Abstract
The human and Old World primate genomes possess conserved endogenous retrovirus sequences that have been implicated in evolution, reproduction, and carcinogenesis. Human endogenous retrovirus (HERV)-K with 5′LTR-gag-pro-pol-env-rec/np9-3′LTR sequences represents the newest retrovirus family that integrated into the human genome 1 to 5 million years ago. Although a high-level expression of HERV-K in melanomas, breast cancers, and teratocarcinomas has been demonstrated, the mechanism of the lineage-specific activation of the long terminal repeat (LTR) remains obscure. We studied chromosomal HERV-K expression in MeWo melanoma cells in comparison with the basal expression in human embryonic kidney 293 (HEK293) cells. Cloned LTR of HERV-K (HML-2.HOM) was also characterized by mutation and transactivation experiments. We detected multiple transcriptional initiator (Inr) sites in the LTR by rapid amplification of complementary DNA ends (5′ RACE). HEK293 and MeWo showed different Inr usage. The most potent Inr was associated with a TATA box and three binding motifs of microphthalmia-associated transcription factor (MITF). Both chromosomal HERV-K expression and the cloned LTR function were strongly activated in HEK293 by transfection with MITF-M, a melanocyte/melanoma-specific isoform of MITF. Coexpression of MITF and the HERV-K core antigen was detected in retinal pigmented epithelium by an immunofluorescence analysis. Although malignant melanoma lines MeWo, G361, and SK-MEL-28 showed enhanced HERV-K transcription compared with normal melanocytes, the level of MITF-M messenger RNA persisted from normal to transformed melanocytes. Thus, MITF-M may be a prerequisite for the pigmented cell lineage-specific function of HERV-K LTR, leading to the high-level expression in malignant melanomas.
Introduction
The human genome contains 2.7 x 106 retroelements that cover 42% of the total chromosomal DNA (3 billion nucleotides) [1]. Unlike other forms of retroelements, endogenous retroviruses (ERVs), which make up 8% of the present-day human genomic sequences [1], are derived from archaic infectious retroviruses that integrated into hominids including humans and great apes. Human ERVs (HERVs) represent ERVs prevailing in the human genome [2]. Studies indicated roles of the envelope protein of HERV-W, syncytin, in placental development [3,4] and in neuroinflammation [5].
HERV-K represents the newest family of HERVs that was integrated into the genome approximately 1 to 5 million years ago [6]. HERV-K proviruses with well-conserved 5′LTR-gag-pro-pol-env-rec/np9-3′LTR structure, in which LTR signifies the long terminal repeat, exist at 30 to 50 loci on the human chromosomes [1,7,8]. HERV-K types I and II encode regulatory proteins, Np9 and Rec, respectively, at their 3′ termini, which was expressed through double splicing [9,10]. Rec and Np9 exert an oncogenic function by interacting with cellular zinc-finger transcription factors including the promyelocytic leukemia zinc finger (PLZF) protein [11–13]. Intriguingly, high-level HERV-K expression was detected in breast cancers, teratocarcinomas, and melanomas [9,14–17]. Very importantly, intensification of HERV-K expression has been implicated in malignant transformation of melanomas [18–20].
HERV-K transcription is enhanced by stepwise incubation with estradiol and progesterone in breast cancer cells [21,22], although it remains unclear whether the LTR function is directly controlled by the nuclear receptors. Involvement of CpG methylation in the LTR-controlling mechanism was also suggested [23]. A model of the U3-R-U5 framework of the LTR has been proposed very recently [24], in which Sp1 and Sp6 were found essential for activation of the TATA-less promoter. However, the mechanism underlying the cancer-specific HERV-K expression remains unknown.
On the other hand, microphthalmia-associated transcription factor (MITF, also termed Mi) is essential for the development of retinal pigmented epithelium (RPE) and neural crest-derived melanocytes [25]. In addition to the genes essential for melanin synthesis and melanosome formation, a number of genes that support cell cycle progression and survival are induced by MITF [26,27]. MITF, having a basic helix-loop-helix, leucine-zipper (bHLH-LZ) structure [28,29], recognizes the E-box, CA(C/T)GTG, as well as the M-box (TCAYRTG or CAYRTGA) sequences in the promoter/enhancer region of the target genes, where Y and R represent pyrimidine and purine, respectively [25,30]. Importantly, the MITF gene (also referred to as MI, WS2A, bHLHe32) is a proposed lineage-specific oncogene, based on the gene amplification and its growth-promoting function in metastatic melanomas [27,31]. Having multiple first exons, MITF gives rise to various isoforms, such as MITF-A, -B, -H and M, having different N-terminal peptides [25,29]. Only the MITF-M isoform (MITF variant 4, NM_000248) undergoes melanocyte/melanoma-specific transcription in response to the α-melanocyte-stimulating hormone signaling [29], whereas other isoforms are controlled by distinct promoters and transcribed in many tissues and cancer cell lines [32].
We studied HERV-K transcription controlling mechanisms by determining the initiator (Inr) sites and enhancer sequences in the LTR. Our results indicate that an arrangement of (MITF motifs)-(TATA box)-(Inr) functions as the core enhancer/promoter inducible by MITF-M.
Materials and Methods
Cell Culture
Adenovirus-transformed human embryonic kidney 293 (HEK293) cells and melanoma lines, MeWo and G361, were supplied by the Japanese Collection of Research Bioresources (Osaka, Japan) and maintained with minimum essential medium supplemented with sodium pyruvate, nonessential amino acids, antibiotics, and 10% fetal bovine serum (FBS). SK-MEL-28 melanoma cells were from the American Type Culture Collection. Human epidermal melanocytes from lightly pigmented adult skin (HEMa-LP) were purchased from Invitrogen (Carlsbad, CA), and were cultured in Medium 254 (calcium/serum-free) with human melanocyte growth supplement (Invitrogen).
Plasmids
The HERV-K LTR sequences (GenBank AF074086.2) were amplified by polymerase chain reaction (PCR) from a Bac clone, RP11-33P21 having approximately 154-kb sequences of chromosome 7. The 5′ and 3′ primers used are NheI-KLTR5-2 (5′-ccgctagctgtggggaaaagcaagagagatca-3′) and BglII-KLTR3-2 (5′-gcagatctcacctgtgggtgtttctcgtaaggt-3′). The amplified LTR sequences (nos. 1–945) were inserted into pGL3-Basic (Promega, Madison, WI) at the NheI/BamHI sites. pGL3-Control (Promega), in which the SV40 early promoter is linked to the Luc gene, was used as a positive control. In vitro mutagenesis was carried out with the Gene Tailor Site-Directed Mutagenesis System and Platinum Taq High Fidelity (Invitrogen). The MITF-M clone, pHMI-9, was provided by Dr Shigeaki Shibahara through the RIKEN Bioresource Center (RDB 1786). The MITF-M-encoding sequences were excised by HindIII/XbaI digestion and recloned into pRc/CMV (Invitrogen) at the HindIII/XbaI sites.
Luciferase Assay
Cells (105 in each well of a 24-well plate) were seeded 24 hours before transfection. Plasmid transfection was with Effectene (Qiagen, Hilden, Germany). Forty-eight hours after the transfection, cells were lysed in Glo-lysis buffer (Promega) for determination of the luciferase activity with the Steady-Glo luciferase assay system (Promega) [33]. For induction with phorbol 12-myristate 13-acetate (PMA), transfected cells (at 16 hours) were starved in the medium containing 0.5% FBS for 8 hours and incubated with or without 20 ng/ml of the drug (Sigma-Aldrich, St Louis, MO) for an additional 24 hours.
Rhesus Monkey Tissues
Tissues of rhesus monkeys, a 13-year-old female and a 6-year-old male, were obtained immediately after killing by exsanguination through the bilateral carotid arteries under deep anesthesia using ketamine hydrochloride and sodium pentobarbital at the Primate Research Institute, Kyoto University, in accordance with the guidelines of the institute.
RNA Purification
Tissues and cultured cells were homogenized in RNAwiz (Ambion, Austin, TX) to purify RNA. Isopropanol-precipitated RNA was further purified with the RNeasy MinElute Cleanup system (Qiagen). DNA fragments possibly contained in the RNA preparations were digested with DNase I (Invitrogen, amplification grade) before reverse transcription (RT)-PCR. The High-Pure RNA Isolation Kit (Roche Diagnostics, Mannheim, Germany) was used for normal melanocytes.
Reverse Transcription-Polymerase Chain Reaction
Semiquantitative RT-PCR was performed with the SuperScript One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen). KP1–2 (targeting nos. 911–933, 5′-ctctcgtcccaccttacgagaaa-3′), KP1 (nos. 916–933, 5′-tcccaccttacgagaaa-3′), KP3 (gag, 5′-gagccattaccggctctgctacat-3′), and KP7 (Rec, 5′-acaaaaccgccatcgtcatcatgg-3′) were newly designed. KP-g3 (5′ end of gag, 5′-tttgccccattatcacccta-3′) and KP-e1 (env, 5′-gtgacatcccgcttaccatgtgataagtg-3′) have been described [9]. For the detection of the MITF isoforms, MITF-A-F (5′-aaagtcaaccgctgaagagcag-3′), MITF-B-F2 (5′-gtatgcattttggttttccca-3′), MITF-H-F (5′-atgttcatgccatgctcctt-3′), and MITF-M-F2(ATG) (5′-atgctggaaatgctagaatataatcact-3′) were used as forward primers. Reverse primers were MITF-b1b-R (5′-actcacgggcactctctgtt) and MITF-R2 (5′-gaatgtgtgttcatgcctgg-3′). Human 18S ribosomal RNA (rRNA) was amplified with Hu-18S-F (5′-GTAACCCGTTGAACCCCATT-3′) and Hu-18S-R (5′-ccatccaatcggtagtagcg-3′). Rhesusmonkey 18S rRNA was tested with Mmula-18S-F (5′-aattccgataacgaacgaga-3′) and Mmula-18S-R (5′-atctaagggcatcacagacc-3′). These primers were synthesized by Invitrogen.
Determination of the Transcriptional Initiation Sites
The 5′ ends of HERV-K full-length RNA were determined by the 5′ rapid amplification of complementary DNA ends (5′ RACE) system (Invitrogen). We used a primer, KP7, targeting the env/rec/np9 region as the gene-specific primer in the RT reaction. KP-g3 (5′-tttgccccattatcacccta-3′) targeting a 5′ terminal gag region and BglII-GSP3-RACE5 (5′-ttccgagctcgctcgagcgtaccttcaccctaga-3′) targeting the leader region were used at the PCR and nested PCR steps, respectively. Products were digested with SpeI and BglII to be cloned into pUC19 and sequenced.
Immunofluorescence Experiments
Human adult normal tissue panels (T8234605) and human fetus tissue arrays (BE01015) were was purchased from Biochain (Hayward, CA) and US Biomax (Rockville, MD), respectively. Immunofluorescence staining was performed as described [34]. After the antigen retrieval by incubation in the LAB solution (Polysciences, Warrington, PA) for 15 minutes at 60°C, cells were permeabilized and incubated with an anti-MITF (rabbit IgG, ab20663; Abcam, Cambridge, United Kingdom) antibody (at a dilution of 1:250) and an anti-HERV-K core (HERM-1831-5, mouse monoclonal; Austral Biologicals, San Ramon, CA) antibody (1:500) for 90 minutes. HERM-1831-5 represents a mouse monoclonal antibody (clone 4D4/F7) obtained by immunization of mice with HERV-K gag antigen (82 kDa). Tissue slides were next incubated with secondary antibodies conjugated with Alexa Fluor 488 or 594 (Invitrogen) for 60 minutes, followed by a 10-minute incubation with Hoechst 33342 (Sigma-Aldrich) for mild nuclear staining. For the control experiment, the anti-HERV-K antibody (1 µg) was neutralized by incubation with the HERV-K gag protein (50 µg) (HERV-K CA; ProteinTech Group, Chicago, IL) in phosphate-buffered saline (500 µl) for 18 hours at 4°C. Fluorescence images were captured by Nikon E600 with the Digital Sight DS-Fi1 system (Nikon, Tokyo, Japan).
Results
Differences in the HERV-K LTR Regulation between HEK293 and MeWo Cells
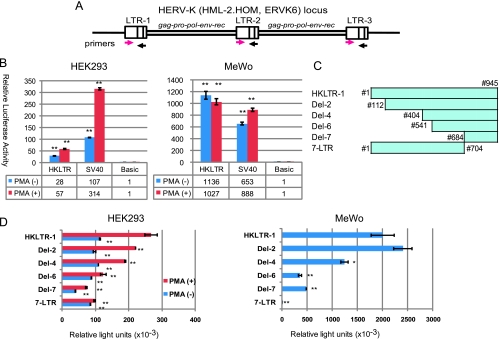
The LTR region (nucleotides nos. 1–945) of HERV-K (HML-2. HOM, human endogenous MMTV-like), also referred to as ERVK6, on chromosome 7 (GenBank AF074086.2) was amplified by PCR from BAC clone RP11-33P21 and inserted into pGL3-Basic. This locus has a tandem repeat of the HERV-K genome with three LTRs: LTR-1, -2, and -3 (Figure 1A) [35]. Our clones HKLTR-1 to -5 were ∼99% identical to the reported sequences. We use the nucleotide numbering of AF074086.2 throughout this report, in which nos. 1 and 968 corresponded to the 5′ and 3′ ends of LTR-1 [35]. On the basis of the human genome sequence data (NT_007819.16 and NT_079592.2) and our sequencing analyses, however, we place an A residue at no. 796 so that nos. 793 to 780 octamer reads ′5′-AATAAATA-3′ instead of ′5′-AATGAATA-3′.
Figure 1.
Detection of the promoter function in cloned HERV-K LTR. (A) The HERV-K locus (HML-2.HOM, ERVK6) on chromosome 7. A tandem repeat of the LTR-gag-pro-pol-env-rec/np9-LTR structure containing LTR-1, -2, and -3 are illustrated as described [35]. The forward and reverse primers used for the LTR sequence amplification are indicated by magenta and black arrows, respectively, at the positions of their target sequences. (B) Luciferase (luc) gene reporter assay with HKLTR-1-Luc in HEK293 and MeWo cells. The pGL3-control plasmid containing the SV40 early promoter (SV40) and pGL3-Basic without a promoter (Basic) were examined as the positive and negative controls of the experiments. Luciferase activity was determined after stimulation with (+) or without (-) PMA (20 ng/ml) in serum reduced (0.5%) medium. Measurements (mean ± SD) are presented in relation to the enzyme activity (=1) obtained by pGL3-Basic. Experiments were carried out in triplicate two times (**P < .01 vs Basic). (C) Deletion mutants examined. Nucleotide numbers (#) contained in each plasmid are indicated. (D) Luc assay with the cloned HERV-K LTR and its deletion mutants. Luciferase activity in transfected HEK293 cells was determined after stimulation with (+) or without (-) PMA (20 ng/ml) in serum-reduced (0.5%) medium. Luciferase activities in MeWo cells incubated with 10% FBS were analyzed. Results (mean ± SD) are shown in relative light units (RLUs). Statistical significance: **P < .01 and *P < .05 versus HKLTR (without PMA stimulation).
In the luciferase gene (Luc) reporter assays, HKLTR-1 was three-fold less active than the control SV40 early promoter (pGL3-control) in human embryonic kidney HEK293 cells without the large T antigen. The strong Luc expression in MeWo (Figure 1B, right) seemed to reflect the high-level chromosomal HERV-K expression. PMA (20 ng/ml) enhanced the HERV-K LTR and the SV40 promoter two-fold and three-fold, respectively, in serum-starved HEK293 cells (Figure 1B, left). Intriguingly, the LTR was not influenced by PMA in serum-starved MeWo cells, whereas the SV40 promoter was 1.4-fold enhanced (Figure 1B, right). The LTR function in MeWo cells may not be further potentiated by protein kinase C pathway stimulation. Other HERV-K LTR clones, HKLTR-2 to -5, were indistinguishable from HKLTR-1 in these and the following assays (data not shown).
Deletion analysis of the HERV-K LTR indicated that the promoter activity is regulated by different regions between HEK293 and MeWo cells (Figure 1, C and D). We were not able to identify a specific region that responded to PMA in HEK293. Most intriguingly, both Del-7 (nos. 684–945) and 7-LTR (nos. 1–704) were able to induce Luc expression in HEK293 cells, whereas Del-7, but not 7-LTR, could exert the promoter function in MeWo. We hypothesized the presence of at least two promoters, one in 7-LTR and the other in Del-7, which are activated in different modes between the two cell lines.
Multiple Initiation Sites in Chromosomal HERV-K LTRs
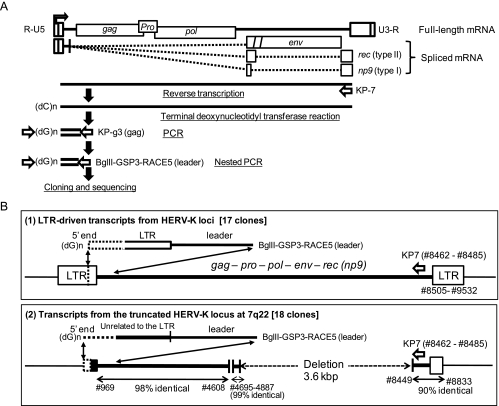
To determine the initiator (Inr) sequences in the HERV-K LTR on the chromosomal DNA, we performed a 5′ RACE experiment with RNA purified from MeWo and HEK293. To determine the 5′ terminus of the full-length viral RNA, we designed gene-specific primers: KP-7 (targeting rec/np9), KP-g3 (gag), and BglII-GSP3-RACE5 (5′ leader) for the RT reaction, PCR, and nested PCR, respectively (Figure 2A). Seventeen of 40 clones obtained from MeWo were derived from the HERV-K LTR-driven viral gene transcripts (Figure 2B, subpanel (1)). Four clones contained unrelated 50 to 80 nucleotide sequences; the other 19 are described in the next section.
Figure 2.
Summary of the 5′ RACE experiments. (A) Strategy for determination of the 5′ end of the viral RNA. HERV-K full-length and spliced RNAs are schematically shown with the gag, PR, pol, env, and rec/np9 genes (top). Experimental processes are illustrated with gene specific primers. (B) Products detected in the experiments. (1) Detection of the transcriptional start points in HERV-K LTR. Twenty-one of 50 clones contained a 3′ terminal region of the 5′ LTR followed by the leader sequences. (2) Expression of a HERV-K-like element rearranged by the loss of 5′ LTR and a 3.6-kb deletion spanning pol and env. The transcripts without a LTR sequence were aligned with the segmented HERV-K locus at 7q22. Twenty-four of fifty 5′ RACE clones contained this structure.
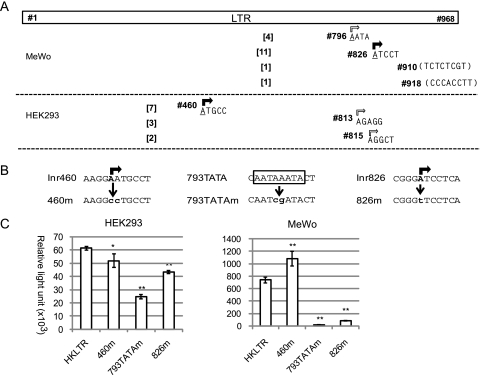
Within the HERV-K LTR-derived 5′ RACE products, we found 11 clones starting at nucleotide no. 826, four clones starting at no. 796, and single clones starting at no. 910 or no. 918 (Figure 3A). All of the detected initiation sites were within the Del-7 region, supporting the Luc assay results in MeWo (Figure 1D). However, nos. 910 (T/U residue) and 918 (C residue) were inappropriate as a conventional initiation nucleotide and were speculated to be 5′ ends of processed RNA. Nucleotide BLAST (Basic Local Alignment Search Tool, National Center for Biotechnology Information) searches indicated that each of the no. 826-initiated transcripts, with an exception of clone-3, was templated by a certain HERV-K locus on chromosome 1, 6, 7, 19, or 22 on the basis of the nucleotide substitutions at a few positions (Figure W1B). We failed to identify the templates for the no. 796-initiated transcript (four clones) having an A-to-T alteration at no. 929 in the human genomic sequences (Figure W1A). Cell line-specific chromosomal alteration and locus-specific transcriptional activation were speculated for the minor initiation at no. 796. Thus, no. 826 seemed to serve as the most potent Inr (Inr826), which was functional at several loci on the human chromosomes. Interestingly, we found, 25 bp upstream of Inr826, octanucleotide 5′-AATAAATA-3′ (nos. 793–800) matching the TATA box consensus sequence TATA(A/T)A(A/T) [36,37] (Figure 3B).
Figure 3.
Multiple Inrs detected in the HERV-K LTR. (A) Results of the 5′ RACE analyses. Positions of the 5′ ends (rectangular arrows) of the HERV-K mRNA from MeWo and HEK293 cells are schematically shown. Number of detected clones (from among the 40 clones) is in brackets. (B) Mutation of Inr460, 793TATA, and Inr826. The original nucleotide sequences and mutated sequences in 460m, 793TATAm, and 826m are indicated. Major and minor initiation sites are indicated by filled and open rectangular arrows, respectively. The 793TATA sequences, 5′-TATA(T/A)A(A/T)(A/G)-3′, corresponding to the TATA box consensus are boxed. (C) Luc assay with HKLTR-1 and the Inr mutants in HEK293 and MeWo cells. Luciferase activity (mean ± SD) is shown in RLUs. Experiments were performed in triplicate two times. Statistical significance: **P < .01 and *P < .05 versus HKLTR-1.
Within the 40 clones of the 5′ RACE products from HEK293 RNA, we identified 7 starting at no. 460, 3 starting at no. 813, and 2 starting at no. 815 (Figures 3A and W2). These were also consistent with the Luc assay result that both 7-LTR and Del-7 acted as an independent promoter in HEK293 cells. Nucleotide no. 460 seemed to be the major initiation site in this cell line and was tentatively termed Inr460. We noted that all of the 5′ RACE clones starting at Inr460 shared 33 nucleotide substitutions when compared with HERV-K (HML-2.HOM) and were derived from a locus on chromosome 3.
Detection of HERV-K-like Transcripts without a 5′ LTR
Nineteen of the 40 clones obtained from MeWo had 5′-terminal unknown sequences preceding the 5′ leader sequences of HERV-K (Figures 2B, subpanel (2), and W1C). We could identify the template sequences at 7q22, which showed a 3.6-kb deletion spanning the pol and env genes (ref: NT_007933.15 and NT_2079596). Because of the long deletion, these transcripts may have been more efficiently reverse transcribed and amplified in the 5′ RACE experiment than the full-length HERV-K messenger RNA (mRNA).
Requirement of the Inr Sequences for the LTR Function
To test whether the major Inr sites (Inr460 and Inr826) and the TATA box-like sequences (793TATA) are necessary for the LTR function, we constructed mutants termed HKLTR-460m, -826m, and 793TATAm (Figure 3B). In MeWo cells, the 460m mutation even increased the luciferase activity 1.4-fold (Figure 3C), suggesting the possibility that no. 460 is involved in the mechanism to render 7-LTR inactive in MeWo (Figure 1D). The 793TATAm and 826m mutations decreased the Luc expression by 97% and 88%, respectively (Figure 3C), indicating that both 793TATA and Inr826 play an essential role in the cloned LTR.
Unlike the result in MeWo, luciferase activity produced from HKLTR-460m was 15%decreased in HEK293 (Figure 3C). Although Inr826-initiated transcription was not detected by 5′ RACE, Luc expression by 793TATAm and 826m were decreased 60% and 30%, respectively. Thus, 793TATA and Inr826 were also vital in HEK293. Inr460 seemed to contribute to the LTR function in HEK293, but less significantly. The predominant appearance of the Inr460-initiated transcript (Figure 3A) may have caused by biased Inr usage at the unique locus (Figure W2A) in the nonproducer cell line.
Activation of the Cloned HERV-K LTR by MITF-M
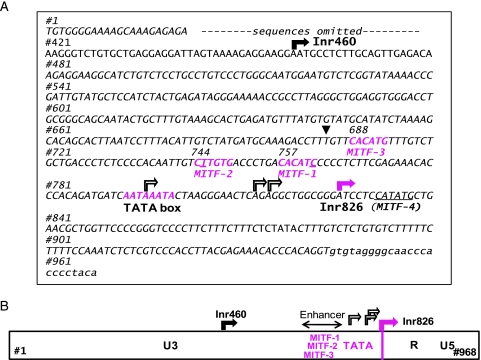
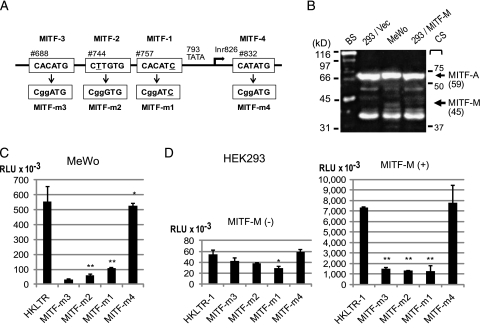
Because HERV-K is efficiently expressed in melanomas and teratocarcinomas, transcription regulatory factor(s) governing the pigmented tissue development could be considered as a potential HERV-K inducer(s). MITF, the MITF-M isoform in particular, is a melanocytic master transcription factor and an oncogene of melanoma [38–40]. Nucleotide sequences corresponding to E-box and related sequences including CACATG, CATGTG, and CATATG serve as the core target sites of MITF to induce gene expression [25]. Intriguingly, three hexanucleotides resembling the MITF binding core motifs were detected upstream of 793TATA within the Del-7 region (Figure 4A): MITF-1 (CACATC starting at no. 757, with one nucleotide mismatch underlined), MITF-2 (CTTGTG at no. 744), and MITF-3 (CACATG at no. 688). An additional motif, MITF-4 (CATATG at no. 832), was found six bases downstream of Inr826 (Figure 5A). We inserted one or two nucleotide mutations into each MITF motif to generate mutants termed MITF-1m (CggATG), MITF-2m (CggGTG), MITF-3m (CggATC), and MITF-4m (CggATG). Results of the Luc assays indicated that MITF-1, -2, and -3, but not MITF-4, contribute significantly to the promoter function of the LTR in MeWo cells (Figure 5C).
Figure 4.
Transcriptional regulatory elements in HERV-K LTR. (A) Nucleotide nos. 1 to 968 of HERV-K LTR are shown according to the numbering in GenBank AF074086.2 (HML-2.HOM). Rectangular arrays indicate the major initiation sites (Inr460 and Inr826) determined by 5′ RACE. MITF-M-responsive sequences (MITF-1, -2, and -3) and 793TATA are highlighted (magenta). MITF-4 is underlined. A list of MITF binding core sequences used in the known MITF-induced genes is provided in reference [25]. The first nucleotide (no. 684) of the Del-7 region is marked by an arrowhead. (B) Proposed framework of HERV-K LTR. Inr826 defines the U3-R junction. Relative positions of Inr826, 793TATA, and the enhancer region formed by MITF-1, -2, and -3 are indicated. Inr460 is also indicated.
Figure 5.
MITF-binding motifs essential for the HERV-K LTR activation by MITF-M. (A) (Top) An array of the MITF-binding motifs (MITF-1, -2, -3, and -4) is graphically shown in relation to the initiation sites (Inr826). (Bottom) Mutated nucleotides in MITF-m1, -m2, -m3, and -m4 are indicated in lower case. (B) Detection of MITF-M by Western blot analysis. Lysates of HEK293 (293), MeWo, and HEK293 transfected with MITF-M (293/MITF-M) were probed with an anti-MITF antibody. Molecular masses (kDa) and positions of the biotinylated protein standards (BS) and prestained color standards (CS) are indicated. MITF-A (520 amino acid residues, 59 kDa; GenBank BAA32288.1; National Center for Biotechnology Information Reference Sequence: NP_937802.1) and 44-kDa MITF-M (419 residues; NM_000248.3) are indicated on the basis of their molecular masses and the RT-PCR analyses shown in Figure 6. (C) Luc assay with the MITF motif mutants in MeWo. Experiments were performed in triplicate two times. Luciferase activity (mean± SD) is shown in RLUs. Statistical significance: **P < .01 and *P < .05 versus HKLTR-1. (D) Transactivation of the HERV-K LTR by MITF-M in HEK293 cells. HKLTR-1 and the mutants were transfected in combination with a CMV promoter-driven MITF-M expression vector (+) or with the empty vector (-) at a weight ratio of 1:1.
The basal-level Luc expression in HEK293 was not greatly altered by any of the MITF motif mutations (Figure 5D). When HKLTR-luc was cotransfected with a CMV promoter-driven MITF-M expression vector, a substantial increase in Luc expression was observed (∼140-fold increase in the luciferase activity). This strong transactivation by MITF-M was cancelled in the MITF-1m, MITF-2m, and MITF-3m mutants, but not in MITF-4m. In Western blot analysis, the MITF-M protein (419 amino acids long) appeared as a minor band of ∼45 kDa in MITF-M-transfected HEK293 (293/MITF-M) cells as well as in MeWo (Figure 5B). In contrast, HEK293 cells transfected with the control vector (293/vec) lacked this protein. The Del-7 region (nos. 684–945) as well as the entire LTR clone exhibited a dose-dependent response to MITF-M in HEK293, whereas 7-LTR was insensitive to MITF-M (Figure W3). Thus, forced expression of MITF-M in HEK293 caused strong HERV-K LTR activation depending on the MITF-1, -2, and -3 motifs.
Induction of Chromosomal HERV-K by MITF-M
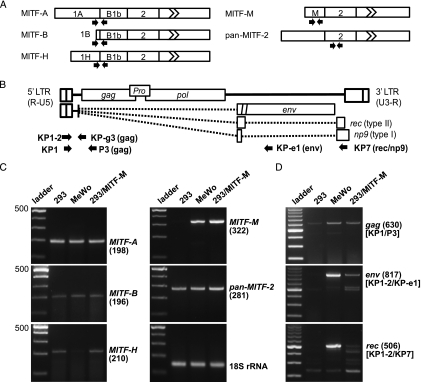
We analyzed MITF gene expression in HEK293 and MeWo by RT-PCR with isoform-specific primers (Figure 6, A and C). MITF-A and MITF-B were transcribed equally in these cell lines. MITF-H was expressed more efficiently in HEK293 than in MeWo. MITF-M was clearly expressed in MeWo but not in HEK293. Total MITF expression did not significantly differ between the cell lines as analyzed with the pan-MITF-2 primer pairs. HEK293 cells transfected with the MITF-M expression vector showed a substantial increase in the MITF-M mRNA without an alteration in other isoforms. These results were consistent with the Western blot analysis results, in which two dominant MITF bands appeared equally in the two cell lines, whereas MITF-M was detectable in MeWo, but not in HEK293 (Figure 5D) as observed in various cancer cell lines [32].
Figure 6.
Induction of chromosomal HERV-K expression by forced expression of MITF-M. (A) MITF variant mRNAs with 5′ terminal isoform-specific exons are shown in ribbon-like structures. Exons 3 to 9 were compressed in the finned box. Positions of the forward and reverse primers are indicated by arrows. The pan-MITF-2 primer pairs were designed to detect exon 2 shared by the known MITF variants. (B) Structure of the gag-encoding full-length HERV-K RNA and spliced mRNAs encoding env and rec/np9. Splicing events are indicated by dotted lines. Arrows show positions of the primers used in RT-PCR. (C) Results of the RT-PCR experiments for MITF mRNAs. HEK293 (293), MeWo, and MITF-M transfected HEK293 (293/MITF-M) were analyzed. Position of the 500-bp marker and sizes (in bp) of the amplified products are indicated in parentheses. The control reaction for 18S ribosomal RNA is also shown. (D) RT-PCR analyzing the gag, env, and rec mRNAs. Primers applied are shown in brackets.
We next examined these cells for production of full-length HERV-K mRNA encoding gag-pol, spliced env mRNA, and doubly spliced rec/np9 mRNA (Figure 6, B and D). Forward primers (KP1, KP1-2) were designed to hybridize sequences downstream of Inr826. Consistent with the previous report [9], high-level expression of the spliced env mRNA and the doubly spliced rec mRNA was evident in MeWo, whereas neither the env nor the rec RT-PCR product was detectable in HEK293. The gag-encoding viral RNA was found in HEK293 at a lower level, whereas it was increased three-fold in MeWo. Interestingly, MITF-M-transfected HEK293 cells significantly enhanced the env and rec mRNA synthesis. The gag-encoding RNA was also increased two-fold by MITF-M. Thus, forced expression of MITF-M caused chromosomal HERV-K activation in HEK293.
Elements Conserved between the Human and Rhesus-type HERV-K LTRs
We detected 113 LTR copies with 95% or higher sequence identity to the HERV-K LTR (HML-2.HOM) in the human genomic sequences (Table W1). MITF-1, MITF-2, MITF-3, 793TATA, and Inr826 were conserved in 108 LTRs, 102 of which had an equivalent MITF motif (5′CACGTG3′) in place of MITF-3 (5′CACATG3′). Inr460 was also conserved in 86 LTRs but inactivated by a nucleotide substitution (A-to-C) in 27 LTRs.
In the rhesus monkey genome, we detected 33 LTR-like elements with 80% or higher nucleotide identity to the HERV-K LTR (Table W2). The GGA(Inr460)ATGC sequence was disrupted in 12 LTRs but existed in other LTRs in a modified form (AGAAGGC), which may possibly retain the Inr function. Twenty-seven LTRs had 793TATA, whereas four showed a single nucleotide substitution in the octanucleotide. None of the rhesus LTRs had an Inr826-like sequence. However, a heptanucleotide matching the consensus of Inr, PyPyA+1N(T/A)PyPy [41,42], was inserted at a position corresponding to Inr826 in 13 LTRs. Three MITF motifs were contained in three LTRs, two MITF motifs in six LTRs, and only one MITF motif in 23 LTRs. The rhesus version of HERV-K could possibly be controlled by mechanisms similar to those found in human HERV-K.
Detection of MITF and HERV-K Expression in RPE
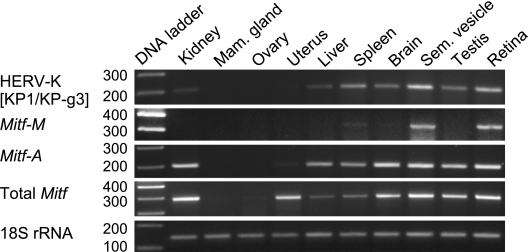
Rhesus monkey tissues were examined for MITF and rhesus-type HERV-K expression by RT-PCR (Figure 7). The human MITF primer pairs shown previously (Figure 6A) were also used in this experiment because the MITF isoform sequences were identical between the two species. The primer pair (KP-1/KP-g3) targeting the sequences shared by the human and rhesus-type HERV-K was used to detect the gag-encoding transcript. Rhesus 18S ribosomal RNA was amplified for normalization.
Figure 7.
Expression of the older version HERV-K and MITF in rhesus monkey tissues. Rhesus monkey (Macaca mulatta) tissues including the mammary gland (Mam. gland) and seminal vesicle (Sem. vesicle) were examined by RT-PCR for expression of the rhesus HERV-K-like elements and the MITF variants. Macaca (Mac) 18S rRNA was also analyzed for standardization of the RNA amounts.
The MITF-A mRNA level was similarly high in the kidney, brain, seminal vesicle, testis, and retina but was undetectable in the mammary gland and ovary under the assay conditions. Enhanced MITF-M expression was observed not only in the retina but also in the seminal vesicle. The spleen and brain seemed to produce MITF-M mRNA less predominantly. Total MITF expression was most intense in the kidney, retina, and seminal vesicle but very weak in the mammary gland and ovary. The MITF expression profiles in the rhesus tissues were consistent with the expression pattern extracted from the human EST database and conventional analyses [32], although the seminal vesicle has not been examined previously.
Rhesus-type HERV-K expression was most active in the pigmented tissues, retina, and seminal vesicle [43], supporting the observation that the rhesus-type LTRs also contain MITF motifs (Table W2). Less predominant but correlated expression of HERV-K and MITF-M were detected in the spleen and brain. Other tissues including the testis and kidney showed a level of HERV-K mRNA, which was not correlated to MITF-M mRNA.
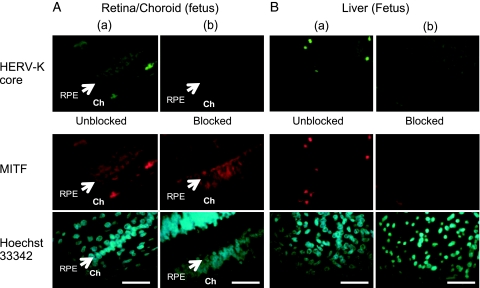
We performed an immunofluorescence analysis with a human fetus tissue array (Figure 8). Because MITF-M-specific antibody was not available, an antibody (ab20663) recognizing the C-terminal domain shared by all of the MITF isoforms was used in combination with an anti-HERV-K core antibody (HERM-1831). In the retina/choroid sections, RPE displayed a strong label by MITF (red, Figure 8A, column a) as reported [44]. The anti-HERV-K antibody also stained RPE strongly (green). The choroid region was poorly reactive with these antibodies. In the control experiment, the anti-HERV-K antibody neutralized with the HERV-K gag peptide failed to stain the RPE (Figure 8A, column b). Other tissues including the liver (Figure 8B) were moderately or poorly reactive with the anti-MITF and anti- HERV-K antibodies. Immunofluorescence images of the seminal vesicle sections were not informative because of an intense autofluorescence of the lipofuscin granules in the epithelium [43] (data not shown).
Figure 8.
Detection of MITF and HERV-K in human RPE. A normal fetus tissue array (BE01015) was labeled for the MITF (red) and HERV-K core (green) proteins. Unblocked (column a) and blocked (column b) anti-HERV-K core (HERM-1831) IgG was used in combination with anti-MITF IgG (ab20663). Uncropped images of the retina/choroid and liver sections are shown. RPE (arrow) and the choroid (Ch) are indicated. Mild nuclear staining with Hoechst 33342 is also shown. Scale bar, 50 µm.
MITF-M and HERV-K Expression in Normal and Transformed Melanocytes
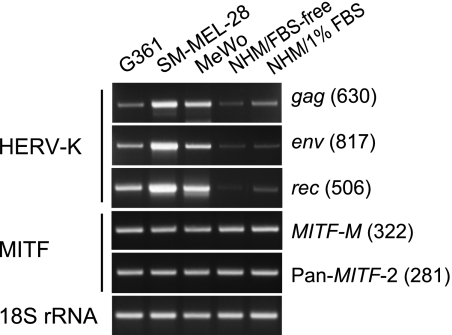
Three malignant melanoma lines, G361, SK-MEL-28, and MeWo, were compared with normal human epidermal melanocytes for the levels of MITF and HERV-K expression by semiquantitative RT-PCR (Figure 9). The levels of total MITF isoforms and MITF-M were constant in the melanoma lines and normal melanocytes. However, G361, SK-MEL-28, and MeWo showed different levels of the gag (full-length) (1:4:2.7), env (1:3:1.4), and rec (1:2:1.5) mRNAs. In normal melanocytes cultured in the serum/calcium-free medium, the amounts of HERV-K mRNAs were only 3% to 10% of those in MeWo. When exposed to 1% FBS for 9 hours, the melanocyte culture increased the gag and rec mRNAs three-fold or higher without a further increase in MITF-M mRNA. Thus, the level of HERV-K transcription was not directly correlated with that of MITF-M in melanoma cells and melanocytes, consistent with the requirement of post-translational modifications including Ser73 phosphorylation by Erk2 for functional activation of MITF-M [25,28,45,46]. MITF-M was thought to determine the melanocyte/melanoma lineage specificity of HERV-K expression.
Figure 9.
Expression of MITF and HERV-K mRNAs in normal and transformed melanocytes. Semiquantitative RT-PCR was performed with RNA purified from normal human melanocytes (NHM/FBS-free) and melanoma lines, G361, SK-MEL-28, and MeWo. NHM cultured with 1% FBS for 9 hours (NHM/1% FBS) was also analyzed. MITF-M, total MITF, and HERV-K mRNAs (gag, env, and rec), and 18S rRNA were amplified with primers shown in Figure 6.
Discussion
We studied transcriptional activation of the HERV-K LTR in melanoma cells. The three MITF binding motifs, the TATA box (793TATA), and the major Inr site (Inr826) seemed to serve as a potent enhancer/promoter unit. In nonmelanoma HEK293 cells, both the cloned LTR function and chromosomal HERV-K expression were strongly activated by forced expression MITF-M, an RPE/melanocyte-specific isoform of MITF. Requirement of MITF-M for the LTR activation seemed to explain the lineage-specific expression of HERV-K.
Endogenous retrovirus expression may undergo varied pathways of transcriptional regulation depending on cell lineages and cellular contexts as reported previously [24,47]. Presence of the multiple Inr sites may reflect the complexity of cellular transcription regulatory mechanisms and the flexibility of retroviral LTRs, allowing ubiquitous basal-level expression [1]. By focusing on the high-level expression of HERV-K in melanoma, we identified MITF-M as an inducer of the HERV-K LTR. The arrangement of MITF binding motifs (MITF-1, -2, and -3), 793TATA, and Inr826 (Figure 4B) may compose a typical enhancer/promoter structure for RNA polymerase II as found in all of the retroviral LTRs studied so far [48]. Because the 5′ end of the R region is defined by the Inr, the U3 region the HERV-K LTR is thought to span from no. 1 to no. 825. This region is long enough to recruit other regulatory proteins, which may cooperate with the initiation factors associated with the core sequences in melanomas.
In normal melanocytes under the calcium/serum-free conditions, MITF-M was highly expressed without inducing HERV-K transcription. Interestingly, serum stimulation enhanced HERV-K transcription in these cells. The transactivating function of MITF-M is known to be enhanced on Ser73 phosphorylation by MAP kinase in response to c-kit [28]. Varied signaling molecules that control MITF-M have been reported [25,45,46]. Melanocytes may gain the constitutive MITF-M-activating functions during the malignant transformation. HEK293 cells would be able to activate it but only if it is synthesized in the cells.
Our results do not rule out the possibility that other MITF isoforms are involved in HERV-K LTR activation by a direct or indirect interaction with MITF-M. Moreover, the TFE family proteins including TFE3, TFEb, and TFE are closely related to the MITF proteins and also interact with the canonical E-box motif, CA(C/T)GTG, and related sequences [29,49]. MITF and melanocyte-specific genes are activated in breast cancers [50], which might contribute to the HERV-K expression in the cell type [21,51].
Retroviruses trigger oncogenesis by varied mechanisms including 1) virus-encoded oncogene(s) and 2) promoter/enhancer insertion upstream of tumor-inducing genes [48]. The rec/np9 genes of HERV-K are expressed through double splicing as found in the tax gene, an oncogene of human T-cell leukemia virus [48]. The Rec/Np9 proteins also seemed to exert an oncogenic function through the interaction with cellular zinc-finger proteins [11–13]. In addition, MITF-M activates not only the 5′ LTR that controls the viral gene expression but also the 3′ LTR, which could activate cell growth/survival genes downstream of the HERV-K loci. Melanoma metastasis involves varied mechanisms including integrin α4 expression [52], chromosomal instability [53], microRNA expression [54], and loss of AP-2 [55]. HERV-K may possibly influence the processes of melanoma progression through the MITF-M-mediated LTR activation and/or Rec/Np9 production. Recent studies on HERV-K also support this hypothesis [18–20].
Supplementary Material
Acknowledgments
The authors thank Shigeaki Shibahara (Tohoku University) for the MITF-M clone and Heui-Soo Kim (Pusan National University) for discussion.
Abbreviations
- MITF
microphthalmia-associated transcription factor
- 5′ RACE
rapid amplification of complementary DNA ends
- RPE
retinal pigmented epithelium
Footnotes
This work was supported in part by the Cooperation Research Program of Primate Research Institute, Kyoto University (H. H.) and by a Grant-in-Aid for Scientific Research (C) 21590434 (I. K.) from MEXT Japan.
This article refers to supplementary materials, which are designated by Figures W1 to W3 and Tables W1 and W2 and are available online at www.neoplasia.com.
References
- 1.Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101(suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tristem M. Identification and characterization of novel human endogenous retrovirus families by phylogenetic screening of the Human Genome Mapping Project Database. J Virol. 2000;74:3715–3730. doi: 10.1128/jvi.74.8.3715-3730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frendo J-L, Olivier D, Cheynet V, Blond J-L, Bouton O, Vidaud M, Rabreau M, Evain-Brion D, Mallet F. Direct involvement of HERV-W env glycoprotein in human trophoblast cell fusion and differentiation. Mol Cell Biol. 2003;23:3566–3574. doi: 10.1128/MCB.23.10.3566-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc Natl Acad Sci USA. 2004;101:1731–1736. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Antony JM, van Marle G, Opii W, Butterfield DA, Mallet F, Yong VW, Wallace JL, Deacon RM, Warren K, Power C. Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nat Neurosci. 2004;7:1088–1095. doi: 10.1038/nn1319. [DOI] [PubMed] [Google Scholar]
- 6.Jha AR, Pillai SK, York VA, Sharp ER, Storm EC, Wachter DJ, Martin JN, Deeks SG, Rosenberg MG, Nixon DF, et al. Cross-sectional dating of novel haplotypes of HERV-K 113 and HERV-K 115 indicate these proviruses originated in Africa before Homo sapiens. Mol Biol Evol. 2009;26:2617–2626. doi: 10.1093/molbev/msp180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dewannieux M, Harper F, Richaud A, Letzelter C, Ribet D, Pierron G, Heidmann T. Identification of an infectious progenitor for the multiplecopy HERV-K human endogenous retroelements. Genome Res. 2006;16:1548–1556. doi: 10.1101/gr.5565706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayer J, Sauter M, Racz A, Scherer D, Mueller-Lantzsch N, Meese E. An almost-intact human endogenous retrovirus K on human chromosome 7. Nat Genet. 1999;21:257–258. doi: 10.1038/6766. [DOI] [PubMed] [Google Scholar]
- 9.Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 10.Armbruester V, Sauter M, Krautkraemer E, Meese E, Kleiman A, Best B, Roemer K, Mueller-Lantzsch N. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin Cancer Res. 2002;8:1800–1807. [PubMed] [Google Scholar]
- 11.Denne M, Sauter M, Armbruester V, Licht JD, Roemer K, Mueller-Lantzsch N. Physical and functional interactions of human endogenous retrovirus proteins Np9 and rec with the promyelocytic leukemia zinc finger protein. J Virol. 2007;81:5607–5616. doi: 10.1128/JVI.02771-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaufmann S, Sauter M, Schmitt M, Baumert B, Best B, Boese A, Roemer K, Mueller-Lantzsch N. Human endogenous retrovirus protein Rec interacts with the testicular zinc-finger protein and androgen receptor. J Gen Virol. 2007;91:1494–1502. doi: 10.1099/vir.0.014241-0. [DOI] [PubMed] [Google Scholar]
- 13.Boese A, Sauter M, Galli U, Best B, Herbst H, Mayer J, Kremmer E, Roemer K, Mueller-Lantzsch N. Human endogenous retrovirus protein cORF supports cell transformation and associates with the promyelocytic leukemia zinc finger protein. Oncogene. 2000;19:4328–4336. doi: 10.1038/sj.onc.1203794. [DOI] [PubMed] [Google Scholar]
- 14.Muster T, Waltenberger A, Grassauer A, Hirschl S, Caucig P, Romirer I, Fodinger D, Seppele H, Schanab O, Magin-Lachmann C, et al. An endogenous retrovirus derived from human melanoma cells. Cancer Res. 2003;63:8735–8741. [PubMed] [Google Scholar]
- 15.Lower R, Lower J, Tondera-Koch C, Kurth R. A general method for the identification of transcribed retrovirus sequences (R-U5 PCR) reveals the expression of the human endogenous retrovirus loci HERV-H and HERV-K in teratocarcinoma cells. Virology. 1993;192:501–511. doi: 10.1006/viro.1993.1066. [DOI] [PubMed] [Google Scholar]
- 16.Patience C, Simpson GR, Colletta AA, Welch HM, Weiss RA, Boyd MT. Human endogenous retrovirus expression and reverse transcriptase activity in the T47D mammary carcinoma cell line. J Virol. 1996;70:2654–2657. doi: 10.1128/jvi.70.4.2654-2657.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bieda K, Hoffmann A, Boller K. Phenotypic heterogeneity of human endogenous retrovirus particles produced by teratocarcinoma cell lines. J Gen Virol. 2001;82:591–596. doi: 10.1099/0022-1317-82-3-591. [DOI] [PubMed] [Google Scholar]
- 18.Reiche J, Pauli G, Ellerbrok H. Differential expression of human endogenous retrovirus K transcripts in primary human melanocytes and melanoma cell lines after UV irradiation. Melanoma Res. 2010;20:435–440. doi: 10.1097/CMR.0b013e32833c1b5d. [DOI] [PubMed] [Google Scholar]
- 19.Schanab O, Humer J, Gleiss A, Mikula M, Sturlan S, Grunt S, Okamoto I, Muster T, Pehamberger H, Waltenberger A. Expression of human endogenous retrovirus K is stimulated by ultraviolet radiation in melanoma. Pigment Cell Melanoma Res. 2011;24:656–665. doi: 10.1111/j.1755-148X.2011.00860.x. [DOI] [PubMed] [Google Scholar]
- 20.Serafino A, Balestrieri E, Pierimarchi P, Matteucci C, Moroni G, Oricchio E, Rasi G, Mastino A, Spadafora C, Garaci E, et al. The activation of human endogenous retrovirus K (HERV-K) is implicated in melanoma cell malignant transformation. Exp Cell Res. 2009;315:849–862. doi: 10.1016/j.yexcr.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 21.Ono M, Kawakami M, Ushikubo H. Stimulation of expression of the human endogenous retrovirus genome by female steroid hormones in human breast cancer cell line T47D. J Virol. 1987;61:2059–2062. doi: 10.1128/jvi.61.6.2059-2062.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golan M, Hizi A, Resau JH, Yaal-Hahoshen N, Reichman H, Keydar I, Tsarfaty I. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia. 2008;10:521–533. doi: 10.1593/neo.07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lavie L, Kitova M, Maldener E, Meese E, Mayer J. CpG methylation directly regulates transcriptional activity of the human endogenous retrovirus family HERV-K(HML-2) J Virol. 2005;79:876–883. doi: 10.1128/JVI.79.2.876-883.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fuchs NV, Kraft M, Tondera C, Hanschmann K-M, Lower J, Lower R. Expression of the human endogenous retrovirus group HML-2/HERV-K does not depend on canonical promoter elements but is regulated by the transcription factors Sp1 and Sp3. J Virol. 2011;85:3436–3448. doi: 10.1128/JVI.02539-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheli Y, Ohanna M, Ballotti R, Bertolotto C. Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res. 2009;23:27–40. doi: 10.1111/j.1755-148X.2009.00653.x. [DOI] [PubMed] [Google Scholar]
- 26.Levy C, Khaled M, Robinson KC, Veguilla RA, Chen PH, Yokoyama S, Makino E, Lu J, Larue L, Beermann F, et al. Lineage-specific transcriptional regulation of DICER by MITF in melanocytes. Cell. 2010;141:994–1005. doi: 10.1016/j.cell.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Levy C, Khaled M, Fisher DE. MITF: master regulator of melanocyte development and melanoma oncogene. Trends Mol Med. 2006;12:406–414. doi: 10.1016/j.molmed.2006.07.008. [DOI] [PubMed] [Google Scholar]
- 28.Hemesath TJ, Price ER, Takemoto C, Badalian T, Fisher DE. MAP kinase links the transcription factor microphthalmia to c-Kit signalling in melanocytes. Nature. 1998;391:298–301. doi: 10.1038/34681. [DOI] [PubMed] [Google Scholar]
- 29.Shibahara S, Takeda K, Yasumoto K, Udono T, Watanabe K, Saito H, Takahashi K. Microphthalmia-associated transcription factor (MITF): multiplicity in structure, function, and regulation. J Investig Dermatol Symp Proc. 2001;6:99–104. doi: 10.1046/j.0022-202x.2001.00010.x. [DOI] [PubMed] [Google Scholar]
- 30.Bertolotto C, Bille K, Ortonne JP, Ballotti R. Regulation of tyrosinase gene expression by cAMP in B16 melanoma cells involves two CATGTG motifs surrounding the TATA box: implication of the microphthalmia gene product. J Cell Biol. 1996;134:747–755. doi: 10.1083/jcb.134.3.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garraway LA, Widlund HR, Rubin MA, Getz G, Berger AJ, Ramaswamy S, Beroukhim R, Milner DA, Granter SR, Du J, et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature. 2005;436:117–122. doi: 10.1038/nature03664. [DOI] [PubMed] [Google Scholar]
- 32.Hershey CL, Fisher DE. Genomic analysis of the microphthalmia locus and identification of the MITF-J/Mitf-J isoform. Gene. 2005;347:73–82. doi: 10.1016/j.gene.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 33.Okuyama T, Kurata S, Tomimori Y, Fukunishi N, Sato S, Osada M, Tsukinoki K, Jin HF, Yamashita A, Ito M, et al. p63(TP63) elicits strong trans-activation of the MFG-E8/lactadherin/BA46 gene through interactions between the TA and DeltaN isoforms. Oncogene. 2008;27:308–317. doi: 10.1038/sj.onc.1210646. [DOI] [PubMed] [Google Scholar]
- 34.Fukunishi N, Katoh I, Tomimori Y, Tsukinoki K, Hata R, Nakao A, Ikawa Y, Kurata S. Induction of DeltaNp63 by the newly identified keratinocyte-specific transforming growth factor beta signaling pathway with Smad2 and IκB kinase α in squamous cell carcinoma. Neoplasia. 2010;12:969–979. doi: 10.1593/neo.101054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reus K, Mayer J, Sauter M, Scherer D, Muller-Lantzsch N, Meese E. Genomic organization of the human endogenous retrovirus HERV-K (HML-2.HOM) (ERVK6) on chromosome 7. Genomics. 2001;72:314–320. doi: 10.1006/geno.2000.6488. [DOI] [PubMed] [Google Scholar]
- 36.Watson JD, Baker TA, Bell SP, Gann A, Levine M, Losick R. In: Molecular Biology of the Gene. 5th ed. Cummings Benjamin., editor. San Francisco, CA: Cold Spring Harbor Laboratory Press; 2004. pp. 347–377. Chapter 2: Mechanisms of Transcription. [Google Scholar]
- 37.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Humer J, Waltenberger A, Grassauer A, Kurz M, Valencak J, Rapberger R, Hahn S, Lower R, Wolff K, Bergmann M, et al. Identification of a melanoma marker derived from melanoma-associated endogenous retroviruses. Cancer Res. 2006;66:1658–1663. doi: 10.1158/0008-5472.CAN-05-2452. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y, Radfar S, Liu S, Riker A, Khong H. Mitf-Mdel, a novel melanocyte/melanoma-specific isoform of microphthalmia-associated transcription factor-M, as a candidate biomarker for melanoma. BMC Med. 2010;8:14. doi: 10.1186/1741-7015-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis IJ, Kim JJ, Ozsolak F, Widlund HR, Rozenblatt-Rosen O, Granter SR, Du J, Fletcher JA, Denny CT, Lessnick SL, et al. Oncogenic MITF dysregulation in clear cell sarcoma: defining the MiT family of human cancers. Cancer Cell. 2006;9:473–484. doi: 10.1016/j.ccr.2006.04.021. [DOI] [PubMed] [Google Scholar]
- 41.Lo K, Smale ST. Generality of a functional initiator consensus sequence. Gene. 1996;182:13–22. doi: 10.1016/s0378-1119(96)00438-6. [DOI] [PubMed] [Google Scholar]
- 42.Javahery R, Khachi A, Lo K, Zenzie-Gregory B, Smale ST. DNA sequence requirements for transcriptional initiator activity in mammalian cells. Mol Cell Biol. 1994;14:116–127. doi: 10.1128/mcb.14.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leung CS, Srigley JR. Distribution of lipochrome pigment in the prostate gland: biological and diagnostic implications. Hum Pathol. 1995;26:1302–1307. doi: 10.1016/0046-8177(95)90293-7. [DOI] [PubMed] [Google Scholar]
- 44.Das G, Choi Y, Sicinski P, Levine E. Cyclin D1 fine-tunes the neurogenic output of embryonic retinal progenitor cells. Neural Dev. 2009;4:15. doi: 10.1186/1749-8104-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shah M, Bhoumik A, Goel V, Dewing A, Breitwieser W, Kluger H, Krajewski S, Krajewska M, Dehart J, Lau E, et al. A role for ATF2 in regulating MITF and melanoma development. PLoS Genet. 2010;6:e1001258. doi: 10.1371/journal.pgen.1001258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sonnenblick A, Levy C, Razin E. Interplay between MITF, PIAS3, and STAT3 in mast cells and melanocytes. Mol Cell Biol. 2004;24:10584–10592. doi: 10.1128/MCB.24.24.10584-10592.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ling J, Pi W, Bollag R, Zeng S, Keskintepe M, Saliman H, Krantz S, Whitney B, Tuan D. The solitary long terminal repeats of ERV-9 endogenous retrovirus are conserved during primate evolution and possess enhancer activities in embryonic and hematopoietic cells. J Virol. 2002;76:2410–2423. doi: 10.1128/jvi.76.5.2410-2423.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Coffin JM, Hughes SH, Varmus HE. Retroviruses. New York, NY: Cold Spring Harbor Laboratory Press; 1997. [PubMed] [Google Scholar]
- 49.Verastegui C, Bertolotto C, Bille K, Abbe P, Ortonne JP, Ballotti R. TFE3, a transcription factor homologous to microphthalmia, is a potential transcriptional activator of tyrosinase and TyrpI genes. Mol Endocrinol. 2000;14:449–456. doi: 10.1210/mend.14.3.0428. [DOI] [PubMed] [Google Scholar]
- 50.Montel V, Suzuki M, Galloy C, Mose ES, Tarin D. Expression of melanocyte-related genes in human breast cancer and its implications. Differentiation. 2009;78:283–291. doi: 10.1016/j.diff.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 51.Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, Piyathilake CJ, Hunt KK, Johanning GL. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rebhun RB, Cheng H, Gershenwald JE, Fan D, Fidler IJ, Langley RR. Constitutive expression of the α4 integrin correlates with tumorigenicity and lymph node metastasis of the B16 murine melanoma. Neoplasia. 2010;12:173–182. doi: 10.1593/neo.91604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silva AG, Graves HA, Guffei A, Ricca TI, Mortara RA, Jasiulionis MG, Mai S. Telomere-centromere-driven genomic instability contributes to karyotype evolution in a mouse model of melanoma. Neoplasia. 2010;12:11–19. doi: 10.1593/neo.91004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Penna E, Orso F, Cimino D, Tenaglia E, Lembo A, Quaglino E, Poliseno L, Haimovic A, Osella-Abate S, De Pitta C, et al. microRNA-214 contributes to melanoma tumour progression through suppression of TFAP2C. EMBO J. 2011;30:1990–2007. doi: 10.1038/emboj.2011.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bar-Eli M. Role of AP-2 in tumor growth and metastasis of human melanoma. Cancer Metastasis Rev. 1999;18:377–385. doi: 10.1023/a:1006377309524. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.