Abstract
We previously developed a fermentation protocol for lipid accumulation in the oleaginous yeast Y. lipolytica. This process was used to perform transcriptomic time-course analyses to explore gene expression in Y. lipolytica during the transition from biomass production to lipid accumulation. In this experiment, a biomass concentration of 54.6 gCDW/l, with 0.18 g/gCDW lipid was obtained in ca. 32 h, with low citric acid production. A transcriptomic profiling was performed on 11 samples throughout the fermentation. Through statistical analyses, 569 genes were highlighted as differentially expressed at one point during the time course of the experiment. These genes were classified into 9 clusters, according to their expression profiles. The combination of macroscopic and transcriptomic profiles highlighted 4 major steps in the culture: (i) a growth phase, (ii) a transition phase, (iii) an early lipid accumulation phase, characterized by an increase in nitrogen metabolism, together with strong repression of protein production and activity; (iv) a late lipid accumulation phase, characterized by the rerouting of carbon fluxes within cells. This study explores the potential of Y. lipolytica as an alternative oil producer, by identifying, at the transcriptomic level, the genes potentially involved in the metabolism of oleaginous species.
Introduction
In a context of increasing concern about global warming and dwindling stocks of fossil fuels, the search for alternative, renewable sources of energy is now a matter of the utmost importance for modern societies. Biodiesel has rapidly become one of the most promising and widely studied alternative sources of energy.
Commonly, biodiesel is produced from refined or edible oils using methanol and an alkaline catalyst. However, the large-scale production of biodiesel requires considerable amounts of these oils, resulting in a sharp increase of their demand over the last decade. The eventual goal is to gain access to a sustainable energy source, as traditional methods of obtaining oils from plants have raised unexpected ecological and sociological issues (e.g. extensive use of arable land, replacement of food crops with fat-producing crops for biofuel production). As a consequence, the development of new production procedures from non-edible oils appears to be an essential prerequisite for a sustainable biodiesel industry. To that end, several esterification processes are currently developed for the utilization of these oils, often containing free fatty acids (FFA) (e.g. two-step esterification process [1], supercritical methanol esterification [2], lipase esterification [3]).
Microorganisms, including yeasts in particular, have long been studied as alternative sources of oils and fats [4], [5]. Under specific conditions, they synthesize and store lipids in the form of triacylglycerols (TAG) and sterol esters (SE) inside a special compartment of the cell, called the lipid body (LB). These neutral lipids serve as energy source for the cell when required. Some species have been reported to accumulate more than 20% of their dry cell mass in the form of lipids, and have been classified as “oleaginous” microorganisms [6], [7]. Oleaginous yeast species (e.g. Rhodotorula glutinis, Lipomyces starkeyi) are particularly promising in this respect, as they can accumulate more than 70% of their dry cell weight as lipids [7]. Additionally, they can present various fatty acid profiles, depending on the species and/or growth conditions. These features, combined with the ease of genetic manipulation and cultivation, make yeasts a target of choice for potential applications like nutritional supplements production (e.g. production of polyunsaturated fatty acids), or as oil providers for biodiesel production [3], [4].
Yarrowia lipolytica is one of the most widely studied “nonconventional” oleaginous yeast species [8], [9]. It has been isolated from various food-related environments (e.g. cheese, sausage), but also from sewage, soils and oil fields [10]. Its classification by the American Food and Drug Administration as “Generally Recognized As Safe” (GRAS) paved the way for the development of various biotechnological applications, including (i) heterologous protein production [11], (ii) organic acids production [12], and (iii) single-cell oil productions from agroindustrial by-products or wastes [13]. Under specific growth conditions, Y. lipolytica accumulates large amounts of lipid, sometimes accounting for more than 50% of its dry cell weight [14]. One of the major advantages of this yeast is its ability to use hydrophobic substrates (e.g. alkanes, fatty acids, oils) efficiently as a sole carbon source [10], [15]. Y. lipolytica cells accumulate large amounts of lipids on these substrates, using specialized protrusions formed on their cell surface to facilitate the uptake of hydrophobic compounds [16]. These characteristics, together with the availability of the complete genome sequence through the work of the Genolevure Consortium [17], render Y. lipolytica a model of choice for investigations of lipid accumulation in oleaginous yeast species. Various studies have already made use of the genome sequence to decipher aspects of lipid metabolism in Y. lipolytica, and some of the genes involved in the bioconversion, synthesis and mobilization of lipids have been described [18]. Furthermore, the availability of genetic information makes this yeast a suitable candidate for genetic and metabolic engineering approaches aiming to develop optimized yeast strains for the production and storage of large amounts of lipid with a specific fatty acid (FA) composition [15]. In addition, lipid composition of Y. lipolytica is mainly consisted of the C16–C18 fatty acid families, making this yeast attractive to biotechnological applications, such as biodiesel production [19].
Oleaginous microorganisms have been studied over decades, but the general mechanisms underlying their metabolic specificities remain unclear. Lipid accumulation has been described as a consequence of the slower growth observed when oleaginous organisms are subject to nutrient shortages (e.g. nitrogen deficiency) while growing on an excess of carbohydrates [7]. It has therefore been suggested that lipid accumulation in oleaginous organisms is the consequence of a stress response or of an adaptation to a nutrient shift [20]. Only a few studies have focused on the proteomics of lipid accumulation in oleaginous yeasts [20], [21]. Some attempts have been made to describe potential set-ups for the large-scale production of single cell oils (SCO) [22]. However, the optimization of these set-ups will require improvements in our understanding of the phenomena involved at the various “omics” levels. In particular, a complete transcriptomic study appears essential to an understanding of the cascade of metabolic processes involved in the transition from growth to lipid accumulation, for insight into regulatory mechanisms and, ultimately, for the identification of targets of interests for further genetic and metabolic engineering.
In this study, a controlled fed-batch culture was carried out to monitor the transition from biomass production to lipid accumulation under defined conditions. Storage lipid and transcriptomic time-course analyses were performed to explore the accumulation and metabolism of lipids in Y. lipolytica. This study constitutes the first attempt to unravel the transcriptomic response of an oleaginous yeast during its metabolic shift from growth to lipid accumulation.
Materials and Methods
Strain, growth conditions, and fed-batch culture strategy
Y. lipolytica mutant strain JMY1346 was used in this study. JMY1346 is a prototroph derivative of strain JMY1202, which was previously obtained by deletion of the GUT2 gene in the auxotrophic strain Po1d (Leu−, Ura−) [18]. One frozen stock culture was used for the inoculation of a two-step preculture, which was further used to inoculate the fed-batch in defined mineral medium (see Table S1) [23].
Fed-batch cultures were performed in a 20 l bioreactor, with the Braun Biostat E fermenting system (Braun, Germany), without oxygen limitation. The temperature was set to 28°C and the pH to 5.8. Custom-built software was used for online acquisition and regulation of the controlled parameters (i.e. stirring rate, pH, temperature, relative pressure, partial pressure of dissolved oxygen, additions of bases and antifoaming agent). Relative pressure in the bioreactor was maintained at 0.3 bar. No more than 0.2 ml of antifoaming agent (Struktol JG73, Schill+Seilacher group, Germany) was added to the culture. During fed-batch culture, the bioreactor was supplied with three sterile feeds (glucose, salt and base, i.e. ammonia or potassium hydroxide), via Masterflex and Gilson peristaltic pumps (Cole-Parmer Instrument Company, USA, Gilson Inc., USA). Glucose feed concentration was 740 g.l−1. The masses of glucose and nitrogen added to the fermentor were estimated online, by weighing (CPA16001S, Sartorius, Germany). Outlet gas composition (after condensation) was analyzed by mass spectrometry (PRIMA 600 s, VG Gas, United Kingdom). O2 consumption and CO2 production rates were calculated from mass balances, taking into account changes in gas volume in the reactor, inlet airflow (as measured with a mass flow meter, Brooks, USA), temperature, humidity and pressure. The glucose concentration within the bioreactor was evaluated with custom-built software based on carbon mass balance and taking into account various data acquired online (i.e. glucose mass, gas analysis and inlet/outlet gas flow).
The fed-batch culture was divided into three phases, based on different carbon and nitrogen feeding strategies [23]: (I) growth phase, (II) transition phase, and (III) nitrogen limitation. During the growth phase (I), glucose flow was exponential, to ensure a constant specific growth rate without nutrient limitation. Nitrogen was supplied via 10 mol.l−1 ammonia solution, which was also used for pH regulation. The transition phase (II) corresponded to nitrogen limitation in the presence of excess carbon. This phase could be divided into two parts: (i) a decrease in the nitrogen concentration of the broth, (ii) a transition of nitrogen input from the pH regulation pump to an independent peristaltic pump. The starting point of phase II may thus be considered to correspond to the beginning of nitrogen depletion, triggered by shifting the pH-regulating solution from ammonia (10 mol.l−1) to potassium hydroxide (10 mol.l−1). Once the nitrogen concentration fell below 10 mmol.l−1, a controlled supply of 5 mol.l−1 ammonia solution was initiated, leading to a stabilization of the Carbon/Nitrogen (C/N) ratio near ca. 30 Cmol.mol−1. The lipid accumulation phase (III) began when the C/N ratio reached ca. 20 Cmol.mol−1, as established by Granger et al. [24]. After 33 h, the glucose supply was limited, to stop lipid accumulation. Throughout the experiment, samples (ca. 300 mg of cell dry weight) were harvested, frozen in liquid nitrogen, and stored at −80°C.
Biomass analyses
Biomass production was determined by measuring A600 and cell dry weight, as estimated for three replicates after filtration and drying (200 mmHg, 60°C, for 48 h, until a constant weight was reached). Ash composition was determined after two complete combustions in a muffle furnace at 550°C for 12 h, in the presence of 200 µl of a 20 g.l−1 NH4NO3 solution. The biomass formula was determined by elemental analysis of C, H, O and N. The total fatty acid content of the dried samples was analyzed as described previously [25].
Supernatant analyses
The sugar and organic acid concentrations of filtered supernatants were determined by HPLC (Ultimate 3000, Dionex, USA) with an Aminex HPX-87H+ column (Bio-Rad, USA), under the following conditions: 50°C, with 5 mM H2SO4 as the eluent (flow rate 0.5 ml.min−1) and dual detection with a refractometer at 50°C (Shodex, Japan) and UV measurement at 210 nm (Dionex, USA). Standards were used for compound identification and quantification. The glucose concentration of culture supernatants was also determined with a YSI Model 27 A glucose analyzer (Yellow Springs Instruments, USA). The residual ammonium concentration in the culture medium was determined with an ammonium ion electrode (PH/ISE meter model 710A+Ammonia Gas-Sensing Electrode Model 95-12, Orion Research Inc., USA). A combination of the various macroscopic analyses was used to calculate carbon mass and redox balances with a maximal error of ca. 4%.
RNA extractions and microarrays
We used 11 sampling points, regularly spaced over the period of fed-batch culture, for transcriptomic analysis. Frozen samples were mechanically disrupted with a bead beater (Microdismenbrator, Braun, Germany) and a tungsten bead (Ø∼7 mm), for 2 min, at 2600 rpm. RNA was extracted from the resulting powder with the RNEasy Midi Kit (Qiagen, The Netherlands). The quality and quantity of RNA were assessed by capillary microelectrophoresis, with an RNA 600 Nano LabChips and a Bioanalyzer 2100 (Agilent, USA). The mRNA obtained was reverse transcribed and labeled using the ChipShot™ Direct Labeling kit (Promega, USA). Each sample was labeled with Cy5, and a mixture of all the RNA samples was labeled with Cy3 and used as a reference. The resulting labelled cDNAs were further purified using the ChipShot™ membrane Clean-up system (Promega, USA). Microarray probes were designed and slides were produced by Eurogentec (Belgium). Samples were hybridized in the Discovery XT System (Ventana Medical Systems Inc., Roche, Switzerland). Microarray slides were prehybridized with a solution of 2× SSC, 0.2% SDS, 1% BSA at 42°C for 30 min. 200 µl of the hybridization solution, containing 20 µL of labeled cDNA (50 pmol of Cy3 reference sample and 50 pmol of Cy5 sample of interest) and 180 µL of hybridization buffer (Chyp Hyb buffer, Ventana Medical System) were added on the printed side of the slide. Arrays were scanned with an Innoscan 700 Microarray Scanner (Innopsys, France) and fluorescence was measured with GenePix Pro v3.0 software (Molecular Devices, USA).
Data filtration, normalization and statistical analyses
Raw transcriptomic data were filtered and normalized with R software [26], and the Limma package of the Bioconductor library [27], [28]. A preliminary filtration of the dataset was carried out with the quality flags provided by GenePix software. Spots with a quality flag value below “0” were removed from the analysis. Local background estimates were corrected by the “normexp+offset” method, using an offset value of 50 [29]. Background levels were subtracted from the data, which were then further normalized by the PrintTip Loess method [30]. Where possible, missing values for the filtration and normalization processes were simulated with the iKNN algorithm [31]. The normalized data were analyzed further, with MeV software [32]. First, the samples were clustered hierarchically on the basis of mean linkage and Euclidean distance. A 100 dendrograms were simulated for the calculation of bootstrap values, and the resulting tree was drawn with the “ape” package for R [33]. Samples were group based on the resolved clusters, and two-class unpaired Significance Analysis of Microarrays (SAM) tests [34] were performed between the resulting groups of samples. Genes were identified as differentially expressed in cases of significant detection, with a false discovery rate lower than 1×10−5 and an absolute fold change between two groups of more than 1.5. K-means clustering was performed on the expression profiles of the identified genes, using Pearson correlation as distance metric, and a maximum number of 50 iterations as convergence criteria. Functional classification of the genes was performed according to Gene Ontology Terms [35] defined during the genome annotation of Y. lipolytica [17], and by comparisons with homologous genes in S. cerevisiae. The data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus [36] and are accessible through GEO Series accession number GSE29046 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE29046).
Results
A fed-batch lipid accumulation process for transcriptomic analysis
In oleaginous microorganisms, the initiation of lipid accumulation during lipid synthesis is caused by the exhaustion of a primary nutrient from the culture medium. Although many nutrients can be limiting, nitrogen limitation is the easiest condition to control and is generally the most efficient type of limitation for inducing lipid accumulation. During the growth phase, the carbon flux is distributed between the four macromolecular pools (carbohydrate, lipid, nucleic acid, protein). When nitrogen becomes unavailable, the catalytic growth rate slows down rapidly, whereas the rate of carbon assimilation slows more gradually [7], [37]. This results in the preferential channelling of carbon flux toward lipid synthesis, leading to an accumulation of triacylglycerols within the lipid body of the cell. When carbon is present in large excess, its uptake is limited only by the substrate transport system of the cell. In this case, limiting concentrations of nitrogen in the medium lead to the induction of lipid accumulation. The critical nitrogen concentration for lipid induction in Y. lipolytica has been found to be about 10−3 mol l−1 [23]. It is important for nitrogen concentration to exceed this threshold value to prevent the production of secondary metabolites (citric acid) that will otherwise affect lipid accumulation.
During the transition between the growth phase (growth with the production of catalytic biomass) and the lipid accumulation phase (decrease in growth rate due to nutrient limitation and the diversion of excess carbon to lipid production), some pathways are repressed (nucleic acid and protein synthesis), whereas others are induced (fatty acid and triacylglycerol synthesis). When non-oleaginous microorganisms are placed in the same nutrient-limiting conditions the available carbohydrate substrate, is diverted into various polysaccharides, including glycogen and various glucans and mannans. Here, we try to identify, at the transcriptomic level, the genes potentially accountable of the oleaginous character of the cell in Y. lipolytica.
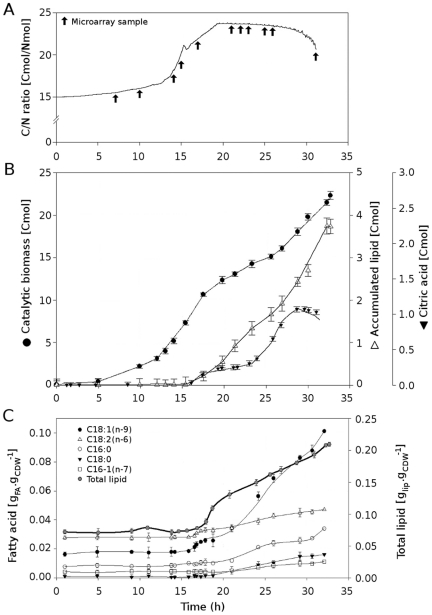
Growth and lipid accumulation were monitored during the culture (Figure 1). Evolution of parameters such as C/N ratio, biomass production and lipid accumulation can be divided into three major phases: (i) a growth phase, from the start of the culture to ca. 15 h, (ii) a short transition phase and (iii) a lipid accumulation phase from ca. 18 to 33 h. The first phase was characterized by an exponential increase of biomass, with a specific growth rate of 0.27 h−1, and a stable C/N consumption ratio of 15.5±1 Cmol/Nmol (Figure 1A and 1B). During this phase, lipid level and composition remained stable (Figure 1C). The second phase can be described as the entry into nitrogen limitation (15 h), resulting in a break in the biomass accumulation curve, with a decrease in specific growth rate from 0.27 h−1 to 0.07 h−1. The cessation of nitrogen feeding was effective from ca. 15 h (Figure 1A), but the nitrogen supply was not exhausted until 18.5 h. This second phase can be considered as an adaptation period, corresponding to the time required for the biomass to use all of the nitrogen present in the medium. During this phase, the C/N consumption ratio increased to 23 Cmol/Nmol (Figure 1A). The third phase began when lipid accumulation effectively increased, starting from 18.7 h (Figure 1B). Once the lipid production phase had begun, lipid content appeared to increase steadily until ca. 33 h. This pattern of lipid accumulation was linked to the linear glucose and nitrogen feed (i.e. constant C/N ratio, as described in Figure 1A). Maximum glucose concentration was below 0.2 g.l−1 during phase III, with the accumulation of less than 1 Cmol of citric acid (Figure 1B), i.e. 2.9 g.l−1. The fatty acid profile of the cells did not change significantly upon entry into the nitrogen limitation phase (Figure 1C). Lipid content began to change significantly after ca. 17.5 h. Lipid accumulation was characterized principally by an increase in C18:1n-9 content, with a mean specific accumulation rate of 0.006 g.gCDW −1.h−1, and by a smaller increase in C18:2n-6,9 and C16:0 content (0.002 g.gCDW −1.h−1). Although not being under optimal lipid accumulation conditions, carbon overflow was avoided since citrate production was very low [8].
Figure 1. Macrokinetic and storage lipid analyses of the fed-batch culture.
(A) changes in C/N ratio, as monitored during the fed-batch culture; (B) catalytic biomass, accumulated lipid and citric acid production; (C) total lipid and fatty acid production. Mean values and error bars were calculated using three replicates.
Identification of global transcriptomic responses during the fed-batch process
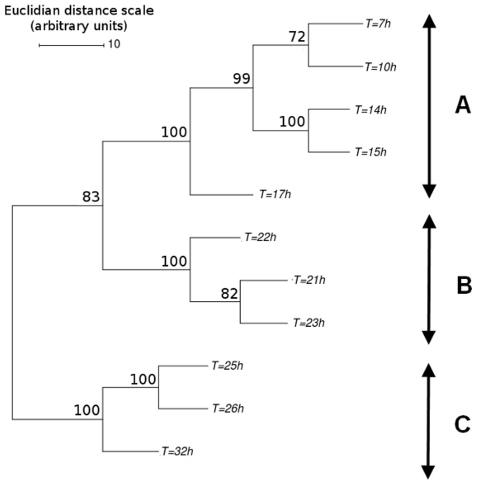
Hierarchical clustering analysis was carried out on the transcriptomic profiles of the samples, based on average linkage, Euclidean distance and 100 bootstrap replicates. The resulting dendrogram (Figure 2) highlights a sequential change in the global transcriptomic response. Different metabolic steps can thus be distinguished, mostly resembling those observed at the macroscopic scale, but with a few important differences. A first group of samples (Figure 2, group A) correspond to the growth phase, as identified in macrokinetic analyses. However, despite the switch to nitrogen-limiting conditions at ca. 15 h, the global transcriptomic response of these samples does not change until much later, with no major changes detected until T = 17 h. The transition phase is probably a transient state, highlighting a progressive transcriptomic response that will affect later stages of the fed batch culture. Finally, the lipid accumulation phase, as described in the macrokinetic approach, could be subdivided into two steps on the basis of transcriptomic analysis: an early phase (from T = 21 h to T = 23 h), and a late phase (from T = 25 h to T = 32 h). This distinction provides the first indication of a dual transcriptomic response to nitrogen-limiting conditions, which will be discussed further below.
Figure 2. Hierarchical clustering of the global transcriptomic profiles obtained during the fed-batch culture.
Bootstrap values were calculated from a 100 tree replicates. Three metabolic phases can be distinguished at the transcriptomic level: (A) biomass production, (B) early lipid accumulation, and (C) late lipid accumulation. Euclidian distance scale, as calculated by MeV software, is given in arbitrary units.
Differential expression during the transition to lipid accumulation
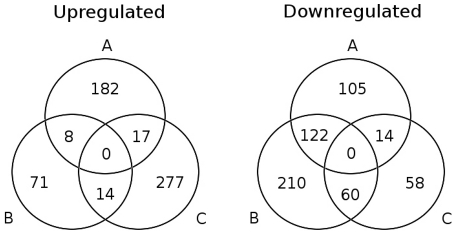
SAM tests were performed between each of the three transcriptomic response groups identified above. 569 different genes were identified as significantly over- or under-expressed in at least two of the three transcriptomic subsets (Table S2). Pairwise comparisons of the identified subsets provided a global overview of the transcriptomic response (Figure 3). We found that 207 genes were overexpressed during the biomass production phase, whereas 93 genes were significantly upregulated during the early accumulation phase and 308 genes were significantly upregulated during the late accumulation phase. Only a limited number of these genes (ca. 7%) appeared to be upregulated during two different phases, highlighting a time-related specificity of the upregulation response. Most of the genes upregulated after the switch to nitrogen-limiting conditions (i.e. genes detected as upregulated during either phase B and/or C) were expressed during the late accumulation phase (ca. 80%). Downregulation was observed for 241 genes during biomass production, 392 genes during the early accumulation phase and 132 genes during the late accumulation phase. The downregulation response appeared to be a slower, transient phenomenon, with large numbers of genes downregulated in consecutive phases. Most repressed genes (ca. 69%) are detected during the early lipid accumulation phase, highlighting the decrease in metabolic activity constituting the primary response to nitrogen starvation.
Figure 3. Distribution of genes identified as differentially expressed during the transition to lipid accumulation.
Three metabolic states are distinguished during fed-batch : (A) biomass production, (B) early lipid accumulation, and (C) late lipid accumulation.
The genes identified as specifically expressed in one of the three transcriptomic phases were further classified into 20 functional categories, based on GO Terms (Table 1). Many metabolic processes were found to be involved in the various phases, but some functional categories were particularly frequently represented. In particular, genes involved in the cell cycle and in cellular component biogenesis and organization were numerous. A large proportion of these genes displayed downregulation after the imposition of nitrogen limitation, reflecting the slower growth observed at the start of the accumulation phase. Genes associated with translation followed a similar pattern. Other major categories identified were linked to the stress response, nucleic acid metabolism and transport. Lipid metabolism accounted for only a small proportion of genes, most of which were overexpressed during the late accumulation phase.
Table 1. Functional classification of the upregulated/downregulated genes identified during the transition to lipid accumulation.
| Number of genes up/downregulated during each transcriptomic phase | |||
| Functional category | A | B | C |
| Amino acid metabolism | 11/6 | 4/12 | 15/3 |
| Carbohydrate metabolism | 8/4 | 4/9 | 10/4 |
| Catabolism | 0/5 | 3/2 | 6/1 |
| Cell cycle | 12/10 | 4/17 | 34/3 |
| Cellular aromatic compound metabolism | 7/1 | 2/5 | 2/0 |
| Cellular component biogenesis | 34/4 | 1/25 | 14/3 |
| Cellular component organization | 52/17 | 11/51 | 64/12 |
| Cellular metabolism | 16/4 | 7/15 | 14/7 |
| Lipid metabolism | 2/3 | 1/12 | 17/0 |
| Nucleic acids metabolism | 48/14 | 5/39 | 46/8 |
| Protein metabolism | 1/1 | 0/1 | 5/0 |
| Protein modification | 13/8 | 4/16 | 19/4 |
| Response to stimulus | 20/12 | 10/24 | 36/5 |
| Signalling | 8/1 | 0/9 | 10/1 |
| Transcription | 7/7 | 3/7 | 20/3 |
| Translation | 74/2 | 1/52 | 10/1 |
| Transport | 29/22 | 14/28 | 42/8 |
| Transposition | 0/0 | 0/0 | 1/0 |
| Vitamin metabolism | 2/0 | 1/1 | 2/2 |
| Unknown | 37/45 | 30/65 | 119/27 |
(A) biomass production, (B) early lipid accumulation, and (C) late lipid accumulation. Functional categories were determined based on GO Terms defined during the annotation of Y. lipolytica, and by comparisons with homologous genes in S. cerevisiae.
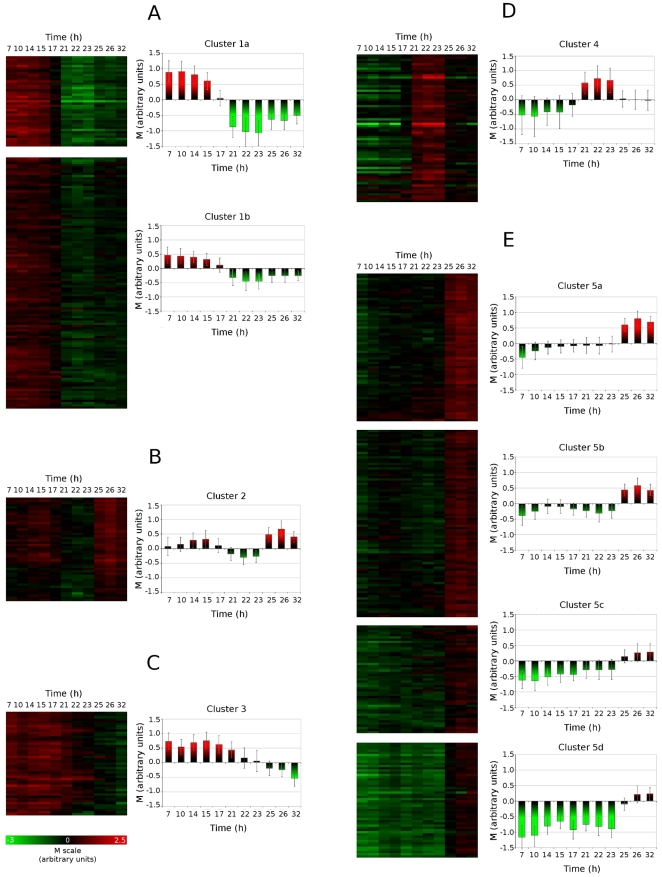
Additional classification by K-means clustering was performed to obtain further insight into the transcriptomic response (particularly as concerns the chain of metabolic events preceding and following nitrogen limitation). The transcriptomic profiles of the 569 differentially expressed genes were resolved into nine clusters, based on mean expression profiles (Figure 4). Some clusters could be further regrouped into similar response patterns, but with different intensities. Clusters 1 to 3 contained 249 genes repressed at one point during the course of the fed-batch process. Clusters 1a and 1b contained 43 and 112 genes, respectively, that were strongly repressed upon entry into the nitrogen limitation phase. Cluster 2 corresponds to a transient repression phenomenon and includes 47 genes downregulated in the early accumulation phase but recovering significant levels of expression in the late stages of lipid production. By contrast, cluster 3 included 47 genes that were strongly downregulated during the late accumulation phase. In parallel, clusters 4 and 5 contained 320 genes overexpressed at one point during the lipid accumulation phase. Cluster 4, in particular, may be considered to correspond to an immediate response to nitrogen limitation, whereas clusters 5a, 5b 5c and 5d contain mostly genes upregulated during the late accumulation phase. The major genes identified in these clusters are presented in Tables 2, 3 and S3, and their role in the chain of metabolic events is discussed below.
Figure 4. K-Mean clustering of the differentially expressed genes identified during the transition to lipid accumulation.
The 569 genes (detailed in Table S2) were resolved into 9 clusters, which could further be classified into 5 response profiles: (A) downregulation upon nitrogen limitation, (B) transient repression, followed by late overexpression, (C) downregulation during late accumulation phase, (D) upregulation upon nitrogen limitation, and (E) upregulation during late accumulation phase. Mean expression values and error bars were calculated, based on the expression profiles of the genes identified in each clusters.
Table 2. Genes involved in energy and lipid metabolism, identified as differentially expressed during the transition to lipid accumulation.
| Fold Change (arbitrary units) | ||||||
| Gene Labels | Gene Names | Description | Cluster | B/A | C/B | C/A |
| Glyoxylate bypass | ||||||
| YALI0C16885g | ICL | Isocitrate lyase | 1b | 0.53 | 1.09 | 0.57 |
| YALI0D19140g | MLS1 | Malate synthase 1, glyoxysomal | 4 | 2.09 | 0.29 | 0.61 |
| TCA cycle | ||||||
| YALI0D16753g | MDH1 | Malate dehydrogenase, mitochondrial | 3 | 0.67 | 0.85 | 0.57 |
| YALI0D09361g | ACO1 | Aconitase | 5a | 1.03 | 1.58 | 1.64 |
| Cellular energy homeostasis | ||||||
| YALI0B00704g | ADK | Adenylate kinase | 5b | 0.83 | 1.55 | 1.29 |
| YALI0F26521g | ADK2 | Adenylate kinase | 4 | 1.2 | 0.64 | 0.77 |
| Oxidative phosphorylation | ||||||
| YALI0A20680g | n.a. | NADH dehydrogenase | 2 | 0.76 | 1.58 | 1.21 |
| YALI0E23089g | n.a. | NADH dehydrogenase | 3 | 0.86 | 0.65 | 0.56 |
| YALI0A02651g | n.a. | NADH dehydrogenase | 5c | 1.15 | 1.75 | 2 |
| YALI0E08617g | VMA13 | Vacuolar H+ ATPase, H subunit | 3 | 1.16 | 0.54 | 0.62 |
| YALI0F09405g | PPA1/VMA16 | Vacuolar H+ ATPase, c” subunit | 5b | 1.1 | 1.86 | 2.05 |
| YALI0B03982g | ATP2 | ATP synthase, beta chain | 3 | 0.68 | 0.77 | 0.52 |
| CoenzymeA synthesis | ||||||
| YALI0F09625g | DPCK | Dephospho-CoA kinase | 4 | 1.71 | 0.89 | 1.52 |
| Pentose phosphate pathway | ||||||
| YALI0B00836g | PRS5 | Ribose-phosphate pyrophosphokinase | 1b | 0.61 | 1.26 | 0.76 |
| YALI0B15598g | GND1 | 6-phosphogluconate dehydrogenase | 1b | 0.76 | 0.82 | 0.62 |
| YALI0F07711g | PGI1 | Glucose-6-phosphate isomerase | 3 | 1.35 | 0.59 | 0.8 |
| Glycolysis/gluconeogenesis | ||||||
| YALI0C06369g | GAPDH | Glyceraldehyde-3-P dehydrogenase | 3 | 0.93 | 0.64 | 0.59 |
| YALI0F07711g | PGI1 | Glucose-6-phosphate isomerase | 3 | 1.35 | 0.59 | 0.8 |
| YALI0B02728g | PGAM1 | Phosphoglycerate mutase | 1b | 0.66 | 0.93 | 0.61 |
| YALI0E00264g | ALD4 | Aldehyde dehydrogenase | 5c | 0.74 | 1.9 | 1.4 |
| YALI0E17787g | n.a. | Alcohol dehydrogenase 2 | 1b | 0.49 | 1.88 | 0.92 |
| Lipid metabolism | ||||||
| YALI0C05951g | FAD1 | Delta-9 fatty acid desaturase | 5c | 0.87 | 1.65 | 1.44 |
| YALI0F06578g | DGA2 | Diacylglycerol acyltransferase | 5a | 1.16 | 1.89 | 2.2 |
| YALI0D05995g | SAC1 | Inositol/phosphatidylinositol phosphatase | 5a | 0.93 | 1.96 | 1.82 |
| YALI0E11561g | LIP15 | Lipase | 5b | 0.73 | 2.13 | 1.54 |
| YALI0C03003g | n.a. | Peroxisomal 2,4-dienoyl-CoA reductase | 5b | 0.9 | 1.68 | 1.51 |
| YALI0E14322g | n.a. | Peroxisomal 2,4-dienoyl-CoA reductase | 5b | 1.14 | 1.54 | 1.76 |
| YALI0F23793g | FALDH1 | Fatty aldehyde dehydrogenase | 5b | 1.13 | 1.45 | 1.65 |
Genes were identified as differentially expressed in cases of significant detection, with a false discovery rate lower than 1×10−5 and an absolute fold change between two groups of more than 1.5. Gene labels and descriptions were attributed according to the Genolevure database (http://www.genolevures.org/). Gene names were given, whenever possible, according to the identification of homologous genes in S. cerevisiae (n.a. : not available).
Table 3. Regulators and transcription factors identified as differentially expressed during the transition to lipid accumulation.
| Fold Change (arbitrary units) | ||||||
| Gene Labels | Gene Names | Description | Cluster | B/A | C/B | C/A |
| YALI0F30173g | TFB2 | TFIIH transcription factor subunit | 1b | 0.61 | 0.9 | 0.54 |
| YALI0F03630g | n.a. | Putative zinc finger transcription factor, Zn(2)-Cys(6) family | 3 | 0.92 | 0.67 | 0.62 |
| YALI0A14542g | TUP1 | General repressor of transcription | 5a | 1.1 | 1.78 | 1.95 |
| YALI0D02673g | PTR3 | Component of the SPS amino-acid sensor complex | 5a | 1.23 | 1.28 | 1.58 |
| YALI0D09647g | n.a. | Putative arginine metabolism regulation protein | 5a | 1.04 | 1.47 | 1.52 |
| YALI0F02783g | NPR2 | Putative nitrogen permease regulator NPR2-like | 5a | 1.1 | 1.54 | 1.7 |
| YALI0F17424g | Hap1 | Putative zinc finger transcription factor, HAP1-like activator | 5a | 1.19 | 1.75 | 2.07 |
| YALI0D20482g | n.a. | Putative nitrogen regulatory protein, AREA-like | 4 | 1.68 | 0.62 | 1.04 |
| YALI0E18986g | UBI4 | Ubiquitin | 4 | 2.87 | 0.67 | 1.92 |
| YALI0D13046g | Otu1 | Putative zinc finger transcription factor, OTU1-like | 5c | 1.53 | 0.99 | 1.51 |
| YALI0C19151g | Cat8 | Putative zinc finger transcription factor, CAT8-like | 5d | 1.16 | 1.71 | 1.99 |
| YALI0A10637g | Hal9 | Putative zinc finger transcription factor, HAL9-like | 4 | 1.51 | 0.95 | 1.44 |
| YALI0E29909g | PDR6 | Pleiotropic drug resistance regulatory protein | 5b | 0.74 | 1.94 | 1.43 |
| YALI0B12716g | HAC1 | Basic leucine zipper transcription factor, involved in the unfolded protein response | 5b | 1.09 | 2.05 | 2.24 |
Genes were identified as differentially expressed in cases of significant detection, with a false discovery rate lower than 1×10−5 and an absolute fold change between two groups of more than 1.5. Gene labels and descriptions were attributed according to the Genolevure database (http://www.genolevures.org/). Gene names were given, whenever possible, according to the identification of homologous genes in S. cerevisiae (n.a. : not available).
Discussion
Lipid accumulation phenotypes have been known for a long time, but the biochemical differences between oleaginous and non oleaginous organisms remain to be elucidated. Despite the development of high-throughput sequencing, transcriptomic and proteomic methods, together with economic and industrial interest in oil production, “-omics” information for oleaginous yeasts remains scarce. Only a limited number of proteomics studies in recent years have focused on the mechanisms of lipid accumulation [20], [21], [38].
Y. lipolytica has been shown to accumulate up to 36% of its dry weight as lipid [7]. However, recent studies on strain ACA-DC 50109 growing on glucose as the sole carbon source highlighted problems achieving lipid accumulation rates of more than 20%, despite strong nitrogen limitation and high C/N ratios [39]. Together with a preferential use of hydrophobic substrates, these results called into question the potential of Y. lipolytica for lipid production an accumulation via a de novo fatty acid biosynthesis pathway [20].
In this study, we used a controlled fed-batch set-up to analyze the behavior of Y. lipolytica during the transition from growth to lipid accumulation. Culture parameters were carefully modulated to control the induction of lipid accumulation. The calculated glucose and nitrogen flows implied that lipid production was suboptimal with respect to the maximum levels obtained in fed-batch mode [23], [40]. In particular, dual nitrogen and glucose limitation was used in later stages of fed-batch culture, to prevent excess citric acid production that might ultimately partly mask the transcriptomic response. Despite these suboptimal conditions, our results clearly demonstrate that Y. lipolytica can accumulate lipids synthesized de novo, using glucose as the sole carbon source.
Transcriptomic response of an oleaginous yeast to nitrogen-limiting conditions
One of the primary consequences of nitrogen limitation is a decrease in cell proliferation. As already observed in the proteomes of other oleaginous yeast species, such as L. starkeyi and R. toruloides [20], [21], much of the transcriptomic response of Y. lipolytica reflects the decrease in growth rate upon nitrogen limitation. Clusters 1a and 1b contain a large number of genes related to cellular metabolism, cell growth and particularly protein synthesis, including 61 genes encoding ribosomal subunits, two translation initiation factors, and seven translation elongation factors (Table S2).
Meanwhile, the assimilated glucose is redirected towards the citric acid cycle to provide lipid biosynthesis, initiated by the fatty acid synthase (FAS), from acetyl-CoA, malonyl-CoA and NADPH [4], [5]. Acetyl-CoA is cleaved from citrate within the cytosol by ATP:citrate lyase (ACL). This protein has been identified as a key enzyme of lipid accumulation in oleaginous organisms, even though some non-oleaginous species exhibit ACL activity [7]. ACL is considered the main provider of Acetyl-CoA for both FAS, and Acetyl-CoA Carboxylase (ACC) that will further provide the FAS with malonyl-CoA [7]. On the other hand, malic enzyme (ME) is considered to be the supplier of NADPH in most oleaginous species. All four enzymes have been found in Y. lipolytica, but neither displays a significant change in transcription level in response to nitrogen limitation. This could be easily interpreted for the ME, as in Y. lipolytica, contrary to most oleaginous yeasts, only a single mitochondrial form of the enzyme is predicted to exist. Therefore it should be unable to provide the NADPH to the cytosolic FAS. Meanwhile, several enzymatic studies in oleaginous yeasts have shown that ACL activity increases during lipid accumulation [41]. Thus, if similar increases in ACL levels occur in Y. lipolytica, they are presumably regulated posttranslationally. As for ACC, transcriptomic data were below our quality threshold, which led us to the incapacity to draw any conclusion for now regarding its expression under nitrogen limiting conditions.
While the main genes governing fatty acid synthesis do not seem to be directly controlled at the transcriptomic level, lipid accumulation could also be a passive consequence of the rerouting of carbon fluxes leading to the production of Acetyl-CoA. Acetyl-CoA production may depend on a sequence of biochemical events, beginning with an increase in AMP deaminase activity shortly after nitrogen limitation [42]. Transcriptomic data tend to highlight a slight increase in expression of the related gene AMD1 (data not shown), but this increase is not statistically significant, suggesting that the sharp increase in AMP deaminase activity, like the one reported for AMD in Rhodosporidium toruloides [7], may also be induced by posttranslational modifications rather than through transcriptional control. Several genes encoding proteins involved in protein modification displayed significant overexpression during the fed-batch process (Table 1, and Table S2) and are therefore potential targets of choice for future investigations of the impact of posttranslational modification on the metabolism of oleaginous yeasts.
Another key regulation point is the inhibition of the AMP-dependent isocitrate dehydrogenase (IDH) in the TCA cycle [4], [7]. In our study, no significant change in IDH expression was detected during lipid accumulation either. However, Morgunov and colleagues [43] isolated IDH from Y. lipolytica subjected to nitrogen starvation and in vitro enzymatic activity tests showed that the enzyme was still present and functional under nitrogen limiting conditions. The decrease in IDH activity could therefore be controlled by cellular AMP content rather than transcriptionally. In parallel, the gene YALI0C16885g, encoding isocitrate lyase (ICL), seems to be strongly and immediately repressed by nitrogen limitation (Table 2). ICL is a peroxisomal enzyme involved in the glyoxylate pathway, in which it converts isocitrate into glyoxylate and succinate [44]. A strong decrease in ICL activity has also been reported in several oleaginous yeasts under nitrogen-limiting conditions [41]. With the presumably low levels of IDH activity [7], and the repression of ICL transcription, isocitrate cannot be metabolized through the TCA or glyoxylate cycles. However, the accumulating isocitrate can be rerouted within the TCA to produce citrate, through the action of aconitase (YALI0D09361g). Interestingly, aconitase would be expected to be equally active under conditions of carbon and nitrogen limitation [7], but its gene was significantly more strongly expressed during the late phase of lipid accumulation, favouring citrate accumulation (Table 2). When put together, our transcriptomic results tend to confirm the re-routing of the TCA towards citrate accumulation. Citrate can be further provided to ACL, via the action of the Citrate/Malate Translocase (CMT) [7]. The gene encoding this particular transport system has not been identified in the genome of Yarrowia so far. Transcriptomic results highlighted several genes encoding transport systems, some of which are over-expressed during lipid accumulation (Table S2). However, none of them appear at this stage as a potential candidate for a mitochondrial translocase system.
While citrate seems to be a key metabolite for lipid accumulation [7], ACL also requires both CoA and ATP for the synthesis of acetyl-CoA. It is interesting to notice that several genes involved in the metabolism of CoA and ATP display significant changes in their expression during the lipid accumulation process. Notably, the gene encoding dephospho-CoA kinase (DPCK, YALI0F09625g), the final enzyme in the CoA biosynthesis pathway, was significantly overexpressed during the late phase of lipid accumulation (Table 2). DPCK plays an important role in regulating CoA biosynthesis [45]. The overexpression of this gene may be an indicator of an increase in the CoA pool during lipid accumulation. As for ATP, its intracellular concentration was studied in various oleaginous yeasts in which no significant changes were highlighted during lipid accumulation [7]. However, ATP, ADP and AMP levels have been reported to fluctuate in Y. lipolytica under nitrogen-limiting conditions [43]. In our study, several genes involved in ATP metabolism have been identified as displaying differential expression in response to nitrogen limitation in Y. lipolytica, such as (i) ADK1 and ADK2, encoding two adenylate kinases involved in cellular energy homeostasis [46], (ii) genes involved in oxidative phosphorylation, (iii) the ATP2 gene, encoding the mitochondrial ATP synthase beta chain [47], (iii) genes coding for ATPases subunits. All these observations suggest that the adenine nucleotide pool in Y. lipolytica during the transition to lipid accumulation may evolve more dynamically than what has been previously reported in other oleaginous yeasts, such as L. starkeyi [7]. As CoA and ATP are essential metabolites in acetyl CoA synthesis, they could ultimately affect lipid synthesis indirectly, via their mobilization and/or their availability for ACL.
Subsequent steps in lipid synthesis, such as elongation and desaturation, appear more likely to be controlled at the transcriptomic level. In particular, the gene encoding the delta-9 fatty acid desaturase [7] was overexpressed during lipid accumulation (Table 2). The expression of this gene was directly correlated with the accumulation of C18:1, as shown by the lipid profile of the fed batch (Figure 1c). Moreover, two genes related to lipid storage are significantly expressed in the late stage of lipid accumulation: (i) SAC1, encoding an inositol/phosphatidylinositol phosphatase previously shown to be a component of lipid particles in Y. lipolytica [38], and (ii) DGA2, encoding a diacylglycerol acyltransferase. More specifically, DGA2 has been recently found to encode a member of the type 1 acyl-CoA:diacylglycerol acyltransferase family (DGAT1), which has not previously been identified in yeasts, but is commonly found in mammals and plants [48]. The enzyme Dga2p has been highlighted as a major contributor to TAG synthesis in Y. lipolytica, via an acyl-CoA-dependent mechanism. Furthermore, expression of this enzyme not only contributes to TAG synthesis, but also affects the size and morphology of lipid bodies. The regulation of these two genes implies an intense lipid storage activity, although the expression rates of the three remaining acyltransferases (i.e. DGA1, LRO1 and ARE1) were not significantly altered during culture. Interestingly, a lipase encoding gene, LIP15, is also over-expressed during lipid accumulation, which could suggest that lipid turnover proceeds in parallel with lipid accumulation. Y. lipolytica possesses many genes coding for lipases, among which (i) two genes coding for intracellular TAG lipases homologous to S. cerevisiae, namely TGL3 and TGL4 [18], and (ii) 16 paralogs of genes coding for lipase, regrouped to form the LIP family [49]. However, little information is known about the role and specificity of each member of this lipolytic arsenal. Only three isoenzymes (Lip2p, Lip7p and Lip8p) have been partly characterized so far [49]. Hence, the exact function of the protein encoded by the LIP15 gene has yet to be fully resolved. It could thus represent a target of choice for future analyses, especially since some genes involved in TAG degradation such as TGL5 have yet to be discovered in Yarrowia [18]. The enhancement of lipid storage capacity combined with the repression of lipid turnover have already proven to be effective strategies to improve lipid accumulation in Y. lipolytica [18], [50]. Genes highlighted in our study, such as DGA2, or LIP15 could be potential targets for genetic manipulation in order to alter the lipid accumulation capacity of Y. lipolytica.
Finally, several glycolysis genes were repressed during lipid accumulation (Table 2). Previous studies have emphasized the negative feedback control of phosphofructokinase (PFK) and pyruvate kinase (PK), the presumed principal regulators of glucose uptake, by citrate accumulation [7]. Our transcriptomic study also highlights the transcriptional regulation of genes involved in glycolysis, indicating a global gene response to the carbon overflow induced by nitrogen limitation [8]. It is therefore probable that repression of glycolysis eventually limits lipid accumulation on the long term. Improvements in our understanding of the regulation mechanisms involved in glucose uptake could lead to the identification of alternative targets for the production of over-accumulating strains.
Adaptation of Y. lipolytica to nitrogen limitation
Yeasts and fungi are able to utilize diverse nitrogen sources, including ammonium, amino acids, urea, nitrogen bases and purine derivatives as nutriment for growth [51], [52]. During culture, specific permeases are synthesized, permitting the incorporation of the nitrogen containing substrate [53]. These nitrogen sources may either be used directly in biosynthetic pathways or catabolized to generate ammonium, glutamate, and glutamine. In the fed-batch process used in this study, nitrogen limitation is induced by the temporary cessation of ammonium supplementation. It is therefore unsurprising that much of the transcriptomic data collected here reflects upon the transition of the cell metabolism in search for an alternative nitrogen source. The genes differentially expressed during this nitrogen-induced transition are presented in Table S3.
The above phenomena, however, are regulated by complex mechanisms involving several interacting and competing regulatory systems, some of which have activities beyond the scope of nitrogen metabolism [51], [52]. Specific transcriptional controls affecting single biosynthetic pathways (e.g. arginine, branched-chain amino acids, lysine, methionine) have been described [54]–[57], but are not yet entirely understood in Y. lipolytica. One gene encoding a protein involved in the regulation of arginine metabolism (YALI0D09647g) was found to be expressed in the late stage of lipid accumulation (Table 3). However, the repression of genes involved in aromatic amino-acid biosynthesis may also reflect the general control of amino-acid biosynthesis (GAAC) [58]. Regulation of the GAAC in S. cerevisiae is reportedly mediated by the transcription factor Gcn4 [52], [58], for which an homolog has been identified in Y. lipolytica (YALI0E27742g), but no significant expression could detected under nitrogen limitation.
A large proportion of the genes identified above are subject to nitrogen catabolite repression (NCR) [51], [52]. A GATA-like transcription factor (YALI0D20482g) appears to become particularly active upon nitrogen limitation (Table 3). Similar factors have been linked to NCR regulation in fungi and yeasts [51], [59], [60]. In S. cerevisiae, NCR involves the inhibition of two transcription factors of the GATA family: Gln3 and Gat1/Nil1 [61], [62]. These transcription factors recognize a 5′-GATA-3′ sequence located upstream from genes subject to NCR. Gln3 and Gat1/Nil1 have high levels of sequence similarity, but differ in their expression patterns. Gln3 is constitutively expressed in S. cerevisiae and is inhibited by Ure2 at high nitrogen levels [60]. By contrast, the expression of Gat1/Nil1 is repressed under normal conditions by another GATA family transcription factor, DEH1/Gzf3 [65]. Under specific nitrogen conditions, a fourth GATA factor, Dal80, has also been shown to inhibit Gat1 [60]–[62]. Three GATA transcription factor genes have been identified in Y. lipolytica: (i) one encoding a homolog of Gzf3 (YALI0C22682g) and (ii) two encoding Gat1/Gln3-like factors (YALI0D20482g, YALI0F17886g). As in S. cerevisiae, these two Gat1/Gln3-like factors identified in Y. lipolytica display high levels of sequence similarity. However, only YALI0D20482g has been shown to be differentially expressed under nitrogen-limiting conditions. Therefore, we would expect this gene to encode a Gat1-like transcription factor, whereas YALI0F17886g encodes a constitutively produced Gln3-like protein.
Finally, additional genes linked to the regulation of nitrogen metabolism were identified from the transcriptomic data (Table 3). One such gene, Ptr3, encodes a subunit of the Ssy1-Ptr3-Ssy5 (SPS) sensor complex. This complex is involved in ammonium source detection and has been linked to the control of various permease-encoding genes [63], [64]. Another, the NPR2 gene, belongs to a family of regulators involved in the posttranslational control of nitrogen permease [65].
Regulation
The general mechanisms regulating lipid accumulation in oleaginous organisms have yet to be completely elucidated, despite the enormous potential they represent. Through transcriptomic analyses of Y. lipolytica, we have highlighted the action of several transcription factors and regulatory proteins, some of which had already been identified as potentially important (Table 3). In particular, YALI0F30173g, which encodes a subunit of the transcription factor TFIIH, appears to be strongly repressed by nitrogen limitation. TFIIH is a known general transcription factor involved in the RNA polymerase II preinitation complex and in nucleotide excision repair [66]. Similarly, YALI0A14542g encodes a TUP1-like general repressor of transcription activated within hours of nitrogen limitation. The regulation of these two factors may be linked to the repression of transcription and protein synthesis observed following nitrogen limitation.
Five genes encoding regulatory proteins of the zinc cluster family were identified as differentially expressed upon nitrogen limitation. Zinc cluster proteins (ZCP) are named after a family of zinc-containing structural motifs generally associated with DNA, RNA or protein-binding properties [67]. As such, ZCP proteins include diverse transcriptional factors involved in the regulation of various metabolic processes [67], [68]. One of these factors, encoded by YALI0F03630g, was also found to be repressed, mostly at the late stage of lipid production. However, the sequence of this gene displays little similarity to those of yeast transcription factors of unknown function.
Four of these five genes encode transcription factors of the zinc cluster family. The four ZCP-like transcription factors identified are putative homologs of Hap1, Otu1, Cat8 and Hal9. The Hap1 gene encodes a yeast heme activator protein that promotes the transcription of many genes in response to heme and oxygen [69]. Otu1 has hydrolase activity responsible for removing conjugated ubiquitin from proteins, thus potentially playing an important role in regulating protein turnover by preventing degradation [70]. It has been shown to participate in the regulation of ubiquitin, which was also significantly expressed (Table 3). Cat8 encodes a known activator of gluconeogenic enzymes [71]. Finally, Hal9 encodes a putative transcription factor involved in halotolerance [72].
Finally, two additional transcription factors, involved in pleiotropic drug resistance and unfolded protein response, were detected in cluster 5b. Interestingly, these two metabolic processes were previously identified as responding to changes in nitrogen sources in S. cerevisiae, despite a lack of direct involvement in nitrogen metabolism [52]. This finding confirms the observations made on the baker's yeast transcriptome, but also highlights the complexity and interconnectivity of regulatory mechanisms in yeast.
Concluding remarks
In conclusion, this study highlights genes potentially involved in the oleaginous characters of the cell at the transcriptomic level. The expression of genes encoding several key enzymes, such as ACL, ME, AMD and IDH, does not seem to be regulated at the transcriptional level. However, a complex cascade of transcriptional events may lead to the increase of the various substrate and cofactor pools necessary for the synthesis of both acetyl-CoA and NADPH. These findings are consistent with the hypothesis that lipid accumulation in oleaginous yeasts is a consequence of the rerouting of carbon fluxes upon nitrogen limitation, rather than specific and/or enhanced lipid metabolism activity.
Supporting Information
Growth conditions, composition of the fed-batch medium, salts and vitamins stock solutions.
(DOC)
Description of the 569 genes identified as differentially expressed during the transition to lipid accumulation.
(XLS)
Genes involved in nitrogen-metabolism identified as differentially expressed during the transition to lipid accumulation.
(DOC)
Acknowledgments
We would like to thank Jérome Montels at ENSIACET (Toulouse) for his help in biomass formula determination. In addition, we would like to express our gratitude to Dr Cécile Neuveglise, Dr Ramdane Haddouche and Dr Thierry Dulermo for the fruitful discussions on Yarrowia lipolytica's metabolism. Finally, we thank Julie Sappa of Alex Edelman and Associates for her excellent help in correcting the English version of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was labellized by Aerospace Valley (a non-profit association) and financially supported in part by CALIN French national research program (DGCIS), by the Lipicaero French national research program (ANR-PNRB) No. 0701C0089 and by the EU in the 7th Framework Programme for Research and Technological Development, under ALFA-BIRD (Alternative Fuels and Biofuels for Aircraft Development) project. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ramadhas AS, Jayaraj S, Muraleedharan C. Biodiesel production from high FFA rubber seed oil. Fuel. 2005;84:335–340. [Google Scholar]
- 2.Minami E, Saka S. Thermodynamic studies on cloud point of biodiesel with its fatty acid composition. Fuel. 2006;85:2479–2483. [Google Scholar]
- 3.Nie K, Xie F, Wang F, Tan T. Lipase catalyzed methanolysis to produce biodiesel: optimization of the biodiesel production. J Mol Catal B: Enzyme. 2006;43:142–147. [Google Scholar]
- 4.Ratledge C. Fatty acid biosynthesis in microorganisms being used for Single Cell Oil production. Biochimie. 2004;86:807–815. doi: 10.1016/j.biochi.2004.09.017. [DOI] [PubMed] [Google Scholar]
- 5.Beopoulos A, Nicaud J-M, Gaillardin C. An overview of lipid metabolism in yeasts and its impact on biotechnological processes. Appl Microbiol Biotechnol. 2011;90:1193–1206. doi: 10.1007/s00253-011-3212-8. [DOI] [PubMed] [Google Scholar]
- 6.Thorpe RF, Ratledge C. Fatty Acid Distribution in Triglycerides of Yeasts Grown onGlucose or n-Alkanes. Microbiol. 1972;72:151–163. [Google Scholar]
- 7.Ratledge C, Wynn JP. The biochemistry and molecular biology of lipid accumulation in oleaginous microorganisms. Adv Appl Microbiol. 2002;51:1–51. doi: 10.1016/s0065-2164(02)51000-5. [DOI] [PubMed] [Google Scholar]
- 8.Beopoulos A, Cescut J, Haddouche R, Uribelarrea J-L, Molina-jouve C, et al. Yarrowia lipolytica as a model for bio-oil production. Prog Lipid Res. 2009;48:375–387. doi: 10.1016/j.plipres.2009.08.005. [DOI] [PubMed] [Google Scholar]
- 9.Beopoulos A, Chardot T, Nicaud J-M. Yarrowia lipolytica: A model and a tool to understand the mechanisms implicated in lipid accumulation. Biochimie. 2009;91:692–696. doi: 10.1016/j.biochi.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 10.Barth G, Gaillardin C. Physiology and genetics of the dimorphic fungus Yarrowia lipolytica. FEMS Microbiol Rev. 1997;19:219–237. doi: 10.1111/j.1574-6976.1997.tb00299.x. [DOI] [PubMed] [Google Scholar]
- 11.Madzak C, Gaillardin C, Beckerich J-M. Heterologous protein expression and secretion in the non-conventional yeast Yarrowia lipolytica: a review. J Biotechnol. 2004;109:63–81. doi: 10.1016/j.jbiotec.2003.10.027. [DOI] [PubMed] [Google Scholar]
- 12.Finogenova TV, Morgunov IG, Kamzolova SV, Chernyavskaya OG. Organic Acid Production by the Yeast Yarrowia lipolytica: A Review of Prospects. Appl Biochem Microbiol. 2005;41:418–425. [PubMed] [Google Scholar]
- 13.Papanikolaou S, Muniglia L, Chevalot I, Aggelis G, Marc I. Accumulation of a cocoa-butter-like lipid by Yarrowia lipolytica cultivated on agro-industrial residues. Curr Microbiol. 2003;46:124–130. doi: 10.1007/s00284-002-3833-3. [DOI] [PubMed] [Google Scholar]
- 14.Ratledge C. Single cell oils for the 21st century. In: Cohen Z, Ratledge C, editors. Single cell oils: microbial and algal oils. AOCS Press, Champaign, IL, USA; 2005. pp. 1–20. [Google Scholar]
- 15.Fickers P, Benetti P-H, Waché Y, Marty A, Mauersberger S, et al. Hydrophobic substrate utilisation by the yeast Yarrowia lipolytica, and its potential applications. FEMS Yeast Res. 2005;5:527–543. doi: 10.1016/j.femsyr.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 16.Mlícková K, Roux E, Athenstaedt K, d'Andrea S, Daum G, et al. Lipid accumulation, lipid body formation, and acyl coenzyme A oxidases of the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2004;70:3918–3924. doi: 10.1128/AEM.70.7.3918-3924.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, et al. Genome evolution in yeasts. Nature. 2004;430:35–44. doi: 10.1038/nature02579. [DOI] [PubMed] [Google Scholar]
- 18.Beopoulos A, Mrozova Z, Thevenieau F, Le Dall M-T, Hapala I, et al. Control of lipid accumulation in the yeast Yarrowia lipolytica. Appl Environ Microbiol. 2008;74:7779–7789. doi: 10.1128/AEM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Steen EJ, Kang Y, Bokinsky G, Hu Z, Schirmer A, et al. Microbial production of fatty-acid-derived fuels and chemicals from plant biomass. Nature. 2010;463:559–562. doi: 10.1038/nature08721. [DOI] [PubMed] [Google Scholar]
- 20.Liu H, Zhao X, Wang F, Jiang X, Zhang S, et al. The proteome analysis of oleaginous yeast Lipomyces starkeyi. FEMS Yeast Res. 2011;11:42–51. doi: 10.1111/j.1567-1364.2010.00687.x. [DOI] [PubMed] [Google Scholar]
- 21.Liu H, Zhao X, Wang F, Li Y, Jiang X, et al. Comparative proteomic analysis of Rhodosporidium toruloides during lipid accumulation. Yeast. 2009;26:553–566. doi: 10.1002/yea.1706. [DOI] [PubMed] [Google Scholar]
- 22.Li Y, Zhao Z, Bai F. High-density cultivation of oleaginous yeast Rhodosporidium toruloides Y4 in fed-batch culture. Enzym Microb Tech. 2007;41:312–317. [Google Scholar]
- 23.Cescut J. Accumulation d'acylglycérols par des espèces levuriennes à usage carburant aéronautique : physiologie et performances de procédés. 2009. PhD thesis, University of Toulouse, France. Available: http://eprint.insa-toulouse.fr/archive/00000289/01/Cescut.pdf. Accessed 2011 Nov 4.
- 24.Granger L-M, Perlot P, Goma G, Pareilleux A. Kinetics of growth and fatty acid production of Rhodotorula glutinis. Appl Microbiol Biotechnol. 1992;37:13–17. [Google Scholar]
- 25.Cescut J, Severac E, Molina-Jouve C, Uribelarrea J-L. Optimizing pressurized liquid extraction of microbial lipids using the response surface method. J Chromatogr. 2011;1218:373–379. doi: 10.1016/j.chroma.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 26.R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. 2011. Available: http://www.r-project.org. Accessed 2011 Nov 4.
- 27.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth GK. Limma: linear models for microarray data. In: R, Carey V, Huber W, Irizarry R, Dudoit S, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. Springer, New York, USA; 2005. pp. 397–420. [Google Scholar]
- 29.Ritchie ME, Silver J, Oshlack A, Holmes M, Diyagama D, et al. A comparison of background correction methods for two-colour microarrays. Bioinformatics. 2007;23:2700–2707. doi: 10.1093/bioinformatics/btm412. [DOI] [PubMed] [Google Scholar]
- 30.Smyth GK, Speed T. Normalization of cDNA microarray data. Methods. 2003;31:265–273. doi: 10.1016/s1046-2023(03)00155-5. [DOI] [PubMed] [Google Scholar]
- 31.Brás LP, Menezes JC. Improving cluster-based missing value estimation of DNA microarray data. Biomol Eng. 2007;24:273–282. doi: 10.1016/j.bioeng.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 32.Saeed AI, Sharov V, White J, Li J, Liang W, et al. TM4: a free, open-source system for microarray data management and analysis. Biotech. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 33.Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 34.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nature genetics. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucl Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ratledge C. Regulation of lipid accumulation in oleaginous micro-organisms. Biochem Soc Trans. 2002;30:1047–1050. doi: 10.1042/bst0301047. [DOI] [PubMed] [Google Scholar]
- 38.Athenstaedt K, Jolivet P, Boulard C, Zivy M, Negroni L, et al. Lipid particle composition of the yeast Yarrowia lipolytica depends on the carbon source. Proteomics. 2006;6:1450–1459. doi: 10.1002/pmic.200500339. [DOI] [PubMed] [Google Scholar]
- 39.Papanikolaou S, Galiotou-Panayotou M, Chevalot I, Komaitis M, Marc I, et al. Influence of glucose and saturated free-fatty acid mixtures on citric acid and lipid production by Yarrowia lipolytica. Curr Microbiol. 2006;52:134–142. doi: 10.1007/s00284-005-0223-7. [DOI] [PubMed] [Google Scholar]
- 40.Costes P, Uribelarrea J-L, Cescut J, Molina-Jouve C. Novel method for cultivating yeasts of the Yarrowia genus. 2010. Patent n° WO/2010/076432.
- 41.Holdsworth JE, Veenhuis M, Ratledge C. Enzyme activities in oleaginous yeasts accumulating and utilizing exogenous or endogenous lipids. J Gen Microbiol. 1988;134:2907–2915. doi: 10.1099/00221287-134-11-2907. [DOI] [PubMed] [Google Scholar]
- 42.Evans CT, Ratledge C. Possible regulatory roles of ATP:citrate lyase, malic enzyme, and AMP deaminase in lipid accumulation by Rhodosporidium toruloides CBS 14. Can J Microbiol. 1985;31:1000–1005. [Google Scholar]
- 43.Morgunov IG, Solodovnikova NY, Sharyshev AA, Kamzolova SV, Finogenova TV. Regulation of NAD(+)-dependent isocitrate dehydrogenase in the citrate producing yeast Yarrowia lipolytica. Biochemistry (Mosc) 2004;69:1391–1398. doi: 10.1007/s10541-005-0086-3. [DOI] [PubMed] [Google Scholar]
- 44.Fukui S, Tanaka A. Yeast peroxisomes. Trends Biochem Sci. 1979;4:246–249. [Google Scholar]
- 45.Daugherty M, Polanuyer B, Farrell M, Scholle M, Lykidis A, et al. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J Biol Chem. 2002;277:21431–21439. doi: 10.1074/jbc.M201708200. [DOI] [PubMed] [Google Scholar]
- 46.Cooper AJ, Friedberg EC. A putative second adenylate kinase-encoding gene from the yeast Saccharomyces cerevisiae. Gene. 1992;114:145–148. doi: 10.1016/0378-1119(92)90721-z. [DOI] [PubMed] [Google Scholar]
- 47.Saltzgaber-Muller J, Kunapuli SP, Douglas MG. Nuclear genes coding the yeast mitochondrial adenosine triphosphatase complex. Isolation of ATP2 coding the F1-ATPase beta subunit. J Biol Chem. 1983;258:11465–11470. [PubMed] [Google Scholar]
- 48.Beopoulos A, Haddouche R, Kabran P, Dulermo T, Chardot T, et al. Identification and characterization of DGA2, an acyltransferase of the DGAT1 acyl-CoA:diacylglycerol acyltransferase family in the oleaginous yeast Yarrowia lipolytica. New insights into the storage lipid metabolism of oleaginous yeasts. Appl Microbiol Biotechnol. 2011 doi: 10.1007/s00253-011-3506-x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fickers P, Marty A, Nicaud JM. The lipases from Yarrowia lipolytica: Genetics, production, regulation, biochemical characterization and biotechnological applications. Biotechnol Adv. 2011;29:632–644. doi: 10.1016/j.biotechadv.2011.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Dulermo T, Nicaud JM. Involvement of the G3P shuttle and β-oxidation pathway in the control of TAG synthesis and lipid accumulation in Yarrowia lipolytica. Metab Eng. 2011;13:482–491. doi: 10.1016/j.ymben.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 51.Marzluf GA. Genetic regulation of nitrogen metabolism in the fungi. Microbiol Mol Biol Rev. 1997;61:17–32. doi: 10.1128/mmbr.61.1.17-32.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Godard P, Urrestarazu A, Vissers S, Kontos K, Bontempi G, et al. Effect of 21 different nitrogen sources on global gene expression in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 2007;27:3065–3086. doi: 10.1128/MCB.01084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kingsbury JM, Goldstein AL, McCusker JH. Role of nitrogen and carbon transport, regulation, and metabolism genes for Saccharomyces cerevisiae survival in vivo. Eukaryot Cell. 2006;5:816–824. doi: 10.1128/EC.5.5.816-824.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dubois E, Hiernaux D, Grennon M, Wiame JM. Specific induction of catabolism and its relation to repression of biosynthesis in arginine metabolism of Saccharomyces cerevisiae. J Mol Biol. 1978;122:383–406. doi: 10.1016/0022-2836(78)90417-5. [DOI] [PubMed] [Google Scholar]
- 55.Kohlhaw GB. Leucine biosynthesis in fungi: entering metabolism through the back door. Microbiol Mol Biol Rev. 2003;67:1–15. doi: 10.1128/MMBR.67.1.1-15.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Becker B, Feller A, el Alami M, Dubois E, Piérard A. A nonameric core sequence is required upstream of the LYS genes of Saccharomyces cerevisiae for Lys14p-mediated activation and apparent repression by lysine. Mol Microbiol. 1998;29:151–163. doi: 10.1046/j.1365-2958.1998.00916.x. [DOI] [PubMed] [Google Scholar]
- 57.Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–532. doi: 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hinnebusch AG. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988;52:248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Christensen T, Hynes MJ, Davis MA. Role of the regulatory gene areA of Aspergillus oryzae in nitrogen metabolism. Appl Environ Micriobiol. 1998;64:3232–3237. doi: 10.1128/aem.64.9.3232-3237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boczko EM, Cooper TG, Gedeon T, Mischaikow K, Murdock DG, et al. Structure theorems and the dynamics of nitrogen catabolite repression in yeast. Proc Natl Acad Sci USA. 2005;102:5647–5652. doi: 10.1073/pnas.0501339102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Coffman J, Rai R, Loprete D, Cunningham T, Svetlov V, et al. Cross regulation of four GATA factors that control nitrogen catabolic gene expression in Saccharomyces cerevisiae. J Bacteriol. 1997;179:3416–3429. doi: 10.1128/jb.179.11.3416-3429.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cooper TG. Transmitting the signal of excess nitrogen in Saccharomyces cerevisiae from the Tor proteins to the GATA factors: connecting the dots. FEMS Microbiol Rev. 2002;26:223–238. doi: 10.1111/j.1574-6976.2002.tb00612.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Forsberg H, Ljungdahl PO. Sensors of extracellular nutrients in Saccharomyces cerevisiae. Curr Genet. 2001;40:91–109. doi: 10.1007/s002940100244. [DOI] [PubMed] [Google Scholar]
- 64.Boles E, André B. Role of transporter-like sensors in glucose and amino acid signalling in yeast. In: Boles E, Reinhard K, editors. Molecular mechanisms controlling transmembrane transport. Springer, Berlin; 2004. pp. 121–153. [Google Scholar]
- 65.Spielewoy N, Guaderrama M, Wohlschlegel JA, Ashe M, Yates JR, et al. Npr2, yeast homolog of the human tumor suppressor NPRL2, is a target of Grr1 required for adaptation to growth on diverse nitrogen sources. Eukaryot Cell. 2010;9:592–601. doi: 10.1128/EC.00192-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drapkin R, Reardon JT, Ansari A, Huang JC, Zawel L, et al. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature. 1994;368:769–772. doi: 10.1038/368769a0. [DOI] [PubMed] [Google Scholar]
- 67.MacPherson S, Larochelle M, Turcotte B. A fungal family of transcriptional regulators: the zinc cluster proteins. Microbiol Mol Biol Rev. 2006;70:583–604. doi: 10.1128/MMBR.00015-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Todd RB, Andrianopoulos A. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet Biol. 1997;21:388–405. doi: 10.1006/fgbi.1997.0993. [DOI] [PubMed] [Google Scholar]
- 69.Pfeifer K, Kim KS, Kogan S, Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989;56:291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- 70.Messick TE, Russell NS, Iwata AJ, Sarachan KL, Shiekhattar R, et al. Structural basis for ubiquitin recognition by the Otu1 ovarian tumor domain protein. J Biol Chem. 2008;283:11038–11049. doi: 10.1074/jbc.M704398200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hedges D, Proft M, Entian KD. CAT8, a new zinc cluster-encoding gene necessary for derepression of gluconeogenic enzymes in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:1915–1922. doi: 10.1128/mcb.15.4.1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mendizabal I, Rios G, Mulet JM, Serrano R, Larrinoa IF de. Yeast putative transcription factors involved in salt tolerance. FEBS Letters. 1998;425:323–328. doi: 10.1016/s0014-5793(98)00249-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Growth conditions, composition of the fed-batch medium, salts and vitamins stock solutions.
(DOC)
Description of the 569 genes identified as differentially expressed during the transition to lipid accumulation.
(XLS)
Genes involved in nitrogen-metabolism identified as differentially expressed during the transition to lipid accumulation.
(DOC)