Abstract
Abstract
Homeostatic processes that regulate electrical activity in neurones are now an established aspect of physiology and rest on a large body of experimental evidence that points to roles in development, learning and memory, and disease. However, the concepts underlying homeostasis are too often summarized in ways that restrict their explanatory power and obviate important subtleties. Here, we present a review of the underlying theory of homeostasis – control theory – in an attempt to reconcile some existing conceptual problems in the context of neuronal physiology. In addition to clarifying the underlying theory, this review highlights the remaining challenges posed when analysing homeostatic phenomena that underlie the regulation of neuronal excitability. Moreover, we suggest approaches for future experimental and computational work that will further our understanding of neuronal homeostasis and the fundamental neurophysiological functions it serves.
Timothy O'Leary is a postdoctoral researcher at the University of Edinburgh, where he received his PhD in 2009, from the School of Informatics. He currently works as an electrophysiologist in the Centre for Integrative Physiology, where he carried out experimental work on neuronal homeostasis. His background is in mathematics, physical sciences and computing and he has moved toward neuroscience, particularly biophysics and neuronal physiology. His wider interests are in systems approaches to understanding fundamental biological processes (such as homeostatic regulation) and methods for acquiring and analysing experimental data. David Wyllie is Professor of Ion Channel Physiology and Pharmacology in the Centre for Integrative Physiology at The University of Edinburgh. His long-standing research interests in the behaviour of single ion channels began during his PhD in the Department of Pharmacology at UCL and extended to studies of synaptic plasticity when a post-doctoral researcher at UCSF. After moving back to the UK, first to UCL and subsequently to Edinburgh his research has focussed on structure–function studies of NMDA receptors and how subtype-dependent biophysical and pharmacological properties sub-serve physiological and pathophysiological functions and investigation of synaptic function, in particular, in models of CNS developmental dysfunction and neurodegeneration.
 |
Introduction
Our central nervous system comprises billions of neurones, all of which are excitable and each of which is coupled to potentially thousands of neighbours. Individually, neurones are exquisitely dynamic, capable of responding to chemical and electrical signals over multiple timescales and modifying their properties and ongoing activity in ways that collectively give rise to our every thought and action. In spite of the inherent complexity of nervous systems, we already understand some of the fundamental principles that govern the patterns of connectivity and signalling activity in neurones. Activity-dependent synaptic plasticity serves as a mechanism for at least two (not necessarily distinct) processes: storing memories and establishing functional networks during development (Buonomano & Merzenich, 1998; Martin et al. 2000). It is also known that the intrinsic properties of cells – properties that dictate how a cell integrates and responds to electrical signals – are themselves capable of being modified in response to particular patterns of activity in ways that facilitate learning and development (Daoudal & Debanne, 2003; Disterhoft & Oh, 2006). Both of these modification processes, synaptic and intrinsic, therefore shape activity and are simultaneously shaped by activity.
Described in this way, it is remarkable that the CNS functions in a stable manner at all: small perturbations in activity can conceivably alter the functional properties of an entire network of cells, which can in turn lead to further modifications elsewhere. What system is in place to keep these modification processes in check? There can be no central, global control mechanism monitoring and adjusting the properties of each individual cell in a coordinated manner. Instead, global control is observed as an emergent feature of the nervous system, arising from the combined effects of a hierarchy of regulatory mechanisms operating on the level of cellular networks, individual cells, subcellular domains and, ultimately, individual genes and proteins.
Homeostatic processes that regulate levels of electrical activity in neurones are currently the subject of intense investigation. Not only do these phenomena contribute to the development and ongoing function of neuronal networks, they also stand as important determinants of the nervous system's response to physiological insult and disease (Turrigiano & Nelson, 2004; Davis, 2006; Marder & Goaillard, 2006; Turrigiano, 2007). Classically, activity-regulating mechanisms are split into two categories: those that exert their effect by modifying synapses and those that modify intrinsic membrane properties (Desai, 2003). Although these categories can be distinguished by their underlying mechanisms – prototypical examples include glutamate receptor insertion and removal in the synaptic case (Turrigiano et al. 1998; but see Echegoyen et al. 2007) and the modification of voltage-gated ion channel density in the intrinsic case (Desai et al. 1999) – it is important to point out that both categories overlap to a degree, and must interact in the system as a whole. Overlap is evident both in terms of the underlying signalling pathways and in terms of their net effect on physiological properties. For example, intracellular calcium concentration has been identified experimentally as an important signal for both synaptic and intrinsic homeostasis (Turrigiano et al. 1994; Yeung et al. 2004; Thiagarajan et al. 2005; O'Leary et al. 2010) and synaptic properties have a substantial impact on intrinsic excitability as a result of ongoing synaptic activity (Chance et al. 2002).
The diversity seen among the mechanisms of neuronal homeostasis is also a characteristic of their effects. We know, for example, that rhythm-generating circuits in crustaceans achieve robust output by modifying the membrane conductance properties of their constituent cells in response to pharmacological manipulation and physical circuit lesion (Turrigiano et al. 1994) and that mammalian central neurones modify their intrinsic membrane properties in a way that counters the effects of imposed changes in excitatory drive (Desai et al. 1999; Brickley et al. 2001; van Welie et al. 2006; Maffei & Turrigiano, 2008; O'Leary et al. 2010). More subtle functional roles for homeostasis of neuronal activity have also been identified, for example, in maintaining the fidelity of long-term memories in networks with cell turnover (Meltzer et al. 2005) and in enhancing the learning performance of canonical synaptic plasticity rules, such as Hebbian learning (Triesch, 2007).
This review focuses on how intrinsic properties of neurones are regulated via homeostatic mechanisms, especially those which regulate membrane conductances. Far from being a simple story, experimental evidence and theory suggest that neurones face and solve an astonishingly complex problem in regulating their electrical properties, and a significant part of how this regulation is achieved remains to be understood. A comprehensive understanding of homeostasis requires a rigorous and general theory. Therefore, a large part of this review is devoted to a critique of the established theories, from which we will introduce the concepts of control theory– a theoretical discipline that was originally developed to describe general homeostatic phenomena, and which is of particular use in understanding neuronal homeostasis (Davis, 2006). We will focus on principles, with the dual aim of illustrating the subtleties of homeostatic control whilst highlighting the complexity of several outstanding questions.
The need for control
The regulation of a neurone's intrinsic properties in part depends on regulating the expression of proteins, such as those that form ion channel subunits. It therefore makes sense to consider how cells regulate protein expression in general before discussing homeostatic processes that control neuronal excitability per se. It is worth pointing out that what follows is a necessarily simplified account in terms of level of biological detail, with the aim of emphasising concepts. Furthermore, intrinsic properties are affected by factors other than ion channel expression – for example, post-translation modification of membrane channels and their interactions with other proteins and ligands – any of which can cause dramatic changes in the biophysical properties of a neurone without necessarily altering its ion channel expression profile (Levitan, 1994 2006; Davis et al. 2001). However, arguments that apply to channel density or expression level apply to other factors so there is nothing to be lost conceptually by restricting our discussion.
We know that the level of expression of a given protein in a cell is controlled – levels do not escalate arbitrarily or disappear at random under non-pathological conditions (Kozak, 1992). In other words, cells have a ‘target’ expression level that is achieved and maintained. Even within a particular cell type, these targets are not fixed: the developmental context will typically determine whether more or less of a particular protein is needed at any given time. For example, the expression of ion pumps such as chloride–potassium co-transporters is triggered at a particular developmental stage (Rivera et al. 1999). Thus, homeostatic systems do indeed track a target, but in general this can be a moving target with heterogeneity in its profile over any population of cells.
The goal of imposing target expression profiles cannot be achieved by merely turning a gene ‘on’ or ‘off’ in a given cell. Existing proteins are constantly being modified, trafficked and degraded; therefore, basal production levels must balance such dissipative processes if stable expression levels are to be maintained. Moreover, biochemical signalling, and gene expression in particular, is stochastic, and therefore noisy. Noise leads to substantial fluctuations in protein expression, as demonstrated by direct measurements in single-cell organisms (Elowitz et al. 2002).
Given these constraints, it is clear that the rates of the processes that govern gene expression need to be modified by some control mechanism that is responsive to changes in demand. In other words, regulatory mechanisms must incorporate a feedback signal even in the absence of the external perturbations that are the focus of most theories and experiments in the field of neuronal homeostasis. In this sense it is evident that many low-level cellular processes must be homeostatic.
The problem of low-level gene regulation has prompted a great deal of experimental and theoretical work in systems biology (Barabasi & Oltvai, 2004). Importantly, it is now known that the genetic regulatory pathways of many individual gene products comprise feedback and feedforward control loops. These loops operate on the level of transcription and translation, and evidence for their existence has been found in all species that have been thus far examined, from prokaryotes to eukaryotes, including humans (Lee et al. 2002; Shen-Orr et al. 2002; Tsang et al. 2007).
All of these findings, which are relevant to generic cellular processes in all organisms and cell types, have important implications for homeostasis in the nervous system. For example, it is possible that certain types of homeostatic compensation that occur in neurones are merely a consequence of the collective, low-level regulation of the cell's basic components, and not something endowed upon neurones due to their role in information processing per se. Indeed, a recent study provides an example of low-level compensation of intrinsic properties that is achieved via post-transcriptional interference of channel-coding genes, rather than being coupled to electrical activity (MacLean et al. 2003). Nonetheless, it is also possible and even likely that neurones have evolved specialized control machinery, particularly where ion channel regulation is concerned. Firstly, their role in information processing means that neurones must regulate their biophysical properties so as to satisfy specific constraints. Secondly, due to their connectivity, neuronal activity is tightly coupled over a multicellular network, so regulatory adjustments within individual cells have knock-on effects that impinge on the organism as a whole (Marder et al. 2010).
Together, these observations suggest that homeostatic mechanisms fit into a hierarchy which operates on the levels of single proteins, protein networks, whole cells, cellular networks, organs and ultimately entire organisms. The question of how mechanisms interact across hierarchical levels is of central importance in neuronal homeostasis. We will return to this problem after defining homeostasis in its most general form on a single level.
Static or homeostatic?
What do we mean by ‘homeostatic regulation’? Definitions differ, but a common theme is that of a system returning to a ‘set-point’, ‘target value’ or ‘previous state’ following some perturbation (Turrigiano, 1999; Turrigiano & Nelson, 2004; Davis, 2006; O'Donnell & Nolan, 2011). Such a definition can have an overly restrictive interpretation: in the most extreme form it could be construed to mean that nothing really changes in homeostatic systems! For example, as O'Donnell & Nolan (2011) have recently pointed out, naïve homeostatic models are hard to reconcile with observed heterogeneities in intrinsic properties of cells over a defined brain region. In principle, this is at odds with one of the seminal computational studies of homeostasis (Abbott & LeMasson, 1993) which identified emergent heterogeneities in the homeostatically regulated conductance profiles of coupled neurones, in spite of their target activity levels being identical. Experimentalists and theoreticians are, of course, acutely aware of the complexity of biological systems, so there is sometimes a need to describe phenomena such as homeostasis in a simplified manner for practical reasons. There is, however, a danger that important subtleties may get lost in the general discussion. This may result in apparent contradictions or the emergence of superfluous concepts such as ‘allostasis’ (‘stability through change’) – a term necessitated by the mistaken view that homeostatic processes imply static systems. This idea is comprehensively critiqued in Day (2005).
Some authors have attempted to solve the problem of finding an adequate yet simple conceptual model of neuronal homeostasis by restricting the range of phenomena that homeostasis applies to (Davis, 2006), or by embellishing a static theory with the notion of ‘customizable set points’ (Turrigiano, 2008). We do not believe the underlying theory requires restriction or embellishment for it to be compatible with physiology, however. In order to motivate a discussion of this theory we have tried to identify three potential sources of its oversimplification:
Targets. The stipulation that the target output of a homeostatic system is fixed in time or in space is unnecessary and restrictive.
Loci. The variable(s) being manipulated and the variable(s) being monitored in a homeostatic system are distinct in general and can be a subset of the total set of available variables.
Hierarchies and emergent behaviour. Homeostatic systems interact in general and individually they can be broken down into underlying regulatory processes that give rise to the apparent regulation of higher-level, collective processes.
If the results of theoretical and experimental studies are to be interpreted in a coherent theory, we need a way to compare varying definitions of homeostasis and the physiological processes they apply to. Fortunately, a rigorous theory of homeostatic control already exists in the wider field of cybernetics, which was originally defined as the study of “control and communication in the animal and the machine” (Wiener, 1965). It is interesting to note that despite having its origins in physiology, cybernetics, and its daughter-field, control theory, together comprise a wealth of valuable theory that is rarely mentioned in contemporary experimental physiology literature.
Control theory: the mother of homeostasis
A typical model of neuronal homeostasis is depicted in Fig. 1A: a neurone has a target activity level which may be perturbed by changes in external excitatory drive. In this example, the cell compensates for transient increases in excitatory input by changing the surface expression of hyperpolarising channels and leak channels. Importantly, the same cell has a target activity level which varies over a period of time, for example, as the cell matures. This entails distinct compensation strategies: the immature cell might favour up-regulation of leak channels, whilst the mature cell selectively up-regulates hyperpolarising channels. The signal that causes changes in channel expression in this example is calcium concentration, which varies as the result of voltage-gated calcium channels. Thus, the variable being controlled (‘average activity’) is distinct from the signal being monitored (‘calcium concentration’). Figure 1B illustrates homeostatic regulation in another general setting. Many cell properties can undergo plastic changes in their properties (such as overall excitability) in response to some signal, which may be a particular activity pattern or the action of a signalling molecule. The archetypal examples are stable potentiation or depression of a property; this stability can be conferred by homeostatic regulation, whilst the net change in the property corresponds to a change in the homeostatic target.
Figure 1. A typical model of neuronal homeostasis in different contexts.

A, a timeline spanning the lifetime of a neurone is depicted, along with a target activity profile which decreases as the cell matures. At both immature and ‘mature’ stages, the cell is capable of homestatically compensating for transient increases in external excitatory input which tend to increase its activity above the target level. Internal calcium concentration, which is often found to be a key ‘error signal’, is seen to correlate with changes in excitatory drive, but on a slower timescale that reflects an accumulated, or integrated, deviation from the target activity level. In this example, compensation is achieved by regulating the expression of different types of membrane conductances in response to changes in the error signal. The strategies for achieving compensation differ between the immature and mature stages (as illustrated by the different ratios of membrane channels), so as to achieve a different target output at the same time as compensating for the same change in external input. B, in a more general context, homeostatic regulation can be reconciled with stable plastic changes (such as those induced by long-term potentiation or depression) by viewing ‘stable changes’ as changes in the homeostatic target.
These hypothetical yet biologically plausible models qualitatively describe many observed instances of neuronal homeostasis whilst exemplifying some of the subtle points we have already mentioned, including the notion that targets need not be fixed, and that signals that are compared to this target (e.g. calcium concentration) can be distinct from the variable being controlled (spiking activity). Moreover, they illustrate some of the key components of an abstract control system, which we will introduce next.
Figure 2A shows a diagram of the canonical feedback control system. The process being controlled (the plant in engineering terminology) merely transforms inputs into outputs. A familiar example of a plant is a single neurone which transforms inputs in the form of synaptic currents into a temporal pattern of neurotransmitter release at its axon terminals. However, in general, the identity of inputs and outputs may be less obvious. For example, in the context of protein regulation, the plant can be regarded as the cell, but the inputs might be intracellular signals that activate transcription factors, whilst the outputs could be the expression level of the protein being regulated, or even the state of other physiological variables such as membrane potential which may be influenced by the protein in question.
Figure 2. Canonical control framework.
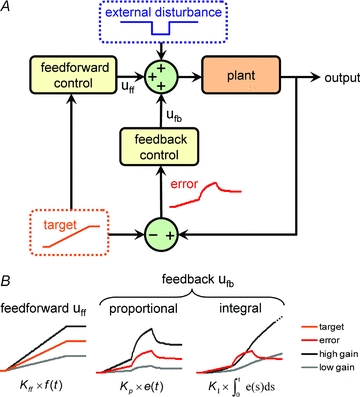
A, uff and ufb represent the feedforward and feedback control signals, respectively; the plant is the process being controlled. Example signals for the target output, the error and the external disturbance are shown. The error is the difference between the output and the target. B, example control signals based on a target signal, f(t), and an error signal, e(t). Kff, KP and KI are the gains for the (proportional) feedforward, proportional feedback and integral feedback control strategies, respectively.
Control systems are faced with the goal of driving the plant's output toward a target output profile by delivering a control signal to the plant's inputs. The inputs may also be subjected to external perturbations. A good example of a ‘target’ might be the characteristic changes in biophysical properties of neurones that occur during cortical development (Ben-Ari & Spitzer, 2010), whilst ‘external disturbances’ in network activity occur during, say, sensory deprivation (Maffei & Turrigiano, 2008). Two generic control paths exist: feedforward and feedback, so-called because the former feeds a control signal to the plant inputs but is blind to the outputs, whilst the latter reads plant outputs and feeds a control signal back to the input. There are numerous options for control strategies with important distinctions that are relevant to their biological plausibility. In order to qualitatively illustrate the behaviour of a generic control system, we have constructed a specific model of a plant with a time-varying target output and step-disturbance (Fig. 2B, Fig. 3); the mathematical details of this example are included in the Appendix.
Figure 3. Example behaviour of a simple linear control system.
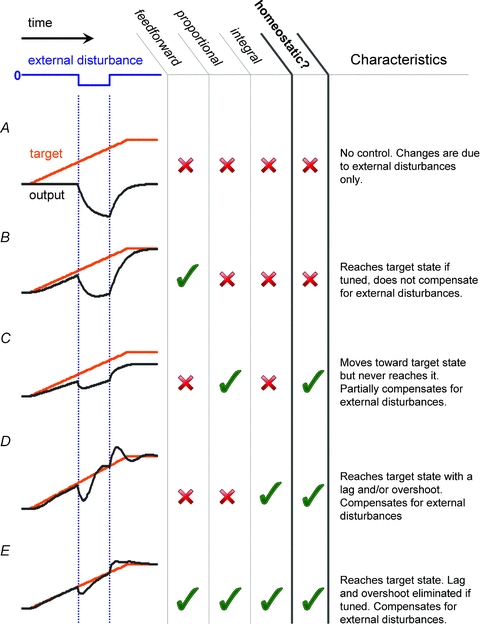
Behaviour of typical linear control systems: A, system with no feedforward or feedback control showing a perturbation in output caused by the external disturbance alone; B, proportional feedforward control; C and D, proportional and integral feedback control; E, combination of all three linear feedforward–feedback controls. The plant is modelled as a linear, first-order system (see Appendix) that exhibits exponential decay of its output to zero in the absence of input. Such a model might be used to describe how the expression level of a protein in a cell is subject to a constitutive degradation processes.
A subset of the plant outputs is accessible to the feedback control process; this subset (which may not encompass the entire set of outputs) is called the set of observables. In our example, observables could be resting membrane potential itself, or some other variable which depends on resting potential such as spiking rate or intracellular calcium concentration. Clearly, observables need to be measurable by biological sensor mechanisms that are known to exist in the cell. Intracellular calcium concentration is often favoured because many calcium-dependent signalling pathways are present in neurones, and calcium buffers are known to interfere with homeostatic compensation in a variety of experimental settings (Turrigiano et al. 1994; Berridge et al. 2003; Davis, 2006).
Observables may be transformed by the feedback controller (time-averaged or amplified, for example) before being compared against a target. The result of this comparison is an error signal, representing the difference between the target state and the observed state (Fig. 2B). This error signal forms the basis of the feedback control signal, which is delivered back to the plant as an input (Fig. 2A). If the variable of interest is resting membrane potential, then the error signal may be coded as intracellular calcium concentration, which varies with membrane potential due to voltage-dependent calcium channels. This example also illustrates a component of time-averaging (integration), since the dynamics of calcium fluctuations are filtered versions of the fluctuations in membrane potential due to calcium channel kinetics, buffering and secondary release systems such as internal stores (Berridge et al. 2003).
Feedback signals can be derived from the error signal in many ways; we will introduce several well-known feedback control paradigms. Proportional control, or P-control, generates a signal that is equal to a scaled version of the error signal (Fig. 2B and Fig. 3). Returning to the example in Fig. 1, the expression level of the various channels at any given time would be proportional to the difference in activity level from the target value at that time. Proportional controllers produce a homeostatic response when the feedback is negative, but suffer from the problem that they do not ever achieve their target value, nor do they provide full homeostatic compensation: they exhibit a constant error, regardless of how large the amplification factor (or gain) is set (Bakshi & Bakshi, 2009). Voltage-gated conductances can be thought of individually as proportional controllers. For example, the hyperpolarization-activated mixed-cation current (Ih) is activated when the membrane potential becomes very negative (<−70 mV) and has a reversal potential of about –30 mV. Thus, Ih tends to drive the membrane potential back to a more positive value when it is hyperpolarised. The ‘set point’ is therefore the reversal potential of the conductance, but the channels themselves can never drive the membrane potential to this value because the channels inactivate and the current they pass tends to zero as the reversal potential is approached.
One way of eliminating the steady error that P-controllers introduce is to make the control signal proportional to the total error accumulated over time, that is, the integral of the error with respect to time. Controllers that use this policy are therefore called integral controllers (I-controllers, Fig. 3D). Owing to the fact that the control signal at any given time depends on the error history, I-controllers are prone to suffering from overshoot and even ringing (successive overshoots followed by undershoots). Nevertheless, they reach their target state eventually and the degree of ringing can be traded off against the speed of response by lowering their gain. An important recent finding relating to bacterial chemotaxis establishes the necessity of integral control in biochemical signalling under a broad set of assumptions (Yi et al. 2000) so it is entirely possible that more complex cells such as neurones possess integral control mechanisms that participate in activity-dependent homeostasis. Indeed, there are many examples of modelling studies that employ integral controllers to explore this possibility (van Rossum et al. 2000; Buonomano, 2005; Zhang & Golowasch, 2007). For many authors, the key issue regarding their plausibility lies in finding a signalling network that implements integration over a suitable timescale for homeostasis – which is normally assumed to be a slow process requiring reliable integration over long timescales (Turrigiano & Nelson, 2000; Desai, 2003; Davis, 2006; Turrigiano, 2007). Several modelling studies have successfully implemented calcium-dependent control of membrane conductances, either by explicitly integrating the calcium concentration over time and varying conductances in proportion to this signal (Gunay & Prinz, 2010; Olypher & Prinz, 2010), or by varying the rate of change of conductance density in proportion to the instantaneous calcium concentration, thereby implicitly integrating the calcium signal (Abbott & LeMasson, 1993; LeMasson et al. 1993). The latter approach offers an attractive solution to the question of plausibility, since its biological interpretation does not require an additional ‘calcium-integrating mechanism’; rather, the integration is performed by a model of the dynamics of channel expression, so a ‘long’ integration time simply corresponds to a ‘slow’ rate of change of channel density.
Proportional and integral control policies are just two examples of linear controllers. That is, their output for two different inputs can be summed to give the output when the two inputs are combined. There is a much wider class of controllers that are non-linear, but nevertheless quite simple to implement and plausible in biological systems, which are highly non-linear in general. Perhaps the most familiar example of a non-linear controller (and one that can be found in most domestic heating systems) is the bang-bang controller. As its name suggests, the bang-bang controller simply switches a control signal ‘on’ (bang!) once the error signal is detectable, then abruptly turns it ‘off’ (bang!) once the error reaches zero (Fig. 4). Although this evidently entails a series of overshoots and undershoots, the controller has the key advantages of being exceptionally simple to implement (the control signal merely needs to be turned on or off) and, perhaps surprisingly, an optimal control under certain conditions where the speed of response is important (Lasalle, 1959). Indeed, if the amount of over/under-shooting is within physiologically tolerable limits, bang-bang controllers provide a way of reaching target values and compensating fully for disturbances (on average) without needing to integrate errors over long timescales.
Figure 4. Bang-bang control and noise.
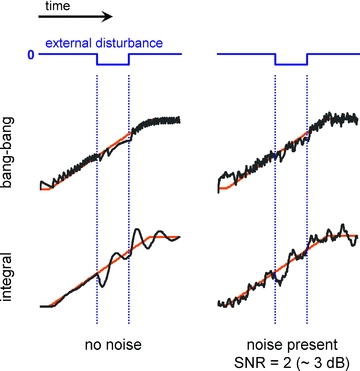
Left, a control system (with plant as in Fig. 3) employing a non-linear bang-bang control (upper plots, details in the Appendix). For comparison, the output of the system employing the integral controller is shown below. Right, the output of these particular bang-bang and integral control systems are similar in the presence of noise.
What might constitute a biological example of a ‘bang-bang’ control? Many physiological processes, such as the activation of a biochemical pathway or the firing of an action potential, exhibit threshold behaviour: beneath threshold, nothing happens, but once the threshold is reached, the chain of events which follows is entirely stereotyped and does not depend on the amount by which the threshold is passed. Bang-bang controllers are characterised by this kind of threshold behaviour and can be used to describe various forms of regulatory control in biology, including the influence of feedback inhibitory interneurons on their target cells and the ‘on–off’ regulation of many genes.
We have not yet confronted the question of what makes the feedback ‘positive’ or ‘negative’, which is an important distinction because homeostatic regulation requires negative feedback. Returning to the example in Fig. 1, negative feedback would down-regulate hyperpolarising channels if the membrane potential were more negative than its target value, and up-regulate them if the membrane potential were positive with respect to the target value. Thus, the sign of a feedback control (whether it is positive or negative) depends on the dynamics of the plant. Interestingly, this means the control mechanism must ‘know’ what effect the regulation of a given variable has on the observables. This built-in ‘knowledge’ of how the plant behaves is an aspect of the internal model principle of control theory – a striking and deep result which states that any effective control system must have an internal representation of the system it is controlling (Francis & Wonham, 1976). How is such knowledge embedded? This is an interesting question; in the case of low-level regulation in individual cells, tuning of the regulatory response probably occurs on an evolutionary timescale. Specifically, the properties of particular gene products (such as the biophysical characteristics of voltage-gated ion channels) and the properties of regulatory signalling networks (the rate constants, interaction sites and identity of the molecules involved) emerge through the trial-and-error nature of evolution as components of a functioning control system. This does, however, introduce a ‘chicken and egg’ dilemma: cellular components perform tasks that are often vital for the survival of an organism, therefore their expression in a cell needs to be regulated – how can appropriate regulatory machinery evolve in anticipation of such components appearing? As with all such questions, the answer presumably comes from considering the simplest, most fundamental forms of regulatory control as starting points for the gradual introduction of additional layers of complexity. These simple control mechanisms might, for example, describe processes that are rate-limited or physically constrained within particular bounds by their nature.
So far, we have focussed on feedback control and ignored the feedforward path. Like feedback control, feedforward control is capable of driving a system toward a target state, but unlike feedback control, it is insensitive to error in the output and therefore cannot underlie homeostasis in isolation. Feedforward controls ‘see’ the target signal and provide an appropriately transformed version of this signal (simple scaling is an example of such a transformation, as illustrated in Fig. 2B) to the plant. Since the feedforward control signal does not depend on measurements of the plant output, delays that are inherent in feedback control strategies can be avoided through their use (this can be seen by comparing the outputs in Fig. 3C and D with that of Fig. 3E). However, in the same way that feedback controllers can be optimised by tuning their parameters according to the dynamics of the plant, feedforward controllers need to ‘know’ how to scale the control signal to avoid overshoot or undershoot. In a neurophysiological context, feedforward controllers could represent processes that lead to stereotyped changes in the functional behaviour of a cell, such changes in intrinsic excitability caused by the action of neuromodulators (see Davis, 2006 for similar examples). More generally, feedforward control has been identified as a fundamental component of gene regulation using model organisms such as yeast (Lee et al. 2002; Mangan & Alon, 2003), whilst at a somewhat higher ‘system’ level, it is an established component of motor control and is integral to the theory of motor learning in humans (Wolpert & Ghahramani, 2000).
One of the motivations we gave for the existence of homeostatic regulation was the presence of noise in biochemical signalling networks. Control mechanisms counteract the perturbations introduced by external disturbances and therefore behave as noise-attenuating filters when the source of the noise is extrinsic (Rao et al. 2002). However, the performance of all models of homeostatic control is also subject to the level of noise present in the control mechanism itself. The abstract ‘sensors’ and ‘control signals’ we have been discussing are ultimately encoded in biochemical signalling networks, which have their own intrinsic noise. Under conditions of high noise, it is possible that the responses due to distinct underlying mechanisms (employing, for example, integral vs. bang-bang control) are effectively indistinguishable in their performance, as illustrated in Fig. 4. As a result, biology may favour simpler control mechanisms than currently anticipated (Yi et al. 2000; Turrigiano, 2008). This has important ramifications for how experimental data are interpreted because several distinct control strategies may be compatible with observed behaviour. Therefore, an important line of investigation – and one which has progressed recently in systems biology (Pedraza & van Oudenaarden, 2005; Rosenfeld et al. 2005) – consists of assessing the levels of noise that regulatory signals are subjected to.
It is hopefully clear why the control-theoretic framework is valuable for discussing neuronal homeostasis (and, indeed, all forms of homeostasis). Most obviously, it provides a consistent way of describing a model: the target, error signal and plant output must be identified explicitly, and the form of the controller can be rigorously categorized. In addition, it reconciles the potential confusion between stable, long-term changes in a property (which correspond to changes in a target) and ongoing changes in the property resulting from a homeostatic response to perturbations (Fig. 1B). This distinction can be lost due to the experimental design: most experiments will take measurements in a regulated property of a ‘treated’ (perturbed) group with respect to a control group. If ongoing plastic changes are present in both, the changes in targets will be invisible since they are subtracted away to reveal the homeostatic component. Potential conceptual problems in interpreting such data can be avoided using the control theory perspective. For example, rather that viewing Hebbian and homeostatic processes as exerting ‘opposing’ effects neuronal properties (e.g. Turrigiano & Nelson, 2000), they can be reconciled as distinct features of the same control system.
In tandem with clarifying homeostasis in general, the control theory perspective also enables an appreciation of the complexity of neuronal homeostasis in particular. Firstly, even if individual control systems can be characterized – such as those that regulate a given conductance in a given cell type – they will interact in the system as a whole. This is indeed observed in recent, elegant work which probes intrinsic, synaptic and circuit-level interactions between regulatory processes in the crab stomatogastric ganglion (Grashow et al. 2010) and at the Drosophila neuromuscular junction (Bergquist et al. 2010). Secondly, the control strategies mentioned so far (and the infinitude of alternative strategies we have omitted) can all be combined together in any individual control system. For example, combinations of feedforward and feedback control can deliver improved performance over the use of one single strategy alone (Kuo, 2002). However, the complexity of these mechanisms is dependent on the sophistication of the biochemical signalling machinery available and on the level of intrinsic noise in the system. These final points are pertinent to issues raised in the next section, where we consider how homeostatic control might exhibit a hierarchy within a single cell.
Distributed vs. central control in single cells
A microscope image of a typical neurone points to an interesting logistical problem for mechanisms that regulate intrinsic membrane properties. Dendritic arbours are complex, with long, thin processes and varicosities that effectively compartmentalise biochemical and electrical signals. Distributions of membrane conductances therefore exert local effects as well as influencing cell-wide electrical properties. Moreover, feedback signals representing the state of physiological variables in such compartments will only reflect local conditions. It is hard to imagine how regulatory control can be orchestrated centrally (at the soma, for example) such that the target intrinsic properties are satisfied in all compartments of the cell simultaneously. This problem, which has received a great deal of attention in the context of synaptic homeostasis (Turrigiano, 2008), has not been addressed in detail for homeostatic control of intrinsic properties (but see Siegel et al. 1994 and comments in Goldberg et al. 2002).
The preceding argument suggests that homeostatic regulation of membrane conductances might operate locally. Indeed, much of the subcellular machinery required for feedback control exists within dendritic compartments. For example, voltage-gated calcium channels, which are well-suited to sensing membrane depolarisation, are distributed throughout the dendrites of many cell types. In addition, mRNAs and polyribosomes are found in neuronal processes and have been demonstrated to mediate local, activity-dependent protein synthesis (Sutton & Schuman, 2006) and regulate dendritic ion channel expression (Raab-Graham et al. 2006). Functional evidence for local regulation of dendritic excitability exists in a variety of contexts and may reflect Hebbian as well as homeostatic modulation (Frick & Johnston, 2005; Kim et al. 2007; Hammond et al. 2008).
What consequences would such distributed regulatory mechanisms have for the functional properties of a neurone? This has not been explored to date, but we would suggest that spatially autonomous homeostatic regulation implies richer emergent behaviour in single cells than global, cell-wide mechanisms. It may also turn out that regulatory control of gross cell properties – such as ‘intrinsic excitability’– result from the combined action of low-level, distributed mechanisms. One global effect of local regulation that was observed in an early computational study (Siegel et al. 1994) is the establishment of non-uniform distributions of voltage-gated conductances across dendritic trees. Such distributions are observed experimentally for a variety of conductances, such as sodium channels (Stuart & Hausser, 1994) and HCN channels (Magee, 1998), whose distribution has indeed been demonstrated to be activity dependent (Shin & Chetkovich, 2007). Local homeostasis therefore provides a potential mechanism for the emergence, maintenance and adaptation of gradients in ion channel expression in single cells.
The issue of noise becomes more important if ionic conductances are indeed locally regulated. Fluctuations in membrane potential resulting from stochastic channel gating are a dominant form of noise in small dendritic compartments (Manwani & Koch, 1999; Faisal et al. 2005; Cannon et al. 2010). As a consequence, local activity-dependent homeostasis might be a situation where simple control strategies (such as the bang-bang controller mentioned earlier) may be more relevant than precisely tuned integral controllers if the noise level leads to equivalent performance. Distinguishing between these possibilities would be extremely difficult experimentally, for it would require long-term, high-resolution monitoring of ion channel distributions in the smallest neuronal processes. Computational models are therefore an attractive medium for investigating putative models of local homeostasis and the effects of noise.
Even if it turns out that many low-level cell properties are regulated by simple mechanisms, there is still room for more complex mechanisms that control emergent intrinsic properties in ways that are relevant to information processing, as has been explored by several authors (Stemmler & Koch, 1999; Triesch, 2005). Furthermore, in order to constrain and control higher-order functional properties (such as spike shape or dendritic integration characteristics), cells must regulate a host of different conductances in concert. This far-from-trivial problem will be discussed next.
Lost in (parameter) space
Neurones express a bewildering array of ion channel types and this diversity reflects the numerous constraints imposed by their biophysical properties (Marder & Goaillard, 2006). For example, pyramidal neurones in the mammalian hippocampus scale their ion channel distribution in a way that sharpens the integration time-window for distal input (Magee, 1998), but also maintain sensitivity to physiologically relevant frequencies in their input such as the theta rhythm (Leung & Yu, 1998) and, of course, need to maintain stable resting membrane potential.
A useful way of describing the diversity of conductances in neurones is to say that neurones have high-dimensional parameter spaces (Taylor et al. 2006; Sobie, 2009). Parameters can be thought of as the functional expression levels of each conductance type, but can also include more general properties such as the kinetics and voltage dependence of each conductance. Allowing for the possibility that each parameter may vary independently leads to the concept of a multidimensional space as illustrated in Fig. 5. Each point in parameter space corresponds to a specification of the conductance properties of an entire cell.
Figure 5. Parameter space and the mapping to intrinsic properties.
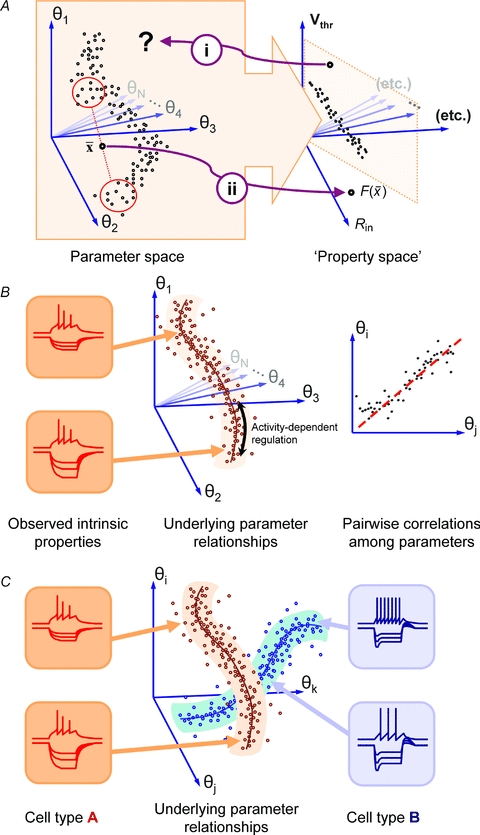
A, the mapping from the parameter space of membrane conductances to the resulting intrinsic properties (in ‘property space’) is complex, and disparate parameter combinations may be mapped to the same point, as illustrated by the squashing of the points onto a plane in property space. (i) As a consequence, the inverse mapping is undefined, so it is difficult to select parameter combinations for a given intrinsic behaviour. (ii) One aspect of the complexity of the mapping is illustrated by the fact that average parameters,  , do not necessarily get mapped to average properties. B, experimental evidence and theoretical work suggest that neurones with stereotyped intrinsic behaviour can have very different underlying conductance parameters. If large numbers of cells are measured, they tend to occupy a connected region of parameter space with a particular shape. Experimental evidence also suggests that many conductance parameters are correlated across different members of a neuronal population; this may indicate that the underlying parameter set is effectively a low-dimensional space within the high-dimensional parameter set –‘submanifold’. Such a submanifold would have fewer effective degrees of freedom, simplifying the problem of activity-dependent regulation. C, the description of conductance parameters in B would predict that neuronal subtypes with distinct electrophysiological properties (fast spiking vs. accommodating, for example) would be distinguished by the shape of their submanifolds. The distinguishing features between types would therefore be most evident in how conductances are co-regulated, rather than in the relative abundance of different conductances within a cell. We have illustrated this by showing that intrinsic properties can vary in certain respects along the submanifold (in input resistance, in the traces shown in the insets) whilst retaining important characteristics (fast spiking vs. accommodating).
, do not necessarily get mapped to average properties. B, experimental evidence and theoretical work suggest that neurones with stereotyped intrinsic behaviour can have very different underlying conductance parameters. If large numbers of cells are measured, they tend to occupy a connected region of parameter space with a particular shape. Experimental evidence also suggests that many conductance parameters are correlated across different members of a neuronal population; this may indicate that the underlying parameter set is effectively a low-dimensional space within the high-dimensional parameter set –‘submanifold’. Such a submanifold would have fewer effective degrees of freedom, simplifying the problem of activity-dependent regulation. C, the description of conductance parameters in B would predict that neuronal subtypes with distinct electrophysiological properties (fast spiking vs. accommodating, for example) would be distinguished by the shape of their submanifolds. The distinguishing features between types would therefore be most evident in how conductances are co-regulated, rather than in the relative abundance of different conductances within a cell. We have illustrated this by showing that intrinsic properties can vary in certain respects along the submanifold (in input resistance, in the traces shown in the insets) whilst retaining important characteristics (fast spiking vs. accommodating).
Two important questions emerge from this description:
How do the points in parameter space map to intrinsic properties such as input resistance, resting membrane potential, firing threshold and so on (‘property space’ in Fig. 5)?
If we could measure the conductance parameters of individual cells among a homogeneous population of real cells – or model cells with stereotyped intrinsic properties – how would they be distributed in parameter space?
Ever since the Hodgkin–Huxley model established a quantitative relationship between membrane conductance and intrinsic properties, both questions have been extensively investigated experimentally and theoretically. Naturally, many of the insights that have been gained about questions (1) and (2) had to wait until the development of sufficiently powerful computer hardware. Early modelling work identified a complex, non-linear mapping between conductance densities of Hodgkin–Huxley type models and the intrinsic properties of model membranes as described by spiking frequency vs. input (FI) curve (Foster et al. 1993). This anticipated later work which has employed substantial computational resources to explore the parameter spaces of cells with a particular intrinsic behaviour and entire networks (Prinz et al. 2003, 2004; Taylor et al. 2006, 2009; Sobie, 2009). An important (and perhaps largely anticipated) finding is that parameter sets that correspond to a given intrinsic behaviour are non-convex. Non-convexity is illustrated in Fig. 5A, where the parameter values of a set of cells sit in a c-shaped set; as a consequence, the average of the conductance parameters of several different neurones does not give rise to average intrinsic properties (Golowasch et al. 2002).
Non-convexity and the resultant failure of averaging pose problems when studying conductance regulation across a whole population of cells: it may not make sense to look at average conductance profiles, since the intrinsic properties they give rise to do not necessarily correspond to these averages (Marder & Taylor, 2011). This issue is acknowledged in recent experimental work that investigates conductance distributions in crustacean rhythm-generating circuits, where multiple conductances were measured in parallel (Schulz et al. 2006; Goaillard et al. 2009).
Moreover, there are experimentally observed instances where distinct conductance profiles map to indistinguishable intrinsic properties (Swensen & Bean, 2005). Although such experimentalists can never exhaustively measure conductance parameters, theoretical work has established that many distinct points in parameter space give rise to identical intrinsic properties (Taylor et al. 2009): that is, the mapping from parameter space to ‘property space’ is many-to-one (Fig. 5A).
Aside from introducing practical problems into experiments, the shapes of parameter sets, and the non-linear, many-to-one nature of the mapping between conductance parameters and intrinsic properties, prompts the more fundamental question of how neurones ‘know’ which way to move through parameter space as they regulate their conductances. For example, we may hypothesise that neurones regulate their resting membrane potential homeostatically. Error signals representing the difference between the actual membrane potential and the desired membrane potential ultimately need to be converted into control signals that regulate, say, the expression densities of the various ion channels in the membrane. If many different combinations of channel densities can lead to the desired change, how does the regulation mechanism settle on any particular combination?
The co-evolution of functional components and constraints in biological systems probably occurs in a modular fashion: new components are integrated into existing metabolic networks along with their functional interactions (Barabasi & Albert, 1999; Jeong et al. 2000; Barabasi & Oltvai, 2004). As a result, regulatory mechanisms are also likely to operate in a modular fashion, implying that groups of conductances are co-regulated (Ball et al. 2010). Evidence for co-regulation already exists in well-characterised preparations such as the crustacean stomatogastric ganglion (MacLean et al. 2003, 2005; Schulz et al. 2006, 2007) and this suggests that although neurones have a potentially high-dimensional parameter space, their effective parameter space is a restricted subset that locally looks like a much lower dimensional space, or in mathematical jargon, a submanifold (Fig. 5B). Parameter submanifolds would explain how neurones solve the problem of regulation, as a given cell type would have a well-defined direction to move in parameter space. In addition, the submanifold description allows a more flexible definition of electrophysiological phenotypes as belonging to distinct submanifolds (Fig. 5C). Such a view is compatible with the large degree of variability observed in conductance parameters of cells with a given identity (Marder, 2011), and may explain a larger amount of variation than can be accounted for by random effects.
Where next?
What are the most promising avenues for further research? This depends on the question being asked. We can identify two main questions: (i) understanding neuronal homeostasis as it happens in specific organisms and (ii) understanding how homeostatic mechanisms can be implemented in biology in general. The former question is important if we want to understand development, function and disease and manipulate the physiology of existing organisms to our own ends. It is also the harder question: probing homeostatic compensation in detail requires many physiological parameters to be measured and controlled simultaneously and data spanning multiple timescales, from milliseconds to months. Optogenetic methods (Gradinaru et al. 2010; Zhang et al. 2010) permit non-invasive measurement and manipulation of electrical signals on the single-cell level over timescales that potentially span the lifetime of an organism. In addition, existing molecular biological techniques such as fluorescent tagging allow the measurement of trafficking and stable expression of important proteins such as ion channels that are the target of homeostatic regulation.
The second question, which addresses homeostasis in a general setting, is already part of an ongoing programme in systems biology. Simple model organisms such as yeast and E. coli provide tractable examples of biological networks that exhibit homeostatic regulation. From an evolutionary viewpoint, the principles learned in these simple organisms will be applicable to more complex organisms and to nervous system function. These principles can be implemented in computational models of single neurones and neuronal networks, providing a bridge between relatively well-understood metabolic networks and the currently obscure details of homeostatic signalling in neurones.
Finally, in order to make sense of current and future findings, we need a coherent theory. Luckily this theory exists, but in a field from which mainstream neuronal physiology has diverged. Our belief is that in this respect, neurophysiology, like some of the homeostatic systems it describes, might benefit from a return to its ‘previous state’.
Acknowledgments
We thank Matthias Hennig, Mark van Rossum, Matt Nolan, Aoife McMahon, Cian O'Donnell and Steph Barnes for helpful discussions and constructive comments on an earlier version of our manuscript. We also thank the anonymous reviewers whose suggestions led to substantial improvements to the final version of this review article.
Appendix: model details
The traces in Fig. 3 and Fig. 4 were obtained by modelling a simple plant (a first-order linear integrator) subject to feedforward and feedback control as specified in the text. Solutions were obtained by numerical integration (backward Euler).
The plant equation and control gains are as follows:
Plant:
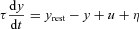
Control:
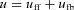
Feedforward:
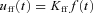
Feedback:
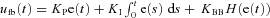
Error:
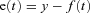
where y is plant output,  is plant time-constant = 1.3 (arbitrary units),
is plant time-constant = 1.3 (arbitrary units), 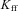 is feedforward gain =
is feedforward gain = 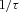 , f is target output,
, f is target output, 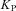 is proportional gain = 2,
is proportional gain = 2,  is the integral gain = 0.4,
is the integral gain = 0.4, 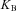 is bang-bang gain = 0.35, H is the Heaviside step-function.
is bang-bang gain = 0.35, H is the Heaviside step-function.
Noise, η,was uniformly distributed with zero autocorrelation and with power equal to −3 dB relative to the target signal.
Author contributions
TO'L conceived and researched the manuscript, created the figures and constructed the model. TO'L and DJAW wrote the manuscript.
References
- Abbott LF, LeMasson G. Analysis of neuron models with dynamically regulated conductances. Neural Computation. 1993;5:823–842. [Google Scholar]
- Bakshi UA, Bakshi MV. Modern Control Theory. Technical Publications; 2009. [Google Scholar]
- Ball JM, Franklin CC, Tobin AE, Schulz DJ, Nair SS. Coregulation of ion channel conductances preserves output in a computational model of a crustacean cardiac motor neuron. J Neurosci. 2010;30:8637–8649. doi: 10.1523/JNEUROSCI.6435-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- Barabasi AL, Oltvai ZN. Network biology: Understanding the cell's functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- Ben-Ari Y, Spitzer NC. Phenotypic checkpoints regulate neuronal development. Trends Neurosci. 2010;33:485–492. doi: 10.1016/j.tins.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergquist S, Dickman DK, Davis GW. A hierarchy of cell intrinsic and target-derived homeostatic signaling. Neuron. 2010;66:220–234. doi: 10.1016/j.neuron.2010.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- Brickley SG, Revilla V, Cull-Candy SG, Wisden W, Farrant M. Adaptive regulation of neuronal excitability by a voltage-independent potassium conductance. Nature. 2001;409:88–92. doi: 10.1038/35051086. [DOI] [PubMed] [Google Scholar]
- Buonomano DV. A learning rule for the emergence of stable dynamics and timing in recurrent networks. J Neurophysiol. 2005;94:2275–2283. doi: 10.1152/jn.01250.2004. [DOI] [PubMed] [Google Scholar]
- Buonomano DV, Merzenich MM. Cortical plasticity: from synapses to maps. Annu Rev Neurosci. 1998;21:149–186. doi: 10.1146/annurev.neuro.21.1.149. [DOI] [PubMed] [Google Scholar]
- Cannon RC, O'Donnell C, Nolan MF. Stochastic ion channel gating in dendritic neurons: morphology dependence and probabilistic synaptic activation of dendritic spikes. PloS Comput Biol. 2010;6:e1000886. doi: 10.1371/journal.pcbi.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance FS, Abbott LF, Reyes AD. Gain modulation from background synaptic input. Neuron. 2002;35:773–782. doi: 10.1016/s0896-6273(02)00820-6. [DOI] [PubMed] [Google Scholar]
- Daoudal G, Debanne D. Long-term plasticity of intrinsic excitability: learning rules and mechanisms. Learn Mem. 2003;10:456–465. doi: 10.1101/lm.64103. [DOI] [PubMed] [Google Scholar]
- Davis GW. Homeostatic control of neural activity: from phenomenology to molecular design. Annu Rev Neurosci. 2006;29:307–323. doi: 10.1146/annurev.neuro.28.061604.135751. [DOI] [PubMed] [Google Scholar]
- Davis MJ, Wu X, Nurkiewicz TR, Kawasaki J, Gui P, Hill MA, Wilson E. Regulation of ion channels by protein tyrosine phosphorylation. Am J Physiol Heart Circ Physiol. 2001;281:H1835–H1862. doi: 10.1152/ajpheart.2001.281.5.H1835. [DOI] [PubMed] [Google Scholar]
- Day TA. Defining stress as a prelude to mapping its neurocircuitry: no help from allostasis. Prog Neuropsychopharmacol Biol Psychiatry. 2005;29:1195–1200. doi: 10.1016/j.pnpbp.2005.08.005. [DOI] [PubMed] [Google Scholar]
- Desai NS. Homeostatic plasticity in the CNS: synaptic and intrinsic forms. J Physiol Paris. 2003;97:391–402. doi: 10.1016/j.jphysparis.2004.01.005. [DOI] [PubMed] [Google Scholar]
- Desai NS, Rutherford LC, Turrigiano GG. Plasticity in the intrinsic excitability of cortical pyramidal neurons. Nat Neurosci. 1999;2:515–520. doi: 10.1038/9165. [DOI] [PubMed] [Google Scholar]
- Disterhoft JF, Oh MM. Learning, aging and intrinsic neuronal plasticity. Trends Neurosci. 2006;29:587–599. doi: 10.1016/j.tins.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Echegoyen J, Neu A, Graber KD, Soltesz I. Homeostatic plasticity studied using in vivo hippocampal activity-blockade: synaptic scaling, intrinsic plasticity and age-dependence. PLoS One. 2007;2:e700. doi: 10.1371/journal.pone.0000700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. doi: 10.1126/science.1070919. [DOI] [PubMed] [Google Scholar]
- Faisal AA, White JA, Laughlin SB. Ion-channel noise places limits on the miniaturization of the brain's wiring. Curr Biol. 2005;15:1143–1149. doi: 10.1016/j.cub.2005.05.056. [DOI] [PubMed] [Google Scholar]
- Foster WR, Ungar LH, Schwaber JS. Significance of conductances in Hodgkin-Huxley models. J Neurophysiol. 1993;70:2502. doi: 10.1152/jn.1993.70.6.2502. [DOI] [PubMed] [Google Scholar]
- Francis BA, Wonham WM. The internal model principle of control theory. Automatica. 1976;12:457–465. [Google Scholar]
- Frick A, Johnston D. Plasticity of dendritic excitability. J Neurobiol. 2005;64:100–115. doi: 10.1002/neu.20148. [DOI] [PubMed] [Google Scholar]
- Goaillard JM, Taylor AL, Schulz DJ, Marder E. Functional consequences of animal-to-animal variation in circuit parameters. Nat Neurosci. 2009;12:1424–1430. doi: 10.1038/nn.2404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J, Holthoff K, Yuste R. A problem with Hebb and local spikes. Trends in Neurosciences. 2002;25:433–435. doi: 10.1016/s0166-2236(02)02200-2. [DOI] [PubMed] [Google Scholar]
- Golowasch J, Goldman MS, Abbott LF, Marder E. Failure of averaging in the construction of a conductance-based neuron model. J Neurophysiol. 2002;87:1129–1131. doi: 10.1152/jn.00412.2001. [DOI] [PubMed] [Google Scholar]
- Gradinaru V, Zhang F, Ramakrishnan C, Mattis J, Prakash R, Diester I, Goshen I, Thompson KR, Deisseroth K. Molecular and cellular approaches for diversifying and extending optogenetics. Cell. 2010;141:154–165. doi: 10.1016/j.cell.2010.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashow R, Brookings T, Marder E. Compensation for variable intrinsic neuronal excitability by circuit-synaptic interactions. J Neurosci. 2010;30:9145–9156. doi: 10.1523/JNEUROSCI.0980-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunay C, Prinz AA. Model calcium sensors for network homeostasis: sensor and readout parameter analysis from a database of model neuronal networks. J Neurosci. 2010;30:1686–1698. doi: 10.1523/JNEUROSCI.3098-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond RS, Lin L, Sidorov MS, Wikenheiser AM, Hoffman DA. Protein kinase a mediates activity-dependent Kv4.2 channel trafficking. J Neurosci. 2008;28:7513–7519. doi: 10.1523/JNEUROSCI.1951-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- Kim J, Jung SC, Clemens AM, Petralia RS, Hoffman DA. Regulation of dendritic excitability by activity-dependent trafficking of the A-type K+ channel subunit Kv4.2 in hippocampal neurons. Neuron. 2007;54:933–947. doi: 10.1016/j.neuron.2007.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Regulation of translation in eukaryotic systems. Annu Rev Cell Biol. 1992;8:197–225. doi: 10.1146/annurev.cb.08.110192.001213. [DOI] [PubMed] [Google Scholar]
- Kuo AD. The relative roles of feedforward and feedback in the control of rhythmic movements. Motor Control. 2002;6:129–145. doi: 10.1123/mcj.6.2.129. [DOI] [PubMed] [Google Scholar]
- Lasalle JP. Time optimal control systems. Proc Natl Acad Sci U S A. 1959;45:573–577. doi: 10.1073/pnas.45.4.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Rinaldi NJ, Robert F, Odom DT, Bar-Joseph Z, Gerber GK, Hannett NM, Harbison CT, Thompson CM, Simon I. Transcriptional regulatory networks in Saccharomyces cerevisiae. Science. 2002;298:799–804. doi: 10.1126/science.1075090. [DOI] [PubMed] [Google Scholar]
- LeMasson G, Marder E, Abbott LF. Activity-dependent regulation of conductances in model neurons. Science. 1993;259:1915–1917. doi: 10.1126/science.8456317. [DOI] [PubMed] [Google Scholar]
- Leung LS, Yu HW. Theta-frequency resonance in hippocampal CA1 neurons in vitro demonstrated by sinusoidal current injection. J Neurophysiol. 1998;79:1592–1596. doi: 10.1152/jn.1998.79.3.1592. [DOI] [PubMed] [Google Scholar]
- Levitan IB. Modulation of ion channels by protein phosphorylation and dephosphorylation. Annu Rev Physiol. 1994;56:193–212. doi: 10.1146/annurev.ph.56.030194.001205. [DOI] [PubMed] [Google Scholar]
- Levitan IB. Signaling protein complexes associated with neuronal ion channels. Nat Neurosci. 2006;9:305–310. doi: 10.1038/nn1647. [DOI] [PubMed] [Google Scholar]
- MacLean JN, Zhang Y, Goeritz ML, Casey R, Oliva R, Guckenheimer J, Harris-Warrick RM. Activity-independent coregulation of IA and Ih in rhythmically active neurons. J Neurophysiol. 2005;94:3601–3617. doi: 10.1152/jn.00281.2005. [DOI] [PubMed] [Google Scholar]
- MacLean JN, Zhang Y, Johnson BR, Harris-Warrick RM. Activity-independent homeostasis in rhythmically active neurons. Neuron. 2003;37:109–120. doi: 10.1016/s0896-6273(02)01104-2. [DOI] [PubMed] [Google Scholar]
- Maffei A, Turrigiano GG. Multiple modes of network homeostasis in visual cortical layer 2/3. J Neurosci. 2008;28:4377–4384. doi: 10.1523/JNEUROSCI.5298-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JC. Dendritic hyperpolarization-activated currents modify the integrative properties of hippocampal CA1 pyramidal neurons. J Neurosci. 1998;18:7613–7624. doi: 10.1523/JNEUROSCI.18-19-07613.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan S, Alon U. Structure and function of the feed-forward loop network motif. Proc Natl Acad Sci U S A. 2003;100:11980–11985. doi: 10.1073/pnas.2133841100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manwani A, Koch C. Detecting and estimating signals in noisy cable structures, I: Neuronal noise sources. Neural Computation. 1999;11:1797–1829. doi: 10.1162/089976699300015972. [DOI] [PubMed] [Google Scholar]
- Marder E. Quantification of Behavior Sackler Colloquium: Variability, compensation, and modulation in neurons and circuits. Proceedings of the National Academy of Sciences of the United States of America. 2011 doi: 10.1073/pnas.1010674108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Goaillard JM. Variability, compensation and homeostasis in neuron and network function. Nat Rev Neurosci. 2006;7:563–574. doi: 10.1038/nrn1949. [DOI] [PubMed] [Google Scholar]
- Marder E, Grashow R, Brookings T. Compensation for variable intrinsic neuronal excitability by circuit-synaptic interactions. J Neurosci. 2010;30:9145–9156. doi: 10.1523/JNEUROSCI.0980-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marder E, Taylor AL. Multiple models to capture the variability in biological neurons and networks. Nat Neurosci. 2011;14:133–138. doi: 10.1038/nn.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin SJ, Grimwood PD, Morris RG. Synaptic plasticity and memory: an evaluation of the hypothesis. Annu Rev Neurosci. 2000;23:649–711. doi: 10.1146/annurev.neuro.23.1.649. [DOI] [PubMed] [Google Scholar]
- Meltzer LA, Yabaluri R, Deisseroth K. A role for circuit homeostasis in adult neurogenesis. Trends Neurosci. 2005;28:653–660. doi: 10.1016/j.tins.2005.09.007. [DOI] [PubMed] [Google Scholar]
- O'Donnell C, Nolan MF. Tuning of synaptic responses: an organizing principle for optimization of neural circuits. Trends Neurosci. 2011;34:51–60. doi: 10.1016/j.tins.2010.10.003. [DOI] [PubMed] [Google Scholar]
- O'Leary T, van Rossum MC, Wyllie DJ. Homeostasis of intrinsic excitability in hippocampal neurones: dynamics and mechanism of the response to chronic depolarization. J Physiol. 2010;588:157–170. doi: 10.1113/jphysiol.2009.181024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olypher AV, Prinz AA. Geometry and dynamics of activity-dependent homeostatic regulation in neurons. J Comput Neurosci. 2010;28:361–374. doi: 10.1007/s10827-010-0213-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedraza JM, van Oudenaarden A. Noise propagation in gene networks. Science. 2005;307:1965. doi: 10.1126/science.1109090. [DOI] [PubMed] [Google Scholar]
- Prinz AA, Billimoria CP, Marder E. Alternative to hand-tuning conductance-based models: construction and analysis of databases of model neurons. J Neurophysiol. 2003;90:3998–4015. doi: 10.1152/jn.00641.2003. [DOI] [PubMed] [Google Scholar]
- Prinz AA, Bucher D, Marder E. Similar network activity from disparate circuit parameters. Nat Neurosci. 2004;7:1345–1352. doi: 10.1038/nn1352. [DOI] [PubMed] [Google Scholar]
- Raab-Graham KF, Haddick PCG, Jan YN, Jan LY. Activity- and mTOR-dependent suppression of Kv1.1 channel mRNA translation in dendrites. Science. 2006;314:144–148. doi: 10.1126/science.1131693. [DOI] [PubMed] [Google Scholar]
- Rao CV, Wolf DM, Arkin AP. Control, exploitation and tolerance of intracellular noise. Nature. 2002;420:231–237. doi: 10.1038/nature01258. [DOI] [PubMed] [Google Scholar]
- Rivera C, Voipio J, Payne JA, Ruusuvuori E, Lahtinen H, Lamsa K, Pirvola U, Saarma M, Kaila K. The K+/Cl– co-transporter KCC2 renders GABA hyperpolarizing during neuronal maturation. Nature. 1999;397:251–255. doi: 10.1038/16697. [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB. Gene regulation at the single-cell level. Science. 2005;307:1962. doi: 10.1126/science.1106914. [DOI] [PubMed] [Google Scholar]
- Schulz DJ, Goaillard JM, Marder E. Variable channel expression in identified single and electrically coupled neurons in different animals. Nat Neurosci. 2006;9:356–362. doi: 10.1038/nn1639. [DOI] [PubMed] [Google Scholar]
- Schulz DJ, Goaillard JM, Marder EE. Quantitative expression profiling of identified neurons reveals cell-specific constraints on highly variable levels of gene expression. Proc Natl Acad Sci U S A. 2007;104:13187–13191. doi: 10.1073/pnas.0705827104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen-Orr SS, Milo R, Mangan S, Alon U. Network motifs in the transcriptional regulation network of Escherichia coli. Nat Genet. 2002;31:64–68. doi: 10.1038/ng881. [DOI] [PubMed] [Google Scholar]
- Shin M, Chetkovich DM. Activity-dependent regulation of h channel distribution in hippocampal CA1 pyramidal neurons. J Biol Chem. 2007;282:33168–33180. doi: 10.1074/jbc.M703736200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel M, Marder E, Abbott LF. Activity-dependent current distributions in model neurons. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:11308–11312. doi: 10.1073/pnas.91.24.11308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobie EA. Parameter sensitivity analysis in electrophysiological models using multivariable regression. Biophys J. 2009;96:1264–1274. doi: 10.1016/j.bpj.2008.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemmler M, Koch C. How voltage-dependent conductances can adapt to maximize the information encoded by neuronal firing rate. Nat Neurosci. 1999;2:521–527. doi: 10.1038/9173. [DOI] [PubMed] [Google Scholar]
- Stuart G, Hausser M. Initiation and spread of sodium action potentials in cerebellar Purkinje cells. Neuron. 1994;13:703–712. doi: 10.1016/0896-6273(94)90037-x. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Swensen AM, Bean BP. Robustness of burst firing in dissociated purkinje neurons with acute or long-term reductions in sodium conductance. J Neurosci. 2005;25:3509–3520. doi: 10.1523/JNEUROSCI.3929-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AL, Goaillard JM, Marder E. How multiple conductances determine electrophysiological properties in a multicompartment model. J Neurosci. 2009;29:5573–5586. doi: 10.1523/JNEUROSCI.4438-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AL, Hickey TJ, Prinz AA, Marder E. Structure and visualization of high-dimensional conductance spaces. J Neurophysiol. 2006;96:891–905. doi: 10.1152/jn.00367.2006. [DOI] [PubMed] [Google Scholar]
- Thiagarajan TC, Lindskog M, Tsien RW. Adaptation to synaptic inactivity in hippocampal neurons. Neuron. 2005;47:725–737. doi: 10.1016/j.neuron.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Triesch J( A gradient rule for the plasticity of a neuron's intrinsic excitability. Artificial Neural Networks: Biological Inspirations. Proceedings of the International Conference on Artificial Neural Networks, 2005. 2005;3696:65–70. [Google Scholar]
- Triesch J. Synergies between intrinsic and synaptic plasticity mechanisms. Neural Computation. 2007;19:885–909. doi: 10.1162/neco.2007.19.4.885. [DOI] [PubMed] [Google Scholar]
- Tsang J, Zhu J, van Oudenaarden A. MicroRNA-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26:753–767. doi: 10.1016/j.molcel.2007.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano G. Homeostatic signaling: the positive side of negative feedback. Curr Opin Neurobiol. 2007;17:318–324. doi: 10.1016/j.conb.2007.04.004. [DOI] [PubMed] [Google Scholar]
- Turrigiano G, Abbott LF, Marder E. Activity-dependent changes in the intrinsic properties of cultured neurons. Science. 1994;264:974–977. doi: 10.1126/science.8178157. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG. Homeostatic plasticity in neuronal networks: the more things change, the more they stay the same. Trends Neurosci. 1999;22:221–227. doi: 10.1016/s0166-2236(98)01341-1. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrigiano GG, Leslie KR, Desai NS, Rutherford LC, Nelson SB. Activity-dependent scaling of quantal amplitude in neocortical neurons. Nature. 1998;391:892–896. doi: 10.1038/36103. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Hebb and homeostasis in neuronal plasticity. Curr Opin Neurobiol. 2000;10:358–364. doi: 10.1016/s0959-4388(00)00091-x. [DOI] [PubMed] [Google Scholar]
- Turrigiano GG, Nelson SB. Homeostatic plasticity in the developing nervous system. Nat Rev Neurosci. 2004;5:97–107. doi: 10.1038/nrn1327. [DOI] [PubMed] [Google Scholar]
- van Rossum MC, Bi GQ, Turrigiano GG. Stable Hebbian learning from spike timing-dependent plasticity. J Neurosci. 2000;20:8812–8821. doi: 10.1523/JNEUROSCI.20-23-08812.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Welie I, van Hooft JA, Wadman WJ. Background activity regulates excitability of rat hippocampal CA1 pyramidal neurons by adaptation of a K+ conductance. J Neurophysiol. 2006;95:2007–2012. doi: 10.1152/jn.00220.2005. [DOI] [PubMed] [Google Scholar]
- Wiener N. Cybernetics: or, Control and Communication in the Animal and the Machine. MIT Press; 1965. [Google Scholar]
- Wolpert DM, Ghahramani Z. Computational principles of movement neuroscience. Nat Neurosci. 2000;3:1212–1217. doi: 10.1038/81497. [DOI] [PubMed] [Google Scholar]
- Yeung LC, Shouval HZ, Blais BS, Cooper LN. Synaptic homeostasis and input selectivity follow from a calcium-dependent plasticity model. Proc Natl Acad Sci U S A. 2004;101:14943–14948. doi: 10.1073/pnas.0405555101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi TM, Huang Y, Simon MI, Doyle J. Robust perfect adaptation in bacterial chemotaxis through integral feedback control. Proc Natl Acad Sci U S A. 2000;97:4649–4653. doi: 10.1073/pnas.97.9.4649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F, Gradinaru V, Adamantidis AR, Durand R, Airan RD, de Lecea L, Deisseroth K. Optogenetic interrogation of neural circuits: technology for probing mammalian brain structures. Nat Protoc. 2010;5:439–456. doi: 10.1038/nprot.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Golowasch J. Modeling recovery of rhythmic activity: hypothesis for the role of a calcium pump. Neurocomputing. 2007;70:1657–1662. doi: 10.1016/j.neucom.2006.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]


