Abstract
Enzymes are increasingly being used in an industrial setting as a cheap and environmentally-friendly alternative to chemical catalysts. In order to produce the ideal biocatalyst, natural enzymes often require optimization to increase their catalytic efficiencies and specificities under a particular range of reaction conditions. A number of enzyme engineering strategies are currently employed to modify biocatalysts, improving their suitability for large-scale industrial applications. These include various directed evolution techniques, semi-rational design techniques, and more recently, the de novo design of novel enzymes. Advances in mutant library design, high-throughput selection processes, and the introduction of powerful computer algorithms have all contributed to the current exponential growth of the field of enzyme engineering. This review article aims to present some of the currently employed strategies for enzyme engineering and attempts to highlight the most recent advances in methodology.
Keywords: Biocatalysts, enzyme engineering, directed evolution, semi-rational design, de novo design
Introduction
Enzymes are excellent catalysts. Millions of years of evolution has afforded enzymes with the ability to accelerate the rate of chemical reactions with a high degree of efficiency, selectivity and specificity. As such, the use of enzymes as biocatalysts for industrial processes is an economically and environmentally attractive option 1. However, natural enzymes are often unable to function in the non-natural conditions dictated by many industrial reactions, which frequently require elevated temperatures, or utilize high substrate concentrations that can inhibit enzyme function. Another important factor that may restrict the use of a biocatalyst is the cost of producing the protein in a stable and pure form. These limitations are currently being addressed by the fast expanding field of enzyme engineering 2. There are already a number of successful reports of natural enzymes being redesigned to a set of specific chemical synthesis requirements, making biocatalysts a more viable option for industrial purposes.
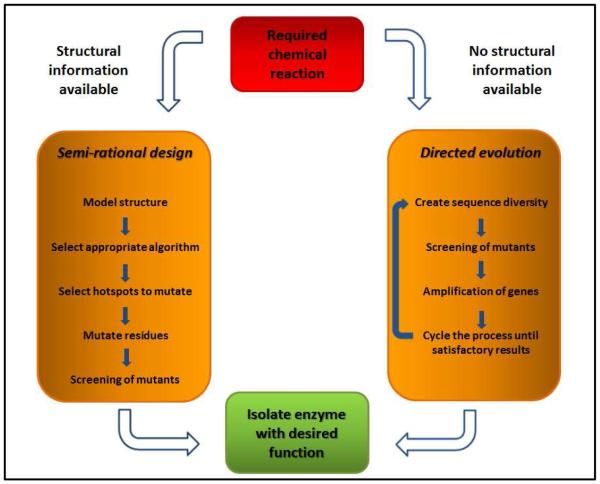
Enzyme engineering is the process by which the natural sequence of an enzyme is altered in order to tailor-design the activity of the enzyme for a particular reaction. Two broad approaches can be undertaken to achieve this – directed evolution and (semi-)rational design (Figure 1). Previously, these two approaches were considered unique and exclusive. However, in recent years there has been an increasing trend in the two approaches being used hand-in-hand to optimize a biocatalyst in a less labor-intensive and more time-efficient manner. Additionally, advances in the field of computational biology and our growing understanding of protein structure have expanded the field of engineering into the realms of de-novo design. We are now creating enzymes capable of catalyzing reactions which cannot be catalyzed by any known naturally occurring enzyme 3-5. Here, the main approaches to enzyme engineering will be discussed in more detail, with specific examples of successful engineering of biocatalysts presented, and recent advances in the field will also be highlighted.
Figure 1.
A summary of the different processes required by semi-rational design and directed evolution.
Directed evolution
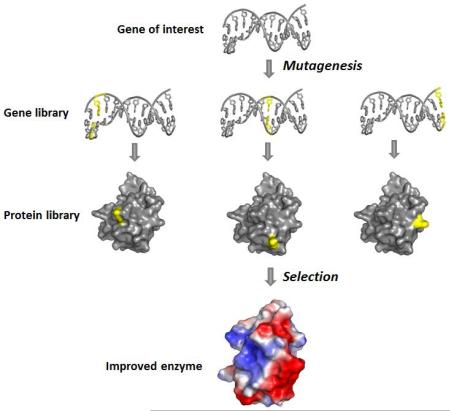
Directed evolution is based upon the principle of natural evolution, whereby the incorporation of random mutations into the sequence of an enzyme allows the creation a large mutant library (103 - 106 mutants) displaying a high level of sequence diversity. This diversity is then explored by high-throughput screening to identify and select for those mutations which produce the desired phenotype or increase the enzyme activity, mimicking the process of natural selection. This selection procedure is repeated several times to produce the final biocatalyst with the desired traits 6.
The challenge of creating the expansive and diverse library of mutants called for by directed evolution has largely been overcome with the development of a number of robust techniques for producing genetic diversity 7. Perhaps the most commonly employed techniques to generate this diversity are error-prone PCR 8, which inserts mutations randomly across genes due to the fact that Taq polymerase lacks 3′ – 5′ exonuclease proofreading activity; and DNA shuffling 9, which involves the recombination of homologous sets of genes. Other techniques to introduce sequence diversity include the use of mutator strains, which lack one or more DNA repair pathways 10; growth of cells harboring a plasmid encoding for the gene of interest in the presence of chemical mutagens such as EMS 11; and sequence saturation mutagenesis (SeSaM), which generates truly random mutations across each nucleotide within a given sequence 12.
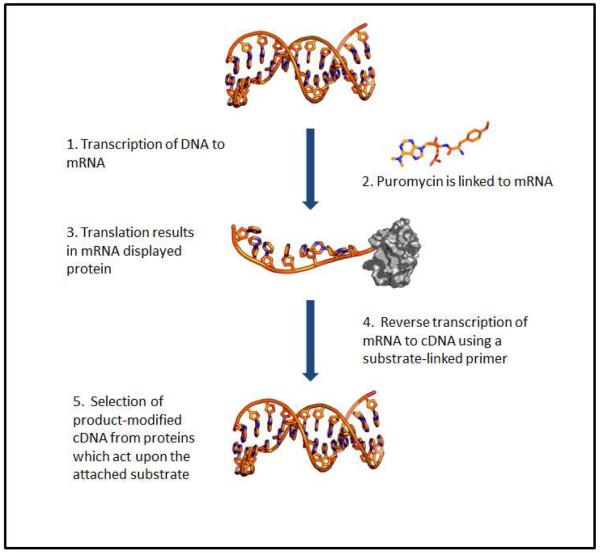
One elegant example in which an enzyme was first selected for its novel activity, and the activity was subsequently catalytically improved by directed evolution, was presented by Seelig and Szostak 13. In order to aid their library screening process, they developed a technique known as mRNA display, which allows for the in vitro selection of enzymes from protein libraries. In this technique a DNA library is first created, and it is then transcribed into mRNA. A modified oligonucleotide containing puromycin (an antibiotic which resembles tRNA) is cross-linked to the 3′ end of the mRNA before in vitro translation, resulting in mRNA-displayed protein. In order to carry out the selection process, the mRNA-displayed protein is linked to the reaction substrate via reverse transcription of the mRNA to cDNA using a substrate-linked primer. Active enzymes can then be selected for as they will convert the substrate into the required product. The cDNA of the active enzymes is then isolated and is used for further rounds of evolution (Figure 2). Using this technique, Seelig and Szostak probed a library which had been prepared by mutating two recognition loops of the DNA binding domain of human retinoid-X-receptor using degenerate primers, preselection of random cassettes for intact open reading frames, and assembly of the final library by an iterative process of restriction and ligation 14, 15. The authors tested the library consisting of 4 × 1012 RNA ligases for a particular novel activity – the ability to catalyze the ligation of a 5′ triphosphorylated RNA oligonucleotide to the 3′ hydroxyl group on a second RNA oligonucleotide. The activity of the resulting isolated RNA ligases was further improved by error-prone PCR. Following several rounds of mutagenesis and selection, 18 novel RNA ligases were found. The 7 most active ligases were expressed in E. coli as part of a maltose binding protein (MBP) fusion to improve stability and solubility of the proteins, and the most active of these fusion proteins was characterized. They found that their evolved RNA ligase was capable of catalyzing this novel reaction 2 × 106 times faster than the uncatalyzed reaction, which is a marked improvement.
Figure 2.
A scheme representing the process of mRNA display of proteins. A library of mRNA displayed proteins is reverse transcribed to cDNA using primers with a substrate covalently attached. This results in a cDNA-mRNA-protein-substrate complex. Proteins that are active against the substrate modify their own cDNA with the resulting reaction product. This cDNA is then selected and is amplified for further rounds of engineering.
One of the greatest advantages of the technique described by Seelig and Szostak, and in fact of directed evolution as a whole, is that no prior structural knowledge of the enzyme is required, permitting the engineering of enzymes whose function is not yet fully understood 16. However, the stochastic nature of directed evolution imposes a serious limitation on this method – that is, the larger the library of mutants screened, the greater the chance of selecting the desired mutant. Consequently, this technique relies heavily on the ability to test the large number of mutants by a high-throughput assay, which is often an extremely labor-intensive process17, 18. The development of techniques such as mRNA display of proteins 13, fluorescence-activated cell-sorting (FACS) of cell-surface displayed mutants 19, and the incorporation of individual bacterial cells into microdroplets as a means of assessing gene expression and enzyme activity 20 have made the screening of large mutant libraries a more practical and achievable process. Nevertheless, the creation of smaller, high-quality libraries containing more mutants displaying the required phenotype, as opposed to larger libraries consisting of a relatively high proportion of non-functional mutants, would be a more practical approach to circumventing the screening bottleneck 21. It is with this aim in mind that researchers have embarked upon the path of semi-rational design of biocatalysts.
Semi-rational design
Semi-rational design is a knowledge-driven process, requiring some degree of understanding of the mechanism by which the enzyme catalyzes a reaction, as well as prior knowledge of either the sequence or the three-dimensional crystal structure of the enzyme. These requirements immediately limit the use of semi-rational design to only those enzymes which have been previously characterized. However, the antithesis of this limitation is that this prior knowledge allows the researcher to design a more specific set of mutations, thereby creating a much smaller library (102 – 103 mutants) with a higher proportion of mutants displaying beneficial traits. Not only does this diminish the task of screening the library, it also allows the researcher to fine-tune the activity of the enzyme more precisely before undertaking any experiments 22.
The typical decision making process that a researcher follows prior to creating a library by semi-rational design is described in Box 1 23. A varied and valuable toolkit is available for semi-rational design, owing to the development of a number of algorithms and programs which simplify the decision making process and allow the researcher to screen libraries in silico 24, 25. One such algorithm is Quantitative Structure-Activity Relationships (QSAR). This multivariate statistical approach establishes a relationship between mutations at given positions in the sequence of an enzyme and the activity of the mutants, providing predictions of the relative activities of the mutants prior to any experimental measurements. This algorithm has been further developed by combining it with ProSAR analysis, which classifies mutations within a given library as neutral, deleterious, or beneficial. The most active mutant is pre-selected using QSAR, and is subsequently used as the template for incorporation of the most beneficial mutations provided by ProSAR, allowing a mutation-activity focused optimization of a biocatalyst 26. Another methodology that is used to predict the value of mutating given amino acids is Combinatorial Active Site Saturation Test (CASTing), which uses the three-dimensional structure of an enzyme to identify key residues surrounding the active site. Several of these positions are then randomized simultaneously in silico, increasing the probability of cooperative effects favorably altering the activity of the enzyme 27.
Box1. The decision process for semi-rational design.
Semi-rational design relies on the previous characterization of the enzyme of interest. There must be either structural or, at the very least, sequence data existing before semi-rational design can be undertaken. An understanding of the mechanism by which the enzyme catalyzes its reaction is also highly beneficial.
Tne first question a researcher must ask is whether the structure has been solved of either the enzyme or a closely related homolog. If the structure of a homolog exists, it is possible to create a three dimensional model of the enzyme of interest, provided that the enzyme sequence is known, thanks to modeling tools such as Swiss Model, available at the ExPASy proteomics server (http://ca.expasy.org).
The next step in the process is to select an appropriate algorithm or program to help select residues to mutate. A wealth of programs are available, and this decision is often dictated by previous experience, or personal preference. The Rosetta suite of programs have revolutionalized the design of enzymes and is widely used (http://rosettadesign.med.unc.edu/). However, other programs exist, such as the HotSpot Wizard, anotherwebserver which can be easily accessed (http://loschmidt.chemi.muni.cz/hotspotwizard/).
In minimal inputs are usually a PDB code or file. Depending on the algorithm used,the program may try to mimimize energy functions, locate evolutionary conserved residues, or assign function to certain residues. The ouput is often in PDB file format, which allows visualization in programs such as PyMOL (http://www.pymol.org/}.
The researcher must then view the results and decide whether the residues selected by the program are suitable formutagenesis. This is a subjective process. Important factors to take into consideration are proximity to the active site, whether the mutation will sterically block the active site or tunnel, and whether the mutated residues will likely alter the chemical reaction taking place. An understanding of the catalytic mechanism is therefore essential.
Finally, the researcher can design a library of residues to mutate based upon their predictions, and undertake their experiments to determine whether their predictions were correct.
Interestingly, one recent study has challenged the widely-accepted belief that active site residues and their neighbors control the specificity of an enzyme 28. The crystal structure of the Baeyer-Villiger monooxygenase PAMO was analyzed to identify randomization hotspots remote to the active site. The aim of mutating a region distant from the active site was to induce allosteric effects, thereby altering the enzyme activity and enantioselectivity. It was predicted that changes in allostery would result in domain shifts, bringing the NADP-binding domain closer to the FAD-binding domain. This structural reorganization would therefore create a new binding pocket for the enzyme. Subsequently, residues Gln93 and Pro94, which are located between the two binding domain interfaces, were selected for saturation mutagenesis studies17. Using the already known crystal structure of the enzyme, Wu et al. 28 were able to design a very specific, high quality mutant library, thereby greatly reducing the screening effort required. After screening of only 400 variants; a meager effort in comparison to the mammoth task of library screening in most cases of directed evolution; two double mutants were found to have altered substrate specificity. Of these, the double mutant Gln93Asn/Pro94Asp was the most active, displaying highly enantioselectivity against cyclohexanone derivatives which are normally not accepted by PAMO 28.
Although semi-rational design allows one to create a smaller, more specific library of mutants with lower redundancy, the question remains – is it even necessary to make a mutant library? Natural evolution has far excelled our current engineering strategies, drawing from millions of years worth of experience in perfecting enzymes for particular processes. Mother Nature has already furnished us with an immense library of diverse sequences. Some researchers have taken advantage of this fact, using preexisting enzymes in their search for the perfect biocatalyst. In a recent report, Höhne and co-workers 29 analyzed the three dimensional crystal structure of (S)-selective transaminase, making predictions of key amino acid mutations which could convert the enzyme to an (R)-selective transaminase. However, rather than create a mutant library, they produced an algorithm to search sequence databases, to ascertain whether any preexisting enzymes already displayed these particular mutations. In doing so, they discovered 17 highly enantiospecific (R)-selective transaminases which had not previously been reported 29. This shift from semi-rational design to rational in silico approaches is gaining more prominence, in part due to our improving knowledge of enzyme catalysis, as well as the development of computer-assisted design methods such as Rosetta 3, 4, 30, 31 which permits the de-novo design of an active site for a particular chemical reaction.
De-novo design
De-novo design of enzymes is modeled on the principle that an enzyme catalyzes a reaction by stabilizing the transition state, therefore lowering the activation energy of the reaction 32. If this principle is taken literally, it should theoretically be possible to arrange a set of atoms in a particular configuration and produce an active enzyme. This is the basis of the computational rational design of de novo enzymes. The first step in designing an enzyme is the in silico modeling of the transition state of a reaction, with active site residues positioned in an optimized geometry to stabilize the transition state using quantum mechanic simulations. Next, the idealized active site is positioned within already existing protein scaffolds and the amino acids are varied to perfectly accommodate the binding pocket using molecular modeling tools such as RosettaMatch 3, 4, 30, 31. Subsequently, this highly challenging method requires a detailed understanding of the catalytic action of the enzyme and a high resolution crystal structure to model the enzyme upon 33.
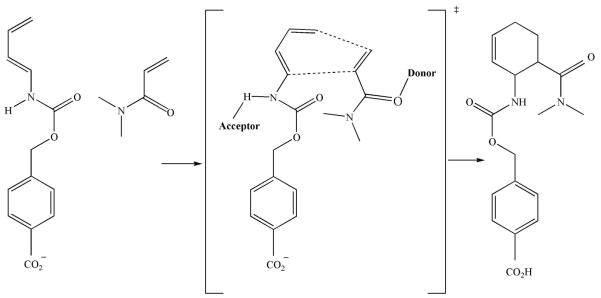
Initial success has been reported by the Baker lab, with the de novo design of a retro-aldolase 3, a Kemp eliminase 4 and most recently, a Diels-Alderase 5. In this latest report, the researchers describe the design of an enzyme capable of forming two carbon-carbon bonds in the cycloaddition of diene and dienophile, forming a cyclohexene ring (Figure 3). While there are reports of enzymes capable of an intramolecular cycloaddition 34, no naturally occurring enzyme capable of catalyzing an intermolecular Diels-Alder reaction is known to exist, making this an important advancement for organic synthesis. First, a model catalytic mechanism was designed, and in silico active site models were designed using Rosetta methodology. Using a carbonyl oxygen from glutamine or asparagine to hydrogen bond to the diene intermediate, and a hydroxyl from serine, threonine or tyrosine to hydrogen bond to the dienophile intermediate, quantum mechanic calculations were used to find the lowest energy transition state. The surrounding protein scaffold was designed using RosettaMatch 31 and RosettaDesign 35, resulting in 84 designs which were tested experimentally. Two of the designed enzymes displayed stereospecific Diels-Alder activity, and their catalytic efficiencies were further improved by mutation of key active site residues to further stabilize the transition state. One of the mutants displayed a catalytic efficiency 20 times greater than catalytic antibodies which undertake the same reaction, perhaps only a modest improvement upon existing biocatalysts for this reaction.
Figure 3.
A reaction scheme of the Diels-Alder reaction carried out by a designed enzyme, which catalyzes a pericyclic [4 + 2] cycloaddition of diene and dienophile to form a chiral cyclohexene ring. An acceptor and donor are required within the active site of the enzyme to activate the two molecules. The carbonyl oxygen from amino acids such as glutamine or asparagine could act as an acceptor for the diene intermediate, while a hydroxyl from amino acids such as serine, threonine or tyrosine could act as a donor for the dienophile intermediate.
The same argument that de novo design does not always produce a highly active biocatalyst can also be applied in the case of the Kemp eliminase 4. Here, the removal of a hydrogen ion from a carbon-hydrogen bond is a key step to initiate the reaction. Using aspartic acid and histidine as potential bases the researchers modeled the active site and tested a number of protein scaffolds to produce 59 energetically stable models. These models were tested experimentally, and eight were found to be active. One of these proteins (KE07) was further improved by mutational analysis, creating an enzyme which could catalyze the reaction a million times faster than the uncatalyzed reaction, which, again equals but does not excel the rates already reported for catalytic antibodies 36. KE07 has since been further improved, first by computational design to perturb the active site backbone geometries, alter the overall active site confirmation, regulate the charge of the active site residues, and to change the length of the loop covering the active site. The residues designated as hotspots by these models were subjected to several rounds of random mutagenesis, resulting in a greater than 400-fold improvement in the catalytic efficiency 37. These reports exemplify the fact that de novo design is not without its drawbacks and difficulties.
An even more challenging case is the de novo design of both a novel active site and the surrounding protein scaffold, from first order principles. One group designed an artificial four-helix bundle di-iron enzyme 38, and then converted it to a phenol oxidase by addition of a phenol binding site. To do this, four bulky leucine residues which blocked the active site required mutation to smaller residues such as glycine. However, any one of these mutations led to a complete destabilization of the fold of the enzyme. By substituting three residues, Val24Thr, Lys25His, and Leu26Asn, which are situated in a loop region distant to the active site, the authors were able to stabilize the protein structure and subsequently solved the structure of the enzyme by NMR 39.
Perhaps the catalytic activity improvements described above for de novo design are not as impressive as some reported for directed evolution, and a dependence upon directed evolution techniques still exists to achieve these improvements. Nevertheless, an important milestone has been reached in enzyme engineering. We are now in a position to tailor-design enzymes to catalyze reactions that do not occur in nature, allowing us to create the ideal biocatalyst 40.
Concluding remarks
Advances in the methodology available to the enzyme engineer have allowed the development of new strategies to design biocatalysts. While most early studies were focused on directed evolution techniques, there has been a shift towards employing directed evolution in conjunction with semi-rational design to produce biocatalysts. This has mainly been thanks to the introduction of a number of excellent computer algorithms, and to the exponential growth in the number of three dimensional structures and protein sequences which are available to the researcher. Smaller, high quality mutant libraries are now commonplace and a range of techniques have been introduced which permit the rapid and straightforward analysis of library members. We are now on the cusp of a new era of complete de novo design of biocatalysts, with successful reports of enzymes being designed to catalyze unnatural reactions already emerging. While these designed enzymes perhaps lag behind evolved enzymes in catalytic efficiencies, they represent an incredible leap forward in our abilities to create “ex nihilo”. They are also important starting points for further improvements using our existing well-proven strategies of directed evolution and semi-rational design. In summary, these exciting advances point towards a number of potentially highly efficient and practical strategies for the production of tailor-designed biocatalysts for industrial purposes.
Acknowledgment
The authors gratefully acknowledge support by the National Institute of Health (grant GM080299, to CSD).
References
- 1.Wohlgemuth R. New Biotechnol. 2009;25:204–13. doi: 10.1016/j.nbt.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Keasling JD. ACS Chem Biol. 2008;3:64–76. doi: 10.1021/cb7002434. [DOI] [PubMed] [Google Scholar]
- 3.Jiang L, Althoff EA, Clemente FR, Doyle L, Rothlisberger D, Zanghellini A, Gallaher JL, Betker JL, Tanaka F, Barbas CF, 3rd, Hilvert D, Houk KN, Stoddard BL, Baker D. Science. 2008;319:1387–91. doi: 10.1126/science.1152692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rothlisberger D, Khersonsky O, Wollacott AM, Jiang L, DeChancie J, Betker J, Gallaher JL, Althoff EA, Zanghellini A, Dym O, Albeck S, Houk KN, Tawfik DS, Baker D. Nature. 2008;453:190–5. doi: 10.1038/nature06879. [DOI] [PubMed] [Google Scholar]
- 5.Siegel JB, Zanghellini A, Lovick HM, Kiss G, Lambert AR, St Clair JL, Gallaher JL, Hilvert D, Gelb MH, Stoddard BL, Houk KN, Michael FE, Baker D. Science. 2010;329:309–13. doi: 10.1126/science.1190239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Labrou NE. Curr Protein Pept Sci. 2010;11:91–100. doi: 10.2174/138920310790274617. [DOI] [PubMed] [Google Scholar]
- 7.Reetz MT. A method for rapid directed evolution. In: Lutz S, Bornscheuer UT, editors. Protein Engineering Handbook. Wiley-VCH; 2011. pp. 409–440. [Google Scholar]
- 8.Zhou YH, Zhang XP, Ebright RH. Nucleic Acids Res. 1991;19:6052. doi: 10.1093/nar/19.21.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stemmer WP. Nature. 1994;370:389–91. doi: 10.1038/370389a0. [DOI] [PubMed] [Google Scholar]
- 10.Bornscheuer UT, Altenbuchner J, Meyer HH. Biotechnol Bioeng. 1998;58:554–9. doi: 10.1002/(sici)1097-0290(19980605)58:5<554::aid-bit12>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 11.Mohan U, Banerjee UC. Chembiochem. 2008;9:2238–43. doi: 10.1002/cbic.200800259. [DOI] [PubMed] [Google Scholar]
- 12.Wong TS, Tee KL, Hauer B, Schwaneberg U. Nucleic Acids Res. 2004;32:e26. doi: 10.1093/nar/gnh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seelig B, Szostak JW. Nature. 2007;448:828–31. doi: 10.1038/nature06032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho G, Keefe AD, Liu R, Wilson DS, Szostak JW. J Mol Biol. 2000;297:309–19. doi: 10.1006/jmbi.2000.3571. [DOI] [PubMed] [Google Scholar]
- 15.Cho GS, Szostak JW. Chem Biol. 2006;13:139–47. doi: 10.1016/j.chembiol.2005.10.015. [DOI] [PubMed] [Google Scholar]
- 16.Dalby PA. Recent Pat Biotechnol. 2007;1:1–9. doi: 10.2174/187220807779813929. [DOI] [PubMed] [Google Scholar]
- 17.Reetz MT. Angew Chem Int Ed Engl. 2011;50:138–74. doi: 10.1002/anie.201000826. [DOI] [PubMed] [Google Scholar]
- 18.Turner NJ. Nat Chem Biol. 2009;5:567–73. doi: 10.1038/nchembio.203. [DOI] [PubMed] [Google Scholar]
- 19.Olsen MJ, Stephens D, Griffiths D, Daugherty P, Georgiou G, Iverson BL. Nat Biotechnol. 2000;18:1071–4. doi: 10.1038/80267. [DOI] [PubMed] [Google Scholar]
- 20.Shim JU, Olguin LF, Whyte G, Scott D, Babtie A, Abell C, Huck WT, Hollfelder F. J Am Chem Soc. 2009;131:15251–6. doi: 10.1021/ja904823z. [DOI] [PubMed] [Google Scholar]
- 21.Lutz S, Patrick WM. Curr Opin Biotechnol. 2004;15:291–7. doi: 10.1016/j.copbio.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Lutz S. Curr Opin Biotechnol. 2010;21:734–43. doi: 10.1016/j.copbio.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kazlauskas RJ, Bornscheuer UT. Nat Chem Biol. 2009;5:526–9. doi: 10.1038/nchembio0809-526. [DOI] [PubMed] [Google Scholar]
- 24.Damborsky J, Brezovsky J. Curr Opin Chem Biol. 2009;13:26–34. doi: 10.1016/j.cbpa.2009.02.021. [DOI] [PubMed] [Google Scholar]
- 25.Bommarius AS, Blum JK, Abrahamson MJ. Curr Opin Chem Biol. 2011;15:194–200. doi: 10.1016/j.cbpa.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 26.Fox RJ, Davis SC, Mundorff EC, Newman LM, Gavrilovic V, Ma SK, Chung LM, Ching C, Tam S, Muley S, Grate J, Gruber J, Whitman JC, Sheldon RA, Huisman GW. Nat Biotechnol. 2007;25:338–44. doi: 10.1038/nbt1286. [DOI] [PubMed] [Google Scholar]
- 27.Reetz MT, Wang LW, Bocola M. Angew Chem Int Ed Engl. 2006;45:1236–41. doi: 10.1002/anie.200502746. [DOI] [PubMed] [Google Scholar]
- 28.Wu S, Acevedo JP, Reetz MT. Proc Natl Acad Sci U S A. 2010;107:2775–80. doi: 10.1073/pnas.0911656107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hohne M, Schatzle S, Jochens H, Robins K, Bornscheuer UT. Nat Chem Biol. 2010;6:807–13. doi: 10.1038/nchembio.447. [DOI] [PubMed] [Google Scholar]
- 30.Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, Davis IW, Cooper S, Treuille A, Mandell DJ, Richter F, Ban YE, Fleishman SJ, Corn JE, Kim DE, Lyskov S, Berrondo M, Mentzer S, Popovic Z, Havranek JJ, Karanicolas J, Das R, Meiler J, Kortemme T, Gray JJ, Kuhlman B, Baker D, Bradley P. Methods Enzymol. 2011;487:545–74. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zanghellini A, Jiang L, Wollacott AM, Cheng G, Meiler J, Althoff EA, Rothlisberger D, Baker D. Protein Sci. 2006;15:2785–94. doi: 10.1110/ps.062353106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garcia-Viloca M, Gao J, Karplus M, Truhlar DG. Science. 2004;303:186–95. doi: 10.1126/science.1088172. [DOI] [PubMed] [Google Scholar]
- 33.Golynskiy MV, Seelig B. Trends Biotechnol. 2010;28:340–5. doi: 10.1016/j.tibtech.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 34.Ose T, Watanabe K, Mie T, Honma M, Watanabe H, Yao M, Oikawa H, Tanaka I. Nature. 2003;422:185–9. doi: 10.1038/nature01454. [DOI] [PubMed] [Google Scholar]
- 35.Kuhlman B, Baker D. Proc Natl Acad Sci U S A. 2000;97:10383–8. doi: 10.1073/pnas.97.19.10383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorn SN, Daniels RG, Auditor MT, Hilvert D. Nature. 1995;373:228–30. doi: 10.1038/373228a0. [DOI] [PubMed] [Google Scholar]
- 37.Khersonsky O, Rothlisberger D, Wollacott AM, Murphy P, Dym O, Albeck S, Kiss G, Houk KN, Baker D, Tawfik DS. J Mol Biol. 2011;407:391–412. doi: 10.1016/j.jmb.2011.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maglio O, Nastri F, Calhoun JR, Lahr S, Wade H, Pavone V, DeGrado WF, Lombardi A. J Biol Inorg Chem. 2005;10:539–49. doi: 10.1007/s00775-005-0002-8. [DOI] [PubMed] [Google Scholar]
- 39.Faiella M, Andreozzi C, de Rosales RT, Pavone V, Maglio O, Nastri F, DeGrado WF, Lombardi A. Nat Chem Biol. 2009;5:882–4. doi: 10.1038/nchembio.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghirlanda G. Nature. 2008;453:164–6. doi: 10.1038/453164a. [DOI] [PubMed] [Google Scholar]