CLOCK is the master transcriptional regulator of the Drosophila circadian clock. Genome-wide analysis is used here to catalog a surprisingly large number of direct target genes of CLOCK and its partner, CYC. This study provides multiple insights into oscillating RNA Pol II occupancy and previously unobserved RNA cycling patterns, as well as temporal binding patterns and gene- and tissue-specific binding of CLOCK/CYC.
Keywords: circadian rhythms, Drosophila, transcription
Abstract
CLOCK (CLK) is a master transcriptional regulator of the circadian clock in Drosophila. To identify CLK direct target genes and address circadian transcriptional regulation in Drosophila, we performed chromatin immunoprecipitation (ChIP) tiling array assays (ChIP–chip) with a number of circadian proteins. CLK binding cycles on at least 800 sites with maximal binding in the early night. The CLK partner protein CYCLE (CYC) is on most of these sites. The CLK/CYC heterodimer is joined 4–6 h later by the transcriptional repressor PERIOD (PER), indicating that the majority of CLK targets are regulated similarly to core circadian genes. About 30% of target genes also show cycling RNA polymerase II (Pol II) binding. Many of these generate cycling RNAs despite not being documented in prior RNA cycling studies. This is due in part to different RNA isoforms and to fly head tissue heterogeneity. CLK has specific targets in different tissues, implying that important CLK partner proteins and/or mechanisms contribute to gene-specific and tissue-specific regulation.
Organisms ranging from cyanobacteria to humans display changes in metabolism, physiology, and behavior that undergo daily oscillations with ∼24-h periods. These oscillations are regulated by core circadian clocks, which function to drive and orchestrate these daily fluctuations. In Drosophila, the core clock is comprised, in part, of two interlocked feedback loops. The principal negative feedback loop includes the basic helix–loop–helix PAS transcription factors CLOCK (CLK) and CYCLE (CYC), which heterodimerize and bind to E-boxes (CACCTG) of the core clock genes period (per) and timeless (tim) to activate their transcription (Yu et al. 2006; Taylor and Hardin 2008). per and tim mRNAs are translated in the cytoplasm; PER and TIM heterodimerize, become phosphorylated, and localize to the nucleus (Hardin et al. 1990; Edery et al. 1994; Curtin et al. 1995; Shafer et al. 2002; Meyer et al. 2006). PER and TIM then repress CLK-mediated transcription, followed by their degradation in the late night/early morning (Edery et al. 1994; Darlington et al. 1998; Ko et al. 2002; Menet et al. 2010; Sun et al. 2010). In the second feedback loop, CLK/CYC directly activates the transcription of vri and pdp1 (Blau and Young 1999; McDonald et al. 2001; Ueda et al. 2002). The resulting proteins, VRI and PDP1, may then regulate clock (Clk) transcription, either negatively (VRI) or positively (PDP1), contributing to rhythmic Clk transcription (Cyran et al. 2003). Another level of regulation is provided by the core clock gene clockwork orange (cwo) (Kadener et al. 2007; Lim et al. 2007; Matsumoto et al. 2007). It is also a CLK/CYC direct target gene and encodes a transcriptional repressor that contributes to the temporal repression of CLK/CYC activity like PER and TIM. These five CLK/CYC direct target genes (per, tim, vri, pdp1, and cwo), along with Clk and cyc, are considered core clock genes and act to maintain robust molecular circadian rhythms of the Drosophila molecular clock.
CLK/CYC and their homologs, CLK/BMAL1, in mammals are considered the master regulators of the molecular circadian clock. For example, ectopic expression of Drosophila Clk in noncircadian locations can induce the formation of ectopic clocks by the criterion of PER expression and cycling (Zhao et al. 2003), and a dominant-negative mutation of Clk strongly diminishes all behavioral and molecular oscillations in flies (clkjrk) (Allada et al. 1998) and mice (CLKΔ19) (King et al. 1997). The circadian period of locomotor activity rhythms is sensitive to Clk gene dose in both organisms (Antoch et al. 1997; Kadener et al. 2008). This central role of CLK/CYC and CLK/BMAL1 suggests a simple model in which the heterodimer directly controls a limited number of key genes. CLK direct target genes in flies like per, tim, vri, pdp1, and cwo—all transcription factors—would then control the cyclical expression of output genes. Consistent with this transcriptional cascade model, studies in Drosophila S2 cells and fly heads identified only 28 CLK direct target genes, including the five transcription factor core clock genes and other transcription factors (Kadener et al. 2007).
To initiate an understanding of the role of CLK in direct target gene regulation, we recently described chromatin immunoprecipitations (ChIPs) for CLK, PER, and RNA polymerase II (Pol II) on per and tim (Menet et al. 2010). CLK is maximally recruited to the promoters of these genes in the early night, Zeitgeber times 14–16 (ZT14–ZT16). At these times, transcription is active, also evident by the presence of Pol II in coding regions. PER binds to per and tim chromatin at ZT18 with a concomitant decrease in transcription and Pol II signal. This is followed by a further decrease in transcription and CLK binding, resulting in minimal transcription and minimal CLK binding at about ZT22–ZT2. The results inspired a model of sequential “ON-DNA” and “OFF-DNA” transcriptional repression. In the “ON-DNA” phase, PER binds to per and tim chromatin, presumably via CLK/CYC, to repress transcription. This is followed by the “OFF-DNA” phase, in which CLK/CYC is mostly absent from chromatin and transcription is minimal.
To identify additional Drosophila direct target genes as well as confirm and extend this model, we expanded on this initial study (Menet et al. 2010) and present here a genome-wide analysis of CLK, PER, CYC, and Pol II binding to chromatin from Drosophila heads. There are ∼1500 CLK-binding peaks, at least 60% of which cycle with maximal CLK binding at ZT14 in early night. At this time, CYC is also present in the same regions that bind CLK, and 4–6 h later, the repressor PER is also bound to CLK direct targets. This suggests that the majority of CLK direct targets are regulated similarly to the core clock genes (Menet et al. 2010). About 30% of target genes show cyclical Pol II binding at promoters or within coding regions, which correlates with active transcription. Many of these CLK direct targets are of interest and have never been previously implicated in circadian transcriptional studies; e.g., in circadian microarray assays focused on identifying cycling mRNAs. A recent study in mice suggests that BMAL1 also binds to a large number of genes in the liver (>2000), only 29% of which had been previously implicated to be under circadian regulation (Rey et al. 2011). In the case of these fly data, we show that the discrepancy with previous cycling RNA studies is due to (1) CLK binding and regulation of specific mRNA isoforms; (2) low mRNA cycling amplitudes for many of these direct target genes, and (3) the tissue complexity of the fly head. Heterogeneity of CLK binding within different head tissues suggests the presence of important CLK partner proteins and mechanisms that contribute to gene-specific and tissue-specific circadian transcriptional regulation.
Results
Identification of CLK direct target genes in Drosophila
To identify CLK direct target genes, we used a strain with two Clk-V5 transgenes (previously described; Kadener et al. 2008) to perform anti-CLK ChIPs at six time points throughout the day (see the Materials and Methods). Anti-V5 ChIPs were performed from Drosophila head extracts, and the CLK-bound DNA fragments were identified using Drosophila Tiling 2.0 arrays (Affymetrix). Peaks of CLK binding were identified using the MAT algorithm (Johnson et al. 2006). One-thousand-five-hundred significant CLK peaks were identified, defined as a site with a P-value of <10−4 in two independent CLK ChIP experiments (see the Materials and Methods; Supplemental Fig. 1).
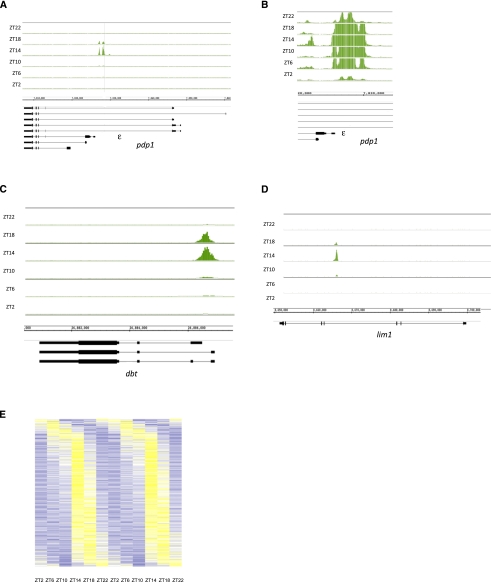
As expected, these significant CLK peaks were enriched for canonical E-boxes as well as degenerate E-boxes (Supplemental Table 2). The top five ClK-binding sites, ranked by statistical significance, are within or adjacent to the known core clock genes vri, tim, pdp1, per, and cwo (Supplemental Table 1). We previously described cycling CLK binding to per and tim, and show here pdp1; there is very strong cycling CLK binding to the middle of the gene, adjacent to the start site of the circadian isoform pdp1ε (Fig. 1A; Zheng et al. 2009), suggesting that this cycling drives circadian transcription of this isoform. Like for per and tim (Taylor and Hardin 2008; Menet et al. 2010), CLK levels increase until the signal peaks at ZT14, after which it decreases back to low levels. However, CLK binding is still above background even at these trough values, suggesting that the chromatin is never completely inaccessible (Fig. 1B).
Figure 1.
CLK binding to direct target genes peaks at ZT14. CLK ChIPs were performed at six different time points throughout the day, and the resulting DNA was analyzed using tiling arrays (Affymetrix). CLK binding is visualized using the IGB. Genes above the genomic coordinates are transcribed from left to right (plus strand), and genes below the genomic coordinates are transcribed from right to left (minus strand). CLK binds rhythmically to the promoters of pdp1 (primarily the ε isoform) (A), of pdp1 (primarily the ε isoform) zoomed in to show binding even at ZT2 (B), of dbt (C), and in the middle of lim1 (D). CLK binding peaks at ZT14 on these three genes. (E) CLK binding cycles on ∼800 genes. Genes were sorted by binding phase, and CLK ChIP signal is portrayed using a heat map in which data for a 24-h period are concatenated to show cycling. CLK ChIP signal ranges from low (dark blue; Z-score between −2 and −0.5; i.e., between 2 and 0.5 standard deviations below the mean) to medium (white; Z-score between −0.5 and +0.5) to highest (yellow; Z-score between 0.5 and 2). For most genes, the highest CLK ChIP signal is at ZT14.
To identify other peaks where CLK binding cycles with a 24-h period, we performed a modified Fourier analysis with an F24 cutoff of 0.7 and a P-value of <0.05 (Wijnen et al. 2005). With these stringent parameters, ∼60% of the 1500 peaks manifest circadian cycling. As observed for the core clock genes (Taylor and Hardin 2008; Menet et al. 2010), maximal CLK binding on most of these genes occurs between ZT14 and ZT16 (Fig. 1E, 2C). Although 40% of CLK peaks were characterized as “noncycling” using our stringent parameters, most oscillate upon visual inspection, suggesting that 60% is a gross underestimate (Supplemental Fig. 2).
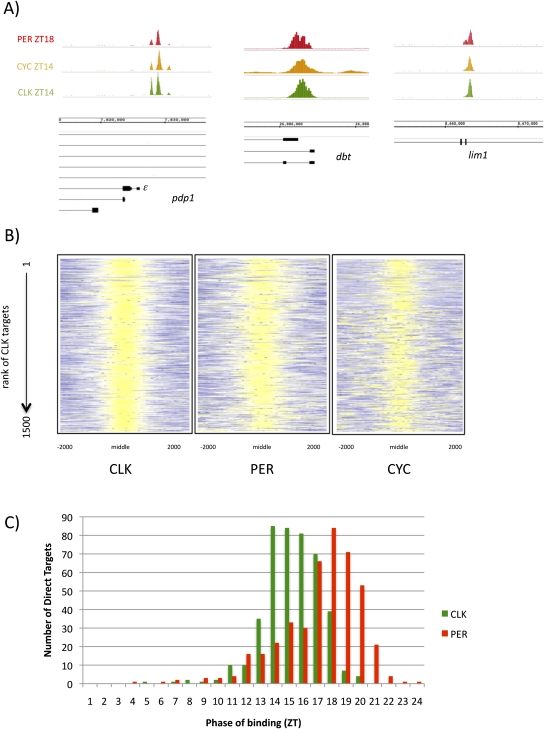
Figure 2.
CLK direct targets also bind PER and CYC. (A) CLK (green; ZT14), CYC (orange; ZT14), and PER (red; ZT18) are all bound in nearly identical locations on pdp1ε, dbt, and lim1. (B) The ChIP signals of CLK, PER, and CYC in regions of CLK binding are represented using a heat map. As expected, CLK ChIP signals are strong (yellow; Z-score between 2 and 6) in the middle of the CLK peaks. The PER ChIP signal is also high (yellow; Z-score between 2 and 6) in these regions, suggesting that PER binds with CLK on the majority of genes. Despite a lower CYC ChIP strength, CYC binding is also enriched where CLK binds (yellow; Z-score between 1.5 and 6). This suggests that the majority of CLK direct targets are also bound by PER and CYC. (C) Histogram showing the phase of cycling CLK (green) and PER (red) binding as determined by Fourier analysis (see the Materials and Methods). CLK binding precedes PER binding by ∼4–6 h.
The cycling CLK-binding sites were visually inspected on the Integrated Genome Browser (IGB; Affymetrix). Three-hundred-fifty-three sites could not be assigned to a single gene, because they were near more than one transcription start site (319 peaks) or in an intergenic region far from any annotated gene (44 peaks). However, ∼500 cycling peaks could be unambiguously mapped to a single gene like the five core clock genes (Fig. 1A; Menet et al. 2010). These genes will henceforth be referred to as the “mapped 500” (see the Materials and Methods; Supplemental Table 1). Two examples are the circadian kinase gene dbt and the homeodomain gene lim1 (Fig. 1C,D; gene #41 and #48, respectively, as shown in Supplemental Table 1).
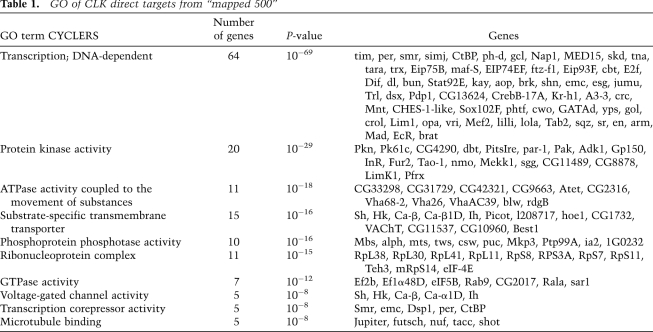
To address the functions of the genes under direct CLK control, we performed a gene ontology (GO) analysis (GO toolbox) (Martin et al. 2004) on the mapped 500. They are highly enriched for transcription factors, ∼10% of the total (64 genes), indicating that CLK sits at the top of a large transcriptional hierarchy. The second most prominent class is protein kinases, including the known circadian kinases dbt, nmo, and sgg (Table 1; Fig. 1C; Martinek et al. 2001; Muskus et al. 2007; Chiu et al. 2011; Yu et al. 2011). The list also includes substrate-specific transmembrane transporters, phosphatases, and ribonucleoprotein complexes.
Table 1.
GO of CLK direct targets from “mapped 500”
CYC is also bound at CLK-binding sites
Given the large number of CLK-binding sites, we asked what fraction is also bound by CYC, the CLK partner. To identify CYC binding, we used a transgenic fly line that contained two copies of a Flag-tagged cyc transgene (see the Materials and Methods). Three independent CYC ChIP–chips were performed at the time of maximal CLK binding, ZT14. Binding sites were identified using the same assay and statistical cutoffs used for CLK binding, with only 575 significant peaks of CYC identified.
As expected, the core circadian genes are the top-ranked CYC targets (five of the top six genes) (Supplemental Table 3). CYC binding is coincident with CLK on pdp1ε (Fig. 2A) as well as on other core clock genes (data not shown). If we restrict our analysis to the top CLK peaks, 92 of the top 100 CLK direct targets also bind CYC significantly. As CLK rank decreases, so does the ability to detect statistically significant levels of CYC via ChIP. To further examine the binding of CYC on all 1500 CLK direct targets, a heat map was used to visualize the binding of CLK and CYC in the region surrounding the CLK peak (Fig. 2B). As expected, maximal CLK ChIP signal is observed in the center of the peak (Z-score between 2 and 6) (Fig. 2B, yellow). CYC ChIP signal is also highest in this location (Z-score between 1.5 and 6) (Fig. 2B, yellow), suggesting that CYC is present at most direct targets despite not being statistically significant. This suggests that the CYC ChIP is relatively weak and the coincidental binding of CYC and CLK at ZT14 is not restricted to the core clock genes; it is found on most if not all CLK direct targets, as shown here for dbt and lim1 (Fig. 2A).
PER binds to CLK-binding sites with a delayed phase
PER binds maximally to the core clock genes per, tim, and pdp1 at ZT18–ZT22, ∼4–8 h after CLK binds, and functions as part of an “ON-DNA” repressive mechanism that both abrogates transcription and removes CLK from the DNA (Supplemental Fig. 3; Menet et al. 2010). To address all CLK target genes, we performed PER ChIP–chips from the same six time points of fly head chromatin used for the CLK ChIP–chip (see the Materials and Methods).
Nearly all CLK direct targets are enriched for PER binding (Fig. 2B), and the phase of maximal PER ChIP signal matches that observed on the core clock genes; PER binds 4–6 h after CLK (ZT18–20) (Fig 2C). To look at PER binding on all 1500 CLK direct targets, we used a heat map to visualize PER ChIP signal at ZT18 in the region surrounding the center of CLK peaks. Nearly all CLK peaks show enriched PER ChIP signal (Z-score 2–6) (Fig 2B, yellow) at regions of high CLK ChIP signal (ZT14) (Fig 2B). When we look more closely at PER binding on individual genes, PER binding at ZT18 almost completely overlaps that of both CLK and CYC at ZT14 (e.g., pdp1ε, lim, and dbt) (Fig. 2A, CLK is in green, CYC is in orange, and PER is in red). This suggests that most CLK direct targets bind the repressor PER at the same location as the CLK/CYC heterodimer but with a delayed phase.
At least 33% of cycling CLK direct targets show oscillating Pol II
To test whether the binding of these three circadian transcription factors also leads to rhythmic transcriptional activation, we examined Pol II occupancy. Pol II ChIP–chips with an antibody recognizing the entire Pol II holoenzyme were performed from the same six time points of Drosophila head chromatin used for CLK and PER ChIP–chips. Using the methods described above, ∼6000 peaks of statistically significant Pol II binding were identified. Most are prominent signals at the 5′ ends of genes and resemble those characterized as stalled or poised Pol II in Drosophila tissue culture cells and in embryos (Muse et al. 2007; Zeitlinger et al. 2007). Visual inspection also revealed signals throughout some abundantly transcribed genes as well as peaks of Pol II at the 3′ ends of some genes (data not shown).
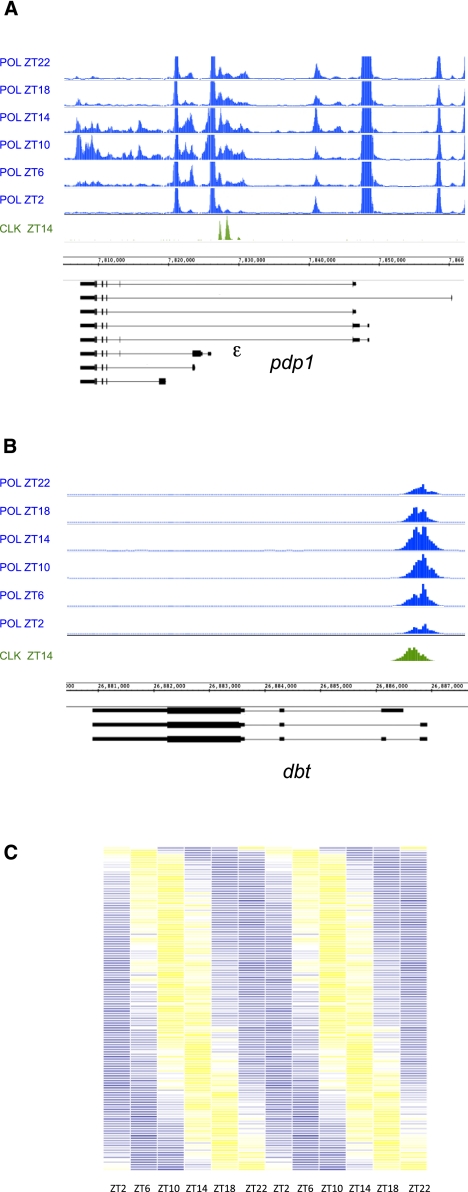
To assess Pol II cycling, we screened all cycling CLK direct target genes (∼800 genes, including both the 500 mapped genes and the ∼300 genes that are mapped to more than one gene) using a combination of visual inspection and computation (see the Materials and Methods). Unlike ChIPs for CLK, PER, and CYC, where signal may reflect binding in specific tissues, Pol II ChIPs may be more similar to assaying mRNA; a cycling signal may be masked by strong Pol II signal in another tissue. For example, if a gene is a CLK target in one tissue but is expressed independently of CLK in another, CLK, PER, and CYC would bind to the gene and visibly cycle in only the first tissue, but the Pol II signal would be a combination of cycling expression in the first tissue and constitutive expression in others (with invisible Pol II cycling). Despite this limitation, 267 CLK direct target genes (33%) had detectable cycling Pol II. Pol II was either present throughout the ORF (47 genes), promoter-proximal (194 genes), or both (26 genes). The 73 genes that show cycling Pol II throughout their ORFs, presumably elongating Pol II, include a number of highly transcribed genes and most core clock genes (e.g., pdp1) (Fig. 3A). As previously shown for tim and per, these profiles show that transcription is relative “OFF” in the late night and early morning (ZT18, ZT22, and ZT2) and then relatively “ON” from ZT6 to ZT10 (Taylor and Hardin 2008; Menet et al. 2010). This phase of Pol II cycling is also nearly identical on most of those genes with promoter-proximal Pol II. For example, pdp1 and dbt show cycling Pol II at their promoters/start sites, which is maximal at ZT10 (Fig. 3A,B). To look at Pol II promoter-proximal cycling more broadly, we identified cycling Pol II peaks that overlap with CLK peaks and double-plotted Pol II ChIP signals across circadian time as a heat map (Fig. 3C). Although Pol II phase is more widely distributed than that of CLK, peak signals (Z-score 1.5–2) (Fig. 3C, yellow) are between ZT6 and ZT14 in most cases. This corresponds to the time of peak circadian transcription (ZT6–ZT12) (So and Rosbash 1997; Menet et al. 2010).
Figure 3.
Approximately 30% of CLK direct target genes have cycling Pol II on their promoters and/or in their coding regions. Pol II ChIP–chips were performed on fly head chromatin collected every 4 h for a total of six time points. (A,B) Pol II binding is visualized on the IGB (Affymetrix). Genes above the genomic coordinates are transcribed from left to right (plus strand), and genes below the genomic coordinates are transcribed from right to left (minus strand). For comparison, CLK binding is shown in green. (A) Pol II occupancy on pdp1 shows both cycling promoter-proximal Pol II binding and cyclical binding of Pol II in the coding region of the circadian-controlled ε isoform. (B) Cyclical promoter-proximal Pol II binding is observed on the circadian kinase dbt. Pol II is always present at the promoter but increases to peak between ZT10 and ZT14. No cyclical Pol II in the coding region is detectable. (C) Heat map showing the Pol II ChIP signal across circadian time on those genes that have overlapping Pol II and CLK peaks (<30% of cycling CLK peaks). Data are double-plotted to aid in the visualization of cycling. Lowest ChIP signal is shown in blue (Z-score −6 to −2), and highest ChIP signal is shown in yellow (Z-score 2–6). Most Pol II peaks oscillate with a phase of ZT10–ZT14. Very few Pol II peaks are maximal at ZT22 or ZT0.
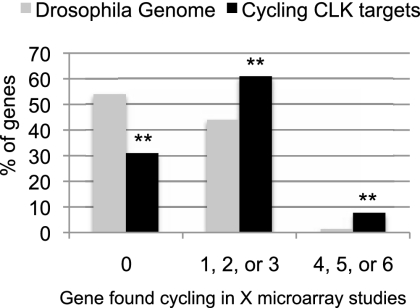
Does CLK binding correlate with mRNA cycling? To address this question, we compared the distribution of cycling mRNAs between the whole Drosophila genome and the mapped 500 CLK direct targets. Only 1.4% of all mRNAs in the genome were consistently identified as cycling; i.e., in at least four of six microarray studies (Fig. 4; McDonald and Rosbash 2001; Wijnen et al. 2006; Kadener et al. 2007). In contrast, 7% of CLK direct targets cycle in at least four of these studies (difference with 1.4%; P-value <0.0001). A much larger fraction of all Drosophila genes, ∼43%, were inconsistently identified as cycling (i.e., identified in one, two, or three microarray studies), whereas an even larger fraction of CLK direct targets, 62%, were in this category (difference with 43%; P-value <0.0001).
Figure 4.
CLK direct targets are enriched for cycling genes. Six different circadian microarray data sets were analyzed and cycling genes were identified. We categorized genes as cycling (identified in four, five, or six independent studies), inconsistently cycling (identified in one, two, or three independent studies) or not cycling (never identified in a study). The graph shows the distribution of all the of genes in the Drosophila genome (gray) or CLK direct target genes (black; only those with cycling binding) into these categories. CLK direct targets are enriched in cycling genes (identified in four or more studies) as well as genes inconsistently identified to cycle (identified in one, two, or three studies). They also show a substantial decrease in the number of genes not identified as cycling in any of the six studies. Double asterisks indicate that the difference between CLK direct targets and the genome as a whole is statistically significant (P-value <10−4).
To directly evaluate the transcription of CLK target genes, we examined mRNAs by quantitative RT–PCR (qRT–PCR) from 10 CLK direct target genes. Their rank of CLK ChIP signal ranged from #7 to #322 (see Supplemental Table 1), and they showed Pol II cycling in either their promoters (seven genes) or in their ORFS (three genes). We also examined 10 CLK direct target genes without detectable Pol II cycling.
Only two of 10 CLK direct targets genes without cycling Pol II showed mRNA cycling (Supplemental Fig. 4). In contrast, eight of the 10 CLK direct target genes with cycling Pol II showed cycling mRNA levels with amplitudes between twofold and threefold and peak expression at approximately ZT14, suggesting that cycling Pol II via ChIP indeed correlates with cycling transcription (Supplemental Fig. 5). Because seven of these eight genes were identified in less than two microarray studies, it is likely that the relatively low cycling amplitude (twofold to threefold), combined with some experimental variability, may make cycling mRNAs more difficult to detect by Affymetrix microarrays than by PCR (see the Discussion). The other two genes, dbt and lim1, showed oscillating mRNA, but with amplitudes just below the cycling cutoff (1.4-fold) (Supplemental Fig. 5).
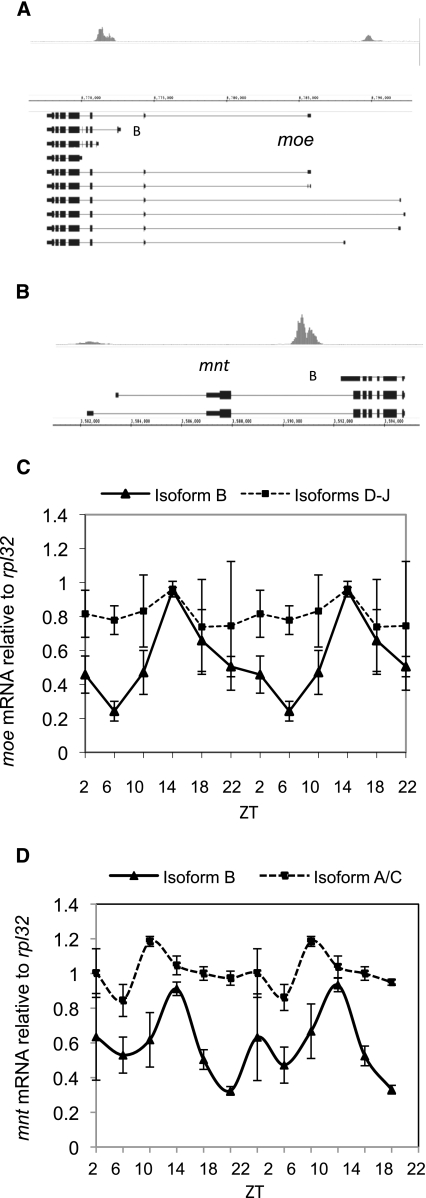
Another possible explanation for the low frequency of microarray-detectable mRNA cycling among CLK direct targets is isoform specificity, as observed for pdp1 (Fig. 1A). Indeed, CLK direct targets are enriched for alternative starts when compared with the whole genome (55% of the mapped 500 vs. 13.75% of the genome; P-value <0.0001), and visual inspection indicates that many CLK-binding sites appear linked to specific transcription start sites. For example, moe and mnt (identified in either one or zero microarray studies, respectively) have apparent isoform-specific CLK binding (Fig. 5A,B). To test for moe and mnt isoform-specific mRNA cycling, RNA was assayed by qRT–PCR. The major CLK-bound isoforms are cycling with amplitudes of approximately threefold, whereas other isoforms are not (Figs. 5C,D). Importantly, this distinction could go undetected in microarray studies where only the 3′ ends of transcripts are assayed.
Figure 5.
CLK binding results in isoform-specific mRNA cycling. Fifty-five percent of CLK direct targets have alternative start sites. (A) CLK binds to both the short (isoform B) and long isoforms of moe at ZT14. (B) CLK binds to the short isoform (isoform B) of mnt at ZT14. (C) mRNA levels at six time points throughout the day are double-plotted and show that the mRNA resulting from isoform B of moe cycles with a peak amplitude at ZT14 (triangles; solid line). In contrast, mRNAs resulting from the longer isoforms (isoforms D–J) of moe do not cycle (squares; dashed line). (D) mRNA levels at six time points throughout the day are double-plotted and show that the mRNA resulting from isoform B of mnt cycles with a peak amplitude at ZT14 (triangles; solid line). In contrast, mRNAs resulting from the longer isoforms (isoforms A and C) of mnt do not cycle (squares; dashed line).
Approximately 1500 CLK direct targets sum CLK binding from multiple tissues
A third possible explanation for the poor mRNA cycling of CLK direct target genes is the tissue heterogeneity of fly heads. Perhaps CLK binding and cycling transcription occurs in one location, whereas much more active but temporally constant transcription takes place in another. To test this possibility, we performed head chromatin CLK ChIP from an eyeless strain. Previous studies have shown that a single copy of GMR-hid causes a complete loss of eye tissue with little or no effect on circadian behavior (Malpel et al. 2004). GMR-hid flies expressing two extra copies of dCLK-V5 were harvested at ZT14 and used for CLK ChIP–chip (see the Materials and Methods). By comparing CLK peaks in control and GMR-hid flies, we were able to identify putative CLK targets only in the eye, predominantly absent from the eye, and in the eye as well as other head tissues.
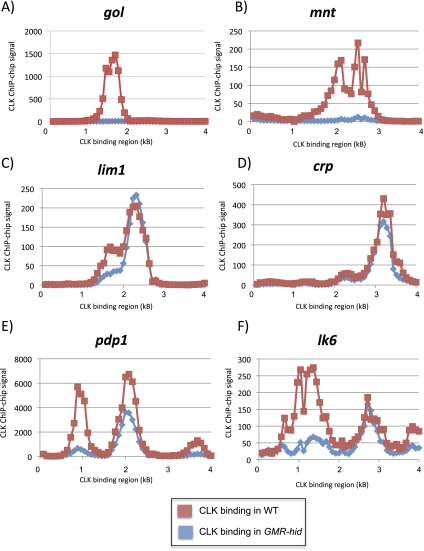
Forty-four percent of CLK direct target genes are no longer detectable in GMR-hid chromatin (see the Materials and Methods). For example, both gol and mnt show dramatically reduced CLK ChIP signals (Fig. 6A,B). Moreover, qRT–PCR shows that both Mnt and Gol mRNA cycles in wild-type flies, but that mRNA levels decrease dramatically in GMR-hid flies, indicating that that these genes are expressed predominantly in the eye (Supplemental Fig. 6; data not shown). However, many apparently eye-specific CLK targets are not expressed solely or even predominantly in the eye (data not shown), suggesting that they are CLK direct targets in the eye but are probably expressed by other transcription factors elsewhere in the head.
Figure 6.
CLK binding can be tissue-specific. CLK ChIP–chips were performed on either wild-type (CLK-V5) or GMR-hid (CLK-V5, GMR-hid) flies at ZT14. In GMR-hid flies, the majority of the eye tissue is ablated. We compare CLK ChIP signal in wild-type (red) and GMR-hid (blue) flies on six different genes. CLK binding is undetectable on gol (A) and mnt (B) when eyes are ablated, suggesting that gol and mnt are CLK direct targets primarily in eye tissue. CLK binding is unaffected on lim1 (C) and crp (D) in GMR-hid, suggesting that these genes are CLK targets in non-eye tissue. Interestingly, on some genes with multiple CLK promoter peaks, the two peaks of CLK binding are differentially affected in GMR-hid. One of the peaks of CLK binding on both pdp1 (E) and lk6 (F) is greatly diminished in GMR-hid, while another peak is much less affected.
In contrast to putative eye CLK-binding genes, ∼20% of CLK-binding sites are unchanged or even increase in GMR-hid compared with the control strain. Two examples are the transcription factors lim1 and crp (Fig. 6C,D), which are therefore putative CLK targets in non-eye tissue only. Interestingly, a previous study showed that lim1 mRNA is highly enriched in a subset of circadian neurons (Kula-Eversole et al. 2010), the l-LNvs, raising the possibility that this is one of the tissues in which CLK binds to lim1.
The remaining ∼40% of direct target genes show intermediate decreases in CLK binding upon eye ablation, suggesting that CLK associates with these genes in the eyes as well as in other head tissues. This category not surprisingly includes the core clock genes, as they are known to undergo mRNA cycling in the eyes as well as in circadian neurons. Intriguingly, the CLK-binding pattern changes between GMR-hid and the control strain on a number of genes; e.g., the core clock gene pdp1 and lk6 (Fig. 6E,F). In both cases, only one CLK peak is clearly decreased upon eye ablation. This suggests that CLK-binding sites can vary qualitatively and/or quantitatively between tissues depending on tissue-specific features, such as the binding of coactivators or repressors or changes in chromatin state. This notion may also help explain the very broad peaks of CLK binding observed on a number of genes, such as pdp1.
Discussion
Previous circadian models in Drosophila suggested a transcriptional cascade in which CLK directly controls a limited number of genes, including core clock genes, which then drive the oscillating expression of many different output genes. The results of this study indicate that CLK directly regulates not only the five core clock genes (i.e., pdp1, vri, tim, per, and cwo), but also many output genes, including ∼60 additional transcription factors. Some of these are tissue-specific; e.g., lim1 and crp (Table 1; Fig. 6). In addition, CLK direct target gene regulation may impact timekeeping in yet unforeseen ways. For example, CLK, PER, and CYC bind to three of the four known circadian kinases; i.e., dbt (rank #41), nmo (rank #48), and sgg (rank #794) (Martinek et al. 2001; Muskus et al. 2007; Chiu et al. 2011; Yu et al. 2011). Although none of these mRNAs have been previously reported to cycle, both dbt and sgg have cycling Pol II (Fig. 3B; data not shown), and dbt shows weak oscillations via qRT–PCR (Supplemental Fig. 5). nmo expression is enriched in circadian neurons and has been shown to cycle in l-LNvs (Kula-Eversole et al. 2010). The data, taken together, indicate that this simple ChIP–chip strategy has uncovered important relationships and suggest that the transcriptional regulation of some of these new target genes warrants further investigation.
Most of the 1500 CLK direct target genes are also bound by two other circadian transcription factors: CYC and PER. Because a previous study from our laboratory showed that there is a progressive, ordered recruitment of CLK, Pol II, and PER on per and tim (Menet et al. 2010), the same basic mechanism is conserved on most CLK direct targets. CLK binding increases in late morning and gives rise to an increase in Pol II signal where detectable (ZT6–ZT10). CLK binding is maximal in the early night (ZT14), and both CLK binding and Pol II occupancy decrease rapidly after the repressor PER is bound to chromatin 4–6 h later, at ZT18 (Fig. 2C; Supplemental Fig. 3). Interestingly, PER binds to nearly all CLK direct targets at the identical CLK/CYC locations, suggesting PER recruitment via protein–protein interactions (Fig. 2A,B; Menet et al. 2010) .
The identical binding sites for CLK, CYC, and PER suggest that binding is not background binding or “sterile” binding with no functional consequence. This is because three components of the circadian transcription machinery are present with proper temporal regulation. Pol II cycling on ∼30% of cycling CLK targets further supports this interpretation. The Pol II signal is maximal from mid- to late morning (ZT6–ZT10), which slightly anticipates the maximal transcription times of core circadian genes like per and tim (So and Rosbash 1997; Menet et al. 2010). Most Pol II signals are promoter-proximal and may reflect poised Pol II complexes often found on genes that respond quickly to environmental stimuli (Rougvie and Lis 1988; Kim et al. 2005; Muse et al. 2007; Saha et al. 2011).
To address RNA cycling, we examined 10 direct target genes with Pol II cycling. Eight of these genes show oscillating mRNA with >1.5-fold amplitude, suggesting that oscillating Pol II indeed reflects cycling transcription. Because this assay may underestimate cycling transcription due to tissue heterogeneity (i.e., masking by noncycling gene expression elsewhere in the head), ∼30% is a minimal estimate of CLK direct targets with cyclical mRNA.
Interestingly, previous microarray studies did not detect many of these genes (Fig. 4). One possibility is that the alternative start sites that characterize 55% of CLK direct targets are not detectable on microarrays; e.g., moe and mnt (Fig. 5). However, several mRNAs that cycle robustly by qRT–PCR are not isoform-specific and are also not consistently identified in microarray studies. A second possibility is that the relatively low cycling amplitude of many target genes—twofold or less compared with the much greater amplitudes of core clock genes such as tim (10-fold), per (15-fold), and pdp1 (20-fold) assayed in parallel (data not shown)—may be more difficult to detect on microarrays.
Low-amplitude cycling may result from relatively stable mRNA, which will dampen the amplitude of cycling transcription. Alternatively, or in addition, low-amplitude cycling may reflect cycling in one head location and noncycling elsewhere within the head, which will dampen head RNA cycling amplitude. This is likely for many eye-specific CLK targets, which appear expressed elsewhere in the head via a CLK-independent mechanism.
A third and arguably more interesting explanation for low-amplitude cycling is that CLK binds on promoters with other transcription factors within single tissues. These could include chromatin modifiers and would function together with CLK in a gene- and tissue-specific fashion. For example, a gene could be constitutively expressed at a basal level by one transcription factor, with temporal CLK binding causing a modest boost to transcription. For example, gol is a CLK target exclusively in the eye, and gol mRNA cycles with a fourfold amplitude (Supplemental Fig. 5). Rather than cycling from “OFF” (no or very low mRNA levels) to “ON,” however, gol mRNA levels are quite high even at the trough or lowest time points (data not shown). This suggests that gol cycles from a substantial basal level in the late night and daytime to an even higher level of expression in the evening and early night. Since mRNA levels decrease by >10-fold in GMR-hid flies (Supplemental Fig. 6), trough transcription levels are not likely from other tissues. Therefore, CLK probably acts on gol and other targets not as an “ON/OFF switch,” but rather in concert with other factors to boost a basal level of gene expression at a particular time of day and cause low-amplitude cycling within a single tissue.
The large number of CLK target genes in fly heads is explained in part by tissue-specific CLK binding. Transcription assays that measure the cycling of mRNA and Pol II binding in one head tissue can be masked by noncycling expression in another. The ChIP assays, in contrast, are not plagued with the same problem. They can identify a gene bound by the cycling circadian transcription machinery even if the same gene is constitutively expressed elsewhere in the head. Surprisingly 44% of CLK direct targets were no longer detected when eyes were ablated with GMR-hid (e.g., Fig. 6). Because many of these mRNAs are not particularly eye-enriched (data not shown), we infer that their genes are constitutively expressed under the control of other transcription factors elsewhere in the head.
The large number of target genes is also explained by the efficiency and sensitivity of the ChIP assay. We infer that it can detect CLK binding from a relatively low number of cells within the fly head. Lim1 is one example and is expressed predominantly in a subset of circadian neurons (l-LNvs; enriched more than four times relative to head) (Kula-Eversole et al. 2010). Preliminary cell-specific CLK ChIP–chip experiments from LNvs confirm that lim1 is an enriched CLK direct target in these cells (data not shown), suggesting that this is the source of a large fraction of the binding signal in the head ChIP–chip experiments. Experiments are under way to more clearly define circadian neuron-specific CLK-binding patterns.
This tissue specificity also suggests the existence of factors and/or chromatin modifications that help regulate CLK-mediated gene expression. They could enable CLK binding to specific genes in one tissue or inhibit binding in another tissue. These tissue-specific factors are strongly indicated by the pdp1 and lk6 CLK-binding patterns, which change so strikingly and specifically in GMR-hid. Although not unprecedented (Slattery et al. 2011), tissue-specific factors that enable or inhibit specific DNA-binding locations are intriguing and warrant further investigation and identification.
Materials and methods
Transgenic fly construction and crosses
The following fly strains were used: yw, yw;; WT dCLK-V5 (Kadener et al. 2008). To generate yw; GMR-hid/cyo; dCLK-V5 flies, yw;;dCLK-V5 flies were crossed to yw;GMR-hid/cyo flies (Grether et al. 1995) (Bloomington Stock Center no. 5771). CYC-Flag transgenic flies were generated by injecting yw embryos with pCasPeR4.0 cyc7.2-3xFlag (BestGene, Inc.). pCasPeR4.0 cyc7.2-3xFlag was generated in several steps using PCR to amplify a 7176-base-pair (bp) sequence of cyc (cyc7.2) from yw genomic DNA. A 5147-bp fragment beginning 2 kb upstream of the cyc transcription start site (+1) and ending 1 kb downstream from the 3′ untranslated region (UTR) (+3147) was amplified in two steps and cloned into the pBS vector. A fragment from −2000 to +1932 was ligated into pBS using SmaI/EcoRI (+1932). A second fragment spanning from +1932 (EcoRI) to +3147 (NotI; 1.2-kb fragment) was ligated into the same vector. The sequence encoding 3XFlag tag was inserted before the stop codon at the C terminus using overlap PCR to generate pBS-cyc5147-3xFlag. This vector was then digested by NcoI (−1635) and NotI to release a 4.7-kb fragment of cyc. Another 2.5-kb fragment (−4030 to −1566) of cyc upstream sequence was amplified by PCR and digested by KpnI/NcoI (−1635). The KpnI/NcoI (2.5 kb) PCR product and the NcoI/NotI (4.7-kb) fragment release from pBS were cloned into pCasPeR4.0, resulting in the pCasPeR4.0 cyc7.2-3xFlag vector. This vector was verified by sequencing. The cyc7.2-3xFlag transgene rescues the arrythmicity of cyc01 (period of 24.0 h in DD [constant darkness]) (data not shown).
ChIP–chips
Yw;;dCLK-V5 flies were entrained for 3 d in 12 h:12 h light:dark cycles and then harvested every 4 h for a total of six time points. ChIPs for CLK, PER, and Pol II were performed from the same chromatin samples as previously described (Menet et al. 2010). Three independent CYC ChIPs were performed at ZT14 as previously described, except that anti-Flag M2 affinity gel (Sigma) was used for the immunoprecipitation (Menet et al. 2010). Tiling arrays were performed as described previously (Menet et al. 2010).
ChIP–chip data analysis
To identify significant peaks of CLK, CYC, PER, and Pol II binding throughout the genome, Affymetrix .CEL files for both input and immunoprecipitation samples from two or more independent experiments were analyzed using MAT (Johnson et al. 2006). This analysis assigns each peak a MAT score that is a statistical value describing the likelihood that a particular genomic region is enriched in the immunoprecipitation relative to the input sample. It is this value that we refer to as the “ChIP signal.” Peaks were considered significant if they have a P-value of <10−4 at any of the six time points (for CLK, PER, and Pol II) or at the time point the experiment was performed (CYC). The resulting peaks were consolidated by grouping overlapping peaks together. Peaks were preliminarily mapped to genes using an algorithm that assigned each peak to the gene (ORF) it was in. If the peak was not in an ORF, it was then assigned to the two nearest genes. This method led to ∼20% of peaks being mapped inaccurately. For example, a peak in the 3′ end of an ORF on the top strand and in the promoter of a gene on the bottom strand would be inappropriately mapped to the ORF on the top strand. To ensure that the peaks were mapped as correctly as possible, all peaks shown to be cycling (see below) were visually inspected and mapped to the nearest promoter or promoters. If a peak was >2000 bp from a promoter, it was considered to be intergenic. The results of the automated mapping as well as the visual mapping are both listed in Supplemental Table 1.
The resulting list of CLK peaks was further analyzed to remove any possible background peaks (Supplemental Fig. 1). First, we removed any peaks that were statistically significant only when both CLK ChIP–chip data sets were analyzed together, but not when they were analyzed independently. Second, we removed any peaks found to be statistically significant when we performed anti-V5 ChIP in a wild-type (yw; no V5 tag present) background. Finally, peaks showing cyclical CLK binding were identified using a Fourier analysis that compares the pattern of CLK binding with a sine wave with a 24-h period and assigns each peak a F24 score, which reflects how well the values match the curve. In this study, a peak was considered cycling if it had a F24 score of >0.7 and a P-value of <0.05 (after 10,000 iterations) (Wijnen et al. 2005). The resulting ∼800 cycling CLK peaks were then inspected visually (see above).
To identify peaks of CYC and PER binding that overlapped with CLK binding, statistically significant peaks of CYC and PER were cross-referenced to the list of statistically significant CLK peaks to identify overlapping peaks (any percentage of overlap was considered as “overlapping”). In addition, the ChIP–chip signals of CLK, CYC, and PER from the region (±2000 bp) from the center of the CLK peaks were extracted, transformed into log2 scale, and plotted as a heat map using heatmap.2 in R (Fig. 2B).
To identify peaks of cycling Pol II, we used two approaches. First, we analyzed those significant peaks of Pol II binding using a Fourier analysis (see above) to identify cycling peaks. This list was then cross-referenced to a list of significant cycling CLK peaks both by overlapping location and by gene name to identify those CLK direct targets that had cycling Pol II. Second, we performed a visual inspection of Pol II on all cycling CLK direct target genes to (1) determine the location of the cycling peak, (2) verify the correct annotation of the Pol II peak (see above), (3) verify the computational cycling analysis, and (4) look for peaks not found in the computational analysis due to peak consolidation, which often made some cycling peaks undetectable via Fourier analysis. To be classified as cycling by visual analysis, a peak had to cycle with an amplitude >2 and have two or more high time points.
To identify how CLK binding changes when eyes are ablated in GMR-hid, CLK ChIP–chips were performed on heads from yw;; WT dCLK-V5 (wt) and GMR-hid; dCLK-V5 (GMR-hid) at ZT14 as previously described (Menet et al. 2010). Affymetrix .CEL files from both yw;; WT dCLK-V5 (wild type) and GMR-hid; dCLK-V5 (GMR-hid) were analyzed using both MAT (Johnson et al. 2006) and Cisgenome 2.0 (Ji et al. 2008). Since the output of MAT is a statistical value and not a linear scale, we used values of CLK ChIP signal generated using Cisgenome2.0 to better compare the amount of CLK binding in wild-type and GMR-hid fly heads. A ratio of CLK ChIP signal was calculated and peaks were classified as either (1) missing in GMR-hid (no detectable peak at all), (2) unchanged in GMR-hid (CLK ChIP signal in GMR-hid was >90% of the signal in wild type), or (3) intermediate effects. To generate Figure 6, CLK ChIP signals (from MAT analysis) from the region (±2000 bp) from the center of the CLK peaks was extracted and plotted to show the difference in CLK ChIP signal between wild type and GMR-hid.
Microarray analysis
To determine whether CLK direct targets are enriched for genes that have cycling mRNAs, data from six different sets of circadian microarray studies from two different laboratories were normalized together using GCRMA in R (McDonald and Rosbash 2001; Wijnen et al. 2006; Kadener et al. 2007). Each of the six studies were analyzed separately, and cycling mRNAs were identified as those that have an F24 of at least 0.7 and an amplitude of at least 1.5-fold. Despite the uniform analysis, there was only limited overlap in the identified cycling transcripts. We categorized genes as cycling (identified in four, five, or six studies), inconsistently cycling (identified in one, two, or three studies), or not cycling (never identified as cycling) in order to examine whether CLK direct targets have cycling mRNAs.
RNA isolation and qRT–PCR
Total RNA was isolated from fly heads using Trizol reagent (Invitrogen) and were DNase-treated using Turbo DNA-free (Ambion). Three micrograms of DNase-treated total RNA was used for RT–PCR using SuperScript II (Invitrogen) and random primers (Promega) following the manufacturers' protocols, including a final RNase H (New England Biolabs) digestion. The resulting cDNA was used in qPCR using the Syber Master Mix (Qiagen) and a Rotorgene qPCR machine (Qiagen). Primers used for qRT–PCR are available in Supplemental Table 4.
Data availability
Affymetrix microarray data for all of the ChIP–chips performed in this study will be available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), accession number GSE32613.
Acknowledgments
Arno Greenleaf generously provided the RNA Pol II polyclonal antibody used in this study. We thank Emi Nagoshi, Sebastian Kadener, Michael Marr, and Paul Hardin for helpful comments and discussion. We thank Krissy Palm Danish for administrative assistance, and Emily Bokser, Sarah Duncan, and Xiao Chen for technical assistance. J.R. was supported in part by a grant from the National Science Foundation (DGE 0549390). A.Z. was supported in part by a grant from the National Institutes of Health (PO1 NS44232). This work was supported in part by a grant from the National Institutes of Health (P01 NS44232) to M.R.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.178079.111.
References
- Allada R, White N, So W, Hall J, Rosbash M 1998. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell 93: 791–804 [DOI] [PubMed] [Google Scholar]
- Antoch MP, Song E-J, Chang A-M, Vitaterna MH, Zhao Y, Wilsbacher LD, Sangoram AM, King DP, Pinto LH, Takahashi JS 1997. Functional identification of the mouse circadian clock gene by transgenic BAC rescue. Cell 89: 655–667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blau J, Young MW 1999. Cycling vrille expression is requiered for a functional Drosophila clock. Cell 99: 661–671 [DOI] [PubMed] [Google Scholar]
- Chiu JC, Ko HW, Edery I 2011. NEMO/NLK phosphorylates PERIOD to initiate a time-delay phosphorylation circuit that sets circadian clock speed. Cell 145: 357–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtin K, Huang ZJ, Rosbash M 1995. Temporally regulated nuclear entry of the Drosophila period protein contributes to the circadian clock. Neuron 14: 365–372 [DOI] [PubMed] [Google Scholar]
- Cyran SA, Buchsbaum AM, Reddy KL, Lin MC, Glossop NR, Hardin PE, Young MW, Storti RV, Blau J 2003. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 112: 329–341 [DOI] [PubMed] [Google Scholar]
- Darlington TK, Wager-Smith K, Ceriani MF, Staknis D, Gekakis N, Steeves TDL, Weitz CJ, Takahashi JS, Kay SA 1998. Closing the circadian loop: CLOCK-induced transcription of its own inhibitors per and tim. Science 280: 1599–1603 [DOI] [PubMed] [Google Scholar]
- Edery I, Zwiebel LJ, Dembinska ME, Rosbash M 1994. Temporal phosphorylation of the Drosophila period protein. Proc Natl Acad Sci 91: 2260–2264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether ME, Abrams JM, Agapite J, White K, Steller H 1995. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev 9: 1694–1708 [DOI] [PubMed] [Google Scholar]
- Hardin PE, Hall JC, Rosbash M 1990. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 343: 536–540 [DOI] [PubMed] [Google Scholar]
- Ji H, Jiang H, Ma W, Johnson DS, Myers RM, Wong WH 2008. An integrated software system for analyzing ChIP–chip and ChIP-seq data. Nat Biotechnol 26: 1293–1300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS 2006. Model-based analysis of tiling-arrays for ChIP–chip. Proc Natl Acad Sci 103: 12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Stoleru D, McDonald M, Nawathean P, Rosbash M 2007. Clockwork Orange is a transcriptional repressor and a new Drosophila circadian pacemaker component. Genes Dev 21: 1675–1686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadener S, Menet JS, Schoer R, Rosbash M 2008. Circadian transcription contributes to core period determination in Drosophila. PLoS Biol 6: e119 doi: 10.1371/journal.pbio.0060119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B 2005. A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King DP, Zhao Y, Sangoram AM, Wilsbacher LD, Tanaka M, Antoch MP, Steeves TDL, Vitaterna MH, Kornhauser JM, Lowrey PL, et al. 1997. Positional cloning of the mouse circadian clock gene. Cell 89: 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko HW, Jiang J, Edery I 2002. Role for Slimb in the degradation of Drosophila Period protein phosphorylated by Doubletime. Nature 420: 673–678 [DOI] [PubMed] [Google Scholar]
- Kula-Eversole E, Nagoshi E, Shang Y, Rodriguez J, Allada R, Rosbash M 2010. Surprising gene expression patterns within and between PDF-containing circadian neurons in Drosophila. Proc Natl Acad Sci 107: 13497–13502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim C, Chung BY, Pitman JL, McGill JJ, Pradhan S, Lee J, Keegan KP, Choe J, Allada R 2007. Clockwork orange encodes a transcriptional repressor important for circadian-clock amplitude in Drosophila. Curr Biol 17: 1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malpel S, Klarsfeld A, Rouyer F 2004. Circadian synchronization and rhythmicity in larval photoperception-defective mutants of Drosophila. J Biol Rhythms 19: 10–21 [DOI] [PubMed] [Google Scholar]
- Martin D, Brun C, Remy E, Mouren P, Thieffry D, Jacq B 2004. GOToolBox: Functional analysis of gene datasets based on gene ontology. Genome Biol 5: R101 doi: 10.1186/gb-2004-5-12-r101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinek S, Inonog S, Manoukian AS, Young MW 2001. A role for the segment polarity gene shaggy/GSK-3 in the Drosophila circadian clock. Cell 105: 769–779 [DOI] [PubMed] [Google Scholar]
- Matsumoto A, Ukai-Tadenuma M, Yamada RG, Houl J, Uno KD, Kasukawa T, Dauwalder B, Itoh TQ, Takahashi K, Ueda R, et al. 2007. A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock. Genes Dev 21: 1687–1700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M 2001. Microarray analysis and organization of circadian gene expression in Drosophila. Cell 107: 567–578 [DOI] [PubMed] [Google Scholar]
- McDonald MJ, Rosbash M, Emery P 2001. Wild-type circadian rhythmicity is dependent on closely spaced E boxes in the Drosophila timeless promoter. Mol Cell Biol 21: 1207–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menet JS, Abruzzi KC, Desrochers J, Rodriguez J, Rosbash M 2010. Dynamic PER repression mechanisms in the Drosophila circadian clock: From on-DNA to off-DNA. Genes Dev 24: 358–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer P, Saez L, Young MW 2006. PER-TIM interactions in living Drosophila cells: An interval timer for the circadian clock. Science 311: 226–229 [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K 2007. RNA polymerase is poised for activation across the genome. Nat Genet 39: 1507–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muskus MJ, Preuss F, Fan JY, Bjes ES, Price JL 2007. Drosophila DBT lacking protein kinase activity produces long-period and arrhythmic circadian behavioral and molecular rhythms. Mol Cell Biol 27: 8049–8064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey G, Cesbron F, Rougemont J, Reinke H, Brunner M, Naef F 2011. Genome-wide and phase-specific DNA-binding rhythms of BMAL1 control circadian output functions in mouse liver. PLoS Biol 9: e1000595 doi: 10.1371/journal.pbio.1000595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rougvie AE, Lis JT 1988. The RNA polymerase II molecule at the 5′ end of the uninduced hsp70 gene of D. melanogaster is transcriptionally engaged. Cell 54: 795–804 [DOI] [PubMed] [Google Scholar]
- Saha RN, Wissink EM, Bailey ER, Zhao M, Fargo DC, Hwang JY, Daigle KR, Fenn JD, Adelman K, Dudek SM 2011. Rapid activity-induced transcription of Arc and other IEGs relies on poised RNA polymerase II. Nat Neurosci 14: 848–856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafer OT, Rosbash M, Truman JW 2002. Sequential nuclear accumulation of the Clock proteins Period and Timeless in the pacemaker neurons of Drosophila melanogaster. J Neurosci 22: 5946–5954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery M, Ma L, Negre N, White KP, Mann RS 2011. Genome-wide tissue-specific occupancy of the Hox protein Ultrabithorax and Hox cofactor Homothorax in Drosophila. PLoS ONE 6: e14686 doi: 10.1371/journal.pone.0014086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So WV, Rosbash M 1997. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J 16: 7146–7155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun WC, Jeong EH, Jeong HJ, Ko HW, Edery I, Kim EY 2010. Two distinct modes of PERIOD recruitment onto dCLOCK reveal a novel role for TIMELESS in circadian transcription. J Neurosci 30: 14458–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P, Hardin PE 2008. Rhythmic E-box binding by CLK-CYC controls daily cycles in per and tim transcription and chromatin modifications. Mol Cell Biol 28: 4642–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda HR, Matsumoto A, Kawamura M, Iino M, Tanimura T, Hashimoto S 2002. Genome-wide transcriptional orchestration of circadian rhythms in Drosophila. J Biol Chem 277: 14048–14052 [DOI] [PubMed] [Google Scholar]
- Wijnen H, Naef F, Young MW 2005. Molecular and statistical tools for circadian transcript profiling. Methods Enzymol 393: 341–365 [DOI] [PubMed] [Google Scholar]
- Wijnen H, Naef F, Boothroyd C, Claridge-Chang A, Young M 2006. Control of daily transcript oscillations in Drosophila by light and the circadian clock. PLoS Genet 2: e39 doi: 10.1371/journal.pgen.0020039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zheng H, Houl JH, Dauwalder B, Hardin PE 2006. PER-dependent rhythms in CLK phosphorylation and E-box binding regulate circadian transcription. Genes Dev 20: 723–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Houl JH, Hardin PE 2011. NEMO kinase contributes to core period determination by slowing the pace of the Drosophila circadian oscillator. Curr Biol 21: 756–761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J, Stark A, Kellis M, Hong JW, Nechaev S, Adelman K, Levine M, Young RA 2007. RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat Genet 39: 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Kilman VL, Keegan KP, Peng Y, Emery P, Rosbash M, Allada R 2003. Drosophila clock can generate ectopic circadian clocks. Cell 113: 755–766 [DOI] [PubMed] [Google Scholar]
- Zheng X, Koh K, Sowcik M, Smith CJ, Chen D, Wu MN, Sehgal A 2009. An isoform-specific mutant reveals a role of PDP1ε in the circadian oscillator. J Neurosci 29: 10920–10927 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Affymetrix microarray data for all of the ChIP–chips performed in this study will be available at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo), accession number GSE32613.