Abstract
DNA-dependent RNA polymerases such as T7 RNA polymerase perform the transcription of DNA into mRNA with high efficiency and high fidelity. Although structural studies have provided a detailed account of the molecular basis of transcription, the relative importance of factors like hydrogen bonds and steric effects remain poorly understood. We report herein the first study aimed at systematically probing the importance of steric and electrostatic effects on the efficiency and fidelity of DNA transcription by T7 RNAP. We used synthetic nonpolar analogues of thymine with size varied in sub-Angström increments to probe the steric requirements of T7 RNAP during the elongation mode of transcription. Enzymatic assays with internal radiolabelling were performed to compare the efficiency of transcription of modified DNA templates with a natural template containing thymine as reference. Furthermore, we analyzed effects on the fidelity by measuring the composition of RNA transcripts by enzymatic digestion followed by 2D-TLC separation. Our results demonstrate that hydrogen bonds play an important role in the efficiency of transcription, but, interestingly, they do not appear to be required for faithful transcription. Steric effects (size and shape variations) are found to be significant both in insertion of a new RNA base and in extension beyond it.
The transcription of DNA is an essential enzymatic process for gene expression and is central to all forms of life. Although studied for decades, it is only more recently that its molecular mechanisms have been revealed (1). This DNA-templated RNA synthesis is mediated by polymerases which operate with high processivity (ca. 100 nucleotides/s) (2) and high selectivity/fidelity (error rate of less than 10−5) (3), thereby averting the appearance of mutagenic or cytotoxic errors (4, 5). Despite the progress of our qualitative understanding of transcription through structural studies (1), a more quantitative understanding of the transcription process at a molecular level is still needed (6). For instance, biochemical studies have demonstrated that hybridization energy does not fully account for the fidelity observed in DNA replication and transcription, thus suggesting that the confinement within the active site of polymerases affects base pair thermodynamics (7-10). Although studies have recently been performed with DNA polymerases to assess the relative importance of hydrogen bonds, sterics, stacking, and solvation during replication (vide infra), the roles of these forces in DNA transcription have not yet been addressed in detail.
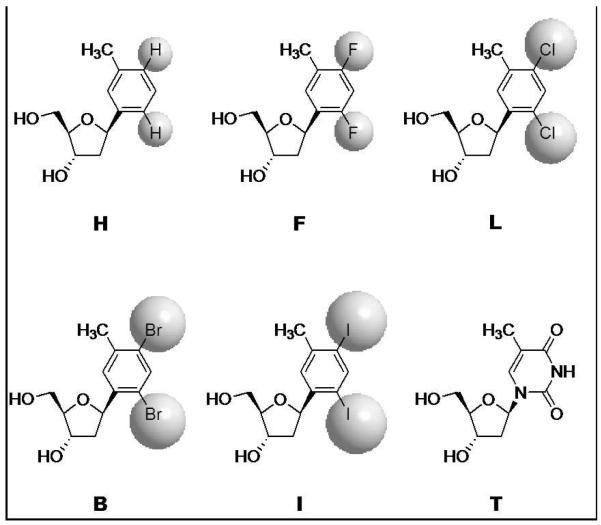
We have introduced nonpolar nucleoside analogues for probing the relative importance of electrostatic and steric effects during DNA replication (11). We have previously shown that E. Coli DNA polymerase I (Klenow fragment) and other A-family DNA polymerases can efficiently and selectively replicate isosteres of thymine (Figure 1) even though they undergo little or no hydrogen bonding (12, 13). A size-exclusion / steric matching model has been proposed which provides a conceptual framework for the role of steric interactions of the base pair within the confined environment of the enzyme’s active site (7, 14, 15). To test steric effects systematically, we recently reported the design and synthesis of a series of nonpolar nucleoside analogues with size varied in sub-Angström increments (Figure 1) (16, 17) and we demonstrated that small size variations can dramatically affect the efficiency and fidelity of the enzymatic replication performed by DNA polymerase I in vitro and by cellular enzymes in E. Coli (18, 19). Overall these results demonstrate that A-family polymerases’ contribution to the stability of base pairs is mainly due to steric interactions within the active site (20). Although all DNA polymerases operate by a chemically similar two-metal-ion mechanism (21), we have shown that these probes can be used to reveal differences in the relative importance of hydrogen bonds and sterics. For instance, hydrogen bonds were found to be important for Y-family error-prone DNA polymerase IV, while steric effects were much less important, suggesting considerable steric flexibility, which may aid in replication beyond damaged bases (19). In line with the size-exclusion model, we have noted a correlation between the structural flexibility of the active site and the fidelity of the enzyme: rigid active site in high-fidelity T7 DNA polymerase I (22) and loose active site in low-fidelity Dpo4 polymerase (23). Here we report the unprecedented use of these nonpolar analogues to probe the roles of steric and electrostatic effects on the efficiency and fidelity of DNA transcription by T7 RNA polymerase.
Figure 1.
Structures and nomenclature of nonpolar nucleoside analogues of thymidine. The van der Waals spheres of the heteroatoms are shown in grey to illustrate the size variations among the series. The interatomic bond lengths (Å) vary as follows: 1.08 (Ar-H), 1.36 (Ar-F), 1.74 (Ar-Cl), 1.90 (Ar-Br), and 2.10 (Ar-I) (18).
Unlike DNA polymerases, RNA polymerases perform a de novo templated synthesis (24). The transcription process starts with the enzyme binding a promoter region and unwinding the upstream DNA duplex, thereby opening a “transcription bubble” (25). The initiation step then involves binding and polymerization of individual nucleoside triphosphates (rNTPs) until a sufficiently long transcript (ca. 8-10 nt) has been formed. Because shorter transcripts can easily dissociate from the template many short products are formed during this “abortive cycling”. The polymerase then undergoes a major conformational change (26-28) – which triggers the release of the promoter – and enters its elongation mode which is characterized by a high processivity (ca. 30-fold increase of the rate of formation of phosphodiester bond in the elongation compared to the initiation phase for T7 RNA polymerase) (29). Structural studies have revealed the different steps of the stepwise nucleotide addition process: 1) rNTP binding at a pre-insertion site, 2) conformational change of the enzyme, 3) chemical reaction, and 4) translocation. Although there are notable structural differences among DNA-templated RNA polymerases, the overall sequence of events is a common feature. The growing RNA transcript remains within the transcription bubble where it is bound to the DNA template. This DNA:RNA heteroduplex adopts an underwound A conformation and its stability is an important factor affecting the efficiency of transcription elongation (30).
Although structural studies have revealed the molecular basis of DNA transcription, our understanding of this process remains essentially qualitative and the detailed balance of forces that determine the efficiency and the fidelity at the molecular level remains elusive. The current studies address this by (a) comparing nonpolar thymine template analogues to the polar thymine, thus addressing the importance of electrostatics in the absence of steric differences, and (b) comparing transcription of variably-sized and shape-altered analogues, thus allowing for the evaluation of the role of sterics (size and shape) in formation of a new RNA-DNA base pair and extension beyond it.
EXPERIMENTAL SECTION
Synthesis of non-polar nucleoside analogues
Compounds H, F, and L were synthesized as previously described (17). Compounds B and I were prepared according to modified procedures (Supporting Information). The synthesis of compound MeL is reported in the Supporting Information. The corresponding DMT-protected and phosphoramidate derivatives were prepared as previously described (16).
Oligonucleotide syntheses
DNA oligonucleotides were synthesized on an Applied Biosystems 394 synthesizer using standard α-cyanoethylphosphoramidite chemistry. DNA oligonucleotides were synthesized in DMT-off mode, deprotected in concentrated NH4OH at 55°C overnight, purified by preparative 20% denaturing polyacrylamide gel electrophoresis, isolated by the crush and soak method, characterized by MALDI-TOF mass spectrometry (Supporting information), and quantitated by absorbance at 260 nm. Molar extinction coefficients were calculated by the nearest neighbor method as previously described (16, 18).
In vitro transcription
1 μL of a 1 μM solution of promoter (final concentration = 100 nM), 1 μL of a 1 μM solution of DNA template (final concentration = 100 nM), and 1 μL of Tris.HCl+NaCl (10 mM each) were annealed by heating to 95°C in 3.5 mins followed by a slow cooling to 4°C in 60 mins. 1 μL of 200 μM of GTP, CTP, and UTP (final concentration = 20 μM) were added, followed by 1 μL of α-32P-ATP (10 μCi, Perkin Elmer). 1 μL of 10x Takara buffer (400 mM Tris-HCl (pH 8.0), 80 mM MgCl2, 20 mM spermidine) and 1 μL of 50 mM DTT were added. After a quick centrifugation of the mixture 1 μL (50 Units (31)) of T7 RNAP (Takara) was added and the mixture was incubated at 37°C. The reaction was quenched by addition of 10 μL of gel loading buffer (10 M urea in 1x TBE) and heating at 75°C for 3 mins and then subjected to denaturing gel electrophoresis (20% polyacrylamide, 7M urea, 1x TBE). Quantification was performed on a PhosphorImager (Molecular Dynamics) with the ImageQuant program.
Transcripts digestion and 2D TLC analyzes
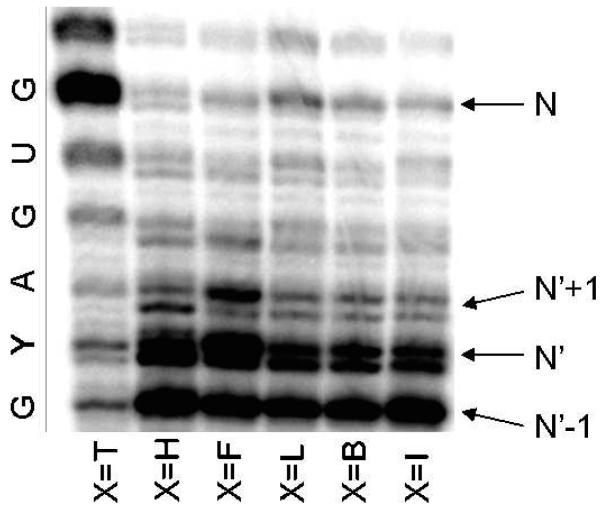
Ribonuclease I (RNase I) was used to digest RNA transcripts and yield a mixture of 3′-monophosphate nucleosides. Because the phosphorus atoms of phosphodiesters remain linked to the 3′ hydroxyl group the 3′-radiolabelled nucleoside monophosphates are those which were connected to the 5′ end of radiolabelled ATP in the RNA transcript. Therefore, we excised band N’+1 (see Figure 3) since the radiolabelled phosphorus atom present at the 3′ end (A) of the transcript will be transferred to the 3′-monophosphate nucleoside Y, thus making the site of interest for the determination of the transcription fidelity visible by radioimaging.
Figure 3.
Enzymatic transcription with different DNA templates (X=T, H, F, L, B, and I). N denotes the band corresponding to the full transcript whereas N’ denotes the band corresponding to the transcript stalled at the position of the artificial modification X. The transcript sequence is noted on the left. Reaction time: 30 min.
The band of interest was located by exposure to film (Kodak X-OmatAR5). It was then excised and the product was eluted with 200 μL of water for 8 hours. 1 μL of 3M sodium acetate, 1 μL of ca. 1 mM tRNA (Sigma Aldrich) and 1.2 mL of ethanol were then added to the solution which was kept overnight in the freezer. After a centrifugation, the supernatant solution was removed. 16 μL of water, 2 μL of 10x TNE buffer, and 1 μL RNAse I (10 U, Epicentre) were successively added and the mixture was incubated at 37°C for 3-4 hours. The reaction was then directly loaded on TLC plates (HPTLC plates, 100 x 100 mm, Merck, Darmstadt, Germany). Elution was performed with the following mixtures: 1st dimension: isobutyric acid/ammonia/H2O: 66/1/33, 2nd dimension: sodium phosphate (0.1 M, pH 6.8)/ammonium sulfate/n-propanol: 100(v)/60(w)/2(v). Quantification was performed on a PhosphorImager (Molecular Dynamics) with the ImageQuant program.
RESULTS
Templates containing analogues
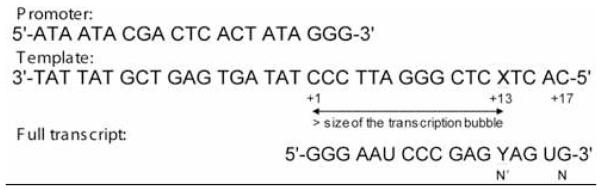
We prepared template DNAs containing nonpolar isostere F or native T to evaluate the importance of hydrogen bonding in synthesis of a new base pair at a position 13 nt downstream from the transcription start site (Figure 2). We also synthesized templates with variably sized thymine analogues H, F, L, B, and I (Figure 1) to measure the role of steric effects in this process. Modified DNAs were characterized by mass spectrometry (see Supporting Information).
Figure 2.
Sequences of the DNA promoter top strand, DNA template, and RNA full transcript. X denotes the position where nonpolar analogues (H, F, L, B, and I) were placed. Y, at position N’ of the RNA transcript, denotes the nucleoside placed opposite X during the enzymatic transcription by T7 RNAP.
In vitro transcription assays
Enzymatic transcription reactions were performed with T7 RNAP on a 35-mer DNA template featuring a) the consensus double-stranded T7 RNAP 18-mer promoter, and b) a 17-mer containing the non-polar nucleoside analogues at position 13 (Figure 2). The modification was placed 13 nucleotides downstream of the initiation position to ensure that the enzyme was in elongation mode once it reached that position. The context of the modified nucleobase (X) was 5′TXC, which should be noted since context can exert influences on RNA polymerase fidelity (32, 33).
Enzymatic transcription reactions were monitored by internal radioisotopic labelling with α-32P-ATP. Since T7 RNAP does not exhibit intrinsic proof-reading (33), we applied the standard model (34) used in running-start reactions with DNA polymerases to quantify the efficiency of transcription which is defined by the ratio of the rate constant for polymerization (kpol) and the rate constant for pausing (koff). The efficiency of transcription at a site m can thus be calculated as follows:
where Im is the intensity of the band corresponding to the stalled product at position m and In is the intensity of each elongated transcript. In the present case, transcripts up to 18-mer were taken into account as longer transcripts were seen to be minor by-products most probably arising from RNA-templated RNA synthesis (35). As defined, the efficiency of transcription represents the ability of the polymerase to pass a site m and produce extended transcripts.
Transcription efficiency
We first assessed the overall transcription efficiency by comparing the amount of full transcript produced with the different templates. A gel electrophoretic analysis of the enzymatic reactions with different DNA templates is shown in Figure 3. Full-length transcript (N) is clearly visible for the thymine template and for the most efficient of the analogues. We noted that several bands appear doubled both for natural and unnatural templates. A composition analysis was performed on the two N’+1 bands excised separately and showed no significant differences in composition at this position (see Supporting Information). In the subsequent data analyses we therefore considered the ensemble of two bands together, since the apparent inhomogeneity occurred at an earlier site in the transcript.
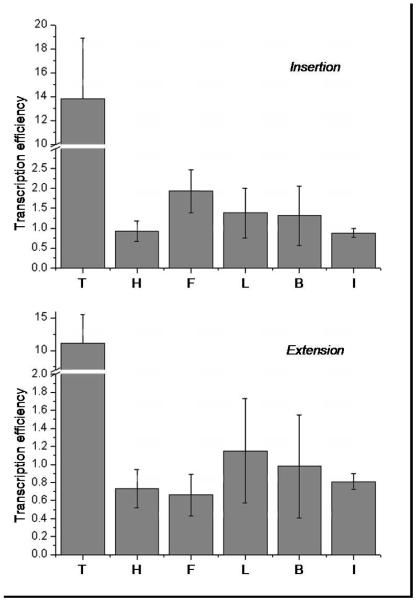
In comparison to the natural thymidine-containing template, the transcription of DNA templates containing nonpolar analogues gave markedly lower yields of full transcript (5-10% relative to the natural template). To gain a more detailed understanding of the origin of this lower efficiency, we determined the efficiency of insertion and elongation with equation (1). The results are represented in Figure 4 below.
Figure 4.
Comparison of the transcription efficiencies of insertion (top) and extension (bottom) for varied DNA templates (X=T, H, F, L, B, and I). Data obtained from equation (1) where m=N’-1 (insertion) or N’ (extension). Reaction time = 30 min (3 replicates). Error bars represent one standard deviation.
When comparing the natural template T with its nonpolar isostere F, the results show that the efficiency of both insertion and extension are considerably decreased (by ca. one order of magnitude). Comparison among the series of nonpolar analogues allows probing the effect of small size variations on the efficiency of transcription and interesting effects were observed. First, the smallest and largest substrates, H and I respectively, display the poorest efficiency of insertion, significantly lower than with analogue F (Student’s t-test between H and F: Prob>∣t∣=0.0538 (p-value=0.0715); Student’s t-test between F and I: Prob>∣t∣=0.0528 (p-value=0.0613)). Second, a small preference for F over L and B is observed. The relationship between efficiency of transcription and size of the analogues thus follows a bell-shaped trend similar to previous observations with DNA polymerase I (18), except that the maximum here lies at F, while previous DNA polymerases showed a slightly larger preference (L). In contrast, the efficiencies of elongation show little dependence upon the size of the analogues, thereby suggesting that steric constraints are lower further into the enzyme active site.
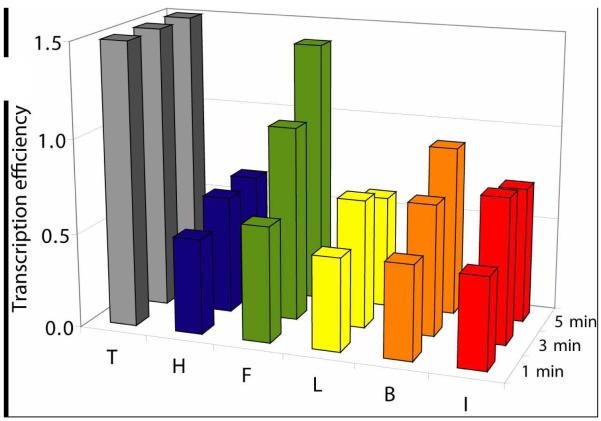
We then monitored the initial phase of transcription and determined the transcription efficiency of insertion at low product conversion (i.e. at short reaction times) (Figure 5 and Supporting Information).
Figure 5.
Comparison of the transcription efficiencies of insertion between different DNA templates (X=T, H, F, L, B, and I) during the initial phase of transcription at low product conversion. Data obtained from equation (1) where m=N’-1. Bars for T are truncated, actual values being 10.1 at 1 min, 9.6 at 3 min, and 10.0 at 5 min. Reaction time noted on the right (1, 3, and 5 min).
Overall this experiment confirms the ca. 10-fold reduction in the efficiency of transcription when thymine is replaced by a nonpolar analogue. However, the results clearly show variations among the series of analogues, F being processed much better (ca. 2 times) than the other size variants, thereby confirming that the correlation between the efficiency of insertion and the size of the nonpolar analogues follows a curve centered on F, which is the same size as thymine.
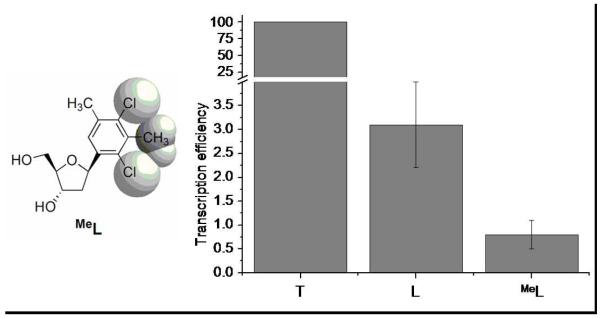
In addition to nucleobase size, another relevant steric property is shape. To assess how the shape of thymine, with its substitution at 2,4 positions, and lack thereof at the 3-position, affect activity, we prepared a previously unknown nonpolar nucleoside analogue having a distorted shape. Compound MeL features an extra methyl group protruding at the 3-position of the phenyl ring (Figure 6), disrupting the thymine-like shape markedly (36). It is a nonpolar mimic of N-3-methylated thymine, a common form of DNA damage (37-40). We tested the effect of this shape modification by comparing enzymatic transcription reactions with DNA templates L and MeL (Figure 6 and Supporting Information).
Figure 6.
/B> Structure of shape-modified nonpolar nucleoside analogue MeL with van der Waals spheres shown in grey (left), and full transcript yields of enzymatic transcription reactions with different DNA templates (X=T, L, and MeL) (right). Normalized yields (template T, 100). Reaction time: 30 min (3 replicates). Error bars represent one standard deviation.
The experiments revealed that the shape-modified analogue MeL is processed less efficiently by T7 RNAP and leads to significantly more stalling. The full transcript yield is 4 times lower with MeL than with L, thereby demonstrating that a thymine-like shape makes a significant contribution to the overall efficiency of base pair formation (presumably opposite adenine, vide infra). More detailed analysis revealed that both the efficiency of insertion and extension are adversely affected when L is replaced by MeL (see Supporting Information), consistent with the notion that the base pair being synthesized must fit into an active site that normally surrounds a canonical pair.
Transcription fidelity
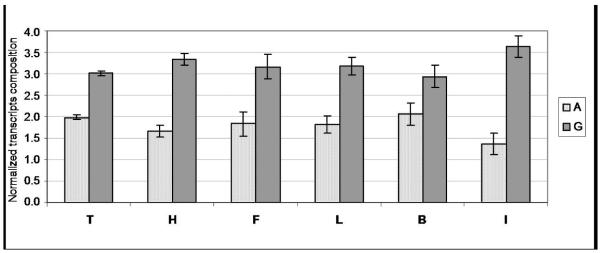
To determine the fidelity of the enzymatic transcription of the thymine analogues, we analyzed the composition of transcripts by performing an enzymatic digestion followed by 2D-TLC separation and quantification by radioimaging (Figure 7 and Supporting Information). This established method has been previously used in similar contexts (41, 42).
Figure 7.
Normalized composition of transcripts N’+1 determined by enzymatic digestion followed by 2D TLC separation and quantification by radioimaging. Error bars represent one standard deviation. Minimum of 3 replicates.
For X=T, the enzymatic digestion of the transcript N’+1 should yield a ratio of 3 radiolabeled GMP mononucleotides to 2 radiolabeled AMP (see Experimental section for analysis). Any deviation from this composition should therefore reflect a decreased fidelity. As shown in Figure 7, the composition analysis of the transcript N’+1 formed from the enzymatic transcription of the natural DNA template where X=T indeed gives the expected ratio of 3 GMP to 2 AMP. When the enzymatic transcription was carried out with the modified DNA template containing the nonpolar isostere F the results showed a similar composition of transcript, thereby indicating a comparable fidelity of transcription. Interestingly, significant differences were observed among the series of nonpolar analogues (Figure 7). The smallest (H) and the largest (I) analogues gave the largest deviation from the ideal fidelity one expects if they were recognized as thymine analogues, showing that they are processed with significantly lower fidelity (the theoretical limit for the case of a 0% fidelity opposite site X being a value of 1.25 for A, Figure 7).
DISCUSSION
Effects on the efficiency of insertion
Our results clearly show an overall reduction in the efficiency of nucleotide insertion by T7 RNAP when hydrogen-bonding groups are missing in the templating base. This can either be due to an inhibition of the required conformational change of the enzyme or to a misalignment of the rNTP within the active site of T7 RNAP. Patel et al. have recently suggested, based on transient state kinetics studies, that the induced-fit conformational change is the rate-determining step of DNA transcription by T7 RNAP (29). It is therefore reasonable to surmise that the variations of transcription efficiency observed here are due to perturbation of the rates or populations of open vs. closed conformations of the enzyme. Crystal structures of the complexes with a complementary rNTP in the preinsertion (43) and insertion (44) sites have shown that the template base and the incoming rNTP adopt more favorable orientations and are in closer contacts – through hydrogen bonds – in the insertion site than in the preinsertion site. Furthermore, previous studies with DNA Pol I have shown that hydrogen bonds play a significant (although not essential) role in stabilizing the closed ternary complex (45). Since DNA Pol I and T7 RNAP share considerable homology (46, 47) it is plausible that the same is true for T7 RNAP. Overall these data suggest that hydrogen bonds are important for triggering the conformational change of T7 RNAP, and that the effect is greater for this RNA polymerase than for A-family DNA polymerases. Specifically which hydrogen bonds play the most important role is discussed below.
Our observation that varying templating base size in very small gradations affects efficiency significantly suggests that the steric accommodation of the incoming NTP within the enzyme active site also contributes to the transcription efficiency, most probably by affecting the positioning of reactive groups (43). This conclusion is in line with biochemical studies which have shown that in T7 RNAP mutations which facilitate the accommodation of incorrect base pairs in the catalytically competent closed conformation increase misincorporation (33, 43). The observation that a template base (MeL) having a shape improper for pairing with any natural base gives a lower efficiency of insertion also supports the idea that steric effects do play a role in transcription efficiency.
Effects on the efficiency of extension
Our results show that the extension step with T7 RNA polymerase depends strongly on hydrogen bonding groups in the templating base. Hydrogen bonds involving nucleobases in polymerase active sites can play a role two ways: via H-bonds between the bases, and between the bases and the minor groove of the enzyme. Hirao et al. (41) and Romesberg et al. (42) have shown that T7 RNAP is capable of efficiently incorporating and extending synthetic base pairs lacking hydrogen bonds between them. This is in contrast with our present observation, where transcription efficiency is apparently lower. However, the compounds used in those prior studies all feature polar moieties pointing toward the minor groove, while the present compounds do not. This suggests that the most important electrostatic effect is not hydrogen bonds between the bases, but rather with the enzyme directly. Biochemical studies have previously shown that mutation H784A results in an enhanced extension rate of misincorpations (33). In addition, examination of recent structural data reveals likely polar contacts between H784 and the minor groove edge of the 3′ end of the RNA transcript (distance of 2.8 Å between the N-3 of H784 and the N-3 of the terminal adenine in the RNA transcript) (44). Our results suggest that moving a nonpolar base opposite such a polar group is destabilizing at the transition state of extension, possibly by desolvating the polar group. Similar effects have been seen with DNA polymerases previously (48). Our results suggest that electrostatic interactions between H784 and the minor groove edge of the base pair is important for the extension (49). Overall the results suggest that H784 plays in crucial role as a checkpoint and that it senses newly-formed base pairs through polar contacts in the minor groove of the 3′ end of the RNA transcript.
Effects on the fidelity
The results obtained here show similar values of fidelity when thymine is replaced by synthetic nonpolar analogues, thereby showing that the enzyme recognizes these substrates as if they were thymine templates. This result demonstrates that in T7 RNAP, Watson-Crick hydrogen bonds are not essential to determine the fidelity of transcription. This is consistent with the results of Hirao et al. (41) and Romesberg et al. (42), who have shown substantial fidelity in novel nonpolar base pairs. Biochemical studies have shown that enzyme mutations that stabilize the catalytically-active closed complex increase misincorporation (33). This gives support to a screening and selection of the appropriate rNTP that takes place when the enzyme is in the catalytically-incompetent open state – the induced-fit conformational change from the open to the closed state being thus powered by the binding energy of the rNTP into the active site opposite the templating DNA nucleobase (43). Since T7 RNAP does not display intrinsic proof-reading (33), the fidelity of the transcription is determined by the selectivity of recognition of the correct rNTP in the preinsertion complex. Our results indicate that fidelity is retained in the absence of hydrogen bonds between the partners. Small size variations, tested with the series of nonpolar analogues varying size in sub-Angström increments, indicate that the preinsertion site is somewhat plastic and can accommodate the smallest size variations reasonably well. However, larger size variations – as seen with analogues H and I – significantly reduced the fidelity of the transcription, thereby indicating the presence of significant steric gating.
Overall, the results show the utility of size-varied nonpolar nucleobase analogues for analyzing mechanisms in a class of polymerases distinct from DNA polymerases. Future work will apply these analogues to the mechanistic study of enzymes involved in eukaryotic transcription.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge the U.S. National Institutes of Health (GM072705) for support. We also ackowledge Dr. Kazuteru Usui for his contribution to the synthesis of dB. S.U. thanks the French-American Fulbright commission and the Region Alsace for financial support.
Funding information. We acknowledge the U.S. National Institutes of Health (GM072705) for support. S.U. thanks the French-American Fulbright commission and the Region Alsace for financial support.
Abbreviations
- TLC
Thin Layer Chromatography
- NTP
Nucleoside TriPhosphate
- DTT
Dithiothreitol
- TBE
Tris-Borate-EDTA
Footnotes
SUPPORTING INFORMATION AVAILABLE. Experimental details on the synthesis and characterization of compounds B, I, and MeL, and additional data on the enzymatic transcription assays can be found in the Supporting Information. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Kornberg RD. The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. USA. 2007;104:12955–12961. doi: 10.1073/pnas.0704138104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nudler E. RNA Polymerase Active Center: The Molecular Engine of Transcription. Annu. Rev. Biochem. 2009;78:335–361. doi: 10.1146/annurev.biochem.76.052705.164655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sydow JF, Cramer P. RNA polymerase fidelity and transcriptional proofreading. Curr. Opin. Struct. Biol. 2009;19:732–739. doi: 10.1016/j.sbi.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 4.Saxowsky TT, Doetsch PW. RNA polymerase encounters with DNA damage: Transcription-coupled repair or transcriptional mutagenesis? Chem. Rev. 2006;106:474–488. doi: 10.1021/cr040466q. [DOI] [PubMed] [Google Scholar]
- 5.Doetsch PW. Translesion synthesis by RNA polymerases: occurrence and biological implications for transcriptional mutagenesis. Mutat. Res. 2002;510:131–140. doi: 10.1016/s0027-5107(02)00258-0. [DOI] [PubMed] [Google Scholar]
- 6.Greive SJ, von Hippel PH. Thinking quantitatively about transcriptional regulation. Nature Rev. Mol. Cell Biol. 2005;6:221–232. doi: 10.1038/nrm1588. [DOI] [PubMed] [Google Scholar]
- 7.Kool ET, Morales JC, Guckian KM. Mimicking the structure and function of DNA: Insights into DNA stability and replication. Angew. Chem. Int. Ed. 2000;39:990–1009. doi: 10.1002/(sici)1521-3773(20000317)39:6<990::aid-anie990>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- 8.Goodman MF. Hydrogen bonding revisited: Geometric selection as a principal determinant of DNA replication fidelity. Proc. Natl. Acad. Sci. USA. 1997;94:10493–10495. doi: 10.1073/pnas.94.20.10493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Echols H, Goodman MF. Fidelity Mechanisms in DNA-Replication. Annu. Rev. Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- 10.Petruska J, Goodman MF, Boosalis MS, Sowers LC, Cheong C, Tinoco I. Comparison between DNA Melting Thermodynamics and DNA-Polymerase Fidelity. Proc. Natl. Acad. Sci. USA. 1988;85:6252–6256. doi: 10.1073/pnas.85.17.6252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krueger AT, Kool ET. Model systems for understanding DNA base pairing. Curr. Opin. Chem. Biol. 2007;11:588–594. doi: 10.1016/j.cbpa.2007.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kool ET. Synthetically modified DNAs as substrates for polymerases. Curr. Opin. Chem. Biol. 2000;4:602–608. doi: 10.1016/s1367-5931(00)00141-1. [DOI] [PubMed] [Google Scholar]
- 13.Moran S, Ren RXF, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc. Natl. Acad. Sci. USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kool ET. Active site tightness and substrate fit in DNA replication. Annu. Rev. Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- 15.Kool ET. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 2001;30:1–22. doi: 10.1146/annurev.biophys.30.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Kim TW, Kool ET. A series of nonpolar thymidine analogues of increasing size: DNA base pairing and stacking properties. J. Org. Chem. 2005;70:2048–2053. doi: 10.1021/jo048061t. [DOI] [PubMed] [Google Scholar]
- 17.Kim TW, Kool ET. A set of nonpolar thymidine nucleoside analogues with gradually increasing size. Org. Lett. 2004;6:3949–3952. doi: 10.1021/ol048487u. [DOI] [PubMed] [Google Scholar]
- 18.Kim TW, Delaney JC, Essigmann JM, Kool ET. Probing the active site tightness of DNA polymerase in subangstrom increments. Proc. Natl. Acad. Sci. USA. 2005;102:15803–15808. doi: 10.1073/pnas.0505113102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silverman AP, Jiang QF, Goodman MF, Kool ET. Steric and electrostatic effects in DNA synthesis by the SOS-induced DNA polymerases II and IV of Escherichia coli. Biochemistry-Us. 2007;46:13874–13881. doi: 10.1021/bi700851z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kool ET, Sintim HO. The difluorotoluene debate - a decade later. Chem. Comm. 2006:3665–3675. doi: 10.1039/b605414e. [DOI] [PubMed] [Google Scholar]
- 21.Steitz TA. A mechanism for all polymerases. Nature. 1998;391:231–232. doi: 10.1038/34542. [DOI] [PubMed] [Google Scholar]
- 22.Kim TW, Brieba LG, Ellenberger T, Kool ET. Functional evidence for a small and rigid active site in a high fidelity DNA polymerase - Probing T7 DNA polymerase with variably sized base pairs. J. Biol. Chem. 2006;281:2289–2295. doi: 10.1074/jbc.M510744200. [DOI] [PubMed] [Google Scholar]
- 23.Mizukami S, Kim TW, Helquist SA, Kool ET. Varying DNA base-pair size in subangstrom increments: Evidence for a loose, not large, active site in low-fidelity Dpo4 polymerase. Biochemistry-Us. 2006;45:2772–2778. doi: 10.1021/bi051961z. [DOI] [PubMed] [Google Scholar]
- 24.Kennedy WP, Momand JR, Yin YW. Mechanism for de novo RNA synthesis and initiating nucleotide specificity by T7 RNA polymerase. J. Mol. Biol. 2007;370:256–268. doi: 10.1016/j.jmb.2007.03.041. [DOI] [PubMed] [Google Scholar]
- 25.Tahirov TH, Temiakov D, Anikin M, Patlan V, McAllister WT, Vassylyev DG, Yokoyama S. Structure of a T7 RNA polymerase elongation complex at 2.9 angstrom resolution. Nature. 2002;420:43–50. doi: 10.1038/nature01129. [DOI] [PubMed] [Google Scholar]
- 26.Durniak KJ, Bailey S, Steitz TA. The structure of a transcribing T7 RNA polymerase in transition from initiation to elongation. Science. 2008;322:553–557. doi: 10.1126/science.1163433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steitz TA. The structural basis of the transition from initiation to elongation phases of transcription, as well as translocation and strand separation, by T7 RNA polymerase. Curr. Opin. Struct. Biol. 2004;14:4–9. doi: 10.1016/j.sbi.2004.01.006. [DOI] [PubMed] [Google Scholar]
- 28.Yin YW, Steitz TA. Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science. 2002;298:1387–1395. doi: 10.1126/science.1077464. [DOI] [PubMed] [Google Scholar]
- 29.Anand VS, Patel SS. Transient state kinetics of transcription elongation by T7 RNA polymerase. J. Biol. Chem. 2006;281:35677–35685. doi: 10.1074/jbc.M608180200. [DOI] [PubMed] [Google Scholar]
- 30.von Hippel PH. Transcription - An integrated model of the transcription complex in elongation, termination, and editing. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 31.One unit is, as defined by the supplier, the amount of the enzyme that incorporates 1 nmol of [3H]GMP into the acid-insoluble products in 1 hour at 37°C and pH 8.0.
- 32.Holmes SF, Erie DA. Downstream DNA Sequence Effects on Transcription Elongation. J. Biol. Chem. 2003;278:35597–35608. doi: 10.1074/jbc.M304496200. [DOI] [PubMed] [Google Scholar]
- 33.Huang JB, Brieba LG, Sousa R. Misincorporation by wild-type and mutant T7 RNA polymerases: Identification of interactions that reduce misincorporation rates by stabilizing the catalytically incompetent open conformation. Biochemistry-Us. 2000;39:11571–11580. doi: 10.1021/bi000579d. [DOI] [PubMed] [Google Scholar]
- 34.Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical Basis of DNA-Replication Fidelity. Crit. Rev. Biochem. Mol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- 35.Cazenave C, Uhlenbeck OC. RNA Template-Directed RNA-Synthesis by T7 RNA-Polymerase. Proc. Natl. Acad. Sci. USA. 1994;91:6972–6976. doi: 10.1073/pnas.91.15.6972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shape effects were tested on the 2,4-dichloro analogue for synthetic reasons - MeL being synthetically more accessible than MeF.
- 37.Falnes PO. Repair of 3-methylthymine and 1-methylguanine lesions by bacterial and human AlkB proteins. Nucleic Acids Res. 2004;32:6260–6267. doi: 10.1093/nar/gkh964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koivisto P, Robins P, Lindahl T, Sedgwick B. Demethylation of 3-methylthymine in DNA by bacterial and human DNA dioxygenases. J. Biol. Chem. 2004;279:40470–40474. doi: 10.1074/jbc.M407960200. [DOI] [PubMed] [Google Scholar]
- 39.Delaney JC, Essigmann JM. Mutagenesis, genotoxicity, and repair of 1-methyladenine, 3-alkylcytosines, 1-methylguanine and 3-methylthymine, in alkB Escherichia coli. Proc. Natl. Acad. Sci. USA. 2004;101:14051–14056. doi: 10.1073/pnas.0403489101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huff AC, Topal MD. DNA Damage at Thymine N-3 Abolishes Base-Pairing Capacity during DNA-Synthesis. J. Biol. Chem. 1987;262:12843–12850. [PubMed] [Google Scholar]
- 41.Hirao I, Kimoto M, Mitsui T, Fujiwara T, Kawai R, Sato A, Harada Y, Yokoyama S. An unnatural hydrophobic base pair system: site-specific incorporation of nucleotide analogs into DNA and RNA. Nature Methods. 2006;3:729–735. doi: 10.1038/nmeth915. [DOI] [PubMed] [Google Scholar]
- 42.Seo YJ, Matsuda S, Romesberg FE. Transcription of an Expanded Genetic Alphabet. J. Am. Chem. Soc. 2009;131:5046–5047. doi: 10.1021/ja9006996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Temiakov D, Patlan V, Anikin M, McAllister WT, Yokoyama S, Vassylyev DG. Structural basis for substrate selection by T7 RNA polymerase. Cell. 2004;116:381–391. doi: 10.1016/s0092-8674(04)00059-5. [DOI] [PubMed] [Google Scholar]
- 44.Yin YW, Steitz TA. The structural mechanism of translocation and helicase activity in T7 RNA polymerase. Cell. 2004;116:393–404. doi: 10.1016/s0092-8674(04)00120-5. [DOI] [PubMed] [Google Scholar]
- 45.Dzantiev L, Alekseyev YO, Morales JC, Kool ET, Romano LJ. Significance of nucleobase shape complementarity and hydrogen bonding in the formation and stability of the closed polymerase-DNA complex. Biochemistry-Us. 2001;40:3215–3221. doi: 10.1021/bi002569i. [DOI] [PubMed] [Google Scholar]
- 46.Sousa R, Mukherjee S. T7 RNA polymerase. Prog. Nucleic Acid Res. Mol. Biol. 2003;73:1–41. doi: 10.1016/s0079-6603(03)01001-8. [DOI] [PubMed] [Google Scholar]
- 47.McAllister WT, Raskin CA. The phage RNA-polymerases are related to DNA-polymerases and reverse transcriptases. Mol. Microbiol. 1993;10:1–6. doi: 10.1111/j.1365-2958.1993.tb00897.x. [DOI] [PubMed] [Google Scholar]
- 48.Morales JC, Kool ET. Minor groove interactions between polymerase and DNA: More essential to replication than Watson-Crick hydrogen bonds? J. Am. Chem. Soc. 1999;121:2323–2324. doi: 10.1021/ja983502+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brieba LG, Sousa R. Roles of histidine 784 and tyrosine 639 in ribose discrimination by T7 RNA polymerase. Biochemistry-Us. 2000;39:919–923. doi: 10.1021/bi992324+. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.