SHQ1 is an essential assembly factor for H/ACA ribonucleoproteins (RNPs). The crystal structures of the yeast Shq1p C-terminal domain, both free and in complex with Cbf5p (the yeast dyskerin/NAP57), reveal how Shq1p acts as a molecular chaperone during H/ACA RNP assembly. Interestingly, Shq1p uses RNA mimicry to bind Cbf5p on its RNA-binding surface, essentially using Cbf5p residues involved in RNA–protein interactions to form specific protein–protein contacts.
Keywords: ribonucleoprotein assembly, H/ACA RNPs, X-linked dyskeratosis congenita, telomerase
Abstract
SHQ1 is an essential assembly factor for H/ACA ribonucleoproteins (RNPs) required for ribosome biogenesis, pre-mRNA splicing, and telomere maintenance. SHQ1 binds dyskerin/NAP57, the catalytic subunit of human H/ACA RNPs, and this interaction is modulated by mutations causing X-linked dyskeratosis congenita. We report the crystal structure of the C-terminal domain of yeast SHQ1, Shq1p, and its complex with yeast dyskerin/NAP57, Cbf5p, lacking its catalytic domain. The C-terminal domain of Shq1p interacts with the RNA-binding domain of Cbf5p and, through structural mimicry, uses the RNA–protein-binding sites to achieve a specific protein–protein interface. We propose that Shq1p operates as a Cbf5p chaperone during RNP assembly by acting as an RNA placeholder, thereby preventing Cbf5p from nonspecific RNA binding before association with an H/ACA RNA and the other core RNP proteins.
H/ACA small nucleolar and small Cajal body ribonucleoproteins (snoRNPs and scaRNPs) are small RNA–protein complexes that are essential for the biogenesis of larger RNPs such as ribosomes, spliceosomes, and telomerase (Yu et al. 1998; Dez et al. 2001; King et al. 2003; Kiss et al. 2010). The mature H/ACA snoRNPs and scaRNPs are composed of a single or double hairpin guide RNA and four evolutionarily conserved proteins (Cbf5p/NAP57, Nop10p/NOP10, Nhp2p/NHP2, and Gar1p/GAR1 in yeast and humans) (Meier 2005; Ye 2007). Most H/ACA particles function as site-specific pseudouridine synthases in an RNA-guided mechanism, the guide RNA providing base-pairing interactions that specify the target uridine (Li 2008). In telomerase, however, the H/ACA domain is not involved in pseudouridylation or telomerase enzymatic activity, but is essential for the accumulation and localization of the mature telomerase RNP (Mitchell et al. 1999; Lukowiak et al. 2001; Zhu et al. 2004). Recent insight into the structure of archaeal H/ACA particles has revealed the molecular basis for the function of this RNP in pseudouridine synthesis (Li and Ye 2006; Duan et al. 2009; Liang et al. 2009). The pseudouridine synthase Cbf5p/NAP57 (also known as dyskerin in humans) harbors the catalytic activity, while the other subunits stabilize the structure of the particle and position the guide and substrate RNAs for catalysis (Henras et al. 1998; Lafontaine et al. 1998; Jin et al. 2007).
The biogenesis of eukaryotic H/ACA snoRNPs and scaRNPs is surprisingly complex, considering that the mature particle only has five components, and involves numerous maturation factors (Kiss 2004; Zhao et al. 2008; Kiss et al. 2010). Two H/ACA-specific factors, called Naf1p and Shq1p in yeast (NAF1 and SHQ1 in humans), are required for the assembly of both snoRNPs and scaRNPs (Dez et al. 2002; Fatica et al. 2002; Yang et al. 2002; Darzacq et al. 2006; Hoareau-Aveilla et al. 2006; Grozdanov et al. 2009a). Naf1p is a structural homolog of Gar1p (Leulliot et al. 2007) and may act as a placeholder during the assembly of the RNP, prior to its replacement with Gar1p for the final maturation and activation of the enzymatic activity (Darzacq et al. 2006; Grozdanov et al. 2009a). The interaction of Naf1p/NAF1 with Cbf5p/NAP57 has been proposed to keep the Cbf5p/NAP57 β7–β10 active site loop in a conformation that prevents the enzyme from being functional (Leulliot et al. 2007).
The function of the second assembly factor, Shq1p/SHQ1, originally identified in yeast (Yang et al. 2002), is unclear. Nonetheless, Shq1p/SHQ1 is essential for the stable expression of H/ACA snoRNAs and scaRNAs and interacts with Cbf5p/NAP57 (Yang et al. 2002; Godin et al. 2009; Grozdanov et al. 2009a). The interaction of NAP57 with SHQ1 excludes that of all of the other core proteins or the second assembly factor, NAF1, and SHQ1 is absent from nucleoli and Cajal bodies, where mature H/ACA particles are concentrated (Grozdanov et al. 2009a). These observations indicate that Shq1p/SHQ1 functions in early steps of the H/ACA RNP biogenesis pathway. Shq1p/SHQ1 contains two structurally independent domains. The N-terminal CS (Chord and Sgt1) domain has structural homology with Hsp90 cochaperones but, surprisingly, does not bind Hsp90 in vitro (Godin et al. 2009; Singh et al. 2009). The structure of the conserved C-terminal domain (Shq1pC) and its function are unknown. Nonetheless, the C-terminal domain mediates the interaction of Shq1p/SHQ1 with Cbf5p/NAP57, whereas the N-terminal domain alone fails to bind to the enzyme (Godin et al. 2009; Grozdanov et al. 2009b).
Recent reports revealed the central role of SHQ1 in human health. In vitro mapping uncovered that the C-terminal domain of SHQ1 interacts with a domain of NAP57 that contains most mutations identified in X-linked dyskeratosis congenita (X-DC) (Grozdanov et al. 2009b). X-DC is a rare bone marrow failure syndrome caused mostly by missense mutations in NAP57, hence also known as dyskerin (Heiss et al. 1998; Meier 2005; Kirwan and Dokal 2008). The molecular mechanism for this disease remains, nonetheless, elusive. X-DC mutations in NAP57 modulate the binding affinity of NAP57 for SHQ1, suggesting that the disruption of the function of SHQ1 in the maturation of H/ACA particles could be a molecular consequence of X-DC mutations (Grozdanov et al. 2009b). In addition, mutations in SHQ1 have been identified in human prostate cancers and indicate that this protein acts as a tumor suppressor (Bullinger et al. 2010; Taylor et al. 2010). SHQ1 has also been identified as a novel target of Notch signaling in T-cell leukemia (Chadwick et al. 2009). Altogether, the putative role of SHQ1 in cancer is consistent with the observed cancer susceptibility of X-DC patients and may be a consequence of the disruption of ribosome and/or telomerase RNP maturation. Obviously, understanding the role of SHQ1 in these pathologies requires the determination of its molecular function.
In order to clarify the link between the function of SHQ1 in H/ACA RNP maturation and X-DC and cancer, we determined the crystal structure of the yeast Shq1p C-terminal domain (Shq1pC), both free and in complex with Cbf5p lacking its catalytic domain (Cbf5pΔcat). These structures show that Shq1pC adopts a new protein fold that binds Cbf5p across the RNA-binding interface, yet in a region that is distinct from the X-DC mutational cluster on the PUA domain. However, a C-terminal extension of Cbf5p that is specific to eukaryal enzymes and that contains further X-DC mutations is specifically bound by Shq1pC. Moreover, the interaction of Shq1p with Cbf5p is essential for viability in yeast and SHQ1 competes with RNA for NAP57 binding, suggesting that Shq1p/SHQ1 acts as an RNA placeholder during RNP assembly.
Results
Shq1pC adopts a novel fold
We determined the structure of Shq1pC (see the Supplemental Material for construct details), both free and in complex with Cbf5pΔcat (see below). The structure of the free protein was solved by single-wavelength anomalous diffraction of selenomethionine-substituted protein (Table 1) and reveals a new protein fold comprised of 17 α helices and a β hairpin arranged in a tightly packed globular structure (Fig. 1A; Supplemental Fig. S1). No structural homologs were detected using structural similarity servers. We propose to name this new fold, composed of a compact structured core wrapped in N-terminal and C-terminal structural elements, the maki fold, in reference to a maki sushi roll.
Table 1.
Data collection and refinement statistics
Figure 1.
Structure of Shq1pC. (A) Stereo cartoon representation of Shq1pC colored from blue (N terminus) to red (C terminus). (B) Residue conservation mapped on the surface of Shq1pC (same orientation as A); residues with more intense green color correspond to high levels of conservation. (C) Electrostatic potential mapped on the surface of Shq1pC (Baker et al. 2001) (same orientation as B); red represents acidic regions of the protein.
In the structure, helices α8–α15 form a three-layered sandwich defining the Shq1pC core that loosely corresponds to the Pfam04925 and InterPro007009 “Shq1-specific” conserved domain identified by sequence analysis (Finn et al. 2008; Hunter et al. 2009). Each layer is formed by a pair of helices arranged in a helix–turn–helix (HtH) motif similar to the repeat units of TPR (tetratrico peptide repeat) folds (HtH1: α8–α9; HtH2: α11–α12; and HtH3: α13–α15, represented in yellow, light green, and dark green, respectively, in Fig. 1A). However, this helical structure is different from the packing observed in TPR superhelices. In Shq1pC, the HtH1 is sandwiched between the HtH2 and HtH3, with the helices of the outer HtHs roughly parallel and the central HtH1 forming a 60° angle with the former. In this arrangement, the loops connecting the helices of each HtH all face the same side of the protein.
The Shq1pC α-helical core sandwich is wrapped by a ring-like structure formed by both the Shq1pC N and C termini, whose secondary structure elements make few interactions with each other and do not form an independent module. The N terminus of Shq1pC is composed of seven α helices (α1–α7, light to dark blue in Fig. 1A) that pack around the entire circumference of the Shq1pC core. The N-terminal helices interact mostly with the side of the HtHs, with the exception of α3 and α4, which interact with the loop of the HtH2 (see below; Fig. 3A, left [below]). The C terminus of Shq1pC that follows its core is composed of two helices (α16 and α17) and a β hairpin inserted between α16 and α17 (red in Fig. 1A). These structural elements interact with the side of the HtH2 and the N-terminal α2 and α3 helices. This ring leaves the loops of the HtHs exposed and places the extended loop of the β hairpin on the same side of the protein.
Figure 3.
Comparison of the Shq1p/Cbf5p and RNA/aCbf5 interfaces. Side-by-side comparison in the same relative orientations of the Shq1p/Cbf5p (left) (this study) and the RNA/aCbf5 (right) (PDB code 3HAY; Duan et al. 2009) complexes. The molecules are represented in the same color code as Figures 1 and 2. (A) The Shq1p HtH1 (yellow) and HtH2 (light green) loops are positioned at the same positions as the guide RNA P1 stem (light blue/orange), and the α4 helix obstructs the ACA-binding site. (B) Shq1p forms salt bridges to basic Cbf5p residues (left) that are involved in binding the RNA P1 stem phosphate backbone (right). (C) Shq1p remodels the C-terminal extension from P347 onward (left), which in RNA-bound aCbf5 completes the ACA-binding pocket (right). (Left) The Cbf5p C-terminal, eukaryal-specific extension binds Shq1p in a V-shaped groove formed by helix α17 and the β hairpin.
Since the C-terminal domain of Shq1p presents a new fold, the bare structure provides little information on the function of this domain in H/ACA biogenesis. Mapping residue conservation on the protein reveals an exposed surface on one face (Fig. 1B) that includes the three loops connecting the helices of the HtH motifs and residues from the first three helices of the N-terminal extension. Interestingly, inspection of the electrostatic potential of Shq1pC reveals that this conserved patch coincides with a negatively charged surface of the protein (Fig. 1C). Taken together, these observations suggest that this surface contains many residues important for the function of Shq1p/SHQ1.
Structure of the Shq1p/Cbf5p complex
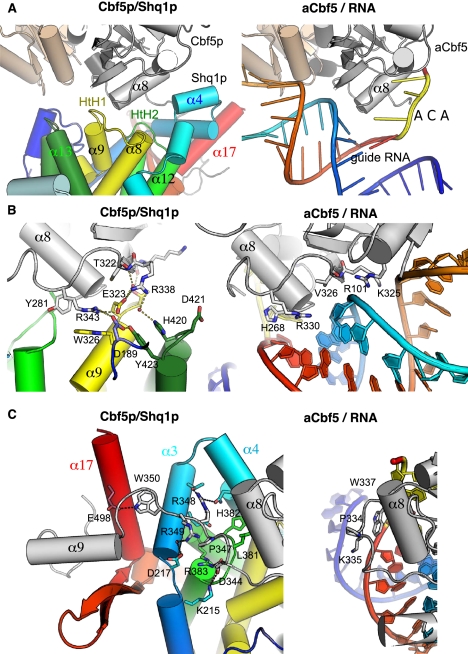
Since Shq1p/SHQ1 binds to Cbf5p/NAP57, and to assign a function to the Shq1p/SHQ1 maki fold, we sought to solve the structure of their complex. In order to circumvent the difficulties in producing soluble recombinant Cbf5p protein (Normand et al. 2006), we generated a Cbf5pΔcat construct lacking the catalytic domain (removal of residues 61–257) (modeled in beige in Fig. 2A), replicating the human NAP57Δcat construct that represents the minimal SHQ1-binding domain (for details of the construct, see Supplemental Fig. S2; Supplemental Material; Grozdanov et al. 2009b). This Cbf5pΔcat construct was also truncated of its 97 C-terminal residues, which are poorly conserved and predicted to be unstructured. This construct was insoluble when expressed alone, but coexpression with Shq1pC yielded a soluble complex amenable to structural studies. The complex crystallized in space group P41212 with two heterodimers in the asymmetric unit, and native data sets were collected at a resolution of 2.8 Å (Table 1). The structure was solved by molecular replacement using our Shq1pC structure and a model of the PUA domain built using the archaeal aCbf5 homolog that was generated using Modeller (Eswar et al. 2008). The structures of free and bound Shq1pC superpose with an RMSD (root-mean-square deviation) of 0.53 Å and are almost identical with the exception of the β hairpin, which is visible in the electron density only in the bound form (red in Fig. 1A).
Figure 2.
Comparison of the structure of the Cbf5p/Shq1p complex with that of the aCbf5/guide RNA complex. (A) Structure of the complex between Shq1pC (same color code as Fig. 1) and the Cbf5pΔcat domain (black and gray for N terminus and PUA domain, respectively). Note the structure of the catalytic domain of yeast Cbf5p was modeled based on that of aCbf5 and is colored beige. (B) Structure of the P. furiosus archaeal aCbf5 protein (same color code as A) in complex with RNA, colored blue (5′) to red (3′) (Protein Data Bank [PDB] code 3HAY) (Duan et al. 2009). The ACA box nucleotides are colored yellow, and, for better comparison, the catalytic domain of aCbf5 is also colored beige.
In the structure, the Shq1pC domain binds to Cbf5pΔcat on the PUA domain and also interacts with the Cbf5p C-terminal residues, up to residue N377 (Fig. 2A). The Shq1pC-binding interface correlates with the conserved surface patch described above (Fig. 1B) and shows a prominent role for the loops of the three HtH structural motifs of the Shq1pC core, but also an involvement of its N-terminal and C-terminal extensions, in the interaction. The Shq1pC/Cbf5pΔcat interaction buries ∼2000 Å2 at the protein–protein interface and involves 11 salt bridges. This high number of salt bridges likely contributes to the stabilization of the complex and explains the observed salt sensitivity of the SHQ1/NAP57 interaction (Grozdanov et al. 2009a).
Structure of Cbf5p, the catalytic subunit of eukaryal H/ACA RNPs
The structure of the Shq1p/Cbf5p complex provides the first representative of a eukaryotic NAP57/dyskerin ortholog, which is noteworthy because of the importance of this protein in X-DC and mammalian telomerase. The PUA domains of archaeal aCbf5 and yeast Cbf5p are almost perfectly superimposable with an RMSD of 1 Å over 92 residues. In addition, our structure of yeast Cbf5p reveals the structure of its C-terminal extension that is absent in archaea (see below; Supplemental Fig. S2) and displays a novel conformation of its N terminus. In aCbf5, the N terminus binds across the PUA domain and forms an additional β strand (β1′) after the PUA β2 strand (dark gray in Fig. 2B), and the N-terminal residues before the β1′ strand form an extended structure that interacts with the β sheet of the catalytic domain. In the yeast protein, the loop following the β1′ strand forms an extended structure that interacts with residues of the α8 helix and has a dramatically different conformation compared with archaea (Fig. 2A). In addition, the residues before the β1′ strand fold in two α helices (αN1 and αN2 in Fig. 2A) and form a compact structure that packs through hydrophobic interactions onto the PUA β sheet.
The interaction of Cbf5p with RNA and with accessory proteins can be accurately predicted using the structures of the archaeal RNPs as templates. In the context of the complete H/ACA RNP, the aCbf5 PUA domain binds the P1 stem of the H/ACA RNA, thereby positioning its pseudouridylation pocket over the active site of the enzyme (Fig. 2B; Li and Ye 2006; Duan et al. 2009). Inspection of the structures reveals that all of the residues involved in salt bridges and hydrogen bonds to the RNA stem are structurally conserved in Cbf5p (R338, R343, R348, K272, and S274; yeast numbering), and simple modeling indicates that yeast Cbf5p would bind to RNA in the same manner as aCbf5. The PUA domain of yeast Cbf5p also contains a preformed ACA-binding pocket (see below). In archaea, this pocket is complemented upon RNA binding by stacking of a C-terminal tryptophan, aCbf5-W337 (W350, in Cbf5p) (see below), that, in the absence of RNA, is disordered in all aCbf5 structures. In the Shq1p/Cbf5p complex, the residues following P347 adopt a novel structure induced by Shq1p binding (see below).
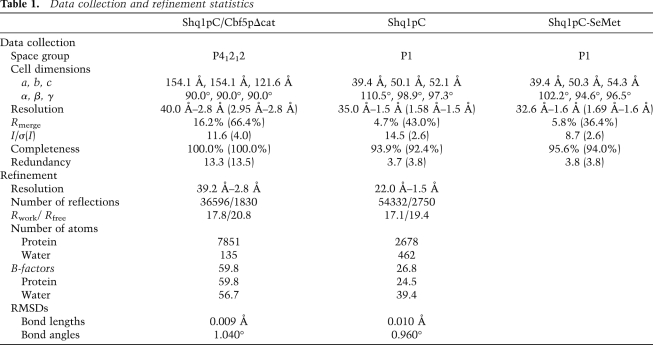
Shq1pC binds the PUA domain of Cbf5p by mimicking contacts between Cbf5p and RNA
The interaction of yeast Cbf5p with Shq1p can be compared with that of archaeal aCbf5 with RNA (Figs. 2, 3). This comparison shows that Shq1p binds to the same region of eukaryotic Cbf5p as RNA binds to the aCbf5 PUA domain (Figs. 2, 3). Furthermore, specific Shq1p structural elements occupy the same position as the RNA phosphate backbone in contacts with aCbf5; namely, the HtH1 and HtH2 loops superpose with the phosphate backbone of the 5′ and 3′ strands of the RNA stem (turquoise and red, respectively, in Fig. 3A, right) and the α4 helix superposes with the ACA motif overhang (yellow in Fig. 3A, right).
Remarkably, there is a nearly perfect overlap of the residues of Cbf5p involved in Shq1p binding with those responsible for RNA binding in aCbf5. Specifically, the contacts formed between Shq1pC and the Cbf5p PUA domain mirror the interactions in the aCbf5/RNA complex: Salt bridges to phosphate backbone groups are replaced by salt bridges to glutamates and aspartates, and hydrogen bonds to the sugar phosphate backbone are replaced by hydrogen bonds to the protein backbone and side chains. An intimate network of interactions is formed around R338 and R343 (Fig. 3B, left), whose equivalent residues in aCbf5 (aCbf5-K325 and R330) (Fig. 3B, right; see Pyrococcus furiosus in Supplemental Fig. S2 for sequence alignment) are major contributors to RNA binding. In our structure, R338 is in the same position as aCbf5-R101 (R111 in yeast), a residue from the catalytic domain also involved in RNA binding, absent in our construct (Fig. 3B, right). R338 and R343 form salt bridges to Shq1p E323 from HtH1 and to D189 from the N-terminal extension. In addition, R343 is stabilized by stacking interactions with W326 (Fig. 3B, left). The stacked side chains of H420 and Y423 from HtH3 contribute to this interaction by hydrogen-bonding to D189 and through water-mediated hydrogen bonds to R338.
In addition to interacting with structural elements of Cbf5p that are important for interactions with the guide RNA, Shq1p also obstructs the ACA-binding pocket of Cbf5p (Fig. 3A–C). This pocket is formed by a cavity between the Cbf5p α8 and α8′ helices, in which the two adenines of the ACA trinucleotide defining every guide RNA are inserted (Fig. 3A, right), and is completely obstructed by the Shq1p α4 helix and the two loops of the first and second HtHs (helices α8–α9 and α11–α12, respectively, yellow and light green in Fig. 3A). No residue-specific interactions are formed between these structural elements, which are mainly stabilized by hydrophobic and van der Waals interactions. Shq1p therefore uses RNA mimicry to form specific protein–protein contacts to the Cbf5p residues involved in protein–RNA interactions and thereby obstructs other regions on the Cbf5p surface important for guide RNA binding.
Shq1pC interacts with the conserved C-terminal extension of Cbf5p
Eukaryal Cbf5 enzymes set themselves apart from their archaeal counterparts by harboring a conserved C-terminal extension corresponding to residues 351–387 in yeast Cbf5p (381–417 in NAP57) (Supplemental Fig. S2). In the archaeal RNP, the C-terminal residues following the PUA domain β17 strand assume an RNA-induced structure, completing the recognition of the ACA motif. Notably, aCbf5-W337 and aCbf5-K335 (W350 and R348, respectively) form stacking, H-bonding, and salt bridges to the RNA phosphate backbone (Fig. 3C, right). In the Cbf5p/Shq1p complex, the conformation of the protein main chain of Cbf5p is almost superimposable with that of aCbf5 up to residue P347 (aCbf5-P334) (Fig. 3C). However, the presence of Shq1p would clash with the following residues in the RNA-bound aCbf5 conformation (Fig. 3A); in particular, Cbf5p-W350 in the RNA-bound conformation would clash with L381 and H382 of the Shq1p HtH2 (Fig. 3C, left). Instead, W350 is inserted in a Shq1p pocket formed by helices α3 and α17 at 15 Å from the position of its archaeal counterpart, and R348 forms hydrogen bonds with residues of the α3 and α4 helices (Fig. 3C, left).
Our structure brings new insight into the function of the C-terminal extension specific to eukaryotic Cbf5/NAP57 proteins. The 30 residues following W350 represent a conserved region of unknown function that is present in eukaryotes but absent in archaea. In the Shq1p/Cbf5p complex, this extension adopts an extended conformation that is stabilized by its interaction with Shq1p and is visible in the electron density up to N377. This region also contains an α-helical (α9) structure that is inserted into a V-shaped groove, formed by the Shq1p β hairpin and the α17 helix (Fig. 3C), and stabilized by significant stacking interactions and six salt bridges. Therefore, this region, specific to eukaryal Cbf5, is bound to conserved residues of Shq1pC. Interestingly, Shq1p lacks homologs in archaea. Therefore, the interaction between Shq1p and Cbf5p extends beyond RNA mimicry, providing a protein-specific binding site that could be accessible in RNA-bound Cbf5p. We propose that this extension functions as a protein–protein interaction site, important for Shq1p association and perhaps that of other protein factors during RNP assembly and maturation.
Shq1p binding to Cbf5p is essential for cell viability
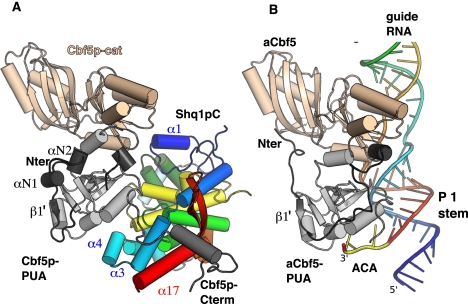
In order to assess the functional importance of the binding of Shq1pC to Cbf5p, we selected evolutionarily conserved Shq1p residues within the Shq1p/Cbf5p interface for mutational analysis. Due to the insolubility of yeast Cbf5p, we used the human homolog NAP57 fused to the maltose-binding protein (MBP-NAP57) for which soluble recombinant protein can be produced. The activity of the SHQ1 mutant proteins were evaluated in vitro by binding to recombinant NAP57, as previously described (Grozdanov et al. 2009a), and in vivo by their ability to complement a shq1 deletion strain, shq1Δ (see the Supplemental Material for strain details). Since we used the human protein homologs in the binding assay, we first tested whether SHQ1 could rescue the lethality of the shq1Δ yeast strain. Consistent with the fact that mammalian NAP57 can functionally replace yeast Cbf5p (Yang et al. 2000), wild-type SHQ1 was fully able to overcome the shq1Δ strain growth defect, suggesting that human SHQ1 interacts with yeast Cbf5p in vivo as Shq1p does (Fig. 4C).
Figure 4.
The Shq1p/Cbf5p interaction is essential for cell viability. (A) Pull-downs of recombinant wild-type and mutant SHQ1 with full-length NAP57 fused to MBP using amylose beads. Residue numbering is shown in italics for the human protein SHQ1 and in parentheses for yeast Shq1p. Input (above) corresponding to 10% of pull-downs (below) of a Coomassie blue-stained SDS-PAGE is shown. (B) Quantification of the amount of SHQ1 mutants pulled down relative to wild type from experiments shown in A and expressed as normalized ratios of SHQ1 constructs over NAP57 (averages of three experiments ± SEM). (C) SHQ1 variants were tested for their ability to rescue shq1Δ strain. Host strain (shq1Δ::TRP1 + pESC-URA[myc-Shq1p]) was transformed by pESC-LEU-based plasmid (empty, encoding myc-Shq1p or myc-SHQ1 variants). Serial dilutions of growing transformants were spotted (left to right; half dilution) on 5FOA-containing medium (1 g/L) and incubated at 30°C. The left column lists proteins encoded by transforming plasmid; mutants refer to myc-SHQ1 variants.
We first tested in vitro the effect of mutations of SHQ1 residues D167 and E296 (D189 and E323 in Shq1p) that are directly involved in salt bridges to Cbf5p R343 and R338 (Fig. 3B) and that are predicted to be major contributors to the stability of the complex. Indeed, mutation to alanine of D189/167 and E323/296 strongly decreases the association of SHQ1 with NAP57 (Fig. 4A,B). In contrast, mutation of E277/249 on the opposite side of SHQ1 and far removed from the Shq1p/Cbf5p interface does not impair the SHQ1/NAP57 interaction. In agreement with the in vitro data, the D189/167A and E323/296A mutants were unable to complement the shq1Δ yeast strain (Fig. 4C), while the control E277/249A mutant retained wild-type growth. Furthermore, SHQ1 substituted with an alanine at W326/299 and at K215/193—which, respectively, stacks on Cbf5p R343 and forms a salt bridge to D344 (Fig. 3C)—was also unable to rescue the shq1Δ strain even though complex formation was less affected in vitro (Fig. 4). The effects of the SHQ1 alanine mutants were reproduced in binding studies to MBP-NAP57 lacking its catalytic domain, MBP-NAP57Δcat (data not shown). These results confirm the interaction of SHQ1 with the PUA domain of NAP57 as a major determinant of the SHQ1/NAP57 interaction and as an essential function of SHQ1.
We also selected a conserved residue from the α13–α14 HtH3 loop, D421/373, which is not in contact with the PUA domain (Fig. 3B). However, in the model with full-length Cbf5p, this aspartate is within salt-bridging distance of R111 (R101 in archaea) (Fig. 3B, right), a residue of the Cbf5p catalytic domain that is involved in binding the phosphate backbone of the guide RNA. Mutation of D421/373 also impairs NAP57 binding in vitro and causes a mild growth defect in vivo, suggesting that hijacking by Shq1p/SHQ1 of Cbf5p/NAP57 residues involved in RNA recognition could extend to the Cbf5p/NAP57 catalytic domain. Surprisingly, the D217/195A mutation, which would only disrupt a water-mediated H bond to R349 in the C-terminal extension of Cbf5p, diminished binding by ∼75% in our in vitro assay, but enhanced growth of the shq1Δ strain. It is possible that the negative impact of this mutant on Shq1 binding is counterbalanced in vivo by its effect on other processes during the assembly and maturation of the RNP. Alternatively, differences between the interactions of the human with yeast proteins in vivo versus that of human with human proteins in vitro account for the differences between the two approaches.
SHQ1 competes with RNA for NAP57 binding
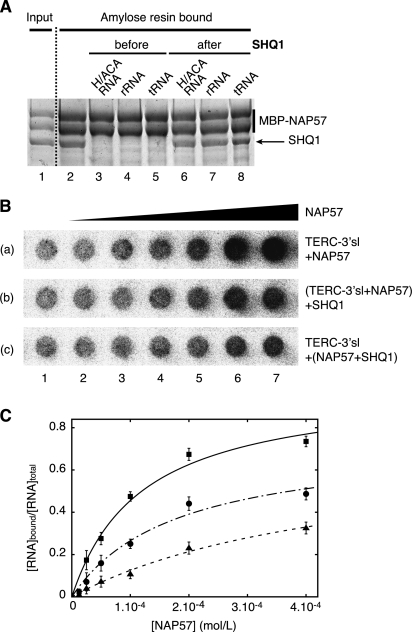
The Shq1p/Cbf5p structure reveals that the interaction of Shq1p with Cbf5p obstructs regions of Cbf5p that are important for binding to H/ACA RNAs, suggesting that binding of Shq1p and RNA to Cbf5p is mutually exclusive. This prediction is consistent with our finding that, within intact cells and in cell lysates, NAP57 associated with SHQ1 is free of H/ACA RNAs (Grozdanov et al. 2009a). Thus, we further investigated the direct interference between Shq1p/SHQ1 and RNA for Cbf5p/NAP57 binding by examining the impact of RNA addition on the binding of SHQ1 to MBP-NAP57 immobilized on amylose resin. Indeed, equimolar amounts of E3 H/ACA RNA abolished SHQ1 binding to NAP57 (Fig. 5A, cf. lanes 2 and 3). Surprisingly, unrelated RNAs, such as a fragment of ribosomal RNA or tRNA, competed equally for binding (Fig. 5A, lanes 4,5; Grozdanov et al. 2009b). Nevertheless, this effect was only observed when the RNAs were added before or simultaneously with SHQ1, but not after SHQ1, indicating a reduced off rate of SHQ1 binding (Fig. 5A, lanes 6–8).
Figure 5.
SHQ1 competes with RNA for NAP57 binding. (A) Pull-downs of SHQ1 with MBP-NAP57 as in Figure 4A: E3 H/ACA RNA, tRNA, and a fragment of 18S rRNA interfere with SHQ1 binding when added before but not after SHQ1. Picture of a Coomassie blue-stained SDS-PAGE. (B) Betascope images of nitrocellulose filters from titrations of NAP57 with human telomerase RNA (TERC-3′sl, 5 fmol per well). Wells from left to right (2–7) contained progressively higher concentrations of NAP57 in twofold steps ranging from 6.25 μM to 0.4 mM. Each strip presents data with different protein mixtures. (Row a) NAP57. (Row b) Same as row a, with SHQ1 (0.425 mM) added to the preformed NAP57/TERC-3′sl mixture. (Row c) NAP57 preincubated 20 min with SHQ1 (0.425 mM) before TERC-3′sl addition. (C) Quantification of bound RNA ratio versus total RNA in each well, and nonlinear curve fitting of the data points. (▪) Interaction between NAP57 and hTERC-3′sl; (•) addition of SHQ1 to a NAP57/hTERC-3′sl solution; (▴) addition of hTERC-3′sl to NAP57/SHQ1 solution.
We next assessed by filter binding the interaction of NAP57 with a radiolabeled H/ACA guide RNA using an in vitro transcribed human telomerase RNA 3′ stem–loop (TERC-3′sl; nucleotides 377–451) (see the Supplemental Material for construct details). This construct was chosen to only present a single NAP57-binding site. NAP57 efficiently binds hTERC-3′sl (Fig. 5B, row a), with an apparent Kd value of 75 μM (Fig. 5C), but, under the same conditions, SHQ1 does not show any significant RNA-binding affinity. This interaction was not displaced by the addition of tRNA or BSA, showing that NAP57 makes a specific interaction with the guide RNA (data not shown). We then incubated SHQ1 with NAP57 before the addition of RNA (Fig. 5B, row c). Under these conditions, the binding of NAP57 to TERC-3′sl decreased (Fig. 5C). This effect is apparently caused by a competition between RNA and SHQ1 for the same binding site of NAP57. In addition, when SHQ1 was added to the premixed NAP57–RNA complexes, TERC-3′sl binding to NAP57 decreased (Fig. 5B [row b], C), indicating that SHQ1 can chase away RNA bound to NAP57 in the absence of the other H/ACA core proteins. Inhibition analysis of the interaction between NAP57 and hTERC-3′sl, assuming a competitive inhibition of SHQ1, revealed a Ki of 110 μM (Fig. 5C). Although the observed binding affinities are low (likely due to competitive binding of RNA copurified with NAP57), the binding affinities of SHQ1 and H/ACA RNA for NAP57 are similar.
Implications for dyskeratosis congenita
Mutations in NAP57 cause X-DC, but only three of these mutations are located in the catalytic domain of the enzyme (full list of mutations and corresponding references can be found on the Telomerase Database, http://telomerase.asu.edu/diseases.html). Most X-DC mutations are found instead in the PUA domain, including the N-terminal residues that complete this domain and the eukaryote-specific C-terminal extension that follows it (Vulliamy et al. 2006). The structure of Cbf5pΔcat in complex with Shq1p allows us to map X-DC mutations in a eukaryal enzyme (Fig. 6), including its N-terminal and C-terminal extensions that are poorly conserved or absent in archaea.
Figure 6.
X-DC mutations cluster in two hot spots. Cbf5pΔcat and Shq1p are represented with the same color code as Figure 2. The Shq1p surface is depicted semitransparently. Residues mutated in X-DC are shown as space-filling models (magenta).
In the PUA domain, residues that are mutated in X-DC (K314, L317, L321, R322, M350, A353, and D359; human numbering) cluster in a patch encompassing residues of the β2′ strand and of the loops around the α8 helix that stacks on the β2′ strand, as in the archaeal structures (Rashid et al. 2006). In the N-terminal extension, the Cbf5p structure places the clusters of mutated residues in different positions compared with those predicted based on models of archaeal proteins. For instance, T19 (T49) is the first residue visible in the electron density (Fig. 6) and is also found mutated in X-DC patients. The equivalent residue in archaea is located 35 Å from the PUA mutation cluster, while in yeast Cbf5p, the new structure of the N terminus places this residue on top of the X-DC mutation cluster of the PUA domain. By extension, the stretch of mutated residues preceding it (Q31, F36-E41, and K43), although not visible in our structure and probably unstructured, should cluster in the same region. The structure of Cbf5p therefore brings all of these reported mutants in one hot spot on the surface of the PUA domain. Interestingly, although some of these X-DC mutations modulate the binding of SHQ1 to NAP57 (Grozdanov et al. 2009b), we found that none of the X-DC mutations in the PUA domain involve residues that interact directly with Shq1pC (Fig. 6). Thus, the effect of these mutations could be linked to local unfolding and destabilization of the Cbf5p protein and/or affect interaction with the full-length Shq1p.
Of the reported X-DC mutations outside the PUA domain, seven are located in the eukaryote-specific C-terminal extension (P384, A386, L398, G402, T408, P409, and S420). The first four of these residues (P354, A356, L368, and G372) are visible in the crystal structure of the complex and are located in the Cbf5p C-terminal extension bound in the V-shaped groove of Shq1pC (Figs. 3C, 6). In contrast to the mutational cluster of the PUA core domain, residues within this second mutational cluster in the C-terminal extension interact directly with Shq1pC. In order to assess the importance of the C-terminal domain in the stability of the interaction, we performed in vitro binding assays with truncated NAP57 constructs. Since this region is rich in basic residues, we first tested the effect of truncation of the poorly conserved charged N and C termini of NAP57 (NAP57 31–422) and found only a small reduction of SHQ1 binding compared with that of full-length NAP57 (FL 1–514) (Fig. 7A, cf. lanes 4 and 5). In contrast, further truncation of the conserved NAP57 C-terminal extension, which is specific to eukaryotes and contains residues mutated in X-DC (NAP57 31–387), reduces binding to full-length SHQ1 (Fig. 7A, lane 6) and to the SHQ1 C-terminal domain (Fig. 7B, lane 6) by an additional 28% and 34%, respectively. Therefore, our structure provides evidence for a second mutational hot spot and suggests that in X-DC patients with such mutations, destabilization of the SHQ1–NAP57 interface in a region distal from the PUA domain could be the underlying mechanism of disease.
Figure 7.
The C-terminal extension of NAP57 that is specific to eukaryal enzymes stabilizes the SHQ1/NAP57 complex. Pull-downs of full-length SHQ1 (A) and of the SHQ1 C terminus (SHQ1-Cter, residues 90–577) (B) with full-length NAP57 (FL 1–514), NAP57 lacking its charged termini (31–422), and NAP57 without the C-terminal extension (31–387) as in Figure 4A. Input (lanes 1–3) and pull-downs (lanes 4–6), including quantifications (below, averages of three experiments ± SEM), are shown and expressed as normalized ratios of SHQ1 constructs over NAP57 variants, with full-length NAP57 set as 1. (Lane 6) Note the reduced binding of the SHQ1 constructs to NAP57 lacking its C-terminal extension.
Discussion
The structure of the Shq1pC/Cbf5pΔcat complex provides a structural framework to understand the role of Shq1p/SHQ1 in the biogenesis of H/ACA particles and the role of Cbf5p/NAP57 mutations in X-DC. We demonstrate that the structure of the C-terminal domain of Shq1p adopts a new fold we refer to as “maki” with no homology with other known protein structures. It exposes a highly conserved surface on one side of the protein that is involved in binding the Cbf5p PUA domain and the C-terminal extension following it. This work also reveals the first structure of a eukaryotic pseudouridine synthase of H/ACA RNPs, going beyond homology-based models and providing experimental evidence for two mutation hot spots on the surface of NAP57 in X-DC. We found that a second set of mutations cluster in the eukaryal-specific C-terminal extension of NAP57 that is directly involved in SHQ1 binding. Deletion of this C-terminal extension impairs binding to SHQ1, which raises the possibility that these mutations affect the cellular SHQ1/NAP57 interaction, destabilize NAP57, and lead to reduced snoRNP and scaRNP biogenesis. In addition, we suggest that mutations in SHQ1 could account for some of the uncharacterized cases of autosomal recessive dyskeratosis congenita (Kirwan and Dokal 2008).
We found that Shq1p achieves specific Cbf5p binding by what is in essence structural mimicry of RNA–protein interactions. RNA mimicry by proteins is relatively rare, and only few structural examples are available. The most noteworthy come from ribosome-associated translation factors, which appear to mimic the shape and charge distribution of tRNA (Brodersen and Ramakrishnan 2003; Nakamura and Ito 2003). However, this molecular mimicry of the isolated protein structures does not necessarily extend when these proteins are part of the ribosome, because these proteins undergo large conformational changes to acquire their functional states on the ribosome (EF-P [Blaha et al. 2009], RF2 [Weixlbaumer et al. 2008], Erf1 [Song et al. 2000], Dom34 [van den Elzen et al. 2010], EF-G [Gao et al. 2009], and Pelota [Kobayashi et al. 2010]). In the case of Shq1p, shape mimicry is not necessary; only mimicry of the precise molecular contacts at the protein–protein interface is important for Cbf5p/NAP57 binding. We further show that binding to Shq1p and RNA is mutually exclusive, which suggests that the Shq1p/Cbf5p interaction is strong enough to prevent loading of the guide RNA. Although Shq1p/SHQ1 does not bind the catalytic domain of Cbf5p/NAP57 alone, Shq1p/SHQ1 might achieve the inhibition of Cbf5p/NAP57 by occupying the RNA-binding platform of the enzyme, a unique function of an RNP assembly factor that has not been demonstrated before. This is reminiscent of the function of some colicin immunity proteins that inhibit the nuclease activity of their toxins by binding an exo site far from the active site and preventing DNA or RNA binding (discussed in Graille et al. 2004). We therefore propose that Shq1p/SHQ1 chaperones Cbf5p/NAP57 in the early steps of H/ACA RNP assembly by stabilizing the inherently unstable Cbf5p/NAP57. Additionally, Shq1p/SHQ1 may prevent Cbf5p/NAP57 from promiscuous association with nucleic acids, thereby preventing spurious pseudouridylation of tRNAs and other RNAs by an enzyme-only mechanism. This could be important, since it has been shown that archaeal aCbf5 has standalone pseudouridine synthase activity on tRNA by recognition of the 3′ CCA nucleotides (Muller et al. 2007) and has high affinity and activity toward other RNA substrates (Zhou et al. 2011).
In summary, through the crystal structure of Shq1pC alone and in complex with Cbf5pΔcat, we established that Shq1p interacts on the RNA-binding surface of Cbf5p and recognizes this protein by employing an RNA mimicry mechanism. Despite the established modulation of SHQ1 binding by X-DC mutations in NAP57, Shq1p/SHQ1 binding occurs distal to the major X-DC mutation cluster, suggesting that structural rearrangements and/or additional interactions are impacted. Future studies will address these possibilities.
Material and methods
Cloning, expression, purification, and pull-downs
Protein samples were prepared as described in the Supplemental Material.
Crystallization and data collection
Both native and SeMet-substituted yeast Shq1pC proteins (5 mg/mL in 20 mM Tris-HCl at pH 7.5, 200 mM NaCl, 10 mM β-mercaptoethanol) were crystallized at 18°C by the sitting drop vapor diffusion method from a 1:1 mixture of protein and precipitant containing 20%–25% PEG 4000, 10% isopropanol, 100 mM HEPES (pH 7.5), and 50 mM MgSO4. Crystals were transferred in the mother liquor containing 35% PEG 4000 prior to flash-freezing in liquid nitrogen. The native Shq1pC/Cbf5pΔcat complex (3.5 mg/mL in 20 mM Tris-HCl at pH 7.5, 100 mM NaCl, 10 mM β-mercaptoethanol) was crystallized at 18°C by the hanging drop vapor diffusion method from 1:1 μL of protein and precipitant containing 4%–8% PEG 2000. Crystals were transferred in the mother liquor containing 30% glycerol prior to flash-freezing in liquid nitrogen.
X-ray diffraction data from crystals of the native Shq1pC, native Shq1pC/Cbf5pΔcat, and SeMet-substituted Shq1pC proteins were collected on the ID14-4 beamline at the European Synchrotron Research Facility (ESRF, Grenoble, France) and Proxima 1 beamline at the SOLEIL synchrotron (Gif-sur-Yvette, France). Diffraction data were processed using MOSFLM (Leslie 1992) and SCALA (Evans 2006). Both crystals of native and SeMet-substituted Shq1pC belong to space group P1, with one molecule per asymmetric unit. Crystals of native Shq1pC/Cbf5pΔcat belong to the P41212 space group, with two heterodimers per asymmetric unit. The cell parameters and data collection statistics are reported in Table 1. Small-angle X-ray scattering (SAXS) data were collected at the ESRF (beamline ID14-EH3) at an electron energy of 13.32 keV and processed with the program pipeline available at the beamline.
Structure solution and refinement
The Shq1pC structure was solved by single anomalous diffraction (SAD) using 1.6 Å data collected at the selenium peak wavelength. The selenium sites were initially found with SHELXD (Schneider and Sheldrick 2002), then phases were determined and improved by density modification with SHELXE (Schneider and Sheldrick 2002). Automatic model building was performed using BUCCANEER (Cowtan,2006), manually completed using COOT (Emsley and Cowtan 2004), and refined against the 1.5 Å resolution native data set using BUSTER (Blanc et al. 2004). The structure of the native Shq1pC/Cbf5pΔcat complex was solved by molecular replacement with PHASER (McCoy et al. 2007) using the free Shq1pC structure and a model of Cbf5p PUA domain based on its archaeal homologs generated by Modeller (Eswar et al. 2008) and refined to 2.8 Å resolution using BUSTER (Blanc et al. 2004).
Refinement statistics are shown in Table 1. In the final models, some regions are not visible in the electron density: residues G143–N163 (N terminus), V345–D361 (α10–α11 loop), S473–P481 (β-hairpin), and P506–Q507 (C terminus) in free Shq1pC; residues M145–G164 (N terminus) and E347–K360 (α10–α11 loop) in bound Shq1pC; and residues M1–D18 (N terminus), P43–S46 (β1′–α1′ loop), and T378–Y386 (C terminus) in bound Cbf5pΔcat. All residues fall within the allowed regions of the Ramachandran plot as defined by MOLPROBITY (Chen et al. 2010), except for Y379 in both free and bound Shq1pC. This residue precedes a cis-proline and is well defined in the electron density. In the crystal of the Cbf5pΔcat/Shq1pC complexes, the Cbf5pΔcat N termini are involved in domain-swapping with each other, resulting in a heterotetrameric complex (Supplemental Fig. S3) such that the N-terminal residues of Cbf5p (dark gray in Fig. 2) belong to different polypeptide chains. Apparent molecular weight determined from gel filtration and SAXS experiments indicate that these heterotetrameric complexes also exist in solution, apparently glued together by domain swapping. Although we cannot exclude the possibility of artifactual domain swapping, it is tempting to speculate that this might occur in mature H/ACA RNPs, which in eukaryotes contain two copies of Cbf5p/NAP57. Figures were prepared using PyMOL (The PyMOL Molecular Graphics System, version 1.3, Schrödinger, LLC)
Accession codes
Protein Data Bank: Coordinates and structure factor amplitudes for Shq1pC and of the Shq1pC/Cbf5pΔcat complex have been deposited with accession codes 3zuz and 3zv0, respectively.
Acknowledgments
We thank P. Legrand and A. Thompson (Soleil synchrotron, Gif-sur-Yvette) for their help in data collection, and Bruno Faivre for cloning Shq1pC. Work at Université Paris-Sud and H.W. were supported by a BIORIB grant (BLAN07-1_194553) from the Agence Nationale de la Recherche (ANR Blanche). Work at Paris Descartes University was supported by a grant from Association pour la Recherche sur le Cancer (ARC). Work at the University of Washington was supported by grant NIH-NIGMS GM-64440 to G.V., and work at the Albert Einstein College of Medicine was supported by NIH HL079566 to U.T.M. H.W. designed the Cbf5p constructs, purified yeast constructs, and solved the crystal structures; R.M.-P. and P.N.G. expressed and purified the recombinant proteins and performed the protein pull-down assays; D.L. performed yeast complementation studies; M.B. and S.R. purified proteins and RNA and carried out filter-binding assays; K.G. performed initial cloning and purifications of yeast proteins; and U.T.M., G.V., and N.L. designed the studies, interpreted data, and wrote the paper with contributions from R.M.-P, M.B., D.L., and H.v.T.
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.176834.111.
References
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA 2001. Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci 98: 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaha G, Stanley RE, Steitz TA 2009. Formation of the first peptide bond: The structure of EF-P bound to the 70S ribosome. Science 325: 966–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G 2004. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr 60: 2210–2221 [DOI] [PubMed] [Google Scholar]
- Brodersen DE, Ramakrishnan V 2003. Shape can be seductive. Nat Struct Mol Biol 10: 78–80 [DOI] [PubMed] [Google Scholar]
- Bullinger L, Krönke J, Schön C, Radtke I, Urlbauer K, Botzenhardt U, Gaidzik V, Carió A, Senger C, Schlenk RF, et al. 2010. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia 24: 438–449 [DOI] [PubMed] [Google Scholar]
- Chadwick N, Zeef L, Portillo V, Fennessy C, Warrander F, Hoyle S, Buckle A-M 2009. Identification of novel Notch target genes in T cell leukaemia. Mol Cancer 8: 35 doi: 10.1186/1476-4598-8-35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC 2010. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66: 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowtan K 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr D Biol Crystallogr 62: 1002–1011 [DOI] [PubMed] [Google Scholar]
- Darzacq X, Kittur N, Roy S, Shav-Tal Y, Singer RH, Meier UT 2006. Stepwise RNP assembly at the site of H/ACA RNA transcription in human cells. J Cell Biol 173: 207–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Henras A, Faucon B, Lafontaine D, Caizergues-Ferrer M, Henry Y 2001. Stable expression in yeast of the mature form of human telomerase RNA depends on its association with the box H/ACA small nucleolar RNP proteins Cbf5p, Nhp2p and Nop10p. Nucleic Acids Res 29: 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dez C, Noaillac-Depeyre J, Caizergues-Ferrer M, Henry Y 2002. Naf1p, an essential nucleoplasmic factor specifically required for accumulation of box H/ACA small nucleolar RNPs. Mol Cell Biol 22: 7053–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan J, Li L, Lu J, Wang W, Ye K 2009. Structural mechanism of substrate RNA recruitment in H/ACA RNA-guided pseudouridine synthase. Mol Cell 34: 427–439 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K 2004. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60: 2126–2132 [DOI] [PubMed] [Google Scholar]
- Eswar N, Eramian D, Webb B, Shen M-Y, Sali A 2008. Protein structure modeling with MODELLER. Methods Mol Biol 426: 145–159 [DOI] [PubMed] [Google Scholar]
- Evans P 2006. Scaling and assessment of data quality. Acta Crystallogr D Biol Crystallogr 62: 72–82 [DOI] [PubMed] [Google Scholar]
- Fatica A, Dlakić M, Tollervey D 2002. Naf1p is a box H/ACA snoRNP assembly factor. RNA 8: 1502–1514 [PMC free article] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz H-R, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. 2008. The Pfam protein families database. Nucleic Acids Res 36: D281–D288 doi: 10.1093/nar/gkm960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y-G, Selmer M, Dunham CM, Weixlbaumer A, Kelley AC, Ramakrishnan V 2009. The structure of the ribosome with elongation factor G trapped in the posttranslocational state. Science 326: 694–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godin KS, Walbott H, Leulliot N, van Tilbeurgh H, Varani G 2009. The box H/ACA snoRNP assembly factor Shq1p is a chaperone protein homologous to Hsp90 cochaperones that binds to the Cbf5p enzyme. J Mol Biol 390: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graille M, Mora L, Buckingham RH, van Tilbeurgh H, de Zamaroczy M 2004. Structural inhibition of the colicin D tRNase by the tRNA-mimicking immunity protein. EMBO J 23: 1474–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov PN, Roy S, Kittur N, Meier UT 2009a. SHQ1 is required prior to NAF1 for assembly of H/ACA small nucleolar and telomerase RNPs. RNA 15: 1188–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozdanov PN, Fernandez-Fuentes N, Fiser A, Meier UT 2009b. Pathogenic NAP57 mutations decrease ribonucleoprotein assembly in dyskeratosis congenita. Hum Mol Genet 18: 4546–4551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiss NS, Knight SW, Vulliamy TJ, Klauck SM, Wiemann S, Mason PJ, Poustka A, Dokal I 1998. X-linked dyskeratosis congenita is caused by mutations in a highly conserved gene with putative nucleolar functions. Nat Genet 19: 32–38 [DOI] [PubMed] [Google Scholar]
- Henras A, Henry Y, Bousquet-Antonelli C, Noaillac-Depeyre J, Gélugne JP, Caizergues-Ferrer M 1998. Nhp2p and Nop10p are essential for the function of H/ACA snoRNPs. EMBO J 17: 7078–7090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoareau-Aveilla C, Bonoli M, Caizergues-Ferrer M, Henry Y 2006. hNaf1 is required for accumulation of human box H/ACA snoRNPs, scaRNPs, and telomerase. RNA 12: 832–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. 2009. InterPro: The integrative protein signature database. Nucleic Acids Res 37: D211–D215 doi: 10.1093/nar/gkn785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Loria JP, Moore PB 2007. Solution structure of an rRNA substrate bound to the pseudouridylation pocket of a box H/ACA snoRNA. Mol Cell 26: 205–215 [DOI] [PubMed] [Google Scholar]
- King TH, Liu B, McCully RR, Fournier MJ 2003. Ribosome structure and activity are altered in cells lacking snoRNPs that form pseudouridines in the peptidyl transferase center. Mol Cell 11: 425–435 [DOI] [PubMed] [Google Scholar]
- Kirwan M, Dokal I 2008. Dyskeratosis congenita: A genetic disorder of many faces. Clin Genet 73: 103–112 [DOI] [PubMed] [Google Scholar]
- Kiss T 2004. Biogenesis of small nuclear RNPs. J Cell Sci 117: 5949–5951 [DOI] [PubMed] [Google Scholar]
- Kiss T, Fayet-Lebaron E, Jády BE 2010. Box H/ACA small ribonucleoproteins. Mol Cell 37: 597–606 [DOI] [PubMed] [Google Scholar]
- Kobayashi K, Kikuno I, Kuroha K, Saito K, Ito K, Ishitani R, Inada T, Nureki O 2010. Structural basis for mRNA surveillance by archaeal Pelota and GTP-bound EF1α complex. Proc Natl Acad Sci 107: 17575–17579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafontaine DLJ, Bousquet-Antonelli C, Henry Y, Caizergues-Ferrer M, Tollervey D 1998. The box H+ACA snoRNAs carry Cbf5p, the putative rRNA pseudouridine synthase. Genes Dev 12: 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW 1992. Recent changes to the MOSFLM package for processing film and image plate data. Joint CCP4 + ESF-EAMCB Newsletter on Protein Crystallography 26 [Google Scholar]
- Leulliot N, Godin KS, Hoareau-Aveilla C, Quevillon-Cheruel S, Varani G, Henry Y, van Tilbeurgh H 2007. The box H/ACA RNP assembly factor Naf1p contains a domain homologous to Gar1p mediating its interaction with Cbf5p. J Mol Biol 371: 1338–1353 [DOI] [PubMed] [Google Scholar]
- Li H 2008. Unveiling substrate RNA binding to H/ACA RNPs: One side fits all. Curr Opin Struct Biol 18: 78–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye K 2006. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307 [DOI] [PubMed] [Google Scholar]
- Liang B, Zhou J, Kahen E, Terns RM, Terns MP, Li H 2009. Structure of a functional ribonucleoprotein pseudouridine synthase bound to a substrate RNA. Nat Struct Mol Biol 16: 740–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowiak AA, Narayanan A, Li ZH, Terns RM, Terns MP 2001. The snoRNA domain of vertebrate telomerase RNA functions to localize the RNA within the nucleus. RNA 7: 1833–1844 [PMC free article] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ 2007. Phaser crystallographic software. J Appl Crystallogr 40: 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier UT 2005. The many facets of H/ACA ribonucleoproteins. Chromosoma 114: 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell JR, Cheng J, Collins K 1999. A box H/ACA small nucleolar RNA-like domain at the human telomerase RNA 3′ end. Mol Cell Biol 19: 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller S, Fourmann J-B, Loegler C, Charpentier B, Branlant C 2007. Identification of determinants in the protein partners aCBF5 and aNOP10 necessary for the tRNA:Ψ55-synthase and RNA-guided RNA:Ψ-synthase activities. Nucleic Acids Res 35: 5610–5624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Ito K 2003. Making sense of mimic in translation termination. Trends Biochem Sci 28: 99–105 [DOI] [PubMed] [Google Scholar]
- Normand C, Capeyrou R, Quevillon-Cheruel S, Mougin A, Henry Y, Caizergues-Ferrer M 2006. Analysis of the binding of the N-terminal conserved domain of yeast Cbf5p to a box H/ACA snoRNA. RNA 12: 1868–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashid R, Liang B, Baker DL, Youssef OA, He Y, Phipps K, Terns RM, Terns MP, Li H 2006. Crystal structure of a Cbf5–Nop10–Gar1 complex and implications in RNA-guided pseudouridylation and dyskeratosis congenita. Mol Cell 21: 249–260 [DOI] [PubMed] [Google Scholar]
- Schneider TR, Sheldrick GM 2002. Substructure solution with SHELXD. Acta Crystallogr D Biol Crystallogr 58: 1772–1779 [DOI] [PubMed] [Google Scholar]
- Singh M, Gonzales FA, Cascio D, Heckmann N, Chanfreau G, Feigon J 2009. Structure and functional studies of the CS domain of the essential H/ACA ribonucleoparticle assembly protein SHQ1. J Biol Chem 284: 1906–1916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H, Mugnier P, Das AK, Webb HM, Evans DR, Tuite MF, Hemmings BA, Barford D 2000. The crystal structure of human eukaryotic release factor eRF1—Mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100: 311–321 [DOI] [PubMed] [Google Scholar]
- Taylor BS, Schultz N, Hieronymus H, Gopalan A, Xiao Y, Carver BS, Arora VK, Kaushik P, Cerami E, Reva B, et al. 2010. Integrative genomic profiling of human prostate cancer. Cancer Cell 18: 11–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Elzen AMG, Henri J, Lazar N, Gas ME, Durand D, Lacroute F, Nicaise M, van Tilbeurgh H, Séraphin B, Graille M 2010. Dissection of Dom34-Hbs1 reveals independent functions in two RNA quality control pathways. Nat Struct Mol Biol 17: 1446–1452 [DOI] [PubMed] [Google Scholar]
- Vulliamy TJ, Marrone A, Knight SW, Walne A, Mason PJ, Dokal I 2006. Mutations in dyskeratosis congenita: Their impact on telomere length and the diversity of clinical presentation. Blood 107: 2680–2685 [DOI] [PubMed] [Google Scholar]
- Weixlbaumer A, Jin H, Neubauer C, Voorhees RM, Petry S, Kelley AC, Ramakrishnan V 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322: 953–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Isaac C, Wang C, Dragon F, Pogacic V, Meier UT 2000. Conserved composition of mammalian box H/ACA and box C/D small nucleolar ribonucleoprotein particles and their interaction with the common factor Nopp140. Mol Biol Cell 11: 567–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang PK, Rotondo G, Porras T, Legrain P, Chanfreau G 2002. The Shq1p.Naf1p complex is required for box H/ACA small nucleolar ribonucleoprotein particle biogenesis. J Biol Chem 277: 45235–45242 [DOI] [PubMed] [Google Scholar]
- Ye K 2007. H/ACA guide RNAs, proteins and complexes. Curr Opin Struct Biol 17: 287–292 [DOI] [PubMed] [Google Scholar]
- Yu YT, Shu MD, Steitz JA 1998. Modifications of U2 snRNA are required for snRNP assembly and pre-mRNA splicing. EMBO J 17: 5783–5795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao R, Kakihara Y, Gribun A, Huen J, Yang G, Khanna M, Costanzo M, Brost RL, Boone C, Hughes TR, et al. 2008. Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J Cell Biol 180: 563–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liang B, Li H 2011. Structural and functional evidence of high specificity of Cbf5 for ACA trinucleotide. RNA 17: 244–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Tomlinson RL, Lukowiak AA, Terns RM, Terns MP 2004. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell 15: 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]