Abstract
PIM1 kinase and MYC are commonly co-expressed in human prostate cancer and synergize to induce rapidly progressing prostate cancer in mouse models. Deficiency of the Pim kinase genes is well tolerated in vivo, suggesting that PIM1 inhibition might offer an attractive therapeutic modality for prostate cancer, particularly for MYC-expressing tumors. Here we examine the molecular consequences of Pim1 and MYC over-expression in the prostate as well as the effects of depleting Pim1 in prostate carcinoma cells with high levels of MYC. Over-expression of Pim1 in the mouse prostate induces several pro-tumorigenic genetic programs including cell cycle genes and Myc-regulated genes prior to the induction of any discernible pathology. Pim1 depletion by RNA interference in mouse and human prostate cancer cells decreased cellular proliferation, survival, Erk signaling, and tumorigenicity even when MYC levels were not significantly altered. These results indicate that PIM1 may be necessary to maintain tumorigenicity, and further support efforts aimed at developing PIM1 inhibitors for prostate cancer therapy.
Keywords: Pim1 kinase, MYC, gene expression, Erk, tumorigenicity
Introduction
The PIM kinase family consists of three constitutively active serine/threonine kinases, PIM1, PIM2 and PIM3, which have been implicated in diverse human malignancies including leukemias and lymphomas, as well as pancreatic, hepatic, oral and prostate cancers (Magnuson et al , Shah et al 2008). PIM1 kinase is an attractive molecular target for cancer pharmacotherapy because it enhances the transforming potential of oncogenes. For instance, Pim1 accelerates Myc-initiated lymphomagenesis and prostate cancer development in mouse models (Verbeek et al 1991, Wang et al 2010). Whereas mice with homozygous deficiency in Myc uniformly exhibit widespread developmental defects and embryonic lethality (Davis et al 1993), mice with homozygous deficiency in all three Pim kinases are viable, fertile, and exhibit only minor phenotypic abnormalities (Mikkers et al 2004). These observations show that inhibition of PIM kinase activity is well-tolerated in vivo, and suggest that drugs targeting PIM kinases might be better tolerated than those targeting MYC.
Using a prostate regeneration system, we have recently shown that Pim1 potently interacts with MYC to accelerate prostate cancer progression although Pim1 alone did not lead to significant pathology (Wang et al 2010). MYC/Pim1 expression in this model led to the development of neuroendocrine prostate cancer, apparently by differentiation via an adenocarcinoma intermediate. The mechanisms underlying cooperativity between Pim1 and Myc in prostate tumorigenesis in vivo have not been clearly elucidated. Studies have suggested that Pim1 may enhance Myc tumorigenesis by increasing Myc protein stability or transcriptional activity (Chen et al 2005, Kim et al 2010, Zhang et al 2008, Zippo et al 2007). We have reported that Pim1 over-expression in the prostate in vivo produced minimal morphological alterations by 6 weeks (Wang et al 2010). These results prompted us to test whether we could discern molecular alterations induced by Pim1 over-expression in the absence of concurrent over-expression of Myc. In this report, we used gene expression profiling to investigate the molecular pathways modulated by expression of Pim1 and MYC alone or together in the mouse prostate. We also knocked down Pim1 expression using RNA interference to test whether Pim1 is necessary to maintain the malignancy of MYC-expressing prostate cancer cells. Our results show that Pim1 knockdown resulted in decreased proliferation, cell survival, Erk signaling, and tumorigenicity even when MYC levels were not significantly altered.
Results
Gene expression programs regulated by Pim1 and c-MYC in prostate cells in vivo
To characterize the effects of Pim1 and MYC on prostate tumorigenesis, we used a tissue recombination model in which lentiviral vectors over-expressing MYC, Pim1, or MYC and Pim1 were used to infect normal mouse prostate cells. Cells were implanted under the renal capsule of SCID mice hosts, and regenerated prostate tissue grafts were harvested after 6 weeks. We have previously shown that at this time point, Pim1 grafts were morphologically indistinguishable from control grafts, consisting of benign prostatic glands (Wang et al 2010). MYC grafts, on the other hand, contained multiple foci of high grade PIN (HGPIN) lesions, (Wang et al 2010). However, MYC/Pim1 grafts had the most severe pathology, consisting of poorly differentiated cancer with evidence of neuroendocrine differentiation. These observations indicate that Pim1 may be dependent on collaborating mutations in genes such as in Myc before it can manifest its tumorigenic effects.
To gain insight into the oncogenic functions of Pim1 in prostate cells in vivo, we compared the gene expression profiles of these regenerated mouse prostate grafts over-expressing human MYC, mouse Pim1, both Pim1 and MYC, or control lentivirus (Wang et al 2010) using Affymetrix microarray analysis. By employing Significance Analysis of Microarrays (SAM) analysis, we were able to identify 4,172 probes (corresponding to 2770 unique genes) that were differentially expressed between control, Pim1, MYC, or MYC/Pim1 grafts (Supplementary Table S1). To identify subgroups of genes with similar expression profiles, we performed hierarchical clustering (Supplementary Fig. S1) for the genes deemed significant in the microarray analysis. The most extreme gene expression changes were observed in three MYC/Pim1 grafts and revealed several distinct clusters of genes with altered expression. Gene expression differences among samples are a probably a reflection of the stages of tumorigenesis in each of the grafts. For example, one MYC/Pim1 array clustered with the MYC arrays (Supplementary Fig. S1), suggesting that RNA isolated from this sample might have come from tissue consisting of mostly HGPIN.
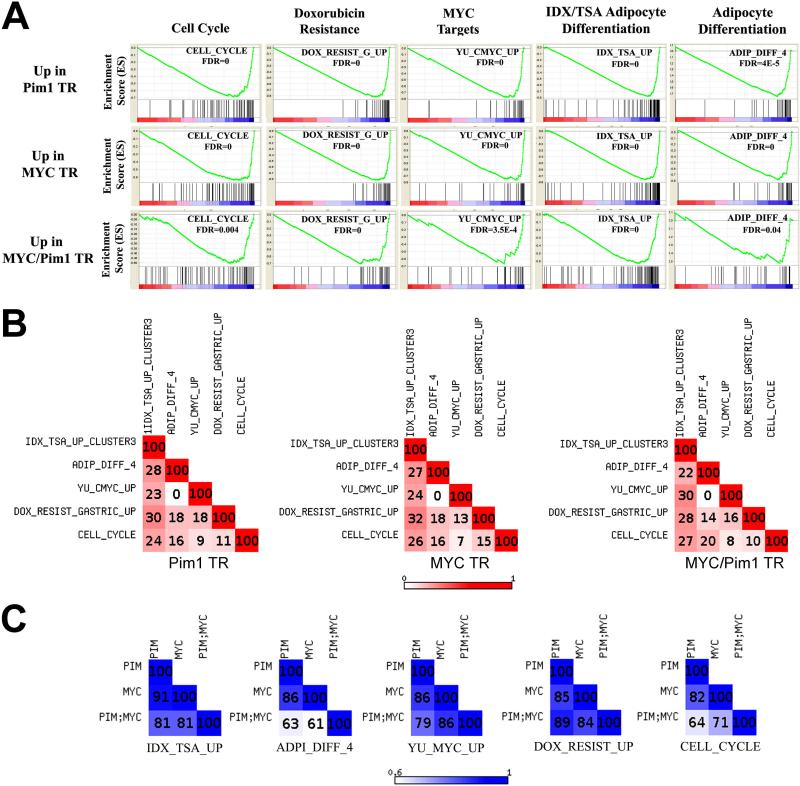
While single analysis of differentially expressed genes can provide insight into the underlying biological differences between samples, it can miss modest but coordinated changes in groups of genes (such as those belonging to the same cellular pathway) which are functionally significant in the tissue or cell type being studied. Gene Set Enrichment Analysis (GSEA) is an approach that identifies pathway-level changes using groups of genes or ‘gene sets’ that are significantly enriched or depleted -as a group- in a pair-wise comparison of experimental vs. control samples (Mootha et al 2003, Subramanian et al 2005). We performed GSEA comparing the control grafts to each of the Pim1, MYC and MYC/Pim1 groups (a total three comparisons). Remarkably, all three groups showed significant enrichment for several gene sets, including cell cycle genes and Myc target genes (Fig. 1, Supplementary Table S2). Other gene sets significantly enriched in Pim1, MYC and MYC/Pim1 grafts include genes involved in differentiation. For example, IDX/TSA cluster 3 refers to a gene set upregulated in a model of adipocyte differentiation of 3T3L1 cells induced by insulin, dexamethasone treatment (Burton et al 2004). Many of the genes in this cluster are also related to the cell cycle as differentiation is associated with exit from the cell cycle. Another notable gene set identified by GSEA analysis consists of genes up-regulated in doxorubicin resistant gastric cancer cells. Therefore the gene sets identified as enriched in Pim1, MYC or MYC/Pim1 grafts consist of genes involved in cell cycle progression, differentiation and drug resistance. The differential expression of several selected genes identified by this analysis was confirmed by qRT-PCR (Figure S1B). While observing these gene expression changes in MYC- or MYC/Pim1-expressign neoplastic prostate grafts is not surprising, it is unexpected that grafts expressing Pim1 alone also show such gene expression changes, considering that Pim1 grafts at 6 weeks show no discernible morphological abnormalities (Wang et al 2010). Thus Pim1 may perturb the expression of cancer-relevant genes before the appearance of any discernible histological abnormalities.
Figure 1. Gene expression profiling identifies pathways dysregulated in Pim1 and MYC-expressing prostate tissue recombinant grafts.
A, GSEA analysis was performed to identify ‘gene sets’ significantly enriched in Pim1, MYC and Pim1/MYC grafts relative to control prostate grafts. The False discovery rates (FDR) are indicated for each gene set. TR (tissue recombinant graft); IDX (Insulin/Dexamethasone/Isobutylmethylxanthine); TSA (Trichostatin A); Adip_Diff (Adipocyte differentiation); Yu_MYC (MYC target genes); Dox_Resist_Gastric (Doxorubicin resistant gastric carcinoma cells). See also Figure S1 and Tables S1 and S2.
B, Limited overlap among leading edge genes across the top gene sets enriched in Pim1, MYC and Pim1/MYC prostate tissue recombinant grafts. The numbers indicate percent overlap of leading edge genes in each gene set. These results indicate that the 5 gene sets are distinct.
C, Overlap of leading edge genes in gene sets enriched by expression of Pim1, MYC or Pim1/MYC. The numbers indicate percent overlap of leading edge genes in each experimental group. These data indicate that Pim1, MYC and MYC/Pim1 regulate a similar set of genes within each gene set.
The enrichment of these gene set modules in the prostate regenerated grafts is not due to the presence of a common set of genes in all modules, as leading edge analysis showed little overlap among them (Fig. 1B, Supplementary Fig. S2). The ‘leading edge’ in a gene set represents the individual genes that are primarily responsible for the observed association. Examination of the ‘leading edge’ could provide important insights into the biological character of the gene set. The similarity between modules enriched in Pim1 expressing grafts to those in MYC-expressing grafts (Figs. 1A,C) is consistent with reports in the literature that Pim1 enhances MYC transcriptional activity (Chen et al 2005, Kim et al 2010, Zhang et al 2008, Zippo et al 2007). Thus our gene expression analysis supports the hypothesis that over-expression of Pim1 stimulates the transcriptional activity of endogenous Myc present in the cells, resulting in the activation of gene programs regulated by Myc.
Establishment of MYC/Pim1 Tumor Cell Lines
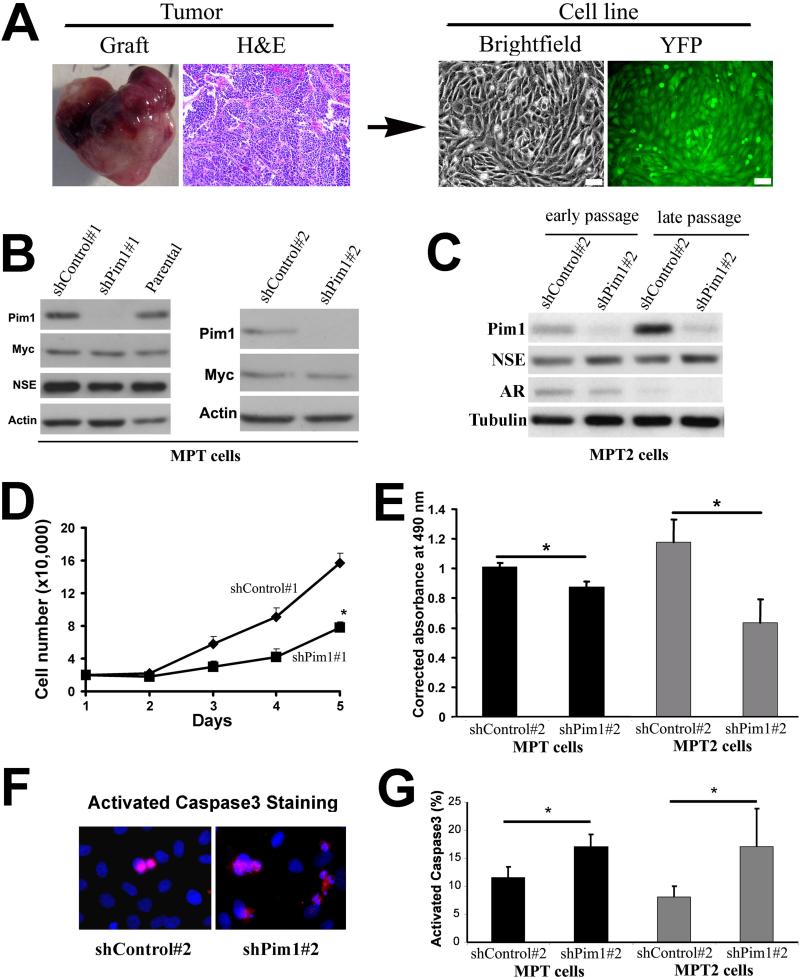
To further investigate the relationship between Myc and Pim1, we tested whether Pim1 plays a role in maintaining the tumorigenic potential of Myc. To examine this possibility, we created a cell line (MPT: MYC and Pim1 over-expressing Tumor cells) from MYC/Pim1 prostate tissue recombinant grafts. As we have shown, the MYC/Pim1 grafts formed aggressive prostate tumors (Fig. 2A and (Wang et al 2010)). MPT cells expressed MYC and Pim1 protein (Fig 2B), yellow fluorescent protein as a marker indicating their derivation from a MYC/Pim1 tumor (Fig. 2A), as well as the neuroendocrine cell marker neuron-specific enolase (NSE) (Fig. 2B), consistent with neuroendocrine differentiation in the original MYC/Pim1 tumors (Wang et al 2010). To study the effects of Pim1 expression, these cells were subsequently infected with shRNAmir against mouse Pim1 (named shPim1#1). To control for off-target effects, additional stable cell line was established expressing shRNA targeting Pim1 (named shPim1#2). Western blot analysis showed that Pim1 protein levels were dramatically reduced by both Pim1 RNAs (Fig. 2B). Furthermore, Pim1 knockdown did not result in a compensatory increase in Pim2 or Pim3 (Figure S4). Notably, Pim1 knock-down did not appreciably change the levels of MYC protein in MPT cells (Fig. 2B).
Figure 2. Knockdown of Pim1 expression in MYC/Pim1-overexpressing prostate tumor derived (MPT) cell lines.
A, Establishment of MYC/Pim1 Tumor (MPT) prostate cell line from a MYC/Pim1-expressing tumor graft. The right panel shows the image of established cell line from 6-week MYC/Pim1 overexpressing tumor. YFP fluorescence can be seen under fluorescence microscope as the cells contain lentivirus construct bicistronically expressing YFP in addition to MYC or Pim1 (Scale bar: 100 μm).
B, Western blot analysis of parental MPT cells or MPT cells stably expressing shRNAmir against Pim1 (shPim1#1), non-silencing shRNAmir (shControl#1), shRNA against Pim1 (shPim1#2) or control (shControl#2). MPT cells express NSE and MYC, the levels of which were not altered by Pim1 knockdown. Actin served as loading control.
C, MPT2 cells were derived from 4-week MYC/Pim1 overexpressing prostate grafts (see Figure S2) and Pim1 expression stably knocked down using shPim1#2. MPT2 cells expressed androgen receptor (AR) in early passage, but not in late passage. NSE was also expressed in MPT2 cells. Tubulin served as loading control.
D, Growth curve were generated by counting shPim1#1 and shControl#1 MPT cells in duplicates. *P<0.05
E, Cell proliferation was measured using the CellTiter 96® AQueous One Solution Cell Proliferation Assay (MTS, Promega). The absorbance is directly proportional to the number of living cells in culture. Results represent the mean ± s.d. of quadruplicate wells after 3 days of incubation of 2000 cells. *P<0.05.
F, Analysis of apoptosis by activated caspase-3 staining (red), with nuclei counterstained with DAPI (blue) (Original magnification: 400×).
G, Quantification of apoptosis rate by counting active capase-3 positive cells. The results represent the mean ± s.d. of two independent experiments *P<0.05.
We also established a second cell line, MPT2 from a 4-week MYC/Pim1 graft (Fig. 2C and Supplementary Fig. S3). Unlike the 6-week grafts reported above, histological and immunohistochemical analysis revealed that 4-week MYC/Pim1 grafts exhibited HGPIN with features of microinvasion. These lesions were androgen receptor (AR) positive and neuroendocrine cell marker synaptophysin negative (Supplementary Fig. S3). We stably knocked down Pim1 expression in MPT2 cells using shPim1#2 (Fig. 2C). We observed that MPT2 cells expressed NSE and initially expressed AR (Fig. 2C). However, AR expression was lost with increasing passage in culture (Fig. 2C).
Further characterization of MPT and MPT2 cells showed that Pim1 knock-down substantially reduced cellular proliferation compared to cells expressing control shRNA (Fig. 2D,E). PIM1 has been implicated in anti-apoptotic functions via interactions with several molecules including BAD (Aho et al 2004) and ASK1 (Gu et al 2009). Accordingly, Pim1 knockdown increased rates of apoptosis as determined by activated Caspase 3 staining in both MPT and MPT2 cells in low serum conditions (Fig. 2F,G).
Pim1 knockdown impairs prostate tumor cell tumorigenicity
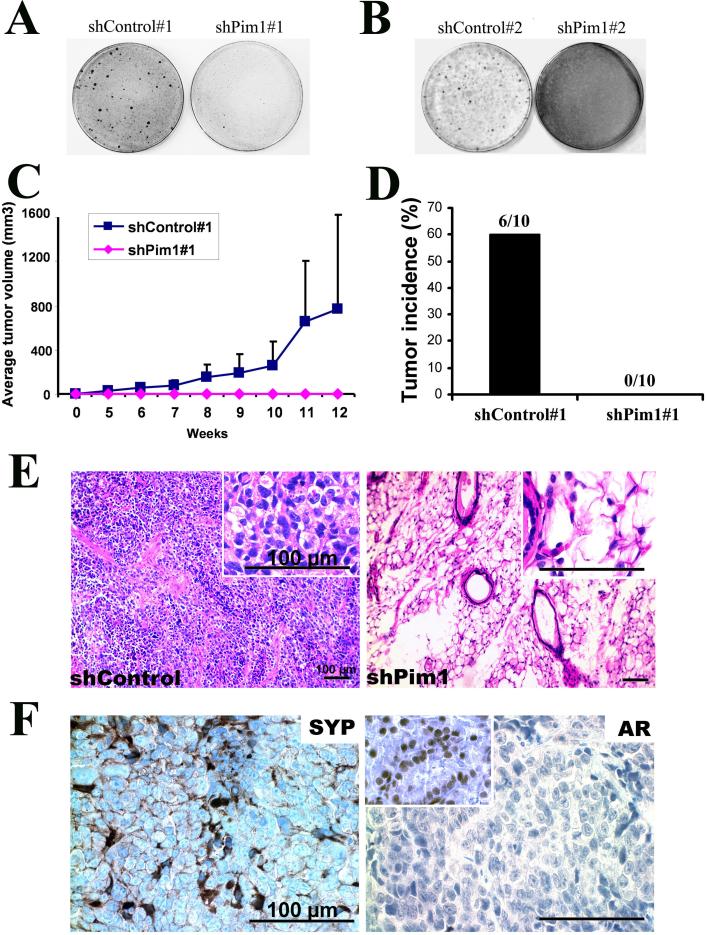
We used a focus-formation assay to test the effects of Pim1 knock-down on prostate cancer cell tumorigenicity in vitro. The results showed that control MPT and MPT2 cells lost contact inhibition and formed foci, which is typical of cancer cells. However, Pim1 knock-down in both cell lines sharply reduced focus-forming ability (Fig. 3A, B). To examine in vivo tumorigenicity, we injected either Pim1 knock-down (shPim1#1) or control (shControl#1) MPT cells subcutaneously into athymic nude mice and monitored tumor growth over 12 weeks. Although no tumors developed in the Pim1 knock-down group (N=10), 60% of mice in the control group (N=10) formed large tumors (Figs. 3C, D). In histology, the control grafts consisted of sheets of tumor cells expressing the neuroendocrine differentiation marker synaptophysin (SYP), but not androgen receptor (AR), recapitulating the features of original MYC/Pim1 tumor from which the cell line was established (Fig. 2A and Figs. 3E,F).
Figure 3. Pim1 expression is required to maintain tumorigenicity of MYC/Pim1 tumor cells.
A and B, Stable knockdown of Pim1 by shRNA reduces colony formation in MPT (A) and MPT2 (B) cells.
C and D, Pim1 knockdown (shPim1#1) abrogates tumorigenicity of MPT cells when injected subcutaneously into nude mice.
E, Representative H&E images of graft sections. Control group showed high-grade tumor consistent with the original MYC/Pim1 tissue recombinant graft tumor while Pim1 knockdown group showed fat and blood vessel cells with absence of tumor cells. Scale bar: 100 μm. Insets: Higher-magnification images.
F, Immunohistochemical staining showed that sections from control grafts were synaptophysin (SYP) positive and androgen receptor (AR) negative. Scale bar: 100 μm. Pim1 knockdown grafts could not be analyzed because no tumor cells were found by H&E staining. Insets: AR positive control.
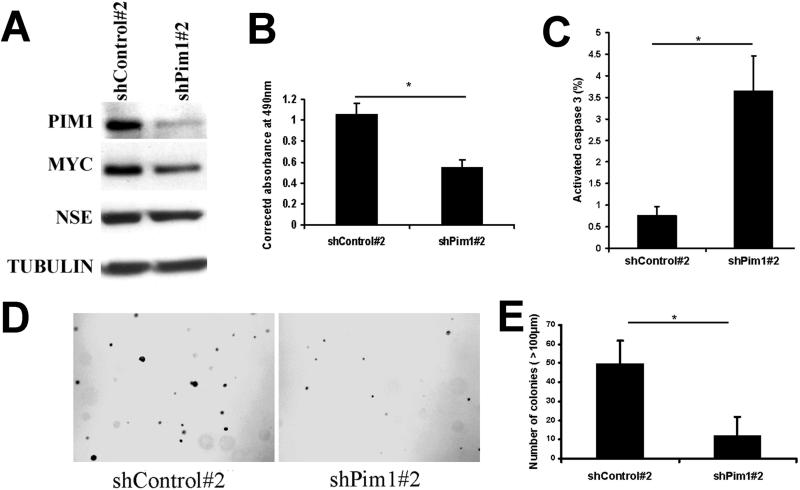
We extended these findings beyond the two MPT cell lines, by examining the effects of PIM1 knockdown in DU145, a human prostate cancer cell line with appreciable endogenous PIM1 and MYC expression. To reduce PIM1 expression in DU145, we used shPim1#2 which targets both human and mouse PIM1 mRNA. We verified that PIM1 was substantially knocked down in DU145 cells (Fig. 4A). Furthermore, unlike MPT cells, endogenous MYC levels were reduced by PIM1 knockdown in DU145 cells (Fig. 4A). NSE was also expressed by DU145 cells, consistent with previous results (Leiblich et al 2007), and its expression was not altered by PIM1 knockdown (Fig. 4A). PIM1 reduction impaired the proliferation and survival of DU145 cells (Fig. 4B, C). In addition, DU145 cell tumorigenicity as determined by growth in soft agar was reduced following PIM1 depletion (Fig. 4D,E). These results, taken together with our previously published results on the effects of Pim1 over-expression on prostate cell transformation (Wang et al 2010), indicate that although Pim1 over-expression alone may not be sufficient to initiate the development of invasive prostate cancer, continued Pim1 expression is required to maintain prostate cancer cell tumorigenicity.
Figure 4. Pim1 knockdown in DU145 cells reduces proliferation, survival and tumorigenicity.
A, Western blot analyses for PIM1, MYC and NSE in DU145 cells with stable PIM1 knockdown by shRNA. Tubulin served as a loading control.
B, Cell proliferation was measured by MTS assay. The results are mean ± s.d. of quadruplicate wells after 3 days of incubation of 4000 cells. *P<0.05.
C, Quantitation of apoptosis rate by counting active capase-3 positive cells after DU145 cells were treated with serum free medium overnight. The results are mean ± s.d. of two independent experiments. *P<0.05.
D, Representative images of soft agar assay show that PIM1 knockdown in DU145 cells decreased the size and number of colonies (Original magnification: 4×).
E, Quantitation of colonies from soft agar assay of Pim1 knockdown and control DU145 cells. The results are mean ± s.d. of triplicates. *P<0.05.
Pim1 knockdown impairs ERK signaling pathway activation
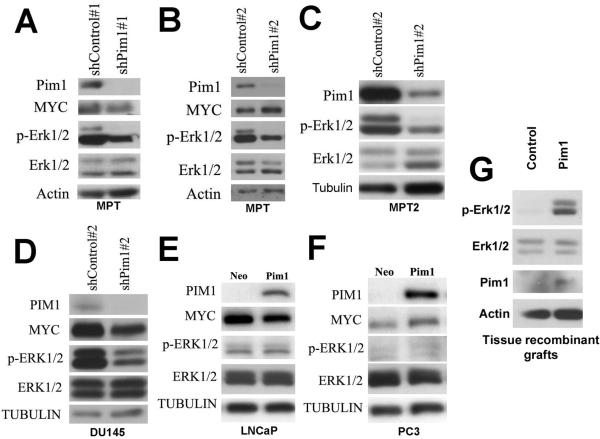
We explored signaling pathways that may be dysregulated in Pim1 knock-down cells, including the PI3-kinase/Akt and MAPK/Erk signaling pathways. While we found no consistent changes in Akt signaling (not shown), Western blot analysis showed that ERK1/2 phosphorylation was consistently reduced by Pim1 knock-down in MPT, MPT2, and DU145 cells compared to corresponding control cells (Fig. 5A-D). We next examined whether Pim1 over-expression is sufficient to induce active ERK in prostate carcinoma cells. We found that stable over-expression of Pim1 in LNCaP and PC3 prostate cancer cells (Kim et al 2010, Roh et al 2003) did not alter ERK phosphorylation (Fig. 5E,F). However, examination of Pim1-expressing tissue recombinant prostate grafts showed evidence of enhanced Erk1/2 phosphorylation (Fig. 5G). As these Pim1-expressing grafts showed no evidence of prostate neoplasia (Wang et al 2010), the results indicate that in prostate tissue in vivo, Pim1 overexpression is associated with enhanced Erk1/2 activity. Overall, these results indicate that PIM1 may be necessary for maintaining ERK signaling in prostate cells.
Figure 5. Pim1 is required to maintain Erk signaling in prostate carcinoma cells.
A-C. Western blots showed that Pim1 knockdown in MPT and MPT2 cells by two different sets of shPim1 constructs decreased phosphorylation of Erk1/2 compared to control shRNA.
D, PIM1 knockdown in DU145 cells decreases ERK1/2 phosphorylation of Erk1/2
E and F, Western blots showed that Pim1 overexpression did not increase ERK phosphorylation in LNCaP and PC3 cells compared to vector control cells (Neo).
G, Western blot analysis of 6-week tissue recombinant grafts generated from cells expressing control or Pim1 lentivirus. Note increased phosphorylation of Erk1/2 in Pim1 graft compared to control.
Discussion
In this report we investigated the molecular changes induced by expression of Pim1 and MYC in the mouse prostate in vivo as well as the functional consequences of depleting Pim1 in prostate tumor cells. We found that Pim1 expression engages multiple gene programs important for tumorigenesis even in the absence of any discernible morphological abnormalities. Several gene modules were disrupted by Pim1 over-expression, including modules involving cell cycle genes, genes altered during differentiation, MYC targets and genes up-regulated in doxorubicin resistant cancer cells. Dysregulation of MYC target genes in Pim1-expressing prostate grafts is consistent with published reports showing Pim1 enhances MYC protein stability and transcriptional activity (Chen et al 2005, Kim et al 2010, Zhang et al 2008, Zippo et al 2007). It has been shown that hypoxia can induce Pim1 which contributes to solid tumor formation (Chen et al 2009a). The gene programs we have shown to be regulated by Pim1 may also be dysregulated when Pim1 is induced in hypoxic tissue and thereby sensitize the tissue to tumorigenesis due to expression of oncogenes such as MYC.
Although Pim1 is weakly oncogenic, it synergizes potently with Myc to induce invasive prostate cancer with features of neuroendocrine differentiation (Wang et al 2010). The basis for this synergism between Pim1 and Myc is not well understood. Pim1 is a constitutively active serine/threonine kinase, so its protein level correlates its kinase activity (Qian et al 2005). To understand the function of Pim1 on Myc/Pim1 cooperation in prostate tumorigenicity, we knocked down Pim1 expression in MYC/Pim1 mouse tumor derived cell lines and a human prostate cancer cell line DU145. We found that, while Pim1 failed to initiate development of invasive prostate adenocarcinoma, depletion of Pim1 expression resulted in decreased growth rate, survival, and tumorigenic potential, indicating that Pim1 is required for maintenance of the tumorigenic phenotype. In MPT cells, Pim1 knockdown reversed their tumorigenic phenotype even when MYC level was not decreased, which suggested that Pim1 and Myc synergism in prostate cancer depend on other mechanisms besides enhancing Myc activity and stability. Unlike in DU145 cells, Pim1 knock-down in MPT and MPT2 cells did not decrease Myc levels perhaps due to the fact that these cells express exogenous MYC. It is important to point out that MPT cells derived from MYC/Pim1 neuroendocrine prostate cancer without AR expression, so it may not reflect the situation in most prostate tumors which are AR positive. However, we found similar results in MPT2 cells which are derived from AR positive HGPIN/cancer lesions. In addition, data from multiple human prostate cancer cell lines consistently showed that depletion of PIM1 attenuates tumor cell proliferation and/or tumorigenicity. Our results indicate that PIM1 shRNA impairs proliferation and tumorigenicity of DU145 cells, consistent with other reports in 22rv1(Morishita et al 2008) and PC3 cells (Hu et al 2009).
We observed reduced Erk phosphorylation in Pim1 knock-down cells. It has been shown that Pim1-depleted and Pim1-inhibitor treated bone marrow cells have impaired Erk phosphorylation (Grundler et al 2009). The Raf–MEK–ERK/MAPK pathway is subject to many levels of regulation. Pim1 may have common targets that affect the MAPK pathway. In Eμ-Myc mice, Tpl2 can compensate for the absence of Pim1 and Pim2 (Berns et al 1999). Tpl2 is also known as Map3k8, and has been reported to be involved in Mek/Erk1/2 activation (Jager et al 2010). It is still unclear whether Pim1 directly or indirectly affects Erk phosphorylation. In the primary mitogen regulated pathway, extracellular stimuli activate Raf-1, which phosphorylates and activates MAPK kinase (Mek), leading to the phosphorylation and activation of the extracellular signal regulated kinases (Erk) (Kolch 2000). Activated Erk/Mapk translocates to the nucleus and activates gene expression by phosphorylation of a series of substrates including MYC. It remains to be determined whether Pim1 affects Raf-1 or Mek phosphorylation, or interacts with other proteins that are involved in this pathway.
Several studies suggest that targeting Pim1 could provide a promising strategy in anti-cancer therapy. Importantly, Pim1 depletion has only subtle effects in normal cells. Pim1 deficient mice are ostensibly normal, healthy and fertile (Laird et al 1993). Pim1, Pim2 and Pim3 compound knockout mice are viable and fertile, but show a reduction in body size, suggesting Pim kinases might act as sensitizers for growth factor signaling pathways (Mikkers et al 2004). Pim kinase inhibition using SGI-1776 in prostate cancer cells or CLL cells (B-cell chronic lymphocytic leukemia) led to a concentration dependent induction of apoptosis (Chen et al 2009b, Mumenthaler et al 2009). A PIM1-specific monoclonal antibody has also shown efficacy in inhibiting the tumorigenicity of PIM1-expressing tumors in mouse models (Hu et al 2009). In conjunction with these studies, our results further support that Pim1 represents an efficient target for prostate cancer therapy.
Materials and Methods
Cell lines and constructs
DU145 human prostate cancer cells (ATCC) were cultured in RPMI medium with 10% fetal bovine serum (FBS). MPT and MPT2 mouse prostate cancer cells were established from MYC/Pim1 prostate tissue recombinant tumors. The generation of MYC/Pim1 graft tumors has been described (Wang et al 2010). Briefly, MYC/Pim1 tumors were minced and digested by collagenase, then plated on collagen coat plate with DMEM/F12/10% FBS medium. After reaching confluence, the cells were split into regular cell culture dish.
Lentivirus preparation, transduction and stable knockdown of Pim1
Lentiviral pGIPZ shRNAmir against mouse Pim1 (V2LMM_46214) and the sequence-scrambled, Non-silencing-GIPZ lentiviral shRNAmir control (RHS4346) were provided by Vanderbilt Functional Genomics shared resource (Open Biosystems). Lentiviral pLKO.1 shRNA targeting human, mouse, rat Pim1 (RHS3979-9631245) and empty vector were obtained from Open Biosystems. Lentiviral shRNAmir or shRNA vectors with Δ8.9 and VSVG (provided by Dr. David Baltimore, Caltech) were transfected into 293FT cells (Invitrogen) to obtain viral supernatants as described (Wang et al 2010). MPT, MPT2 or DU145 cells were infected by virus in the presence of 8μg/ml polybrene. 10 or 5μg/ml Puromycin were added to select stably transduced cells after 2 days of infection.
Transcriptional profiling analysis
Total RNA was isolated with TRIzol (Invitrogen) and RNAeasy kit from Qiagen. RNA quality was checked on an Agilent Bioanalyzer. All samples used for microarray analysis have high quality score (RIN >7). RNA (1 μg) was reverse transcribed with T7-oligo(dT) primer and labeled with biotin using Affymetrix One Cycle Target Labeling kit following manufacturer's protocol. Three to four replicates of each group were prepared, labeled, and hybridized to Affymetrix Mouse Gene 1.0 ST v1.r4 arrays and scanned on Affymetrix GeneChip scanner 3000. Data were collected using Affymetrix GCOS software and pre-processed in Expression Console (version 1.1)(Affymetrix, Santa Clara CA) using the RMA-Sketch method. Normalized data were imported to Microsoft Excel for SAM (Significance Analysis of Microarrays) (Tusher et al 2001). Multiclass SAM was performed (delta = 0.43, FDR < 0.0152) without array centering. Genes deemed significant in the multiclass SAM were ordered based on their similarity in expression patterns with hierarchical clustering in R. Distance was calculated using the Euclidean model and clusters were generated using the complete linkage method. GSEA v2.0.6 (Subramanian et al 2005) was used to test for enrichment in Collections C2 – C5 of the Molecular Signatures Database (MSigDB version 2.5). Comparisons were made three ways: control vs. Pim1 over-expressing, control vs. MYC over-expressing, and control vs. MYC/Pim1 over-expressing.
Western blot analysis
Cells were washed with PBS, RIPA buffer with protease inhibitors was added, and cells were harvested with cell scraper, briefly sonicated and spun down. For analysis of tissues, prostate grafts from control and Pim1-expressing tissue recombinants grown for 6 weeks (Wang et al 2010) were used to make lysates. Lysate was run on SDS-PAGE, transferred to PDVF membranes. Membranes were blotted with the following antibodies: c-MYC, Pim1, AR and Actin (Santa Cruz Biotechnology); p44/42 MAPK (Erk1/2), phospho-p44/42 MAPK (Thr202/Tyr204) (Cell signaling); NSE (Neomarker); β-actin (Santa Cruz Biotechnology); β-tubulin (Sigma).
Proliferation assay
MPT Cells were seeded at 20,000 cells per well in a six-well plate. Viable cell number was determined by hemacytometer counts of trypan blue-excluding cells for 4 days. For MTS assay, 2000 MPTs cells or 4000 DU145 cells were plated in quadruplicate per well in 96-well plates. Three days later, CellTiter 96® Aqueous One Solution (Promega) was added to each well and results were read at 490 nm in a plate reader according to the manufacturer's instruction.
Active Caspase 3 staining
Cells grown were plated on glass coverslips. Cells were fixed in 4% paraformaldehyde for 10 min followed by permeabilization in 1% Triton X-100 for 10 min. After washing in PBS, cells were blocked in 10% goat serum, stained with activated caspase-3 (Cell signal) at 1:500. Coverslips were mounted on slides using Vectashield mounting medium (Vector Laboratories). An average of 1000 cells was counted per coverslip.
Colony Formation Assay
5,000 of MPT cells were plated in 10 cm dishes. Triplicate experiments were performed for each cell line. The medium was changed every 3-4 days. After 10 days, the cells were fixed and stained with crystal violet.
Soft Agar Assay
A 2 ml of lower layer of 0.6% agar in RPMI-10% FBS was placed into each well of 6 well plate. After agar solidified, 2 ml of 0.3% top agarose in RPMI-10% FBS containing 10,000 cells was added to each well. The cells were fed every 3-4 days with RPMI-10% FBS. The plates were incubated at 37°C, in 5% CO2 incubator for 2 weeks. The colonies that are larger than 100μm were counted.
In vivo tumorigenicity assay
1×105 control MPT cells or Pim1 knockdown cells were mixed with 15μl of Matrigel (Becton Dickinson Labware) and injected subcutaneously in both flanks of 8-week-old male athymic nude mice (BALB/c strain; Harlan Sprague Dawley). Grafts were measured weekly since they were visible. All mice were sacrificed by 12 weeks after injection. Animal care and experiments were carried out according to the protocols approved by the Institutional Animal Care and Use Committees at Vanderbilt University. Grafts were paraffin-embedded, sectioned, stained with H&E, and analyzed by light microscopy. A tumor was defined as a palpable mass that contained carcinoma cells upon histological examination. Immunostaining was performed as described (Abdulkadir et al 2000, Abdulkadir et al 2001) using anti-synaptophysin (BD Biosciences), anti-androgen receptor (Santa Cruz Biotechnology).
Supplementary Material
Acknowledgements
We thank Drs. Robert Matusik, Simon Hayward, Vito Quaranta, Xiuping Yu, and Fritz Parl for helpful discussions. Micrographs were taken from Cell Imaging Core at Vanderbilt University Medical Center. This work was supported by grant RO1CA123484 from the NCI to S.A.A. while D.G. is supported by R01CA152601 from the NCI and BC093803 from the DOD. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest: The authors declare no conflict of interest.
References
- Abdulkadir SA, Carvalhal GF, Kaleem Z, Kisiel W, Humphrey PA, Catalona WJ, et al. Tissue factor expression and angiogenesis in human prostate carcinoma. Hum Pathol. 2000;31:443–447. doi: 10.1053/hp.2000.6547. [DOI] [PubMed] [Google Scholar]
- Abdulkadir SA, Carbone JM, Naughton CK, Humphrey PA, Catalona WJ, Milbrandt J. Frequent and early loss of the EGR1 corepressor NAB2 in human prostate carcinoma. Hum Pathol. 2001;32:935–939. doi: 10.1053/hupa.2001.27102. [DOI] [PubMed] [Google Scholar]
- Aho TL, Sandholm J, Peltola KJ, Mankonen HP, Lilly M, Koskinen PJ. Pim-1 kinase promotes inactivation of the pro-apoptotic Bad protein by phosphorylating it on the Ser112 gatekeeper site. FEBS Lett. 2004;571:43–49. doi: 10.1016/j.febslet.2004.06.050. [DOI] [PubMed] [Google Scholar]
- Berns A, Mikkers H, Krimpenfort P, Allen J, Scheijen B, Jonkers J. Identification and characterization of collaborating oncogenes in compound mutant mice. Cancer Res. 1999;59:1773s–1777s. [PubMed] [Google Scholar]
- Burton GR, Nagarajan R, Peterson CA, McGehee RE., Jr. Microarray analysis of differentiation-specific gene expression during 3T3-L1 adipogenesis. Gene. 2004;329:167–185. doi: 10.1016/j.gene.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Chen J, Kobayashi M, Darmanin S, Qiao Y, Gully C, Zhao R, et al. Hypoxia-mediated up-regulation of Pim-1 contributes to solid tumor formation. Am J Pathol. 2009a;175:400–411. doi: 10.2353/ajpath.2009.080972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LS, Redkar S, Bearss D, Wierda WG, Gandhi V. Pim kinase inhibitor, SGI-1776, induces apoptosis in chronic lymphocytic leukemia cells. Blood. 2009b;114:4150–4157. doi: 10.1182/blood-2009-03-212852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen WW, Chan DC, Donald C, Lilly MB, Kraft AS. Pim family kinases enhance tumor growth of prostate cancer cells. Mol Cancer Res. 2005;3:443–451. doi: 10.1158/1541-7786.MCR-05-0007. [DOI] [PubMed] [Google Scholar]
- Davis AC, Wims M, Spotts GD, Hann SR, Bradley A. A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 1993;7:671–682. doi: 10.1101/gad.7.4.671. [DOI] [PubMed] [Google Scholar]
- Grundler R, Brault L, Gasser C, Bullock AN, Dechow T, Woetzel S, et al. Dissection of PIM serine/threonine kinases in FLT3-ITD-induced leukemogenesis reveals PIM1 as regulator of CXCL12-CXCR4-mediated homing and migration. J Exp Med. 2009;206:1957–1970. doi: 10.1084/jem.20082074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu JJ, Wang Z, Reeves R, Magnuson NS. PIM1 phosphorylates and negatively regulates ASK1-mediated apoptosis. Oncogene. 2009;28:4261–4271. doi: 10.1038/onc.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XF, Li J, Vandervalk S, Wang Z, Magnuson NS, Xing PX. PIM-1-specific mAb suppresses human and mouse tumor growth by decreasing PIM-1 levels, reducing Akt phosphorylation, and activating apoptosis. J Clin Invest. 2009;119:362–375. doi: 10.1172/JCI33216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager J, Gremeaux T, Gonzalez T, Bonnafous S, Debard C, Laville M, et al. Tpl2 kinase is upregulated in adipose tissue in obesity and may mediate interleukin-1beta and tumor necrosis factor-{alpha} effects on extracellular signal-regulated kinase activation and lipolysis. Diabetes. 2010;59:61–70. doi: 10.2337/db09-0470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Roh M, Abdulkadir SA. Pim1 promotes human prostate cancer cell tumorigenicity and c-MYC transcriptional activity. BMC Cancer. 2010;10:248. doi: 10.1186/1471-2407-10-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolch W. Meaningful relationships: the regulation of the Ras/Raf/MEK/ERK pathway by protein interactions. Biochem J 351 Pt. 2000;2:289–305. [PMC free article] [PubMed] [Google Scholar]
- Laird PW, van der Lugt NM, Clarke A, Domen J, Linders K, McWhir J, et al. In vivo analysis of Pim-1 deficiency. Nucleic Acids Res. 1993;21:4750–4755. doi: 10.1093/nar/21.20.4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiblich A, Cross SS, Catto JW, Pesce G, Hamdy FC, Rehman I. Human prostate cancer cells express neuroendocrine cell markers PGP 9.5 and chromogranin A. Prostate. 2007;67:1761–1769. doi: 10.1002/pros.20654. [DOI] [PubMed] [Google Scholar]
- Magnuson NS, Wang Z, Ding G, Reeves R. Why target PIM1 for cancer diagnosis and treatment? Future Oncol. 2010;6:1461–1478. doi: 10.2217/fon.10.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkers H, Nawijn M, Allen J, Brouwers C, Verhoeven E, Jonkers J, et al. Mice deficient for all PIM kinases display reduced body size and impaired responses to hematopoietic growth factors. Mol Cell Biol. 2004;24:6104–6115. doi: 10.1128/MCB.24.13.6104-6115.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Morishita D, Katayama R, Sekimizu K, Tsuruo T, Fujita N. Pim kinases promote cell cycle progression by phosphorylating and down-regulating p27Kip1 at the transcriptional and posttranscriptional levels. Cancer Res. 2008;68:5076–5085. doi: 10.1158/0008-5472.CAN-08-0634. [DOI] [PubMed] [Google Scholar]
- Mumenthaler SM, Ng PY, Hodge A, Bearss D, Berk G, Kanekal S, et al. Pharmacologic inhibition of Pim kinases alters prostate cancer cell growth and resensitizes chemoresistant cells to taxanes. Mol Cancer Ther. 2009;8:2882–2893. doi: 10.1158/1535-7163.MCT-09-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian KC, Wang L, Hickey ER, Studts J, Barringer K, Peng C, et al. Structural basis of constitutive activity and a unique nucleotide binding mode of human Pim-1 kinase. J Biol Chem. 2005;280:6130–6137. doi: 10.1074/jbc.M409123200. [DOI] [PubMed] [Google Scholar]
- Roh M, Gary B, Song C, Said-Al-Naief N, Tousson A, Kraft A, et al. Overexpression of the oncogenic kinase Pim-1 leads to genomic instability. Cancer Res. 2003;63:8079–8084. [PubMed] [Google Scholar]
- Shah N, Pang B, Yeoh KG, Thorn S, Chen CS, Lilly MB, et al. Potential roles for the PIM1 kinase in human cancer - a molecular and therapeutic appraisal. Eur J Cancer. 2008;44:2144–2151. doi: 10.1016/j.ejca.2008.06.044. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verbeek S, van Lohuizen M, van der Valk M, Domen J, Kraal G, Berns A. Mice bearing the E mu-myc and E mu-pim-1 transgenes develop pre-B-cell leukemia prenatally. Mol Cell Biol. 1991;11:1176–1179. doi: 10.1128/mcb.11.2.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Kim J, Roh M, Franco OE, Hayward SW, Wills ML, et al. Pim1 kinase synergizes with c-MYC to induce advanced prostate carcinoma. Oncogene. 2010;29:2477–2487. doi: 10.1038/onc.2010.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wang Z, Li X, Magnuson NS. Pim kinase-dependent inhibition of c-Myc degradation. Oncogene. 2008;27:4809–4819. doi: 10.1038/onc.2008.123. [DOI] [PubMed] [Google Scholar]
- Zippo A, De Robertis A, Serafini R, Oliviero S. PIM1-dependent phosphorylation of histone H3 at serine 10 is required for MYC-dependent transcriptional activation and oncogenic transformation. Nat Cell Biol. 2007;9:932–944. doi: 10.1038/ncb1618. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.