Abstract
Cell polarity is typically oriented by external cues such as cell-cell contacts, chemoattractants, or morphogen gradients. In the absence of such cues, however, many cells can spontaneously polarize in a random direction, suggesting the existence of an internal polarity-generating mechanism whose direction can be spatially biased by external cues. Spontaneous “symmetry-breaking” polarization is likely to involve an autocatalytic process set off by small random fluctuations [1]. Here we review recent work on the nature of the autocatalytic process in budding yeast and on the question of why polarized cells only develop a single “front”.
Polarized cells have a clear front-to-back axis, generally orient cytoskeletal elements along that axis, and localize many cell constituents asymmetrically. A common feature that defines the cell’s front is the local accumulation of the active, GTP-bound, form of a polarity-regulating Rho-family GTPase at the cell cortex. The GTPase is Cdc42 or Rac in animals and fungi, or Rop in plants: hereafter we use Cdc42 for simplicity. This master regulator interacts with “effectors” (proteins that bind specifically to GTP-Cdc42) to orient the cytoskeleton along the front-back axis. Although Cdc42 remains stably concentrated at the front of the cell, studies measuring fluorescence recovery after photobleaching (FRAP) indicated that individual Cdc42 molecules only reside at the front for a few seconds at a time, and other polarity regulators display similarly brief residence times [2]. Thus, cells develop and maintain a dynamically renewed patch of concentrated GTP-Cdc42. How does a random cortical location (and not the rest of the cortex) accumulate GTP-Cdc42?
Disagreement on the requirements for symmetry breaking in yeast
Wild-type yeast cells normally select a front from among a limited number of predetermined sites that contain pre-localized “landmark” proteins [3]. Thus, to study symmetry breaking, investigators forced cells to ignore the predetermined sites and pick a random site instead. This was achieved in two different ways. One involved removal of the “bud site selection” protein Rsr1, without which the landmarks can no longer stimulate local Cdc42 activation. The other involved bypassing the landmarks altogether by inducing the expression of Cdc42Q61L, a mutant that cannot hydrolyze GTP and is thus constitutively activated. In both cases, clusters of concentrated Cdc42 (or Cdc42Q61L) spontaneously formed at random locations. However, whereas rsr1Δ mutants did not require F-actin in order to polarize, the Cdc42pQ61L expressors did [4, 5]. These differences led to some controversy regarding the mechanism(s) of symmetry breaking.
What role does F-actin play in symmetry breaking?
One hypothesis to reconcile the findings from different systems is that yeast have two parallel symmetry-breaking mechanisms, one actin-mediated and the other actin-independent [2]. In rsr1Δ mutants, both would be redundantly operative, so F-actin would not be required. In Cdc42Q61L expressors, the actin-independent mechanism (which probably requires localized Cdc42 activation: see below) no longer works on the constitutively active Cdc42Q61L, allowing the system to uncover the existence of a second, actin-mediated, pathway. In this view, the different model systems each provide insight into one of two normal symmetry-breaking mechanisms.
This hypothesis does not account for several features of the Cdc42Q61L system. First, Cdc42Q61L clustering was only observed when Cdc42Q61L was overexpressed: near-endogenous levels of Cdc42Q61L did not promote clustering [5]. Second, overexpression (but not near-endogenous expression) of Cdc42Q61L caused cell lysis, presumably due to cell-wall weakening [5, 6]. Third, unlike wild-type or rsr1Δ mutant cells, which made one and only one front, Cdc42Q61L overexpressors often generated two or three [4, 6].
These features raise the possibility that the primary symmetry-breaking event in Cdc42Q61L overexpressors is the generation of weak spots in the yeast cell wall. Clustering of F-actin and Cdc42Q61L at the weak spots may occur as an attempt to repair the damage. We note that lethal mutants unrelated to Cdc42 regulation can also cause F-actin clustering followed by lysis [7]. Moreover, laser-induced cell-wall wounding triggers clustering of F-actin at the wound site (Kono and Pellman, personal communication). Either the Cdc42Q61L–induced wounding (whose molecular basis is not understood) or the subsequent clustering response could be F-actin dependent, explaining the actin requirement for Cdc42Q61L clustering. Multiple clusters could develop as Cdc42Q61L overexpression leads to increasing numbers of wounds, and sub-lethal levels of Cdc42Q61L that do not induce wounds would not induce clustering. This plausible hypothesis explains the unexpected features of the Cdc42Q61L system. If it is correct, then actin-mediated mechanisms may have little relevance to normal polarity establishment. Thus, in the remainder of the review, we focus on what has been learned from studies using the rsr1Δ system, in which cells proliferate rather than lyse and F-actin is not required for symmetry breaking.
Autocatalytic clustering of GTP-Cdc42 at the cell cortex
Analysis of the genetic requirements for polarization in rsr1Δ cells led to the identification of the scaffold protein Bem1 as an essential factor for symmetry breaking [5]. Although Bem1 binds to many proteins, its role in breaking symmetry appears to involve linking just two of its binding partners: the guanine nucleotide exchange factor (GEF) that activates Cdc42, and a Cdc42 effector called p21-activated kinase (PAK) [8]**. An artificial GEF-PAK fusion protein was able to bypass the requirement for Bem1 in symmetry breaking, and similar GEF-PAK complexes have been discovered in other fungal and animal systems [9, 10], suggesting that this mechanism may be widely employed.
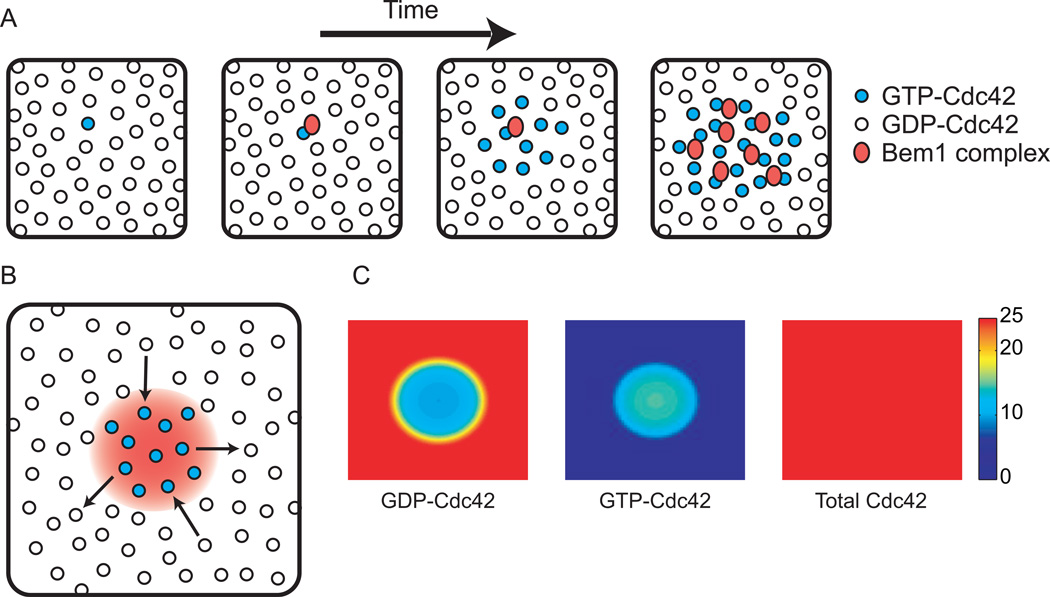
Based on these findings, Kozubowski et al. [8]** proposed a model in which the Bem1 complex mediates autocatalytic clustering of GTP-Cdc42 (Fig. 1A). In this model, the PAK effector domain allows stochastically-arising cortical GTP-Cdc42 to capture a cytoplasmically-diffusing Bem1 complex. The complex-associated GEF then activates neighboring GDP-Cdc42, leading to further Bem1 complex recruitment and Cdc42 activation in a positive feedback loop. Left unchecked, this mechanism would spread GTPCdc42 all over the plasma membrane. However, if Bem1 complexes are limiting, then spreading stops once the complexes are depleted from the cytoplasm. A dynamic polarized steady state arises in which diffusion of GTP-Cdc42 away from the cluster (and subsequent GTP hydrolysis) is balanced by diffusion of GDP-Cdc42 into the cluster (and subsequent activation by the locally-enriched Bem1 complexes)(Fig. 1B).
Figure 1. Autocatalytic clustering of GTP-Cdc42 by the Bem1 complex.
(A) A stochastically-activated GTP-Cdc42 on the membrane (viewed as en face snapshots) can recruit a Bem1 complex from the cytoplasm. The Bem1-associated GEF catalyzes conversion of neighboring GDP-Cdc42 into GTP-Cdc42, which can recruit more cytoplasmic Bem1 complexes, generating a positive feedback loop that “grows” a cluster of GTP-Cdc42. (B) Lateral diffusion of GTP-Cdc42 away from the cluster of Bem1 complexes (red gradient) followed by GTP hydrolysis, produces GDP-Cdc42. Conversely, lateral diffusion of GDP-Cdc42 into the cluster provides fresh substrate for the GEF, dynamically renewing the cluster. Because GTP-Cdc42 diffuses out at the same rate that GDP-Cdc42 diffuses in, the total (GTP- plus GDP-bound) Cdc42 concentration is uniform. (C) Simulation of the dynamic steady state illustrated in B. Color denotes concentration of the indicated species. See supplement for equations and methods.
This simple system resembles a number of theoretical reaction-diffusion models developed by mathematicians to explain the emergence of a spatial pattern from homogeneous starting conditions [1, 11, 12]. A key feature of such models is that diffusion of the polarity “activator” (here, GTP-Cdc42) must be slow compared to diffusion of the polarity “substrate” (here, the cytoplasmic Bem1 complex). Slow diffusion of Cdc42 (and bound Bem1 complexes) keeps the autocatalysis localized, and fast diffusion of free Bem1 complexes allows their rapid capture by the growing GTP-Cdc42 cluster, depriving distant locations of autocatalytic potential. Indeed, mathematical modeling shows that for well-chosen parameters (see supplementary data), the Bem1 complex can generate and sustain a cluster of GTP-Cdc42 in a sea of GDP-Cdc42 (Fig. 1C). In yeast cells, the difference between cytoplasmic and plasma-membrane protein mobility is particularly stark (cytoplasm, 4–11 µm2/s [13, 14]; membrane 0.0025–0.036 µm2/s [15, 16]: the unusually slow diffusion in the yeast plasma membrane may stem from its intimate apposition to the rigid cell wall). Thus, it is probable that a combination of local activation (due to slow membrane diffusion) and long-range inhibition (due to rapid cytosolic diffusion) underlies symmetry breaking.
Concentrating Cdc42: exploiting the difference in membrane/cytoplasmic diffusion
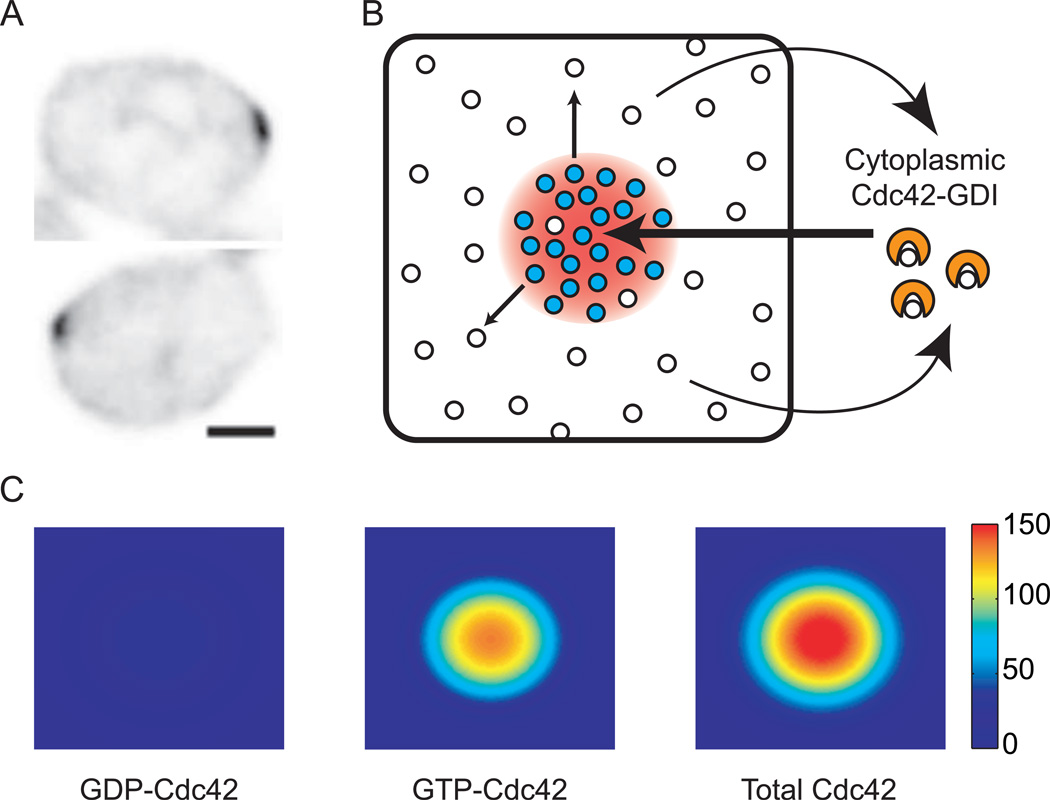
Even if the Bem1 complex promotes local clustering of GTP-Cdc42, it could not raise the local GTP-Cdc42 concentration above that of the surrounding GDP-Cdc42. Thus, the total (GDP + GTP) Cdc42 concentration would remain uniform at the cortex (Fig. 1C). However, immunofluorescence and GFP-tagging studies show that yeast cells do dramatically concentrate total Cdc42 at the polarization site (Fig. 2A) [17, 18]. How does this local enrichment take place?
Figure 2. Concentrating total Cdc42 by GDI-mediated mobilization of GDP-Cdc42.
(A) Inverted images of polarized GFP-Cdc42 in pre-budding yeast cells. Scale bar = 2 µm. Images taken from [28]**. (B) GDI extracts GDP-Cdc42 from the membrane, and rapid cytoplasmic diffusion promotes uniform re-deposition of GDP-Cdc42 all over the membrane. Conversion of GDP-Cdc42 to GTP-Cdc42 within the cluster depletes local GDP-Cdc42, so the rate of GDI-mediated extraction is lower inside the cluster than outside it. Thus, the GDI promotes a net "lux of GDP-Cdc42 from outside to inside the cluster (arrows), resulting in the accumulation of a higher concentration GTP-Cdc42 than the surrounding GDP-Cdc42. (C) Simulation of the dynamic steady state illustrated in B. Color denotes concentration of the indicated species. There is a small depletion of GDP-Cdc42 in the peak that is hard to see with this color bar. See supplement for equations and methods.
A possible answer to this question exploits another known Cdc42 regulator, the GDP-dissociation inhibitor (GDI). Cdc42 binds tightly to membranes because of a hydrophobic C-terminal geranylgeranyl modification [19]. Upon binding to Cdc42, a hydrophobic pocket in the GDI buries the geranylgeranyl group, thereby extracting Cdc42 into the cytoplasm. Goryachev and Pokhilko [20]** pointed out that if the GDI binds GDPCdc42 but not GTP-Cdc42 (an idea with considerable biochemical support [21]**), then the GDP-Cdc42 would be much more mobile than GTP-Cdc42, due to the rapid cytoplasmic diffusion of the Cdc42-GDI complex. As the Bem1 complex grows a cluster of GTP-Cdc42, it simultaneously depletes local GDP-Cdc42. Then, rapid influx of GDP-Cdc42 from cytoplasmic GDI complexes into the cluster outpaces the slow outward diffusion of GTPCdc42p along the membrane, resulting in local enrichment of GTP-Cdc42 to a level far above that of the surrounding GDP-Cdc42 (Fig. 2B).
Mathematical modeling demonstrated that in combination, the Bem1 complex and the GDI would lead to effective growth and maintenance of a concentrated patch of GTP-Cdc42 [20]** (Fig. 2C). The patch would be stable despite the rapid turnover of individual Cdc42 molecules as the GDI brings in fresh GDP-Cdc42 from the cytoplasm to replace the loss of GTP-Cdc42 by lateral diffusion. Experimental support for an important role of the GDI in promoting such Cdc42 dynamics came from a FRAP study showing that the rapid turnover of GFP-Cdc42 in polarized yeast cells (t1/2 4–5 s) was greatly reduced in cells lacking the GDI (t1/2 20 s) [22]**. By selectively speeding the diffusion of GDP-Cdc42, the GDI promotes formation and dynamic maintenance of a concentrated GTP-Cdc42 cluster.
In principle, the GDI mechanism suffices to concentrate Cdc42 at the polarization site. However, there is only one GDI recognizable by sequence homology in the yeast genome, and deletion of the gene is neither lethal [23] nor prohibitive of Cdc42 polarization [22]**[24]. Thus, there must be another way to concentrate Cdc42. Because a cytoplasmic pool of Cdc42 was still (unexpectedly) detected in cells lacking the known GDI [25, 26], yeast may harbor an unknown protein with GDI-like properties. However, a fundamentally distinct mechanism, discussed next, has also been proposed that would enable Cdc42 concentration even if it remained stuck to membranes [16].
Concentrating Cdc42: exploiting polarized vesicle trafficking
GTP-Cdc42 orients actin cables towards the polarization site. Myosin V delivers secretory vesicles along actin cables, leading to vesicle fusion with the plasma membrane at that site [27]. Secretory vesicles are likely to carry Cdc42 [4], raising the possibility that polarized vesicle delivery could lead to local enrichment of Cdc42 at the front.
A key feature of an operational vesicle-delivery model is that the concentration of Cdc42 on secretory vesicles must exceed that at the polarization site; otherwise, vesicle fusion would dilute Cdc42 at that site. Mathematical modeling recently showed that in order for vesicle trafficking of Cdc42 to contribute to, and not destroy, polarization, the Cdc42 must be actively trapped in forming endocytic vesicles [28]** (see Box 1). At present, we do not know whether endocytic trapping actually occurs for Cdc42.
The degree to which Cdc42 could be polarized by vesicle trafficking depends on the quantitative balance between the vesicle delivery rate and the diffusion of Cdc42 at the membrane. If diffusion is too fast (or vesicle delivery too slow) then the Cdc42-concentrating effect of each secretory vesicle fusion event would be largely dissipated by diffusion before the next vesicle arrives [28]**. With current estimates of the Cdc42 diffusion constant [16] and vesicle trafficking rate [28]**, the polarity generated by this mechanism is insufficient to account for the Cdc42 polarization observed in cells lacking the known GDI (Box 1). Thus, in addition to the proposed GDI and vesicle trafficking mechanisms, there is yet another Cdc42-concentrating mechanism in yeast that remains to be discovered.
Making one and only one front
Autocatalytic clustering and concentration of GTP-Cdc42 explains how a local stochastic fluctuation in GTP-Cdc42 concentration can develop a highly concentrated focus of Cdc42. But why does only a single focus of Cdc42 develop? Why wouldn’t autocatalytic amplification occur at many sites? One could imagine that the probability of initiating an autocatalytic loop is small, possibly because cells have a small absolute number of molecules of some key polarity factor [29]. In that case, it would be highly improbable for a cell to develop more than one cluster within a relevant timeframe. However, this mechanism carries the significant cost that a cell might have to wait a long time for even a single polarity cluster to develop.
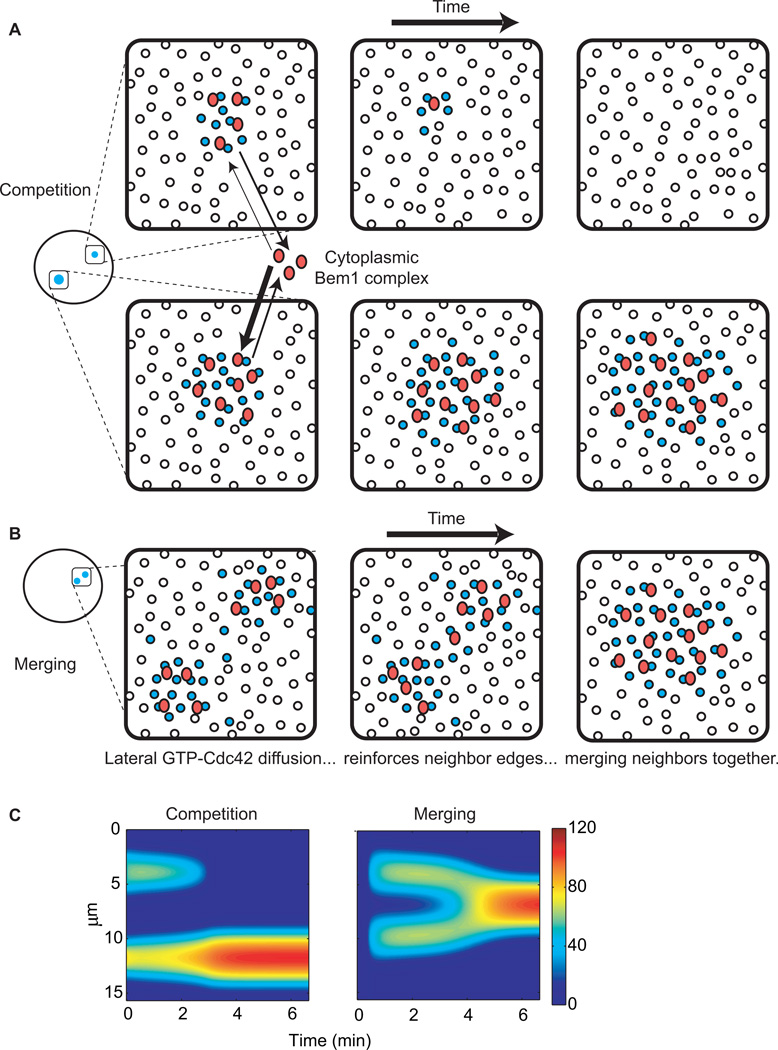
Alternatively, it could be that multiple clusters do in fact develop on a frequent basis, but there is some way to ensure that all but one cluster is eliminated. One way that this could occur is through competition: mathematical modeling of the Bem1 complex/GDI mechanism predicted that if two clusters formed, then as Bem1 complexes were depleted from the cytoplasm, the clusters would compete with each other for those complexes that were released back into the cytoplasm [20]**[30]**. Because larger clusters have more GTP-Cdc42, they would have more opportunities to capture free Bem1 complexes, so the largest cluster would grow at the expense of smaller clusters (Fig. 3A,C). Additionally, clusters near each other would tend to merge together, as diffusion of GTP-Cdc42 away from one cluster would reinforce the nearest edge of the neighboring cluster, biasing the autocatalytic process in that direction and “moving” the cluster centroids together (Fig. 3B,C). Over time, competition and merging would result in a single “winning” cluster, which would contain most of the Bem1 complexes, preventing new clusters from getting off the ground.
Figure 3. Competition between clusters and merging of nearby clusters leads to a single winning cluster.
(A) If two clusters of unequal size form at distant cortical locations, both will grow until the Bem1 complex is depleted from the cytoplasm. However, Bem1 complexes can return to the cytoplasm following dissociation from GTP-Cdc42. Rapid cytoplasmic diffusion allows each complex to randomly sample the cortex. Larger clusters have more GTP-Cdc42 available to “catch” such cytoplasmic Bem1 complexes, resulting in the gradual accumulation of more Bem1 complex (and GTP-Cdc42) at the expense of the smaller cluster, which would disappear. (B) If two clusters form near each other, outward diffusion raises the GTP-Cdc42 concentration between the clusters, which spatially biases recruitment of Bem1 complexes and GTP-Cdc42 activation. This bias would gradually shift each cluster’s centroid until both clusters merged into one. (C) Simulation of competition and merging. Color denotes concentration of GTP-Cdc42. Y axis indicates the perimeter of a cell, and X axis indicates time. See supplement for equations and methods.
The competition hypothesis predicts that polarity establishment would often proceed via a multi-cluster intermediate. Two clusters were observed in a small number of cells (6%), providing limited support for this hypothesis [30]**. Because cells were imaged at 1.5 min intervals, it remains possible that multi-cluster intermediates occurred at higher frequency, but that competition was so fast as to be totally resolved between timepoints, escaping detection. If that is the case, then it may be that cells develop one and only one front because nascent polarity clusters engage in a winner-takes-all competition.
Conclusions and open questions
Recent studies suggest that symmetry breaking in yeast relies on a simple positive feedback loop in which stochastically-arising cortical GTP-Cdc42 captures GEF-containing complexes from the cytoplasm, promoting GTP-loading of neighboring Cdc42 molecules to grow a GTP-Cdc42 cluster. Further concentration of GTP-Cdc42 at the polarization site occurs through the action of GDI as a GDP-Cdc42 mobilization factor, and perhaps by vesicular recycling of Cdc42. However, the single known GDI is non-essential for polarity establishment, and mathematical modeling predicts that the vesicular pathway would be ineffective given current estimates of vesicle trafficking rates and the Cdc42 diffusion constant. Thus, other Cdc42-concentrating mechanisms probably remain to be discovered.
A symmetry-breaking mechanism that relies on stochastically-arising GTP-Cdc42 should often lead to formation of multiple clusters, but yeast cells only ever make one bud. Co-existence of multiple clusters has been observed in cells undergoing polarity establishment, but only in a surprisingly small minority of cells. This may result from the technical challenges of detecting faint and fleeting intermediate states, or an unknown mechanism may restrict most cells to initiate only one Cdc42 cluster. We need faster and more sensitive movies of polarity establishment to differentiate between these possibilities.
When multiple clusters do form, competition between them assures that only a single cluster emerges victorious. Mathematical modeling of known biochemical reactions has recapitulated this competition (though not its apparent speed), providing some hope that the known processes may suffice to explain the establishment of one and only one front. However, it is worth noting that occasional cells appear to dismantle a perfectly good polarization site and relocate it elsewhere [30]**. Moreover, a remarkable Cdc42 mutant manages to polarize and survive (albeit only in a narrow range of growth conditions), despite the fact that the mutant fails to couple budding with the cell cycle and can grow several sequentially-emerging buds within one cycle [31]. This mutant does not require the normal Cdc42-directed GEF, and its mechanism of polarization remains mysterious.
Other open questions concern the mechanisms whereby relevant spatial stimuli, like bud site selection proteins and pheromone gradients, can accurately bias the symmetry-breaking positive feedback to occur at desirable locations. Moreover, it is clear that Cdc42 polarization does not always produce the same downstream consequence: pheromone-treated cells generate projections that do not resemble buds, and the basis for these differences in growth pattern remains to be determined.
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
The Supplementary information provides details, the mathematical model assumptions, equations, and parameters used to simulate Cdc42 distribution in the various Figures. See Supplementary information in the online version, at doi: xxxxxxx.
References
- 1.Turing A. The Chemical Basis of Morphogenesis. Philos Trans R Soc Lond B Biol Sci. 1952;237:37–72. doi: 10.1098/rstb.2014.0218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wedlich-Soldner R, Wai SC, Schmidt T, Li R. Robust cell polarity is a dynamic state established by coupling transport and GTPase signaling. The Journal of cell biology. 2004;166:889–900. doi: 10.1083/jcb.200405061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Park HO, Bi E. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol Mol Biol Rev. 2007;71:48–96. doi: 10.1128/MMBR.00028-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wedlich-Soldner R, Altschuler S, Wu L, Li R. Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science (New York, N.Y. 2003;299:1231–1235. doi: 10.1126/science.1080944. [DOI] [PubMed] [Google Scholar]
- 5.Irazoqui JE, Gladfelter AS, Lew DJ. Scaffold-mediated symmetry breaking by Cdc42p. Nature Cell Biology. 2003;5:1062–1070. doi: 10.1038/ncb1068. [DOI] [PubMed] [Google Scholar]
- 6.Gulli MP, Jaquenoud M, Shimada Y, Niederhauser G, Wiget P, Peter M. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 2000;6:1155–1167. doi: 10.1016/s1097-2765(00)00113-1. [DOI] [PubMed] [Google Scholar]
- 7.Tang HY, Cai M. The EH-domain-containing protein Pan1 is required for normal organization of the actin cytoskeleton in Saccharomyces cerevisiae. Molecular and Cellular biology. 1996;16:4897–4914. doi: 10.1128/mcb.16.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kozubowski L, Saito K, Johnson JM, Howell AS, Zyla TR, Lew DJ. Symmetry-Breaking Polarization Driven by a Cdc42p GEF-PAK Complex. Curr Biol. 2008;18:1719–1726. doi: 10.1016/j.cub.2008.09.060. Dissection of the role of Bem1 in symmetry-breaking. Point mutations that block PAK or GEF binding to Bem1 can be by-passed by fusing Bem1 to the respective binding partner, but only if both the PAK and the GEF can associate with a single Bem1 molecule. Suprisingly, a GEF-PAK fusion bypasses the requirement for Bem1 entirely, arguing that Bem1's role in symmetry-breaking is to link the GEF and a PAK into one complex.
- 9.Schiller MR, Chakrabarti K, King GF, Schiller NI, Eipper BA, Maciejewski MW. Regulation of RhoGEF activity by intramolecular and intermolecular SH3 domain interactions. The Journal of biological chemistry. 2006;281:18774–18786. doi: 10.1074/jbc.M512482200. [DOI] [PubMed] [Google Scholar]
- 10.Feng Q, Albeck JG, Cerione RA, Yang W. Regulation of the Cool/Pix proteins: key binding partners of the Cdc42/Rac targets, the p21-activated kinases. The Journal of biological chemistry. 2002;277:5644–5650. doi: 10.1074/jbc.M107704200. [DOI] [PubMed] [Google Scholar]
- 11.Gierer A, Meinhardt H. A theory of biological pattern formation. Kybernetik. 1972;12:30–39. doi: 10.1007/BF00289234. [DOI] [PubMed] [Google Scholar]
- 12.Jilkine A, Edelstein-Keshet L. A comparison of mathematical models for polarization of single eukaryotic cells in response to guided cues. PLoS computational biology. 2011;7:e1001121. doi: 10.1371/journal.pcbi.1001121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maeder CI, Hink MA, Kinkhabwala A, Mayr R, Bastiaens PI, Knop M. Spatial regulation of Fus3 MAP kinase activity through a reaction-diffusion mechanism in yeast pheromone signalling. Nature cell biology. 2007;9:1319–1326. doi: 10.1038/ncb1652. [DOI] [PubMed] [Google Scholar]
- 14.Slaughter BD, Schwartz JW, Li R. Mapping dynamic protein interactions in MAP kinase signaling using live-cell fluorescence fluctuation spectroscopy and imaging. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:20320–20325. doi: 10.1073/pnas.0710336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Valdez-Taubas J, Pelham HR. Slow diffusion of proteins in the yeast plasma membrane allows polarity to be maintained by endocytic cycling. Curr Biol. 2003;13:1636–1640. doi: 10.1016/j.cub.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 16.Marco E, Wedlich-Soldner R, Li R, Altschuler SJ, Wu LF. Endocytosis optimizes the dynamic localization of membrane proteins that regulate cortical polarity. Cell. 2007;129:411–422. doi: 10.1016/j.cell.2007.02.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziman M, Preuss D, Mulholland J, O'Brien JM, Botstein D, Johnson DI. Subcellular localization of Cdc42p, a Saccharomyces cerevisiae GTP- binding protein involved in the control of cell polarity. Molecular biology of the cell. 1993;4:1307–1316. doi: 10.1091/mbc.4.12.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Richman TJ, Sawyer MM, Johnson DI. Saccharomyces cerevisiae Cdc42p localizes to cellular membranes and clusters at sites of polarized growth. Eukaryot Cell. 2002;1:458–468. doi: 10.1128/EC.1.3.458-468.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Johnson DI. Cdc42: An essential Rho-type GTPase controlling eukaryotic cell polarity. Microbiol Mol Biol Rev. 1999;63:54–105. doi: 10.1128/mmbr.63.1.54-105.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Goryachev AB, Pokhilko AV. Dynamics of Cdc42 network embodies a Turing-type mechanism of yeast cell polarity. FEBS letters. 2008;582:1437–1443. doi: 10.1016/j.febslet.2008.03.029. Mathematical modeling of a system incorporating the known interactions and activities of Cdc42, the Bem1 complex, and the GDI in 8 reaction-diffusion equations demonstrates a Turing-like ability to polarize. The authors propose that competition between clusters explains the eventual dominance of one and only one cluster. This model differs from many previous models in taking a "bottom-up" approach and showing that polarity emerges from known biochemical reactions.
- 21. Johnson JL, Erickson JW, Cerione RA. New insights into how the Rho guanine nucleotide dissociation inhibitor regulates the interaction of Cdc42 with membranes. The Journal of biological chemistry. 2009;284:23860–23871. doi: 10.1074/jbc.M109.031815. Shows that GDI preferentially extracts GDP-bound Cdc42 from membranes, which was not appreciated from previous work in which GDI bound equally well to detergent-solubilized GDP-Cdc42 or GTP-Cdc42.
- 22. Slaughter BD, Das A, Schwartz JW, Rubinstein B, Li R. Dual modes of cdc42 recycling fine-tune polarized morphogenesis. Developmental cell. 2009;17:823–835. doi: 10.1016/j.devcel.2009.10.022. This paper presents a truly impressive collection of Cdc42 FRAP studies in various conditions. Perturbing the actin cytoskeleton mildly slows Cdc42 dynamics, while perturbing the GDI greatly slows Cdc42 dynamics. Even upon elimination of the known GDI and F-actin there is still a reasonably rapid Cdc42 exchange, but it cannot maintain a polarized Cdc42 distribution indefinitely. The data are fit to a mathematical model that assumes the GDI and actin provide parallel routes for Cdc42 traffic to the polarization site. However, aspects of this model were challenged in [28]**.
- 23.Masuda T, Tanaka K, Nonaka H, Yamochi W, Maeda A, Takai Y. Molecular cloning and characterization of yeast rho GDP dissociation inhibitor. The Journal of biological chemistry. 1994;269:19713–19718. [PubMed] [Google Scholar]
- 24.Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nature cell biology. 2010;12:477–483. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koch G, Tanaka K, Masuda T, Yamochi W, Nonaka H, Takai Y. Association of the Rho family small GTP-binding proteins with Rho GDP dissociation inhibitor (Rho GDI) in Saccharomyces cerevisiae. Oncogene. 1997;15:417–422. doi: 10.1038/sj.onc.1201194. [DOI] [PubMed] [Google Scholar]
- 26.Tiedje C, Sakwa I, Just U, Hofken T. The Rho GDI Rdi1 Regulates Rho GTPases by Distinct Mechanisms. Molecular biology of the cell. 2008;19:2885–2896. doi: 10.1091/mbc.E07-11-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pruyne D, Bretscher A. Polarization of cell growth in yeast. J. Cell Sci. 2000;113:571–585. doi: 10.1242/jcs.113.4.571. [DOI] [PubMed] [Google Scholar]
- 28. Layton AT, Savage NS, Howell AS, Carroll SY, Drubin DG, Lew DJ. Modeling vesicle traffic reveals unexpected consequences for cdc42p-mediated polarity establishment. Curr Biol. 2011;21:184–194. doi: 10.1016/j.cub.2011.01.012. The first mathematical model that explicitly incorporates membrane traffic by exocytosis and endocytosis during Cdc42 polarization. Inclusion of membrane traffic changes the model's behavior and shows that (applying other previously used assumptions) polarity is no longer sustainable. In contrast, actively endocytosed transmembrane proteins can become polarized by directed vesicle traffic. Thus, either Cdc42 traffic unexpectedly resembles that of such membrane proteins, or directed vesicle traffic is not an effective mechanism to polarize Cdc42.
- 29.Altschuler SJ, Angenent SB, Wang Yand, Wu LF. On the spontaneous emergence of cell polarity. Nature. 2008;454:886–889. doi: 10.1038/nature07119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Howell AS, Savage NS, Johnson SA, Bose I, Wagner AW, Zyla TR, Nijhout HF, Reed MC, Goryachev AB, Lew DJ. Singularity in polarization: rewiring yeast cells to make two buds. Cell. 2009;139:731–743. doi: 10.1016/j.cell.2009.10.024. Yeast cells are artificially re-wired to use an actin-dependent feedback loop rather than the Bem1-mediated reaction-diffusion mechanism to polarize Cdc42. "Rewired" cells often formed two Cdc42 clusters, which competed slowly. Cluster pairs that did not resolve by the time of bud emergence resulted in simultaneous growth of two buds. This paper provides strong evidence that singularity (one-and-only-one front) is an intrinsic property of the polarization positive feedback mechanism.
- 31.Caviston JP, Tcheperegine SE, Bi E. Singularity in budding: a role for the evolutionarily conserved small GTPase Cdc42p. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12185–12190. doi: 10.1073/pnas.182370299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.