Abstract
Summary: We introduce ProfileChaser, a web server that allows for querying the Gene Expression Omnibus based on genome-wide patterns of differential expression. Using a novel, content-based approach, ProfileChaser retrieves expression profiles that match the differentially regulated transcriptional programs in a user-supplied experiment. This analysis identifies statistical links to similar expression experiments from the vast array of publicly available data on diseases, drugs, phenotypes and other experimental conditions.
Availability: http://profilechaser.stanford.edu
Contact: abutte@stanford.edu
Supplementary Information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
The introduction of the DNA microarray and other genome-level technologies has provided an unprecedented, systems-level view of cellular transcription. This detailed characterization of gene expression provides a unique opportunity for data-driven discovery of connections between biological conditions and phenotypes. Past work has demonstrated the utility of gene expression-based discovery in predicting the mechanisms of genetic and chemical perturbations (Hughes et al., 2000) and linking drugs and diseases (Hassane et al., 2008; Lamb et al., 2006), even when integrating data across different platforms and cell types (Dudley et al., 2009).
While these studies have demonstrated the power of gene expression-based approaches to biological hypothesis generation, few tools exist to exploit the primary molecular data cataloged in repositories such as the Gene Expression Omnibus (GEO) (Barrett et al., 2009; Chen et al., 2008). Here we introduce ProfileChaser, a web server that allows for content-based gene expression search with a user-supplied experiment. Our tool mines GEO DataSets for experiments that differentially regulate the same transcriptional programs. ProfileChaser provides an accessible and powerful interface for leveraging public data to inform new experiments and predict novel associations between diseases, drugs, genotypes and phenotypes.
2 METHODS
ProfileChaser aims to link biological conditions that have similar patterns of differential gene expression. Conceptually, if the differentially expressed genes in a ‘breast cancer versus normal tissue’ experiment match the differentially expressed genes in a ‘Mock versus Drug X’ experiment, we might predict that drug X could match biological processes involved in breast cancer. We expand this approach beyond drug–disease relationships to include all cellular phenotypes, perturbations and genetic modulations: we leverage GEO DataSet (GDS) annotations as previously described (Morgan et al., 2009) to automatically generate and search 14 974 combinatorial comparisons between experimental conditions comprising 37 710 microarrays from 1815 GEO DataSets.
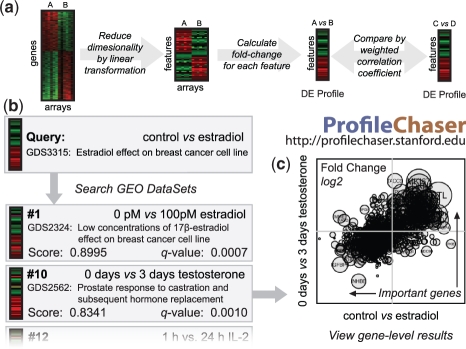
ProfileChaser indexes and searches GEO DataSets using a combination of previously developed techniques (Fig. 1a) (Chen et al., 2007; Engreitz et al., 2010a, b). To begin, we apply independent component analysis to a compendium of human gene expression data, obtaining a reduced set of gene expression features that describe biologically relevant transcriptional programs (Engreitz et al., 2010a). We call these features fundamental components of gene expression. We reduce the dimensionality of GEO DataSet experiments by mapping genes to their unique human homologs and projecting the resulting data into the feature space identified by these fundamental components. This 50-fold reduction in complexity allows for speedier and more accurate retrieval of relevant experiments, even across tissue types, platforms and species (Engreitz et al., 2010b). Next, we create a differential expression (DE) profile for each of the 14 875 experimental comparisons, including the fold-change and P-value of DE for each fundamental component. These DE profiles summarize the changes in the expression of conserved transcriptional programs, and together form ProfileChaser's main database.
Fig. 1.
Graphical overview of ProfileChaser. (a) To index differential expression comparisons in GEO DataSets, each experiment is processed according to the flowchart. (b) The user supplies a query experiment and retrieves matching GEO DataSet comparisons. (c) For each ranked result with q<0.05, the user may view a gene-level comparison of the query and retrieved experiment. Genes that are differentially expressed in both experiments may prove important to the shared function or mechanism. Additional details are available in the Supplementary Materials.
To query this collection of DE profiles, the user supplies an experiment that compares two conditions. ProfileChaser generates a DE profile in feature space as described above, then searches its database to find similar experiments (Fig. 1b). As opposed to Gene Set Enrichment Analysis (GSEA)-based approaches that require arbitrarily chosen gene-set cutoffs, we score the similarity between two DE profiles using a weighted Pearson's correlation: fundamental-component fold-changes are weighted by their P-value of DE (Engreitz et al., 2010b). A null distribution for this measure is computed from the correlation coefficients of all 14 875 experimental comparisons, allowing for the calculation of a conservative false discovery rate for each retrieved result.
For the top results, users can view the genes that contributed most to the similarity between the query and retrieved comparisons. The Gene Results page displays a weighted scatterplot of the most significant genes, as well as a table of genes ranked by their contribution to the P-value-weighted correlation measure. This ranking highlights the genes that might play an important role in the biology shared by the two experiments (Fig. 1c).
3 RESULTS
To illustrate the utility of our method, we queried ProfileChaser to find experiments that resemble the treatment of MCF7 breast cancer cells with 17β-estradiol, the natural ligand of estrogen receptor (GDS3315). The top matching comparisons included several variations of this experiment performed by separate groups (Fig. 1b, Supplementary Fig. S1). In addition, ProfileChaser identified other experimental contexts involving hormone-induced proliferation, including cytotoxic T cell response to interleukin-2 (GDS3222) and primary prostate response to testosterone treatment (GDS2562). An examination of significant genes confirmed that proliferation linked these biological conditions: in the comparison of GDS3315 with GDS2562, for instance, the second most significant gene encoded Ki-67, the canonical proliferation marker (Supplementary Fig. S2).
Next, we used the comparison of tumorigenic and non-tumorigenic breast cancer cells (GDS2618) to identify compounds that might specifically target these cell subpopulations (Supplementary Fig. S3). The comparison of dasatinib-resistant versus dasatinib-sensitive prostate cancer cell lines scored significantly, suggesting that tumorigenic breast cancer cells might be sensitive to this Src inhibitor. Treating basal breast cancer cell lines with dasatinib significantly decreases the proportion of ALDH1-positive tumorigenic cells, supporting this hypothesis (Kurebayashi et al., 2010).
Finally, we recently used these methods to search GEO for datasets that resembled the comparison of neural stem cells from wild-type and FoxO3−/− mice (Engreitz et al., 2010b). We identified four matching profiles that linked FoxO3 to hypoxia, a relationship validated by recent experimental work with our collaborators (Renault et al., 2009). Thus, experiments retrieved by ProfileChaser provide statistical evidence for associations that may be verified by further experimental or computational analyses.
ProfileChaser comparisons are defined by GEO annotations, and thus interpretation of results may not be straightforward. In particular, experimental designs with multiple factors may confound analysis (Supplementary Fig. S4). Subset names assigned by GEO may also be misleading. To properly interpret results, users should examine the design of each experiment using the GEO web interface and references therein. ProfileChaser cannot take advantage of potential paired-sample information, and DE profiles generated from the same experiment are not independent, since they contain overlapping samples. Finally, users should note that the absence of a positive result does not imply that two conditions are unrelated.
The ability to query GEO based on differential expression will allow for the identification of new associations between diseases, drugs, genotypes and phenotypes. Future work will expand ProfileChaser to incorporate other sources of gene expression data, including European Bioinformatics Institute ArrayExpress (Parkinson et al., 2011).
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Alex Skrenchuk for technical support.
Funding: National Library of Medicine (R01 LM009719 to A.J.B., T15 LM007033 to A.A.M. and J.T.D.); Howard Hughes Medical Institute; Hewlett Packard Foundation and the Lucile Packard Foundation for Children's Health.
Conflict of interest: R.M. was employed at Optra Systems, hired to build the database-backed website and tools.
REFERENCES
- Barrett T., et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., et al. AILUN: reannotating gene expression data automatically. Nat. Methods. 2007;4:879. doi: 10.1038/nmeth1107-879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R., et al. GeneChaser: identifying all biological and clinical conditions in which genes of interest are differentially expressed. BMC Bioinformatics. 2008;9:548. doi: 10.1186/1471-2105-9-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudley J.T., et al. Disease signatures are robust across tissues and experiments. Mol. Syst. Biol. 2009;5:307. doi: 10.1038/msb.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J.M., et al. Independent component analysis: mining microarray data for fundamental human gene modules. J. Biomed. Inform. 2010a;43:932–944. doi: 10.1016/j.jbi.2010.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engreitz J.M., et al. Content-based microarray search using differential expression profiles. BMC Bioinformatics. 2010b;11:603. doi: 10.1186/1471-2105-11-603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassane D.C., et al. Discovery of agents that eradicate leukemia stem cells using an in silico screen of public gene expression data. Blood. 2008;111:5654–5662. doi: 10.1182/blood-2007-11-126003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes T.R., et al. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- Kurebayashi J., et al. Preferential antitumor effect of the Src inhibitor dasatinib associated with a decreased proportion of aldehyde dehydrogenase 1-positive cells in breast cancer cells of the basal B subtype. BMC Cancer. 2010;10:568. doi: 10.1186/1471-2407-10-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb J., et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- Morgan A., et al. Dynamism in Gene Expression Across Multiple Studies. Physiol. Genomics. 2009;40:128–140. doi: 10.1152/physiolgenomics.90403.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson H., et al. ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renault V.M., et al. FoxO3 regulates neural stem cell homeostasis. Cell Stem Cell. 2009;5:527–539. doi: 10.1016/j.stem.2009.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.