Abstract
Cytokinins are hormones that are involved in various processes of plant growth and development. The model of cytokinin signalling starts with hormone perception through membrane-localized histidine kinase receptors. Although the biochemical properties and functions of these receptors have been extensively studied, there is no solid proof of their subcellular localization. Here, cell biological and biochemical evidence for the localization of functional fluorophor-tagged fusions of Arabidopsis histidine kinase 3 (AHK3) and 4 (AHK4), members of the cytokinin receptor family, in the endoplasmic reticulum (ER) is provided. Furthermore, membrane-bound AHK3 interacts with AHK4 in vivo. The ER localization and putative function of cytokinin receptors from the ER have major impacts on the concept of cytokinin perception and signalling, and hormonal cross-talk in plants.
Keywords: AHK3, cytokinin perception, endoplasmic reticulum, FIDSAM
Introduction
Cytokinins, a class of adenine-derived plant hormones, have been implicated in almost every aspect of plant growth and development, including root and shoot growth, vasculature differentiation, photomorphogenesis, senescence, fertility, and seed development (Muller and Sheen, 2007a; Werner and Schmulling, 2009) as well as in responses to cold and osmotic stress (Tran et al., 2007; Jeon et al., 2010). It is well established that cytokinin perception and signalling is mediated by a multistep two-component circuitry. In Arabidopsis thaliana three transmembrane histidine kinases, namely AHK2, AHK3, and AHK4, serve as cytokinin receptors (Inoue et al., 2001; Suzuki et al., 2001; Ueguchi et al., 2001). Cytokinin binding to their CHASE domain is proposed to initiate autophosphorylation of the receptors at a conserved histidine residue in the transmitter domain (Pas et al., 2004; Muller and Sheen, 2007b). The phosphoryl group is then transferred to a conserved aspartate residue in the receptor's receiver domain. Histidine phosphotransfer proteins (AHPs) finally transmit the signal to response regulators (ARRs), which then regulate the cellular responses (Muller and Sheen, 2007b; Werner and Schmulling, 2009; Kieber and Schaller, 2010).
Although the cytokinin receptors have been extensively studied regarding their specific functions, biochemical properties, and expression patterns (Higuchi et al., 2004; Nishimura et al., 2004; Riefler et al., 2006; Romanov et al., 2006), their subcellular localization and molecular function are still not fully determined. It has been assumed that they reside in the plasma membrane, and a green fluorescent protein (GFP) fusion of AHK3 appears to localize to the plasma membrane of protoplasts (Kim et al., 2006). However, further attempts to ascertain this localization led to the observation that they show a more diverse localization pattern (Dortay et al., 2008).
In this report, cell biological and biochemical evidence for the localization of functional fluorescent protein fusions of AHK3—a representative of the cytokinin receptor family—in the endoplasmic reticulum (ER) is provided. This localization is detected not only in transiently transformed tobacco (Nicotiana benthamiana) and Arabidopsis cells but also in stably transgenic Arabidopsis plants. The present observation entails a reconsideration of the current model of cytokinin signal perception and, as other hormone receptors are also located in the ER, opens new perspectives for hormonal cross-talk at this cellular compartment in plant cells.
Materials and methods
Construction of cDNA fusions
To generate the fusion proteins, attB sites were added via PCR-mediated ligation to the coding regions of AHK1 (AT2G17820), AHK3 (AT1G27320), AHK4 (AT2G01830), ERS1 (AT2G40940), and NHL3 (AT5G06320) with or without a STOP codon and recombined into pDONR™201 according to the manufacturer's protocol (Invitrogen). The cDNA was then transferred via LR reaction (Invitrogen) into the destination vectors pH7WGF2, pH7FWG2, or pB7WGR2 (Karimi et al., 2002) and pABindmCherry (Bleckmann et al., 2010).
For constructs under the control of the ubiquitin 10 (UBQ10) promoter, a gateway cassette (reading frame A) was inserted into the vectors pUGT1kan+ and pUGT2kan+ (Karin Schumacher, unpublished) at the SmaI site in the multiple cloning site. The AHK3 coding sequence was then inserted in the destination vectors by LR reaction.
For the fusion construct with internal GFP (AHK3intGFP), linker sequences (coding for GGGGS/T) were added via PCR to the coding sequence of GFP using the primers GFP-BcuI-S and GFP-BcuI-A (Supplementary Table S1 available at JXB online). For ligation into the AHK3 entry clone an appropriate restriction site was produced via site-directed mutagenesis in the AHK3 coding sequence at position 123 (corresponding to amino acid 41) where the linker–GFP–linker sequence was introduced. The AHK3intGFP cDNA was then recombined into pMDC32 (Curtis and Grossniklaus, 2003) by LR reaction. For mating-based split-ubiquitin system (mbSUS) assay, the AHK4 cDNA was transferred by LR reaction to pMetYC-Dest and the AHK3 and ERS1 cDNAs to pXNubA22 (Grefen et al., 2009).
Transient gene expression in Nicotiana benthamiana leaves and Arabidopsis seedlings
Transient transformations of N. benthamiana leaves with the Agrobacterium tumefaciens strain GV3101 pMP90 containing the expression constructs were carried out as described in Schutze et al. (2009). The transformed leaves were assayed for fluorescence by confocal laser scanning microscopy (CLSM) 2–3 d post-infiltration. The transgene expression from the estradiol-inducible promoter was induced 2–3 d after infiltration with 20 μM β-estradiol supplemented with 0.1% Tween-20. For transient expression in Arabidopsis seedlings, the Agrobacterium strains containing the fusion constructs and the marker constructs used were grown as described (Marion et al., 2008), prior to infiltration diluted in 5% sucrose, 200 μM acetosyringone to an OD600 of 2.0, and mixed 1:1. 3–4 d old Arabidopsis efr1 seedlings (Zipfel et al., 2006) were transformed via vacuum infiltration as described by Marion et al. (2008) and the seedlings were examined for fluorescence 3 d post-infiltration.
CLSM and fluorescence intensity decay shape analysis microscopy (FIDSAM)
CLSM and FIDSAM as well as the used spectromicroscopic systems and measurement protocols have been described previously (Elgass et al., 2009; Schleifenbaum et al., 2010).
Construction of transgenic Arabidopsis lines
The transgenes were transformed into Arabidopsis ahk2-2ahk3-3 plants (ahk2ahk3, Higuchi et al., 2004) via the floral dip method and selected by phenotype (complemented dwarf phenotype of the ahk2ahk3 mutant background). Twenty independent ahk2-2ahk3-3 lines complemented by AHK3-GFP and 10 independent lines complemented by GFP-AHK3 were isolated. After verification of the transgene integration, the lines were analysed for the GFP fluorescence signal using CLSM and FIDSAM. The line with the most intense GFP signal was used for imaging and endogylcosidase H (EndoH) assays.
EndoH assay
The EndoH assay was performed according to the manufacturer's manual (New England BioLabs) by using crude protein extracts of transiently transformed tobacco or Arabidopsis leaves. The proteins were analysed by SDS–PAGE and western blot using a GFP antibody.
Root growth and yeast mbSUS assays
For the root elongation assay, seedlings were grown vertically on 0.5× MS plates supplemented with different concentrations of kinetin. The root length was measured 6 d post-germination. The yeast mbSUS assays using AHK4-Cub-PLV and the NubA fusions of AHK3 and ERS1 as constructs were carried out as described previously (Grefen et al., 2009; Caesar et al., 2011).
Results and Discussion
FP fusions of AHK3 localize to the ER
In order to examine the subcellular localization of AHK3, C- and N-terminal GFP fusions of the receptor were transiently expressed in leaf epidermal cells of N. benthamiana and in cotyledon cells of Arabidopsis seedlings under the control of either the 35S Cauliflower mosaic virus (35S) or the Arabidopsis UBQ10 promoter. Both AHK3–GFP and GFP–AHK3 showed an ER-like localization pattern in tobacco and Arabidopsis independent of the promoter used (Fig. 1A–D, Supplementary Fig. S1 at JXB online). The identity of the endomembrane system as ER was verified by the co-localization of the GFP fusions of AHK3 with the mCherry-tagged ER marker ER-rk CD3-959 (Nelson et al., 2007; Fig. 1A–D, Supplementary Fig. S1B, C). In addition, there was a co-localization of the GFP fusion proteins of AHK3 with the ER-localized red fluorescent protein (RFP) fusion of the ethylene receptor ERS1 (Grefen et al., 2008; Fig. 2). In contrast, no co-localization of these AHK3 fusion proteins was found with the mCherry-tagged Golgi marker G-rk CD2-967 (Nelson et al., 2007; Supplementary Fig. S2) and the RFP fusion of the plasma membrane protein NHL3 (Varet et al., 2003; Fig. 3). Furthermore, the GFP fusion of the plasma membrane-bound AHK1, which is a positive regulator of drought and salt stress response and functions as an osmosensor in yeast (Urao et al., 1999; Tran et al., 2007; Wohlbach et al., 2008), also did not co-localize with an mCherry fusion of AHK3 (Fig. 4). The yellow colour, partially visible in the merged images of Supplementary Fig. S2D, and Figs 3A, C, 4, results from the very strong GFP signal in this area and the physically restricted resolution of light microscopy below Abbe's diffraction limit (250 nm). However, the magnified images of these yellow domains showed an incomplete overlap of the GFP and RFP or mCherry fluorescence (Fig. 4; Supplementary Fig. S3), indicating that the fusion proteins localize to different membrane compartments.
Fig. 1.
The Arabidopsis cytokinin receptor AHK3 localizes to the ER in transiently transformed tobacco leaf cells and Arabidopsis seedlings. (A–D) and (F) Confocal images of transiently transformed tobacco epidermal leaf cells co-expressing the indicated AHK3 fusion protein under the control of the 35S promoter or the UBQ10 promoter with the ER marker ER-rk CD3-959. (E) Confocal images of transiently transformed tobacco epidermal leaf cells expressing an AHK3–mCherry fusion protein under the control of the estradiol-inducible promoter (XVE). Images were recorded 2 h (I), 4 h (II), and 24 h (III) after application of 20 μM β-estradiol. Images (I) and (II) were recorded at the highest sensitivity settings of the microscope at which the mCherry fluorescence was just detectable. (G) Confocal images of transiently transformed Arabidopsis cotyledon cells co-expressing the indicated AHK3 fusion protein with the ER marker ER-rk CD3-959. Bars represent 10 μm.
Fig. 2.
AHK3–GFP and GFP–AHK3 fusion proteins co-localize with ERS1–RFP. (A–D) Confocal images of transiently transformed tobacco epidermal leaf cells co-expressing the indicated AHK3 fusion protein under the control of the 35S or the UBQ10 promoter and an RFP fusion of the ethylene receptor ERS1 (ERS1–RFP). Bars represent 10 μm.
Fig. 3.
AHK3–GFP and GFP–AHK3 fusion proteins do not co-localize with the plasma membrane-localized fusion protein NHL3–RFP. (A–E) Confocal images of transiently transformed tobacco epidermal leaf cells co-expressing the indicated AHK3 fusion protein under the control of the 35S promoter or the UBQ10 promoter and the plasma membrane-localized fusion protein NHL3–RFP. (F) Confocal images of transiently transformed Arabidopsis cotyledon cells co-expressing the indicated AHK3 fusion protein and the plasmalemma-localized fusion protein NHL3–RFP. Bars represent 10 μm.
Fig. 4.
AHK3–mCherry does not co-localize with a GFP fusion of the plasma membrane-bound Arabidopsis histidine kinase 1 (AHK1–GFP). Confocal images of different magnification of transiently transformed tobacco epidermal leaf cells co-expressing AHK3–mCherry under the control of the estradiol-inducible promoter (XVE) and AHK1–GFP under control of the 35S promoter. The image series of the second row represents a magnified detail of the images of the first row. The image series of the third row derives from an independent cell. Images were recorded 4 h after application of 20 μM β-estradiol. Bars represent 10 μm.
It has been reported that strong expression of fusion proteins under the control of promoters such as 35S or UBQ10 might lead to mislocalization artefacts (Bleckmann et al., 2010). A C-terminal mCherry fusion of AHK3 (AHK3–mCherry) expressed from an estradiol-controlled promoter system was therefore used to trigger the expression level of the histidine kinase (Bleckmann et al., 2010). After transformation of the construct into N. benthamiana leaves, the expression was induced by brushing the leaves with β-estradiol. AHK3–mCherry fluorescence was detectable 2 h after β-estradiol application at the earliest (Fig. 1E). Already at this early time point the AHK3–mCherry fluorescence displayed a net-like ER localization pattern, which did not change within the next 22 h (Fig. 1E). The localization of β-estradiol-induced AHK3–mCherry was never observed in the plasma membrane.
The amino acid sequence of AHK3 contains potential signals for the secretory pathway and ER export, respectively [Fig. 6A; iPsort Prediction, http://ipsort.hgc.jp/ (Bendtsen et al., 2004); YLoc Prediction, www.multiloc.org/YLoc (Hanton et al., 2005; Langhans et al., 2008; Briesemeister et al., 2010)]. To exclude a possible mislocalization of the fusion proteins due to masking of potential sorting signals, a construct was generated where GFP is inserted between the first and the second predicted transmembrane domain of AHK3 (AHK3intGFP; see Fig. 6A for the details of the insertion site). AHK3intGFP showed the identical ER localization pattern to AHK3–GFP and GFP–AHK3 when transiently expressed in tobacco leaves as well as in Arabidopsis seedlings, and co-localized with the mCherry-tagged ER marker (Fig. 1F, G) but not with the plasma membrane marker NHL3–RFP (Fig. 3E, F).
Fig. 6.
The GFP fusion proteins of AHK3 show EndoH sensitivity and are glycosylated in vivo. (A) Amino acid sequence of AHK3. The transmembrane domains are shown in blue, the histidine kinase domain in red, and the pseudo receiver domain and the receiver domain in purple. N-X-S/T sequons are framed. The predicted signal peptide is italicized, and the putative ER export signals are underlined. The green triangle marks the site where GFP was inserted into AHK3intGFP. (B) Representations of AHK3, AHK1, and the ethylene receptor ERS1. Putative glycosylation sites are indicated with asterisks. (C) The electrophoretic mobility of AHK3 is endoglycosidase H (EndoH) sensitive. Equal volumes of protein extracts from transiently transformed tobacco leaves expressing the indicated fusion proteins were treated with EndoH (+) or mock treated (–), followed by western blot analysis and immunodetection using an anti-GFP antibody. The fusion proteins are indicated by arrowheads.
Signal-induced translocation of receptors to the plasma membrane is reported for animal systems (Shuster et al., 1999; Song et al., 2004). Therefore, assays were carried out to determine whether the application of kinetin, a synthetic cytokinin, has an influence on the subcellular localization of the N- and C-terminal GFP fusions of AHK3. However, 4 h of kinetin treatment did not alter the ER localization of AHK3–GFP and GFP–AHK3 (Supplementary Fig. S4 at JXB online).
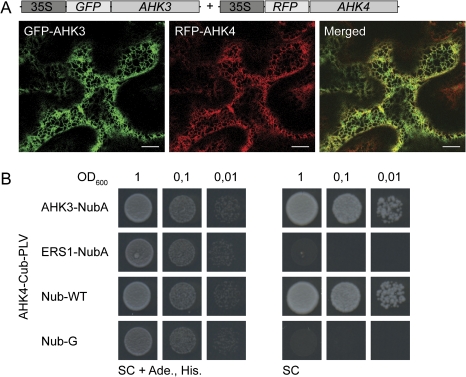
Furthermore, it was tested whether the intracellular location of AHK3 changed when it was co-expressed with an RFP fusion of its sister receptor, AHK4. As shown in Fig. 5A, both fusion proteins co-localized in the ER. To determine whether AHK3 is able to interact with AHK4 in the membrane, in vivo interaction studies were performed using the yeast mbSUS (Grefen et al., 2009). The mbSUS experiments revealed that AHK3 not only forms homo-oligomers (data not shown) but also interacts with AHK4 in vivo (Fig. 5B). No interaction was observed with ERS1 (Fig. 5B), which also localized to the ER (Fig. 2; Grefen et al., 2008). These data suggest that the cytokinin receptors are able specifically to homo- and heterodimerize in the ER and that their interaction has no influence on their subcellular localization.
Fig. 5.
The cytokinin receptor AHK4 co-localizes with AHK3 in the ER and interacts with AHK3 in vivo (yeast). (A) Confocal images of transiently transformed tobacco epidermal leaf cells co-expressing RFP–AHK4 and GFP–AHK3 under control of the 35S promoter. Bars represent 10 μm. (B) Yeast mbSUS protein–protein interaction analysis. The AHK4-Cub-PLV construct was transformed in yeast strain THY.AP4 (MATa), and the Nub constructs of AHK3 and ERS1 were transformed in yeast strain THY.AP5 (MATα). After mating, activation of the reporter gene was determined by growth of the transformants in a dilution series (OD600nm from 1 to 0.01) on SC medium (SC). The presence of the plasmid was assayed by growth on SC medium supplemented with adenine and histidine (SC+Ade., His.). Co-transformations of the AHK4–Cub-PLV fusion with NubG served as negative control and co-transformation with NubWT served as positive control.
In conclusion, all tested fluorescent protein fusions of AHK3—no matter whether GFP is tagged to the C-terminus, N-terminus, or internally—show an ER localization. A mislocalization of the fusion proteins due to overexpression or masking of potential sorting signals is unlikely. Furthermore, neither cytokinin application nor the co-expression of AHK3 and 4 or their potential in planta interaction are capable of altering the ER localization of AHK3.
The AHK3 protein is EndoH sensitive
To substantiate the ER localization of the AHK3 fusion proteins, a biochemical survey was conducted applying an EndoH assay. EndoH is a glycosidase which cleaves asparagine-linked oligomannose and hybrid, but not complex oligosaccharides from glycoproteins (Maley et al., 1989). EndoH, therefore, enables, by electrophoretic mobility shift, the differentiation of ER-localized glycoproteins from glycoproteins in the plasma membrane, whose asparagine-linked glycans are further modified in the secretory pathway and are no longer substrates for the glycolytic enzyme (Hong et al., 2008). In the AHK3 amino acid sequence, five potential N-X-S/T glycosylation sites were identified; one N-terminally of and three within the CHASE domain, and one in the receiver domain close to the C-terminus (Fig. 6A, B). Therefore, a mobility shift of EndoH-treated AHK3 would be expected on condition that AHK3 is located in the ER. As controls, AHK1–GFP, which is a plasma membrane-localized protein (Fig. 4) and has nine potential N-X-S/T glycosylation sites (Fig. 6B), and ERS1–GFP, which is, like ERS1–RFP, bound to the ER (Grefen et al., 2008), were used. The ERS1 single N-X-S/T site is predicted not to be glycosylated due to its C-terminal location (Fig. 6B; Gavel and von Heijne, 1990). Total crude protein extracts of tobacco leaves expressing the GFP fusion proteins were exposed to EndoH or mock treated. After SDS–PAGE and western blot using a GFP-specific antibody, the fusion proteins were analysed for changes in their electrophoretic mobility. There was no mobility shift and, thus, no EndoH sensitivity of plasma membrane-bound AHK1–GFP or of ER-bound ERS1–GFP detected, indicating that AHK1–GFP is not retained in the ER and ERS1 is not glycosylated in tobacco cells (Fig. 6C). The unaltered pattern of AHK1–GFP in particular also proves that the reaction mixture conditions per se have no influence on the electrophoretic mobility of the fusion proteins. In contrast, the EndoH-treated AHK3 fusion proteins showed a significant mobility shift compared with the non-treated control (Fig. 6C). Most importantly, there was no high mobility band in the non-treated AHK3 preparations.
The results of the EndoH assays thus support the cell biological observations that the GFP fusions of AHK3 localize to the ER. Furthermore, there appears to be no subfraction of AHK3 in the plasmalemma because the entire population of the cytokinin receptor carries EndoH-sensitive mannose structures typical for ER-resident glycoproteins.
AHK3–GFP and GFP–AHK3 rescue the cytokinin-insensitive phenotype of the ahk2ahk3 receptor mutant
To determine the functionality of the GFP fusions of AHK3, their capability to complement the dwarf and cytokinin-insensitive root growth phenotype of the ahk2-2ahk3-3 (ahk2ahk3) mutant was analysed. The ahk2-2 or the ahk3-3 single mutants were not used for the complementation analysis as they show the wild-type phenotype (Higuchi et al., 2004). Those ahk2ahk3 plants were selected whose dwarf and cytokinin-insensitive root growth phenotypes were complemented by UBQ10-driven expression of AHK3-GFP or GFP-AHK3 demonstrating that both fusion proteins are functional receptors (Fig. 7A–C). Next the AHK3-GFP-complemented transgenic line was studied for the accumulation and subcellular localization of the fusion protein using standard CLSM. Weak fluorescence signals were detected in epidermal and stomatal cotyledon cells. The fluorescence signal appeared in a net-like and discontinuous pattern as well as in the perinuclear space (Fig. 7D). This observation suggests that AHK3–GFP is predominantly localized in the ER. To be sure that background autofluorescence was not recorded, FIDSAM was applied. FIDSAM enhances the contrast of fluorescence images due to efficient background fluorescence repression (Schleifenbaum et al., 2010). In the FIDSAM images, AHK3–GFP again became visible as a discontinuous fluorescence pattern with a net-like structure that is typical for ER-localized fusion proteins (Supplementary Fig. S5 at JXB online) but atypical for plasma membrane proteins (Grefen et al., 2008). These data demonstrate that the observed fluorescence actually derives from the GFP of AHK3–GFP. The low accumulation of AHK3–GFP fusion protein was surprising as the AHK3 fusion constructs were under the control of the constitutive UBQ10 promoter, which usually provides for a high level of accumulation of the corresponding fusion protein. This suggests that the transgenic plants must keep the AHK3 protein amount at a level which is similarly low as that of wild-type Arabidopsis.
Fig. 7.
The GFP fusions of AHK3 are able to complement the mutant phenotype of the ahk2ahk3 receptor mutant and locate to the ER in the complemented lines. (A) Adult plants of the wild type (Col 0), the ahk2ahk3 mutant, and plants expressing AHK3–GFP or GFP–AHK3 from the UBQ10 promoter in the ahk2ahk3 background (AHK3–GFP and GFP–AHK3). (B) Genotypic analysis of AHK3–GFP and GFP–AHK3 lines by PCR using the indicated primer pairs. (C) Inhibition of root elongation by increasing amounts of exogenously applied cytokinin (kinetin). Seedlings of the wild type (Col 0, white columns), the ahk2ahk3 mutant (black columns), and the complemented, heterozygous AHK3–GFP and GFP–AHK3 lines (dark and light grey columns) were grown on 0.5× MS agar plates supplemented with the indicated concentrations of kinetin. Mean values (n=18; standard errors) relative to the root length of non-treated controls and P-values for statistical significance are given. (D) AHK3–GFP fluorescence is detectable in the ER of the AHK3-GFP-complemented Arabidopsis line. CLSM images of epidermis and stomatal cells from cotyledons of etiolated seedlings are shown. N indicates the nucleus. Bars represent 10 μm. (E) EndoH sensitivity of the AHK3–GFP fusion protein in the AHK3-GFP-complemented line. Equal amounts of protein extract from seedlings of the indicated lines were treated or not with endoglycosidase H (EndoH), followed by western blot analysis and immunodetection with an anti-GFP antibody. The fusion proteins are indicated by arrowheads.
To substantiate the ER localization, EndoH assays were performed, using extracts from the AHK3–GFP line, the ERS1–GFP line, and the ahk2ahk3 mutant. Again, an EndoH-caused mobility shift of the AHK3 fusion protein was observed, but not a clear shift of ERS1–GFP (Fig. 7E). Again, there was no high mobility band in the mock-treated AHK3 preparations (Fig. 7E) and no free GFP (data not shown). Thus, the observed complementation of the ahk2ahk3 mutant phenotype was not due to post-translational cleavage of the GFP and release of non-tagged AHK3 or a translocation of an AHK3 subpopulation to the plasma membrane.
Summarizing, the results of the EndoH assay and the CLSM/FIDSAM analysis suggest that AHK3–GFP localizes to the ER not only in transiently transformed tobacco and Arabidopsis cells but also in transgenic plants. As AHK3–GFP complements the cytokinin-insensitive phenotype of the ahk2ahk3 mutant and as there is no indication that a subpopulation of AHK3 targets to the plasma membrane, the receptor appears to function from the ER. However, the possibility that minuscule amounts of AHK3 are transferred to the plasma membrane, which are detectable neither by CLSM nor by western blot after EndoH treatment, cannot be entirely excluded.
Conclusion
The ER localization of AHK3 (and AHK4) has major consequences for the concept of cytokinin perception and signalling in plants. The present data indicate that the cytokinin-binding CHASE domain is not oriented to the apoplast, as previously assumed, but exposed to the ER lumen, whereas the C-terminal kinase domain, that, upon activation, transfers the phosphoryl residues to the nucleocytoplasmic histidine phosphotransfer proteins, is exposed to the cytoplasm. This topology of the receptor is in agreement with the observation that the binding of the cytokinin zeatin to AHK3 and AHK4 has a pH optimum of ∼6.5 (Romanov et al., 2006)—a pH found in the ER lumen (Kim et al., 1998). At pH values of ∼5.5—as reported for the apoplast (Li et al., 2005)—the binding of zeatin to AHK3 is almost abolished (Romanov et al., 2006). Thus, when the cytokinin receptors are located in the ER and expose the CHASE domain to the lumen, they bind their ligand with much higher affinity. Furthermore, although the subcellular distribution of active cytokinins and their ability to permeate the cell membrane (Laloue et al., 1981) have not yet been examined in detail, many enzymes involved in cytokinin biosynthesis, such as the isopentenyl transferases (IPTs) and lonely guys (LOGs), and in catabolism, such as cytokinin oxidases (CKXs), are not only found in plastids (IPTs; Kasahara et al., 2004) but also in the cytoplasm and nucleus (LOGs; Kuroha et al., 2009) and other organelles such as the vacuole and the ER (CKXs; Werner et al., 2003). These observations suggest intracellular mechanisms which distribute the hormone and its derivates within the cell. In addition, several plasma membrane-bound carriers have been identified which are able to transport cytokinin into the cell (Burkle et al., 2003; Wormit et al., 2004; Hirose et al., 2005; Cedzich et al., 2008) where it could be distributed further. So apparently the current model of cytokinin signal perception at the plasma membrane needs to be reconsidered.
Recent analyses showed that other hormone perception, signalling, and distribution compounds as well as hormone metabolic enzymes are also found at the ER (Friml and Jones, 2010). For instance, ethylene perception by the five ethylene receptors and their interaction with central downstream signalling elements such as constitutive triple response 1 (CTR1) and ethylene insensitive 2 (EIN2) occur at the ER (Chen et al., 2002; Gao et al., 2003; Grefen et al., 2008; Bisson et al., 2009; Bisson and Groth, 2010). Furthermore, the auxin-binding protein 1 (ABP1) and the PIN-formed 5 (PIN5) auxin efflux carrier localize to the ER (Tian et al., 1995; Chen et al., 2006; Mravec et al., 2009), where they are discussed to be involved not only in auxin homeostasis and metabolism but also in auxin signalling (Friml and Jones, 2010). Hormonal cross-talk decisively contributes to the final physiological and developmental output of hormone action (Benkova and Hejatko, 2009). It is, therefore, intriguing to speculate that the ER might represent the intracellular site for hormonal cross-talk.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. AHK3GFP fusion proteins localize to the ER in transiently transformed Arabidopsis cotyledon cells.
Figure S2. GFP fusion proteins of AHK3 do not co-localize with the Golgi marker G-rk CD3-967.
Figure S3. AHK3–GFP fusion proteins do not co-localize with the plasma membrane-localized fusion protein NHL3–RFP.
Figure S4. The ER localization of AHK3–GFP and GFP–AHK3 does not change upon cytokinin treatment.
Figure S5. AHK3–GFP fluorescence is detectable in the ER of the AHK3–GFP-expressing Arabidopsis line.
Table S1. Oligonucleotides used in the study.
Acknowledgments
We thank G. Felix for the efr1 seeds, A. Kakimoto for the ahk2-2ahk3-3 seeds, and K. Schumacher for vectors. We are also grateful to F. de-Courcy for proofreading the manuscript. This work was supported by a DFG grant to KH (HA 2146/10-1) and PhD fellowships of the University of Tübingen to KC and KE.
References
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. Journal of Molecular Biology. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Benkova E, Hejatko J. Hormone interactions at the root apical meristem. Plant Molecular Biology. 2009;69:383–396. doi: 10.1007/s11103-008-9393-6. [DOI] [PubMed] [Google Scholar]
- Bisson MM, Bleckmann A, Allekotte S, Groth G. EIN2, the central regulator of ethylene signalling, is localized at the ER membrane where it interacts with the ethylene receptor ETR1. Biochemical Journal. 2009;424:1–6. doi: 10.1042/BJ20091102. [DOI] [PubMed] [Google Scholar]
- Bisson MM, Groth G. New insight in ethylene signaling: autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Molecular Plant. 2010;3:882–889. doi: 10.1093/mp/ssq036. [DOI] [PubMed] [Google Scholar]
- Bleckmann A, Weidtkamp-Peters S, Seidel CA, Simon R. Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiology. 2010;152:166–176. doi: 10.1104/pp.109.149930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briesemeister S, Rahnenfuhrer J, Kohlbacher O. Going from where to why—interpretable prediction of protein subcellular localization. Bioinformatics. 2010;26:1232–1238. doi: 10.1093/bioinformatics/btq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkle L, Cedzich A, Dopke C, Stransky H, Okumoto S, Gillissen B, Kuhn C, Frommer WB. Transport of cytokinins mediated by purine transporters of the PUP family expressed in phloem, hydathodes, and pollen of Arabidopsis. The Plant Journal. 2003;34:13–26. doi: 10.1046/j.1365-313x.2003.01700.x. [DOI] [PubMed] [Google Scholar]
- Caesar K, Elgass K, Chen Z, Huppenberger P, Witthöft J, Schleifenbaum F, Blatt MR, Oecking C, Harter K. A fast brassinolide-regulated response pathway in the plasma membrane of Arabidopsis thaliana. The Plant Journal. 2011;66:528–540. doi: 10.1111/j.1365-313X.2011.04510.x. [DOI] [PubMed] [Google Scholar]
- Cedzich A, Stransky H, Schulz B, Frommer WB. Characterization of cytokinin and adenine transport in Arabidopsis cell cultures. Plant Physiology. 2008;148:1857–1867. doi: 10.1104/pp.108.128454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JG, Wang SC, Lazarus CM, Napier RM, Jones AM. Altered expression of auxin-binding protein 1 affects cell expansion and auxin pool size in tobacco cells. Journal of Plant Growth Regulation. 2006;25:69–78. [Google Scholar]
- Chen YF, Randlett MD, Findell JL, Schaller GE. Localization of the ethylene receptor ETR1 to the endoplasmic reticulum of Arabidopsis. Journal of Biological Chemistry. 2002;277:19861–19866. doi: 10.1074/jbc.M201286200. [DOI] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiology. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dortay H, Gruhn N, Pfeifer A, Schwerdtner M, Schmulling T, Heyl A. Toward an interaction map of the two-component signaling pathway of Arabidopsis thaliana. Journal of Proteomic Research. 2008;7:3649–3660. doi: 10.1021/pr0703831. [DOI] [PubMed] [Google Scholar]
- Elgass K, Caesar K, Schleifenbaum F, Stierhof Y-D, Meixner AJ, Harter K. Novel application of fluorescence lifetime and fluorescence microscopy enables quantitative access to subcellular dynamics in plant cells. PLoS One. 2009;4:e5716. doi: 10.1371/journal.pone.0005716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Jones AR. Endoplasmic reticulum: the rising compartment in auxin biology. Plant Physiology. 2010;154:458–462. doi: 10.1104/pp.110.161380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chen YF, Randlett MD, Zhao XC, Findell JL, Kieber JJ, Schaller GE. Localization of the Raf-like kinase CTR1 to the endoplasmic reticulum of Arabidopsis through participation in ethylene receptor signaling complexes. Journal of Biological Chemistry. 2003;278:34725–34732. doi: 10.1074/jbc.M305548200. [DOI] [PubMed] [Google Scholar]
- Gavel Y, von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Engineering. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grefen C, Obrdlik P, Harter K. The determination of protein–protein interactions by the mating-based split-ubiquitin system (mbSUS) Methods in Molecular Biology. 2009;479:217–233. doi: 10.1007/978-1-59745-289-2_14. [DOI] [PubMed] [Google Scholar]
- Grefen C, Stadele K, Ruzicka K, Obrdlik P, Harter K, Horak J. Subcellular localization and in vivo interactions of the Arabidopsis thaliana ethylene receptor family members. Molecular Plant. 2008;1:308–320. doi: 10.1093/mp/ssm015. [DOI] [PubMed] [Google Scholar]
- Hanton SL, Renna L, Bortolotti LE, Chatre L, Stefano G, Brandizzi F. Diacidic motifs influence the export of transmembrane proteins from the endoplasmic reticulum in plant cells. The Plant Cell. 2005;17:3081–3093. doi: 10.1105/tpc.105.034900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Pischke MS, Mahonen AP, et al. In planta functions of the Arabidopsis cytokinin receptor family. Proceedings of the National Academy of Sciences, USA. 2004;101:8821–8826. doi: 10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose N, Makita N, Yamaya T, Sakakibara H. Functional characterization and expression analysis of a gene, OsENT2, encoding an equilibrative nucleoside transporter in rice suggest a function in cytokinin transport. Plant Physiology. 2005;138:196–206. doi: 10.1104/pp.105.060137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z, Jin H, Tzfira T, Li J. Multiple mechanism-mediated retention of a defective brassinosteroid receptor in the endoplasmic reticulum of Arabidopsis. The Plant Cell. 2008;20:3418–3429. doi: 10.1105/tpc.108.061879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Higuchi M, Hashimoto Y, Seki M, Kobayashi M, Kato T, Tabata S, Shinozaki K, Kakimoto T. Identification of CRE1 as a cytokinin receptor from Arabidopsis. Nature. 2001;409:1060–1063. doi: 10.1038/35059117. [DOI] [PubMed] [Google Scholar]
- Jeon J, Kim NY, Kim S, et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. Journal of Biological Chemistry. 2010;285:23371–23386. doi: 10.1074/jbc.M109.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Inze D, Depicker A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends in Plant Science. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- Kasahara H, Takei K, Ueda N, Hishiyama S, Yamaya T, Kamiya Y, Yamaguchi S, Sakakibara H. Distinct isoprenoid origins of cis- and trans-zeatin biosyntheses in Arabidopsis. Journal of Biological Chemistry. 2004;279:14049–14054. doi: 10.1074/jbc.M314195200. [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE. The perception of cytokinin: a story 50 years in the making. Plant Physiology. 2010;154:487–492. doi: 10.1104/pp.110.161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Ryu H, Hong SH, Woo HR, Lim PO, Lee IC, Sheen J, Nam HG, Hwang I. Cytokinin-mediated control of leaf longevity by AHK3 through phosphorylation of ARR2 in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2006;103:814–819. doi: 10.1073/pnas.0505150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Johannes L, Goud B, Antony C, Lingwood CA, Daneman R, Grinstein S. Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proceedings of the National Academy of Sciences, USA. 1998;95:2997–3002. doi: 10.1073/pnas.95.6.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroha T, Tokunaga H, Kojima M, Ueda N, Ishida T, Nagawa S, Fukuda H, Sugimoto K, Sakakibara H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. The Plant Cell. 2009;21:3152–3169. doi: 10.1105/tpc.109.068676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laloue M, Pethe-Terrine C, Gruen J. Uptake and metabolism of cytokinins in tobacco cells: studies in relation to the expression of their biological activities. In: Guern J, Peaud-Lenoel C, editors. Metabolism and molecular activities of cytokinins. Berlin: Springer; 1981. pp. 80–96. [Google Scholar]
- Langhans M, Marcote MJ, Pimpl P, Virgili-Lopez G, Robinson DG, Aniento F. In vivo trafficking and localization of p24 proteins in plant cells. Traffic. 2008;9:770–785. doi: 10.1111/j.1600-0854.2008.00719.x. [DOI] [PubMed] [Google Scholar]
- Li J, Yang H, Peer WA, et al. Arabidopsis H+-PPase AVP1 regulates auxin-mediated organ development. Science. 2005;310:121–125. doi: 10.1126/science.1115711. [DOI] [PubMed] [Google Scholar]
- Maley F, Trimble RB, Tarentino AL, Plummer Jr., TH Characterization of glycoproteins and their associated oligosaccharides through the use of endoglycosidases. Analytical Biochemistry. 1989;180:195–204. doi: 10.1016/0003-2697(89)90115-2. [DOI] [PubMed] [Google Scholar]
- Marion J, Bach L, Bellec Y, Meyer C, Gissot L, Faure JD. Systematic analysis of protein subcellular localization and interaction using high-throughput transient transformation of Arabidopsis seedlings. The Plant Journal. 2008;56:169–179. doi: 10.1111/j.1365-313X.2008.03596.x. [DOI] [PubMed] [Google Scholar]
- Mravec J, Skupa P, Bailly A, et al. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- Muller B, Sheen J. Arabidopsis cytokinin signaling pathway. Science Signaling STKE. 2007 a doi: 10.1126/stke.4072007cm5. 2007, cm5. [DOI] [PubMed] [Google Scholar]
- Muller B, Sheen J. Cytokinin signaling pathway. Science Signaling STKE. 2007 b doi: 10.1126/stke.4072007cm4. 2007, cm4. [DOI] [PubMed] [Google Scholar]
- Nelson BK, Cai X, Nebenfuhr A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. The Plant Journal. 2007;51:1126–1136. doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- Nishimura C, Ohashi Y, Sato S, Kato T, Tabata S, Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. The Plant Cell. 2004;16:1365–1377. doi: 10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pas J, von Grotthuss M, Wyrwicz LS, Rychlewski L, Barciszewski J. Structure prediction, evolution and ligand interaction of CHASE domain. FEBS Letters. 2004;576:287–290. doi: 10.1016/j.febslet.2004.09.020. [DOI] [PubMed] [Google Scholar]
- Riefler M, Novak O, Strnad M, Schmulling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. The Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romanov GA, Lomin SN, Schmülling T. Biochemical characteristics and ligand-binding properties of Arabidopsis cytokinin receptor AHK3 compared to CRE1/AHK4 as revealed by a direct binding assay. Journal of Experimental Botany. 2006;57:4051–4058. doi: 10.1093/jxb/erl179. [DOI] [PubMed] [Google Scholar]
- Schleifenbaum F, Elgass K, Sackrow M, Caesar K, Berendzen K, Meixner AJ, Harter K. Fluorescence intensity decay shape analysis microscopy (FIDSAM) for quantitative and sensitive live-cell imaging: a novel technique for fluorescence microscopy of endogenously expressed fusion-proteins. Molecular Plant. 2010;3:555–562. doi: 10.1093/mp/ssp110. [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C. Bimolecular fluorescence complementation (BiFC) to study protein–protein interactions in living plant cells. Methods in Molecular Biology. 2009;479:189–202. doi: 10.1007/978-1-59745-289-2_12. [DOI] [PubMed] [Google Scholar]
- Shuster SJ, Riedl M, Li X, Vulchanova L, Elde R. Stimulus-dependent translocation of kappa opioid receptors to the plasma membrane. Journal of Neuroscience. 1999;19:2658–2664. doi: 10.1523/JNEUROSCI.19-07-02658.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song RX, Barnes CJ, Zhang Z, Bao Y, Kumar R, Santen RJ. The role of Shc and insulin-like growth factor 1 receptor in mediating the translocation of estrogen receptor alpha to the plasma membrane. Proceedings of the National Academy of Sciences, USA. 2004;101:2076–2081. doi: 10.1073/pnas.0308334100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Miwa K, Ishikawa K, Yamada H, Aiba H, Mizuno T. The Arabidopsis sensor His-kinase, AHK4, can respond to cytokinins. Plant and Cell Physiology. 2001;42:107–113. doi: 10.1093/pcp/pce037. [DOI] [PubMed] [Google Scholar]
- Tian H, Klambt D, Jones AM. Auxin-binding protein 1 does not bind auxin within the endoplasmic reticulum despite this being the predominant subcellular location for this hormone receptor. Journal of Biological Chemistry. 1995;270:26962–26969. doi: 10.1074/jbc.270.45.26962. [DOI] [PubMed] [Google Scholar]
- Tran LS, Urao T, Qin F, Maruyama K, Kakimoto T, Shinozaki K, Yamaguchi-Shinozaki K. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2007;104:20623–20628. doi: 10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi C, Koizumi H, Suzuki T, Mizuno T. Novel family of sensor histidine kinase genes in Arabidopsis thaliana. Plant and Cell Physiology. 2001;42:231–235. doi: 10.1093/pcp/pce015. [DOI] [PubMed] [Google Scholar]
- Urao T, Yakubov B, Satoh R, Yamaguchi-Shinozaki K, Seki M, Hirayama T, Shinozaki K. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. The Plant Cell. 1999;11:1743–1754. doi: 10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varet A, Hause B, Hause G, Scheel D, Lee J. The Arabidopsis NHL3 gene encodes a plasma membrane protein and its overexpression correlates with increased resistance to Pseudomonas syringae pv. tomato DC3000. Plant Physiology. 2003;132:2023–2033. doi: 10.1104/pp.103.020438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Motyka V, Laucou V, Smets R, Van Onckelen H, Schmulling T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. The Plant Cell. 2003;15:2532–2550. doi: 10.1105/tpc.014928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner T, Schmulling T. Cytokinin action in plant development. Current Opinion in Plant Biology. 2009;12:527–538. doi: 10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- Wohlbach DJ, Quirino BF, Sussman MR. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. The Plant Cell. 2008;20:1101–1117. doi: 10.1105/tpc.107.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wormit A, Traub M, Florchinger M, Neuhaus HE, Mohlmann T. Characterization of three novel members of the Arabidopsis thaliana equilibrative nucleoside transporter (ENT) family. Biochemical Journal. 2004;383:19–26. doi: 10.1042/BJ20040389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, Caniard A, Jones JD, Boller T, Felix G. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.