Abstract
The chromatin organization modifier domain (chromodomain) was first identified as a motif associated with chromatin silencing in Drosophila. There is growing evidence that chromodomains are evolutionary conserved across different eukaryotic species to control diverse aspects of epigenetic regulation. Although originally reported as histone H3 methyllysine readers, the chromodomain functions have now expanded to recognition of other histone and non-histone partners as well as interaction with nucleic acids. Chromodomain binding to a diverse group of targets is mediated by a conserved substructure called the chromobox homology region. This motif can be used to predict methyllysine binding and distinguish chromodomains from related Tudor “Royal” family members. In this review, we discuss and classify various chromodomains according to their context, structure and the mechanism of target recognition.
Keywords: Lysine Methylation, Histone Code, Chromatin Signaling, Chromatin Remodeling, Nucleosome Recognition, Binary Switches in Chromatin, Tudor Royal Family
Introduction
A genomic revolution was launched with the completion of the human genome project a decade ago that led to new paradigms for understanding gene expression (Schmutz et al., 2004, Consortium, 2004). Due to complex processes ranging alternative splicing and post □translational modifications, there are estimated to be over 1 million different human protein forms among 21,000 genes. Central to these processes are distinct chromatin transactions yielding epigenetic phenomena (Bonasio et al., 2010). The unit particle of chromatin is the nucleosome consisting of conserved histones that wrap a DNA superhelix (Luger et al., 1997, Khorasanizadeh, 2004). Signaling to nucleosomes occurs via post-translational modifications of histones and DNA cytosine methylations (Bannister and Kouzarides, 2011, Feng et al., 2010b).
A histone code has been proposed that states distinct modifications of histones act sequentially or in combination to recruit proteins, and bring about downstream regulatory events of gene expression (Strahl and Allis, 2000). Chromodomains are the founding class of histone code methyllysine “readers”. The methyllysine histone codes are also subjects of other “readers” through modular interactions of Tudor, MBT, PWWP, PHD finger, Ankyrin repeats, and WD repeats (Taverna et al., 2007). These effector modules are integral to control of transcription that responds to epigenetic initiators from environmental and developmental cues. Epigenetic effects influence common human diseases such as cancers, heart disease, diabetes and psychiatric disorders from patient to patient. A new study shows the most well-known chromodomain for reading a methyllysine in repressive chromatin, the Polycomb chromodomain, when fused to transcription factors can synthetically activate transcription in human cells (Haynes and Silver, 2011). A better understanding of functions ascribed to chromodomains should facilitate translational research and the discovery of therapeutic or diagnostic tools.
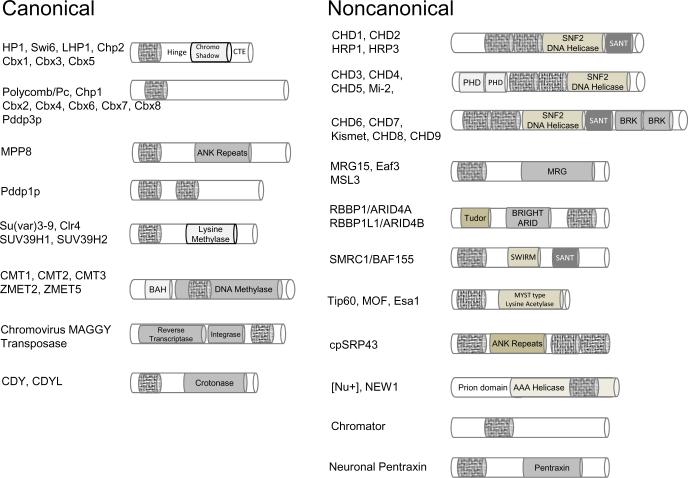
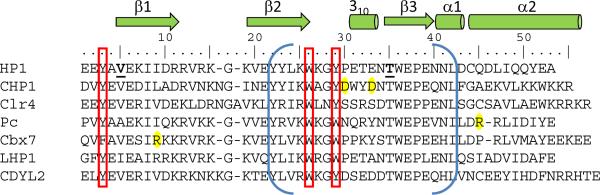
Historically, the chromodomain was discovered as a motif responsible for imprinting a determined state into the chromatin of Drosophila, present in the Heterochromatin protein 1 (HP1) and Polycomb (Pc) proteins (James and Elgin, 1986, Paro, 1990, Paro and Hogness, 1991, Aasland and Stewart, 1995). Subsequently, the three-dimensional structures corresponding to the HP1 chromodomain and chromo shadow domain revealed a conserved fold consisting of three anti-parallel β-strands followed by an α-helix (Ball et al., 1997a, Brasher et al., 2000). Their modularity facilitated the prediction of chromodomain and chromo shadow domain from amino acid sequences using SMART or InterPro tools (Letunic et al., 2009, Hunter et al., 2009) (Figure 1). The chromodomain is a hallmark of eukaryotic species, with the exception of a few cyanobacteria that encode one chromodomain-containing protein. According to SMART database, the human and Drosophila genomes encode 55 and 25 chromodomain-containing polypeptides, respectively (Table 1) (Letunic et al., 2009). For certain chromodomain proteins there are multiple variants (e.g., 5 Drosophila HP1s, HP1a, HP1b, HP1c, HP1d and HP1e (Vermaak et al., 2005) vs. 3 mammalian HP1 variants HP1α, HP1β and HP1γ).
Figure 1.
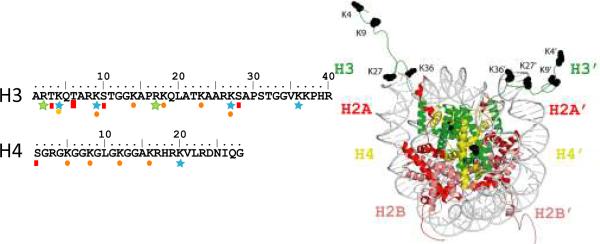
Diverse chromodomains recognize methyl-lysine marks in H3 and H4 tails. a) Composition of chromodomain proteins, and their classification into canonical and noncanonical polypeptides. Chromodomain module is represented with a hatched rod. (b) Schematic of the absolutely conserved histone H3 and H4 tails and their locations in the nucleosome core particle. Sites of modifications with methylation of lysine (blue star), methylation of arginine (green star), phosphorylation of threonine (red square) and acetylation of lysines (orange circle) are labeled in histone tail sequences.
Table 1.
Prevalence of chromodomain proteins in selected eukaryotic species derived from SMART database.
| Species | Total number of chromodomain proteins |
|---|---|
| Nostoc punctiforme | 1 |
| Saccharomyces cerevisiae | 4 |
| Schizosaccharomyces pombe | 9 |
| Tetrahymena thermophile | 18 |
| Caenorhabditis elegans | 22 |
| Arabidopsis thaliana | 23 |
| Drosophila melanogaster | 25 |
| Homo sapiens | 55 |
A range of functions is associated with chromodomains. A majority specifically associates with sites of lysine-methylation along the absolutely conserved histone H3 and H4 tails (histone methyl-marks) (Fig. 1 and Table 2). The interaction of a chromodomain with a histone methyl-mark is sensitive to the degree of methylation as well as to the presence of adjacent histone modifications such as phosphorylation of serines/threonine, methylation of arginine or acetylation of lysine. These are referred to as binary switches in histone biology (Fischle et al., 2003a). In addition, chromodomains can associate with RNA and DNA in a sequence-selective manner, whereas the chromo shadow domain binds peptides with a central valine (Smothers and Henikoff, 2000). The dissociation constants measured display a wide μM range, suggesting these effectors can mediate versatile functions. Positive and negative regulations of these interactions have been associated with phosphorylation sites in chromodomain proteins as discussed below.
Table 2.
Peptide targeting functions reported for chromodomain proteins.
| Recruiting Histone Mark | Localization/Function | Chromodomain |
|---|---|---|
| H3K4me3 | Transcription start sites, transcription activity (Barski et al., 2007, Santos-Rosa et al., 2002, Schneider et al., 2004) | Human CHD1 (Flanagan et al., 2005), CHD2, CHD8 (Flanagan et al., 2007, Rodriguez-Paredes et al., 2009), Eaf3 (Xu et al., 2008), CHD7 (Takada et al., 2007) |
| H3K4me2 | Active or poised chromatin, transcription activity (Bernstein et al., 2005, Liang et al., 2004) | |
| H3K4me1 | Gene enhancers, transcription activity (Heintzman et al., 2007) | CHD7 (Bajpai et al.,2010, Schnetz et al., 2009) |
| H3K9me2/3 | Pericentric heterochromatin, transcription activity or silencing (Barski et al., 2007, Bernstein et al., 2005, Liang et al., 2004) | HP1 (Bannister et al., 2001, Jacobs and Khorasanizadeh, 2002), LHP1 (Zhang et al., 2007), MPP8 (Quinn et al.), Chp1 (Schalch et al., 2009), Swi6 (Nakayama et al., 2001, Sadaie et al., 2008), Chp2 (Sadaie et al., 2008), Clr4 (Zhang et al., 2008), MAGGY (Kordis, 2005), Tip60 (Sun et al., 2009), CHD7 (Takada et al., 2007), Pdd1/3p (Taverna et al., 2002), Cbx2/4/7 (Bernstein et al., 2006), Su(var)3–9 (Jacobs et al., 2004), CMT3 (Lindroth et al., 2004) |
| H3K9me1 | Pericentric heterochromatin, transcription activity(Peters et al., 2003) | CDYL, CDY (Fischle et al., 2008) |
| H3K27me2/3 | Body of genes, transcription silencing (Cao et al., 2002) | LHP1 (Zhang et al., 2007), Pc (Fischle et al., 2003b, Min et al., 2003), CBX2/7 (Bernstein et al., 2006) |
| H3K27me1 | Pericentric heterochromatin, transcription silencing (Heintzman et al., 2007) | - |
| H3K36me2/3 | Body and 3' end of genes, transcription activity (Lee et al., 2007) | MRG15 (Zhang et al., 2006), Eaf3 (Sun et al., 2008, Xu et al., 2008), MSL3 (Sural et al., 2008) |
| H4K20me2/3 | Chromosomal integrity, transcription silencing (Lan and Shi, 2009, Mikkelsen et al., 2007) | - |
| H4K20me1 | Body of genes, transcription elongation or silencing (Karachentsev et al., 2007, Vakoc et al., 2006) | MSL3 (Kim et al., 2010) |
| Antagonizing Histone Mark | Localization/Function | Chromo |
|---|---|---|
| H3T3ph | Mitotic centromeres, mitosis regulation (Dai et al., 2005, Rosasco-Nitcher et al., 2008) | Human CHD1 (Flanagan et al., 2005) |
| H3R2me2asym | Body and 3' end of genes(Guccione et al., 2007) | Human CHD1 (Flanagan et al., 2005) |
| H3S10ph | Transcription activity, chromosome condensation and segregation during mitosis (Hendzel, 1997, Johansen and Johansen, 2006, Wei et al., 1999) | HP1 (Fischle et al., 2005, Jacobs and Khorasanizadeh, 2002) |
| H4K16ac | Genomewide distribution, transcription activity (Wang et al., 2008) | MSL3 (Kim et al., 2010) |
| Recruiting Chromatin Factor | Localization/Function | ChSh |
|---|---|---|
| EMSY | DNA-damage response and chromatin remodeling (Hughes-Davies et al., 2003) | HP1(Ekblad et al., 2005, Huang et al., 2006) |
| CAF-1 | Nucleosome assembly during DNA replication and repair (Gaillard et al., 1996, Kaufman et al., 1995, Murzina et al., 1999, Smith and Stillman, 1989) | HP1 (Thiru et al., 2004) |
| PIWI | RNA silencing complex/binds to piRNAs (Brennecke et al., 2007, Vagin et al., 2006) | HP1 (Brower-Toland et al., 2007) |
| DIM-2 | Heterochromatin/DNA methylation (Kouzminova and Selker, 2001, Tamaru and Selker, 2001) | N. crassa HP1 (Honda and Selker, 2008) |
| Histone H3 | Activation of JAK2 by chromosomal translocations or point mutations in haematological malignancies (Dawson et al., 2009) | HP1a (Dawson et al., 2009) |
Here we classify chromodomains based on their structure and mechanism of target recognition (Fig. 1a). Those with high sequence homology to the HP1 chromodomain will be referred to as canonical. They form selective interactions with a conserved lysine-methylated motif (ARKS) originally found in the H3 tail at H3K9 and H3K27 sites (Fig. 1b), but also present within other proteins (Table 3). Canonical chromodomains are abundantly encoded in mammals (17 polypeptides in total), whereas lower eukaryotes use far fewer (Letunic et al., 2009). In budding yeast, the methylations of H3K9 and H3K27 are both absent (Millar and Grunstein, 2006), hence HP1 protein is not among the total of four chromodomain proteins encoded. In contrast, the fission yeast implements H3K9 methylation, and encodes four polypeptides (Clr4, Swi6, Chp1 and Chp2) that are able to bind this mark among a total of 8 chromodomain polypeptides encoded (Zhang et al., 2008, Schalch et al., 2009).
Table 3.
Human proteins that contain an ARKS/T motif are classified according to function with their accession codes listed in parenthesis. These may be putative physiological targets of a variety of lysine methyltransferases.
| Zinc finger | Transcription regulation | Histone methyltransferase/demethylase | Adapter | Chromosomal stability/DNA replication | Catalytic activity | Polymeraseassociated | |
|---|---|---|---|---|---|---|---|
| Nuclear functions | |||||||
| ZFHX4 (Q86UP3) | ADNP (Q9H2P0) | NSD1 (Q96L73) | TRRAP (Q9Y4A5) | MCM5 (P33992) | BMP2K (Q9NSY1) | BN51 (P05423) | |
| ZNF231 (Q9UPA5) | TSHZ3 (Q63HK5) | NSD3 (Q9BZ95) | ATF-IP (Q6VMQ6) | DPOG1 (P54098) | PRP4 (Q13523) | RPA194 (O95602) | |
| ZNF449 (Q6P9G9) | PHTF2 (Q8N3S3) | G9a (Q96KQ7) | ACRC (Q96QF7) | LIG1 (P18858) | EXDL2 (Q9NVH0) | POLR1A (B7ZKR9) | |
| ZNF484 (Q5JVG2) | Sp110 (Q9HB58) | MLL2 (O14686) | CLIP4 (P57075) | GTBP (P52701) | NTHL1 (P78549) | ||
| ZNF518A (Q6AHZ1) | MR (P08235) | MLL5 (Q8IZD2) | WDR62 (O43379) | FAC1 (Q9NVI1) | EST1A (Q86US8) | ||
| ZNF644 (Q9H582) | GR (P04150) | EHMT1 (Q9H9B1) | Repo-Man (Q69YH5) | ATAD2B (Q9ULI0) | |||
| ZNF707 (Q96C28) | BBX (Q8WY36) | UTX (O15550) | RBM12 (Q9NTZ6) | CSTF-77 (Q12996) | |||
| ZNF839 (A8KOR7) | BCL6 (E3UVQ2) | UTY (O14607) | NEIL3 (Q8TAT5) | ||||
| ZBED6 (P86452) | BCOR (Q6W2J9) | ||||||
| CTCF (P49711) | |||||||
| SPT16 (Q9Y5B9) | |||||||
| PAS3 (Q8IXF0) | |||||||
| RORα (P35398–2) | |||||||
| Catalytic activity/ Signalling | Membraneassociated | Cell migration/Adhesion/Cytoscheleton/Apoptosis | Transport | Cell cycle/Microtubuleassociated | Development/Differentiation/Disease |
|---|---|---|---|---|---|
| CPRK (Q8IWU2) | DMXL2 (Q8TDJ6) | LAMA4 (Q16363) | LTC4 (P33527) | MAP-1A (P78559) | CHRD (Q9H2X0) |
| VIP1 (Q6PFW1) | FAM171A1 (Q5VUB5) | BOC (Q9BWV1) | AFTPH (Q6ULP2) | KIF17 (Q9P2E2) | BMP-1 (P13497) |
| DHTKD1 (Q96HY7) | EMILIN-4 (Q13201) | ATP13A4 (Q4VNC1) | GTSE-1 (Q9NYZ3) | SAP-97 (Q12959) | |
| DPDE3 (Q08499) | PRTG (Q2VWP7) | ADB2 (Q13349) | GABRG2 (P18507) | CLASP2 (O75122) | NRK (Q7Z2Y5) |
| PPIP5K2 (O43314) | GCC1 (Q96CN9) | MAC2BP (Q08380) | GABRG3 (Q99928) | DP-150 (Q14203) | LNP (Q9C0E8) |
| MTR (Q99707) | C9orf144B (Q6ZU69) | PC-LKC (Q9BYE9) | Ru2 (Q9UPZ3) | DNM1 (Q05193) | CASP-14 (P31944) |
| XDH (P47989) | 5-HT-7 (P34969) | N-RAP (Q86VF7) | LYST (Q99698) | RYR-3 (Q15413) | EDF (P08476) |
| AP-A (Q07075) | CIP150 (Q4ADV7) | CK-73 (Q86Y46) | CrAT (P43155) | Munc13–3 (Q8NB66) | |
| PIK3C2B (B7ZM86) | MUC-16 (Q8WXI7) | SYNC (Q9H7C4) | Synip (Q6ZWJ1) | AP-N (P15144) | |
| ART3 (Q13508) | DCST2 (Q5T1A1) | ARPC5 (O15511) | VATB (P15313) | GVINP1 (Q7Z2Y8) | |
| JAK-3 (P52333) | COG2 (Q86U99) | DAAM1 (Q9Y4D1) | 40-2-3 (Q96QD9) | NF-1 (P21359) | |
| SE (Q14534) | TMC3 (Q7Z5M5) | Acinus (Q9UKV3) | SNX19 (Q92543) | mHag HA-1 (Q92619) | |
| USP31 (Q70CQ4) | TMEM68 (Q96MH6) | Carma 3 (Q9BWT7) | CAB4 (O00305) | GRASP-1 (Q4V328) | |
| DEPTOR (Q8TB45) | LPPR5 (Q32ZL2) | NLRP6 (P59044) | PN5 (Q9UI33) | MECl-1 (P40306) | |
| LEI (P30740) | GTSCR1 (Q86UQ5) | PDCD6IP (Q8WUM4) | KCTD6 (Q8NC69) | Cyclin-F (P41002) | |
| TULP4 (Q9NRJ4) | OST267 (Q96RD1) | GADD34 (O75807) | SKD2 (Q9UN37) | ||
| FBXO41(Q8TF61) | TMEM215 (Q68D42) | PRG-2 (Q6T4P5) | HGS (O14964) | ||
| USPL1 (Q5W0Q7) | JP-2 (Q9BR39) | SMG7 (Q92540) | RIM 2 (Q9UQ26) | ||
| MR-GEF (Q92565) | DHHC-22 (Q8N966) | DOCK1 (B2RUU3) | IQGAP2 (Q13576) | ||
| EPN2 (O95208) | DOCK5 (Q9H7D0) | ||||
| MMR (P22897) |
The ARKS/T binding chromodomains have been found in a subset of so-called chromoviruses (a form of transposases) that target the H3K9 methyl-mark. Similar chromodomain-containing transposases are encoded in plants, but none have been reported in mammals (Gorinsek et al., 2004, Gorinsek et al., 2005). Numerous other chromodomains have been reported to recognize different targets along the histone tails, e.g., the H3K4, H3K36 and H4K20 methyl-marks (Table 3). Often these chromodomains have amino and carboxyl-terminal extensions and/or internal inserts. A significant number of chromodomains display no interaction with methylated peptides. Some are associated with nucleic acid recognition, and others may recognize unmethylated peptides. We will refer to all these chromodomains as noncanonical (Fig. 1a).
The chromobox homology motif harbors critical aromatic residues for methyllysine recognition. We discuss the use of this motif as a prediction tool for the identification of chromodomains and their functions. We conclude with a comparison of chromodomains with Tudor, MBT, PWWP and Agenet domains collectively coined as the Royal family (Maurer-Stroh et al., 2003). As noncanonical chromodomains often fold like the Tudor domain, we conclude by evaluating attributes of Tudor domains.
CANONICAL CHROMODOMAINS
HP1 and Polycomb chromodomains
In Drosophila, HP1 and Polycomb chromodomains direct the localization of distinct chromatin repressive complexes to the H3K9 and H3K27 methyl-marks, respectively (Jacobs et al., 2001, Fischle et al., 2003b). In mammals, the Polycomb protein has expanded to five (Cbx2, Cbx4, Cbx6, Cbx7 and Cbx8) proteins; the Cbx1, Cbx3 and Cbx5 correspond to the HP1 variants. The five Polycomb variants have roles in assembly and function of the various forms of the polycomb repressive complex 1 (PRC1) that regulates tissue-specific development and disease (Simon and Kingston, 2009). A common mechanism of methylated histone tail recognition is achieved by the HP1 and Polycomb chromodomains (Fig. 2).
Figure 2.
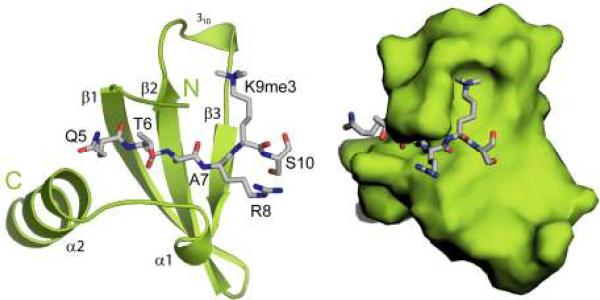
Structural properties of canonical chromodomains. a) Ribbon and surface representation of the HP1 chromodomain (green) bound to the H3K9me3 peptide (grey). (b) Differentially methylated peptides (yellow) form the similar cation-π interactions with the aromatic cage of the HP1 chromodomain (green). The free hydrogens in mono- and dimethyllysine form water-mediated hydrogen bonds (dotted lines) with the chromodomain. c) Sequence alignment of HP1 chromodomain with other well-studied canonical chromodomains. The red bars depict the aromatic residues of the cage. The blue brackets correspond to the chromobox homology motif. In bold is the threonine that resides in the Casein Kinase II consensus sequence. Highlighted in yellow are the residues that uniquely contribute to structure of complexes.
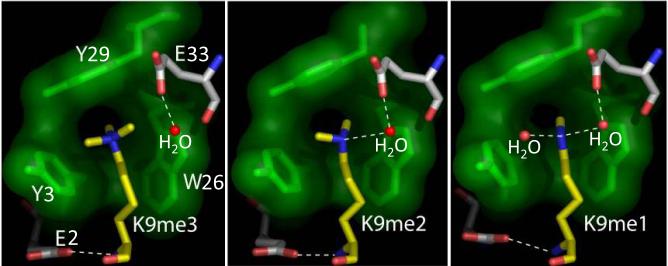
Canonical chromodomains accommodate the histone H3 peptide in a groove between β1 and β3 strands and complete a β-sandwich architecture (Jacobs et al., 2001, Nielsen et al., 2002, Jacobs and Khorasanizadeh, 2002, Kaustov et al., 2011) (Fig. 2a). The binding involves the insertion of the methyllysine moiety into a groove lined by three aromatic residues coined as an aromatic cage (Jacobs and Khorasanizadeh, 2002). In this manner, the disordered histone H3 tail adopts an ordered conformation upon binding to the chromodomain, whereas the structure and dynamics of the chromodomain do not change upon binding. Their interaction requires the presence of methyllysine at H3K9 and formation of cation-π interactions with delocalized electrons of three conserved aromatic residues (Fig. 2b). HP1 forms robust interactions with mono, di and trimethylated H3K9 (Fischle et al., 2003b, Jacobs and Khorasanizadeh, 2002), but the binding to di and trimethylation is physiologically-relevant (Grewal and Elgin, 2007). Substitution with a neutral trimethyllysine analog, the tert-butylnorleucine, results in 30-fold weaker binding to the H3K9 methyl-mark (Hughes et al., 2007). Interestingly, a superimposable aromatic residue arrangement has been found in the human phosphatidylcholine transfer protein binding to the quaternary amine of dilinoleoyl-phosphatidylcholine (Roderick et al., 2002).
Electrostatic interactions dominate in the binding of the HP1 chromodomains to the target peptide (Jacobs and Khorasanizadeh, 2002, Kaustov et al., 2011). Acidic residues of the peptide groove or adjacent surfaces form polar interactions. In contrast, the Polycomb and mammalian Cbx variants rely on hydrophobic side chains at their surface to clasp the target peptide (Fischle et al., 2003b, Min et al., 2003, Kaustov et al., 2011) (Fig. 2c). Subtle amino acid substitutions in the peptide groove can lead to conformational heterogeneity in the backbone, and inability to bind peptide (Jacobs et al., 2001, Kaustov et al., 2011). Sequence variations among these chromodomains result in a spectrum of binding affinity towards the H3K9 and H3K27 methyl-marks (Bernstein et al., 2006, Kaustov et al., 2011) (Fig. 2c). For example, Cbx7 binds the H3K9 and H3K27 methyl-marks with micromolar affinity whereas Cbx6 and Cbx8 associate using millimolar affinity. Sequence variation has also led to bimodal chromatin interactions. An arginine present in the β1 of Cbx7 chromodomain (Fig. 2c) is crucial for non-coding RNA recognition near where H3K27 methyl-mark binds (Yap et al., 2010). Another novel interaction is the oligomerization by the chromodomain of yeast Swi6 when interacting with H3K9 methylated nucleosomal substrates (Canzio et al., 2011). Point mutations of the Swi6 chromodomain that increase its dimerization ability were shown to enhance yeast chromatin silencing in vivo (Canzio et al., 2011) in agreement with a chromodomain role in spreading of heterochromatin (Nakayama et al., 2001).
Chromodomain phosphorylation regulates peptide binding. Canonical chromodomains have a conserved threonine in β3 (Fig. 2c) that resides in a Casein Kinase II consensus sequence. This site is phosphorylated as a response to DNA damage, and disrupts HP1 binding (Ayoub et al., 2008). Phosphorylation may mediate disruption of the π-cation bonding in the aromatic cage by increasing conformational heterogeneity or electrostatic interference (Fig. 2a,c). In contrast, the phosphorylation of acidic residues near the HP1 chromodomain improves peptide binding (Hiragami-Hamada et al., 2011). Many Cbx proteins as well as the plant HP1/LHP1 protein do not discriminate between methylated H3K9 and H3K27 peptides. In these, phosphorylation is believed to regulate targeting (Hatano et al., 2010, Zhang et al., 2007, Kaustov et al., 2011).
Other Canonical Chromodomains
In fission yeast, a Cbx-like chromodomain resides in the Chp1 protein that tethers the RNA-induced initiation of transcription gene silencing (RITS) complex to centromeres. Chp1 binds the H3K9 methyl-mark 10-fold tighter than the Swi6/HP1 chromodomain (Schalch et al., 2009). There is a larger interface of the Chp1 complex (625 Å2) as compared to Drosophila HP1 (521 Å2) (Schalch et al., 2009), and a zinc ion is coordinated near the aromatic cage by two aspartic acids (Fig. 2c). Binding to zinc may improve conformational order and serve advantageous to peptide binding.
The mammalian M-phase phosphoprotein 8 (MPP8) affects tumor cell proliferation and invasiveness by regulating E-cadherin expression (Matsumoto-Taniura et al., 1996, Bua et al., 2009, Quinn et al., 2010, Kokura et al., 2010). MPP8 interacts with GLP and ESET, two lysine methyltransferases and the DNA methyltransferase DNMT3A (Kokura et al., 2010). Its canonical chromodomain forms a specific interaction with the H3K9 methyl-mark (Chang et al., 2011). Phosphorylation of Tyr and Ser/Thr sites within the chromodomain of MPP8 are thought to control interactions with the H3K9 methyl-mark (Chang et al., 2011).
The canonical chromodomains in Tetrahymena reside in Pdd1p and Pdd3p proteins, which play a crucial role in programmed DNA elimination. Pdd1p contains two chromodomains and the first chromodomain is selective for both H3K9me3 and H3K27me3 (Taverna et al., 2002, Liu et al., 2007). Pdd3p has a single chromodomain and binds H3K9me3 (Taverna et al., 2002, Liu et al., 2007). These function in the macronucleus where mega base pairs of DNA, often repeat sequences, are eliminated from the genome (Chalker, 2008).
Binary Switch for Canonical Chromodomains
The interactions of canonical chromodomains with the ARKS/T-methylated peptides are significantly reduced by Ser/Thr phosphorylation in this motif, e.g., the H3S10 or H3S28 phosphorylation (Fischle et al., 2005, Niessen et al., 2009). Both of these marks correlate with chromodomain protein dissociation from chromatin (Niessen et al., 2009). A phosphorylation near the ARKS/T, the H3T6 was recently discovered (Garske et al., 2010). A significantly reduced H3K9 methyl-mark binding by mammalian HP1 variants was observed in the presence of H3T6 phosphorylation (Kaustov et al., 2011). The acetylation of H3K4 may also influence peptide binding in certain cases. Extensive contacts between the Chp1 chromodomain and H3K4 were detected for the binding to the H3K9 methylated peptide (Schalch et al., 2009). In yeast, the acetylation of H3K4 signals the blockage of Chp1 interaction and prevents the spreading of the RITS complex (Xhemalce and Kouzarides, 2010). The canonical chromodomain interaction with a methyllysine is therefore modulated according to multitude of adjacent modifications that may help protect these remarkably stable methylated lysine marks (Bannister et al., 2002).
Enzymes with a Canonical Chromodomain
The suppressor of variegation 3–9 (Su(var)3–9) protein contains a SET domain that deposits the H3K9 methyl-mark in heterochromatin (Rea et al., 2000, Grewal and Jia, 2007). Deletion of the chromodomain leads to catalytic inefficiency and compromises fidelity (Chin et al., 2006, Eskeland et al., 2004). Its homolog in fission yeast is Clr4, the sole H3K9 methyltransferase that is recruited to heterochromatin by the RITS complex (Zhang et al., 2008, Motamedi et al., 2008, Chen et al., 2008). Initial targeting of Clr4 relies on siRNAs and methylated H3K9 recognition. Clr4 chromodomain binding to H3K9me3 initiates downstream K9 methylation events allowing for the stable association of the RITS complex through the Chp1 chromodomain. This allows the RNAi machinery to process more siRNAs, which reinforce a looping mechanism that further propagates heterochromatin formation (Zofall and Grewal, 2006). This machinery is sensitive to H3K4 acetylation as Clr4 and Chp1 chromodomains could not bind to the H3K9 methyl-mark via the binary methyl/acetyl switch (Xhemalce and Kouzarides, 2010).
In Arabidopsis, various DNA methylation strategies are used to silence genes (Feng et al., 2010a). A subset of these enzymes is the three chromomethyltransferase variants, of which only CMT3 is transcribed and CMT1 and CMT2 are considered pseudogenes. These also include maize ZMET2 and ZMET5 (Papa et al., 2001) and rice CMT1, CMT2 and CMT3 (Sharma et al., 2009). The chromodomain is embedded between motifs I and IV of the DNA methyltransferase enzyme, and is suspected of contributing to catalysis (Fig. 1a). The CMT3 chromodomain forms a specific interaction with the lysine 9-methylated histone H3 in the context of long peptides (Lindroth et al., 2004). CMT3-related chromodomains have a characteristic insertion in the β1 strand suggestive of another protein interaction surface (Lindroth et al., 2004). As DNA methylation has been shown to target nucleosomes (Chodavarapu et al., 2010), the CMT3 chromodomain can provide a discriminatory handle and target CMT3 to selectively function in distinct heterochromatic regions.
In a rice blast fungus Magnaportha grisea, an enzyme with reverse transcriptase and integrase functions called the chromovirus MAGGY contains the canonical chromodomain (Fig. 1a) (Kordis, 2005, Gao et al., 2008). To verify binding to the H3K9 methyl-mark in vivo, this chromodomain was fused to the Tf1 retrotransposon of fission yeast. As the wild-type Tf1 does not localize to heterochromatic regions, the presence of the chimeric retrotransposon was attributed to MAGGY localization to heterochromatin. This chromovirus and others may hijack host mechanisms to integrate into the host genome using natural histone methylation marks.
The chromodomain on the Y chromosome (CDY) family of proteins are crotonases or enoyl-CoA hydratases (Hamed et al., 2008) with a canonical chromodomain (Lahn and Page, 1997). Early observation that CDY catalytic motif is a histone acetyltransferase specific for the histone H4 tail has not been reproduced (Lahn et al., 2002), and the modification created by this family remains elusive. Despite a high sequence identity, the CDY, CDYL, and CDYL2 chromodomains differ in their binding preferences for H3K9 and H3K27 methyl-marks as well as for methylated non-histone peptides (Fischle et al., 2008, Franz et al., 2009, Kim et al., 2006).
Putative targets of canonical chromodomains
We discussed a variety of biological scenarios mediating the canonical chromodomain interaction with histone H3 tail. In the last few years, evidence has accumulated for the interaction of these chromodomains with other proteins via a methylated ARKS/T motif (Huang and Berger, 2008, Daujat et al., 2005, Sampath et al., 2007). Moreover, the consensus sequence of numerous lysine methyltransferases in the mammalian genome is the ARKS/T motif (Allis et al., 2007). These observations suggest a wider role for the canonical chromodomain in cellular signaling. We prepared Table 2 to list human polypeptides containing the ARKS/T motif, and categorized them according to presumed function. We suggest these become methylated and subject of chromodomain recognition for a variety of nuclear control mechanisms (Table 2 upper row). In addition, we found many proteins that function outside of the nucleus and may be involved in signaling, membrane association, transport or microtubule organization (Table 2 lower row).
NONCANONICAL CHROMODOMAINS
CHD double chromodomains cooperate to bind the H3K4 methyl-mark
Chromo-ATPase/helicase-DNA-binding (CHD) proteins have double chromodomains N-terminal to a SNF2-type helicase and are vital for chromatin remodeling during development (Woodage et al., 1997, Flaus et al., 2006, Ho and Crabtree, 2010). S. cerevisiae encodes a single CHD family member, CHD1. In metazoans the family has expanded to four CHD proteins in Drosophila and nine in humans, highlighting the importance of the CHD proteins to acquire new functions in more complex organisms such as transcription elongation, DNA damage repair, osteogenic differentiation, and neurogenesis (Lutz et al., 2006, Rodriguez-Paredes et al., 2009, Srinivasan et al., 2005, Shur et al., 2006a, Shur et al., 2006b, Surapureddi et al., 2006, Takada et al., 2007, Bajpai et al., 2010, Hurd et al., 2010). These proteins were divided into four phylogenetic classes based on their chromodomain sequences (Flanagan et al., 2007).
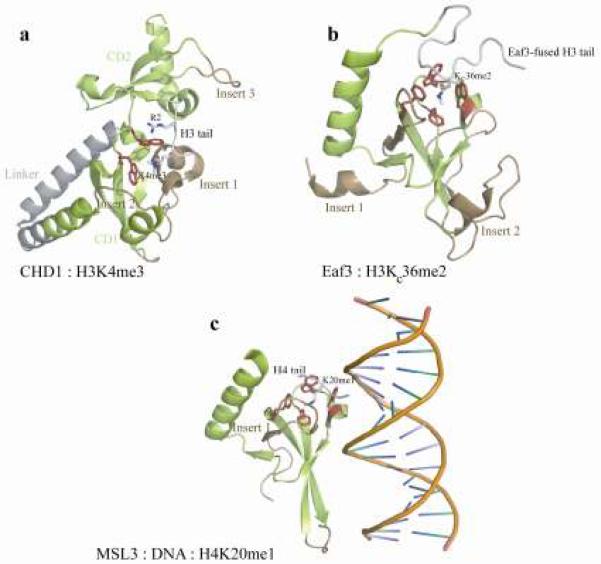
In CHD1 (and CHD2) protein the modules position orthogonally using a helical linker (Flanagan et al., 2005, Flanagan et al., 2007) (Fig. 3a). The unconserved sequence inserts block the canonical peptide binding groove in chromodomain 1, and peptide binding is redirected to the junction of the two chromodomains. Two conserved aromatic residues in chromodomain 1 form the cage for methyllysine recognition and support binding to mono, di and trimethylated H3K4 mark in metazoan CHD1 proteins (Flanagan et al., 2005, Morettini et al., 2011). The peptide affinity is substantially reduced with the presence of asymmetric dimethylation of H3R2 or phosphorylation of H3T3 despite the formation of identical chromodomain-peptide complexes with these dually-modified peptides (Flanagan et al., 2005). Therefore, a methyl/methyl and a methyl/phos switch can influence CHD1 interactions. The phosphorylation of CHD proteins such as CHD2 also may affect affinity or specificity for peptides (Flanagan et al., 2007). The antagonistic effect of H3T3 on H3K4me3 recognition has also been observed for TAF3 and TFIID transcription factors that do not contain a chromodomain, suggesting a broader role of binary switches in regulating chromatin structure and transcription (Varier et al.).
Figure 3.
Sequence inserts in noncanonical chromodomains modulate binding activity. The canonical architecture of chromodomains is displayed in green, inserts are in brown, aromatic cage residues are drawn in ball and stick, and peptides are in gray. a) The double chromodomains of human CHD1 linked by a novel helix-turn-helix (gray). (b) The solution structure of the yeast Eaf3 chromodomain with its C-terminal extension containing a methyllysine analog mimicking the binding of the H3K36 methyl-mark to its aromatic cage. (c) The MSL3 chromodomain in complex with a DNA duplex promoting its selectivity for the H4K20me1 peptide. The peptide sits on the DNA minor groove, with its methyllysine inserted in the four-residue aromatic cage.
Both human and yeast CHD1 are essential for transcription elongation (Simic et al., 2003, Sims et al., 2007), and the mammalian CHD1 is essential for the pluripotency of embryonic stem cells (Gaspar-Maia et al., 2009). In yeast, the lack of a key aromatic residue and changes in the folding of insert 1 prohibit the binding to the H3K4 methyl-mark or other histone peptides (Flanagan et al., 2007, Okuda et al., 2007, Sims et al., 2005) (Fig. 3a). Biochemical and structural studies on the yeast CHD1 recently revealed a function for chromodomain in catalysis (Hauk et al., 2010). The second chromodomain was shown to contact lobe 1 while the inter-chromodomain linker blocked the DNA-binding surface of lobe 2 of the SNF2-helicase. It was further shown that chromodomains enhance substrate specificity of CHD1 for nucleosomes as compared to naked DNA (Hauk et al., 2010). Due to the structural diversity noted for CHD proteins, chromodomains may mediate diverse regulatory functions.
Interestingly, the chromodomains of CHD7 have a unique specificity for the monomethylated H3K4 mark, and ChIP-chip studies have shown that CHD7 tracks H3K4 monomethylation patterns at enhancer motifs (Bajpai et al., 2010, Schnetz et al., 2009). Another study suggested CHD7 chromodomains bind the trimethylated H3K9 mark when CHD7 is recruited for osteoblastogenesis (Takada et al., 2007), suggesting CHD7 has bimodal histone specificity. CHD8 chromodomains have also been shown to bind the H3K4 methyl-mark (Rodriguez-Paredes et al., 2009). By contrast, there is a group of CHD proteins related to the Mi-2 protein, which use chromodomains for DNA recognition (Bouazoune et al., 2002, Kunert and Brehm, 2009). Deletion of the Mi-2 chromodomains impairs ATPase activity and nucleosome mobilization (Bouazoune et al., 2002). To compensate for the lack of histone tail binding by this class, Plant Homeodomain (PHD) fingers are shown to interact with the histone H3 tail selectively (Mansfield et al., 2011, Musselman et al., 2009, Shi et al., 2006).
Versatility of MRG chromodomains in Eaf3/MRG15 and MSL3
Members of the chromodomain containing Morf-related gene (MRG) family are critical components of HAT and HDAC complexes that regulate global acetylation levels. The human MRG15 protein is the founding member of the MRG family, and has been associated with development, DNA repair, and chromatin remodeling (Biswas et al., 2008, Cai et al., 2003, Chen et al., 2010, Hayakawa et al., 2010, Pena and Pereira-Smith, 2007). It is a stable component of the Tip60 HAT complex and also interacts with the mSin3A HDAC complex (Cai et al., 2003, Doyon et al., 2004, Yochum and Ayer, 2002). The MRG15 homolog in budding yeast, Eaf3, is a member of the NuA4 HAT and Rpd3 HDAC complexes, which have opposing functions in transcriptional regulation (Eisen et al., 2001, Gavin et al., 2002). In S. pombe, Altered polarity protein 13 (Alp13) is a member of the Clr6 HDAC complex. Alp13 chromodomain mutants show increased acetylation levels genome-wide implicating that similar to Eaf3, the chromodomain is critical for regulating the deacetylase activity of the complex (Nakayama et al., 2003). A protein microarray screen did not identify any methyllysine substrates for chromodomains of the MRG class (Kim et al., 2006), as they form very weak interactions with histone peptides.
The MRG15/Eaf3 protein associates with two opposing HAT and HDAC complexes. Although the interaction with the H3K36 methyl-mark is physiologically significant, in vitro affinity is in millimolar range, and the chromodomain does not discriminate among peptides with different sequences (Sun et al., 2008, Xu et al., 2008, Zhang et al., 2006). The binding to the H3K4 methyl-mark may also be functionally relevant (Joshi and Struhl, 2005, Reid et al., 2004, Sun et al., 2008, Xu et al., 2008). Deletion of Eaf3 chromodomain prevents Rpd3S HDAC from distinguishing methylated nucleosome substrates, and hindering deacetylation (Li et al., 2007). In another context, Eaf3 chromodomain is critical for NuA4 HAT activity (Joshi and Struhl, 2005, Reid et al., 2004). The MRG15 protein and the recognition of the H3K36 methyl-mark have also been noted to be crucial to alternative pre-mRNA splicing (Luco et al., 2010, Luco et al., 2011).
The Eaf3 and MRG15 chromodomains are structurally related and contain a four-residue aromatic cage in addition to an N-terminal β-strand, which blocks the canonical peptide recognition site (Fig. 3b) (Zhang et al., 2006). The methyllysine moiety was found inserted into a four-residue aromatic cage. However, the surface of interaction between the chromodomain and the peptide is not as extensive as in the canonical chromodomain (Xu et al., 2008). This accounts for the weak affinity and inability to distinguish between methylated peptides in vitro (Xu et al., 2008). Additionally, Eaf3 contains a unique insert between β2 and β3 that does not affect methyllysine recognition and is not present in the MRG15 chromodomains.
A phylogenetic analysis of the MRG family of chromodomains indicated that the human MRG15 and yeast Eaf3 chromodomains are closely related, whereas the metazoan-specific Male-Specific Lethal 3 (MSL3) is diverged. The MSL3 protein is a member of the MSL histone acetyltransferase complex responsible for H4K16 acetylation and transcription dosage compensation (Straub and Becker, 2007, Lucchesi, 2009). Initially, in vitro studies suggested MSL3 protein preferentially binds nucleosomes harboring the H3K36me3 methyl-mark using its chromodomain (Sural et al., 2008). Subsequent studies identified MSL3 specificity for the H4K20 methyl-mark (Kim et al., 2010, Moore et al., 2011). MSL3 chromodomain also binds to naked DNA or nucleosome DNA (Buscaino et al., 2006, Sural et al., 2008). In support of this the MSL3 chromodomain binds to GA-rich MSL recognition elements (MREs) (Alekseyenko et al., 2008) with sub-micromolar affinity (Kim et al., 2010).
The structure of the MSL3 chromodomain revealed two adjacent minor groove binding surfaces that support a cooperative DNA binding. The binding to DNA generated specificity for a histone methyl-mark, H4K20me1 peptide that bound to DNA and a four-residue aromatic cage in chromodomain (Fig. 3c). The canonical binding groove found in the HP1 chromodomain is blocked with a unique sequence insert, and peptide binds within the minor groove of the DNA. The presence of H4K16 acetylation disrupts MSL3 peptide binding, indicating the usage of a methyl/acetyl switch (Kim et al., 2010). A key arginine in MSL3 chromodomain helps recognize the DNA minor groove in a manner related to an arginine in the distantly-related protein Sac7d (Robinson et al., 1998).
Chromodomain similarity with the MRG family (Bertram and Pereira-Smith, 2001) is a hallmark of the ARID4A/B (AT Rich Interactive Domain 4A/B) also known as RBBP-1 (Retinoblastoma Binding Protein 1) protein, a member of the mSin3A chromatin remodeling complex (Fleischer et al., 2003, Lai et al., 2001). ARID4A is involved in the repression of E2F-dependent transcription and cellular proliferation through its interaction with the retinoblastoma protein (Lai et al., 1999a, Lai et al., 1999b). It is a transcription factor important for osteoblast differentiation, genomic imprinting, and leukemic transformation (Monroe et al., 2010, Wu et al., 2008, Wu et al., 2006). Deletion of the ARID genes regulates histone methylation levels, but chromodomain specificity has not been reported (Wu et al., 2008).
Another potential MRG-related chromodomain resides in Baf (BRG-1 Associated Factor (BAF) protein, which is a member in some SWI/SNF chromatin remodeling complexes (Phelan et al., 1999). The BAF protein contributes to heterochromatin formation and chromatin remodeling during differentiation and transcription regulation (Schaniel et al., 2009).
MYST family chromodomains
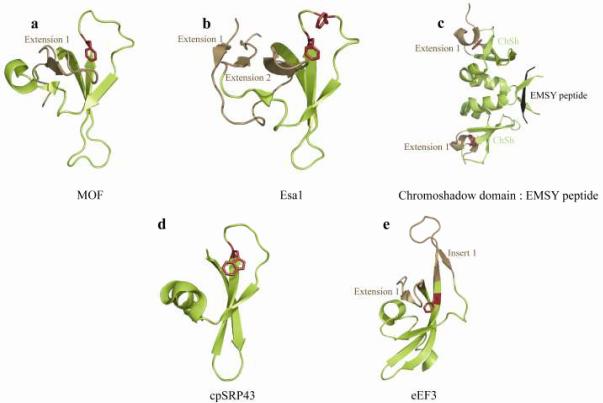
The MYST proteins are lysine acetlytransferases named after the founding members MOZ, YBF2/SAS3, SAS2 and Tip60 (Sapountzi and Cote, 2010, Neal et al., 2000). A protein microarray screen did not identify any methyllysine substrates for chromodomains of the MYST class (Kim et al., 2006). Instead, some MYST chromodomains may be involved in nucleic acid binding (Akhtar et al., 2000, Shimojo et al., 2008). The Drosophila Males-absent on the first (MOF) and the S. cerevisiae Essential Sas-related acetyltransferase 1 (Esa1) proteins acetylate histone H4 genomewide (Hilfiker et al., 1997, Akhtar and Becker, 2000). Loss of Mofresults in a global reduction of H4K16 acetylation, cell cycle arrest and defects in DNA damage repair (Li et al., 2010b, Sharma et al., 2010). Drosophila MOF has an unusual chromodomain structure with short β-strands (Fig. 4a). Aromatic cage residues are not conserved in MOF chromodomain and the canonical peptide binding groove is blocked by an N-terminal extension (Nielsen et al., 2005). Although MOF chromodomain module did not stably bind RNA, the full-length protein requires chromodomain for binding to a non-coding RNA target (Akhtar et al., 2000, Nielsen et al., 2005).
Figure 4.
Noncanonical chromodomains that lack an aromatic cage. The color-coding is similar to that in Figure 3. (a) The Drosophila MOF chromodomain also called a chromobarrel domain. (b) Esa1 chromodomain, also called a knotted-tudor domain. (c) The human chromo shadow dimer in complex with the EMSY peptide. (d) Chromodomain 1 of cpSRP43. (e) The structurally-observed chromodomain-like domain in the eEF3.
Another unusual chromodomain fold is observed for the yeast Esa1 (Shimojo et al., 2008). Two β-strands extend from the amino and carboxyl terminal ends of the Esa1 chromodomain, forming an anti-parallel β sheet, which is crucial for the stability of this module (Shimojo et al., 2008) (Fig. 4b). This structure led to the coining of an inaccurate term the knotted-tudor domain in the Esa1 protein (Shimojo et al., 2008). We found a protein knot detection algorithm (Kolesov et al., 2007) applied to the Esa1 coordinates 2RO0 did not detect any knot. Moreover, SMART and InterPro both predict presence of a chromodomain, but not a tudor domain in Esa1 protein. Interestingly, the Esa1 chromodomain has been shown to interact with single strand RNA, but this is non-specifically and weak. The surface residues important for binding RNA have been shown to have a significant role for the biological function of the Esa1 protein (Shimojo et al., 2008) (Figure 4b).
Tip60 (Tat-interacting protein 60 kDa) chromodomain forms another unique structure (pdb accession 2EKO) closely related to the canonical architecture, and has a two-residue aromatic cage. However, no methyllysine binding is detected for the chromodomain module (Kim et al., 2006). The intact Tip60 was shown to selectively interact with the H3K9 methyl-mark in a manner dependent on the aromatic residues in its chromodomain (Sun et al., 2009). Tip60 interaction with this histone mark is proposed to initiate a Tip60-dependent DNA damage response that enhances acetyltranferase activity (Sun et al., 2009).
The chromo shadow domain: a homodimer with critical carboxy terminal extension
The chromo shadow domain is found C-terminal to the chromodomain in HP1 proteins (Aasland and Stewart, 1995) (Fig. 1a). The chromo shadow domain can also occur without the presence of a chromodomain, e.g. in Drosophila Umbrea protein, which associates with the HP1 protein (Joppich et al., 2009). A number of structures reported for this domain show it forms a symmetric homodimer stabilized by a C-terminal α-helix (Cowieson et al., 2000, Brasher et al., 2000, Mendez et al., 2011). It exhibits a dimerization constant in low μM range (Mendez et al., 2011) (Fig. 4c). The structure of each monomer is highly related to that of the HP1 chromodomain. Two of the three aromatic residues associated with the chromodomain cage are substituted with hydrophobic residues. These block the canonical peptide-binding groove by forming hydrophobic interactions with a helical turn at the N-terminus. The peptide binds to the junction of dimers by forming hydrophobic interactions with a tryptophan in the C-terminus of the chromo shadow domain (Fig. 4c).
The chromo shadow domain mediates HP1 association with a large list of proteins. It is known to drive the initial localization of the HP1 protein to chromatin harboring the H3K9 methyl-mark (Smothers and Henikoff, 2001, Meehan et al., 2003, Lechner et al., 2005). In certain regions of chromatin, it mediates assembly of DNA methylation machinery (Smallwood et al., 2007, Honda and Selker, 2008). The HP1α chromo shadow domain interacted with an H3K36-specific demethylase, and suggested H3K36 demethylation is linked with heterochromatin assembly (Lin et al., 2008). The HP1α chromo shadow domain also interacted with the histone H3 core region near residue 41, and binding was neutralized with JAK2 phosphorylation of tyrosine 41 (Dawson et al., 2009). In plant LHP1, it interacts with LIF2 protein involved in RNA processing (Latrasse et al., 2011) or binds to transcription factors SVP and SCARECROW to repress the activity of their target genes (Cui and Benfey, 2009, Liu et al., 2009).
Partner selectivity for the chromo shadow domain requires the presence of PxVxL type motifs found by a phage display library screen (Smothers and Henikoff, 2000). The three-dimensional structures of the mammalian HP1β chromo shadow domain with PxVxL peptides in CAF155 and EMSY proteins show these peptides associate with a limited surface in the C-terminus of HP1β (Table 2, Figure 4) (Thiru et al., 2004, Huang et al., 2006, Ekblad et al., 2005). Dimerization is required for the binding to a peptide, where a conserved tryptophan sandwiches the central valine of the peptide (Brasher et al., 2000, Thiru et al., 2004, Huang et al., 2006). The affinity of the Drosophila HP1 chromo shadow domain for protein partners was compared (Mendez et al., 2011). In addition to the PxVxL motif, a novel LCVKI was discovered (Mendez et al., 2011). Affinity tightened by 50-fold, suggesting LCVKI-binding drives homodimerization of the HP1 protein. A novel function was attributed to the C-terminal extension (CTE). CTE is essential for stability of dimers and discriminatory binding of HP1 to a partner (Mendez et al., 2011). Therefore, HP1 variants also target chromatin by relying on unique CTE regions.
Chromodomains of Signal Recognition Particle in Chloroplast
Plants rely on chloroplast signal recognition particles cpSRP54 and cpSRP43 for photosynthesis (Schuenemann et al., 1998). The cpSRP43 protein contains three chromodomains with similar architecture (Sivaraja et al., 2005), none forms the aromatic cage (Fig. 4d). The first chromodomain is important for integrating light-harvesting chlorophyll-binding proteins (LHCPs) into thylakoids (Goforth et al., 2004). The C-terminal α-helix of the second chromodomain in cpSRP43 facilitates binding to cpSRP54 with nanomolar affinity (Hermkes et al., 2006, Sivaraja et al., 2005). The second and third chromodomains interact with the Alb3 translocase protein, which is necessary for LHCP insertion into the thylakoid membrane (Falk et al., 2010, Lewis et al., 2010). We note that cpSRP43 protein is the only chromodomain protein with function outside of the nuclear boundary.
Chromodomain of ABC-type ATPase
Elongation factor eEF3 belongs to a family of ATP-binding cassette (ABC) proteins that functions in transporting proteins across membranes, DNA repair and translation (Andersen et al., 2006). Interestingly in yeast, this protein interacts with ribosomes for efficient tRNA release. It is speculated the chromodomain of this protein plays a role in this process by stabilizing an open ribosomal conformation. A chromodomain is not predicted, but the structural characterization of eEF3 revealed a chromodomain-like region. We performed a superposition of this region with the canonical chromodomain and identified extensive insertions in this domain (Fig. 4e) (Andersen et al., 2006). By contrast, SMART and InterPro predict the presence of an embedded chromodomain in a few other AAA helicase modules such as that in the [Nu+] prion formation protein 1 of yeast (Fig. 1a). Therefore, bona fide chromodomains may also contribute to accessing substrates for other ABC proteins.
Distinctions of chromodomain from Tudor clan modules
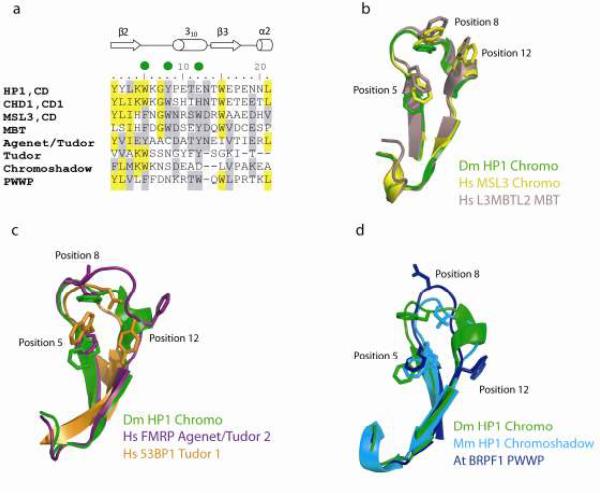
The chromodomain was originally noted to form a similar architecture to a small chromosomal protein in archaea called Sac7d (Ball et al., 1997b). Sac7d forms a module that binds the minor groove for any DNA sequence at micromolar affinity to bend DNA conformation for increased thermal stability (Robinson et al., 1998). Additional work showed chromodomains form consistently smaller distinct modules with structural homology to a group of other eukaryotic domains collectively called the tudor clan. A hallmark of the chromodomains is the presence of a 21-residue chromobox homology motif (Fig. 5a), originally noted when CHD double chromodomains were characterized (Flanagan et al., 2007). This region forms a highly superimposable unit of architecture among chromodomains and participates in partner recognition. Sequence insertions or deletions within this region rarely occur except in cpSRP43 chromoboxes (Fig. 4d). At position 5 of a chromobox there is always an aromatic residue. Other aromatic residues at positions 8 and/or 12 are required for methyllysine binding capability of chromodomains; the position 12 aromatic residue is only present in the MRG class (Fig. 5a).
Figure 5.
The chromobox homology motif is a toolkit for distinguishing the tudor clan modules from the chromodomain. a) A sequence alignment of the chromobox from a selection of chromodomains and related domains: chromo shadow domain, tudor, MBT, PWWP and Agenet. b–d) Ribbon diagram show superposition of the relevant regions of homology and point to key aromatic residues. The superposition of the Drosophila HP1 chromobox with related regions from other domains produced the following rmsd over the backbone residues: 1.21 Å with MBT, 0.44 Å with MSL3, 1.5 Å with 53BP1 Tudor, 2.7 Å with FRMP Agenet, 0.96 Å with chromo shadow and 1.45 Å with PWWP.
Despite low overall sequence identity between MBT, PWWP, Tudor, and Agenet domains, they superimpose well on the canonical chromobox (Fig. 5). There is variability in inter-strand contacts, the folding of the turn and conservation of key aromatic residues associated with these other motifs. Tudor clan modules were coined a “Royal family” (Maurer-Stroh et al., 2003), because they are often shown to be capable of methyllysine recognition by assembling related cages near the chromobox homology motif (Guo et al., 2009, Vezzoli et al., 2010, Min et al., 2007, Botuyan et al., 2006, Ramos et al., 2006, Maurer-Stroh et al., 2003). As compared to chromodomains, there are far less MBT, PWWP and Tudor domains in metazoan genomes, and the Agenet domain is a hallmark of plant genomes (Maurer-Stroh et al., 2003).
Among the members of the tudor clan, the MBT domain forms a chromobox closest to that of chromodomains in the MRG class (Fig. 5a,b). MBT domains are shown to be selective for monomethylated lysine, e.g., the human L3MBTL2 protein recognizes the H4K20 methyl-mark using a canonical chromobox related to the MSL3 chromodomain (Guo et al., 2009). The 53BP1 tudor domains also bind the H4K20 methyl-mark using aromatic residues in positions 5 and 12 (Figure 5a, c) (Botuyan et al., 2006). Lack of an aromatic residue at position 8 is also seen for the Agenet motif in the FMRP protein (Figure 5a, c), which interacts with several trimethylated lysine marks (Ramos et al., 2006). For the PWWP motif of BRFP1, the aromatic residue at position 5 cooperates with residues outside the chromobox to interact with H3K36 methyl-mark, and aromatic residue at position 12 is not used (Figure 5d) (Vezzoli et al.). Therefore, the Tudor, Agenet and PWWP modules each contain a functionally related motif that can be distinguished from chromodomains (Fig. 5).
Dimethylarginine Recognition by Tudor domains
The tudor domain is also associated with recognition of methylated arginine, symmetrically-dimethylated (sDMA) or asymmetrically-dimethylated (aDMA). Their interactions with such glycine-rich peptides are closely related to the representative SMN tudor important for splicing. They exhibit millimolar affinity for binding methylated peptides of a variety of partner proteins. Recently, a surprising micromolar affinity interaction was described for a tudor domain binding to a sDMA partner peptide, and the structure of the complex suggested an asparagine-gated aromatic cage (Liu et al., 2010). In the same protein, Drosophila Tudor, there are 10 other tudor domains, but those lack structural elements for methylated arginine recognition at high affinity.
With regard to the recognition of histone arginine methyl-marks, the mammalian TDRD3 tudor domain exhibits specificity for aDMA in histone H3 and H4 tails (Yang et al., 2010). The same tudor domain is also thought to be the effector for a novel aDMA found in the carboxy terminal domain of RNA Polymerase II (Sims et al., 2011). A protein microarray screen had originally identified TDRD3 as an effector for sDMA peptides (Kim et al., 2006). To what degree TDRD3 discriminates between sDMA and aDMA containing peptides has not been reported. To address this question, we prepared the construct used in protein microarray screen (Kim et al., 2006), and used it in fluorescence polarization peptide binding assays as described (Jacobs et al., 2004). We found TDRD3 binds to aDMA of H3R17 peptide with 50 μM affinity, whereas the binding to a sDMA of H3R17 exhibited a factor of 7 weaker affinity. For a structural basis for this selectivity, we inspected the solution structure deposited in the protein data bank for the human TDRD3 (pdb accession code 2D9T), and identified an unconserved tryptophan that buttresses TDRD3 aromatic cage, potentially enabling selectivity through van der Waals and π-cation interactions with the aDMA.
By contrast to the tudor domain, the chromodomain has never been noted for methylarginine recognition. This is likely due to the high conservation in the chromobox homology motif as well as key aromatic residues. These prohibit the insertion of a methyl arginine moiety in chromodomain aromatic cages. Therefore, the tudor domain sequences have evolved significantly to allow methyllysine or methylarginine selectivity. Interestingly, no chromodomain has been identified to recognize the methylated lysine in the core of histone H3. However, a protein microarray screen has identified a putative interaction between the tudor domain of C20orf104 (also called human PHF20) and the H3K79 methyl-mark (Kim et al., 2006). Subsequent studies are needed to verify the interaction and identify the basis for the unique recognition of this methyl-mark.
Perspective
In less than a decade enormous literature has been published on versatile functions of chromodomains in epigenetic regulations (a selection listed in Table 2). These have often described individual chromodomain modules, and the in vitro targeting potential for histone and other disordered peptides. However, NMR studies that characterized hydrogen exchange properties of the H3 tail in nucleosome arrays revealed a majority of the amide protons were protected from exchange, suggestive of a folded H3 tail (Kato et al., 2009). The methylation of key residues in the H3 tail may thus increase its dynamics, and promote accessibility of individual nucleosomes in diverse gene expression mechanisms. Investigating the interaction of chromodomains with higher order chromatin structure was hampered by challenges in the reconstitution of an appropriate chromatin environment in vitro. However, recent efforts have verified physiologically-relevant chromatin fibers can be reconstituted (Li et al., 2010a). A recombinant method for preparation of histones harboring methyllysine analogs has also been validated for the study of HP1 chromodomain interaction with the H3K9 methyl-mark (Simon et al., 2007).
Recently, a nucleosome-bridging mechanism for the HP1 adaptor protein was described (Canzio et al., 2011). Binding studies between Swi6 and the H3K9-methylated nucleosome arrays led to chromodomain dimerization, and featured array organization related to the spreading of heterochromatin (Canzio et al., 2011). These studies need further development to incorporate the crucial roles of the chromo shadow domain, the hinge and CTE regions (Mendez et al., 2011, Smothers and Henikoff, 2001), which are required for heterochromatin assembly. Therefore, HP1 proteins stabilize a network of modification-dependent interactions. MSL3 chromodomain was also proposed to function in bridging of nucleosomes. The crystal structure of the complex of MSL3 chromodomain with DNA and the H4K20me1 peptide superimposes closely on the tetranucleosome structure at the junction of two nucleosomes (Kim et al., 2010, Schalch et al., 2005). As H4K16 acetylation disrupts MSL3 binding, the chromodomain was suggested to guide the spreading of the MSL complex to nucleosomes lacking acetylation.
Thus far, functions attributed to the majority of chromodomains have been linked to histones and nucleosome regulation. However, a wider usage of lysine methyltransferase activity has been noted for chromatin signaling pathways (Huang and Berger, 2008). The presence of lysine-methylated motifs in non-histone proteins suggests further research on chromodomains will broaden their role in mediating a variety of modification-dependent signaling networks (Table 2).
Acknowledgments
Work in our laboratory was supported by NIH grant 1R01GM070558 and AHA grant 0740058N to S.K.. K.W. was a recipient of Ruth L. Kirschstein National Research Service Award.
Footnotes
Declaration of interest: The authors report no conflicts of interest. The authors alone are responsible for the content and writing of this paper.
References
- AASLAND R, STEWART AF. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3168–73. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AKHTAR A, BECKER PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–75. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- AKHTAR A, ZINK D, BECKER PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–9. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- ALEKSEYENKO AA, PENG S, LARSCHAN E, GORCHAKOV AA, LEE OK, KHARCHENKO P, MCGRATH SD, WANG CI, MARDIS ER, PARK PJ, KURODA MI. A sequence motif within chromatin entry sites directs MSL establishment on the Drosophila X chromosome. Cell. 2008;134:599–609. doi: 10.1016/j.cell.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ALLIS CD, BERGER SL, COTE J, DENT S, JENUWIEN T, KOUZARIDES T, PILLUS L, REINBERG D, SHI Y, SHIEKHATTAR R, SHILATIFARD A, WORKMAN J, ZHANG Y. New nomenclature for chromatin-modifying enzymes. Cell. 2007;131:633–6. doi: 10.1016/j.cell.2007.10.039. [DOI] [PubMed] [Google Scholar]
- ANDERSEN CB, BECKER T, BLAU M, ANAND M, HALIC M, BALAR B, MIELKE T, BOESEN T, PEDERSEN JS, SPAHN CM, KINZY TG, ANDERSEN GR, BECKMANN R. Structure of eEF3 and the mechanism of transfer RNA release from the E-site. Nature. 2006;443:663–8. doi: 10.1038/nature05126. [DOI] [PubMed] [Google Scholar]
- AYOUB N, JEYASEKHARAN AD, BERNAL JA, VENKITARAMAN AR. HP1-beta mobilization promotes chromatin changes that initiate the DNA damage response. Nature. 2008;453:682–6. doi: 10.1038/nature06875. [DOI] [PubMed] [Google Scholar]
- BAJPAI R, CHEN DA, RADA-IGLESIAS A, ZHANG J, XIONG Y, HELMS J, CHANG CP, ZHAO Y, SWIGUT T, WYSOCKA J. CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature. 2010;463:958–62. doi: 10.1038/nature08733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALL LJ, MURZINA NV, BROADHURST RW, RAINE AR, ARCHER SJ, STOTT FJ, MURZIN AG, SINGH PB, DOMAILLE PJ, LAUE ED. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. Embo J. 1997a;16:2473–81. doi: 10.1093/emboj/16.9.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BALL LJ, MURZINA NV, BROADHURST RW, RAINE ARC, ARCHER SJ, STOTT FJ, MURZIN AG, SINGH PB, DOMAILLE PJ, LAUE ED. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 1997b;16:2473–2481. doi: 10.1093/emboj/16.9.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANNISTER AJ, KOUZARIDES T. Regulation of chromatin by histone modifications. Cell Res. 2011;21:381–95. doi: 10.1038/cr.2011.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BANNISTER AJ, SCHNEIDER R, KOUZARIDES T. Histone methylation: dynamic or static? Cell. 2002;109:801–6. doi: 10.1016/s0092-8674(02)00798-5. [DOI] [PubMed] [Google Scholar]
- BANNISTER AJ, ZEGERMAN P, PARTRIDGE JF, MISKA EA, THOMAS JO, ALLSHIRE RC, KOUZARIDES T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–4. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- BARSKI A, CUDDAPAH S, CUI K, ROH TY, SCHONES DE, WANG Z, WEI G, CHEPELEV I, ZHAO K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–37. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- BERNSTEIN BE, KAMAL M, LINDBLAD-TOH K, BEKIRANOV S, BAILEY DK, HUEBERT DJ, MCMAHON S, KARLSSON EK, KULBOKAS EJ, 3RD, GINGERAS TR, SCHREIBER SL, LANDER ES. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–81. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- BERNSTEIN E, DUNCAN EM, MASUI O, GIL J, HEARD E, ALLIS CD. Mouse polycomb proteins bind differentially to methylated histone H3 and RNA and are enriched in facultative heterochromatin. Mol Cell Biol. 2006;26:2560–9. doi: 10.1128/MCB.26.7.2560-2569.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BERTRAM MJ, PEREIRA-SMITH OM. Conservation of the MORF4 related gene family: identification of a new chromo domain subfamily and novel protein motif. Gene. 2001;266:111–21. doi: 10.1016/s0378-1119(01)00372-9. [DOI] [PubMed] [Google Scholar]
- BISWAS D, TAKAHATA S, STILLMAN DJ. Different genetic functions for the Rpd3(L) and Rpd3(S) complexes suggest competition between NuA4 and Rpd3(S) Mol Cell Biol. 2008;28:4445–58. doi: 10.1128/MCB.00164-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BONASIO R, TU S, REINBERG D. Molecular signals of epigenetic states. Science. 2010;330:612–6. doi: 10.1126/science.1191078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOTUYAN MV, LEE J, WARD IM, KIM JE, THOMPSON JR, CHEN J, MER G. Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell. 2006;127:1361–73. doi: 10.1016/j.cell.2006.10.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BOUAZOUNE K, MITTERWEGER A, LANGST G, IMHOF A, AKHTAR A, BECKER PB, BREHM A. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. Embo J. 2002;21:2430–40. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRASHER SV, SMITH BO, FOGH RH, NIETLISPACH D, THIRU A, NIELSEN PR, BROADHURST RW, BALL LJ, MURZINA NV, LAUE ED. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. Embo J. 2000;19:1587–97. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BRENNECKE J, ARAVIN AA, STARK A, DUS M, KELLIS M, SACHIDANANDAM R, HANNON GJ. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell. 2007;128:1089–103. doi: 10.1016/j.cell.2007.01.043. [DOI] [PubMed] [Google Scholar]
- BROWER-TOLAND B, FINDLEY SD, JIANG L, LIU L, YIN H, DUS M, ZHOU P, ELGIN SC, LIN H. Drosophila PIWI associates with chromatin and interacts directly with HP1a. Genes Dev. 2007;21:2300–11. doi: 10.1101/gad.1564307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUA DJ, KUO AJ, CHEUNG P, LIU CL, MIGLIORI V, ESPEJO A, CASADIO F, BASSI C, AMATI B, BEDFORD MT, GUCCIONE E, GOZANI O. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BUSCAINO A, LEGUBE G, AKHTAR A. X-chromosome targeting and dosage compensation are mediated by distinct domains in MSL-3. EMBO Rep. 2006;7:531–8. doi: 10.1038/sj.embor.7400658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAI Y, JIN J, TOMOMORI-SATO C, SATO S, SOROKINA I, PARMELY TJ, CONAWAY RC, CONAWAY JW. Identification of new subunits of the multiprotein mammalian TRRAP/TIP60-containing histone acetyltransferase complex. J Biol Chem. 2003;278:42733–6. doi: 10.1074/jbc.C300389200. [DOI] [PubMed] [Google Scholar]
- CANZIO D, CHANG EY, SHANKAR S, KUCHENBECKER KM, SIMON MD, MADHANI HD, NARLIKAR GJ, AL-SADY B. Chromodomain-mediated oligomerization of HP1 suggests a nucleosome-bridging mechanism for heterochromatin assembly. Mol Cell. 2011;41:67–81. doi: 10.1016/j.molcel.2010.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CAO R, WANG L, WANG H, XIA L, ERDJUMENT-BROMAGE H, TEMPST P, JONES RS, ZHANG Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43. doi: 10.1126/science.1076997. [DOI] [PubMed] [Google Scholar]
- CHALKER DL. Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochim Biophys Acta. 2008;1783:2130–6. doi: 10.1016/j.bbamcr.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHANG Y, HORTON JR, BEDFORD MT, ZHANG X, CHENG X. Structural insights for MPP8 chromodomain interaction with histone H3 lysine 9: potential effect of phosphorylation on methyl-lysine binding. J Mol Biol. 2011;408:807–14. doi: 10.1016/j.jmb.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHEN ES, ZHANG K, NICOLAS E, CAM HP, ZOFALL M, GREWAL SI. Cell cycle control of centromeric repeat transcription and heterochromatin assembly. Nature. 2008;451:734–7. doi: 10.1038/nature06561. [DOI] [PubMed] [Google Scholar]
- CHEN M, TOMINAGA K, PEREIRA-SMITH OM. Emerging role of the MORF/MRG gene family in various biological processes, including aging. Ann N Y Acad Sci. 2010;1197:134–41. doi: 10.1111/j.1749-6632.2010.05197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CHIN HG, PATNAIK D, ESTEVE PO, JACOBSEN SE, PRADHAN S. Catalytic properties and kinetic mechanism of human recombinant Lys-9 histone H3 methyltransferase SUV39H1: participation of the chromodomain in enzymatic catalysis. Biochemistry. 2006;45:3272–84. doi: 10.1021/bi051997r. [DOI] [PubMed] [Google Scholar]
- CHODAVARAPU RK, FENG S, BERNATAVICHUTE YV, CHEN PY, STROUD H, YU Y, HETZEL JA, KUO F, KIM J, COKUS SJ, CASERO D, BERNAL M, HUIJSER P, CLARK AT, KRAMER U, MERCHANT SS, ZHANG X, JACOBSEN SE, PELLEGRINI M. Relationship between nucleosome positioning and DNA methylation. Nature. 2010;466:388–92. doi: 10.1038/nature09147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- CONSORTIUM IHGS. Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–45. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- COWIESON NP, PARTRIDGE JF, ALLSHIRE RC, MCLAUGHLIN PJ. Dimerisation of a chromo shadow domain and distinctions from the chromodomain as revealed by structural analysis. Curr Biol. 2000;10:517–25. doi: 10.1016/s0960-9822(00)00467-x. [DOI] [PubMed] [Google Scholar]
- CUI H, BENFEY PN. Interplay between SCARECROW, GA and LIKE HETEROCHROMATIN PROTEIN 1 in ground tissue patterning in the Arabidopsis root. Plant J. 2009;58:1016–27. doi: 10.1111/j.1365-313X.2009.03839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAI J, SULTAN S, TAYLOR SS, HIGGINS JM. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 2005;19:472–88. doi: 10.1101/gad.1267105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DAUJAT S, ZEISSLER U, WALDMANN T, HAPPEL N, SCHNEIDER R. HP1 binds specifically to Lys26-methylated histone H1.4, whereas simultaneous Ser27 phosphorylation blocks HP1 binding. J Biol Chem. 2005;280:38090–5. doi: 10.1074/jbc.C500229200. [DOI] [PubMed] [Google Scholar]
- DAWSON MA, BANNISTER AJ, GOTTGENS B, FOSTER SD, BARTKE T, GREEN AR, KOUZARIDES T. JAK2 phosphorylates histone H3Y41 and excludes HP1alpha from chromatin. Nature. 2009;461:819–22. doi: 10.1038/nature08448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOYON Y, SELLECK W, LANE WS, TAN S, COTE J. Structural and functional conservation of the NuA4 histone acetyltransferase complex from yeast to humans. Mol Cell Biol. 2004;24:1884–96. doi: 10.1128/MCB.24.5.1884-1896.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- EISEN A, UTLEY RT, NOURANI A, ALLARD S, SCHMIDT P, LANE WS, LUCCHESI JC, COTE J. The yeast NuA4 and Drosophila MSL complexes contain homologous subunits important for transcription regulation. J Biol Chem. 2001;276:3484–91. doi: 10.1074/jbc.M008159200. [DOI] [PubMed] [Google Scholar]
- EKBLAD CM, CHAVALI GB, BASU BP, FREUND SM, VEPRINTSEV D, HUGHES-DAVIES L, KOUZARIDES T, DOHERTY AJ, ITZHAKI LS. Binding of EMSY to HP1beta: implications for recruitment of HP1beta and BS69. EMBO Rep. 2005;6:675–80. doi: 10.1038/sj.embor.7400415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ESKELAND R, CZERMIN B, BOEKE J, BONALDI T, REGULA JT, IMHOF A. The N-terminus of Drosophila SU(VAR)3–9 mediates dimerization and regulates its methyltransferase activity. Biochemistry. 2004;43:3740–9. doi: 10.1021/bi035964s. [DOI] [PubMed] [Google Scholar]
- FALK S, RAVAUD S, KOCH J, SINNING I. The C terminus of the Alb3 membrane insertase recruits cpSRP43 to the thylakoid membrane. J Biol Chem. 2010;285:5954–62. doi: 10.1074/jbc.M109.084996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FENG S, COKUS SJ, ZHANG X, CHEN PY, BOSTICK M, GOLL MG, HETZEL J, JAIN J, STRAUSS SH, HALPERN ME, UKOMADU C, SADLER KC, PRADHAN S, PELLEGRINI M, JACOBSEN SE. Conservation and divergence of methylation patterning in plants and animals. Proc Natl Acad Sci U S A. 2010a;107:8689–94. doi: 10.1073/pnas.1002720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FENG S, JACOBSEN SE, REIK W. Epigenetic reprogramming in plant and animal development. Science. 2010b;330:622–7. doi: 10.1126/science.1190614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FISCHLE W, FRANZ H, JACOBS SA, ALLIS CD, KHORASANIZADEH S. Specificity of the chromodomain Y chromosome family of chromodomains for lysinemethylated ARK(S/T) motifs. J Biol Chem. 2008;283:19626–35. doi: 10.1074/jbc.M802655200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FISCHLE W, TSENG BS, DORMANN HL, UEBERHEIDE BM, GARCIA BA, SHABANOWITZ J, HUNT DF, FUNABIKI H, ALLIS CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–22. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- FISCHLE W, WANG Y, ALLIS CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003a;425:475–9. doi: 10.1038/nature02017. [DOI] [PubMed] [Google Scholar]
- FISCHLE W, WANG Y, JACOBS SA, KIM Y, ALLIS CD, KHORASANIZADEH S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003b;17:1870–81. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLANAGAN JF, BLUS BJ, KIM D, CLINES KL, RASTINEJAD F, KHORASANIZADEH S. Molecular implications of evolutionary differences in CHD double chromodomains. J Mol Biol. 2007;369:334–42. doi: 10.1016/j.jmb.2007.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLANAGAN JF, MI LZ, CHRUSZCZ M, CYMBOROWSKI M, CLINES KL, KIM Y, MINOR W, RASTINEJAD F, KHORASANIZADEH S. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–5. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- FLAUS A, MARTIN DM, BARTON GJ, OWEN-HUGHES T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FLEISCHER TC, YUN UJ, AYER DE. Identification and characterization of three new components of the mSin3A corepressor complex. Mol Cell Biol. 2003;23:3456–67. doi: 10.1128/MCB.23.10.3456-3467.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FRANZ H, MOSCH K, SOEROES S, URLAUB H, FISCHLE W. Multimerization and H3K9me3 binding are required for CDYL1b heterochromatin association. J Biol Chem. 2009;284:35049–59. doi: 10.1074/jbc.M109.052332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAILLARD PH, MARTINI EM, KAUFMAN PD, STILLMAN B, MOUSTACCHI E, ALMOUZNI G. Chromatin assembly coupled to DNA repair: a new role for chromatin assembly factor I. Cell. 1996;86:887–96. doi: 10.1016/s0092-8674(00)80164-6. [DOI] [PubMed] [Google Scholar]
- GAO X, HOU Y, EBINA H, LEVIN HL, VOYTAS DF. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008;18:359–69. doi: 10.1101/gr.7146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GARSKE AL, OLIVER SS, WAGNER EK, MUSSELMAN CA, LEROY G, GARCIA BA, KUTATELADZE TG, DENU JM. Combinatorial profiling of chromatin binding modules reveals multisite discrimination. Nat Chem Biol. 2010;6:283–90. doi: 10.1038/nchembio.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GASPAR-MAIA A, ALAJEM A, POLESSO F, SRIDHARAN R, MASON MJ, HEIDERSBACH A, RAMALHO-SANTOS J, MCMANUS MT, PLATH K, MESHORER E, RAMALHO-SANTOS M. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature. 2009 doi: 10.1038/nature08212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAVIN AC, BOSCHE M, KRAUSE R, GRANDI P, MARZIOCH M, BAUER A, SCHULTZ J, RICK JM, MICHON AM, CRUCIAT CM, REMOR M, HOFERT C, SCHELDER M, BRAJENOVIC M, RUFFNER H, MERINO A, KLEIN K, HUDAK M, DICKSON D, RUDI T, GNAU V, BAUCH A, BASTUCK S, HUHSE B, LEUTWEIN C, HEURTIER MA, COPLEY RR, EDELMANN A, QUERFURTH E, RYBIN V, DREWES G, RAIDA M, BOUWMEESTER T, BORK P, SERAPHIN B, KUSTER B, NEUBAUER G, SUPERTI-FURGA G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–7. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- GOFORTH RL, PETERSON EC, YUAN J, MOORE MJ, KIGHT AD, LOHSE MB, SAKON J, HENRY RL. Regulation of the GTPase cycle in post-translational signal recognition particle-based protein targeting involves cpSRP43. J Biol Chem. 2004;279:43077–84. doi: 10.1074/jbc.M401600200. [DOI] [PubMed] [Google Scholar]
- GORINSEK B, GUBENSEK F, KORDIS D. Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol. 2004;21:781–98. doi: 10.1093/molbev/msh057. [DOI] [PubMed] [Google Scholar]
- GORINSEK B, GUBENSEK F, KORDIS D. Phylogenomic analysis of chromoviruses. Cytogenet Genome Res. 2005;110:543–52. doi: 10.1159/000084987. [DOI] [PubMed] [Google Scholar]
- GREWAL SI, ELGIN SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399–406. doi: 10.1038/nature05914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GREWAL SI, JIA S. Heterochromatin revisited. Nat Rev Genet. 2007;8:35–46. doi: 10.1038/nrg2008. [DOI] [PubMed] [Google Scholar]
- GUCCIONE E, BASSI C, CASADIO F, MARTINATO F, CESARONI M, SCHUCHLAUTZ H, LUSCHER B, AMATI B. Methylation of histone H3R2 by PRMT6 and H3K4 by an MLL complex are mutually exclusive. Nature. 2007;449:933–7. doi: 10.1038/nature06166. [DOI] [PubMed] [Google Scholar]
- GUO Y, NADY N, QI C, ALLALI-HASSANI A, ZHU H, PAN P, ADAMS-CIOABA MA, AMAYA MF, DONG A, VEDADI M, SCHAPIRA M, READ RJ, ARROWSMITH CH, MIN J. Methylation-state-specific recognition of histones by the MBT repeat protein L3MBTL2. Nucleic Acids Res. 2009;37:2204–10. doi: 10.1093/nar/gkp086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAMED RB, BATCHELAR ET, CLIFTON IJ, SCHOFIELD CJ. Mechanisms and structures of crotonase superfamily enzymes--how nature controls enolate and oxyanion reactivity. Cell Mol Life Sci. 2008;65:2507–27. doi: 10.1007/s00018-008-8082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HATANO A, MATSUMOTO M, HIGASHINAKAGAWA T, NAKAYAMA KI. Phosphorylation of the chromodomain changes the binding specificity of Cbx2 for methylated histone H3. Biochem Biophys Res Commun. 2010;397:93–9. doi: 10.1016/j.bbrc.2010.05.074. [DOI] [PubMed] [Google Scholar]
- HAUK G, MCKNIGHT JN, NODELMAN IM, BOWMAN GD. The chromodomains of the Chd1 chromatin remodeler regulate DNA access to the ATPase motor. Mol Cell. 2010;39:711–23. doi: 10.1016/j.molcel.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAYAKAWA T, ZHANG F, HAYAKAWA N, OHTANI Y, SHINMYOZU K, NAKAYAMA J, ANDREASSEN PR. MRG15 binds directly to PALB2 and stimulates homology-directed repair of chromosomal breaks. J Cell Sci. 2010;123:1124–30. doi: 10.1242/jcs.060178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HAYNES KA, SILVER PA. Synthetic reversal of epigenetic silencing. J Biol Chem. 2011;286:27176–82. doi: 10.1074/jbc.C111.229567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HEINTZMAN ND, STUART RK, HON G, FU Y, CHING CW, HAWKINS RD, BARRERA LO, VAN CALCAR S, QU C, CHING KA, WANG W, WENG Z, GREEN RD, CRAWFORD GE, REN B. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–8. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- HENDZEL MJ. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- HERMKES R, FUNKE S, RICHTER C, KUHLMANN J, SCHUNEMANN D. The alpha-helix of the second chromodomain of the 43 kDa subunit of the chloroplast signal recognition particle facilitates binding to the 54 kDa subunit. FEBS Lett. 2006;580:3107–11. doi: 10.1016/j.febslet.2006.04.055. [DOI] [PubMed] [Google Scholar]
- HILFIKER A, HILFIKER-KLEINER D, PANNUTI A, LUCCHESI JC. mof, a putative acetyl transferase gene related to the Tip60 and MOZ human genes and to the SAS genes of yeast, is required for dosage compensation in Drosophila. Embo J. 1997;16:2054–60. doi: 10.1093/emboj/16.8.2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HIRAGAMI-HAMADA K, SHINMYOZU K, HAMADA D, TATSU Y, UEGAKI K, FUJIWARA S, NAKAYAMA J. N-terminal phosphorylation of HP1{alpha} promotes its chromatin binding. Mol Cell Biol. 2011;31:1186–200. doi: 10.1128/MCB.01012-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HO L, CRABTREE GR. Chromatin remodelling during development. Nature. 2010;463:474–84. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HONDA S, SELKER EU. Direct interaction between DNA methyltransferase DIM-2 and HP1 is required for DNA methylation in Neurospora crassa. Mol Cell Biol. 2008;28:6044–55. doi: 10.1128/MCB.00823-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUANG J, BERGER SL. The emerging field of dynamic lysine methylation of nonhistone proteins. Curr Opin Genet Dev. 2008;18:152–8. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- HUANG Y, MYERS MP, XU RM. Crystal structure of the HP1-EMSY complex reveals an unusual mode of HP1 binding. Structure. 2006;14:703–12. doi: 10.1016/j.str.2006.01.007. [DOI] [PubMed] [Google Scholar]
- HUGHES-DAVIES L, HUNTSMAN D, RUAS M, FUKS F, BYE J, CHIN SF, MILNER J, BROWN LA, HSU F, GILKS B, NIELSEN T, SCHULZER M, CHIA S, RAGAZ J, CAHN A, LINGER L, OZDAG H, CATTANEO E, JORDANOVA ES, SCHUURING E, YU DS, VENKITARAMAN A, PONDER B, DOHERTY A, APARICIO S, BENTLEY D, THEILLET C, PONTING CP, CALDAS C, KOUZARIDES T. EMSY links the BRCA2 pathway to sporadic breast and ovarian cancer. Cell. 2003;115:523–35. doi: 10.1016/s0092-8674(03)00930-9. [DOI] [PubMed] [Google Scholar]
- HUGHES RM, WIGGINS KR, KHORASANIZADEH S, WATERS ML. Recognition of trimethyllysine by a chromodomain is not driven by the hydrophobic effect. Proc Natl Acad Sci U S A. 2007;104:11184–8. doi: 10.1073/pnas.0610850104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HUNTER S, APWEILER R, ATTWOOD TK, BAIROCH A, BATEMAN A, BINNS D, BORK P, DAS U, DAUGHERTY L, DUQUENNE L, FINN RD, GOUGH J, HAFT D, HULO N, KAHN D, KELLY E, LAUGRAUD A, LETUNIC I, LONSDALE D, LOPEZ R, MADERA M, MASLEN J, MCANULLA C, MCDOWALL J, MISTRY J, MITCHELL A, MULDER N, NATALE D, ORENGO C, QUINN AF, SELENGUT JD, SIGRIST CJ, THIMMA M, THOMAS PD, VALENTIN F, WILSON D, WU CH, YEATS C. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–5. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- HURD EA, POUCHER HK, CHENG K, RAPHAEL Y, MARTIN DM. The ATP-dependent chromatin remodeling enzyme CHD7 regulates pro-neural gene expression and neurogenesis in the inner ear. Development. 2010;137:3139–50. doi: 10.1242/dev.047894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JACOBS SA, FISCHLE W, KHORASANIZADEH S. Assays for the determination of structure and dynamics of the interaction of the chromodomain with histone peptides. Methods Enzymol. 2004;376:131–48. doi: 10.1016/S0076-6879(03)76009-1. [DOI] [PubMed] [Google Scholar]
- JACOBS SA, KHORASANIZADEH S. Structure of the HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- JACOBS SA, TAVERNA SD, ZHANG Y, BRIGGS SD, LI J, EISSENBERG JC, ALLIS CD, KHORASANIZADEH S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. Embo J. 2001;20:5232–41. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JAMES TC, ELGIN SC. Identification of a nonhistone chromosomal protein associated with heterochromatin in Drosophila melanogaster and its gene. Mol. Cell. Biol. 1986;6:3862–3872. doi: 10.1128/mcb.6.11.3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOHANSEN KM, JOHANSEN J. Regulation of chromatin structure by histone H3S10 phosphorylation. Chromosome Res. 2006;14:393–404. doi: 10.1007/s10577-006-1063-4. [DOI] [PubMed] [Google Scholar]
- JOPPICH C, SCHOLZ S, KORGE G, SCHWENDEMANN A. Umbrea, a chromo shadow domain protein in Drosophila melanogaster heterochromatin, interacts with Hip, HP1 and HOAP. Chromosome Res. 2009;17:19–36. doi: 10.1007/s10577-008-9002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JOSHI AA, STRUHL K. Eaf3 chromodomain interaction with methylated H3-K36 links histone deacetylation to Pol II elongation. Mol Cell. 2005;20:971–8. doi: 10.1016/j.molcel.2005.11.021. [DOI] [PubMed] [Google Scholar]
- KARACHENTSEV D, DRUZHININA M, STEWARD R. Free and chromatin-associated mono-, di-, and trimethylation of histone H4-lysine 20 during development and cell cycle progression. Dev Biol. 2007;304:46–52. doi: 10.1016/j.ydbio.2006.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KATO H, GRUSCHUS J, GHIRLANDO R, TJANDRA N, BAI Y. Characterization of the N-Terminal Tail Domain of Histone H3 in Condensed Nucleosome Arrays by Hydrogen Exchange and NMR. J Am Chem Soc. 2009 doi: 10.1021/ja9070078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUFMAN PD, KOBAYASHI R, KESSLER N, STILLMAN B. The p150 and p60 subunits of chromatin assembly factor I: a molecular link between newly synthesized histones and DNA replication. Cell. 1995;81:1105–14. doi: 10.1016/s0092-8674(05)80015-7. [DOI] [PubMed] [Google Scholar]
- KAUSTOV L, OUYANG H, AMAYA M, LEMAK A, NADY N, DUAN S, WASNEY GA, LI Z, VEDADI M, SCHAPIRA M, MIN J, ARROWSMITH CH. Recognition and specificity determinants of the human cbx chromodomains. J Biol Chem. 2011;286:521–9. doi: 10.1074/jbc.M110.191411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KHORASANIZADEH S. The nucleosome: from genomic organization to genomic regulation. Cell. 2004;116:259–72. doi: 10.1016/s0092-8674(04)00044-3. [DOI] [PubMed] [Google Scholar]
- KIM D, BLUS BJ, CHANDRA V, HUANG P, RASTINEJAD F, KHORASANIZADEH S. Corecognition of DNA and a methylated histone tail by the MSL3 chromodomain. Nat Struct Mol Biol. 2010;17:1027–9. doi: 10.1038/nsmb.1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KIM J, DANIEL J, ESPEJO A, LAKE A, KRISHNA M, XIA L, ZHANG Y, BEDFORD MT. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOKURA K, SUN L, BEDFORD MT, FANG J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. Embo J. 2010;29:3673–87. doi: 10.1038/emboj.2010.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KOLESOV G, VIRNAU P, KARDAR M, MIRNY LA. Protein knot server: detection of knots in protein structures. Nucleic Acids Res. 2007;35:W425–8. doi: 10.1093/nar/gkm312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KORDIS D. A genomic perspective on the chromodomain-containing retrotransposons: Chromoviruses. Gene. 2005;347:161–73. doi: 10.1016/j.gene.2004.12.017. [DOI] [PubMed] [Google Scholar]
- KOUZMINOVA E, SELKER EU. dim-2 encodes a DNA methyltransferase responsible for all known cytosine methylation in Neurospora. Embo J. 2001;20:4309–23. doi: 10.1093/emboj/20.15.4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KUNERT N, BREHM A. Novel Mi-2 related ATP-dependent chromatin remodelers. Epigenetics. 2009;4:209–11. doi: 10.4161/epi.8933. [DOI] [PubMed] [Google Scholar]
- LAHN BT, PAGE DC. Functional coherence of the human Y chromosome. Science. 1997;278:675–80. doi: 10.1126/science.278.5338.675. [DOI] [PubMed] [Google Scholar]
- LAHN BT, TANG ZL, ZHOU J, BARNDT RJ, PARVINEN M, ALLIS CD, PAGE DC. Previously uncharacterized histone acetyltransferases implicated in mammalian spermatogenesis. Proc Natl Acad Sci U S A. 2002;99:8707–12. doi: 10.1073/pnas.082248899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI A, KENNEDY BK, BARBIE DA, BERTOS NR, YANG XJ, THEBERGE MC, TSAI SC, SETO E, ZHANG Y, KUZMICHEV A, LANE WS, REINBERG D, HARLOW E, BRANTON PE. RBP1 recruits the mSIN3-histone deacetylase complex to the pocket of retinoblastoma tumor suppressor family proteins found in limited discrete regions of the nucleus at growth arrest. Mol Cell Biol. 2001;21:2918–32. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI A, LEE JM, YANG WM, DECAPRIO JA, KAELIN WG, JR., SETO E, BRANTON PE. RBP1 recruits both histone deacetylase-dependent and -independent repression activities to retinoblastoma family proteins. Mol Cell Biol. 1999a;19:6632–41. doi: 10.1128/mcb.19.10.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LAI A, MARCELLUS RC, CORBEIL HB, BRANTON PE. RBP1 induces growth arrest by repression of E2F-dependent transcription. Oncogene. 1999b;18:2091–100. doi: 10.1038/sj.onc.1202520. [DOI] [PubMed] [Google Scholar]
- LAN F, SHI Y. Epigenetic regulation: methylation of histone and non-histone proteins. Sci China C Life Sci. 2009;52:311–22. doi: 10.1007/s11427-009-0054-z. [DOI] [PubMed] [Google Scholar]
- LATRASSE D, GERMANN S, HOUBA-HERIN N, DUBOIS E, BUI-PRODHOMME D, HOURCADE D, JUUL-JENSEN T, LE ROUX C, MAJIRA A, SIMONCELLO N, GRANIER F, TACONNAT L, RENOU JP, GAUDIN V. Control of flowering and cell fate by LIF2, an RNA binding partner of the polycomb complex component LHP1. PLoS One. 2011;6:e16592. doi: 10.1371/journal.pone.0016592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LECHNER MS, SCHULTZ DC, NEGOREV D, MAUL GG, RAUSCHER FJ., 3RD The mammalian heterochromatin protein 1 binds diverse nuclear proteins through a common motif that targets the chromoshadow domain. Biochem Biophys Res Commun. 2005;331:929–37. doi: 10.1016/j.bbrc.2005.04.016. [DOI] [PubMed] [Google Scholar]
- LEE MG, VILLA R, TROJER P, NORMAN J, YAN KP, REINBERG D, CROCE LD, SHIEKHATTAR R. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science. 2007;318:447–50. doi: 10.1126/science.1149042. [DOI] [PubMed] [Google Scholar]
- LETUNIC I, DOERKS T, BORK P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–32. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LEWIS NE, MARTY NJ, KATHIR KM, RAJALINGAM D, KIGHT AD, DAILY A, KUMAR TK, HENRY RL, GOFORTH RL. A dynamic cpSRP43-Albino3 interaction mediates translocase regulation of chloroplast signal recognition particle (cpSRP)-targeting components. J Biol Chem. 2010;285:34220–30. doi: 10.1074/jbc.M110.160093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI B, GOGOL M, CAREY M, LEE D, SEIDEL C, WORKMAN JL. Combined action of PHD and chromo domains directs the Rpd3S HDAC to transcribed chromatin. Science. 2007;316:1050–4. doi: 10.1126/science.1139004. [DOI] [PubMed] [Google Scholar]
- LI G, MARGUERON R, HU G, STOKES D, WANG YH, REINBERG D. Highly compacted chromatin formed in vitro reflects the dynamics of transcription activation in vivo. Mol Cell. 2010a;38:41–53. doi: 10.1016/j.molcel.2010.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LI X, CORSA CA, PAN PW, WU L, FERGUSON D, YU X, MIN J, DOU Y. MOF and H4 K16 acetylation play important roles in DNA damage repair by modulating recruitment of DNA damage repair protein Mdc1. Mol Cell Biol. 2010b;30:5335–47. doi: 10.1128/MCB.00350-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIANG G, LIN JC, WEI V, YOO C, CHENG JC, NGUYEN CT, WEISENBERGER DJ, EGGER G, TAKAI D, GONZALES FA, JONES PA. Distinct localization of histone H3 acetylation and H3-K4 methylation to the transcription start sites in the human genome. Proc Natl Acad Sci U S A. 2004;101:7357–62. doi: 10.1073/pnas.0401866101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIN CH, LI B, SWANSON S, ZHANG Y, FLORENS L, WASHBURN MP, ABMAYR SM, WORKMAN JL. Heterochromatin protein 1a stimulates histone H3 lysine 36 demethylation by the Drosophila KDM4A demethylase. Mol Cell. 2008;32:696–706. doi: 10.1016/j.molcel.2008.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LINDROTH AM, SHULTIS D, JASENCAKOVA Z, FUCHS J, JOHNSON L, SCHUBERT D, PATNAIK D, PRADHAN S, GOODRICH J, SCHUBERT I, JENUWEIN T, KHORASANIZADEH S, JACOBSEN SE. Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J. 2004;23:4146–55. doi: 10.1038/sj.emboj.7600430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU C, XI W, SHEN L, TAN C, YU H. Regulation of floral patterning by flowering time genes. Dev Cell. 2009;16:711–22. doi: 10.1016/j.devcel.2009.03.011. [DOI] [PubMed] [Google Scholar]
- LIU H, WANG JY, HUANG Y, LI Z, GONG W, LEHMANN R, XU RM. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev. 2010;24:1876–81. doi: 10.1101/gad.1956010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LIU Y, TAVERNA SD, MURATORE TL, SHABANOWITZ J, HUNT DF, ALLIS CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–45. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUCCHESI JC. The structure-function link of compensated chromatin in Drosophila. Curr Opin Genet Dev. 2009;19:550–6. doi: 10.1016/j.gde.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUCO RF, ALLO M, SCHOR IE, KORNBLIHTT AR, MISTELI T. Epigenetics in Alternative Pre-mRNA Splicing. Cell. 2011;144:16–26. doi: 10.1016/j.cell.2010.11.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUCO RF, PAN Q, TOMINAGA K, BLENCOWE BJ, PEREIRA-SMITH OM, MISTELI T. Regulation of alternative splicing by histone modifications. Science. 2010;327:996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUGER K, MAEDER AW, RICHMOND RK, SARGENT DF, RICHMOND TJ. X-ray structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–259. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- LUTZ T, STOGER R, NIETO A. CHD6 is a DNA-dependent ATPase and localizes at nuclear sites of mRNA synthesis. FEBS Lett. 2006;580:5851–7. doi: 10.1016/j.febslet.2006.09.049. [DOI] [PubMed] [Google Scholar]
- MANSFIELD RE, MUSSELMAN CA, KWAN AH, GARSKE AL, DAVRAZOU F, DENU JM, KUTATELADZE TG, MACKAY JP. The plant homeodomain (PHD) fingers of CHD4 are histone H3-binding modules with preference for unmodified H3K4 and methylated H3K9. J Biol Chem. 2011;286:11779–91. doi: 10.1074/jbc.M110.208207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MATSUMOTO-TANIURA N, PIROLLET F, MONROE R, GERACE L, WESTENDORF JM. Identification of novel M phase phosphoproteins by expression cloning. Mol Biol Cell. 1996;7:1455–69. doi: 10.1091/mbc.7.9.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAURER-STROH S, DICKENS NJ, HUGHES-DAVIES L, KOUZARIDES T, EISENHABER F, PONTING CP. The Tudor domain `Royal Family': Tudor, plant Agenet, Chromo, PWWP and MBT domains. Trends Biochem Sci. 2003;28:69–74. doi: 10.1016/S0968-0004(03)00004-5. [DOI] [PubMed] [Google Scholar]
- MEEHAN RR, KAO CF, PENNINGS S. HP1 binding to native chromatin in vitro is determined by the hinge region and not by the chromodomain. Embo J. 2003;22:3164–74. doi: 10.1093/emboj/cdg306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MENDEZ DL, KIM D, CHRUSZCZ M, STEPHENS GE, MINOR W, KHORASANIZADEH S, ELGIN SC. The HP1a Disordered C Terminus and Chromo Shadow Domain Cooperate to Select Target Peptide Partners. Chembiochem. 2011;12:1084–96. doi: 10.1002/cbic.201000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MIKKELSEN TS, KU M, JAFFE DB, ISSAC B, LIEBERMAN E, GIANNOUKOS G, ALVAREZ P, BROCKMAN W, KIM TK, KOCHE RP, LEE W, MENDENHALL E, O'DONOVAN A, PRESSER A, RUSS C, XIE X, MEISSNER A, WERNIG M, JAENISCH R, NUSBAUM C, LANDER ES, BERNSTEIN BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–60. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MILLAR CB, GRUNSTEIN M. Genome-wide patterns of histone modifications in yeast. Nat Rev Mol Cell Biol. 2006;7:657–66. doi: 10.1038/nrm1986. [DOI] [PubMed] [Google Scholar]
- MIN J, ALLALI-HASSANI A, NADY N, QI C, OUYANG H, LIU Y, MACKENZIE F, VEDADI M, ARROWSMITH CH. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14:1229–30. doi: 10.1038/nsmb1340. [DOI] [PubMed] [Google Scholar]
- MIN J, ZHANG Y, XU RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–8. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MONROE DG, HAWSE JR, SUBRAMANIAM M, SPELSBERG TC. Retinoblastoma binding protein-1 (RBP1) is a Runx2 coactivator and promotes osteoblastic differentiation. BMC Musculoskelet Disord. 2010;11:104. doi: 10.1186/1471-2474-11-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOORE SA, FERHATOGLU Y, JIA Y, AL-JIAB RA, SCOTT MJ. Structural and biochemical studies on the chromo-barrel domain of male specific lethal 3 (MSL3) reveal a binding preference for mono- or dimethyllysine 20 on histone H4. J Biol Chem. 2011;285:40879–90. doi: 10.1074/jbc.M110.134312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MORETTINI S, TRIBUS M, ZEILNER A, SEBALD J, CAMPO-FERNANDEZ B, SCHERAN G, WORLE H, PODHRASKI V, FYODOROV DV, LUSSER A. The chromodomains of CHD1 are critical for enzymatic activity but less important for chromatin localization. Nucleic Acids Res. 2011;39:3103–15. doi: 10.1093/nar/gkq1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MOTAMEDI MR, HONG EJ, LI X, GERBER S, DENISON C, GYGI S, MOAZED D. HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell. 2008;32:778–90. doi: 10.1016/j.molcel.2008.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MURZINA N, VERREAULT A, LAUE E, STILLMAN B. Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell. 1999;4:529–40. doi: 10.1016/s1097-2765(00)80204-x. [DOI] [PubMed] [Google Scholar]
- MUSSELMAN CA, MANSFIELD RE, GARSKE AL, DAVRAZOU F, KWAN AH, OLIVER SS, O'LEARY H, DENU JM, MACKAY JP, KUTATELADZE TG. Binding of the CHD4 PHD2 finger to histone H3 is modulated by covalent modifications. Biochem J. 2009;423:179–87. doi: 10.1042/BJ20090870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAKAYAMA J, RICE JC, STRAHL BD, ALLIS CD, GREWAL SI. Role of histone H3 lysine 9 methylation in epigenetic control of heterochromatin assembly. Science. 2001;292:110–3. doi: 10.1126/science.1060118. [DOI] [PubMed] [Google Scholar]
- NAKAYAMA J, XIAO G, NOMA K, MALIKZAY A, BJERLING P, EKWALL K, KOBAYASHI R, GREWAL SI. Alp13, an MRG family protein, is a component of fission yeast Clr6 histone deacetylase required for genomic integrity. Embo J. 2003;22:2776–87. doi: 10.1093/emboj/cdg248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NEAL KC, PANNUTI A, SMITH ER, LUCCHESI JC. A new human member of the MYST family of histone acetyl transferases with high sequence similarity to Drosophila MOF. Biochim Biophys Acta. 2000;1490:170–4. doi: 10.1016/s0167-4781(99)00211-0. [DOI] [PubMed] [Google Scholar]
- NIELSEN PR, NIETLISPACH D, BUSCAINO A, WARNER RJ, AKHTAR A, MURZIN AG, MURZINA NV, LAUE ED. Structure of the chromo barrel domain from the MOF acetyltransferase. J Biol Chem. 2005;280:32326–31. doi: 10.1074/jbc.M501347200. [DOI] [PubMed] [Google Scholar]
- NIELSEN PR, NIETLISPACH D, MOTT HR, CALLAGHAN J, BANNISTER A, KOUZARIDES T, MURZIN AG, MURZINA NV, LAUE ED. Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature. 2002;416:103–7. doi: 10.1038/nature722. [DOI] [PubMed] [Google Scholar]
- NIESSEN HE, DEMMERS JA, VONCKEN JW. Talking to chromatin: post-translational modulation of polycomb group function. Epigenetics Chromatin. 2009;2:10. doi: 10.1186/1756-8935-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- OKUDA M, HORIKOSHI M, NISHIMURA Y. Structural Polymorphism of Chromodomains in Chd1. J Mol Biol. 2007;365:1047. doi: 10.1016/j.jmb.2006.10.039. [DOI] [PubMed] [Google Scholar]
- PAPA CM, SPRINGER NM, MUSZYNSKI MG, MEELEY R, KAEPPLER SM. Maize chromomethylase Zea methyltransferase2 is required for CpNpG methylation. Plant Cell. 2001;13:1919–28. doi: 10.1105/TPC.010064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARO R. Imprinting a determined state into the chromatin of Drosophila. Trends Genet. 1990;6:416–21. doi: 10.1016/0168-9525(90)90303-n. [DOI] [PubMed] [Google Scholar]
- PARO R, HOGNESS DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc. Natl. Acad. Sci. USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PENA AN, PEREIRA-SMITH OM. The role of the MORF/MRG family of genes in cell growth, differentiation, DNA repair, and thereby aging. Ann N Y Acad Sci. 2007;1100:299–305. doi: 10.1196/annals.1395.031. [DOI] [PubMed] [Google Scholar]
- PETERS AH, KUBICEK S, MECHTLER K, O'SULLIVAN RJ, DERIJCK AA, PEREZ-BURGOS L, KOHLMAIER A, OPRAVIL S, TACHIBANA M, SHINKAI Y, MARTENS JH, JENUWEIN T. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–89. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- PHELAN ML, SIF S, NARLIKAR GJ, KINGSTON RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol Cell. 1999;3:247–53. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- QUINN AM, BEDFORD MT, ESPEJO A, SPANNHOFF A, AUSTIN CP, OPPERMANN U, SIMEONOV A. A homogeneous method for investigation of methylation-dependent protein-protein interactions in epigenetics. Nucleic Acids Res. 2010;38:e11. doi: 10.1093/nar/gkp899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAMOS A, HOLLINGWORTH D, ADINOLFI S, CASTETS M, KELLY G, FRENKIEL TA, BARDONI B, PASTORE A. The structure of the N-terminal domain of the fragile X mental retardation protein: a platform for protein-protein interaction. Structure. 2006;14:21–31. doi: 10.1016/j.str.2005.09.018. [DOI] [PubMed] [Google Scholar]
- REA S, EISENHABER F, O'CARROLL D, STRAHL BD, SUN ZW, SCHMID M, OPRAVIL S, MECHTLER K, PONTING CP, ALLIS CD, JENUWEIN T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–9. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- REID JL, MOQTADERI Z, STRUHL K. Eaf3 regulates the global pattern of histone acetylation in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:757–64. doi: 10.1128/MCB.24.2.757-764.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROBINSON H, GAO YG, MCCRARY BS, EDMONDSON SP, SHRIVER JW, WANG AH. The hyperthermophile chromosomal protein Sac7d sharply kinks DNA. Nature. 1998;392:202–5. doi: 10.1038/32455. [DOI] [PubMed] [Google Scholar]
- RODERICK SL, CHAN WW, AGATE DS, OLSEN LR, VETTING MW, RAJASHANKAR KR, COHEN DE. Structure of human phosphatidylcholine transfer protein in complex with its ligand. Nat Struct Biol. 2002;9:507–11. doi: 10.1038/nsb812. [DOI] [PubMed] [Google Scholar]
- RODRIGUEZ-PAREDES M, CEBALLOS-CHAVEZ M, ESTELLER M, GARCIA-DOMINGUEZ M, REYES JC. The chromatin remodeling factor CHD8 interacts with elongating RNA polymerase II and controls expression of the cyclin E2 gene. Nucleic Acids Res. 2009;37:2449–60. doi: 10.1093/nar/gkp101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ROSASCO-NITCHER SE, LAN W, KHORASANIZADEH S, STUKENBERG PT. Centromeric Aurora-B activation requires TD-60, microtubules, and substrate priming phosphorylation. Science. 2008;319:469–72. doi: 10.1126/science.1148980. [DOI] [PubMed] [Google Scholar]
- SADAIE M, KAWAGUCHI R, OHTANI Y, ARISAKA F, TANAKA K, SHIRAHIGE K, NAKAYAMA J. Balance between distinct HP1 family proteins controls heterochromatin assembly in fission yeast. Mol Cell Biol. 2008;28:6973–88. doi: 10.1128/MCB.00791-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SAMPATH SC, MARAZZI I, YAP KL, SAMPATH SC, KRUTCHINSKY AN, MECKLENBRAUKER I, VIALE A, RUDENSKY E, ZHOU MM, CHAIT BT, TARAKHOVSKY A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- SANTOS-ROSA H, SCHNEIDER R, BANNISTER AJ, SHERRIFF J, BERNSTEIN BE, EMRE NC, SCHREIBER SL, MELLOR J, KOUZARIDES T. Active genes are tri-methylated at K4 of histone H3. Nature. 2002;419:407–11. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- SAPOUNTZI V, COTE J. MYST-family histone acetyltransferases: beyond chromatin. Cell Mol Life Sci. 2010;68:1147–56. doi: 10.1007/s00018-010-0599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHALCH T, DUDA S, SARGENT DF, RICHMOND TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–41. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- SCHALCH T, JOB G, NOFFSINGER VJ, SHANKER S, KUSCU C, JOSHUA-TOR L, PARTRIDGE JF. High-affinity binding of Chp1 chromodomain to K9 methylated histone H3 is required to establish centromeric heterochromatin. Mol Cell. 2009;34:36–46. doi: 10.1016/j.molcel.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHANIEL C, ANG YS, RATNAKUMAR K, CORMIER C, JAMES T, BERNSTEIN E, LEMISCHKA IR, PADDISON PJ. Smarcc1/Baf155 couples self-renewal gene repression with changes in chromatin structure in mouse embryonic stem cells. Stem Cells. 2009;27:2979–91. doi: 10.1002/stem.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHMUTZ J, WHEELER J, GRIMWOOD J, DICKSON M, YANG J, CAOILE C, BAJOREK E, BLACK S, CHAN YM, DENYS M, ESCOBAR J, FLOWERS D, FOTOPULOS D, GARCIA C, GOMEZ M, GONZALES E, HAYDU L, LOPEZ F, RAMIREZ L, RETTERER J, RODRIGUEZ A, ROGERS S, SALAZAR A, TSAI M, MYERS RM. Quality assessment of the human genome sequence. Nature. 2004;429:365–8. doi: 10.1038/nature02390. [DOI] [PubMed] [Google Scholar]
- SCHNEIDER R, BANNISTER AJ, MYERS FA, THORNE AW, CRANE-ROBINSON C, KOUZARIDES T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004;6:73–7. doi: 10.1038/ncb1076. [DOI] [PubMed] [Google Scholar]
- SCHNETZ MP, BARTELS CF, SHASTRI K, BALASUBRAMANIAN D, ZENTNER GE, BALAJI R, ZHANG X, SONG L, WANG Z, LAFRAMBOISE T, CRAWFORD GE, SCACHERI PC. Genomic distribution of CHD7 on chromatin tracks H3K4 methylation patterns. Genome Res. 2009;19:590–601. doi: 10.1101/gr.086983.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHUENEMANN D, GUPTA S, PERSELLO-CARTIEAUX F, KLIMYUK VI, JONES JD, NUSSAUME L, HOFFMAN NE. A novel signal recognition particle targets light-harvesting proteins to the thylakoid membranes. Proc Natl Acad Sci U S A. 1998;95:10312–6. doi: 10.1073/pnas.95.17.10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHARMA GG, SO S, GUPTA A, KUMAR R, CAYROU C, AVVAKUMOV N, BHADRA U, PANDITA RK, PORTEUS MH, CHEN DJ, COTE J, PANDITA TK. MOF and histone H4 acetylation at lysine 16 are critical for DNA damage response and double-strand break repair. Mol Cell Biol. 2010;30:3582–95. doi: 10.1128/MCB.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHARMA R, MOHAN SINGH RK, MALIK G, DEVESHWAR P, TYAGI AK, KAPOOR S, KAPOOR M. Rice cytosine DNA methyltransferases - gene expression profiling during reproductive development and abiotic stress. Febs J. 2009;276:6301–11. doi: 10.1111/j.1742-4658.2009.07338.x. [DOI] [PubMed] [Google Scholar]
- SHI X, HONG T, WALTER KL, EWALT M, MICHISHITA E, HUNG T, CARNEY D, PENA P, LAN F, KAADIGE MR, LACOSTE N, CAYROU C, DAVRAZOU F, SAHA A, CAIRNS BR, AYER DE, KUTATELADZE TG, SHI Y, COTE J, CHUA KF, GOZANI O. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–9. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SHIMOJO H, SANO N, MORIWAKI Y, OKUDA M, HORIKOSHI M, NISHIMURA Y. Novel structural and functional mode of a knot essential for RNA binding activity of the Esa1 presumed chromodomain. J Mol Biol. 2008;378:987–1001. doi: 10.1016/j.jmb.2008.03.021. [DOI] [PubMed] [Google Scholar]
- SHUR I, SOCHER R, BENAYAHU D. In vivo association of CReMM/CHD9 with promoters in osteogenic cells. J Cell Physiol. 2006a;207:374–8. doi: 10.1002/jcp.20586. [DOI] [PubMed] [Google Scholar]
- SHUR I, SOLOMON R, BENAYAHU D. Dynamic interactions of chromatin-related mesenchymal modulator, a chromodomain helicase-DNA-binding protein, with promoters in osteoprogenitors. Stem Cells. 2006b;24:1288–93. doi: 10.1634/stemcells.2005-0300. [DOI] [PubMed] [Google Scholar]
- SIMIC R, LINDSTROM DL, TRAN HG, ROINICK KL, COSTA PJ, JOHNSON AD, HARTZOG GA, ARNDT KM. Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 2003;22:1846–56. doi: 10.1093/emboj/cdg179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMON JA, KINGSTON RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- SIMON MD, CHU F, RACKI LR, DE LA CRUZ CC, BURLINGAME AL, PANNING B, NARLIKAR GJ, SHOKAT KM. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell. 2007;128:1003–12. doi: 10.1016/j.cell.2006.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMS RJ, 3RD, CHEN CF, SANTOS-ROSA H, KOUZARIDES T, PATEL SS, REINBERG D. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem. 2005;280:41789–92. doi: 10.1074/jbc.C500395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMS RJ, 3RD, MILLHOUSE S, CHEN CF, LEWIS BA, ERDJUMENT-BROMAGE H, TEMPST P, MANLEY JL, REINBERG D. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell. 2007;28:665–76. doi: 10.1016/j.molcel.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIMS RJ, 3RD, ROJAS LA, BECK D, BONASIO R, SCHULLER R, DRURY WJ, 3RD, EICK D, REINBERG D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SIVARAJA V, KUMAR TK, LEENA PS, CHANG AN, VIDYA C, GOFORTH RL, RAJALINGAM D, ARVIND K, YE JL, CHOU J, HENRY R, YU C. Three-dimensional solution structures of the chromodomains of cpSRP43. J Biol Chem. 2005;280:41465–71. doi: 10.1074/jbc.M507077200. [DOI] [PubMed] [Google Scholar]
- SMALLWOOD A, ESTEVE PO, PRADHAN S, CAREY M. Functional cooperation between HP1 and DNMT1 mediates gene silencing. Genes Dev. 2007;21:1169–78. doi: 10.1101/gad.1536807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SMITH S, STILLMAN B. Purification and characterization of CAF-I, a human cell factor required for chromatin assembly during DNA replication in vitro. Cell. 1989;58:15–25. doi: 10.1016/0092-8674(89)90398-x. [DOI] [PubMed] [Google Scholar]
- SMOTHERS JF, HENIKOFF S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- SMOTHERS JF, HENIKOFF S. The hinge and chromo shadow domain impart distinct targeting of HP1-like proteins. Mol Cell Biol. 2001;21:2555–69. doi: 10.1128/MCB.21.7.2555-2569.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SRINIVASAN S, ARMSTRONG JA, DEURING R, DAHLSVEEN IK, MCNEILL H, TAMKUN JW. The Drosophila trithorax group protein Kismet facilitates an early step in transcriptional elongation by RNA Polymerase II. Development. 2005;132:1623–35. doi: 10.1242/dev.01713. [DOI] [PubMed] [Google Scholar]
- STRAHL BD, ALLIS CD. The language of covalent histone modifications. Nature. 2000;403:41–5. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- STRAUB T, BECKER PB. Dosage compensation: the beginning and end of generalization. Nat Rev Genet. 2007;8:47–57. doi: 10.1038/nrg2013. [DOI] [PubMed] [Google Scholar]
- SUN B, HONG J, ZHANG P, DONG X, SHEN X, LIN D, DING J. Molecular basis of the interaction of Saccharomyces cerevisiae Eaf3 chromo domain with methylated H3K36. J Biol Chem. 2008;283:36504–12. doi: 10.1074/jbc.M806564200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SUN Y, JIANG X, XU Y, AYRAPETOV MK, MOREAU LA, WHETSTINE JR, PRICE BD. Histone H3 methylation links DNA damage detection to activation of the tumour suppressor Tip60. Nat Cell Biol. 2009;11:1376–82. doi: 10.1038/ncb1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SURAL TH, PENG S, LI B, WORKMAN JL, PARK PJ, KURODA MI. The MSL3 chromodomain directs a key targeting step for dosage compensation of the Drosophila melanogaster X chromosome. Nat Struct Mol Biol. 2008;15:1318–25. doi: 10.1038/nsmb.1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SURAPUREDDI S, VISWAKARMA N, YU S, GUO D, RAO MS, REDDY JK. PRIC320, a transcription coactivator, isolated from peroxisome proliferator-binding protein complex. Biochem Biophys Res Commun. 2006;343:535–43. doi: 10.1016/j.bbrc.2006.02.160. [DOI] [PubMed] [Google Scholar]
- TAKADA I, MIHARA M, SUZAWA M, OHTAKE F, KOBAYASHI S, IGARASHI M, YOUN MY, TAKEYAMA K, NAKAMURA T, MEZAKI Y, TAKEZAWA S, YOGIASHI Y, KITAGAWA H, YAMADA G, TAKADA S, MINAMI Y, SHIBUYA H, MATSUMOTO K, KATO S. A histone lysine methyltransferase activated by non-canonical Wnt signalling suppresses PPAR-gamma transactivation. Nat Cell Biol. 2007;9:1273–85. doi: 10.1038/ncb1647. [DOI] [PubMed] [Google Scholar]
- TAMARU H, SELKER EU. A histone H3 methyltransferase controls DNA methylation in Neurospora crassa. Nature. 2001;414:277–283. doi: 10.1038/35104508. [DOI] [PubMed] [Google Scholar]
- TAVERNA SD, COYNE RS, ALLIS CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell. 2002;110:701–11. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- TAVERNA SD, LI H, RUTHENBURG AJ, ALLIS CD, PATEL DJ. How chromatin-binding modules interpret histone modifications: lessons from professional pocket pickers. Nat Struct Mol Biol. 2007;14:1025–40. doi: 10.1038/nsmb1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- THIRU A, NIETLISPACH D, MOTT HR, OKUWAKI M, LYON D, NIELSEN PR, HIRSHBERG M, VERREAULT A, MURZINA NV, LAUE ED. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. Embo J. 2004;23:489–99. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VAGIN VV, SIGOVA A, LI C, SEITZ H, GVOZDEV V, ZAMORE PD. A distinct small RNA pathway silences selfish genetic elements in the germline. Science. 2006;313:320–4. doi: 10.1126/science.1129333. [DOI] [PubMed] [Google Scholar]
- VAKOC CR, SACHDEVA MM, WANG H, BLOBEL GA. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol Cell Biol. 2006;26:9185–95. doi: 10.1128/MCB.01529-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VARIER RA, OUTCHKOUROV NS, DE GRAAF P, VAN SCHAIK FM, ENSING HJ, WANG F, HIGGINS JM, KOPS GJ, TIMMERS HT. A phospho/methyl switch at histone H3 regulates TFIID association with mitotic chromosomes. Embo J. 29:3967–78. doi: 10.1038/emboj.2010.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VERMAAK D, HENIKOFF S, MALIK HS. Positive selection drives the evolution of rhino, a member of the heterochromatin protein 1 family in Drosophila. PLoS Genet. 2005;1:96–108. doi: 10.1371/journal.pgen.0010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VEZZOLI A, BONADIES N, ALLEN MD, FREUND SM, SANTIVERI CM, KVINLAUG BT, HUNTLY BJ, GOTTGENS B, BYCROFT M. Molecular basis of histone H3K36me3 recognition by the PWWP domain of Brpf1. Nat Struct Mol Biol. 2010;17:617–9. doi: 10.1038/nsmb.1797. [DOI] [PubMed] [Google Scholar]
- WANG Z, ZANG C, ROSENFELD JA, SCHONES DE, BARSKI A, CUDDAPAH S, CUI K, ROH TY, PENG W, ZHANG MQ, ZHAO K. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WEI Y, YU L, BOWEN J, GOROVSKY MA, ALLIS CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- WOODAGE T, BASRAI MA, BAXEVANIS AD, HIETER P, COLLINS FS. Characterization of the CHD family of proteins. Proc Natl Acad Sci U S A. 1997;94:11472–7. doi: 10.1073/pnas.94.21.11472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU MY, ELDIN KW, BEAUDET AL. Identification of chromatin remodeling genes Arid4a and Arid4b as leukemia suppressor genes. J Natl Cancer Inst. 2008;100:1247–59. doi: 10.1093/jnci/djn253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WU MY, TSAI TF, BEAUDET AL. Deficiency of Rbbp1/Arid4a and Rbbp1l1/Arid4b alters epigenetic modifications and suppresses an imprinting defect in the PWS/AS domain. Genes Dev. 2006;20:2859–70. doi: 10.1101/gad.1452206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XHEMALCE B, KOUZARIDES T. A chromodomain switch mediated by histone H3 Lys 4 acetylation regulates heterochromatin assembly. Genes Dev. 2010;24:647–52. doi: 10.1101/gad.1881710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- XU C, CUI G, BOTUYAN MV, MER G. Structural Basis for the Recognition of Methylated Histone H3K36 by the Eaf3 Subunit of Histone Deacetylase Complex Rpd3S. Structure. 2008;16:1740–50. doi: 10.1016/j.str.2008.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YANG Y, LU Y, ESPEJO A, WU J, XU W, LIANG S, BEDFORD MT. TDRD3 is an effector molecule for arginine-methylated histone marks. Mol Cell. 2010;40:1016–23. doi: 10.1016/j.molcel.2010.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YAP KL, LI S, MUNOZ-CABELLO AM, RAGUZ S, ZENG L, MUJTABA S, GIL J, WALSH MJ, ZHOU MM. Molecular interplay of the noncoding RNA ANRIL and methylated histone H3 lysine 27 by polycomb CBX7 in transcriptional silencing of INK4a. Mol Cell. 2010;38:662–74. doi: 10.1016/j.molcel.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- YOCHUM GS, AYER DE. Role for the mortality factors MORF4, MRGX, and MRG15 in transcriptional repression via associations with Pf1, mSin3A, and Transducin-Like Enhancer of Split. Mol Cell Biol. 2002;22:7868–76. doi: 10.1128/MCB.22.22.7868-7876.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG K, MOSCH K, FISCHLE W, GREWAL SI. Roles of the Clr4 methyltransferase complex in nucleation, spreading and maintenance of heterochromatin. Nat Struct Mol Biol. 2008;15:381–8. doi: 10.1038/nsmb.1406. [DOI] [PubMed] [Google Scholar]
- ZHANG P, DU J, SUN B, DONG X, XU G, ZHOU J, HUANG Q, LIU Q, HAO Q, DING J. Structure of human MRG15 chromo domain and its binding to Lys36-methylated histone H3. Nucleic Acids Res. 2006;34:6621–8. doi: 10.1093/nar/gkl989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZHANG X, GERMANN S, BLUS BJ, KHORASANIZADEH S, GAUDIN V, JACOBSEN SE. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- ZOFALL M, GREWAL SI. RNAi-mediated heterochromatin assembly in fission yeast. Cold Spring Harb Symp Quant Biol. 2006;71:487–96. doi: 10.1101/sqb.2006.71.059. [DOI] [PubMed] [Google Scholar]