Human mitochondrial long noncoding RNAs (lncRNAs) have not been described to date. By analysis of deep-sequencing data, the authors identified three lncRNAs generated from the mitochondrial genome and confirmed their expression by northern blotting and strand-specific qRT-PCR. They show that the abundance of these lncRNAs is comparable to their complementary mRNAs and that nuclear-encoded mitochondrial proteins involved in RNA processing regulate their expression. Finally, they show that mitochondrial lncRNAs form intermolecular duplexes and that their abundance is cell- and tissue-specific, suggesting a functional role in the regulation of mitochondrial gene expression.
Keywords: noncoding RNA, mitochondrial RNA, RNA processing
Abstract
Human mitochondrial long noncoding RNAs (lncRNAs) have not been described to date. By analysis of deep-sequencing data we have identified three lncRNAs generated from the mitochondrial genome and confirmed their expression by Northern blotting and strand-specific qRT–PCR. We show that the abundance of these lncRNAs is comparable to their complementary mRNAs and that nuclear-encoded mitochondrial proteins involved in RNA processing regulate their expression. We also identify the 5′ and 3′ transcript ends of the three lncRNAs and show that mitochondrial RNase P protein 1 (MRPP1) is important for the processing of these transcripts. Finally, we show that mitochondrial lncRNAs form intermolecular duplexes and that their abundance is cell- and tissue-specific, suggesting a functional role in the regulation of mitochondrial gene expression.
INTRODUCTION
Eukaryotic transcriptomes have been shown to be extremely complex. The discovery of noncoding RNAs (Okazaki et al. 2002) has expanded the regulatory repertoire of gene expression and our view beyond the central dogma where RNA couples gene expression to protein synthesis (Mattick 2003). An extraordinary range of noncoding RNAs are generated from sense or antisense interspersed intronic or genomic regions overlapping with coding and noncoding sequences, and their expression and function are tightly linked to developmental processes (Carninci et al. 2005; Amaral and Mattick 2008; Mercer et al. 2009). Long ncRNAs (lncRNAs) are diverse in sequence and structure, and genome-wide technologies allow the classification and analysis of their functional relevance (Carninci et al. 2005; Kapranov et al. 2005, 2007). The function of lncRNAs includes the epigenetic control of chromatin structure (Mattick et al. 2009) as well as transcriptional and post-transcriptional gene regulation (Mercer et al. 2009). There are also many types of small regulatory RNAs found in the nucleus and cytoplasm of animal cells (Mattick and Makunin 2005; Taft et al. 2009, 2010). Interestingly, to date only six small ncRNAs have been identified in mitochondria (Lung et al. 2006) as well as short inverted repeats that form extensions of the mitochondrial 16S rRNA (Villegas et al. 2007). However, lncRNAs generated from the mammalian mitochondrial genome have not been identified.
The mitochondrial genome, unlike the complex nuclear genome, is a compact, circular, double-stranded DNA encoding only 13 proteins, which are all subunits of the electron transport chain, as well as two rRNAs and 22 tRNAs required for their translation (Smeitink et al. 2001). Mitochondrial genes for proteins and tRNAs are located on both the heavy and light strands of the genome, which are transcribed as large polycistronic transcripts covering almost the entire length of each strand (Aloni and Attardi 1971; Murphy et al. 1975; Montoya et al. 1981; Mercer et al. 2011). A third transcript covering the start of the heavy strand and the two rRNA genes is also produced (Christianson and Clayton 1988). These long precursor mitochondrial transcripts undergo processing to form functional RNAs (Ojala et al. 1981). In nearly all cases, coding genes are interspersed by one or more tRNAs, which act as “punctuation” marks for processing by RNase P at the 5′ end of tRNAs (Holzmann et al. 2008) and by the mitochondrial RNase Z, elaC homology 2 (ELAC2), at the 3′ end of tRNAs (Takaku et al. 2003; Brzezniak et al. 2011; Lopez Sanchez et al. 2011). A CCA triplet is added to the 3′ ends of tRNAs and specific bases within both tRNAs and rRNAs are often modified, while mRNAs are generally polyadenylated at their 3′ ends. Recently, using a deep-sequencing approach to characterize the 5′ and 3′ ends of all 22 mitochondrial tRNAs, we have found that the regulation of the processing of mitochondrial tRNAs has profound effects on mitochondrial gene expression (Lopez Sanchez et al. 2011). We found that knockdown of the four nuclear-encoded mitochondrial proteins ELAC2, mitochondrial RNase P proteins 1 and 3 (MRPP1 and MRPP3), and pentatricopeptide repeat domain protein 1 (PTCD1) affects the levels of mitochondrial RNAs and their final processing sites (Lopez Sanchez et al. 2011).
Here we have used deep-sequencing data to discover RNAs generated from noncoding sequences of the mitochondrial genome. We have identified three abundant mitochondrial lncRNAs and have found that their expression is regulated by nuclear-encoded mitochondrial processing proteins, in particular, those that comprise the mitochondrial RNase P complex. We show that all three lncRNAs form intermolecular duplexes and their abundance varies in different cell lines and tissues, suggesting that mitochondrial lncRNAs may have functional significance that contributes to the regulation of mitochondrial gene expression.
RESULTS/DISCUSSION
The mitochondrial genome generates three stable lncRNAs
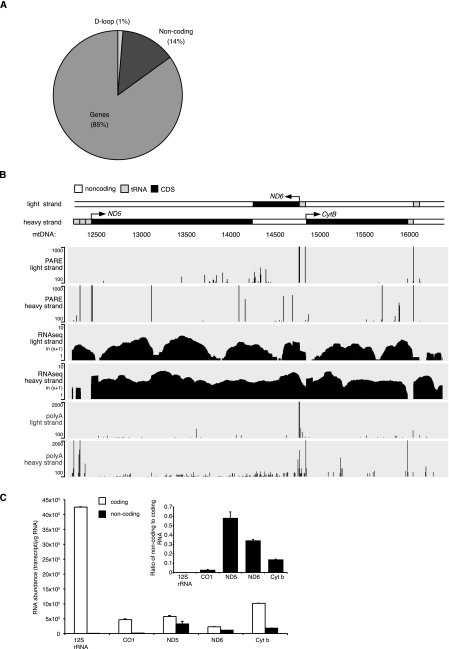
The mitochondrial polycistronic transcript encoding the heavy-strand genes has little noncoding sequence. In contrast, the light-strand polycistronic transcript only encodes seven tRNAs and the ND6 mRNA that are separated by long stretches of noncoding sequences. It is not entirely clear whether the noncoding sequences are degraded or whether any of them are abundant and functionally significant. We used data sets from strand-specific deep sequencing to analyze the presence of lncRNAs in the transcriptome of HeLa mitochondria. We observed that a significant proportion (15.02%, excluding rRNA and tRNA) of reads that uniquely aligned to the mitochondrial genome correspond to noncoding DNA (Fig. 1A). The regions of the mitochondrial genome complementary to the genes that encode ND5, ND6, and Cyt b mRNAs were found to have high levels of lncRNAs (Fig. 1B). The region on the heavy strand that is complementary to the ND6 mRNA is known to be retained as the 3′ untranslated region (UTR) of the mature ND5 mRNA; however, we also found that it is a lncRNA in its own right (see below). The three mitochondrial lncRNAs are punctuated by numerous stop codons and there are no significant open reading frames present (Supplemental Fig. 1). The longest opening reading frame is only 67 amino acids, located in the middle of lncND6 RNA (Supplemental Fig. 1). Therefore, we identified three mitochondrial lncRNAs, lncND5, lncND6, and lncCyt b RNA within the mitochondrial transcriptome (Fig. 1; Supplemental Fig. 1). In addition, the parallel analyses of RNA ends (PARE) and poly(A) analyses of RNA isolated from HeLa cells revealed the canonical 5′ and 3′ ends of mitochondrial coding transcripts (Ojala et al. 1981) as well as potential transcript ends of the lncRNAs (Fig. 1B), which we further investigated here.
FIGURE 1.
Long noncoding RNAs are generated from the light strand of the mitochondrial genome. (A) Noncoding RNAs make up 15% of the human mitochondrial transcriptome, excluding rRNAs and tRNAs. (B) Deep sequencing detected the existence of mitochondrial ND5, ND6, and Cyt b lncRNAs. PARE analyses detect the 5′ ends of mitochondrial transcripts and poly(A) analyses detect the polyadenylated 3′ ends of mitochondrial transcripts. (C) Abundance of the noncoding ND5, ND6, and Cyt b RNAs compared with coding mRNAs and the 12S rRNA determined by strand-specific qRT–PCR.
We used strand-specific qRT–PCR (ss qRT–PCR) to measure the abundance of these lncRNAs and compared them with their complementary transcripts, which encode functional mRNAs. We found that these three lncRNAs encoded by the light strand are 58%, 34%, and 14% as abundant as their complementary coding ND5, ND6, and Cyt b mRNAs, respectively (Fig. 1C). We observed that there was a 1867-fold higher abundance of sense to antisense RNA from the 12S rRNA region, indicating that the ssqRT–PCR provides at least three orders of magnitude higher sensitivity for the target strand of interest over its complementary sequence (Fig. 1C).
Nuclear-encoded proteins regulate the levels of mitochondrial lncRNAs
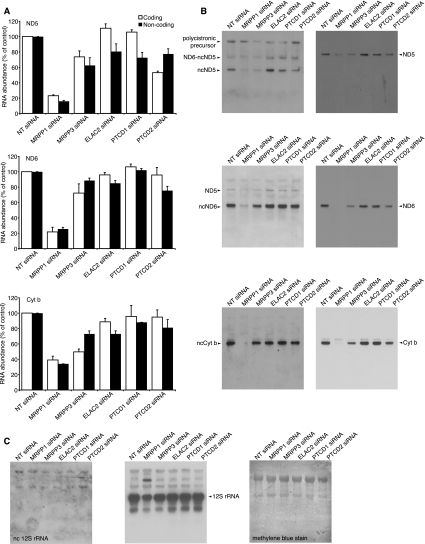
Recently, we have shown that ELAC2, MRPP1, MRPP3, and PTCD1 affect the levels of mature tRNA, rRNA and mRNA expression, and the accumulation of precursor transcripts at the RNA processing level (Lopez Sanchez et al. 2011). Pentatricopeptide repeat (PPR) domain protein 2 (PTCD2) belongs to the family of PPR RNA-binding proteins, which includes PTCD1 and MRPP3, and are involved in the regulation of mammalian mitochondrial gene expression (Rackham and Filipovska 2011), and has been found to affect the processing of the Cyt b mRNA (Xu et al. 2008). We performed ss qRT–PCR on RNA isolated from purified mitochondria of HeLa cells where ELAC2, MRPP1, MRPP3, PTCD1, or PTCD2 were knocked down after 6 d to 3.5% ± 0.4, 1.6% ± 0.5, 1.9% ± 0.6, 5.3% ± 0.8, and 2.2% ± 0.5, respectively, by siRNAs and used nontargeting (NT) siRNAs as a control to determine the contribution of these proteins to the regulation and abundance of lncRNA compared with coding mRNA expression (Fig. 2A). In addition, by using antibodies against ELAC2, MRPP1, MRPP3, PTCD1, or PTCD2, we show that their protein abundance is significantly decreased (Supplemental Fig. 2A). We show knockdown of MRPP1 caused the most dramatic decrease in the abundance of the three lncRNAs (Fig. 2A). Knockdown of MRPP3 decreased the levels of the lncRNAs, although the effect was less dramatic compared with MRPP1. This may be a result of the stability of the MRPP3 protein compared with MRPP1, since their respective mRNAs were knocked down to similar levels. Decrease in ELAC2 levels caused a more subtle decrease in all lncRNAs, whereas PTCD1 knockdown caused a specific decrease in the ND5 lncRNA (Fig. 2A). PTCD2 has been shown to affect Cyt b mRNA processing in the heart (Xu et al. 2008). Therefore, we investigated the effect of PTCD2 knockdown on the mitochondrial lncRNAs. Interestingly, we found that PTCD2 decreased the levels of ND5 lncRNA and the lncND6 RNA, which can form the 3′ UTR of the ND5 mRNA (Fig. 2A). These observed changes suggest an important contribution of the five nuclear-encoded proteins to the regulation of mitochondrial gene expression, possibly reflecting their roles in processing the precursor transcripts from which both coding and noncoding RNAs are derived.
FIGURE 2.
Mitochondrial lncRNAs are regulated by nuclear-encoded proteins involved in mitochondrial RNA processing. (A) Strand-specific qRT–PCR showing the effects of mitochondrial RNA processing proteins on lncRNAs. The effects of ELAC2, MRPP1, MRPP3, PTCD1, and PTCD2 knockdown in HeLa cells were analyzed on mitochondrial transcripts by ss qRT–PCR. (B) RNA isolated from cells transfected with ELAC2, MRPP1, MRPP3, PTCD1, or PTCD2 siRNA and nontargeting (NT) siRNA was analyzed for mitochondrial noncoding and complementary coding RNA abundance by Northern blotting. (C) The 12S rRNA and the complementary noncoding region of 12S rRNA were probed as controls. The data are typical of results repeated on four separate RNA preparations. Each Northern blot was probed for all transcripts by stripping the blot and reprobing. The methylene blue-stained blot shows equal loading of RNA to each well.
MRPP1 is necessary for the accumulation of mitochondrial lncRNAs
We used Northern blotting to detect and compare the abundance of the mitochondrial ncRNAs and their complementary mRNAs and rRNA (Fig. 2B). We show that the lncND5, lncND6, and lncCyt b RNAs are abundant in mitochondria. In contrast, overexposure of the blot probed for the noncoding transcript of the 12S rRNA showed that there were no abundant RNAs corresponding to this region (Fig. 2C), indicating that the ND5, ND6, and Cyt b lncRNAs are specifically enriched in mitochondria. We further investigated the effects of knocking down the RNA processing proteins on the levels of ND5, ND6, and Cyt b lncRNAs and their complementary mRNAs. We found that MRPP1, and in most cases, MRPP3, are important for the abundance of the lncRNAs (Fig. 2B). Knockdown of PTCD2 caused a specific decrease of the ND5 lncRNA. We used a probe that was complementary to the 3′ UTR alone to detect the ND6 lncRNA. We were surprised to detect that the 3′ UTR of ND5 was also present independent of the mature ND5 mRNA, which we describe as lncND6 (Fig. 2B). We show that lncND6 is an abundant transcript that has previously not been annotated or investigated in Northern blots, most likely because probes that were complementary to this region of the ND5 mRNA have not been used. We observe that in addition to the lncND6, we also detect the mature ND5 mRNA that includes the 3′ UTR or lncND6. Interestingly, we do not observe the presence of an ND5 mRNA that is missing the 3′ UTR, suggesting that it may be required for the mature ND5 mRNA (Fig. 2B). MRPP1 or MRPP3 knockdown lowers the abundance of the ND5, ND6, and Cyt b mRNAs and lncRNAs in a similar manner to that observed with ss qRT–PCR (Supplemental Fig. 2B). In addition, MRPP1 knock down decreases the abundance of the 12S rRNA and causes the accumulation of a larger mitochondrial precursor transcript containing the 12S rRNA (Fig. 2C).
Mitochondrial ncRNAs are specifically processed at defined sites
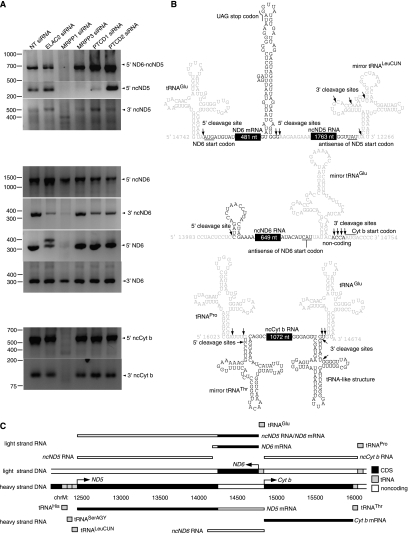
We investigated the effects of protein knockdown on the processing of the 5′ and 3′ ends of the mitochondrial lncRNAs. We used rapid amplification of cDNA ends (RACE) to show that MRPP1 is responsible for processing of the 5′ and 3′ ends of the lncRNAs, with the exception of the 5′ end of the ND6 lncRNA (Fig. 3A). In addition, we observed that MRPP3 and PTCD1 knockdown cause a decrease in the processing of the 5′ end of the ND5 lncRNA, whereas ELAC2 has a minor effect on the processing of the 3′ end of the ND6 lncRNA (Fig. 3A). We observed the enrichment of an additional transcript end when ELAC2 is depleted in cells, which is that of a preprocessed ND6-tRNAGlu transcript, further supporting our previous findings that ELAC2 processes the 3′ end of mitochondrial tRNAs (Lopez Sanchez et al. 2011). Furthermore, we observed that the lncND5 RNA was occasionally found as a 3′ UTR of the ND6 mRNA, as well as an independent lncND5 RNA (Fig. 3A–C). This was also observed in the Northern blot of the lncND5 RNA (Fig. 2B). However, it remains to be determined whether these two isoforms provide an opportunity to differentially regulate ND6 expression.
FIGURE 3.
Identification of the 5′ and 3′ sites of mitochondrial lncRNAs and how they are affected by knockdown of mitochondrial RNA processing proteins. (A) The effects of ELAC2, MRPP1, MRPP3, PTCD1, and PTCD2 knockdown in HeLa cells were analyzed on mitochondrial lncRNAs by rapid amplification of cDNA ends. (B) We show the transcript ends and their position for the mitochondrial RNAs determined by Sanger sequencing (numbering is according to the human mitochondrial DNA reference sequence). (C) Schematic of the identified coding and noncoding ND5, ND6, and Cyt b RNAs.
Next, we identified the exact transcript ends of the mitochondrial lncRNAs by Sanger sequencing of the RACE products (Fig. 3B). We found that the mitochondrial lncRNAs have a defined length and transcript ends (Fig. 3C). Here we have observed cleavage by the mitochondrial RNase P adjacent to antisense or mirror tRNAs (Fig. 3B), indicating that this enzyme may recognize them as substrates, suggesting that mirror tRNAs may have a functional role as previously suggested (Seligmann 2010a,b). Interestingly, we observed several different transcript ends for some of the lncRNA ends (Fig. 3B), indicating the occasional promiscuity of the mitochondrial RNase P. Recently, it has been shown that the mitochondrial RNase P is an unconventional RNase P in that it lacks an RNA component (Holzmann et al. 2008). Our findings that the mitochondrial RNase P cleaves adjacent to mirror tRNA sequences with imprecise substrate selection at certain sites provides further evidence of the enzyme's unconventional characteristics and may explain why mammalian mitochondrial RNase P has two additional protein subunits (Holzmann et al. 2008) relative to plants, where the mitochondrial RNase P fulfills a more canonical role in tRNA processing similar to nuclear RNase P (Gobert et al. 2010). Some of the different consequences of MRPP1 and MRPP3 knockdown might reflect the distinct activities of each RNase P protein. For example, in addition to its requirement for RNase P activity, MRPP1 has been shown to have tRNA methyltransferase activity (Lopez Sanchez et al. 2011).
Mitochondrial ncRNAs form intermolecular duplex structures
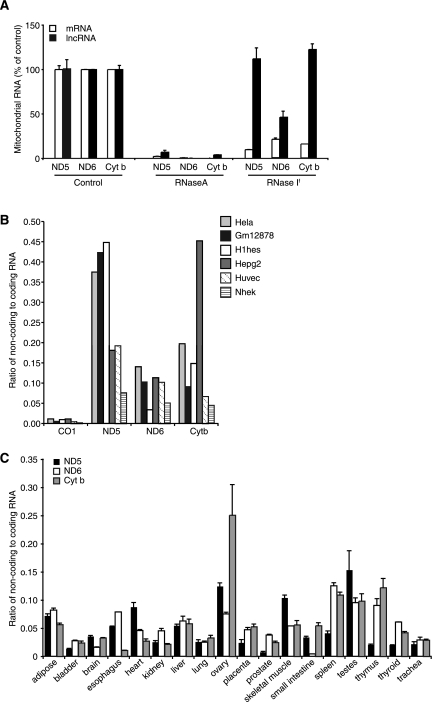
We investigated whether the three mitochondrial lncRNAs were single or double stranded by ss qRT–PCR using mitochondria isolated from cells, lysed, and treated either with RNase A, an RNA endonuclease that cleaves after cytosine and uracil residues, or with RNase If, a single-strand-specific RNA endonuclease. RNase A degraded both the sense and lncRNAs almost completely and to similar extents (Fig. 4A). We show that the lncRNAs primarily form intermolecular duplexes, as they are resistant to RNase If cleavage, unlike their complementary ND5, ND6, and Cyt b mRNAs (Fig. 4A). This indicates that the lncRNAs may be protected by the binding of their respective complementary mRNAs, which are in excess compared with the lncRNAs. Furthermore, the protection of these three lncRNAs suggests that they may have a functional role in mitochondria to either stabilize ND5, ND6, and Cyt b mRNAs or regulate their expression. The ND6 mRNA is the only mitochondrial mRNA that is not polyadenylated (Temperley et al. 2010; Mercer et al. 2011), and the presence of the lncND6 transcript may play a role in stabilizing this mRNA. Moreover, the double-stranded nature of these lncRNAs may impair the translation of their corresponding mRNAs, potentially explaining why the ND6 protein is the least abundant mitochondria encoded protein.
FIGURE 4.
Mitochondrial ncRNAs are predominantly found as duplexes and show cell-specific expression. (A) Mitochondria isolated from HeLa cells were lysed and treated with either RNase A or RNase If, and lncRNAs and mRNAs were measured by ss qRT–PCR. Mitochondria not treated with RNases were used as a control. Differential abundance of lncRNAs compared with their coding counterparts in different cell lines determined from deep-sequencing data sets (B) and in 18 different human tissues (C) determined by ss qRT–PCR, suggesting that they may have a role in the regulation of mitochondrial gene expression.
Cell and tissue-specific expression of mitochondrial lncRNAs
Lastly, we investigated whether the abundance of mitochondrial lncRNAs varies in different cells in strand-specific deep-sequencing data sets and by ss qRT–PCR of RNA from tissues. We observed a large variation in the abundance of the three lncRNAs in different cell lines (Fig. 4B). In addition, we found that there was variation in the abundance of the three lncRNAs across 18 different human tissues, and lncND5 RNA was the most abundant of the three ncRNAs (Fig. 4C). Interestingly, the lncRNAs are most abundant in reproductive tissues such as ovaries and testes. To investigate whether mitochondrial lncRNAs might have a regulatory role, we tested the ratios of lncRNAs compared with adjacent mRNAs from opposite strands and found that the lncRNA ratios are different compared with the mRNAs that are on the opposite strand, suggesting that the regulation of the lncRNAs may be independent from that of coding transcripts (Supplemental Fig. 3). These data further suggest that the regulation of mitochondrial gene expression varies depending on specific cell-type requirements, and implies that the identified lncRNAs may contribute to the regulation of mitochondrial gene expression.
MATERIALS AND METHODS
Cell culture
HeLa human cervical cancer cells were cultured at 37°C under humidified 95% air/5% CO2 in Dulbecco's modified Eagle's medium (DMEM, Invitrogen) containing glucose (4.5 g.l−1), 1 mM pyruvate, 2 mM glutamine, penicillin (100 U.mL−1), streptomycin sulfate (100 μg.mL−1), and 10% fetal bovine serum (FBS).
Transfections
Cells were plated at 60% confluence in 6-well plates or 10-cm dishes and transfected with annealed siRNAs in OptiMEM media (Invitrogen). A total of 125 nM (for 6-well plates) or 145 nM (for 10-cm dishes) of ELAC2, MRPP1, MRPP3, PTCD1, PTCD2, or control off-target siRNAs (Dharmacon) were transfected using Lipofectamine 2000 (Invitrogen). Cell incubations were carried out for up to 3 d and the cells were retransfected and incubated for a further 3 d following transfection.
Mitochondrial isolation and RNase treatments
Mitochondria were prepared from 15 × 106 cells grown overnight in 15-cm dishes. Cells in PBS were sedimented (150g for 5 min at 4°C), resuspended in 4 mL of ice-cold 10 mM NaCl, 1.5 mM MgCl2, and 10 mM Tris-HCl (pH 7.5). Cells were allowed to swell for 5 min on ice and briefly homogenized with a 7-mL glass homogenizer. The sucrose concentration was then adjusted to 250 mM by adding 2 M sucrose in 10 mM Tris-HCl and 1 mM EDTA (pH 7.6) (T10E20 buffer). The nuclei from this suspension were sedimented (1300g for 3 min at 4°C), and the centrifugation step was repeated for the supernatant once more. Mitochondria were sedimented from the supernatant by centrifugation (15,000g for 15 min at 4°C). The mitochondrial suspension was layered on a discontinuous sucrose gradient (1.0 and 1.7 M) in T10E20 buffer and centrifuged at 70,000g for 40 min at 4°C. The mitochondrial fraction was recovered from the interface between the two sucrose cushions, diluted with 400 μL of 250 mM sucrose in T10E20 buffer, and washed once in the same solution. The final mitochondrial pellet was resuspended in 100 μL of 250 mM sucrose in T10E20 buffer and protein concentration as determined by the bicinchoninic acid (BCA) assay (Smith et al. 1985) using bovine serum albumin (BSA) as a standard. For RNase sensitivity assays, mitochondria were lysed in ShortCut reaction buffer (NEB) containing 20 mM MnCl2 and 0.1% Triton X-100 for 15 min at 37°C in the presence or absence of either 100 μg.mL−1 RNase A or 50 U of RNase If.
RNA isolation and strand-specific quantitative RT–PCR
RNA was isolated from highly purified, mitochondria using the miRNeasy Mini kit according to the manufacturer's instructions (Qiagen). RNA from tissues was purchased from Ambion. Contaminating genomic DNA was eliminated by an on-column RNase-free DNase treatment. To quantify the levels of mitochondrial RNA produced from a specific strand of mitochondrial DNA, total cellular RNA was denatured at 65°C for 5 min and reverse transcribed using SuperScript III (Invitrogen) at 55°C for 1 h in the presence of a gene-specific primer that incorporates additional adaptor sequence. The reverse transcriptase was inactivated by incubation at 70°C for 15 min. qRT–PCR was performed with a gene-specific primer designed to anneal at the adjacent side of the cDNA of interest and another primer complementary to the additional adaptor sequence incorporated in the primer used for reverse transcription. PCR was performed with a Corbett Rotorgene 6000 using Platinum UDG SYBR Green mastermix (Invitrogen) and normalized to 18S rRNA. Control reactions were preformed by omitting the reverse transcriptase in the cDNA synthesis; qRT–PCR using this material as a template revealed no detectable amplification.
Deep-sequencing analyses
RNA seq data were downloaded from ENCODE and used in strict accordance with the ENCODE Data Release policy (http://genome.ucsc.edu/ENCODE/index.html). Directional RNA sequenced reads were used to identify sites of nontemplate poly(A) additions to the mitochondrial transcriptome using the following approach. RNA sequencing data from Hela and cell lines data were generated as part of the ENCODE project (Birney et al. 2007) and used in strict accordance with the Data Release policy (http://genome.ucsc.edu/ENCODE/index.html). Raw data reads are available from: http://hgdownload.cse.ucsc.edu/goldenPath/hg18/encodeDCC/wgEncodeCaltechRnaSeq. Sequenced reads were aligned to the human genome requiring exact matching. Those reads that did not map exactly to the genome were then trimmed of three or more −A nucleotides from the 3′ end. Trimmed reads longer than 15 nt were then realigned to the human genome again with those reads that mapped uniquely and exactly following 3′ poly(A) trimming were indicative of polyadenylation sites. Nucleotide sites were additionally required to have evidence of 10 or more aligned reads for consideration in analysis. PARE sequencing data were generated from Hela cell lines (Shin et al. 2010). Raw data reads are accessible from GEO (Accession ID GSE22068): http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE22068. Sequenced reads were initially trimmed of adaptor sequence and aligned uniquely and exactly to the human mitochondrial genome with ZOOM (Lin et al. 2008) with up to two mismatches, permitting sequenced reads to overlap SNPs or modified nucleotides. Read mapping was converted into .bed and .wig format for visualization on the UCSC genome browser (genome.ucsc.edu).
Rapid amplification of cDNA ends (RACE)
We polyadenylated the 3′ ends of RNAs isolated from HeLa cells where MRPP1, MRPP3, ELAC2, PTCD1, and PTCD2 were knocked down with Escherichia coli poly(A) polymerase (NEB) ligated RNA adaptors to the RNA 5′ ends, and reverse transcribed with an oligo d(T) primer. Rapid amplification of cDNA ends (RACE) was then performed to identify the 5′ or 3′ ends of the cDNA of interest.
Northern blotting
RNA (4 μg) was resolved on 1.2% agarose formaldehyde gels, then transferred to 0.45 μm of Hybond-N+ nitrocellulose membrane (GE Lifesciences) and hybridized with biotinylated oligonucleotide probes specific to mitochondrial RNAs. The hybridizations were carried out overnight at 50°C in 5x SSC, 20 mM Na2HPO4, 7% SDS, and 100 μg.mL−1 heparin, followed by washing. The signal was detected using a streptavidin-linked horse radish peroxidase (diluted 1: 2000 in 3x SSC, 5% SDS, 25 mM Na2HPO4 at pH 7.5) by enhanced chemiluminescence (GE Lifesciences).
Immunoblotting
Specific proteins were detected using rabbit αELAC2, αPTCD1 (Sigma, diluted 1:500), αMRPP1, αMRPP3, αPTCD2 (Abcam, diluted 1:500) antibodies, and mouse αporin antibody (MitoSciences, diluted 1:2000) in 1% skim milk powder in PBS (4.3 mM sodium phosphate, dibasic, 137 mM sodium chloride, 2.7 mM potassium chloride, 1.4 mM potassium phosphate, monobasic, PBS). The primary antibodies were detected using goat anti-mouse or goat anti-rabbit horse radish peroxidase (Biorad, diluted 1: 10,000).
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
ACKNOWLEDGMENTS
This work was supported by grants and fellowships from the Australian Research Council (FT0991008 and FT0991113), the National Health and Medical Research Council (572513, 1005030 and Australia Fellowship 631668), and the University of Western Australia.
Footnotes
Article published online ahead of print. Article and publication date are at http://www.rnajournal.org/cgi/doi/10.1261/rna.029405.111.
REFERENCES
- Aloni Y, Attardi G 1971. Symmetrical in vivo transcription of mitochondrial DNA in HeLa cells. Proc Natl Acad Sci 68: 1757–1761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Mattick JS 2008. Noncoding RNA in development. Mamm Genome 19: 454–492 [DOI] [PubMed] [Google Scholar]
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezniak LK, Bijata M, Szczesny RJ, Stepien PP 2011. Involvement of human ELAC2 gene product in 3′ end processing of mitochondrial tRNAs. RNA Biol 8: 616–626 [DOI] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Christianson TW, Clayton DA 1988. A tridecamer DNA sequence supports human mitochondrial RNA 3′-end formation in vitro. Mol Cell Biol 8: 4502–4509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gobert A, Gutmann B, Taschner A, Gossringer M, Holzmann J, Hartmann RK, Rossmanith W, Giege P 2010. A single Arabidopsis organellar protein has RNase P activity. Nat Struct Mol Biol 17: 740–744 [DOI] [PubMed] [Google Scholar]
- Holzmann J, Frank P, Loffler E, Bennett KL, Gerner C, Rossmanith W 2008. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135: 462–474 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Drenkow J, Cheng J, Long J, Helt G, Dike S, Gingeras TR 2005. Examples of the complex architecture of the human transcriptome revealed by RACE and high-density tiling arrays. Genome Res 15: 987–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Lin H, Zhang Z, Zhang MQ, Ma B, Li M 2008. ZOOM! Zillions of oligos mapped. Bioinformatics 24: 2431–2437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez Sanchez MIG, Mercer TR, Davies SM, Shearwood A-MJ, Nygård KKA, Richman TR, Mattick JS, Rackham O, Filipovska A 2011. RNA processing in human mitochondria. Cell Cycle 10: 1–13 [DOI] [PubMed] [Google Scholar]
- Lung B, Zemann A, Madej MJ, Schuelke M, Techritz S, Ruf S, Bock R, Huttenhofer A 2006. Identification of small non-coding RNAs from mitochondria and chloroplasts. Nucleic Acids Res 34: 3842–3852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattick JS 2003. Challenging the dogma: the hidden layer of non-protein-coding RNAs in complex organisms. Bioessays 25: 930–939 [DOI] [PubMed] [Google Scholar]
- Mattick JS, Makunin IV 2005. Small regulatory RNAs in mammals. Hum Mol Genet 14: R121–R132 [DOI] [PubMed] [Google Scholar]
- Mattick JS, Amaral PP, Dinger ME, Mercer TR, Mehler MF 2009. RNA regulation of epigenetic processes. Bioessays 31: 51–59 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Mercer TR, Neph S, Crawford J, Dinger ME, Smith MA, Shearwood A-MJ, Haugen E, Bracken CP, Rackham O, Stamatoyannopoulos JA, et al. 2011. The human mitochondrial transcriptome. Cell 146: 645–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoya J, Ojala D, Attardi G 1981. Distinctive features of the 5′-terminal sequences of the human mitochondrial mRNAs. Nature 290: 465–470 [DOI] [PubMed] [Google Scholar]
- Murphy WI, Attardi B, Tu C, Attardi G 1975. Evidence for complete symmetrical transcription in vivo of mitochondrial DNA in HeLa cells. J Mol Biol 99: 809–814 [DOI] [PubMed] [Google Scholar]
- Ojala D, Montoya J, Attardi G 1981. tRNA punctuation model of RNA processing in human mitochondria. Nature 290: 470–474 [DOI] [PubMed] [Google Scholar]
- Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. 2002. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature 420: 563–573 [DOI] [PubMed] [Google Scholar]
- Rackham O, Filipovska A 2011. The role of mammalian PPR domain proteins in the regulation of mitochondrial gene expression. Biochim Biophys Acta (in press) [DOI] [PubMed] [Google Scholar]
- Seligmann H 2010a. Mitochondrial tRNAs as light strand replication origins: similarity between anticodon loops and the loop of the light strand replication origin predicts initiation of DNA replication. Biosystems 99: 85–93 [DOI] [PubMed] [Google Scholar]
- Seligmann H 2010b. Undetected antisense tRNAs in mitochondrial genomes? Biol Direct 5: 39 doi: 10.1186/1745-6150-5-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin C, Nam JW, Farh KK, Chiang HR, Shkumatava A, Bartel DP 2010. Expanding the microRNA targeting code: functional sites with centered pairing. Mol Cell 38: 789–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smeitink J, van den Heuvel L, DiMauro S 2001. The genetics and pathology of oxidative phosphorylation. Nat Rev Genet 2: 342–352 [DOI] [PubMed] [Google Scholar]
- Smith PK, Krohn RI, Hermanson GT, Mallia AK, Gartner FH, Provenzano MD, Fujimoto EK, Goeke NM, Olson BJ, Klenk DC 1985. Measurement of protein using bicinchoninic acid. Anal Biochem 150: 76–85 [DOI] [PubMed] [Google Scholar]
- Taft RJ, Glazov EA, Lassmann T, Hayashizaki Y, Carninci P, Mattick JS 2009. Small RNAs derived from snoRNAs. RNA 15: 1233–1240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE, et al. 2010. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat Struct Mol Biol 17: 1030–1034 [DOI] [PubMed] [Google Scholar]
- Takaku H, Minagawa A, Takagi M, Nashimoto M 2003. A candidate prostate cancer susceptibility gene encodes tRNA 3′ processing endoribonuclease. Nucleic Acids Res 31: 2272–2278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temperley RJ, Wydro M, Lightowlers RN, Chrzanowska-Lightowlers ZM 2010. Human mitochondrial mRNAs-like members of all families, similar but different. Biochim Biophys Acta 1797: 1081–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villegas J, Burzio V, Villota C, Landerer E, Martinez R, Santander M, Pinto R, Vera MI, Boccardo E, Villa LL, et al. 2007. Expression of a novel non-coding mitochondrial RNA in human proliferating cells. Nucleic Acids Res 35: 7336–7347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Ackerley C, Maj MC, Addis JB, Levandovskiy V, Lee J, Mackay N, Cameron JM, Robinson BH 2008. Disruption of a mitochondrial RNA-binding protein gene results in decreased cytochrome b expression and a marked reduction in ubiquinol-cytochrome c reductase activity in mouse heart mitochondria. Biochem J 416: 15–26 [DOI] [PubMed] [Google Scholar]