Abstract
Cellular membranes are composed of proteins and glyco- and phospholipids and play an indispensible role in maintaining cellular integrity and homeostasis by physically restricting biochemical processes within cells and providing protection. Membrane proteins perform many essential functions, which include operating as transporters, adhesion-anchors, receptors, and enzymes. Recent advancements in proteomic mass spectrometry have resulted in substantial progress towards the determination of the plasma membrane (PM) proteome, resolution of membrane protein topology, establishment of numerous receptor protein complexes, identification of ligand–receptor pairs, and the elucidation of signaling networks originating at the PM. Here we discuss the recent accelerated success of discovery-based proteomic pipelines for the establishment of a complete membrane proteome.
Application of mass spectrometry to study membrane proteins
The water insoluble nature of transmembrane proteins renders them challenging, but not impossible, to investigate by traditional biochemical approaches in conjunction with mass spectrometry (MS)(1). In this review, we focus on the eminence of shotgun MS for accelerating the identification and study of membrane proteins. Specifically, we briefly cover recent MS advancements to determine the complete membrane proteome, as a way to better understand membrane protein topology membrane protein–protein interactions, and signaling networks that originate from the membrane surface. Indeed, recent technological and methodological advancements have reduced the barriers which previously impeded membrane protein analysis by MS.
Membrane proteins have been investigated by a gamut of approaches at many stages of sample preparation and peptide MS analysis (for an in-depth discussion see (2)). Membrane proteins are chemically tractable entities and represent ~30% of the molecular targets for currently available pharmaceuticals, largely because they often possess ligand binding domains which can be therapeutically targeted (3). Thus, the application of MS to membrane protein drug discovery represents a powerful new approach (reviewed in (4)). The importance of membrane proteins is evident and MS continues to edge us closer toward the determination of a complete membrane proteome.
Shotgun MS proteomics to determine the complete membrane proteome
The relatively low abundance of membrane proteins in un-fractionated samples has undoubtedly resulted in their under-representation in large-scale proteomic datasets. However, PM proteins are not completely absent in these datasets; they are just more challenging to identify. Several recent technological advancements, including improved sample preparation, instrumentation and better liquid chromatographic (LC) performance, have led to a substantial increase in PM protein representation in large-scale data sets (5). Although the majority of membrane proteins are not accessible by traditional sample preparation techniques, it is a topic is of great importance to this discussion which has previously been reviewed in great detail (2). To fully characterize membrane proteins, we must first identify the complete collection; shotgun MS is well suited for this challenge and can identify thousands of proteins in a single analysis. The determination of a complete membrane protein catalog would represent a substantial development because it could provide a global overview of all the proteins at the PM.
Because membrane proteins associate with lipid bilayers in various ways, a conclusive localization assignment is highly complex (6–7). Membrane proteins are categorized based on how they associate with membranes; integral (membrane penetrating), peripheral (attached via non-covalent bonds), or lipid-anchored (attached through covalent bonds). Integral membrane proteins are further categorized based on the secondary structure of the membrane-spanning domain: the majority cross PMs in an alpha-helical arrangement (e.g., the insulin receptor) whereas a few form beta-barrels (e.g., maltoporin). Alpha-helical transmembrane proteins are further classified into 4 basic types based on which terminus of the protein resides in the lumen, and the number of times the protein traverses the membrane. Peripheral membrane proteins can attach to the PM in several ways, such as an in-plane α-helix (e.g., microtubule-affinity-regulating-kinase or via electrostatic interactions (e.g., diphtheria toxin). Lastly, lipid-anchored PM proteins are attached by covalent bonding directly to lipids (e.g., G proteins) through attachment to a fatty acid, prenyl- group, or a GPI anchor. If we also consider protein–protein interactions with PM proteins, it becomes clear that proteins associate with the PM in many complex arrangements. Thus it is not trivial to determine if a given protein is a “membrane protein” by simple computational examination of the amino acid sequence. Indeed, traditional biochemical approaches to characterize membrane proteins involve isolation of these hydrophobic and insoluble proteins-- a process that is both laborious and technically challenging. Consequently, discovery-based approaches such as shotgun MS are particularly well suited for the elucidation of membrane proteins.
Analysis of membrane proteins is complicated by their hydrophobicity, complex post-translational modifications (PTMs), and the fact that they are present at low abundance. Improved instrument performance boosts membrane protein identification compared to previous analyses such that a more in-depth analysis of complex protein mixtures is now easily attainable. Optimization of peptide chromatography and processing can also facilitate the analysis of low abundance proteins in complex mixtures; this has helped in the deep probing of the PM proteome and represents an active area of research which holds great promise. Raising the temperature from 20 to 60 degrees during micro LC was recently shown to boost the number of membrane proteins identified in a standard analysis and should become standard practice with the commercial availability of column ovens (8). The development of MS-compatible detergents has also aided the identification of PM proteins and has been shown to increase PM protein identifications and sequence coverage from the insoluble fraction of rat brain homogenate (9–10).
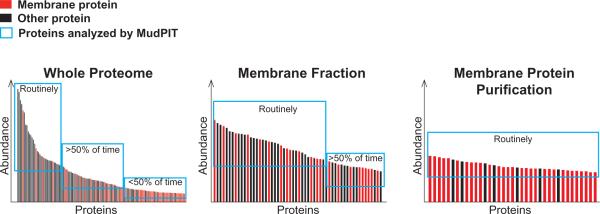
Membrane proteins are analyzed at several “degrees” in shotgun proteomics (Box 1). The coarsest, yet most unbiased, level of membrane protein analysis is within the whole proteome, as in a crude cell lysate or tissue homogenate. Multidimensional Protein Identification Technology (MudPIT) facilitates proteome-level analysis of proteins present across many orders of magnitude in abundance by separating peptides based on their charge and hydrophobicity (11). Membrane proteins are readily identified by MudPIT in crude, complete proteome samples. In theory, whole proteome-level analysis is least likely to be biased by differences in sample preparations between scientists and labs; therefore it should yield the most comparable data. The removal of highly abundant cytoskeletal proteins also can increase PM protein identifications; however the reproducibility of these depletions will likely complicate the comparison of multiple independent analyses (12). Furthermore, all types of membrane proteins will be available for analysis in the comprehensive protein identification given that there is minimal sample manipulation.
Membrane proteins can be easily enriched by centrifugation. The question of which proteins are associated with membranes in specific cell types has been addressed many times by various biochemical enrichments followed by MS analysis. However, the complex arrangement and connectivity of cellular membranes make isolation of perfectly pure PM fractions nearly impossible. Fractionation and enrichment procedures are simple and useful approaches to analyze membrane proteins by reducing the protein complexity and in turn increasing the chances that less abundant PM proteins are identified. The mammalian brain represents a tissue of long-standing interest which is vastly enriched for membranous structures. Within the central nervous system, neurons are connected in complex arrangements to form circuits. Specialized neuron–neuron junctions called chemical synapses represent key membrane-enriched structures because they facilitate molecular communication between neurons. A shining example of the usefulness of MS for analysis of biochemical membrane fractions comes from the Post-Synaptic Density (PSD) which has been performed multiple times with great, but likely incomplete, determination (13–14).
It has been predicted that 20–35% of the mammalian genome encodes membrane proteins, and this represents a bench-mark for any purification comparison (15). A recent MudPIT analysis of the insoluble fraction of rodent brain homogenate found that 35% of the identified proteins were annotated by Gene Ontology as PM proteins and that 22% contained at least one transmembrane domain (10). No fractionation is absolute, and highly abundant soluble non-PM membrane proteins (nuclear and endoplasmic reticulum [ER]) often cannot be completely removed from PM fractions. However, isotopic labeling allows relative quantification of the degree of enrichment of PM proteins, and represents a substantial improvement from the vast majority of the previous studies performed without any quantitation (16). Comparison of the PM and endomembrane fractions has revealed that as many as 25% of the proteins found in the PM fraction are indeed biological contaminants rather than true PM proteins (16). In other similar bodies of work, absolute membrane protein quantitation by isobaric tag for relative and absolute quantitation (iTRAQ) tagging was successfully used to identify previously uncharacterized membrane proteins in pancreatic zymogen granules and HeLa cells (17–18).
MS has also proven to be effective for analyzing the protein components of detergent-resistant membrane domains, microdomains or so-called “rafts”, which have been proposed to be enriched with proteins responsible for a concentrated function, and which constitute a very active area of membrane research (19–20). In hopes of comprehensively analyzing the membrane proteomes of micro-organisms and rodent lung endothelial cells, multiplexed fractionation approaches have been used with some success, but require increased MS data acquisition durations (21–22). Many 1-D and 2-D gel-based approaches have also been effectively employed and represent a valuable method to separate intact proteins by molecular weight (reviewed in (5)). Another angle is to couple membrane fractionation with quantitative comparison of wild type and disease states for differential membrane protein analysis (23–25). Lastly, membrane capture followed by PM protein “shaving” (peptides are cleaved from intact membrane protein fractions) holds promise given that peptides are often removed into solution away from the membrane (reviewed in (26)).
Finally, purified samples facilitate the deepest analysis because the protein and peptide complexity is significantly reduced, thereby affording the mass spectrometer the opportunity to analyze many peptides and proteins of interest given that there are far fewer proteins present. Additionally, highly enriched samples are still required to investigate PTMs or protein complex stoichiometry. The investigation of PM protein PTMs represents yet another active area of research in which MS-based approaches are making substantial contributions (see (5)). Another successful approach is to isolate membrane proteins by affinity capture of a specific PTM; this approach has been useful for studying both glycosylated and palmitoylated proteins (27–28). Antibody-based purification of specific target membrane proteins provides the highest degree of enrichment and can facilitate analysis at substantially increased resolution.
Proteomic methods to determine membrane protein topology
Mass spectrometry is one of the most powerful tools available to study proteins; MS now can easily identify and quantify proteins, determine PTMs, and also explore protein structure (29–30). PM protein topology can be complex and is a critical determinant of function which can be effectively investigated with MS. Membrane protein structure is notoriously difficult to study by traditional high-resolution methods such as X-ray crystallography and NMR spectroscopy. Recently, however, MS in conjunction with hydrogen/deuterium (H/D-MS) exchange, hydroxyl radical –or/oxidative probing, or covalent tagging with regents such as carbodiimide diisopropylcarbodiimide (DiPC-MS) have become more widely applied; these techniques provide powerful approaches to investigate protein structure, folding and topology in membranes at the submolecular level (Figure 1a & b) (31–33). The pioneering MS studies to probe membrane protein topology were accomplished by identifying protease-sensitive regions: these amino acids likely represent surface-exposed and protease-accessible stretches that are excluded from membranes (32). Initial studies of membrane protein topology by MS were small in scale and focused on well characterized proteins of interest, e.g. bacteriorhodopsin (34–36). One key attribute of MS-based topology approaches is that they can be potentially applied to any membrane protein given that this technique requires no structural information.
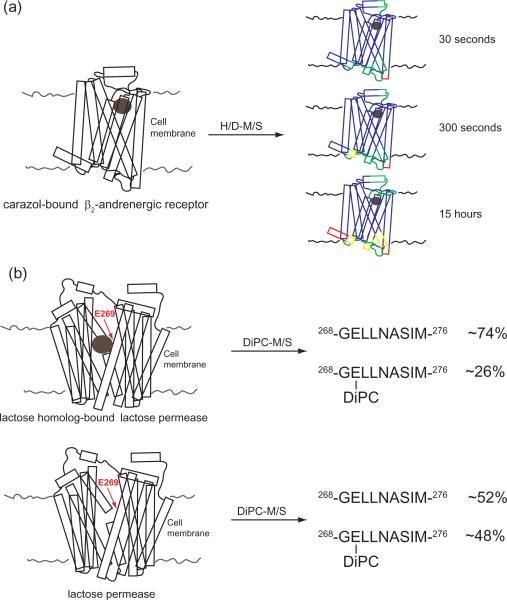
Figure 1. Mass spectrometry in conjunction with hydrogen/deuterium (H/D-MS) exchange and carbodiimide diisopropylcarbodiimide (DiPC-MS) chemical probing reveals membrane protein structural characteristics.
(a) Upon ligand binding, PM receptors can undergo structural rearrangements. Changes in domain labeling of the ligand-bound GPCR β2-andrenergic receptor (PDB:2RH1), after various exchange durations, is shown as an example of how H/D-MS can be used to probe the molecular details of ligand activation (41). Blue regions indicate protein domains which exchange less than 20%, green regions show 20–60% exchange, yellow 70–80%, and red 90–100%. From these results, one can conclude that some regions of integral membrane proteins are more dynamic than others of this PM protein (41). (b) Specific binding pocket amino acid residues can play an important role in ligand binding of PM receptors. Differential DiPC labeling of lactose permease (PDB:1PV6) in the ligand-bound and ligand-free state revealed a key role for aspartic acid (E)-269 (44). Shown are the ligand-bound (i) and ligand-free (ii) states and the percentage of the 268-GELLNASIM-276 peptide tagged. This analysis revealed a dramatic change in the distribution of E-269 which was modified, from a nearly equal proportion in the free state to a nearly three-fold increase of the unmodified in the bound state.
Most MS-based PM protein structure studies take advantage of unequal labeling of PM protein domains, which are dictated by the physical accessibility of the protein. Typically, two or more experimental conditions are compared in order to probe structure topology. H/D-MS is based on the chemistry of hydrogen atom exchange between proteins and the surrounding aqueous solution (37). For transmembrane proteins, the rate of H/D exchange is dictated by the extent of solvent accessibility through the lipid bilayer, the degree of participation of amide hydrogens in secondary and tertiary structural H-boding, and can be complicated by electrostatic effects (38–39). In combination with structure-function mutational analysis, H/D-MS has revealed that most H-bonding interactions in membrane proteins only moderately stabilize the folded state; this finding was unexpected and exemplifies the strength of the approach (40). Additionally, HD-MS analysis of the β2-adrenergic receptor with increasing deuterium incubation durations revealed several dynamic regions within this G protein-coupled receptor (GPCR) (41).Overall, H/D-MS can readily reveal structural characteristics at medium-resolution (on average 10 amino acids) and is capable of moderate-throughput analyses(38).
Oxidative ˙OH labeling and DiPC covalent chemical tagging with MS have revealed native membrane protein structural details at high resolution (42). ˙OH labeling occurs exclusively at Methionine (Met) residues, which represents a key advantage over many other chemical tagging approaches which can be promiscuous in the residues which they modify. Interestingly, Met residues have been artificially introduced into membrane proteins and with ˙OH labeling effectively serve as conformational probes (43). DiPC is also fairly specific in the residues which modifies and preferentially hits Aspartic acid (Asp) and Glutamic acid (Glu). DiPC-M/S has been applied to PM proteins and revealed an important role of Glu269 in substrate binding of the membrane protein lactose permease (44). The labeling specificity of these approaches provides structural information at higher resolution than H/D-MS, but is restricted by the amino acid identities in a protein domain of interest.
A new and exciting approach is the application of phospholipid bilayer nanodiscs with H/D-MS to the study of membrane proteins. Phospholipid bilayer nanodiscs provide a stable and controllable native-like membrane environment which aids solubility issues and allows protein analysis in native conformations (45). A new approach, although not yet applied to membrane proteins, couples laser temperature jumping with fast photochemical oxidation, enabling MS-based folding analysis at sub-millisecond time resolution (46). This novel technique could hold promise for the investigation of transmembrane protein folding and membrane insertion dynamics in reconstituted systems. Perhaps the most exciting possibility for determination of protein topology by MS and chemical labeling is to determine membrane protein topology in living cells; this goal, however, remains a daunting endeavor.
Shotgun MS facilitates the mapping of membrane protein–protein interactions
MS applications are making progress toward the comprehensive identification of all PM proteins, and probe their topology. MS has also proven to be particularly useful to discover PM protein–protein interactions; such experiments can reveal which proteins physically (and often functionally) interact, thereby representing an essential step towards elucidating the molecular function of PM proteins. Most investigations fall into one of two general approaches: isolation of membrane protein complexes by antibody purification or in vitro binding experiments followed by MS. Often in vitro binding experiments are most suitable for finding direct high affinity interactions such as ligand and receptor pairs whereas antibody purifications are best for the identification of multi-protein complexes. With current shotgun MS technologies, it is now possible to identify nearly every protein in purified protein samples of low complexity.
Antibody purifications have revealed the molecular composition of receptor complexes with unexpected and exciting results. The N-methyl-D-aspartate (NMDA) and 5-hydroxytryptamine (5-HT2C)receptors were early examples characterized by MS, but recently many more have followed, including the α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR), β-Aminobutyric acid (GABA), and kainite receptors (Figure 2a) (47–50). Furthermore, it has become increasingly evident that most cell surface receptor complexes contain auxiliary subunits (such as transmembrane AMPA receptor regulatory proteins and receptor activity-modifying proteins), which can modulate functional properties and membrane insertion patterns. Membrane extracts from rodent brains often serve as input material for affinity purification. High quality antibodies for endogenous proteins are required, but tagged proteins also can be utilized. In one exceptional case, an epitope-tagged glutamate receptor was selectively expressed in the cerebellum and used to determine a brain region-specific receptor complex (51). To ensure accurate identification of specific versus non-specific interactors the inclusion of negative control mice which lack the target protein are of high value: any proteins found in these purifications are likely irrelevant (52). It is important to note that endogenous antibodies can compete for protein binding interfaces and that the addition of epitope tags could artificially disrupt protein–protein interactions. Although there is no easy way to counter these challenges, utilizing polyclonal antibodies which recognize multiple epitopes or inserting the recombinant tag at a different position has proven to be useful. Recently, blue-native PAGE was used to further characterize affinity-purified receptor complexes by determining the molecular weight of the intact complex (53–54). Thus, by summing the molecular weight of the components one can show that all the masses can be accounted for by the identified molecules present in the complex.
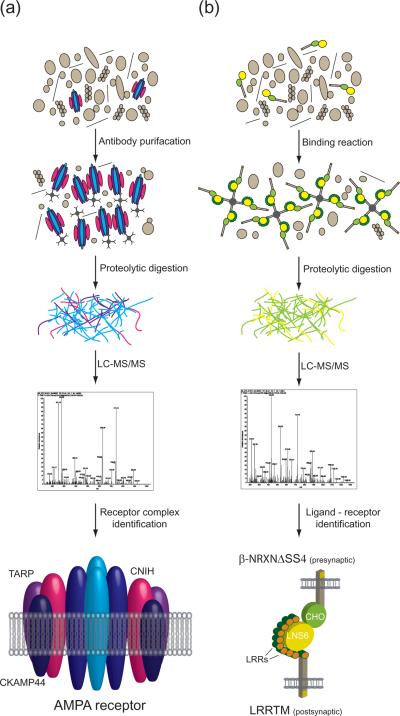
Figure 2. Membrane protein–protein interactions are identified by biochemical approaches in combination with shotgun proteomics.
(a) Antibody purification effectively isolates intact PM receptor complexes from membrane extracts. Purified protein complexes are digested into peptides and subsequently analyzed by LC-MS/MS. The AMPA receptor complex, which has been successfully characterized by antibody purification followed by MS analysis, is shown to illustrate the importance of this approach. AMPA receptor core complexes consist of heterotetramers of glutamate receptor 2 and either glutamate receptor 1, glutamate receptor 3, or glutamate receptor 4 (shown in sky and royal blue). MS-based protein analyses have identified the auxiliary AMPA receptor subunits transmembrane AMPA receptor regulatory proteins (TARPs shown in purple, cornichons (CNIH; fuschia), and cysteine-knot AMPAR modulating protein (CKAMP44, navy blue) (47). (b) In vitro binding reactions with the extracellular domain of receptors as bait and membrane fractions as prey in conjunction with MS can effectively identify ligand–receptor pairs. To illustrate this approach, the ligand–receptor pair (presynaptic neurexin (NRXN) and leucine rich repeat transmembrane protein (LRRTM)) is shown; their interaction was identified by in vitro binding assays followed by MS protein analysis(55). LRRs = Leucine Rich Repeats, CHO = O-linked sugar domain, LNS6 = laminin neurexin and sex hormone-binding protein domain-6.
Ligand–receptor pairs are of critical biological importance given that their binding is directly responsible for important signaling and adhesion events. Shotgun proteomics has proven to be a powerful tool to investigate these events in great detail. The “bait protein” is often produced in heterologous cells and generated as a fusion for easy isolation, and the “prey proteins” can be of any origin which can be extracted from membranes under conditions which disrupt endogenous protein–protein interactions. Using these fusion proteins, the binding event can be potentially recapitulated in vitro. After binding, interacting proteins are easily identified by MS and in a well performed experiment, the interacting “prey protein” should be among the most abundant proteins identified. For some “bait proteins”, removal of background, non-specific “prey protein” binders is required to identify true interacting proteins. This works perfectly for some ligand–receptor pairs; however, there is no guarantee that all endogenous receptor–ligand pairs are amenable to this approach. Thus, numerous false negatives can be encountered and must be considered in any dataset generated through the use of this approach. Another potential concern with this approach is that proteins which never interact in endogenous biological contexts (e.g., because they localize to opposite sides of membranes), now have the opportunity to bind. A recent example of the value of in vitro binding approaches comes from two reports which both successfully identified neurexins as the pre-synaptic ligands for the newly identified synaptogenic molecule leucine rich repeat transmembrane protein 2 LRRTM2 (55–56) (Figure 2b).
In addition to these commonly used approaches, several groups have successfully used less standard approaches to study membrane protein interactions. Because high quality antibodies are not always available and some proteins are not easily expressed, purifying and analyzing native membrane protein complexes would be a substantial development. Most of the current work in this area relies on front-end multidimensional biochemical separations, and although still in its infancy, it certainly holds promise (57). Progress has also been made in analyzing intact membrane protein complexes directly in the mass spectrometer, in a process termed “top-down native MS” (58). In theory, top-down native MS is superior to all shotgun approaches given that the entire polypeptide chain is analyzed in the mass spectrometer. Indeed, an intact membrane protein complex ionized in a micellar solution could be maintained and detected in the gas phase of the mass spectrometer (59). Although there are substantial challenges in all top-down MS approaches, the potential payoff is great given that proteins are analyzed intact without “dark regions” which can be missed when using shotgun approaches.
Shotgun MS identifies signaling networks across membranes
Once PM protein interaction networks have been elucidated, it is critically important to determine how they are integrated in order to transmit signals across PMs. The PM is a critical cellular location for the integration of signaling events and is enriched for surface receptors (for example, GPCRs, receptor tyrosine kinases (RTKs), adhesion signaling molecules, and channels). MS analysis of membrane protein signaling has recently gained attention and is revealing signaling pathways at unprecedented levels (60–61). Indeed, membrane protein signaling has been investigated by increasingly sophisticated methodologies which have revealed the intricate complexity present in physiological signaling networks.
To investigate membrane signaling, cell cultures are typically treated with a ligand (such as a growth factor) and changes in protein abundance or PTM (e.g. phosphorylation) are determined by MS. This approach generates high quality data from hundreds to tens of thousands of proteins per analysis. Although most studies have utilized very similar experimental designs, the importance of thoughtful controls is critical if meaningful conclusions are to be derived. In order to investigate brassinosteroid (BR) membrane signaling, Arabidopsis thaliana seedlings were treated with brassinosteroids and PM fractions were analyzed on 2-D gels followed by MS (62). These analyses identified several proteins whose abundance changed in response to BR signaling; later analyses identified these proteins as belonging to a family of membrane-associated kinases downstream of BR transmembrane receptors. Although this gel-based approach proved successful in plants, such analysis has not proven to be highly applicable to mammalian systems.
Stable isotope labeling facilitates relative peptide and protein measurements and have greatly accelerated the investigation of membrane proteins. In a seminal study, stable isotope labeling was used with affinity capture of phosphorylated proteins to investigate epidermal growth factor signaling (63–65). More recently, similar analysis has been performed to investigate ephrin B (EphB) and brain-derived neurotrophic signaling (66–67). These reports successfully identified hundreds of proteins with changed abundances in ligand treated samples when compared to controls. However, one caveat of these reports, and many others like them, is that phosphopeptides were not used for the quantitation of signaling events. Rather, the changes were assumed by calculating the changes in unmodified peptides (and later protein) levels in the phospho-enriched sample. Most recently, similar experiments were performed to investigate T cell receptors, EphB, and Flt3; these investigations have uncovered hundreds to thousands of regulated phosphorylation events and quantitated these changes at the phosphopeptide level (68–70). The next challenge will be to extend such analyses to the phospho-site level in vivo with a rigorous analytical approach.
Other interesting, less orthodox studies of membrane signaling have been reported and include a chemical proteomic approach in which kinases were enriched in samples of drug-treated cells through the use of immobilized nonselective kinase inhibitors and then identified by MS (71). This approach could prove valuable for drug development given that it enables the quantification of the abundance of hundreds of kinases in drug versus control treated cells (71). Another report of note investigated post-synaptic signaling at the PSD after treatment of organotypic brain slice cultures with NMDA (72). In this work, phosphorylation was found to be increased in 127 and decreased in 101 proteins. These results confirm the existence of combinatorial signaling programs at synapses which likely confer complex information-processing and signaling diversity. Overall, whereas the analysis of PM protein signaling networks by MS is relatively straightforward, the biology which it reveals is highly complex.
Concluding remarks
MS has proven to be a powerful approach to accelerate our understanding of membrane proteins by facilitating discovery-based investigations. These unexpected findings have had a significant effect on the membrane protein field and have propelled it in new and exciting directions. In summary, MS comprehensively identifies PM proteins, probes PM topology, maps PM protein–protein interactions, and elucidates PM signaling networks. The recent advancements in shotgun proteomic instrumentation and experimental design ensure that MS will remain the method of choice to study membrane protein structure, signaling, and molecular interactions in the near and distant future.
Text Box 1. Discovery-based shotgun analysis of the membrane proteome.
A complete catalog of membrane proteins is currently lacking, and data-dependent shotgun proteomics is the best suited approach for discovery-based investigations of the membrane proteome (Figure I). It is currently impossible to conclude that all membrane proteins have been analyzed. The membrane proteome is dynamic and certainly varies between cell types; thus it is impossible to completely determine in a single analysis (this is true for any proteome type). In the best case, MS can accomplish a comprehensive membrane proteome analysis under one set of biological conditions. To determine that an analysis is truly comprehensive, however, repeated analyses of the same sample should show no new protein identifications. Another way to gain confidence that a proteomic analysis is in fact comprehensive is to compare the proteome to the transcriptome mRNA profiles. Although this mRNA-protein comparison might not be entirely fair, it should provide a useful quantitative benchmark of the portion of the protein encoding genome which could potentially be identified in a MS experiment (73).
Recently, the prospects and advantages of single ion-reaction monitoring (SRM) on triple quadrapole mass spectrometers have been highlighted (74). In SRM experiments the mass spectrometer is set up to monitor specific intact peptides and fragment ions (called transitions). This is in contrast to the data-dependent approach commonly used in shotgun experiments in which nearly all peptides will be potentially available for analysis. Although targeted proteomics might hold some promise for accurate protein abundance determination, it is not without its limitations. First, ion suppression critically limits the ability to detect low abundance ions in complex mixtures (75). Enrichment or depletion is required to detect membrane proteins in complex mixtures such as whole brain homogenate. Second, confidence in the identity of the observed peptide is not measured relative to ALL possible digestion products (incomplete and non-specific digestion), and the false positive rates are unknown and most likely high unless a large number of transitions are performed. Lastly, targeted analysis requires a priori knowledge of what is present [sc1]and therefore is a limited discovery tool. SRM is, however, a valuable tool to verify results as long as knowledge of its limits are understood.
Box 1, Figure I.
Views of the membrane proteome based on different enrichment strategies.
Acknowledgements
We would like to thank Guoan Zhang, Thomas Neubert, Joris de Wit and Terunaga Nakagawa for useful discussions regarding the determination of signaling networks, and membrane protein interactions. Additionally, Albert Heck provided critical reading of the review, and Emily J. Larrimer gave valuable editorial advice. Many of the ideas presented here were developed through scientific discussion with current members of the Yates laboratory. The authors would like to acknowledge funding support from NIH grants P41 RR011823 and 5R01 MH067880 to JY.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Eichacker LA, et al. Hiding behind hydrophobicity. Transmembrane segments in mass spectrometry. J Biol Chem. 2004;279(49):50915–50922. doi: 10.1074/jbc.M405875200. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 2.Speers AE, Wu CC. Proteomics of integral membrane proteins--theory and application. Chem Rev. 2007;107(8):3687–3714. doi: 10.1021/cr068286z. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 3.Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002;1(9):727–730. doi: 10.1038/nrd892. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 4.Weinglass AB, Whitelegge JP, Kaback HR. Integrating mass spectrometry into membrane protein drug discovery. Curr Opin Drug Discov Devel. 2004;7(5):589–599. Translated from eng. in eng. [PubMed] [Google Scholar]
- 5.Wu CC, Yates JR., 3rd The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003;21(3):262–267. doi: 10.1038/nbt0303-262. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 6.Alberts B. Molecular biology of the cell. 4th Ed. Garland Science; New York: 2002. p Varied pagination. [Google Scholar]
- 7.Bethani I, Skanland SS, Dikic I, Acker-Palmer A. Spatial organization of transmembrane receptor signalling. EMBO J. 2010;29(16):2677–2688. doi: 10.1038/emboj.2010.175. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Speers AE, Blackler AR, Wu CC. Shotgun analysis of integral membrane proteins facilitated by elevated temperature. Anal Chem. 2007;79(12):4613–4620. doi: 10.1021/ac0700225. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 9.Chen EI, Cociorva D, Norris JL, Yates JR., 3rd Optimization of mass spectrometry-compatible surfactants for shotgun proteomics. J Proteome Res. 2007;6(7):2529–2538. doi: 10.1021/pr060682a. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen EI, McClatchy D, Park SK, Yates JR., 3rd Comparisons of mass spectrometry compatible surfactants for global analysis of the mammalian brain proteome. Anal Chem. 2008;80(22):8694–8701. doi: 10.1021/ac800606w. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Washburn MP, Wolters D, Yates JR., 3rd Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat Biotechnol. 2001;19(3):242–247. doi: 10.1038/85686. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 12.Moebius J, et al. The human platelet membrane proteome reveals several new potential membrane proteins. Mol Cell Proteomics. 2005;4(11):1754–1761. doi: 10.1074/mcp.M500209-MCP200. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 13.Walikonis RS, et al. Identification of proteins in the postsynaptic density fraction by mass spectrometry. J Neurosci. 2000;20(11):4069–4080. doi: 10.1523/JNEUROSCI.20-11-04069.2000. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jordan BA, et al. Identification and verification of novel rodent postsynaptic density proteins. Mol Cell Proteomics. 2004;3(9):857–871. doi: 10.1074/mcp.M400045-MCP200. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 15.Stevens TJ, Arkin IT. Do more complex organisms have a greater proportion of membrane proteins in their genomes? Proteins. 2000;39(4):417–420. doi: 10.1002/(sici)1097-0134(20000601)39:4<417::aid-prot140>3.0.co;2-y. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 16.Nelson CJ, Hegeman AD, Harms AC, Sussman MR. A quantitative analysis of Arabidopsis plasma membrane using trypsin-catalyzed (18)O labeling. Mol Cell Proteomics. 2006;5(8):1382–1395. doi: 10.1074/mcp.M500414-MCP200. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 17.Chen X, Ulintz PJ, Simon ES, Williams JA, Andrews PC. Global topology analysis of pancreatic zymogen granule membrane proteins. Mol Cell Proteomics. 2008;7(12):2323–2336. doi: 10.1074/mcp.M700575-MCP200. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han CL, et al. A multiplexed quantitative strategy for membrane proteomics: opportunities for mining therapeutic targets for autosomal dominant polycystic kidney disease. Mol Cell Proteomics. 2008;7(10):1983–1997. doi: 10.1074/mcp.M800068-MCP200. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 19.Jia JY, et al. Quantitative proteomics analysis of detergent-resistant membranes from chemical synapses: evidence for cholesterol as spatial organizer of synaptic vesicle cycling. Mol Cell Proteomics. 2006;5(11):2060–2071. doi: 10.1074/mcp.M600161-MCP200. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 20.Gupta N, et al. Quantitative proteomic analysis of B cell lipid rafts reveals that ezrin regulates antigen receptor-mediated lipid raft dynamics. Nat Immunol. 2006;7(6):625–633. doi: 10.1038/ni1337. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 21.Che FY, et al. Comprehensive Proteomic Analysis of Membrane Proteins in Toxoplasma gondii. Mol Cell Proteomics. 2011;10(1):M110. doi: 10.1074/mcp.M110.000745. Translated from eng. 000745 (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y, et al. Enhancing identifications of lipid-embedded proteins by mass spectrometry for improved mapping of endothelial plasma membranes in vivo. Mol Cell Proteomics. 2009;8(6):1219–1235. doi: 10.1074/mcp.M800215-MCP200. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hofmann A, et al. Proteomic cell surface phenotyping of differentiating acute myeloid leukemia cells. Blood. 2010;116(13):e26–34. doi: 10.1182/blood-2010-02-271270. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 24.Boyd RS, et al. Protein profiling of plasma membranes defines aberrant signaling pathways in mantle cell lymphoma. Mol Cell Proteomics. 2009;8(7):1501–1515. doi: 10.1074/mcp.M800515-MCP200. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang H, et al. Quantitative analysis of cell surface membrane proteins using membrane-impermeable chemical probe coupled with 18O labeling. J Proteome Res. 2010;9(5):2160–2169. doi: 10.1021/pr9009113. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Griffin NM, Schnitzer JE. Overcoming key technological challenges in using mass spectrometry for mapping cell surfaces in tissues. Mol Cell Proteomics. 2010 doi: 10.1074/mcp.R110.000935. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yount JS, et al. Palmitoylome profiling reveals S-palmitoylation-dependent antiviral activity of IFITM3. Nat Chem Biol. 2010;6(8):610–614. doi: 10.1038/nchembio.405. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun B, et al. Shotgun glycopeptide capture approach coupled with mass spectrometry for comprehensive glycoproteomics. Mol Cell Proteomics. 2007;6(1):141–149. doi: 10.1074/mcp.T600046-MCP200. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 29.Cravatt BF, Simon GM, Yates JR., 3rd The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450(7172):991–1000. doi: 10.1038/nature06525. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 30.Xiao H, et al. Mapping protein energy landscapes with amide hydrogen exchange and mass spectrometry: I. A generalized model for a two-state protein and comparison with experiment. Protein Sci. 2005;14(2):543–557. doi: 10.1110/ps.041001705. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chandra V, et al. Structure of the intact PPAR-gamma-RXR-alpha nuclear receptor complex on DNA. Nature. 2008:350–356. doi: 10.1038/nature07413. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore CR, et al. Proteolytic fragments of the nicotinic acetylcholine receptor identified by mass spectrometry: implications for receptor topography. Biochemistry. 1989;28(23):9184–9191. doi: 10.1021/bi00449a034. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 33.Kiselar JG, Chance MR. Future directions of structural mass spectrometry using hydroxyl radical footprinting. J Mass Spectrom. 2010;45(12):1373–1382. doi: 10.1002/jms.1808. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hufnagel P, Schweiger U, Eckerskorn C, Oesterhelt D. Electrospray ionization mass spectrometry of genetically and chemically modified bacteriorhodopsins. Anal Biochem. 1996;243(1):46–54. doi: 10.1006/abio.1996.0480. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 35.Ball LE, et al. Mass spectrometric analysis of integral membrane proteins: application to complete mapping of bacteriorhodopsins and rhodopsin. Protein Sci. 1998;7(3):758–764. doi: 10.1002/pro.5560070325. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Whitelegge JP, Gundersen CB, Faull KF. Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein Sci. 1998;7(6):1423–1430. doi: 10.1002/pro.5560070619. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Busenlehner LS, et al. Stress sensor triggers conformational response of the integral membrane protein microsomal glutathione transferase 1. Biochemistry. 2004;43(35):11145–11152. doi: 10.1021/bi048716k. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 38.Englander SW, Sosnick TR, Englander JJ, Mayne L. Mechanisms and uses of hydrogen exchange. Curr Opin Struct Biol. 1996;6(1):18–23. doi: 10.1016/s0959-440x(96)80090-x. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shaw BF, et al. Neutralizing Positive Charges at the Surface of a Protein Lowers Its Rate of Amide Hydrogen Exchange without Altering Its Structure or Increasing Its Thermostability. J Am Chem Soc. 2010 doi: 10.1021/ja9067035. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Joh NH, et al. Modest stabilization by most hydrogen-bonded side-chain interactions in membrane proteins. Nature. 2008;453(7199):1266–1270. doi: 10.1038/nature06977. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, et al. Dynamics of the beta2-adrenergic G-protein coupled receptor revealed by hydrogen-deuterium exchange. Anal Chem. 2010;82(3):1100–1108. doi: 10.1021/ac902484p. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pan Y, Stocks BB, Brown L, Konermann L. Structural characterization of an integral membrane protein in its natural lipid environment by oxidative methionine labeling and mass spectrometry. Anal Chem. 2009;81(1):28–35. doi: 10.1021/ac8020449. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 43.Pan Y, Brown L, Konermann L. Site-directed mutagenesis combined with oxidative methionine labeling for probing structural transitions of a membrane protein by mass spectrometry. J Am Soc Mass Spectrom. 2010;21(11):1947–1956. doi: 10.1016/j.jasms.2010.08.004. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 44.Weinglass AB, et al. Elucidation of substrate binding interactions in a membrane transport protein by mass spectrometry. EMBO J. 2003;22(7):1467–1477. doi: 10.1093/emboj/cdg145. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hebling CM, et al. Conformational analysis of membrane proteins in phospholipid bilayer nanodiscs by hydrogen exchange mass spectrometry. Anal Chem. 2010;82(13):5415–5419. doi: 10.1021/ac100962c. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen J, Rempel DL, Gross ML. Temperature jump and fast photochemical oxidation probe submillisecond protein folding. J Am Chem Soc. 2010;132(44):15502–15504. doi: 10.1021/ja106518d. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Engelhardt J, et al. CKAMP44: a brain-specific protein attenuating short-term synaptic plasticity in the dentate gyrus. Science. 2010;327(5972):1518–1522. doi: 10.1126/science.1184178. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 48.Kreienkamp HJ. Organisation of G-protein-coupled receptor signalling complexes by scaffolding proteins. Curr Opin Pharmacol. 2002;2(5):581–586. doi: 10.1016/s1471-4892(02)00203-5. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 49.Schwenk J, et al. Native GABA(B) receptors are heteromultimers with a family of auxiliary subunits. Nature. 2010;465(7295):231–235. doi: 10.1038/nature08964. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 50.Zhang W, et al. A transmembrane accessory subunit that modulates kainate-type glutamate receptors. Neuron. 2009;61(3):385–396. doi: 10.1016/j.neuron.2008.12.014. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Selimi F, Cristea IM, Heller E, Chait BT, Heintz N. Proteomic studies of a single CNS synapse type: the parallel fiber/purkinje cell synapse. PLoS Biol. 2009;7(4):e83. doi: 10.1371/journal.pbio.1000083. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Muller CS, et al. Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc Natl Acad Sci U S A. 2010;107(34):14950–14957. doi: 10.1073/pnas.1005940107. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Helbig AO, Heck AJ, Slijper M. Exploring the membrane proteome--challenges and analytical strategies. J Proteomics. 2010;73(5):868–878. doi: 10.1016/j.jprot.2010.01.005. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 54.Schagger H, Cramer WA, von Jagow G. Analysis of molecular masses and oligomeric states of protein complexes by blue native electrophoresis and isolation of membrane protein complexes by two-dimensional native electrophoresis. Anal Biochem. 1994;217(2):220–230. doi: 10.1006/abio.1994.1112. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 55.de Wit J, et al. LRRTM2 interacts with Neurexin1 and regulates excitatory synapse formation. Neuron. 2009;64(6):799–806. doi: 10.1016/j.neuron.2009.12.019. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ko J, Fuccillo MV, Malenka RC, Sudhof TC. LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron. 2009;64(6):791–798. doi: 10.1016/j.neuron.2009.12.012. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maddalo G, et al. A systematic analysis of native membrane protein complexes in Escherichia coli. J Proteome Res. 2011;10:1848–1859. doi: 10.1021/pr101105c. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 58.Barrera NP, et al. Mass spectrometry of membrane transporters reveals subunit stoichiometry and interactions. Nat Methods. 2009;6(8):585–587. doi: 10.1038/nmeth.1347. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321(5886):243–246. doi: 10.1126/science.1159292. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 60.Dengjel J, Kratchmarova I, Blagoev B. Receptor tyrosine kinase signaling: a view from quantitative proteomics. Mol Biosyst. 2009;5(10):1112–1121. doi: 10.1039/b909534a. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 61.Huang PH, White FM. Phosphoproteomics: unraveling the signaling web. Mol Cell. 2008;31(6):777–781. doi: 10.1016/j.molcel.2008.09.001. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tang W, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321(5888):557–560. doi: 10.1126/science.1156973. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Blagoev B, et al. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21(3):315–318. doi: 10.1038/nbt790. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 64.Blagoev B, Ong SE, Kratchmarova I, Mann M. Temporal analysis of phosphotyrosine-dependent signaling networks by quantitative proteomics. Nat Biotechnol. 2004;21(3):315–318. doi: 10.1038/nbt1005. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 65.Olsen JV, et al. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127(3):635–648. doi: 10.1016/j.cell.2006.09.026. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 66.Zhang G, Fenyo D, Neubert TA. Screening for EphB signaling effectors using SILAC with a linear ion trap-orbitrap mass spectrometer. J Proteome Res. 2008;7(11):4715–4726. doi: 10.1021/pr800255a. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Spellman DS, Deinhardt K, Darie CC, Chao MV, Neubert TA. Stable isotopic labeling by amino acids in cultured primary neurons: application to brain-derived neurotrophic factor-dependent phosphotyrosine-associated signaling. Mol Cell Proteomics. 2008;7(6):1067–1076. doi: 10.1074/mcp.M700387-MCP200. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mayya V, et al. Quantitative phosphoproteomic analysis of T cell receptor signaling reveals system-wide modulation of protein-protein interactions. Sci Signal. 2009;2(84):ra46. doi: 10.1126/scisignal.2000007. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 69.Jorgensen C, et al. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science. 2009;326(5959):1502–1509. doi: 10.1126/science.1176615. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 70.Choudhary C, et al. Mislocalized activation of oncogenic RTKs switches downstream signaling outcomes. Mol Cell. 2009;36(2):326–339. doi: 10.1016/j.molcel.2009.09.019. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 71.Bantscheff M, et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol. 2007;25(9):1035–1044. doi: 10.1038/nbt1328. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 72.Coba MP, et al. Neurotransmitters drive combinatorial multistate postsynaptic density networks. Sci Signal. 2009;2(68):ra19. doi: 10.1126/scisignal.2000102. Translated from eng. in eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Koskenniemi K, et al. Proteomics and Transcriptomics Characterization of Bile Stress Response in Probiotic Lactobacillus rhamnosus GG. Mol Cell Proteomics. 2011;10(2):M110. doi: 10.1074/mcp.M110.002741. Translated from eng. 002741 (in eng) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ahrens CH, Brunner E, Qeli E, Basler K, Aebersold R. Generating and navigating proteome maps using mass spectrometry. Nat Rev Mol Cell Biol. 2010;11(11):789–801. doi: 10.1038/nrm2973. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]
- 75.Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5(4):573–588. doi: 10.1074/mcp.M500331-MCP200. Translated from eng. in eng. [DOI] [PubMed] [Google Scholar]