Abstract
The mapping of environment, through variation in individuals' life histories, to dynamics can be complex and often poorly known. Consequently, it is not clear how important it is dynamically. To explore this, I incorporated lessons from an empirical system, a soil mite, into an individual-based model. Individuals compete for resource and allocate this according to eight ‘genetic’ rules that specify investment in growth or reserves (which influences survival or fecundity), size at maturation and reproductive allocation. Density dependence, therefore, emerges from competition for food, limiting individual's growth and fecundity. We use this model to examine the role that genetic and phenotypically plastic variation plays in dynamics, by fixing phenotypes, by allowing phenotypes to vary plastically and by creating genetic variation between individuals. Variation, and how it arises, influences short- and long-run dynamics in a way comparable in magnitude with halving food supply. In particular, by switching variation on and off, it is possible to identify a range of processes necessary to capture the dynamics of the ‘full model’. Exercises like this can help identify key processes and parameters, but a concerted effort is needed across many different systems to search for shared understanding of both process and modelling.
Keywords: phenotypic plasticity, population dynamics, selection, individual-based model, evolution, systems ecology
1. Introduction
Given the unprecedented rate of environmental change, and the increasing recognition that ecological systems provide extremely valuable services to society, there is a pressing need to understand how biological systems respond to environmental change in order to be able to manage and protect the services they provide. Predicting the way a complex ecological system will behave requires some form of modelling approach. Models are simplifications of reality; so choices need to be made at the outset as to what processes to include. For some general, strategic questions, models can be made very simple and then be solved analytically. However, for population prediction, such models may be inappropriate because they miss important biological details that influence the way that the system responds to any environmental change. These ‘important biological details’ include (i) individual differences caused by age, stage or previous history [1], (ii) the mechanism by which density influences demographic rates, and that different rates may have different functions with density [2,3], (iii) the extent to which the environment fluctuates over time and space [2,4,5], and (iv) evolutionary dynamics that arise from genetic and phenotypic dynamics [6–8]. Although these factors are probably ubiquitous, they are sometimes not modelled for two good reasons: either the data are unavailable, or because more complexity in processes leads to more cumbersome models.
Empirical data were, until recently, largely unavailable. However, in the past two decades, empirical systems have been studied in great depth, and the biological processes underpinning phenotypic, population and evolutionary dynamics have begun to be elucidated [3,6,7,9–11]. Field-based systems have been illuminating, and much has been performed to disentangle different processes contributing to the population and evolutionary dynamics by dissecting detailed population time series coupled with pedigree analysis. Laboratory ecological systems have the particular benefit of allowing experimental dissection of the processes that underlie the dynamics. For one amenable empirical model (the soil mite, Sancassania berlesei), we have experimentally dissected the relationship between phenotypic and dynamical variation. This mite is small enough to conduct replicated free-running population experiments, yet large enough to undertake detailed investigations on individuals [12]. One over-riding conclusion that emerges from our laboratory experiments is that individuals are plastic and vary in resource allocation decisions according to the food they gain. Thus, per capita food (PCF) determines growth rate, which determines age and size at maturity [13]. A female's size and resources determine reproduction by affecting the number and size of eggs; the latter also influences the offspring life history in a context-dependent way [14,15]. Long-running population experiments show that these phenotypic relationships evolve in response to environmentally imposed selection pressures [16], and therefore have a genetic component; these experiments also demonstrate that population dynamics evolve [16]. Population responses to perturbations in food supply are therefore the integration across the population of the individual responses to changing resource levels, and, through delayed effects, previous environments including those of parents [13–15,17].
Therefore, data are increasingly available to inform the mechanistic drivers of dynamical change. The issue of modelling the system then comes down to the choice of processes to model. The traditional modelling approach, that values analytical simplicity greatly, can be characterized by statistical analogy as a ‘forwards selection’ approach, as typically the simplest possible model is taken as the starting point and single processes are added. A priori, however, it is not clear the extent to which different biological details really matter in determining dynamical properties, and therefore which processes are necessary and sufficient to capture the dynamics of any biological system. The choice of processes is particularly important for models aiming to be used as a predictive tool for management. To inform these choices, we need to undertake some exercises where we build models rich in biological detail and investigate their properties, to identify the extent to which biological detail affects the details of the model result. Continuing the statistical analogy, the requirement in such cases may be calls for a ‘backwards selection’ approach from a maximal model.
To explore the dynamical importance of between-individual variation (arising from variation in genes plus variation in resources, leading to phenotypic plasticity), I have incorporated many of the biological drivers of the dynamics we observe in the mite experiments into an individual-based model. The mapping of environment to dynamics is complex, and depends on many indirect pathways, even crossing generations via the resources allocated to eggs, as a main mechanism driving parental effects. The purpose of this model is not to fit it specifically to empirical time series (although methods are available to do this, e.g. [18]), but to build an informed model that captures a meaningful range of biological mechanisms translating environment into dynamics. Using this biologically realistic ‘system model’, we explore the extent to which different endogenous and exogenous processes are particularly important in capturing the overall dynamics.
2. Methods
The model description follows the 7-step overview, design and details (ODD) protocol for describing individual- and agent-based models [19]. The model used for this paper is coded in R v. 2.10.0 [20]. Code is available on request.
(a). Purpose
The purpose of the model is to model an ecological, single-species system using a range of factors that have been identified as key drivers of demography from experiments on the mite laboratory system. This model can then be used to explore the relationship between phenotypic, population and evolutionary dynamics, with a particular view towards identifying which of the processes driving phenotypic dynamics are important for transient and long-term dynamics.
(b). State variables and scales
The agents in the model are individuals that reproduce clonally. Individuals take in resources and allocate them to growth, reserves or reproduction according to eight rules (genes), which are passed on to offspring (with potential for mutation). Each individual's state is tracked via its size (Si), age (Ai), reserves (Ri) and its maturation status. Reproduction is clonal and dependent only on resources. Individuals are followed from birth to death, and the time step is daily (to match our experimental programme). Space is implicit. Population size and structure are only constrained by the resource input.
(c). Process overview and scheduling
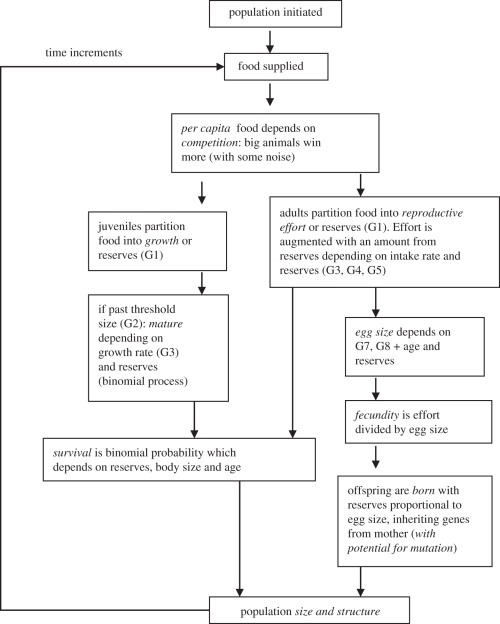
The model is summarized in figure 1. Briefly, each time step begins with food being supplied. This is shared out between individuals according to a competition function (see §3). An individual's PCF is spent in a way determined by its variants (‘alleles’) of its life-history rules (‘genes’). In particular, juveniles invest PCF in growth or reserves, or adults in reproduction or reserves. Juveniles mature once past a (genetic) size threshold and at a probability determined by their growth rate and reserves. Adults allocate food and reserves to reproductive effort, which is then partitioned into eggs of a size determined by age, size and reserves. Survival is a binomial process depending on size, age and reserves, and occurs at the end of each time step.
Figure 1.
Schematic layout of the structure of the individual-based model. The heritable allocation rules, ‘genes’ 1–8 are specified by G1 … G8. Sequencing runs from top to bottom.
(d). Design concepts
(i). Emergence
Individuals' life histories emerge from their patterns of resource investment in growth, reserves and fecundity. These investment decisions are either genetically fixed or are gene × environment interactions that are plastic in response to food supply and internal state (age, size, etc.). Density dependence occurs only in the sharing of food between individuals, and thereafter density-dependent relationships between demographic rates and density arise from investment decisions. Population dynamics emerges from individual rules. Differential survival and reproduction of individuals with different variants of the genes (‘alleles’) is natural selection. Mutation (a small per capita chance of a change in an allele from parent to offspring) allows new genetic variation to emerge leading to sustained evolutionary dynamics.
(ii). Individual characteristics
Individuals do not interact, mate, sense or move.
(iii). Stochasticity
Stochasticity occurs in daily food supply, the maturation decision and daily survival. Food supply is a stochastic environmental variable around a deterministic mean. For a given food level, the deterministic per capita share is multiplied by a random deviate to proxy variation created by encounter rate, spacing of food and contest competition. Maturation and survival decisions are binomial deviates around a deterministic mean.
(iv). Observation
At each time step, individuals' states are recorded (size, age and reserves), along with the number of eggs laid and their sizes, their age at maturation and death. Each individual's alleles are recorded at birth, along with parental identity. At each time step, the population's genome, pedigree and demography are known, along with population size and structure.
(e). Initialization
The population is initiated with a number of individuals (throughout 1000, selected from a random uniform distribution of sizes from 0.1 to 4.9). Genetic variation was initiated by drawing values from uniform distributions (see appendix A for details). Simulations were typically run for 1000 time steps. For each scenario, five simulation runs at each set of starting values were performed.
(f). Input
Resource is supplied to the population at a specified amount (number of units) per time step. Here, we use two regimes, constant food (with the baseline being 200 units d−1) and periodic food (with the deterministic signal being a sine wave of amplitude 150 units, centred around 200 units and a 100 day period). ‘Constant’ and ‘variable’ food supplies were chosen to reflect empirical investigations. To reflect small-scale environmental stochasticity, the daily food is multiplied by a normal deviate of mean 1 ± 0.3 s.d.
(g). Submodels
(i). Competition
Resource is divided up among individuals so that large individuals get proportionally more. Two competition rules are used. (i) Logistic competition, such there is an asymptote above which larger animals do not consume more. The competitive ability (C) of each individual (i) of size (S) is defined by
(ii) Linear sharing of food according to the body size of each individual. These rules correspond to empirical results associated with different competitive scenarios. When food is clumped and easily defendable, the biggest individuals can eat continuously, whereas when food is less defendable, patches get depleted via scramble competition, leading to a size-asymptote in intake (G. Truelove & T. G. Benton 2006, unpublished data). Noise is added each time step to each Ci by multiplying it by a normal deviate of mean 1 ± 0.15 s.d. The daily food is then shared out in proportion to the distribution of Ci across the population, giving each individual its PCF.
(ii). Genes
There are eight genes that specify individuals' life histories. Variation in individuals' genes (‘alleles’) creates phenotypic differences.
G1, Reserves. Empirically, juveniles can choose to grow slowly but have higher adult fecundity [14], implying the existence of investment in reserves. G1 is the percentage of resources that go into reserves (the rest is invested in growth in juveniles or reproductive effort in adults).
G2 and G3, Maturation. In mites, maturation is consistent with Day & Rowe's (2002) threshold maturation model: well-resourced individuals growing fast continue to grow past the threshold maturation size [13]. We assume that the probability of an individual maturing at time t is a linear function of the individual's size, Si, above its genetic threshold, G2i, a quadratic function of its reserves, Ri (such that maturation probability increases fast with well-resourced animals), and inversely proportional to its current growth increment. G3i is then a scaling factor for this relationship.
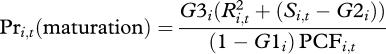 |
Individuals that mature at one time step can reproduce at the next.
G4–G6 specify the rate at which ‘reserves’ are withdrawn to complement current food supply and determine reproductive effort E. The results of Plaistow et al. [14] imply both capital and income breeding, and that the balance between these varies between individuals developed under low, medium and high food. Hence, we choose three different rules, ‘expressed’ in different contexts. If reserves are low (less than 10% body size), reserves are withdrawn at a rate of 0, 50 or 100 per cent (G4 takes three values); if reserves are not low and the intake rate is high (less than 10× reserves), reserves are withdrawn at a rate of 0, 50 or 100 per cent (G5 takes three values); otherwise, reserves are withdrawn at a rate between 0 and 100 per cent (G6 is a continuous proportion).
G7 and G8 determine an individual's investment in each offspring [15,21]. G7 is the slope of a regression of egg size, ϕ, on maternal age. G8 modifies the intercept with respect to the overall effort being invested, such that
Fecundity is then a function of the total reproductive effort and the specified egg size. We assume that energetic reserves contribute to half the egg (with the other half being, for example, water), and that individuals are born with some reserves contributed via yolk (30% of their initial size). Eggs hatch as time increments (so contribute to competition at the next time step).
(iii). Survival
An individual's probability of survival at time t is related to its size (Si), reserves (Ri) and age (Ai).
Pr(survival) was bounded at zero and unity. The parameter values were arbitrarily chosen, but provide realistic patterns of size-based survival.
(h). Modelling experiments
Three scenarios were used to examine the role of variation in the life history on dynamics:
— individuals had their alleles assigned at random, providing a genetically heterogeneous population;
— individuals had their alleles for genes fixed at the same value. This provided a genetically homogeneous population but allowed phenotypic variation between individuals in response to their state (resources, age, size, etc.). To assess the role of variation in each gene, one or more genes were allowed to vary, while the rest were held constant. To assess the role of selection, genes were fixed at the initial population mean genotype, or at the mean genotype following 1000 steps of selection; and
— individuals had their phenotype fixed and therefore genes were neutral. The phenotypes constrained were size at maturity, the percentage of resources invested in reserves and egg size (so all individuals matured at the same size, although age varied, and laid the same size eggs, although fecundity varied).
To assess the ‘necessary and sufficient processes’ that replicated the full dynamics (all eight genes variable), a ‘stepwise backwards’ process was undertaken by fixing genes in increasing rank order of their single gene effects (figure 2) until an ANOVA of the five model runs showed significant differences from the baseline (all eight genes variable).
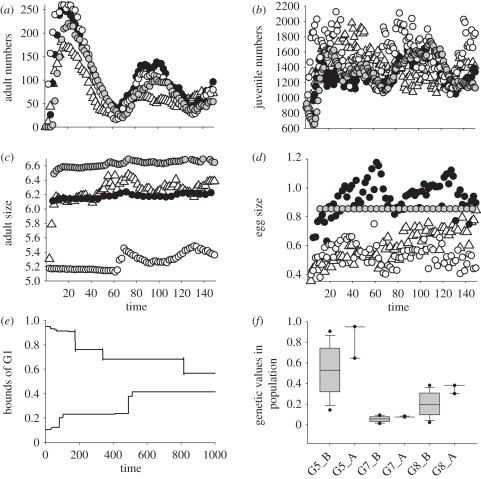
Figure 2.
Transient dynamics and the role of genetic and phenotypic variation between individuals. The eco-evolutionary dynamics of a single run of the IBM where initial genetic variation occurs in all genes (G, triangles), or where the values of the genes are fixed at the mean value of five runs of the variable model (GF, black circles), the mean value of the initial conditions (NOG, white circles) or where the phenotype is fixed (PF, adults have a fixed threshold maturation size, a fixed egg size and a fixed investment rate in reserves; grey circles). (a) Adult dynamics, (b) juvenile dynamics, (c) mean adult size at maturation, (d) egg size, (e) the upper and lower limits of G1, illustrating the narrowing of genetic variability owing to selection; (f) shows the initial genetic variation for G5, G7 and G8 and the final variation after 1000 time steps. Values for GF are G1 = 0.4797552, G2 = 6.25119, G3 = 0.8206453, G4 = 0.00305499, G5 = 0.5, G6 = 0.7390198, G7 = 0.0982889, G8 = 0.361522; values for NOG are G1 = 0.5050849, G2 = 5.024989, G3 = 0.5050392, G4 = 0.5, G5 = 0.5, G6 = 0.5050981, G7 = 0.05050591, G8= 0.2050123; values for the fixed-phenotypes are adult size = 6.414548, investment in reserves 0.4798, egg size = 0.8547742.
3. Results
The model captures some of the patterns we see in the experimental observations. Population dynamics is density-dependent, typically with decaying oscillations (compare figure 2 with time series in earlier studies [17,21]). Individual's growth rates are inversely correlated with population density, such that age at maturity is a positive correlate of total population density at birth (r = 0.52, n = 884 individuals tracked throughout life over 1000 time steps) and maturity (r = 0.98); similarly, lifetime fecundity is a function of density at birth, maturity and death (r = 0.51, 0.29 and 0.4, respectively). In the model, most mortality happens to juveniles, such that the median age of death is 2 days (mean 6); this is consistent with our empirical understanding [22]. The mean age of maturity of a sample of individuals (884 out of 50 000 individuals tracked) is 20 days, s.d. 6, range: 11–45; again the patterns were not inconsistent with our observations [13,23]. The model suggests survival to maturity is only approximately 2 per cent, but once there, adults live for an average of 16 days (s.d. 13). Genetics leads to mother–daughter similarity, as do maternal environmental effects: mothers laying large eggs give a competitive advantage to offspring. There are therefore correlations between mother and offspring in: egg size laid (r = 0.67, n = 432), fecundity (r = 0.49), longevity (r = 0.42) and age at maturity (r = 0.95).
Variation between individuals in their phenotypes, and how it arises, has marked effects on the population dynamics. Figure 2 contrasts the transient phenotypic, genetic and population dynamics between the three modelling scenarios: (i) phenotypically fixed population (PF), (ii) genetically fixed populations (where phenotypic plasticity remains, but all individuals have the same genotype). We illustrate two genetically fixed populations: one where the genes are fixed at the mean of the initial conditions (GFi), one where they are fixed at the mean values at the end of 1000 time steps (GFe), after selection has operated, and (iii) genetic variation in all genes (G, allowing different individuals to respond to the same environment in different ways). Overall, the population dynamics of the three ‘fixed’ populations is considerably more variable than the population with genetic variability, and cohort cycles are sustained for longer (figure 2a,b). The phenotype-fixed treatment shows little phenotypic variation as expected. The average size at maturity tracks resource availability in the genetically variable population, to a lesser extent in the GFi population and not at all in the GFe population (figure 2c). Conversely, egg size instead tracks resource availability in the GFe population, to a lesser extent in the GFi population and not at all in the genetically variable population (figure 2d).
That size at maturity is strongly density-dependent in the genetically variable population, coupled with a small egg size (and thus high juvenile mortality) tends to dampen the oscillations in adult numbers faster than in the fixed treatments (figure 2a), such that between time 75 and time 125, adult numbers are considerably lower (means of 56 for G, 68 for GFi, 105 for GFe, 77 for PF). Fewer adults result in more PCF for each, and a greater size and per capita fecundity, leading to more juveniles. However, these are born smaller, compete heavily, grow slowly and suffer greater mortality (figure 2b). The sum of these two effects is marked, with the genetically variable population averaging 68 or 65 per cent of the biomass of the fixed-phenotype or GFe populations during the initial phase of 150 days (here, biomass is the size-weighted total population size, approximated by the sum of all individuals' body sizes).
Genetic variation not only alters mean phenotypes but also creates opportunities for selection, with the response to selection creating phenotypic (and population) dynamics. The upward drift in egg size (figure 2d) arises from strong selection, as reflected in the distribution of alleles in the population (figure 2f): after 1000 time steps, selection on genes 7 and 8 has moved the mean genotype to the top end of the initial range, leading to the production of large eggs.
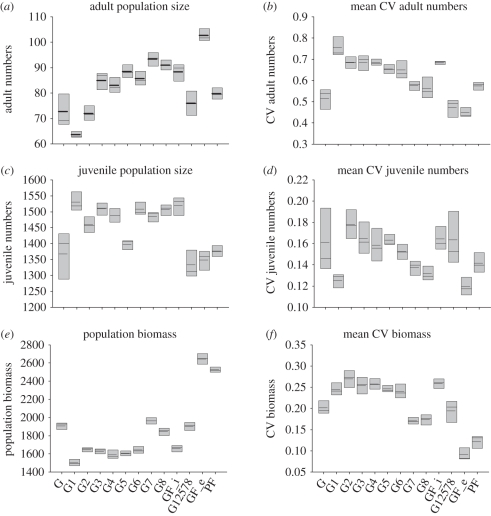
The way variation in different genes contributes to the mean and variability in transient dynamics is summarized in figure 3. Fixing all genes at the mean initial conditions (i.e. GFi), but allowing one to vary shows, for example, that variation in G1 (resources into reserves or growth/fecundity) decreases the average adult population size but increases its variance, and vice versa with juvenile numbers. This pattern arises because investment in reserves affects growth rate, age- and size-at-maturity decisions and reproductive effort, and individuals with high values of G1 invest more in reserves, and so have a slower growth, smaller size at maturity but higher fecundity. Removing genetic variation by fixing 7 or 8 of the genes increases the relative variability in adult numbers (reinforcing the results of figure 2, indicating that genetically variable population dynamics decline in variability more quickly). Using a systematic stepwise backwards procedure to find a minimal model, the behaviour of which was consistent with the model including variation in all genes, identified one that included variation in G1, G2, G5, G7 and G8 (figure 3). This suggests that key processes in the dynamics are resource allocation to growth/fecundity or reserves, allocation of reserves to reproductive effort and apportionment of that effort between offspring.
Figure 3.
The impact of genetic variation on population size. Boxplots showing the mean (dashed line), median and interquartile ranges of variables for the initial 250 time steps of five replicate runs, using baseline conditions (see appendix A), and varying the source of genetic variation. The initial population had variation in all genes (G), no genes (NOG, fixed at mid range) or single genes (G1, G2…). In this last case, seven genes were set at midrange, and one was varied. G12578 is variation in G1, G2, G5, G7 and G8. GF refers to genes fixed at the mean values at the end of 1000 steps of G, and PF refers to where the phenotype is fixed using means from the end of G (see legend to figure 2 for values). Biomass is the sum of all individuals' sizes across the population. NB: the long-run averages (time steps 501 : 1000) are qualitatively similar but differ in some instances; for example, where genes are fixed (NOG), age and size at maturity are less variable, so cohort cycles tend to be maintained leading to the highest CVs.
Genetic variation also changes the predictability of the dynamics (contrast the height of the grey bars, representing the way the mean population size varies over the five replicated simulations). It also changes the variation in the dynamics over time measured by the mean coefficients of variation (CVs; variation in single genes tends to increase temporal variance in adult dynamics). Finally, compare the three fixed treatments (GFi and GFe—the mean genotypes before and after selection, and PF—the fixed-phenotype treatment). The PF versus GFe comparison shows the difference caused by plasticity alone (which can be considerable; see adult numbers). The GFe versus GFi indicates the role of selection over 1000 time steps. This is most noticeable with respect to the evolution of biomass, where GFi and GFe differ considerably. This is due to selection to increase egg size, reducing their mortality and therefore preventing their loss from the system.
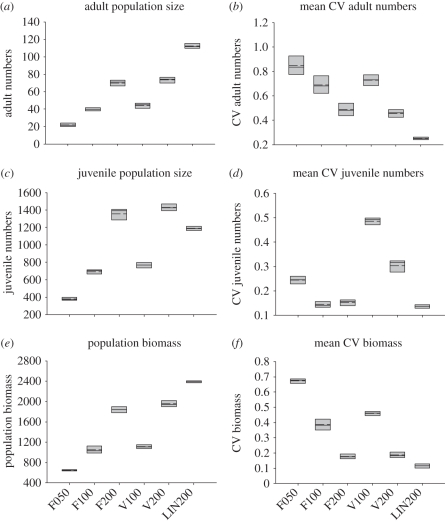
Having examined the sensitivity of dynamics to genetic and phenotypic variation, we can now ask how the magnitude of effect compares with the impact of external processes, such as the amount and variance in food supply. Using the same baseline conditions but varying the food amount, or its variance, has a strong effect on the dynamics (figure 4). Changing food levels affects population size in a nonlinear way (quartering the food results in a third of the biomass). Over the first 250 time steps, the baseline adult population size is 70 ± 4 s.d., at half the food this falls to 40 ± 2.3 s.d. and at a quarter to 22 ± 2.2 s.d. Adding variability to the food gives 74 ± 4 s.d. at baseline food and 44 ± 3 s.d. at half food, and therefore it does not have much effect on mean population size; however, it does impact on the population variability (especially of juveniles) as the dynamics follow the food (figure 4d). Changing the competition function, allowing individuals to access food proportional to their body size, also has a very strong effect on population size (figure 4a,c,e). This is because logistic competition creates an upper asymptote in the PCF an individual can gain (giving a type III functional response), reducing the selection to continue to grow over a certain size. A linear share (type I functional response) creates a continuing advantage to growing, selecting for larger individuals and thus changing stage structure, as more biomass is partitioned into larger adults and less into juveniles. The average number of adults across the first 250 time steps changes from 70 ± 4 s.d. to 113 ± 4 s.d. with the change in competition from type III to type I. The absolute change in population biomass caused by different levels of genetic variation (figure 3e) is similar in magnitude to decreasing food supply to the population by 50 per cent, or changing the competition function (figure 4e).
Figure 4.
The effect of changing the food supply and competition on population parameters over the initial 250 time steps. Categories are F200 (baseline, see appendix A), F100 (half food), F050 (quarter food), FV200 (baseline food, but varying about the mean as a sine wave), FV100 (half food, variable), LIN200 (linear share of food proportional to body size, cf. baseline where there is a maximum amount an individual can gain). Descriptive statistics for first 250 days.
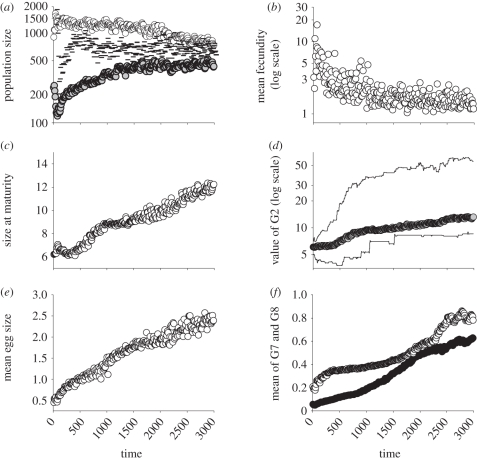
Within the simulations reported earlier, there was no mutation; so selection occurs on the initial genetic variation. Allowing mutation allows evolutionary change to be sustained (figure 5). Under many parameter values, size at maturity is driven upwards by increasing selection on egg size as K-selection selects for increased competitive ability. The change in life histories that results impacts on the population dynamics in different ways at different temporal scales (after the initial transients, adult population size increases over 500 time steps before decreasing again). Long-running laboratory experiments typically document similar changes in dynamics [12], and the empirical evolution of dynamics is addressed in [16].
Figure 5.
Evolution with mutation. (a) Population dynamics (grey circles = biomass × 0.1, open circles = juveniles, dashes = adults (×10), (b) mean per capita fecundity at each time step declines as egg size increases, (c) mean size at maturity, (d) mean value of G2 (on log scale) with upper and lower population bounds, (e) average egg size, and (f) mean values of G7 (black circles) and G8 (white circles). Initial and final means for eight genes are G1 = 0.5170865, 0.3324225; G2 = 6.0492818, 13.0910140; G3 = 0.5276286, 0.6085805; G4 = 0.4960000, 0.5550122; G5 = 0.4840000, 0.6894866; G6 = 0.5080018, 0.5774387; G7 = 0.0546385, 0.6193221; G8 = 0.2066729, 0.7998085. Data are plotted every 20th time step.
4. Discussion
The purpose of this modelling exercise was to explore the way in which variability between individuals, created by genetic and plastic responses to resources, impacts on short-term population dynamics, and therefore our ability to predict ecological responses to environmental change. Our model incorporates the sorts of biological mechanisms underlying phenotypic dynamics that may be broadly applicable across many systems. Population dynamics arises from interactions between individuals over resources, rather than being imposed as a density-dependent function. Summarizing the results, even when life is simple, with ecologically rational rules, we perhaps still need to measure most things in order to create predictive models.
The long-running mite laboratory study has highlighted that individuals vary because life histories are highly responsive to current [13] and past environments, whether the individual's own or via parental effects across generations [17,23], and also that life histories change rapidly in response to selection in populations [16]. This latter result indicates that allocation rules describing reaction norms are heritable. Resource-related growth rates, plasticity in maturation, variation in number and resources allocated to each offspring, and heritability in reaction norms have been found in a range of well-studied organisms (such as ungulates [7,24], plants [25], isopods [26], Daphnia [27] and fishes [28]), and so these sorts of processes may well be general. On the basis of such results, the IBM assumes some simple mechanisms underlying individual phenotypic dynamics, and although the specifics are mite-informed, the model may be more general in terms of the processes included.
The IBM allows the investigation of the dynamical effects of different biological processes that cannot easily be modified empirically. Specifically, we can use it to switch off phenotypic and genetic variation, and, by comparing the results, we can identify processes, and sources of variation, that contribute most to the results of the ‘full model’. Thus, sensitivity analysis of the model can provide insight into the processes or parameters most influential for the dynamics, and so can act as a guide to future research effort in order to reduce process and parameter uncertainty [29,30]. The results described here indicate that (i) variation between individuals arising from phenotypically plastic allocation rules interacts with genetic variation to influence dynamics. Hence, variation in population dynamical responses to a perturbation is a necessary result of variation between individuals in life histories [1]; (ii) that genetic variation in a range of processes throughout the animal's life was needed to approximate the full model: the rate at which an individual allocates resources to growth or reserves, the maturation threshold size (correlated with age at maturity), the way it allocates resources to reproductive effort and the way the effort is partitioned between propagules. These relationships therefore are necessary to describe the whole life history and how it responds to environmental resources, as well as its evolutionary fitness.
This model further suggests two important lessons: firstly, as is increasingly being recognized, it is not possible to separate ecological and evolutionary timescales [6,31,32]. Therefore, under a stationary environment, the dynamical attractor is not a population dynamical one, but a population and evolutionary one. Population growth rate, or carrying capacity, can evolve significantly over a small handful of generations ([16,33], figure 5). Secondly, although selection is a powerful force, indirect effects may prevent selection from driving phenotypic traits in response to it. For example, on average, maturation size is selected to increase under the majority of conditions explored in this paper (figure 5c); however, during periods of high competition, the average adult body size may nonetheless decrease in the population (figure 2c, black points, time 50–100). This is similar to Soay sheep on St Kilda, where body size is selected to increase, but average body size has decreased in recent decades in response to changes in age structure and thus competition [7].
In the model, density dependence works through resource competition, with the available resources being partitioned into growth (or reproduction) and reserves. The effects of density therefore impact on all demographic traits and do not just influence survival or fecundity [34]. They also lead to complex lag structures in the dynamics, as generation time itself varies with density. The model indicates that changing the amount of food, or the way it is competed for, can have significant impacts on the population dynamics. For example, the spatial aggregation of food can change competition from scramble to contest, changing individual growth rates, body sizes and population dynamics [12]. The way density dependence operates is therefore context-dependent, making identification of the correct process more difficult, especially in the field [35–37]. Despite the focus in the literature on the effects of different mechanisms of density dependence, it is interesting to note that the impacts of individual variation are not dissimilar in magnitude to halving the food supply, or changing the model for competition from scramble to contest.
So, what conclusions can be drawn from this approach and this specific model? Ecological systems are obviously complex, but this exercise in understanding the behaviour of the system has pinpointed some positive ways to identifying processes that are particularly important for predicting dynamical responses. The key message is that variation between individuals (driven by genetic variation and plastic allocation rules) determines transient dynamics, owing to complex direct and indirect effects in the way that individuals, and thus the population, respond to environmental change [24]. Furthermore, this model reinforces empirical findings that population dynamics (as well as phenotypic dynamics) are likely to carry signatures of responses to selection. That the biological processes included in the model may be quite widespread suggests the general result that the predictive models need to incorporate more biological details to be realistic. This is a challenge for ecology for two reasons: one is that collecting data on the way individuals vary and its causes is complicated. Secondly, we need to develop a framework for numerical modelling in terms of how to analyse, report and develop analytical approximations, as well as identifying parameter and process uncertainty. It is possible to have a science based on numerical models with widespread community support, as exemplified by the earth systems' community [30,38].
Acknowledgements
Thanks to Andrew Beckerman, an anonymous referee and Matthew Evans for comments; and the mite laboratory, past and present, for inputs. The funding for the mite programme comes from NERC. Esa Ranta first stimulated me to make an IBM, and he remains much missed.
Appendix A
(a). Baseline initial settings
- n < −1000
# original pop size
- itime < −1000
# number of time steps
- Xo < -runif(n,0.1,4.9);
# X is vector of sizes,
- food < −200*rnorm(1,1,0.3)
# food supplied per day, 200* random normal deviate
- pfood < −(1/(1 + exp(−1 × (Xo − 4)))) × rnorm(n,1,0.15)
# competition and sharing of food
share < −food*(pfood)/sum(pfood).
(b). Genetic variation (G)
G1 varies from a random uniform distribution between 0.1 and 0.95
G2 varies from a random uniform distribution between 5.00 and 7.05
G3 varies from a random uniform distribution between 0.1 and 0.95
G4 takes three equally likely values of 0.0, 0.5 or 1.0
G5 takes three equally likely values of 0.0, 0.5 or 1.0
G6 varies from a random uniform distribution between 0.1 and 0.95
G7 varies from a random uniform distribution between 0.01 and 0.100
G8 varies from a random uniform distribution between 0.01 and 0.400
(c). If genes fixed at midrange of baseline genetic variation (NOG)
G1 varies from a random uniform distribution between 0.50 and 0.51
G2 varies from a random uniform distribution between 5.00 and 5.05
G3 varies from a random uniform distribution between 0.50 and 0.51
G4 is 0.5
G5 is 0.5
G6 varies from a random uniform distribution between 0.50 and 0.51
G7 varies from a random uniform distribution between 0.05 and 0.051
G8 varies from a random uniform distribution between 0.20 and 0.21
(d). Mutation
G1 mutants come from a random uniform distribution between 0.1 and 0.95
G2 is initially specified by a lower and upper range. Mutations in G2 are from random uniform deviates within this range, modified by a random deviate in the range 1–1.05 × upper limit, and 0.95–1.0 × lower limit.
G3 mutants come from a random uniform distribution between 0.0 and 1.0
G4 mutants take three equally likely values of 0, 0.5 or 1.0
G5 mutants take three equally likely values of 0.0, 0.5 or 1.0
G6 mutants come from a random uniform distribution between 0.0 and 1.0.
G7 is initially specified by a lower and upper range. Mutations in G7 are from random uniform deviates within this range, modified by a random deviate in the range 1–1.05 × upper limit, and 0.95–1.0 × lower limit.
G8 is initially specified by a lower and upper range. Mutations in G2 are from random uniform deviates within this range, modified by a random deviate in the range 1–1.05 × upper limit, and 0.95–1.0 × lower limit.
References
- 1.Benton T. G., Plaistow S. J., Coulson T. N. 2006. Complex population dynamics and complex causation: devils, details and demography. Proc. R. Soc. B 273, 1173–1181 10.1098/rspb.2006.3495 (doi:10.1098/rspb.2006.3495) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sinclair A. R. E., Pech R. P. 1996. Density dependence, stochasticity, compensation and predator regulation. Oikos 75, 164–173 10.2307/3546240 (doi:10.2307/3546240) [DOI] [Google Scholar]
- 3.Coulson T., Catchpole E. A., Albon S. D., Morgan B. J. T., Pemberton J. M., Clutton-Brock T. H., Crawley M. J., Grenfell B. T. 2001. Age, sex, density, winter weather, and population crashes in Soay sheep. Science 292, 1528–1531 10.1126/science.292.5521.1528 (doi:10.1126/science.292.5521.1528) [DOI] [PubMed] [Google Scholar]
- 4.Ovaskainen O., Cornell S. J. 2006. Space and stochasticity in population dynamics. Proc. Natl Acad. Sci. USA 103, 12 781–12 786 10.1073/pnas.0603994103 (doi:10.1073/pnas.0603994103) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Greenman J. V., Benton T. G. 2005. The impact of environmental fluctuations on structured discrete time population models: resonance, synchrony and threshold behaviour. Theor. Popul. Biol. 68, 217–235 10.1016/j.tpb.2005.06.007 (doi:10.1016/j.tpb.2005.06.007) [DOI] [PubMed] [Google Scholar]
- 6.Ellner S. P., Geber M. A., Hairston N. G. 2011. Does rapid evolution matter? Measuring the rate of contemporary evolution and its impacts on ecological dynamics. Ecol. Lett. 14, 603–614 10.1111/j.1461-0248.2011.01616.x (doi:10.1111/j.1461-0248.2011.01616.x) [DOI] [PubMed] [Google Scholar]
- 7.Ozgul A., Tuljapurkar S., Benton T. G., Pemberton J. M., Clutton-Brock T. H., Coulson T. 2009. The dynamics of phenotypic change and the shrinking sheep of St. Kilda. Science 325, 464–467 10.1126/science.1173668 (doi:10.1126/science.1173668) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yoshida T., Jones L. E., Ellner S. P., Fussmann G. F., Hairston N. G. 2003. Rapid evolution drives ecological dynamics in a predator–prey system. Nature 424, 303–306 10.1038/nature01767 (doi:10.1038/nature01767) [DOI] [PubMed] [Google Scholar]
- 9.Coulson T., Tuljapurkar S., Childs D. Z. 2010. Using evolutionary demography to link life history theory, quantitative genetics and population ecology. J. Anim. Ecol. 79, 1226–1240 10.1111/j.1365-2656.2010.01734.x (doi:10.1111/j.1365-2656.2010.01734.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desharnais R. A., Costantino R. F., Cushing J. M., Dennis B., Henson S. M., King A. 2005. Population dynamics and laboratory ecology. San Diego, CA: Elsevier. See http://www.elsevierdirect.com/ISBN/9780120139378/Population-Dynamics-and-Laboratory-Ecology [Google Scholar]
- 11.Zheng C. Z., Ovaskainen O., Hanski I. 2009. Modelling single nucleotide effects in phosphoglucose isomerase on dispersal in the Glanville fritillary butterfly: coupling of ecological and evolutionary dynamics. Phil. Trans. R. Soc. B 364, 1519–1532 10.1098/rstb.2009.0005 (doi:10.1098/rstb.2009.0005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Benton T. G., Beckerman A. P. 2005. Population dynamics in a noisy world: lessons from a mite experimental system. Adv. Ecol. Res. 37, 143–181 [Google Scholar]
- 13.Plaistow S. J., Lapsley C. T., Beckerman A. P., Benton T. G. 2004. Age and size at maturity: sex, environmental variability and developmental thresholds. Proc. R. Soc. Lond. B 271, 919–924 10.1098/rspb.2004.2682 (doi:10.1098/rspb.2004.2682) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plaistow S. J., Lapsley C. T., Benton T. G. 2006. Context-dependent intergenerational effects: the interaction between past and present environments and its effect on population dynamics. Am. Nat. 167, 206–215 10.1086/499380 (doi:10.1086/499380) [DOI] [PubMed] [Google Scholar]
- 15.Plaistow S. J., St Clair J. J. H., Grant J., Benton T. G. 2007. How to put all your eggs in one basket: empirical patterns of offspring provisioning throughout a mother's lifetime. Am. Nat. 170, 520–529 10.1086/521238 (doi:10.1086/521238) [DOI] [PubMed] [Google Scholar]
- 16.Cameron T. C., O'Sullivan D., Reynolds A., Piertney S. B., Benton T. G. Evolution of population dynamics in response to selection on life-histories. doi: 10.1111/ele.12107. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plaistow S. J., Benton T. G. 2009. The influence of context-dependent maternal effects on population dynamics: an experimental test. Phil. Trans. R. Soc. B 364, 1049–1058 10.1098/rstb.2008.0251 (doi:10.1098/rstb.2008.0251) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wood S. N. 2010. Statistical inference for noisy nonlinear ecological dynamic systems. Nature 466, 1102–1113 10.1038/nature09319 (doi:10.1038/nature09319) [DOI] [PubMed] [Google Scholar]
- 19.Grimm V., et al. 2006. A standard protocol for describing individual-based and agent-based models. Ecol. Model. 198, 115–126 10.1016/j.ecolmodel.2006.04.023 (doi:10.1016/j.ecolmodel.2006.04.023) [DOI] [Google Scholar]
- 20.Team RDC. 2009. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing [Google Scholar]
- 21.Benton T. G., Plaistow S. J., Beckerman A. P., Lapsley C. T., Littlejohns S. 2005. Changes in maternal investment in eggs can affect population dynamics. Proc. R. Soc. B 272, 1351–1356 10.1098/rspb.2005.3081 (doi:10.1098/rspb.2005.3081) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benton T. G., Cameron T. C., Grant A. 2004. Population responses to perturbations: predictions and responses from laboratory mite populations. J. Anim. Ecol. 73, 983–995 10.1111/j.0021-8790.2004.00859.x (doi:10.1111/j.0021-8790.2004.00859.x) [DOI] [Google Scholar]
- 23.Beckerman A. P., Benton T. G., Lapsley C. T., Koesters N. 2003. Talkin' 'bout my generation: environmental variability and cohort effects. Am. Nat. 162, 754–767 10.1086/381056 (doi:10.1086/381056) [DOI] [PubMed] [Google Scholar]
- 24.Reuter H., Jopp F., Holker F., Eschenbach C., Middelhoff U., Breckling B. 2008. The ecological effect of phenotypic plasticity: analyzing complex interaction networks (COIN) with agent-based models. Ecol. Inform. 3, 35–45 10.1016/j.ecoinf.2007.03.010 (doi:10.1016/j.ecoinf.2007.03.010) [DOI] [Google Scholar]
- 25.Donohue K., Polisetty C. R., Wender N. J. 2005. Genetic basis and consequences of niche construction: plasticity-induced genetic constraints on the evolution of seed dispersal in Arabidopsis thaliana. Am. Nat. 165, 537–550 10.1086/429162 (doi:10.1086/429162) [DOI] [PubMed] [Google Scholar]
- 26.Hassall M., Helden A., Benton T. 2003. Phenotypic plasticity and interpopulation differences in life history traits of Armadillidium vulgare (Isopoda: Oniscidae). Oecologia 137, 85–89 10.1007/s00442-003-1325-1 (doi:10.1007/s00442-003-1325-1) [DOI] [PubMed] [Google Scholar]
- 27.LaMontagne J. M., McCauley E. 2001. Maternal effects in Daphnia: what mothers are telling their offspring and do they listen? Ecol. Lett. 4, 64–71 10.1046/j.1461-0248.2001.00197.x (doi:10.1046/j.1461-0248.2001.00197.x) [DOI] [Google Scholar]
- 28.Persson L., de Roos A. M., Claessen D., Bystrom P., Lovgren J., Sjogren S., Svanback R., Wahlstrom E., Westman E. 2003. Gigantic cannibals driving a whole-lake trophic cascade. Proc. Natl Acad. Sci. USA 100, 4035–4039 10.1073/pnas.0636404100 (doi:10.1073/pnas.0636404100) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hawkins E., Sutton R. 2011. Estimating climatically relevant singular vectors for decadal predictions of the Atlantic Ocean. J. Climatol. 24, 109–123 10.1175/2010jcli3579.1 (doi:10.1175/2010jcli3579.1) [DOI] [Google Scholar]
- 30.Hawkins E., Sutton R. 2009. The potential to narrow uncertainty in regional climate predictions. Bull. Am. Meteorol. Soc. 90, 1095–1107 10.1175/2009bams2607.1 (doi:10.1175/2009bams2607.1) [DOI] [Google Scholar]
- 31.Stockwell C. A., Hendry A. P., Kinnison M. T. 2003. Contemporary evolution meets conservation biology. Trends Ecol. Evol. 18, 94–101 10.1016/S0169-5347(02)00044-7 (doi:10.1016/S0169-5347(02)00044-7) [DOI] [Google Scholar]
- 32.Olsen E. M., Heino M., Lilly G. R., Morgan M. J., Brattey J., Ernande B., Dieckmann U. 2004. Maturation trends indicative of rapid evolution preceded the collapse of northern cod. Nature 428, 932–935 10.1038/nature02430 (doi:10.1038/nature02430) [DOI] [PubMed] [Google Scholar]
- 33.Zanesse A. 2010. Ecological and evolutionary consequences of environmental perturbations for population persistence. PhD thesis, University of Leeds, Leeds, UK [Google Scholar]
- 34.De Roos A. M., Persson L., McCauley E. 2003. The influence of size-dependent life-history traits on the structure and dynamics of populations and communities. Ecol. Lett. 6, 473–487 10.1046/j.1461-0248.2003.00458.x (doi:10.1046/j.1461-0248.2003.00458.x) [DOI] [Google Scholar]
- 35.Freckleton R. P., Watkinson A. R., Green R. E., Sutherland W. J. 2006. Census error and the detection of density dependence. J. Anim. Ecol. 75, 837–851 10.1111/j.1365-2656.2006.01121.x (doi:10.1111/j.1365-2656.2006.01121.x) [DOI] [PubMed] [Google Scholar]
- 36.Hixon M. A., Pacala S. W., Sandin S. A. 2002. Population regulation: historical context and contemporary challenges of open vs. closed systems. Ecology 83, 1490–1508 10.1890/0012-9658(2002)083[1490:prhcac]2.0.co;2 (doi:10.1890/0012-9658(2002)083[1490:prhcac]2.0.co;2) [DOI] [Google Scholar]
- 37.Dennis B., Taper M. L. 1994. Density-dependence in time-series observations of natural-populations: estimation and testing. Ecol. Monogr. 64, 205–224 10.2307/2937041 (doi:10.2307/2937041) [DOI] [Google Scholar]
- 38.Collins M., Booth B. B. B., Harris G. R., Murphy J. M., Sexton D. M. H., Webb M. J. 2006. Towards quantifying uncertainty in transient climate change. Clim. Dyn. 27, 127–147 10.1007/s00382-006-0121-0 (doi:10.1007/s00382-006-0121-0) [DOI] [Google Scholar]