Abstract
Total internal reflection fluorescence microscopy (TIRFM) has been proven to be an extremely powerful technique in animal cell research for generating high contrast images and dynamic protein conformation information. However, there has long been a perception that TIRFM is not feasible in plant cells because the cell wall would restrict the penetration of the evanescent field and lead to scattering of illumination. By comparative analysis of epifluorescence and TIRF in root cells, it is demonstrated that TIRFM can generate high contrast images, superior to other approaches, from intact plant cells. It is also shown that TIRF imaging is possible not only at the plasma membrane level, but also in organelles, for example the nucleus, due to the presence of the central vacuole. Importantly, it is demonstrated for the first time that this is TIRF excitation, and not TIRF-like excitation described as variable-angle epifluorescence microscopy (VAEM), and it is shown how to distinguish the two techniques in practical microscopy. These TIRF images show the highest signal-to-background ratio, and it is demonstrated that they can be used for single-molecule microscopy. Rare protein events, which would otherwise be masked by the average molecular behaviour, can therefore be detected, including the conformations and oligomerization states of interacting proteins and signalling networks in vivo. The demonstration of the application of TIRFM and single-molecule analysis to plant cells therefore opens up a new range of possibilities for plant cell imaging.
Keywords: Evanescent, epifluorescence, fluorescent protein, microscopy, single-molecule total internal reflection fluorescence (TIRF) microscopy
Introduction
Accurate intracellular localization of proteins is a critical part of understanding their function. The development of green fluorescent protein (GFP) and its derivative fluorescent proteins (FPs) has provided scientists with invaluable tools to study the location and dynamics of plant proteins (Goodin et al., 2007; Mathur, 2007). By coupling the use of FPs with the remarkable advances in imaging techniques over the last few decades, it is now possible to visualize and study biological processes at the subcellular level, and even at the single-molecule level in a living plant.
Several fluorescence microscopy techniques are available for the study of proteins in living plant cells (Shaw, 2006). At its simplest, the sample is wide-field illuminated through the objective (epifluorescence, Fig. 1a), and the resultant fluorescence is detected by a camera or viewed through the eyepiece. However, fluorescent objects outside the focal plane contribute high levels of background to the detected signal. In order to reduce or even eliminate this out-of-focus component, optical sectioning techniques have been developed.
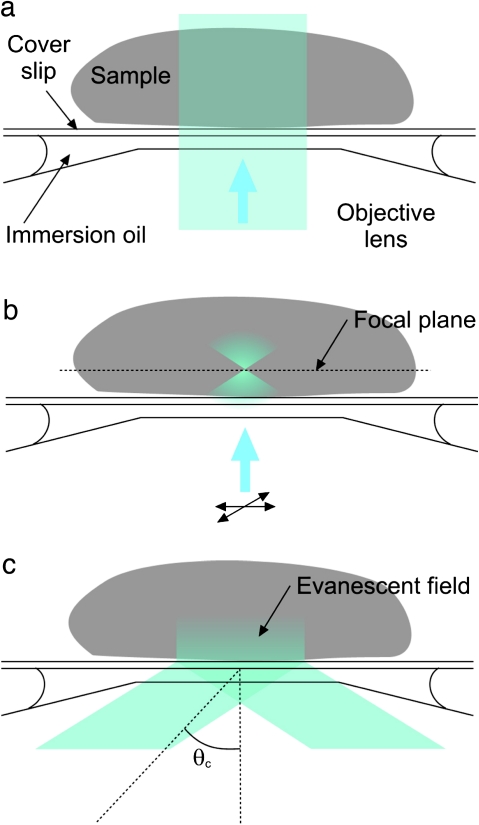
Fig. 1.
Comparisons between different fluorescence microscopy techniques. (a) Epifluorescence (EPI). The beam is incident along the central axis of the objective and illuminates the entire sample. Both in- and out-of-focus fluorescence signal is collected. (b) Laser confocal scanning microscopy (LSCM). The laser beam is focused in the plane of interest, reducing fluorescence excitation in other planes, and scanned across the sample. A pinhole (not shown) rejects most of the out-of-focus excited fluorescence and further confines the image to the focal plane of interest. (c) Total internal reflection fluorescence microscopy (TIRFM). A beam incident on the interface between two mediums (e.g. coverslip glass and water/sample) with different refractive indices at an angle greater than the critical angle θc is totally internally reflected. An evanescent field is generated, the intensity of which decays exponentially over a few hundred nanometres. (This figure is available in colour at JXB online.)
One such technique is laser confocal scanning microscopy (LCSM), which significantly reduces the background caused by out-of-focus and scattered light. This gives a high signal-to-background ratio (SBR) and improves the image resolution considerably compared with wide-field fluorescence microscopy, particularly in the axial direction (Conchello and Lichtman, 2005). It should be pointed out that SBRs are sometimes erroneously equated with signal-to-noise ratios (SNRs). The former is a measure of contrast in an image (i.e. the ratio between the intensities of pixels in the focal and deeper planes), whereas the SNR describes the variability in the intensity of a single pixel. LCSM uses point illumination, which is scanned across the sample to build up an image (Fig. 1b). A confocal pinhole in front of the detector spatially filters the fluorescence to eliminate out-of-focus information. The increased resolution is at the cost of decreased signal intensity since much of the sample fluorescence signal is blocked (at the pinhole). However, the resultant increased exposures, which can cause sample photobleaching, and slow image acquisition times are the major drawbacks of this technique. Image acquisition speeds can be increased using spinning-disk and line-scanning confocal techniques, but there will always be some loss of image resolution (Shaw, 2006).
An alternative approach to optical sectioning that has proven to be extremely powerful in animal cell research for generating images with a high SBR is total internal reflection fluorescence microscopy (TIRFM). If a laser beam strikes the interface between two materials of high and low refractive index at an angle greater than the critical angle (given by Snell’s Law), the incident light will undergo total internal reflection (Fig. 1c). In the microscope, these materials are the lens/immersion oil/coverslip and sample/water, respectively. Although the excitation beam does not pass through the sample, an ‘evanescent’ electromagnetic field is generated whose intensity decays exponentially with distance from the interface into the sample. The field penetrates for a few hundred nanometres (<400 nm) into the sample in the z-direction (Axelrod, 2001; Schneckenburger, 2005; Wang et al., 2006; Konopka and Bednarek, 2008a). Consequently, only the fluorophores nearest to the glass surface (within the evanescent field) are selectively excited and their fluorescence collected by the microscope optics. This limitation on the excitation depth, which could be considered as a disadvantage of this technique, is precisely its main advantage; TIRFM therefore generates images with the highest SBR, where background fluorescence is nearly absent and photobleaching is dramatically reduced. Total internal reflection illumination can be achieved using either a prism-based or, with a high numerical aperture objective lens (NA >1.4), an objective-based configuration (for reviews, see (Axelrod, 2001; Toomre and Manstein, 2001; Schneckenburger, 2005).
TIRFM has been utilized very effectively for imaging membrane proteins in animal systems, for example epidermal growth factor (Webb et al., 2006), single-channel calcium microdomains (Demuro and Parker, 2006), and the dynamics of the yeast cytoskeleton (Chan et al., 2009). Until recently, however, TIRFM had only been applied in plant research to the in vitro study of the actin cytoskeleton and microtubule dynamics, and endoplasmic reticulum (ER) dynamics in protoplasts, which lack a cell wall (Michelot et al., 2005, 2006; Goodin et al., 2007; Vidali et al., 2009; Ye et al., 2009). The thinness of the plasma membrane in animal cells makes them ideal for TIRFM. In contrast, plant cells have a rigid cell wall surrounding the plasma membrane, which varies in thickness depending upon the tissue, growth conditions, and developmental stage, but can be >100 nm. This thickness could, in principle, prevent the penetration of the evanescent field to the plasma membrane and beyond. The cell wall could also lead to scattering of the illumination, resulting in excitation of fluorophores beyond the evanescent field. The consequent view that TIRFM is of limited use with intact plant cells for the visualization of biological processes in and beyond the plasma membrane (Shaw, 2006) has caused plant scientists to show little interest in the technique.
An alternative to epifluorescence is variable-angle epifluorescence microscopy (VAEM); instead of an evanescent field, this uses a narrow band of illumination that passes through the sample almost parallel to the coverslip, yielding a high SNR for visualizing events at or near the plasma membrane of intact cells. As with TIRFM, little has been published in plant research using VAEM. However, it has been used to image in vivo dynamics of secretory vesicles in pollen tubes (Wang et al., 2006) and to study the dynamics and function of dynamin-related proteins (DRPs), which are required for cytokinesis and cell expansion (Konopka and Bednarek, 2008a, b; Konopka et al., 2008). VAEM showed a significant improvement over epifluorescence illumination for actin filaments in growing root hairs (Konopka and Bednarek, 2008b), increasing the resolution of individual actin cables. It was also used with plasma membrane markers for the study of plant endocytosis (Konopka and Bednarek, 2008a, b; Konopka et al., 2008). Using VAEM in pollen tubes and expanding hypocotyl epidermal cells and TIRF techniques in protoplasts, the dynamics of actin turnover have also been examined (Staiger et al., 2009; Blanchoin and Staiger, 2010; Smertenko et al., 2010; Zhang et al., 2010); these approaches have facilitated a previously hitherto unknown level of understanding of the dynamic control of actin filament turnover in plant cells indicating the potential that such techniques can offer.
Here, by performing a comparative study with other wide-field fluorescence microscopies, it is demonstrated for the first time that TIRFM is a valuable tool for in vivo analysis of fluorescent proteins in intact plant cells and can be used for single-molecule analysis in plants. It is shown that TIRF imaging of intact cells is possible, not only at the plasma membrane, but also, due to the physiology of plant cells and the presence of large vacuoles, in organelles situated in the cytoplasm of the plant cell. How to distinguish between TIRFM and VAEM (Konopka and Bednarek, 2008b), also known as highly inclined laminated optical sheet microscopy (HILO; Tokunaga et al., 2008), which can be difficult, is also described, and it is demonstrated that the high SBR that is seen is indeed due to genuine TIRF. It is then demonstrated for the first time in plant cells that TIRFM can be used for single-molecule analysis in plant cells. These techniques, which are becoming routine in mammalian cells, are a major advance for plant biology. For example, the new super-resolution imaging methods such as photoactivated localization microscopy (PALM; capable of nanometre resolution) depend on the ability to detect single molecules. The particular advantages of TIRF illumination and single-molecule analysis, which include the ability to determine the stoichiometry of protein complexes and in situ protein conformation as derived from single-molecule polarization and single-molecule fluorescence resonance energy transfer (FRET), are discussed.
Materials and methods
Plant material
Arabidopsis thaliana plants, ecotype Colombia-0, were used as negative controls and the GFP–MAP4 line (Marc et al., 1998) was used to visualize cortical microtubules. The rest of the GFP lines showing subcellular localization in A. thaliana were a generous gift from Dr S. Kurup (Rothamsted Research, Harpenden, UK): mGFP5–ER (Haseloff et al., 1997), LTI6b, N84725, N84727, N84728, and N84733 (Cutler et al., 2000).
Plant growth conditions
For visualization of intact root cells, seeds were surface sterilized in 10% (v/v) household bleach for 5 min, washed three times with sterile water, and plated on 10 cm×10 cm square plates with 0.5 M MS agar [2.2 g l−1 Murashige and Skoog medium, 1% (w/v) agar, pH 6.2, with KOH]. Plated seed were stratified for 2 d, in the dark at 4 °C, to achieve the highest germination synchronization. The seedlings were then allowed to germinate and grow under continuous light (125 μmol m−2 s−1) at 22 °C for 7 d.
Wide-field fluorescence and TIRF microscopy
To visualize intact cells, a ∼7 mm fragment of root tip was transferred to a 35 mm glass-bottomed culture dish (no. 1.5, uncoated, γ-irradiated, Mat Tek). A small drop of sterile water was applied to a coverslip, which was deposited on top of the root fragment. Without further manipulation of the coverslip, more water was applied to the edge of the coverslip to ensure that no air bubbles were trapped while keeping the root in direct contact with the glass bottom of the culture dish. A drop of immersion oil was applied to the microscope objective and the culture dish was placed on top of it.
The microscope is a custom-built objective-type inverted TIRF microscope, incorporating a ×100 objective lens (Zeiss, α-plan fluar, NA=1.45). Excitation light enters the rear port of the microscope via a single mode optical fibre and spatial filter assembly, the pinhole of which is imaged onto the back focal plane of the objective lens. A micrometer allows continuous lateral adjustment of the spatial filter assembly, and hence the position of the beam at the back aperture of the objective, to switch between TIRF and epifluorescence modes. It is possible to verify that the microscope is in TIRF mode when a sample is on the microscope because only then does the excitation beam return through the objective and become clearly visible on a piece of sticky tape temporarily, partially blocking the optical path at the filter cube. The excitation source is a 491 nm solid state laser (Cobolt, Calypso), the filter cube contained the filters HQ480/40X, Q505LP, and HQ525/50M (Chroma), and the detector was an electron multiplication CCD (DV887, Andor). The frame rate used is a trade-off between the intensity SNR, the photobleaching rate, and the laser power’s effects on the cell. It was decided to integrate full frames for 100 ms, acquired at 10 Hz, and the laser power incident on the sample was 1.5 mW, but the optimum parameters could be determined by further study.
The angle of the beam at the sample was measured using a semi-circular glass block protractor with a diameter of 230 mm and equal refractive index to the objective lens (Comar). By placing the block centrally on the objective with a drop of immersion oil to bridge the gap, TIR is prevented. The location and divergence of the beam are marked on the outside of the block and the angles measured.
For single-molecule studies, the sample was bleached until individual spots, rather than structures, were visible (∼75 min). Spots were determined to be single molecules if their size was equal to the point-spread function of the microscope, their intensity was consistent with that expected from single enhanced GFP (EGFP) molecules, and they exhibited single-step photobleaching characteristics. Data were analysed using custom-written software (D Rolfe et al., unpublished results).
Results
TIRFM can be used to visualize fluorescent proteins in intact plant cells
The use of TIRFM in whole plant tissues was investigated to determine whether fluorescently labelled proteins could be imaged in intact cells and in organelles beyond the cell surface. As mentioned previously, there have been major concerns within the scientific community that the plant cell wall would disturb the evanescent field, or simply impede the observation of deeper structures because of its thickness. Initially fluorescent proteins in Arabidopsis protoplasts were imaged using TIRFM (data not shown); however, the principal interest was in using TIRFM on intact plant tissues. Transient transformation by bombardment of onion epidermal cells was conducted; however, the frequency of transformation was relatively low and the imaging was difficult because of the lack of homogeneity in tissue thickness and the difficulty in getting the tissue to lie completely flat. Therefore, the focus of study changed to imaging Arabidopsis roots from stably transformed transgenic lines carrying a variety of fluorescently labelled proteins, and it was possible to detect fluorescence in the plasma membrane and in various organelles, including microtubules, vacuolar membrane, ER, and nuclei, located at the periphery of the intact plant cells due to the presence of the central vacuole (Figs 2, 3). Sample mounting was critical for optimal imaging; a ∼7 mm fragment of the root tip was transferred to a 35 mm glass-bottomed culture dish and a drop of sterile water was applied to a coverslip which was deposited on top of the root fragment without air bubbles. Ensuring that the root was touching the bottom of the culture dish was imperative, not only to obtain focused images, but also to guarantee that the sample to be analysed was within the reach of the evanescent field. In order to be able to compare epifluorescence and TIRF techniques, illumination intensity was kept constant for both illumination modes but the CCD gain was reduced when the epifluorescence images were saturated.
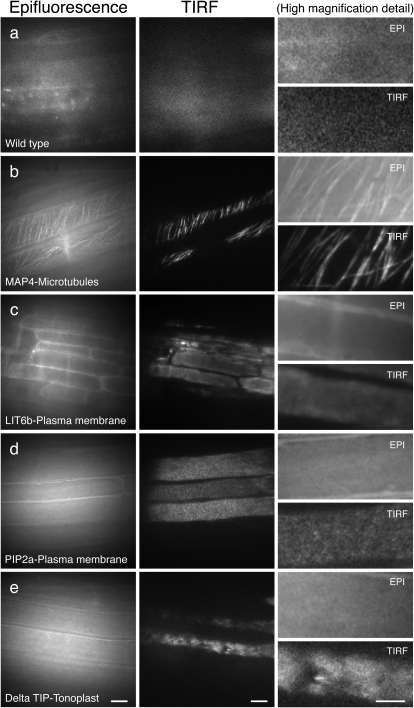
Fig. 2.
Analysis of plasma membrane and cytoskeleton markers by epifluorescence and TIRF microscopy in roots. Arabidopsis roots imaged using epifluorescence and TIRF. (a) Wild-type control (Col-0); (b) MAP4–GFP (microtubule marker); (c) LIT6b–GFP (plasma membrane marker); (d) N84725 (PIP2a; plasma membrane marker); (e) N84727 (Delta TIP; vacuolar membrane marker). Scale bars in columns 1 and 2=10 μm. Scale bars in details=5 μm
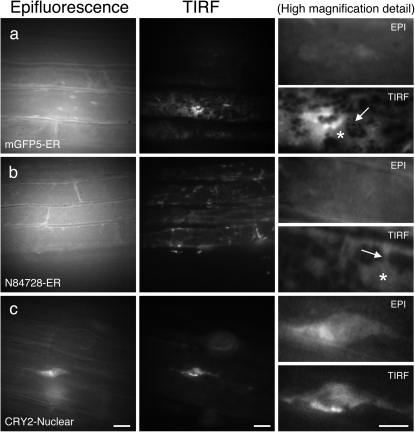
Fig. 3.
Analysis of subcellular organelle markers by epifluorescence and TIRFM in roots. Arabidopsis roots imaged using epifluorescence and TIRFM. (a) mGFP5–ER (ER marker); (b) N84728 (ER marker). Individual ER structures and the presence of extended cisternal lamellae connected to a tubular ER network can only be distinguished in the TIRF images (asterisks, ER cisternae; arrow, ER tubules); (c) N84733 (chromosome marker). Scale bars=10 μm. Scale bars in details=5 μm
Wild-type Arabidopsis roots were used to evaluate the initial levels of autofluorescence and light scatter (Fig. 2a). Regions where the root tissue was in closest contact with the glass of the culture dish were pre-selected for imaging. In contrast to the epi-image, the TIRF image shows almost no fluorescence background. Figure 2b shows GFP-labelled microtubules (MAP4) in the root epidermis. The TIRF image shows a clearer image of the microtubule arrangement, such that individual or small groups of microtubules can be distinguished, with a higher SBR compared with the same cells imaged with epifluorescence illumination (Fig. 2b, detail).
To visualize localization within the plasma membrane, two different markers were used—LTI6b (Kurup et al., 2005) (Fig. 2c) and PIP2a (Line N84725; Cutler et al., 2000) (Fig. 2d). While the epi-images showed a blur of fluorescence, the TIRF images provided a more detailed and localized signal, with the detected fluorescence restricted to the plasma membrane of individual cells. It is also evident from these images that TIRFM enhances the visualization of cell structures by removing most of the out-of-focus fluorescence signal. These images revealed a distinct fluorescent pattern for each plasma membrane marker; while the LIT6b marker has a very diffuse fluorescence distribution, PIP2a presents a discrete punctated fluorescence pattern (Fig, 2c, d, detail). This specific pattern of expression can be used to infer valuable information about the localization and dynamics of these labelled proteins.
The tonoplast marker, Delta TIP, was used to image the vacuolar membrane of root epidermal cells (Line N84727; Cutler et al., 2000) (Fig. 2e). TIRF images showed discrete localization of the fluorescence signals in particular areas of the tonoplast, with regions partially depleted of fluorescent signal, indicating specific spatial localization of the protein marker. In contrast, images obtained with epifluorescence could not differentiate between areas with or without fluorescence, showing reduced resolution and signal evenly distributed along the vacuolar membrane (Fig. 2e, detail). Similar signal localization of the tonoplast marker (DIP aquaporin) was previously observed using VAEM and was proposed as corresponding to regions of vacuole membrane invagination (Konopka and Bednarek, 2008b).
To determine whether TIRFM can be used to image fluorescent proteins targeted to organelles within the cell, an array of protein markers were used that are localized to the ER and the nucleus. Figure 3a (mGFP5–ER; Haseloff et al., 1997) and 3b (Line N84728, GFP fusion to an unknown protein to the ER surface; Cutler et al., 2000) shows GFP localization in the ER of root epidermal cells. The GFP fluorescent signal presented a very distinctive pattern, accumulating in certain areas such as the edges of the cell, whilst other areas did not show any GFP signal. The epi-illumination images appeared blurred and lacked structural definition, whilst the images obtained using TIR illumination presented a specific distribution of GFP fluorescence, allowing the resolution and localization of GFP-positive structures, due to their much higher SBRs. The ER presents two characteristic forms: cisternae or lamellae and tubular elements. Only in the TIRF images can the different ER structures and the presence of extended cisternal lamellae all along the cytoplasm connected to a tubular ER network be resolved (Fig. 3a, b, detail).
Interesting results were obtained using the nuclear-targeted marker, CRY2–GFP (Line N84733; Cutler et al., 2000) (Fig. 3c), with this marker enabling visualization of the nuclei in root epidermal cells using epifluorescence and TIRFM. Most importantly, TIRF images were of higher resolution with superior SBRs compared with epifluorescence, allowing enhanced visualization of intranuclear structures (Fig. 3c, detail).
TIRFM is distinguishable from VAEM
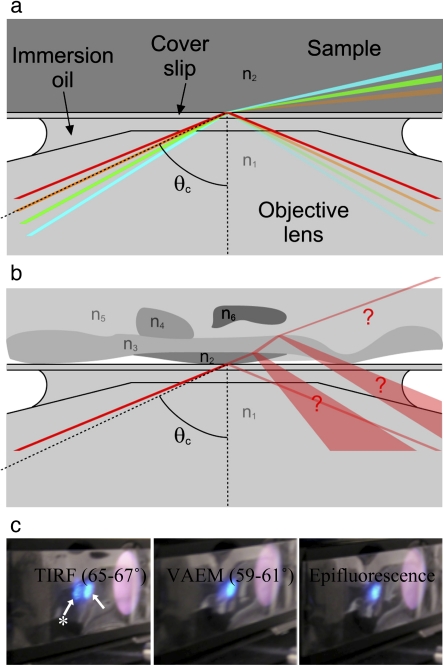
In VAEM, a narrow band of illumination (not an evanescent field) is generated which penetrates into the sample and passes through it almost parallel to the coverslip, yielding a high SBR for visualizing events at or near the plasma membrane of intact cells. In a simple sample, with materials of two refractive indices, it is easy to predict whether the angle at which the beam leaves the objective will result in TIRFM or VAEM (Fig. 4a). In a more complex sample, such as a plant cell, there may be multiple (and not quantified) refractive indices present, and the microscopy mode is uncertain (Fig. 4b). However, it is only in TIRFM that the excitation beam returns through the objective and the intensity of the reflection increases by orders of magnitude, thus providing an easy method of determining the microscopy mode by visualization of the returning excitation beam (Fig. 4c) (this visualization may not be possible in integrated commercial light-tight microscopes unless they are equipped with a Bertrand lens). The low divergence and brightness of this returned beam also show that this is not due to scattering by the sample or back-reflection from the coverslip.
Fig. 4.
Distinguishing TIRFM and VAEM. (a) The angle at which the beam is incident on the refractive index interface determines whether the microscope operates in TIRFM or VAEM mode. The mode is easy to predict when only two refractive indices n1 and n2 are present. At angles greater than the critical angle θc (red line), the beam is totally internally reflected. If the angle is less than θc (green and blue lines), the beam is refracted and the microscope operates in VAEM mode. A tiny proportion of the beam, dependent on the angle, is reflected. If the beam is incident at the critical angle (orange line), then the width of the beam causes the microscope to operate in mixed modes. (b) It is difficult to predict the microscopy mode when multiple refractive indices are present within the complex geometry of a plant cell. (c) Visualizing the difference between TIRFM, VAEM, and epifluorescence modes using an Arabidopsis root. By temporarily placing a strip of sticky tape in the excitation light path it is possible to visualize the incident (arrow) and totally internally reflected beams simultaneously (arrow with star). In TIRFM the excitation beam returns strongly through the objective and is clearly visible on the sticky tape. In VAEM and standard epifluorescence microscopy, no more than a weak reflection returns through the objective.
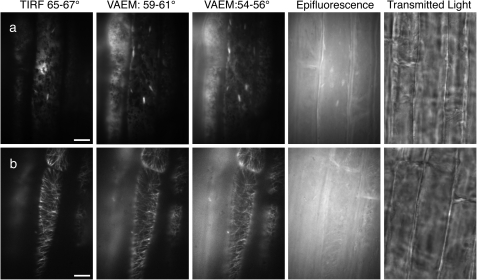
As with TIRFM, little has been published in plant research using VAEM. It has been used to image in vivo dynamics of secretory vesicles in pollen tubes (Wang et al., 2006) and to study the dynamics and function of DRPs, which are required for cytokinesis and cell expansion (Konopka and Bednarek, 2008a, b; Konopka et al., 2008). VAEM showed a significant improvement over epifluorescence illumination for actin filaments in growing root hairs (Konopka and Bednarek, 2008b), increasing the resolution of individual actin cables. It was also used with plasma membrane markers for the study of plant endocytosis (Konopka and Bednarek, 2008a, b; Konopka et al., 2008). The difference between TIRFM and VAEM when imaging the ER or microtubules in plant cells is illustrated in Fig. 5. In both cases, the angle at which the beam exited the objective was adjusted to four positions corresponding to the four angles 66, 60, 55, and 0 °. If the sample is water (refractive index=1.33) and the coverslip, immersion oil, and objective all have a refractive index of 1.51, then 60 ° is approximately the critical angle. Because the beam is not perfectly collimated, but has a spread of ∼2 °, such images are a combination of TIRFM and VAEM modes; 66 ° is pure TIRFM and 55 ° is pure VAEM, while 0 ° is standard epifluorescence. Visually, it is clear that the ER marker (Fig. 5a) has the highest SBR with TIRFM and that the ratio progressively degrades as the angle is reduced. With the microtubules (Fig. 5b), the ratio also degrades with the angle; however, excitation at 66 ° and 60 ° will not necessarily identify the same microtubules within the tissues because of the changes in the depth of field. This shows the use of the two modes, TIRFM for the most superficial structures, VAEM for deeper ones.
Fig. 5.
Comparison of fluorescence techniques in root cells. Comparison of TIRFM, VAEM, epifluorescence, and white light transmission of EGFP in root cells. The angle of incident light was measured to determine the technique used and hence compare them. Scale bars=10 μm. (a) Endoplasmic reticulum (ER) marker (mGFP5–ER). (b) Microtubule marker (MAP4–GFP).
While this discussion has assumed a simple dual refractive index system, the fact that the reflected beam could or could not be observed as per Fig. 4c suggests that it is a good approximation. In any case, the existence of other components (such as the cell wall) with different refractive indices does not prevent TIR, but merely introduces ambiguity as to the interface where it occurs (Axelrod, 2001). The refractive indices of plant cell components have been little characterized, but in the few examples given all are comparable with glass or water (Gausman et al., 1974; Margalit et al., 2010). It is therefore possible that TIR takes place either at the coverslip–water interface, or at the cell wall–cytoplasm interface, which might account for the increased depth of field that has been observed. However, single-molecule sensitivity in superficial structures will be achieved whether TIR occurs at the coverslip–water or the cell wall–cytoplasm interface.
Single-molecule fluorescence microscopy can be achieved in intact plant cells
The superior SBR of TIRF images also enables single-molecule fluorescence microscopy studies of plant proteins. Ensemble fluorescence images provide a bulk measurement from multiple proteins, masking rare and asynchronous events. At the single-molecule level, however, it is possible to track the path of individual protein molecules in space and time while simultaneously recording the stoichiometry of protein clusters.
The low concentration of fluorescing molecules required for single-molecule detection can be achieved either through using an ultra low initial concentration or by bleaching a higher concentration. Although the first strategy is more difficult biochemically with expressed proteins, it may be necessary when the diffusion rate is high enough to maintain a bulk level of fluorescing molecules in the evanescent field.
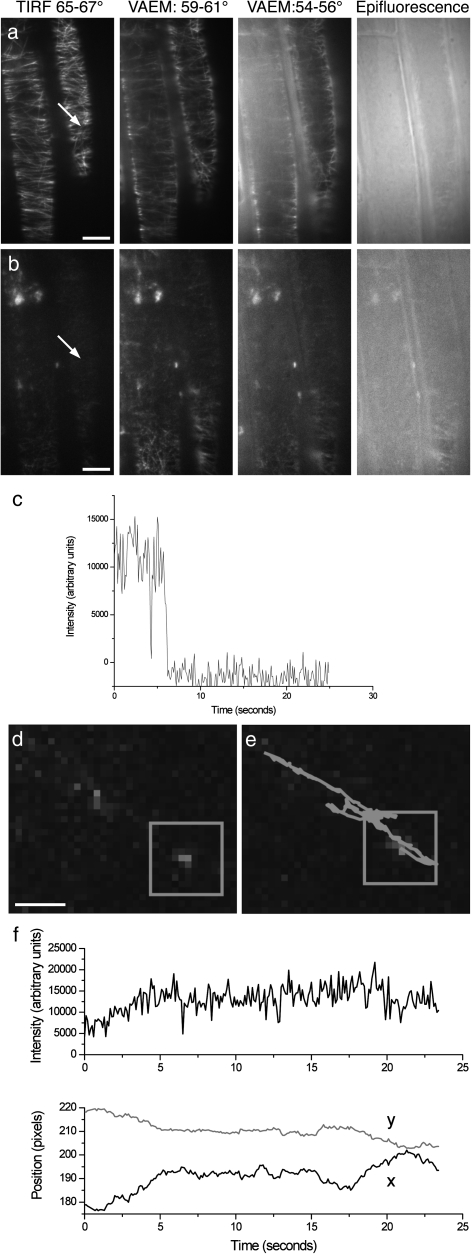
In order to demonstrate that single-molecule fluorescence microscopy is achievable in intact plant cells, attempts were made to bleach MAP4–EGFP, the microtubule marker shown in Fig. 5b. After 75 min bleaching with epi-illumination, parts of the sample showed small spots consistent with the fluorescence intensity time courses of individual EGFP molecules and displaying the characteristic blinking of a single molecule, of which an example is shown in Fig 6c; Fig. 2a shows that there is no fluorescence when there is no GFP present. Figure 6b shows the same field of view as Fig. 6a, but following bleaching. After bleaching, cytoplasmic streaming could still be seen in the cells and cell turgor was unaffected, indicating that viability had not been compromised. As before, the background intensity is lowest for TIRF illumination. Analysis of one typical molecule in this image is shown in Fig. 6d–f. By collecting a long series of images, it is possible to extract dynamic information on molecules in the sample. The intensity and co-ordinates of the molecule were determined for each image in the series, thereby revealing the trace followed by the molecule (Supplementary Movie S1 availablke at JXB online). Figure 6d shows that this particular molecule displays a back and forth motion constrained to a single line, which from observations of the pre-bleached images may coincide with a microtubule.
Fig. 6.
Single-molecule fluorescence microscopy in root cells. Single-molecule analysis of EGFP in root cells using the microtubule marker (MAP4–GFP). (a) The microtubule marker (MAP4–GFP) imaged under TIRFM, VAEM, and epifluorescence illumination. Scale bars=10 μm. (b) The same field of view as (a), after 75 min bleaching under epi-illumination. (c) Total intensity of a single MAP4–GFP marker through time, showing the typical intensity and blinking of a single GFP molecule. (d) Detail of one frame from the time series. The box indicates the single molecule. The molecule analysed here was located as indicated by the arrow in 6a and b. Scale bars=1 μm. (e) As (d), but with the path of the molecule superimposed. The molecule’s initial position was at the top left end. (f) Total intensity and position of the fluorescent spot through time.
Discussion
TIRFM has become a widespread technique for imaging structures at the cell surface, but its application to plant research has been limited due to the depth of the evanescence field and the perceived problems associated with the plant cell wall. There have only been a few reports of the use of TIRFM in fungi and plants, all of which involve analysis of fluorescent molecules at the cell surface, in protoplasts, or protein extracts. These include the study of the cytoskeleton and microtubule dynamics in Neurospora crassa hyphae and germ tubes (Uchida et al., 2008), ER dynamics in protoplasts (Goodin et al., 2007), and in vitro actin filament elongation in moss (Vidali et al., 2009) and tobacco protoplasts (Smertenko et al., 2010). TIRFM has also been used to characterize in vitro actin filament polymerization in Arabidopsis (Michelot et al., 2005; Ye et al., 2009), indicating that TIRFM provides a robust tool for the analysis of highly dynamic cellular processes such as actin dynamics.
Herein the wider application of TIRFM for plant developmental studies in subcellular locations within living plant cells has been demonstrated. It has been shown for the first time that it is possible to analyse fluorescently labelled proteins by TIRFM in organelles within the plant cell, possibly due to their localization close to the plasma membrane, and as such a wealth of opportunities for plant science researchers to increase our understanding significantly over a wide range of biological processes have been identified. The thickness of the plant cell wall can vary significantly depending upon the growth conditions and particular tissue/cell type; however, in general, the thickness of Arabidopsis root epidermal cell walls is between 100 nm and 200 nm although the inner cell walls in the root are thinner (∼70 nm) (Kramer et al., 2007). The images presented demonstrate that this does not limit the penetration of the evanescent field and therefore does not restrict the application of TIRFM to intact plant tissues.
The ability to image fluorescent proteins targeted to organelles located within the cytoplasm, for example nuclei, is probably due to the central vacuole that is present in plant cells. When the cells are hydrated, the vacuole is fully expanded and displaces organelles and the cytoplasm outwards towards the plasma membrane and cell wall. In doing so, in contrast to animal cells, it brings cell structures and organelles closer to the cell surface and consequently within the evanescent field, opening up a new range of possibilities for this technique for plant research. In addition, although the evanescent field is much weaker at greater depths, it is still sufficiently intense to excite bright fluorescence if the laser intensity and fluorophore concentrations are both high, as here.
Several authors have used VAEM to study plant biological processes associated with the cell surface, and have generated enhanced images compared with epifluorescence (Konopka and Bednarek, 2008a, b; Konopka et al., 2008); however, it has been shown that TIRF images have a superior SBR to VAEM. Indeed, due to the excellent ratio, TIRFM is compatible with single-molecule microscopy. Since TIRFM can be performed in many cell organelles and it has been demonstrated that single-molecule analysis is possible, it is likely that these techniques will provide valuable tools for a diverse range of plant imaging studies. Rare protein events, which would otherwise be masked by the average molecular behaviour, can therefore be detected. In this study, for example, it was demonstrated how individual protein molecules in living plant cells can be tracked in space and time. Furthermore, the low protein densities required for single-molecule studies are those commonly found in physiological conditions. As an example, FRET between single molecules has been used to find the distribution of intramolecular distances in membrane proteins on the nanometre scale (1–10 nm) in animal systems (Webb et al., 2008a). A particular advantage of the TIR excitation mode is that the polarization of the evanescent field is well defined (Webb et al., 2008b) and can hence be used to measure the relative orientation of fluorescent probes. The combination of FRET and fluorescence polarization at the single-molecule level can be used to report on the conformation and oligomerization state of interacting proteins and signalling networks in vivo. The fluorescence SBRs in the images shown herein are comparable with those observed in mammalian cells, suggesting that the combination of TIRFM and single-molecule microscopy could equally be applied to the analysis of protein oligomerization and conformation in living plant cells.
TIRFM provides an opportunity to study protein dynamics because of its fast acquisition rates (even faster than spinning-disk confocal microscopes), selective excitation fields, decreased background levels, and low levels of bleaching. For example, in the study of slow vacuolar channels (Perez et al., 2008), the authors appreciate the necessity of implementing high-resolution techniques such as TIRFM that would allow sufficient temporal and spatial resolution to address tonoplast dynamics. Although its application to plant imaging is still in development, TIRFM has the resolution and precision necessary to help address new and exciting biological questions relating to protein conformation, stoichiometry, and dynamics in living plant cells.
This work therefore demonstrates for the first time that TIRFM can be used to generate high contrast images, that are superior to other approaches, from fluorescently labelled proteins in intact plant cells. It is also shown that TIRF imaging is possible not only at the plasma membrane level, but also in organelles, for example the nucleus. These TIRF images show the highest SBR and it is shown that they can be used for single-molecule microscopy. This demonstration of the application of TIRFM and single-molecule analysis to plant cells opens up a new range of possibilities for plant cell imaging.
Supplementary data
Supplementary data are available at JXB online.
Movie S1. Single-molecule fluorescence microscopy in root cells. Single-molecule analysis of EGFP in root cells using the microtubule marker (MAP4–GFP), showing the intensity and location of a single fluorescent molecule through time (the video is at 2.5× acquisition speed, created with the Cinepak codec).
Acknowledgments
We would like to thank Dr S. Kurup (Rothamsted Research, Harpenden, UK) for the generous gift of fluorescently labelled marker lines used in this work. We would also like to thank the STFC for providing access to the Central Laser Facility and the TIRF microscope, and Dr D Rolfe for assistance with single-molecule data analysis. Financial support was provided by the Biotechnology and Biological Science Research Council, and the Science and Technology Facilities Council.
References
- Axelrod D. Total internal reflection fluorescence microscopy in cell biology. Traffic. 2001;2:764–774. doi: 10.1034/j.1600-0854.2001.21104.x. [DOI] [PubMed] [Google Scholar]
- Blanchoin L, Staiger CJ. Plant formins: diverse isoforms and unique molecular mechanism. Biochimica et Biophysica Acta. 2010;1803:201–206. doi: 10.1016/j.bbamcr.2008.09.015. [DOI] [PubMed] [Google Scholar]
- Chan C, Beltzner CC, Pollard TD. Cofilin dissociates Arp2/3 complex and branches from actin filaments. Current Biology. 2009;19:537–545. doi: 10.1016/j.cub.2009.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conchello JA, Lichtman JW. Optical sectioning microscopy. Nature Methods. 2005;2:920–931. doi: 10.1038/nmeth815. [DOI] [PubMed] [Google Scholar]
- Cutler SR, Ehrhardt DW, Griffitts JS, Somerville CR. Random GFP::cDNA fusions enable visualization of subcellular structures in cells of Arabidopsis at a high frequency. Proceedings of the National Academy of Sciences, USA. 2000;97:3718–3723. doi: 10.1073/pnas.97.7.3718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demuro A, Parker I. Imaging single-channel calcium microdomains. Cell Calcium. 2006;40:413–422. doi: 10.1016/j.ceca.2006.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gausman HW, Allen WA, Escobar DE. Refractive-index of plant-cell walls. Applied Optics. 1974;13:109–111. doi: 10.1364/AO.13.000109. [DOI] [PubMed] [Google Scholar]
- Goodin MM, Chakrabarty R, Banerjee R, Yelton S, Debolt S. New gateways to discovery. Plant Physiology. 2007;145:1100–1109. doi: 10.1104/pp.107.106641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haseloff J, Siemering KR, Prasher DC, Hodge S. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proceedings of the National Academy of Sciences, USA. 1997;94:2122–2127. doi: 10.1073/pnas.94.6.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Backues SK, Bednarek SY. Dynamics of Arabidopsis dynamin-related protein 1C and a clathrin light chain at the plasma membrane. The Plant Cell. 2008;20:1363–1380. doi: 10.1105/tpc.108.059428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY. Comparison of the dynamics and functional redundancy of the Arabidopsis dynamin-related isoforms DRP1A and DRP1C during plant development. Plant Physiology. 2008a;147:1590–1602. doi: 10.1104/pp.108.116863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopka CA, Bednarek SY. Variable-angle epifluorescence microscopy: a new way to look at protein dynamics in the plant cell cortex. The Plant Journal. 2008b;53:186–196. doi: 10.1111/j.1365-313X.2007.03306.x. [DOI] [PubMed] [Google Scholar]
- Kramer EM, Frazer NL, Baskin TI. Measurement of diffusion within the cell wall in living roots of Arabidopsis thaliana. Journal of Experimental Botany. 2007;58:3005–3015. doi: 10.1093/jxb/erm155. [DOI] [PubMed] [Google Scholar]
- Kurup S, Runions J, Kohler U, Laplaze L, Hodge S, Haseloff J. Marking cell lineages in living tissues. The Plant Journal. 2005;42:444–453. doi: 10.1111/j.1365-313X.2005.02386.x. [DOI] [PubMed] [Google Scholar]
- Marc J, Granger CL, Brincat J, Fisher DD, Kao Th, McCubbin AG, Cyr RJ. A GFP–MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. The Plant Cell. 10:1927–1940. doi: 10.1105/tpc.10.11.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit O, Sarafıs V, Zalevsky Z. The effect of grana and inter-grana components of chloroplasts on green light transmission: a preliminary study. Optik-International Journal for Light and Electron Optics. 2010;121:1439–1442. [Google Scholar]
- Mathur J. The illuminated plant cell. Trends in Plant Science. 2007;12:506–513. doi: 10.1016/j.tplants.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Michelot A, Derivery E, Paterski-Boujemaa R, Guerin C, Huang S, Parcy F, Staiger CJ, Blanchoin L. A novel mechanism for the formation of actin-filament bundles by a nonprocessive formin. Current Biology. 2006;16:1924–1930. doi: 10.1016/j.cub.2006.07.054. [DOI] [PubMed] [Google Scholar]
- Michelot A, Guerin C, Huang S, Ingouff M, Richard S, Rodiuc N, Staiger CJ, Blanchoin L. The formin homology 1 domain modulates the actin nucleation and bundling activity of Arabidopsis FORMIN1. The Plant Cell. 2005;17:2296–2313. doi: 10.1105/tpc.105.030908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez V, Wherrett T, Shabala S, Muniz J, Dobrovinskaya O, Pottosin I. Homeostatic control of slow vacuolar channels by luminal cations and evaluation of the channel-mediated tonoplast Ca2+ fluxes in situ. Journal of Experimental Botany. 2008;59:3845–3855. doi: 10.1093/jxb/ern225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneckenburger H. Total internal reflection fluorescence microscopy: technical innovations and novel applications. Current Opinion in Biotechnology. 2005;16:13–18. doi: 10.1016/j.copbio.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Shaw SL. Imaging the live plant cell. The Plant Journal. 2006;45:573–598. doi: 10.1111/j.1365-313X.2006.02653.x. [DOI] [PubMed] [Google Scholar]
- Smertenko AP, Deeks MJ, Hussey PJ. Strategies of actin reorganisation in plant cells. Journal of Cell Science. 2010;123:3019–3028. doi: 10.1242/jcs.071126. [DOI] [PubMed] [Google Scholar]
- Staiger CJ, Sheahan MB, Khurana P, Wang X, McCurdy DW, Blanchoin L. Actin filament dynamics are dominated by rapid growth and severing activity in the Arabidopsis cortical array. Journal of Cell Biology. 2009;184:269–280. doi: 10.1083/jcb.200806185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nature Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- Toomre D, Manstein DJ. Lighting up the cell surface with evanescent wave microscopy. Trends in Cell Biology. 2001;11:298–303. doi: 10.1016/s0962-8924(01)02027-x. [DOI] [PubMed] [Google Scholar]
- Uchida M, Mourino-Perez RR, Freitag M, Bartnicki-Garcia S, Roberson RW. Microtubule dynamics and the role of molecular motors in Neurospora crassa. Fungal Genetics and Biology. 2008;45:683–692. doi: 10.1016/j.fgb.2007.10.013. [DOI] [PubMed] [Google Scholar]
- Vidali L, van Gisbergen PA, Guerin C, Franco P, Li M, Burkart GM, Augustine RC, Blanchoin L, Bezanilla M. Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proceedings of the National Academy of Sciences, USA. 2009;106:13341–13346. doi: 10.1073/pnas.0901170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Teng Y, Wang Q, Li X, Sheng X, Zheng M, Samaj J, Baluska F, Lin J. Imaging of dynamic secretory vesicles in living pollen tubes of Picea meyeri using evanescent wave microscopy. Plant Physiology. 2006;141:1591–1603. doi: 10.1104/pp.106.080168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb SE, Needham SR, Roberts SK, Martin-Fernandez ML. Multidimensional single-molecule imaging in live cells using total-internal-reflection fluorescence microscopy. Optics Letters. 2006;31:2157–2159. doi: 10.1364/ol.31.002157. [DOI] [PubMed] [Google Scholar]
- Webb SED, Roberts SK, Needham SR, Tynan CJ, Rolfe DJ, Winn MD, Clarke DT, Barraclough R, Martin-Fernandez ML. Single molecule imaging and FLIM show different structures for high and low-affinity EGFRs in A431 cells. Biophysics Journal. 2008a;94:1–17. doi: 10.1529/biophysj.107.112623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb SE, Rolfe DJ, Needham SR, Roberts SK, Clarke DT, McLachlan CI, Hobson MP, Martin-Fernandez ML. Simultaneous widefield single molecule orientation and FRET microscopy in cells. Optics Express. 2008b;16:20258–20265. doi: 10.1364/oe.16.020258. [DOI] [PubMed] [Google Scholar]
- Ye J, Zheng Y, Yan A, Chen N, Wang Z, Huang S, Yang Z. Arabidopsis formin3 directs the formation of actin cables and polarized growth in pollen tubes. The Plant Cell. 2009;21:3868–3884. doi: 10.1105/tpc.109.068700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Qu X, Bao C, et al. Arabidopsis VILLIN5, an actin filament bundling and severing protein, is necessary for normal pollen tube growth. The Plant Cell. 2010;22:2749–2767. doi: 10.1105/tpc.110.076257. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.