SUMMARY
The ability to sequence genomes has far outstripped approaches for deciphering the information they encode. Here we present a suite of techniques, based on ribosome profiling (the deep-sequencing of ribosome-protected mRNA fragments), to provide genome-wide maps of protein synthesis as well as a pulse-chase strategy for determining rates of translation elongation. We exploit the propensity of harringtonine to cause ribosomes to accumulate at sites of translation initiation together with a machine learning algorithm to define protein products systematically. Analysis of translation in mouse embryonic stem cells reveals thousands of strong pause sites and novel translation products. These include amino-terminal extensions and truncations and upstream open reading frames with regulatory potential, initiated at both AUG and non-AUG codons, whose translation changes after differentiation. We also define a new class of short, polycistronic ribosome-associated coding RNAs (sprcRNAs) that encode small proteins. Our studies reveal an unanticipated complexity to mammalian proteomes.
INTRODUCTION
In the ten years since the publication of draft human genomes (Lander et al., 2001; Venter et al., 2001), extraordinary advances in DNA sequencing technology (Bentley et al., 2008) have made it possible to obtain comprehensive genomic information rapidly and at low cost. Decoding the information contained in these genomes represents a central challenge for the biological community. Protein-coding regions have been defined according to simple rules about the nature of translation--for example, that open reading frames (ORFs) have a minimum length, biased codon usage and start at the first AUG in a transcript (Brent, 2005). Yet there are many exceptions to these rules, including internal ribosome entry sites, initiation at non-AUG codons, leaky scanning, translational reinitiation and translational frame shifts (Atkins and Gesteland, 2010). Additionally, an abundant class of large intergenic non-coding RNAs (lincRNAs) that do not contain canonical ORFs has been recently been described (Guttman et al., 2009; Guttman et al., 2010). Many of these newly identified transcripts are likely to be functional RNAs, but there are well-documented cases of biologically important short coding regions. For example, the Drosophila tarsal-less/polished rice gene, was originally thought to be a lincRNA (Tupy et al., 2005) but actually encodes a series of short peptides that modulate the activity of the shavenbaby transcription factor (Kondo et al., 2010). The question of which of the potential lincRNAs are actually translated remains largely unaddressed.
We also know that the rate of translation is not constant across a message and translation pauses can regulate synthesis (Darnell et al., 2011; Morris and Geballe, 2000), folding (Kimchi-Sarfaty et al., 2007; Zhang et al., 2009), and localization of a protein (Mariappan et al., 2010) or mRNA (Yanagitani et al., 2011). These pauses can results from codon usage (Irwin et al., 1995), mRNA structure (Namy et al., 2006), or peptide sequence (Nakatogawa and Ito, 2002; Tenson and Ehrenberg, 2002), but little information exists on how generally they occur, let alone their functional impact.
Recently, we described a strategy, termed ribosome profiling, based on deep-sequencing of ribosome-protected mRNA fragments, that makes it possible to monitor translation with a depth, speed and accuracy that rivals existing approaches for following mRNA levels (Guo et al., 2010; Ingolia et al., 2009). By revealing the precise location of ribosomes on each mRNA, ribosome profiling also has the potential to identify protein-coding regions. However, initiation from multiple sites within a single transcript makes it challenging to define all open reading frames, especially in complex transcriptomes. Additionally, ribosome profiling provides a snapshot of ribosome positions but does not report directly on the kinetics of translational elongation or distinguish stalled ribosomes from those engaged in active elongation.
Here we describe a simplified, robust protocol for ribosome profiling in mammalian systems. We have used this technique to determine the kinetics of translation by following run-off elongation after stalling new initiation using the drug harringtonine (Fresno et al., 1977; Huang, 1975; Robert et al., 2009; Tscherne and Pestka, 1975). We further employ harringtonine, which causes ribosomes to accumulate precisely at initiation codons, together with a machine learning algorithm, to define the sites of translation initiation genome-wide. Application of our approach to mouse embryonic stem cells reveals a wide range of novel or modified ORFs, including highly translated short ORFs in the majority of annotated lincRNAs. We now classify these atypical protein-coding transcripts as short, polycistronic ribosome-associated RNAs (sprcRNAs). Additionally, we identify over a thousand strong translational pauses that could act as key regulatory sites. Our approach is readily applicable to other cells and organisms and as such provides a general scheme for decoding complex genomes, monitoring rates of proteins production and exploring the molecular mechanisms used to regulate translation.
RESULTS
A Simplified Mammalian Ribosome Profiling Assay
We first describe a simplified ribosome profiling strategy suitable for the analysis of mammalian cells. In general terms, the assay involves three distinct steps, each of which has been refined. (i) Generation of cell extracts in which ribosomes have been faithfully halted along the mRNA they are translating in vivo. (ii) Nuclease digestion of RNAs that are not protected by the ribosome followed by recovery of the ribosome-protected mRNA fragments. (iii) Quantitative conversion of the protected RNA fragments into a DNA library that can be analyzed by deep sequencing.(Ingolia, 2010; Ingolia et al., 2009)(Lau et al., 2001; Pfeffer et al., 2005) After nuclease treatment, we purified ribosomes and the associated mRNA footprints by ultracentrifugation through a sucrose cushion rather than by sucrose density gradient fractionation, which is a more specialized technique. Protected mRNA fragments from single ribosomes were purified by PAGE, as fragments that derive from other ribosomal complexes are longer—tightly packed ribosome pairs protect 58-62 nt of mRNA (Wolin and Walter, 1988) and 48s pre-initiation complexes are reported to protect 50 nt or 70 nt under different conditions (Lazarowitz and Robertson, 1977; Pisarev et al., 2008).(Ule et al., 2003)(Chi et al., 2009; Lunde et al., 2007) We generated libraries from these purified fragments using our previous published protocol (Ingolia, 2010; Ingolia et al., 2009), modified to use RNA ligation to attach a linker to the 3′ end of the protected RNA fragment (Lau et al., 2001; Pfeffer et al., 2005). Additionally, we used subtractive hybridization to substantially deplete the majority of contaminating ribosomal RNA fragments.
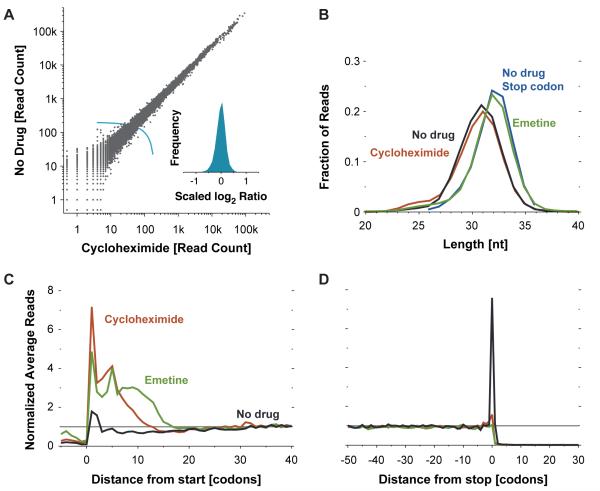
We explored the effects of stabilizing ribosome-mRNA interactions with elongation inhibitors before cell lysis. We compared cycloheximide (Schneider-Poetsch et al., 2010) and emetine pre-treatment to a “no drug” approach in which unperturbed cells were lysed in a buffer that should not support continued elongation. The density of ribosome footprints on each coding sequence, which measures the translation of the gene, agreed well across the three approaches (cycloheximide versus no drug, std dev of log2 ratio (sdlr) 0.20, corresponding to a typical 15% inter-replicate difference; cycloheximide versus emetine, sdlr 0.40; emetine versus no drug, sdlr 0.41) (Figure 1A). We concluded that brief treatment of cells with elongation inhibitors did not significantly change which transcripts were associated with ribosomes and did not distort translation measurements made by ribosome profiling. Thus, pre-treatment can be chosen based on experimental constraints. For example, elongation inhibitors would preserve the cellular state of translation during manipulations such as FACS sorting, whereas flash-freezing and cryogenic lysis would enable the analysis of tissues where infusion of translation inhibitors is challenging.
Figure 1.
Ribosome Profiling in Mouse Embryonic Stem Cells
(A) Effect of elongation inhibitors on ribosome density. The number of ribosome footprint reads that align to the body of each coding sequence (Methods) is plotted for cells that were either untreated or pretreated with cycloheximide (Spearman r = 0.99). The inset shows a histogram of log2 ratios for genes with at least 200 total reads (the threshold shown by the light blue line) normalized by the median ratio (N = 10045, s.d. = 0.20, corresponding to 15 percent difference in measurements).
(B) Ribosome-protected fragment lengths. Plotted is the length distribution of ribosome footprints over the body of messages prepared from cells treated as indicated, as well as for footprints centered on the stop codon for the untreated cells.
(C) Metagene analysis of translation initiation. Average ribosome read density profiles over 4994 well-expressed genes (Table S1), aligned at their start codon, are shown for untreated and drug-treated samples.
(D) Metagene analysis of translation termination. As in (C) but alignment was from stop codons.
Nonetheless, elongation inhibitors do alter the pattern of ribosome footprints along transcripts. Footprints derived from emetine-treated cells are slightly longer than those from untreated or cycloheximide-treated cells (Figure 1B and Figures S1A and S1B), suggesting that emetine stabilizes a different ribosome conformation that protects more mRNA. Furthermore, a metagene analysis, in which many gene profiles are aligned and then averaged, revealed global differences in ribosome density at the beginning and ends of ORFs. The excess of ribosomes at the initiation site and extending over the first five to ten codons is essentially absent from untreated cells (Figure 1C). Such an excess would result from the inhibition of translation elongation in the presence of continuing initiation. Beyond the initial five to ten-codon window we saw no global variation in ribosome density along coding sequences in any sample. An earlier analysis had suggested that the excess ribosomes extending over ~100 codons at the beginning of Saccharomyces cerevisiae ORFs reflected a broadly conserved “ramping” strategy that minimized ribosome stacking and collisions later in the messages (Tuller et al., 2010). While it is possible that such a ramping effect occurs in S. cerevisiae, it does not appear to occur mammalian cells.
Drug pre-treatment also eliminates the excess of ribosomes seen at the stop codon in untreated cells (Figure 1D). The accumulation is still seen when cells are lysed in the presence of a non-hydrolyzable GTP analogue, suggesting that it does not result from continued elongation in the lysate (Stern-Ginossar and Weissman, unpublished data). Interestingly, we saw longer footprints at stop codons (Figure 1B and S1B), suggesting that the accumulating ribosomes are in a different conformation, as has been seen during termination in vitro (Alkalaeva et al., 2006). In summary, while drug pre-treatment does not distort measurements of the overall level of translation of a given message (Figure 1A), caution should be used in interpreting position-specific information.
We characterized translation in a mouse embryonic stem cell line (E14 mESCs), with matched ribosome profiling and mRNA-seq data. We used ribosome footprint density within a coding sequence as a measure of protein synthesis and determined levels of gene expression genome-wide (Figure S1C and Tables S1A and S1B). We also compared protein synthesis with mRNA abundance and showed that there was a broad distribution encompassing over a 10-fold range in the amount of protein produced per transcript (Figure S1D and Tables S1C and S1D). This distribution is asymmetric, suggesting a maximal rate of protein production from an mRNA and substantial dynamic range for decreased translational efficiency. Our data are consistent with recent work that indirectly infers translation levels from absolute mRNA and protein abundance measurements (Schwanhausser et al., 2011). Notably, they found that translation was the single largest contributor to protein abundance, highlighting the value of direct measurements of protein synthesis.
Widespread Presence of Strong Ribosomal Pauses
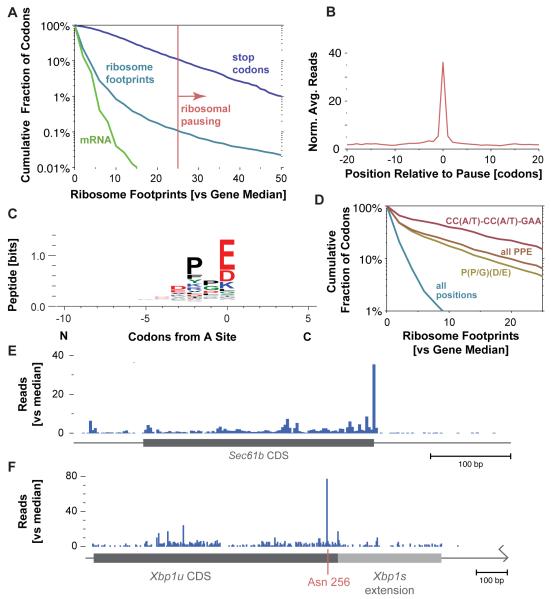
The density of ribosome footprint reads varies substantially at different codons within an individual message (Figure 2A). The footprint count on a codon should be proportional to the average ribosome dwell time there, so this density variation represents differences in the speed of the ribosome. Position specific variability is pervasive in both yeast and mESC ribosome profiling data (Ingolia et al., 2009), but in mammalian translation we find more pronounced pauses where ribosome density is 25-fold or greater than the median density observed across the body of the gene. Based on a typical elongation rate of ~6 codons per second (see below) the pauses we see last for several seconds (Figure 2A), which is enough time for the paused ribosome-nascent chain (RNC) complex to bind co-translational chaperones.
Figure 2.
Analysis of Translational Stall Sites
(A) Distribution of per-codon ribosome footprint counts. The cumulative distribution of footprint counts at each codon, relative to the median density across the gene, is plotted and the 25× median threshold used to identify ribosomal stall sites is indicated. The distribution of density at stop codons, which are excluded from the overall distribution, is shown as well, along with the read densities in randomly-fragmented mRNA, which controls for library generation.
(B) Metagene analysis of translational stalling. Ribosome footprint densities were averaged after aligning gene density profiles at internal translational stall positions (Table S2B).
(C) Peptide motif associated with internal translational stalling.
(D) Ribosome footprints over peptide motif enriched in stall sites. The cumulative distribution of relative ribosome footprint counts for the all Pro-Pro-Glu sites and for those encoded by CC(A/T)-CC(A/T)-GAA are shown along with the more lenient Pro-(Pro/Gly)-(Asp/Glu) sites and the overall data from (A).
(E) Ribosome footprint profile on the Sec61b transcript (median 22.5 footprints per codon).
(F) Ribosome footprint profile on the Xbp1 transcript (median 1.0 footprint per codon). Xbp1 undergoes a nonconventional splicing event (Calfon et al., 2002). The unspliced (Xbp1u) coding sequence is indicated, along with the site of translational stalling at Asn 256 and the extended coding sequence in the spliced (Xbp1s) message.
We find thousands of novel pauses in the body of genes (1500 pauses in 1100 genes found in a set of 4994 well-expressed genes; Tables S2A and S2B) and at termination codons (420 pauses, Table S2C). Interestingly, we see no evidence that pausing causes secondary ribosome accumulation ~10 codons upstream, where a following ribosome would collide with the stalled one (Wolin and Walter, 1988), nor a depletion of ribosomes within the 10 codon “shadow” resulting from paused ribosomes (Figure 2B). The lack of packed ribosomes at pause sites suggests that ribosome density is typically too low to cause frequent encounters between upstream elongating ribosomes and a transiently stalled downstream ribosome (Arava et al., 2003). Alternately, such a collision might relieve ribosomal stalling, allowing for the continual presence of a ribosome at a pause site while minimizing ribosome sequestration. The absence of downstream depletion also argues that the majority of ribosomes continue elongation following these pause sites.
Analysis of the sequence around the pause sites reveals a consensus peptide motif (Figure 2C). There is strong enrichment of glutamate or asparate in the A site at strong pauses, preceded by a proline or glycine and then another proline, with an additional bias towards the GAA glutamate codon and CC(A/T) proline codons. Importantly, we see no enrichment for residues or codons downstream of the A-site, which are not yet being decoded. We also see no evidence that the pause sites are enriched for rare codons. Sites that match the full three-residue consensus have dramatically reduced elongation rates overall (Figure 2D). Translation in E. coli is stalled by similar peptide motifs with a terminal Pro-Pro peptide, in some cases with an Asp codon in the A site (Tanner et al., 2009). Our findings suggest that tRNA identity and nascent peptide sequence can influence the kinetics of elongation, whereas even for rare codons, tRNA recruitment is not rate-limiting.
Our analysis also provides insights into the limited number of previously documented translational pauses. A recent study observed slow termination of two tail-anchored (TA) proteins (Sec61b and Vamp) during in vitro translation (Mariappan et al., 2010). Pausing at the termination codon of TA proteins has been proposed to provide time for the recruitment of the insertion machinery before the release of the C-terminal transmembrane domain from the ribosome exit channel. Our data confirmed termination pausing during the Sec61b and Vamp translation in vivo (Figure 2E), but we found no evidence for this phenomenon in the majority of other TA proteins (3 / 32 have pauses), nor was it restricted to TA proteins (stop codon ribosome density does not differ significantly, Kruskal-Wallis p ~ 0.25). Instead, pausing at termination codons is a common feature of translation.
A second prominent example of a translation pause follows a hydrophobic sequence in the Xbp1 transcription factor (Yanagitani et al., 2011). This hydrophobic domain interacts with the ER membrane and recruits the Xbp1 message ribosome-nascent chain (RNC) complex (Yanagitani et al., 2009). Ribosome pausing facilitates this co-translational localization by delaying the dissociation of the RNC. We confirmed the presence of this pause and identified its precise position as residue Asn 256, which is the last codon required for translational arrest (Yanagitani et al., 2011) (Figure 2F). The biological roles of the pauses we identify remains to be established, but many mRNAs are localized to specific subcellular regions (Martin and Ephrussi, 2009), including a number of mRNAs found on the ER surface that, like Xbp1, do not enter the secretory pathway (Kraut-Cohen and Gerst, 2010), and so the mechanism described for Xbp1 localization may be more general.
Monitoring Kinetics of Translation
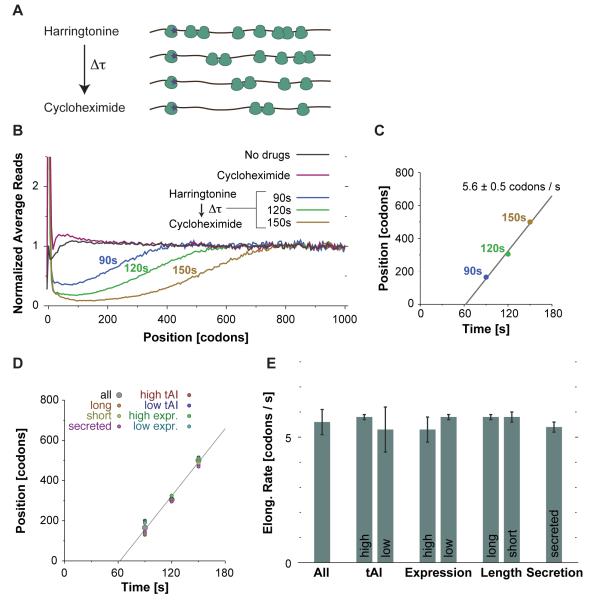
Our knowledge of the kinetics of protein synthesis in vivo has been based on a limited number of specific messages (Bostrom et al., 1986). We reasoned that we could monitor the kinetics of in vivo translation directly by tracing run-off elongation using ribosome profiling. We first stopped new translation using harringtonine, which effectively blocks initiation by inhibiting elongation during the first rounds of peptide bond formation following subunit joining (Fresno et al., 1977; Robert et al., 2009). We then allowed a short time for run-off elongation before adding cycloheximide to halt translation by all active ribosomes. We varied the time allowed for run-off elongation to generate a series of snapshots that could be assembled into a moving picture of translation in vivo (Figure 3A). Metagene analyses revealed a progressive depletion of ribosomes from the 5′ to the 3′ of the messages after harringtonine treatment. Following a delay of ~60 seconds, which presumably reflects the time required for engagement of harringtonine, ribosomes progress from the 5′ ends of transcripts at a rate of 5.6 amino acids per second (Figures 3B and 3C), which is consistent with values from previous single-gene measurements (Bostrom et al., 1986).
Figure 3.
A Pulse-chase Strategy for Measuring Translation Elongation Rates
(A) Schematic of the in vivo run-off elongation experiment.
(B) Metagene analysis of run-off elongation. Ribosome read density was averaged across 5-codon windows for samples prepared with the indicated drug treatments.
(C) Rate of ribosome depletion. The codon position of 50% ribosome depletion is plotted as a function of harringtonine treatment time. Linear fit is x(t) = ax + b, a = 5.6 ± 0.5 codons / s, b = −347 ± 65 codons, r.m.s.d. 22.5.
(D) Ribosome depletion on subsets of genes. Data from (C) is plotted, along with comparable measurements made from the indicated gene subsets.
(E) Elongation rates on subsets of genes. Elongation rates, inferred from linear fit as described in (C), are plotted along with the standard error of the regressed coefficient.
The rate of translation is remarkably consistent between different classes of messages (Figures 3D and 3E). The kinetics of elongation are independent of length and protein abundance and are the same in secreted proteins, whose translation occurs on the ER surface. Translation speed is also independent of codon usage, which is consistent with the absence of pauses at rare codons. This is surprising as it is often assumed that codons corresponding to low abundance tRNAs are decoded more slowly than those read by abundant tRNAs. While this may be the case for specific examples, we find no evidence for a large effect on the overall rate of elongation. An important practical implication for the universality of the average rate of elongation is that ribosome footprint density provides a reliable measure of protein synthesis independent of the particular gene being translated.
Defining Translation Start Sites
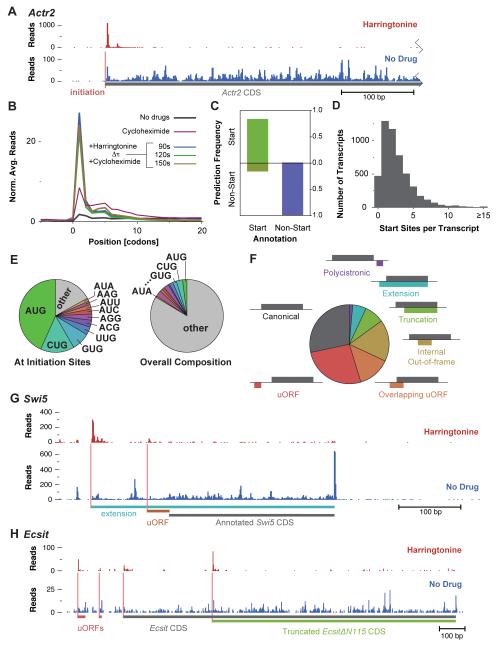
We found that harringtonine treatment also leads to a profound accumulation of ribosomes at the sites of translation initiation (Figures 4A and 4B). This effect likely occurs because harringtonine binds to free 60s subunits, but not those that are joined into an 80s ribosome. Thus, elongating ribosomes are immune to harringtonine, whereas a 60s subunit bound by harringtonine will form an 80s at a start site that does not move forward (Fresno et al., 1977; Robert et al., 2009). We reasoned that this accumulation of ribosomes could serve as a basis for objectively detecting translation initiation. Accordingly we used a support vector machine (SVM)-based machine learning strategy (Joachims, 1999; Noble, 2006) to comprehensively identify initiation sites from harringtonine-treated ribosome footprint profiles, using a “vector” of footprint counts around a candidate translation start site. The SVM model was trained on a set of annotated genes to identify features of footprint profiles that distinguish the start codon from other positions. These profiles capture not just the accumulation of ribosomes at the start codon, but also the distinctive asymmetric pattern of reads across flanking codons. Analysis of a distinct testing set of transcripts not used for training established that this model recognized 86 percent of annotated start codons as sites of translation initiation in comparison to only ~1 percent of other positions (Figures 4C and S2A). Actual false negative and false positive rates may be considerably lower, as not all annotated start sites are correct and there is a substantial rate of translation initiation from non-canonical start sites.
Figure 4.
Harringtonine Enables Automated Identification of Translation Initiation Sites.
(A) Effect of harringtonine on ribosome density for a typical gene. Ribosome footprint read count is shown prior to and following harringtonine treatment (150 s) along the 5′ UTR and the beginning of the coding sequence of Actr2.
(B) Metagene analysis of ribosome footprints surrounding start codons after harringtonine treatment. As in Figure 3B, focusing specifically on the site of translation initiation and the surrounding codons.
(C) Evaluation of start site prediction analysis. Plotted is the fraction of positive and negative initiation site predictions for start and selected non-start codons that were excluded from the training set.
(D) Histogram of initiation sites predicted per transcript.
(E) Distribution of AUG codons and near-AUG codons at predicted sites of translation initiation (left), compared with the overall codon distribution (right).
(F) Classification of reading frames at predicted initiation sites relative to the annotated CDS.
(G) Pattern of initiation and translation on the Swi5 transcript. As in Figure 4A, with the two detected initiation sites shown along with the respective reading frames, one of which produces a conserved amino-terminal extension on the Swi5 protein.
(H) Pattern of initiation and translation on the Ecsit transcript. Four AUG initiation sites are present, two associated with uORFs and two with alternate protein isoforms of Ecsit.
We applied the SVM approach to identify 13454 candidate translation start sites within ~5000 transcripts that were well-expressed in our mouse ES cells (Table S2A). The majority (65%) of these transcripts contain more than one detectable site of translation initiation, with 16 percent containing four or more sites (Figure 4D and Table S3). While the analysis examined all potential translation start sites, we observed a dramatic enrichment for AUG (23-fold; Figure 4E), which provides an independent line of evidence for the accuracy of the SVM approach. We also found a strong enrichment for a specific subset of the near-cognate codons (i.e., codons that differ from AUG by a single nucleotide) at initiation sites (Figure 4E). Initiation at near-cognate sites is sometimes resistant to harringtonine (Starck et al., 2008); Stern-Ginossar and Weissman, unpublished data), so our analysis may underestimate the true prevalence of near-cognate initiation.
Characterization of Alternate Open Reading Frames
We classified the reading frames downstream of the initiation sites we identified based on their relationship to the annotated ORF (Figure 4F). Nearly half (44%) of the AUG initiation sites that we found are unannotated, and the majority of these were downstream of the annotated start and were predicted to produce N-terminally truncated proteins or ORFs encoded in alternate reading frames (Figure S2B). In many cases, the annotated AUG was also used and the alternate protein may not be the primary translation product. However, 280 of the genes with N-terminal truncations lacked detectable initiation on the annotated AUG, either because the annotated start codon is skipped in favor of the internal start site that we identified, or the transcript is truncated and the annotated start codon is absent.
A substantial fraction (14%) of the initiation sites we observed are predicted to produce alternate protein isoforms of known genes (Figure 4F). We identified 570 genes with potential N-terminal extensions and 870 with N-terminal truncations in the 4994 genes we analyzed. Extensions most often resulted from near-cognate initiation (Figure S2B), probably because computational gene annotation selects the first in-frame AUG, though conservation has been used to identify N-terminal extensions from near-cognate initiation (Ivanov et al., 2011). We found an N-terminal extension on the DNA repair protein Swi5 (Figure 4G); its protein sequence is conserved, and there is experimental evidence that endogenous mouse Swi5 is larger than the annotated 89 amino acid protein (Akamatsu and Jasin, 2010). Our data also revealed information about the protein products resulting from alternative splicing, which are often difficult to annotate. For instance, the growth factor Igf2 has two 5′ UTR variants with the same reading frame annotated in both transcripts, but we observed an isoform-specific N-terminal extension (Figure S2C).
The N-terminal truncations are of particular note as they can produce functionally distinct protein isoforms that lack an entire amino-terminal domain. For example, alternate start codons in the Cebpa gene can result in either a full-length transcription factor or in a truncated dominant-negative isoform that contains the DNA-binding domain but not the full transactivation domain (Lin et al., 1993). We observe clear evidence of novel N-terminal truncations that could produce similar antagonistic products. Internal initiation in the Ets family transcription factor Etv5 produces a product that lacks the predicted activation domain (Monte et al., 1996) but contains the domain that mediates DNA binding (Monte et al., 1994) (Figure S2D). This mechanism is not limited to transcription factors--internal initiation in the signaling scaffold Ecsit produces a protein nearly identical to a dominant negative form created by designed N-terminal deletion (Figure 4H) (Kopp et al., 1999).
Exploring Translation of sprcRNAs
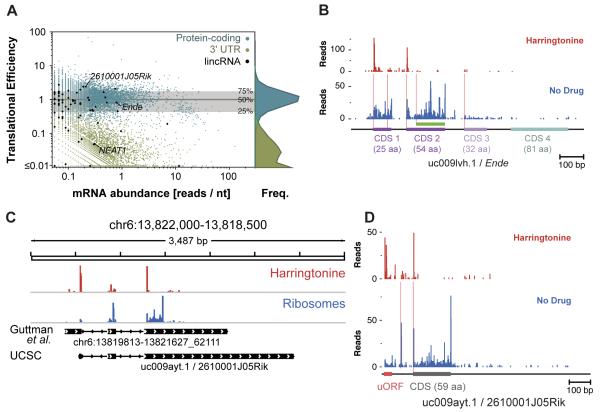
The above analysis focuses on known coding transcripts, but recently an abundant class of RNAs, referred to as lincRNAs, have been identified that lack the characteristics of conventional protein-coding genes. A limited number of lincRNAs such as Xist and HotAir have been shown to act at the RNA level in the nucleus (Brockdorff et al., 1992; Khalil et al., 2009), but the extent to which putative lincRNAs are translated is not known. Accordingly, we searched for translated regions within candidate lincRNAs (Guttman et al., 2009) (Guttman et al., 2010) by finding the most highly ribosome-occupied 90 nt window within the lincRNA and determining its translational efficiency as the ratio of ribosome footprint and mRNA-seq reads.(Guttman et al., 2010)(Guttman et al., 2009) This analysis was very effective at distinguishing between traditional translated coding sequences and their 3′ UTRs, which are poorly translated (Figure 5A).
Figure 5.
Translation of sprcRNAs
(A) Translational efficiency of putative lincRNAs. The translational efficiency, a normalized ratio of ribosome footprint density to mRNA-seq read density, is plotted for the most highly occupied 90 nt window of each lincRNA, protein-coding gene, and coding transcript 3′ UTR, along with a histogram of translational efficiency values for CDSes and 3′UTRs and the median and quartile values for protein-coding genes.
(B) Ribosome footprint profile of the uc009lvh.1 transcript. This RNA is annotated as a non-coding RNA, but we identify two short (25 and 54 amino acids) well-translated ORFs, and see little translation from a longer (81 amino acid) downstream CDS hypothesized to encode a protein (Hassan et al., 2010).
(C) Ribosome footprint profile of the 2610001J05Rik genomic locus. The profile includes transcript-aligned reads mapped to corresponding genomic positions and genomic-aligned reads with no transcript alignment. The annotated non-coding uc009ayt.1 transcript is shown along with the reconstructed transcript (Guttman et al., 2010).
(D) Ribosome footprint profile of the uc009ayt.1 transcript.
Remarkably, the majority of putative lincRNAs contain regions of high translation comparable to protein-coding genes (Figure 5A and Table S4). We saw specific start sites marked by harringtonine followed by ribosome footprints extending to the first in-frame stop codon (Figures 5B-D). (Clemson et al., 2009)These data establish that the majority of lincRNAs are exported to the cytoplasm and effectively engaged by the protein translation machinery. We classify these RNAs as short, polycistronic, ribosome-associated coding RNAs (sprcRNAs) based on our observation that they contain small coding sequences that are bound by elongating ribosomes, and frequently contain multiple ORFs. We also identify a significant subset of true lincRNAs that are not translated, including the well-documented RNA element NEAT1, which regulates mRNA export (Clemson et al., 2009). The extent to which various RNAs act through their translation products and/or directly through their transcript remains a central open question that our dataset should provide a critical resource for addressing.
Widespread Translation of uORFs
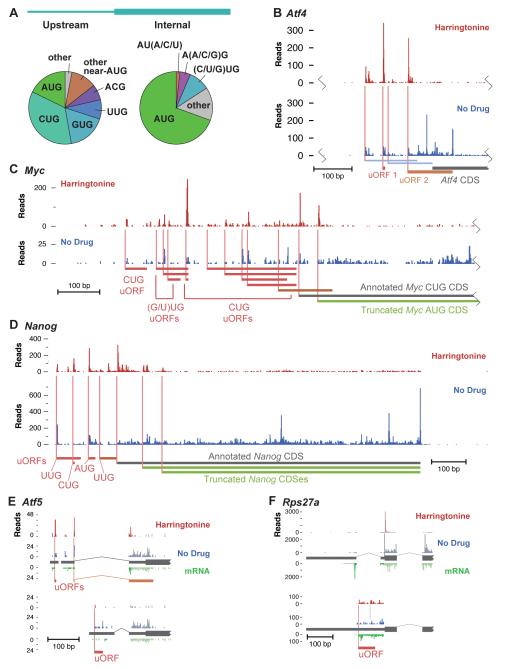
The majority of novel near-cognate initiation sites we detected drive the translation of uORFs (Figure 6A and Figure S4B). This is consistent with the high level of translation that we observe on many 5′ UTRs as opposed to 3′ UTRs, which are almost devoid of ribosomes. These uORF initiation sites are accompanied by elongating ribosome footprints in the untreated sample that are depleted during harringtonine treatment, indicating that they are involved in active translation (Figure 4B). In a few well-studied examples, uORFs have been shown to affect translation of downstream genes. The first uORF in the Atf4 transcript is constitutively translated and ribosomes then reinitiate at either the second uORF or the CDS (Calvo et al., 2009; Lu et al., 2004; Morris and Geballe, 2000) (Figure 6B). This exemplifies two roles of uORFs—some permit downstream reinitiation, whereas others capture some fraction of scanning pre-initiation complexes and decrease CDS translation. There are a small number of well-documented uORFs with near-cognate start codons (Ivanov et al., 2008), but there are no effective computational approaches for identifying them. Our observations suggest that near-cognate uORFs are quite common. The ribosome footprint profiles of Myc and Nanog, two genes that play a critical role in pluripotency, illustrate the complexity of translation; both have multiple uORFs and alternate translation products initiating at both AUG and near-cognate sites (Figures 6C and 6D).
Figure 6.
Translation of Regulatory uORFs and Alternatively Processed Transcripts
(A) Codon distribution at upstream (left) and internal (right) translation initiation sites. Internal sites are only taken from codons 15 through 300, as internal sites further downstream are affected by incomplete ribosome run-off during short harringtonine treatment.
(B) Patterns of initiation and translation on the Atf4 transcript. The two characterized regulatory uORFs, initiated by AUG codons, are highlighted. Two weak non-AUG reading frames are shown in blue.
(C) As in (B) on the Myc transcript. Several near-cognate sites of upstream initiation are shown, along with the annotated CUG initiation codon and the alternate AUG initiation codon.
(D) As in (C) on the Nanog transcript. Upstream open reading frames are shown, along with the CDS and two in-frame AUG initiation sites within the CDS.
(E) Patterns of initiation and translation on the 5′ end of two transcripts of the Atf5 gene. The exon structure is shown with thin gray rectangles for the 5′ UTR and thick gray rectangles for the annotated coding sequence. An mRNA-seq read profile is shown on an inverted y axis. Isoform-specific transcript positions are shown in dark colors and non-isoform-specific positions are shown with faint colors. The major isoform (top) has two uORFs that confer translational regulation on the coding sequence; a distinct uORF is observed in the minor embryonic isoform (bottom).
(F) As (E), for the 5′ end of the Rpl27a transcripts. Only the isoform-specific positions are shown for the minor isoform (bottom), scaled 10x.
Due to the prevalence of alternative transcription start sites and alternative splicing, many genes have multiple 5′ UTR isoforms, potentially including distinct regulatory information (Hughes, 2006). Many novel initiation sites occurred in alternative UTRs; we found 1800 genes showing differential initiation of uORFs in distinct 5′ UTR isoforms. We additionally observed that at least 30% of these genes showed a significant difference in the ratio of ribosome footprint to mRNA-seq reads between the distinct 5′ UTRs of different isoforms. Thus, alternative splicing generates transcripts with different upstream initiation sites and results in different uORF translation. For example, the transcription factor Atf5 is regulated by well-characterized uORFs in one mRNA isoform that are missing from a less-abundant isoform expressed in early development (Hansen et al., 2002). We observe robust translation initiation at a distinct uORF in this second isoform (Figure 6E). Alternative inclusion of uORFs was also seen in ribosomal proteins, including Rps27a, where a small fraction of transcripts had a retained 5′ UTR intron that introduced a uORF (Figure 6F). In the particular case of isoforms where an alternative UTR splice junction is quite close to the shared start codon, ribosome footprints from initiation at the start codon can include enough distinct upstream sequence to distinguish the effect of different UTRs. The gene Pih1d1 has two 5′ UTR variants with distinct uORFs. Strong initiation of the uORF in one isoform led to 50% less initiation of its protein-coding reading frame as compared to initiation of the same protein-coding reading frame in the second isoform (Figure S3). This effect demonstrates the potential impact of the widespread upstream initiation we observe in both alternative and constitutive 5′ UTRs.
Changes in Translation During Embryoid Body Formation
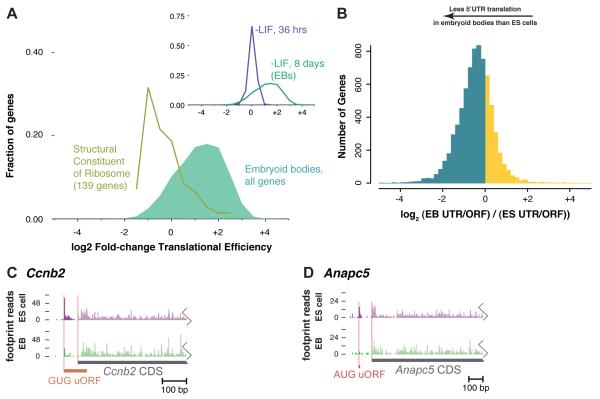
We next asked how the landscape of translation changes when proliferative, pluripotent ES cells undergo differentiation into embryoid bodies (EBs). Withdrawal of leukemia inhibitory factor (LIF) induced differentiation (Figure S4A), which we assessed visually and by the down-regulation of the direct LIF target Klf4 (Niwa et al., 2009), followed by loss of Oct4 expression and the induction of developmental and lineage-specific genes (Figures S4B and S4C and Tables S5A-S5F). We then looked for translational control of gene expression during differentiation and observed strong repression of ribosomal proteins (RPs) in EBs relative to ES cells (Figure 7A, Figure S5D and Table S5F). Although these genes were still highly expressed in embryoid bodies, they were translated 3- to 4-fold less efficiently than the typical transcript (Tables S5D-S5F). The translation of RPs is regulated in response to proliferation and nutrient status (Hamilton et al., 2006), and here we show that this response is a notable feature of EB formation. Polysome profiling experiments have suggested a global increase in cellular translation during early ES cell differentiation, and we see a modest upregulation of RPs in our early timepoint (Sampath et al., 2008). This might lead to a surfeit of ribosomes at the later stage of EB formation. Intriguingly, Akt/mTOR signaling, controls RP expression and may regulate translation during differentiation more generally (Di Cristofano et al., 1998; Sampath et al., 2008). We also observed a modest but quite significant increase in the translational efficiency of integral membrane proteins in EBs (Figure S4E and Table S5F), which could result directly from a redirection of ribosomes to the rough ER, or indirectly through regulatory programs whose targets are enriched for membrane proteins.
Figure 7.
Changes in Upstream Translation During Differentiation
(A) Translational regulation following LIF withdrawal. The distribution of log2 fold-changes of translational efficiency (ratio of sample-normalized ribosome footprint density to mRNA-seq density) is shown for all genes and for those with the GO annotation “structural constituent of ribosome” (see Table S5D). Inset: distributions for all genes, 36 hours and 8 days after LIF withdrawal (see Tables S5A and S5D).
(B) Changes in relative upstream translation in EBs versus ES cells. The ratio of footprints between the 5′UTR and the ORF was computed for each gene and the distribution of log2 change in the 5′UTR/ORF ratio is plotted, with decreases in EB shown in blue and increases in EB shown in yellow.
(C-D) Patterns of translation on the Ccnb5 (C) and Anapc5 (D) transcripts. Ribosome footprints that map to the 5′UTR are in dark colors and the CDS in faint colors. The average, sample-normalized ribosome footprint density on the CDS is slightly higher in the EB sample than the ES cell sample for both.
Translation of uORFs also declined substantially during differentiation. We measured the level of upstream translation using the ratio of ribosome footprint reads in the 5′UTR to the coding sequence of each gene and found that the typical transcript showed a ~25% decrease in 5′UTR translation during differentiation (Figure 7B). This shift can be observed on the 5′UTR of individual genes with defined uORFs (Figures 7C and 7D). It reflects a broad change in the translational apparatus with the potential to impact gene expression genome-wide. Reduced upstream translation might reflect a relative decrease in cap-dependent versus cap-independent initiation, as cap-dependent initiation would be expected to favor upstream sites near the cap. Such a shift has been associated with proliferation in tumorigenesis, and has been linked to the translational control of RPs (Mamane et al., 2006; Ruggero and Sonenberg, 2005). This tumor cell translational program may also be active in ES cells.
DISCUSSION
Here we present a range of ribosome profiling techniques, based on deep sequencing of ribosome-protected fragments, that dramatically expand our ability to define and quantitatively monitor mammalian proteomes. Our approaches provide experimentally based maps of the protein coding potential of complex genomes, and reveal in depth information about the kinetics and mechanism of translation elongation and coupled co-translational events. Finally, ribosome profiling allows high precision, genome-wide measures of the rate of protein synthesis from the density of ribosome footprints, much as RNA-seq experiments measure mRNA abundance from read density; such gene expression measurements may represent the most frequent application of ribosome profiling even after the proteome is fully defined. While there have been remarkable advances in quantitative mass spectrometry (Nilsson et al., 2010), it is difficult to match the large dynamic range and comprehensive nature of deep sequencing. More generally mass spectrometry and ribosome profiling represent highly complementary approaches; for example, comparison between changes in rate of synthesis measured by ribosome profiling and abundance measured by mass spectrometry should reveal examples of regulated degradation of proteins.
A number of novel features of mammalian proteomes emerge from our studies, including the ubiquitous use of alternate initiation sites that drive the production of extended or truncated isoforms of known proteins as well as the translation of sprcRNAs, whose protein-coding potential was not initially apparent. We also observe widespread translation upstream of mammalian protein-coding genes, similar to but more extensive than upstream translation that we observed in yeast (Ingolia et al., 2009). Translation of a uORFs can modulate the expression of the downstream protein-coding gene in response to global (Sonenberg and Hinnebusch, 2009) or gene-specific regulatory signals (Medenbach et al., 2011). We have shown that upstream translation decreases as ES cells undergo differentiation, indicating that it is subject to regulation and may be part of a major program of translational control.
Our studies also establish that many sites of translation initiation, especially upstream initiation, occur at non-AUG codons. While most productive protein synthesis starts at a classical AUG codon, initiation at CUG and GUG codons is widespread and is likely to have broad biological significance. An important open question is how this non-AUG initiation differs mechanistically from AUG initiation and what factors regulate initiation site selection. The bias towards upstream non-AUG initiation seems to conflict with a pure scanning model for start codon recognition, as a pre-initiation complex that bypasses the annotated AUG is no less likely to recognize a subsequent CUG, though the difference could reflect heterogeneous stringencies in scanning complexes.
Non-AUG initiation clearly impacts many aspects of translation. The extensive upstream non-AUG initiation we observe is likely to regulate protein synthesis from specific transcripts in response to global changes in initiation. It is also regulated during EB formation, suggesting a global link with growth and proliferation, and is involved in the synthesis of functional proteins, including the well-studied oncogene and pluripotency factor Myc (Hann et al., 1988). More broadly, it has been implicated in the production of peptides for immune surveillance (Malarkannan et al., 1999), and additional roles will likely emerge as we understand more about which non-AUG codons are used and how this selection is regulated.
EXPERIMENTAL PROCEDURES
Ribosome Footprinting
E14 mESCs were propagated in standard culture feeder-free conditions (Tremml et al., 2008) and differentiation was induced by transferring cells to media lacking LIF in low adhesion dishes. Cells were pre-treated with harringtonine (2 μg/ml), cycloheximide (100 μg/ml), and/or emetine (20 μg/ml) as indicated and detergent lysis was performed in the dish. The lysate was DNase-treated and clarified, and a sample was taken for mRNA-Seq analysis. Lysates were subject to ribosome footprinting by nuclease treatment. Footprint fragments were purified and deep sequencing libraries were generated from these fragments, as well as from poly(A) mRNA purified from untreated lysate. These libraries were analyzed by sequencing on the Illumina GAII and HiSeq.
Footprint Sequence Alignment
Sequences were aligned to a library of transcripts derived from the UCSC Known Genes data set (Hsu et al., 2006) and the reconstructed mESC transcriptome of Guttman et al. (Guttman et al., 2010), and those with no acceptable transcript alignment were then aligned against the genome. Because sequencing reads comprise a variable-length RNA fragment followed by a linker sequence, the first 26 nucleotides were aligned against the reference database using Bowtie and this alignment was extended until it reached the known linker sequence. Alignments were accepted with up to two mismatches, and multiple alignments were allowed for a single sequence but alignments with fewer mismatches were preferred.
For most analyses, footprint alignments were assigned to specific A site nucleotides by using the position and total length of each alignment, calibrated from footprints at the beginning and the end of CDSes (Figures S1A and S1B) as previously described (Ingolia et al., 2009).
Footprint Profile Analysis
Profiles of ribosome footprints across a transcript were constructed by quantifying the number of footprints assigned to each nucleotide position. A set of well-expressed genes was selected based on median footprint density across the coding sequence, excluding the first 15 and last 5 codons due to the accumulation of ribosomes (Figures 1C and 1D). To construct metagene density profiles, individual gene profiles were scaled by their footprint density in the untreated control and all were averaged with equal weight.
Harringtonine Depletion Profile Analysis
Metagene profiles from harringtonine run-off were further normalized by the median value over codons 800-1000, which appeared undepleted at harringtonine treatment times used in this study, and smoothed by averaging disjoint 5-codon windows. The extent of depletion was defined as the earliest codon position, beyond the first 40, that retained at least 50% of the full ribosome density. Subsets of genes for elongation rate analysis were: 1) lowest and highest quintile of tAI, computed according to dos Reis et al. (dos Reis et al., 2004); 2) lowest and highest quintile of ribosome footprint density; 3) short genes, 750-1000 codons, and long genes, over 1000 codons; 4) secreted proteins were identified using SignalP data from Ensembl.
Initiation Site Prediction
Initiation site predictions for each nucleotide position were based on a vector of footprint read counts over 15 codons around the position for each harringtonine sample, concatenated to produce an overall vector. The SVMlight pattern recognition tool (Joachims, 1999) was trained on an arbitrary set of 3200 transcripts, using the annotated start codon as a positive example and ten other positions as negative examples.
Initiation sites were defined as one or more consecutive nucleotide positions that passed an SVM score threshold as well as a minimum of 50 harringtonine footprints total amongst all samples. These consecutive blocks were typically (91%) three or fewer nucleotides long and in no case longer than six nucleotides (Table S3). Initiation sites that contained an AUG codon were assigned to that codon, or if none was present, to any near-cognate codons, and the reading frame was predicted from that codon. Sites with no recognizable initiation codon or with multiple potential near-cognate codons could not be assigned to a specific reading frame and were eliminated from further analyses. The preferential assignment of initiation sites to AUG codons may lead to a modest bias against detecting near-cognate initiation.
LincRNA Analysis
LincRNAs were collected from reconstructed transcripts (Guttman et al., 2010) that lay entirely within the lincRNA chromatin signatures identified by Guttman et al. (Guttman et al., 2009), which excluded known protein-coding genes. Footprint density profiles from the untreated sample were analyzed to identify the 90 nt window with the most positions occupied by at least one ribosome footprint amongst all transcripts in the chromatin region. For annotated protein-coding transcripts, the coding sequence and the 3′ UTR were analyzed separately. The mRNA abundance was calculated as the density of mRNA-seq reads in the window and the translational efficiency was calculated as the ratio between the ribosome footprint and the mRNA-seq read density in the window.
Supplementary Material
ACKNOWLEDGMENTS
We thank M. Barna, S. Luo, D. Ruggero G. Schroth, P. Brown and members of the Weissman lab including A. Frost and S. Churchman for advice and helpful discussions; C. Chu for help with sequencing; S. Rouskin for help developing the ligation protocol; G. Brar for help developing rRNA depletion; J. Hunkapiller and J. Reiter for E14 mESCs. This investigation was supported by the NIH P01 (AG10770) and a Ruth L. Kirschstein NRSA (GM080853) (N.T.I.), the Damon Runyon Cancer Research Foundation (DRG-2033-09) (L.F.L), and by the Howard Hughes Medical Institute (J.S.W.)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession Numbers
Sequencing data were deposited in the GEO database with accession number GSE30839.
REFERENCES
- Akamatsu Y, Jasin M. Role for the mammalian Swi5-Sfr1 complex in DNA strand break repair through homologous recombination. PLoS Genet. 2010;6:e1001160. doi: 10.1371/journal.pgen.1001160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alkalaeva EZ, Pisarev AV, Frolova LY, Kisselev LL, Pestova TV. In vitro reconstitution of eukaryotic translation reveals cooperativity between release factors eRF1 and eRF3. Cell. 2006;125:1125–1136. doi: 10.1016/j.cell.2006.04.035. [DOI] [PubMed] [Google Scholar]
- Arava Y, Wang Y, Storey JD, Liu CL, Brown PO, Herschlag D. Genome-wide analysis of mRNA translation profiles in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2003;100:3889–3894. doi: 10.1073/pnas.0635171100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkins JF, Gesteland RF. Recoding: expansion of decoding rules enriches gene expression. New York, Springer: 2010. [Google Scholar]
- Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom K, Wettesten M, Boren J, Bondjers G, Wiklund O, Olofsson SO. Pulse-chase studies of the synthesis and intracellular transport of apolipoprotein B-100 in Hep G2 cells. The Journal of biological chemistry. 1986;261:13800–13806. [PubMed] [Google Scholar]
- Brent MR. Genome annotation past, present, and future: how to define an ORF at each locus. Genome Res. 2005;15:1777–1786. doi: 10.1101/gr.3866105. [DOI] [PubMed] [Google Scholar]
- Brockdorff N, Ashworth A, Kay GF, McCabe VM, Norris DP, Cooper PJ, Swift S, Rastan S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell. 1992;71:515–526. doi: 10.1016/0092-8674(92)90519-i. [DOI] [PubMed] [Google Scholar]
- Calfon M, Zeng H, Urano F, Till JH, Hubbard SR, Harding HP, Clark SG, Ron D. IRE1 couples endoplasmic reticulum load to secretory capacity by processing the XBP-1 mRNA. Nature. 2002;415:92–96. doi: 10.1038/415092a. [DOI] [PubMed] [Google Scholar]
- Calvo SE, Pagliarini DJ, Mootha VK. Upstream open reading frames cause widespread reduction of protein expression and are polymorphic among humans. Proc Natl Acad Sci U S A. 2009;106:7507–7512. doi: 10.1073/pnas.0810916106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van†Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, et al. FMRP Stalls Ribosomal Translocation on mRNAs Linked to Synaptic Function and Autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Cristofano A, Pesce B, Cordon-Cardo C, Pandolfi PP. Pten is essential for embryonic development and tumour suppression. Nature genetics. 1998;19:348–355. doi: 10.1038/1235. [DOI] [PubMed] [Google Scholar]
- dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–5044. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fresno M, Jimenez A, Vazquez D. Inhibition of translation in eukaryotic systems by harringtonine. Eur J Biochem. 1977;72:323–330. doi: 10.1111/j.1432-1033.1977.tb11256.x. [DOI] [PubMed] [Google Scholar]
- Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–840. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton TL, Stoneley M, Spriggs KA, Bushell M. TOPs and their regulation. Biochemical Society transactions. 2006;34:12–16. doi: 10.1042/BST20060012. [DOI] [PubMed] [Google Scholar]
- Hann SR, King MW, Bentley DL, Anderson CW, Eisenman RN. A non-AUG translational initiation in c-myc exon 1 generates an N-terminally distinct protein whose synthesis is disrupted in Burkitt’s lymphomas. Cell. 1988;52:185–195. doi: 10.1016/0092-8674(88)90507-7. [DOI] [PubMed] [Google Scholar]
- Hansen MB, Mitchelmore C, Kjaerulff KM, Rasmussen TE, Pedersen KM, Jensen NA. Mouse Atf5: molecular cloning of two novel mRNAs, genomic organization, and odorant sensory neuron localization. Genomics. 2002;80:344–350. doi: 10.1006/geno.2002.6838. [DOI] [PubMed] [Google Scholar]
- Hassan AS, Hou J, Wei W, Hoodless PA. Expression of two novel transcripts in the mouse definitive endoderm. Gene Expr Patterns. 2010;10:127–134. doi: 10.1016/j.gep.2010.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Huang MT. Harringtonine, an inhibitor of initiation of protein biosynthesis. Mol Pharmacol. 1975;11:511–519. [PubMed] [Google Scholar]
- Hughes TA. Regulation of gene expression by alternative untranslated regions. Trends Genet. 2006;22:119–122. doi: 10.1016/j.tig.2006.01.001. [DOI] [PubMed] [Google Scholar]
- Ingolia NT. Genome-wide translational profiling by ribosome footprinting. Methods Enzymol. 2010;470:119–142. doi: 10.1016/S0076-6879(10)70006-9. [DOI] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin B, Heck JD, Hatfield GW. Codon pair utilization biases influence translational elongation step times. J Biol Chem. 1995;270:22801–22806. doi: 10.1074/jbc.270.39.22801. [DOI] [PubMed] [Google Scholar]
- Ivanov IP, Firth AE, Michel AM, Atkins JF, Baranov PV. Identification of evolutionarily conserved non-AUG-initiated N-terminal extensions in human coding sequences. Nucleic acids research. 2011;39:4220–4234. doi: 10.1093/nar/gkr007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov IP, Loughran G, Atkins JF. uORFs with unusual translational start codons autoregulate expression of eukaryotic ornithine decarboxylase homologs. Proc Natl Acad Sci U S A. 2008;105:10079–10084. doi: 10.1073/pnas.0801590105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joachims T, Schölkopf B, Burges C, Smola A. Advances in Kernel Methods - Support Vector Learning. MIT Press; Cambridge, MA: 1999. Making large-Scale SVM Learning Practical. [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci U S A. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimchi-Sarfaty C, Oh JM, Kim IW, Sauna ZE, Calcagno AM, Ambudkar SV, Gottesman MM. A "silent" polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–528. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- Kondo T, Plaza S, Zanet J, Benrabah E, Valenti P, Hashimoto Y, Kobayashi S, Payre F, Kageyama Y. Small peptides switch the transcriptional activity of Shavenbaby during Drosophila embryogenesis. Science. 2010;329:336–339. doi: 10.1126/science.1188158. [DOI] [PubMed] [Google Scholar]
- Kopp E, Medzhitov R, Carothers J, Xiao C, Douglas I, Janeway CA, Ghosh S. ECSIT is an evolutionarily conserved intermediate in the Toll/IL-1 signal transduction pathway. Genes Dev. 1999;13:2059–2071. doi: 10.1101/gad.13.16.2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraut-Cohen J, Gerst JE. Addressing mRNAs to the ER: cis sequences act up! Trends Biochem Sci. 2010;35:459–469. doi: 10.1016/j.tibs.2010.02.006. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science. 2001;294:858–862. doi: 10.1126/science.1065062. [DOI] [PubMed] [Google Scholar]
- Lazarowitz SG, Robertson HD. Initiator regions from the small size class of reovirus messenger RNA protected by rabbit reticulocyte ribosomes. J Biol Chem. 1977;252:7842–7849. [PubMed] [Google Scholar]
- Lin FT, MacDougald OA, Diehl AM, Lane MD. A 30-kDa alternative translation product of the CCAAT/enhancer binding protein alpha message: transcriptional activator lacking antimitotic activity. Proc Natl Acad Sci U S A. 1993;90:9606–9610. doi: 10.1073/pnas.90.20.9606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PD, Harding HP, Ron D. Translation reinitiation at alternative open reading frames regulates gene expression in an integrated stress response. J Cell Biol. 2004;167:27–33. doi: 10.1083/jcb.200408003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malarkannan S, Horng T, Shih PP, Schwab S, Shastri N. Presentation of out-of-frame peptide/MHC class I complexes by a novel translation initiation mechanism. Immunity. 1999;10:681–690. doi: 10.1016/s1074-7613(00)80067-9. [DOI] [PubMed] [Google Scholar]
- Mamane Y, Petroulakis E, LeBacquer O, Sonenberg N. mTOR, translation initiation and cancer. Oncogene. 2006;25:6416–6422. doi: 10.1038/sj.onc.1209888. [DOI] [PubMed] [Google Scholar]
- Mariappan M, Li X, Stefanovic S, Sharma A, Mateja A, Keenan RJ, Hegde RS. A ribosome-associating factor chaperones tail-anchored membrane proteins. Nature. 2010;466:1120–1124. doi: 10.1038/nature09296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin KC, Ephrussi A. mRNA localization: gene expression in the spatial dimension. Cell. 2009;136:719–730. doi: 10.1016/j.cell.2009.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medenbach J, Seiler M, Hentze MW. Translational Control via Protein-Regulated Upstream Open Reading Frames. Cell. 2011;145:902–913. doi: 10.1016/j.cell.2011.05.005. [DOI] [PubMed] [Google Scholar]
- Monte D, Baert JL, Defossez PA, de Launoit Y, Stehelin D. Molecular cloning and characterization of human ERM, a new member of the Ets family closely related to mouse PEA3 and ER81 transcription factors. Oncogene. 1994;9:1397–1406. [PubMed] [Google Scholar]
- Monte D, Coutte L, Dewitte F, Defossez PA, Le Coniat M, Stehelin D, Berger R, de Launoit Y. Genomic organization of the human ERM (ETV5) gene, a PEA3 group member of ETS transcription factors. Genomics. 1996;35:236–240. doi: 10.1006/geno.1996.0345. [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatogawa H, Ito K. The ribosomal exit tunnel functions as a discriminating gate. Cell. 2002;108:629–636. doi: 10.1016/s0092-8674(02)00649-9. [DOI] [PubMed] [Google Scholar]
- Namy O, Moran SJ, Stuart DI, Gilbert RJ, Brierley I. A mechanical explanation of RNA pseudoknot function in programmed ribosomal frameshifting. Nature. 2006;441:244–247. doi: 10.1038/nature04735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson T, Mann M, Aebersold R, Yates JR, 3rd, Bairoch A, Bergeron JJ. Mass spectrometry in high-throughput proteomics: ready for the big time. Nat Methods. 2010;7:681–685. doi: 10.1038/nmeth0910-681. [DOI] [PubMed] [Google Scholar]
- Niwa H, Ogawa K, Shimosato D, Adachi K. A parallel circuit of LIF signalling pathways maintains pluripotency of mouse ES cells. Nature. 2009;460:118–122. doi: 10.1038/nature08113. [DOI] [PubMed] [Google Scholar]
- Noble WS. What is a support vector machine? Nature biotechnology. 2006;24:1565–1567. doi: 10.1038/nbt1206-1565. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. Identification of microRNAs of the herpesvirus family. Nat Methods. 2005;2:269–276. doi: 10.1038/nmeth746. [DOI] [PubMed] [Google Scholar]
- Pisarev AV, Kolupaeva VG, Yusupov MM, Hellen CU, Pestova TV. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert F, Carrier M, Rawe S, Chen S, Lowe S, Pelletier J. Altering chemosensitivity by modulating translation elongation. PLoS One. 2009;4:e5428. doi: 10.1371/journal.pone.0005428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruggero D, Sonenberg N. The Akt of translational control. Oncogene. 2005;24:7426–7434. doi: 10.1038/sj.onc.1209098. [DOI] [PubMed] [Google Scholar]
- Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–460. doi: 10.1016/j.stem.2008.03.013. [DOI] [PubMed] [Google Scholar]
- Schneider-Poetsch T, Ju J, Eyler DE, Dang Y, Bhat S, Merrick WC, Green R, Shen B, Liu JO. Inhibition of eukaryotic translation elongation by cycloheximide and lactimidomycin. Nat Chem Biol. 2010;6:209–217. doi: 10.1038/nchembio.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starck SR, Ow Y, Jiang V, Tokuyama M, Rivera M, Qi X, Roberts RW, Shastri N. A distinct translation initiation mechanism generates cryptic peptides for immune surveillance. PLoS One. 2008;3:e3460. doi: 10.1371/journal.pone.0003460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner DR, Cariello DA, Woolstenhulme CJ, Broadbent MA, Buskirk AR. Genetic identification of nascent peptides that induce ribosome stalling. The Journal of biological chemistry. 2009;284:34809–34818. doi: 10.1074/jbc.M109.039040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenson T, Ehrenberg M. Regulatory nascent peptides in the ribosomal tunnel. Cell. 2002;108:591–594. doi: 10.1016/s0092-8674(02)00669-4. [DOI] [PubMed] [Google Scholar]
- Tremml G, Singer M, Malavarca R. Culture of mouse embryonic stem cells. Curr Protoc Stem Cell Biol. 2008 doi: 10.1002/9780470151808.sc01c04s5. Chapter 1, Unit 1C 4. [DOI] [PubMed] [Google Scholar]
- Tscherne JS, Pestka S. Inhibition of protein synthesis in intact HeLa cells. Antimicrob Agents Chemother. 1975;8:479–487. doi: 10.1128/aac.8.4.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Tupy JL, Bailey AM, Dailey G, Evans-Holm M, Siebel CW, Misra S, Celniker SE, Rubin GM. Identification of putative noncoding polyadenylated transcripts in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2005;102:5495–5500. doi: 10.1073/pnas.0501422102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wolin SL, Walter P. Ribosome pausing and stacking during translation of a eukaryotic mRNA. EMBO J. 1988;7:3559–3569. doi: 10.1002/j.1460-2075.1988.tb03233.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagitani K, Imagawa Y, Iwawaki T, Hosoda A, Saito M, Kimata Y, Kohno K. Cotranslational targeting of XBP1 protein to the membrane promotes cytoplasmic splicing of its own mRNA. Mol Cell. 2009;34:191–200. doi: 10.1016/j.molcel.2009.02.033. [DOI] [PubMed] [Google Scholar]
- Yanagitani K, Kimata Y, Kadokura H, Kohno K. Translational pausing ensures membrane targeting and cytoplasmic splicing of XBP1u mRNA. Science. 2011;331:586–589. doi: 10.1126/science.1197142. [DOI] [PubMed] [Google Scholar]
- Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–280. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.