Abstract
Most proteins do not function on their own but as part of large signaling complexes that are arranged in every living cell in response to specific environmental cues. Proteins interact with each other either constitutively or transiently and do so with different affinity. When identifying the role played by a protein inside a cell, it is essential to define its particular cohort of binding partners so that the researcher can predict what signaling pathways the protein is engaged in. Once identified and confirmed, the information might allow the interaction to be manipulated by pharmacological inhibitors to help fight disease.
Key words: cell signaling, protein complexes, protein-protein interactions, affinity tagging, co-immunoprecipitation, peptide array technology
In this review, we discuss protein-protein interactions and how they are essential to propagate signals in signaling pathways. We examine some of the high-throughput screening methods and focus on the methods used to confirm specific protein-protein interactions including; affinity tagging, co-immunoprecipitation, peptide array technology and fluorescence microscopy.
Introduction to Protein-Protein Interactions
Signaling cascades are intrinsic to virtually every biological process. Deciphering these signaling cascades is fundamental to generating a comprehensive and complete understanding of the biological process. This information is essential for novel therapeutic and diagnostic strategies targeted to the diseased state. Central to every signaling cascade is protein complex formation which is based on the ability of proteins to interact with other proteins, nucleic acids (DNA and RNA), small molecules and metals.
The human genome codes for over 500,000 different proteins and it is estimated that over 80% of these proteins function as part of protein complexes in signaling cascades and not as individual proteins.1 In Saccharomyces cerevisiae, over 70% of proteins in the cell have been shown to have interacting partners.2,3 Proteins have enzymatic, structural and regulatory functions and when they bind to other proteins they establish highly regulated functional protein complexes. In untangling these protein complexes, it is essential that we gain an understanding of how proteins are assembled and apply systematic and unbiased approaches to define how proteins in complexes are regulated. When protein complexes are formed in signaling cascades, protein interactions enable one protein to modify another protein in the complex and thus transmit biological information. Often, the recruitment of one protein into a cascade facilitates the recruitment of the next, thus building systematic linear processes.
Central to signaling cascades is the recruitment of signaling hubs. The term ‘hub’ is now widely used to describe a protein that participates in several different signaling pathways and can bind several different proteins. Hub proteins are highly expressed and their expression pattern usually follows that of their binding partners. They function as scaffolding/adaptor proteins and generally do not possess enzymatic activity. These proteins usually have the ability to interact with several different proteins at the same time. The proteins that interact with signaling hubs fall into two broad categories: constitutively bound proteins, and those that are stimulus-dependent or transient. Constitutive protein-protein interactions are predominantly strong interactions found in the M range and usually have a larger interaction interface than transient interactions (reviewed in ref. 4). Transient interactions are usually weak (µM range) and usually arise as a result of posttranslational modification of one or both of the proteins involved in the interaction. Transient interactions are critical to signal propagation. In a transient protein-protein interaction, one or both of the proteins in the interaction undergo conformational change which may reveal a binding site for the next interacting protein. The most important feature of protein-protein interactions is that they are dynamic. They are components of specific signaling complexes and are assembled in response to environmental cues. In this way, the cell deciphers information about conditions outside the cell so that it can generate a precise response. When bound to each other either transiently or constitutively, proteins have the opportunity to modify each other and transmit biological information in a flow pattern that usually (but not always) converge at the cell nucleus whereby they terminate by coding for expression of particular gene sets.
The objective here is to discuss several different high throughput screens and discuss the procedures involved in designing the appropriate verification methods which can be applied to investigate protein-protein interactions.
Protein-Protein Interactions in Signaling Cascades
Protein-protein interactions and signaling pathways are highly conserved. For example, several hundred protein-protein interactions in yeast have also been identified for the corresponding protein orthologs in worm,5,6 and the signaling pathways that control chemotaxis in bacteria have very clear similiarities to the processes that control cell migration in immune cells in higher species.7–10 As well as the signaling pathways, the rules dictating protein-protein interactions and complex formation are highly conserved. The past 15 years has seen the development of a large number of methods which have propelled our understanding of signaling cascades by helping us identify and characterize protein-protein interactions and signaling networks. These methods are practiced or accessed in laboratories all around the world. In deciphering signaling pathways and in establishing the cohort of proteins that interact with a specific protein, there are several approaches that one can take. Computational methods can help predict protein interactions, however most investigators start by using high-throughput proteomics approaches to define protein-protein interaction sets. The logical approach is to then employ assays to confirm protein-protein interactions in several different ways before specific protein-protein interactions can be refined to binding interfaces. These methods rely heavily on the genetics and biology of bacteria, yeast (in particular S. cerevisiae and S. pombe), C. elegans, Drosophila, mice and human cell lines. When reporting protein-protein interactions, it is important that the interactions are characterized by more than one method, an essential step in validating the specific protein-protein interaction. As we will discuss, there are several pitfalls and areas where false-positive results can be obtained. Once an interaction is defined, several further functional experiments such as mutational analysis and knockdown of protein expression by RNA interference must be performed to determine the hierarchy of the particular protein interaction to determine which direction the signal is conveyed in the signaling pathway. From a pharmacological perspective, it is essential that we develop a comprehensive understanding of the particular protein-protein interaction and develop a clear understanding of what the interaction means in the context of cellular function. Achieving this for specific protein-protein interactions may take several years, even decades.
There are several different strategies that one can use in identifying protein-protein interactions. The best are those that employ several different methods to distil and refine a particular protein-protein interaction. In this review, we will focus on the combination of methods best suited to identify protein-protein interactions paying particular attention to the best approaches to take to verify them. We will also focus on peptide array technologies, a powerful and relatively accessible tool used to decipher the specific interfaces on the proteins involved in protein-protein interactions.
Things to Consider before Planning a Protein-Protein Interaction Study
The presumption is that we can elucidate the function of a protein if we identify the particular cohort of proteins that interact with it at any one time. In most cases this is true because there is enough information in the literature to predict with a high degree of certainty the signaling pathways that your test protein is involved in based on the composition of the binding cohort. However, no individual experimental approach can sufficiently reveal all the critical components of protein complexes in signaling pathways. You must first consider what system best supports the function of your protein. For example, if your test protein is predicted to be a nuclear protein or embedded in a lipid bilayer, you must ensure that the buffers you choose are sufficient to adequately expose these complexes. In a protein complex, most proteins will be linked by non-covalent protein-protein interactions and as mentioned earlier, there are varying degrees of stability in this binding. Buffers and techniques used to lyse cells such as sonication, freeze-thaw action etc. are inherently disruptive to complexes so it is very important that methods are chosen that best protect these complexes to ensure that the screen is as comprehensive as possible. One might also be looking for proteins that interact with a test protein after a particular stimulus, such as growth factor stimulation, or the investigator might be trying to identify the proteins that interact with a test protein after treatment with a pharmacological compound. In any screen, it is critical that a library is chosen that provides the broadest and most complete coverage possible of the expressed genes or proteins. For example, when searching broadly for binding partners using yeast-two-hybrid (Y2H) assays, researchers might employ cDNA derived from mouse embryonic stem cells or human fetal brain. However, if a researcher wants a more refined screen and has an idea that their test protein functions as a membrane protein, a membrane protein library might be used. When planning large screens, costing and time factors must be considered too.
There are a series of computational approaches that can be employed to help predict protein-protein interactions (reviewed in ref. 11–13) and identify protein-protein interaction motifs.14 Several databases are in place that store information obtained in high throughput methods such as Y2H and Tandem Affinity Purification followed by Mass Spectrometry (TAP-MS) and information obtained by mining through the literature. Databases, such as the Biomolecular Interaction Network Database15,16 and the Database of Interacting Proteins (DIP) 17,18 store full descriptions of molecular complexes and pathways, and store very useful experimentally determined information on protein-protein interactions. They can also help predict transient protein-protein interactions which often elude biochemical methods.4,19,20 A full list and analysis of protein interaction databases has been compiled by Shoemaker et al.12,21,22 Databases have now expanded to include information on protein-protein interaction interfaces (reviewed in refs. 22–25). Consulting databases like these are well worth the effort and the networks that are deciphered by these computational routes can save precious time during further functional analyses.
Detecting Protein-Protein Interactions and Complexes
Before we discuss some of the high throughput systems, it is important to point out that there are multiple processes that can be employed to obtain information about the complexes and signaling pathways that a protein contributes to (reviewed in refs. 1 and 12). For example, DNA microarray systems are commonly used. These microarrays display genome wide expression patterns as DNA spots immobilized on a solid support.26 The idea for DNA microarrays grew from a desire to compare genes that are activated or suppressed (‘up or down’) between two populations of cells and the technique is now being used as a system to examine co-expression of genes. This is based on studies which have shown that proteins that are involved in the same complex are co-expressed and so identifying co-expressed genes is a good predictor of permanent protein interactions.27–30 The system has now evolved to support protein arrays31 which is a major expansion to the array technology. With protein arrays we now have the opportunity to study protein interactions and determine interaction interfaces as well as studying the enzymatic activity of proteins. This has broken down several barriers that existed in protein complex analysis because the system lends itself to automation and miniaturisation (saving costs), but also supports the investigation of protein-DNA, protein-lipid and protein-protein interactions. The potential of this system is now being harnessed by several pharmaceutical companies because it can be used to screen for compounds that both bind to proteins but also for compounds that inhibit protein-protein interactions, and to search for biomarkers for disease.32 The system has its drawbacks however, for example, the immobilization chemistry results in a random orientation of the displayed proteins and so they may not properly represent protein interfaces and may in fact compromise biological activity.33–35 It is also very difficult to replicate post-translational modifications in the system which results in many transient protein-protein interactions being missed. Several adaptations of the protein array system, such as suspension bead arrays, have the potential to circumnavigate this problem36 and studies investigating post-translational modifications of particular residues such as lysine have been successful and are being used to study ubiqutin (Ub) ligases, SUMOylation E3 ligases and acetyltransferases.37 Advances in peptide synthesis on nitrocellulose support have also helped overcome some of these challenges and is discussed later in the review. Many laboratories still rely on more classical approaches such as synthetic lethality,38 Electrophoretic Mobility Shift Assays (EMSAs) and Far-protein gel blotting. These are all excellent systems and many are mounting a comeback as a first stop tool to analyze protein interactions as revamped techniques. For example, fluorescence EMSAs are now generating information on the binding affinity in protein-protein and protein-nucleic acid interactions (reviewed in refs. 39 and 40).
The composition of signaling complexes can also be studied by X-ray crystallography which is quickly helping to build a clearer picture of how proteins interact. This powerful method has been very influential in placing specific proteins in complex with ribosomes and X-ray crystal structures have now been obtained for complete eukaryotic ribosomal subunits.41,42 This powerful method has its limitations, primarily because X-ray crystallography does not define a unique protein-protein interface, and distinguishing the absolute biological interface in the structure is difficult (discussed in ref. 43). However, in combination with computational methods, protein structure prediction and other bioinformatics tools, protein-protein interaction interfaces can be predicted with a higher degree of certainty. Recent advances and applications of these methods emphasize the benefit of lateral thinking and the application of hybrid approaches that combine a variety of different tools to achieve better efficiency, accuracy, resolution and more comprehensive screens. The remainder of this discussion is focused on describing some of the ‘workhorses’ and validation procedures used to study protein-protein interactions, while focusing on the advantages and disadvantages of each technique. A summary table of the advantages and pitfalls associated with the discussed procedures is included (Table 1).
Table 1.
Comparison of some of the different experimental methods used to study and verify protein-protein interactions
| Experimental Method | Advantages | Problems/pitfalls |
| Protein microarrays | High throughput. Lends itself to automation. Can be miniaturized. Supports protein-DNA studies. Supports protein-drug studies. |
Protein immobilization. Replication of post-translational events. Loss of transient protein interactions. In vitro assay. |
| X-Ray Crystallography | High Resolution. Reveals information on specific atoms. |
Defining the absolute interaction interface is difficult. |
| Yeast Two-hybrid | Robust and relatively accessible. Inexpensive. High throughput. Excellent verification tool. In vivo assay. |
Can't be applied to all interactions. Bait proteins can be toxic. Bait proteins can auto-activate genes. Interactions based on post-translation modifications are missed. High percentage of false positives. Detection of transient interactions. |
| Affinity tagging and pull down assays | Several robust tags are available Several antibody combinations can be used. Can be applied to bacterial, yeast and mammalian systems. Very robust. Compatible with several biological processes such as MS. Can be used to identify protein complexes. High throughput. |
Affinity based. Success based on the quality of the antibodies used. Loss of weakly bound proteins. Tags can interfere with protein function. High percentage of false positives. Expensive in comparison to other methods. In vitro assay. |
| Co-immunoprecipitation | Excellent verification procedure. Proteins are in their native state. Proteins are in their native concentration. Suitable for overexpression assays. Relatively inexpensive. Very reproducible. Can be combined with affinity tagging. Supports studies addressing catalytic activity. Compatible with several biological processes such as MS. |
Several different control combinations are required. Success highly dependent on cell stimulus and lysis procedure. Only high affinity interactions are detected. Most transient interactions are lost. Does not differentiate between direct and indirect interactions. Can be expensive if used as a screening system. |
| Peptide Array Technology | Can be used to map protein interactions to the specific amino acids involved. Peptide support systems are robust. Protein surfaces can be accurately represented. Direct synthesis of peptide by SPOT synthesis and photolithography. |
At least one protein must be available as a recombinant protein. Expensive. Very specific equipment required. Peptide purity. |
| Fluorescence Microscopy | Very flexible, can accommodate many different flurophore combinations. Supports in vivo studies. Very compatible with affinity tagging. Will yield information on the cellular location of the interaction. Can be used to analyze tissue samples. |
Expensive. Very specific equipment required. Success based on the availability of highly specific antibodies. Specific expertise required. |
Two-Hybrid Methods
Yeast two-hybrid (Y2H) methods/reporter assays are one of the most common techniques used to screen for and identify protein-protein interactions.44–46 The system is based on the properties of GAL4 and other site-specific transcription factors in yeast,47 and has been extensively reviewed in reference 48–50. In the assay, two putative interacting proteins X and Y are fused to the DNA binding domain (BD) and transcriptional activation domain (AD) of the transcription factor. The proteins are then co-expressed in a yeast strain which contains the specific DNA-binding site of the transcriptional activator upstream of a reporter gene. The reporter gene of choice is usually the Escherichia coli lacZ gene because its activation is easily detected by the growth of blue colonies when the cells are grown in growth media containing 5-bromo-4-chloroindol-3yl β-D-galactopyranoside (X-gal). The reporter gene is only transcribed when the BD and AD of the transcription factor come together to reconstitute the activator and transcribe the reporter gene. When employing a Y2H screen, the “bait protein” (test protein) is attached to the BD to generate BD-X and the “prey” protein is attached to the AD to generate AD-Y. The BD-X bait-protein is cloned into an expression plasmid which is transfected into a yeast cell that contains a “library” of cDNA clones. These clones contain the AD-Y chimeric protein which has been generated in the same way. Most laboratories now use commercially available Y2H kits with pretransformed libraries. There are several variations of the Y2H system (reviewed in ref. 51) such as one-hybrid screens which are designed to investigate protein-DNA interactions, and three-hybrid screens which have been developed to identify proteins that interact with RNA.52–55 Some successful attempts have also been made to adopt the three-hybrid system to investigate ternary protein complex formation that involves three proteins, the concept being that it is possible to trap the interaction of two proteins which occurs only in the presence of a third protein.55,56 In all two-hybrid screens, either a matrix approach or a library approach can be taken (reviewed in ref. 12).
Y2H screens cannot be applied to all protein-protein interaction screens and even when it can be applied, it has several limitations. A critical first step in the process is to check that the bait protein; (A) is not toxic when expressed in yeast and (B), that it does not autonomously activate the reporter gene (autoactivation). Either of these is a severe handicap, and may result in designing new strategies based around using a truncated form of the bait protein. Truncated proteins obviously lack important sequences of the parent protein and will reduce the ‘coverage’ and efficacy of the system. Having a known positive interacting protein for the bait is an advantage but is not always possible. Other disadvantages to the system are that the bait protein will not be post-translationally modified in the yeast and the bait protein may not fold properly. Again this is an insurmountable challenge, however very exciting developments are taking place in the area of mammalian two-hybrid screening which has been adapted for high throughput screening.57 These screens have a particular advantage in that the expressed proteins are subjected to the essential post-translational modifications which are very important for transient protein-protein interactions.58,59 The main problem with Y2H screens (as with most high throughput screens) is the percentage of false positives where estimates of up to 80% false negatives and up to 60% false positives are associated with this high throughput screening.60–65 To address this, it is essential that once two interacting proteins are identified by Y2H, the interaction is verified by several other biological methods. Two-hybrid systems are themselves an excellent verification system for a specific protein-protein interaction if the interacting protein is cloned directly into the prey AD vector. In addition, the system supports the use of specific point mutations to determine the critical amino acids required for an interaction (reviewed in refs. 49 and 66–68).
Affinity Tagging and Pull Down Assays
Affinity tags are used to purify proteins from bacteria, yeast and mammalian cells. The system is based on transfecting cells with plasmids encoding a bait protein and a tag sequence resulting in the expression of a protein with a tag fused at either the N- or C-terminus of the bait protein. The tagged protein is expressed in the cells, often controlled by an induction system based on nutritional selection,69,70 or the presence of molecular compounds such as Isopropyl β-D-1-thiogalactopyranoside (IPTG),71 Tetracycline72 or temperature.73,74 When the proteins are expressed, the cells are lysed and the tagged protein is “pulled down” or “pulled out” using antibodies against the tag, antibodies against the protein or by using solid supports that bind the tag.
Before describing pull down assays, it is important to mention some of the tagging systems that can be used. Most tags are easily available, and even if they are not readily available, they can be generated by routine polymerase chain reaction (PCR) and simple DNA cloning techniques. Tagging a protein using tags is essential if there is no antibody raised against the studied protein. Using tags like the tags discussed below, have proven essential in deciphering protein function particularly in mammalian expression systems. There are several different tags that can be used. The FLAG-Tag (N-DYK DDD DK-C) is a short tag consisting of 8 amino acids that can be added to either the N or the C-terminus of a protein using recombinant DNA technology. This particular tag was one of the first functional tags to be used in protein biochemistry75–78 and was patented by Hopp et al. (US Patent Number 4,703,004) in 1987. The Myc-Tag (N-EQK LIS EED L-C) is a polypeptide protein tag that can be fused to either the N-terminus or the C-terminus of a protein which was derived from the c-myc gene and the HA-Tag (N-YPY DVP DYA-C) is derived from haemagglutinin and again can be inserted at either the N-terminus or the C-terminus of a protein. All of these tags are excellent for use in mammalian expression systems and are easily applied to the N- and C-terminus of genes in expression vectors. The FLAG-Tag is hydrophobic in nature giving it a distinct advantage over other tags because it is less likely to disrupt the proteins that it is attached to. However, Myc-Tags and HA-Tags are the tag of choice by most laboratories because there are a series of excellent antibodies that recognize them cleanly by immunofluorescence and by protein gel blotting after SDS PAGE. The tags also ensure that the tagged proteins can be immunoprecipitated easily from total cell lysates in pull down assays. There are other tags that are more suitable for recombinant proteins and affinity chromatography. The His-Tag is a polyhistidine-tag that consists of at least five Histidine (His) residues, which can be applied at the N- or C-terminus of the protein. It is also known as the 6xHis-tag. In this tag, it is possible to vary the number of histidine residues and insert a suitable amino acid sequence that facilitates removal of the polyhistidine-tag using endopeptidases after protein pull down. Other tags suitable for isolating recombinant proteins from bacterial expression systems include Maltose Binding Protein tag (MBP-Tag) and Glutathione-S-Transferase tag (GST-Tag). There are other tagging systems that are often used for real time visualization of proteins in cells using microscopy. These fluorescent protein tags exhibit bright fluorescence when exposed to specific wavelengths of light. The green fluorescent protein (GFP) tag was first isolated from the jellyfish Aequorea victoria79 and several derivatives are now being used including cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP). The GFP tag has had a dramatic effect on protein visualization in mammalian cells, plants, Drosophilia, yeast and prokaryotic cells (reviewed in refs. 80–84) and is discussed in more detail later when we describe the use of fluorescence microscopy as a validation tool in protein-protein interactions.
In affinity assays, the protein can be pulled down for analysis or for purification, for example, a GST-protein. However, in most cases the tagged protein can be used as a tool to ‘fish out‘ other proteins that might interact with the test protein by affinity chromatography. A typical experiment might be that the protein is expressed in bacteria, the bacteria lysed and the protein purified and bound in a column to a solid support or ligand linked to a solid support (for example, 6xHis-Tag binds to nickel beads, GST-Tag binds to Glutathione beads). Then a lysate from mammalian cells might be passed through the column. Proteins that have a high affinity for the immobilized protein will bind, while non-specific interacting proteins will be washed through. Bound proteins are then eluted and separated by gel-electrophoresis and identified by protein gel blotting (if antibodies are available in the lab which recognize candidate proteins) or by Mass Spectrometry (MS). MS is providing quantitative identification of protein-protein interactions and is making a dramatic contribution to deciphering protein-protein interactions and mapping signaling proteins when coupled to an appropriate pull down system (reviewed in ref. 85). Expressing the tagged protein in mammalian systems has advantages because post translational modifications of the bait protein are more likely to occur and binding will be performed directly in the cell. It is also very likely (though not certain) that the tagged protein will be directed to the correct cellular location where it can interact with its physiological target. If this system is employed, then capturing the affinity tag with solid support or ligand linked to a solid support is done to wash away non-specific interactions. Bound proteins will be eluted and identified as described above.
Affinity tagging has the distinct advantage over the Y2H system in that it can detect interactions involving more than two proteins and can be used to detect protein complexes if followed by MS.1,86 However, it too has limitations. Depending on the lysis procedure used, nonphysiological targets might be exposed and may be artificially incorporated into the complex and identified as positive interacting proteins. Affinity tagging is also biased toward high affinity interactions and there is always the risk that the Tag will interfere with protein folding and function. False positives can be reduced by washing the immobilized complex with buffers with increased concentrations of detergents and salts but stringent verification techniques must be applied. The Tandem Affinity Purification (TAP) method of purification of protein complexes is now the preferred method of choice for identification of native protein complexes. The method comprises of overexpressing a dual-tagged target protein in host cells, isolation of the fusion protein using two binding steps and then identifying co-bound proteins by MS.1,87–90 Because two tags are expressed with the protein, there are lower levels of non-specific binding and increased complex recovery. The system also benefits from being compatible with several protein identification processes such as native-PAGE, 2D-electrophoresis, protein gel blotting and MS.91–93 Limitations in the process include loss of weakly bound interacting proteins and there is always the possibility that the tag will interfere with folding and function of the protein disrupting how the protein interacts with other proteins.
When using affinity tags, there are several things to consider which may overcome some of the limitations associated with the study and help improve the results. For example, designing baits where the tag is inserted at the N-terminus as opposed to the C-terminus of the protein often addresses the problem of altered stability of the fusion protein. One might also consider generating different tagged versions of a protein and compare the sub cellular location of each. Crosslinking agents are also useful. There are several types of crosslinking agents including chemical crosslinkers, thermoreactive crosslinkers and photo-reactive crosslinkers. These agents can assist the capture of protein-protein interactions and protein complexes by covalently bonding the proteins together when they interact. Crosslinking reagents are used extensively and in combination with MS, are a very powerful tool.1,94–96
High throughput screens provide an excellent start point for determining what cohort of proteins interact with the test protein and in combination with bioinformatics and computational tools they will allow the researcher to predict with a high degree of certainty what signaling pathways the test protein is involved in. It is essential then to move to a series of validation systems to confirm an interaction and also to identify the binding interface between the two proteins. Once the interaction is confirmed, it will then be possible to investigate how the interaction responds to environmental cues and determine how the specific protein-protein interaction is altered in the diseased state.
Verification of Protein-Protein Interactions
Co-immunoprecipitation.
Co-immunoprecipitation (co-IP) is the term given to the process where a protein is extracted from a whole cell lysate using a precipitating antibody which is conjugated to some form of resin (usually Protein A or Protein G). The process is based on the principal of immunoprecipitation (IP) and it can be used as a first stop in detecting protein-protein interactions but is best used as a verification procedure after a high throughput screening process such as two-hybrid analysis, affinity chromatography or MS.86,97
The technique is used to enrich a whole cell lysate for a specific protein, and in the process, ‘pull down’ any other proteins that are associated with that protein. In this technique, the protein being isolated is the antigen and it is essential to have access to an antibody that binds to that antigen in its native form. If antibodies against the test protein are not available, an alternative strategy is to generate a tagged form of the protein (described above) and the protein can be immunoprecipitated with antibodies directed against the tag. To form the antigen-antibody complex, whole cell extracts are prepared which are then incubated with antibodies that recognize the protein being isolated. After some time (for example; 1 h at room temperature or 3 h at 4°C), protein A or protein G resin is added to capture the antibody-antigen complex. The resin binds to the antibody when the mixture is incubated at 4°C with gentle agitation. Centrifugation will capture the resin-antibody-antigen complex and the immunoprecipitate can be washed with buffers with varying salt concentrations to reduce non-specific binding of proteins to the immunoprecipitated complex. The complex is eluted from the beads and the proteins can be identified by protein gel blotting after SDS PAGE, or alternatively, if the co-IP procedure is being used as a first step in the protein-protein interaction study, the elution can be analyzed by MS.
During the entire process, the proteins are maintained in their native state and in their native concentration (unless tagged version of a protein are introduced) which maximizes trapping the cohort of interacting proteins. The process is relatively inexpensive and is very reproducible and the technique can be applied to purify protein complexes from whole cell lysates, tissue preparations and serum samples. The process can be adapted into an assay to assess the binding of two recombinant proteins. The co-IP process is an excellent verification tool. If two proteins are found to interact by a high throughput screen, then acquiring antibodies to both proteins and setting up a co-IP can validate the interaction. The co-IP can then be used to map the interaction of two proteins. For example, if a tagged version of one of the proteins is generated and the interaction confirmed, then truncation mutations and specific point mutations of the tagged protein can be expressed to assess whether the interaction is lost or amplified by the mutation. This is also an excellent approach to take to assess whether the interaction between two proteins is dependent on catalytic activity of one protein in the complex. The co-IP method does have disadvantages however, and like most of the techniques described in this review, the main problem is with contaminants and false positive interactions. Control tests have to be performed to assess antibody specificity and antibody affinity, and controls must be included in every experiment to identify and characterize non-specific binding events. It is also likely that some proteins may interact with the resin used to purify the complex which can be assessed with the appropriate controls.86,97 During the co-IP experiment, it is essential to protect the proteins from digestion and degradation and protect phosphorylation events from decay to maximize the potential to trap transient interactions. For this reason, it is very important to carry out detailed strategic planning of the experiment and to consider the appropriate lysis buffers, protease and phosphatase inhibitors and antibody concentrations to be used.
Peptide array technology.
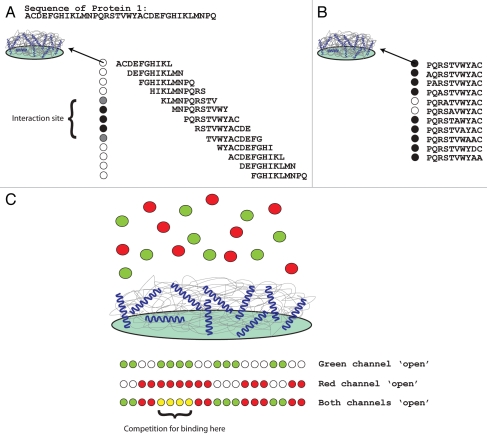
Peptide array technology is growing in popularity as a verification procedure for protein-protein interactions. Here we will discuss the application of peptide arrays to validate and map protein-protein interactions, however peptide arrays can be used for antibody purification and epitope mapping, affinity purification of proteins, analysis of enzyme substrates and protease inhibitors and to screen for drugs that interact with a particular protein. The concept of peptide synthesis is not new98 however advances in the chemistry and synthesis methods have significantly improved peptide folding, peptide presentation and peptide purity and have helped support automation of the technique. Protein-protein interactions are very dependent on the exposed surface which ultimately presents a ‘binding interface’ to the partner protein. Correctly representing this interface in the context of a peptide is the challenge in peptide synthesis. There are several solid support systems used to immobilize peptides including cellulose sheets and glass (reviewed in refs. 99 and 100). Peptides can be synthesized and then covalently attached to a solid support, or peptides can be synthesized directly on the solid support. SPOT synthesis and photolithography are the two main processes which are employed to generate peptide arrays directly on the solid support although SPOT synthesis has become the method of choice for most researchers.100 There are several commercial companies that supply protein arrays or deal in the instrumentation to make protein arrays.100 During SPOT synthesis, a mini reaction chamber is created when a droplet containing the modified amino acid is placed onto the support. Subsequent amino acids are added to the same area of the support and the peptide chain grows using Fmoc solid phase synthesis.99 The result is chains of peptides synthesized on the solid support that have a high chance of folding like they do in the native protein. Protein interactions can be verified by immobilizing one protein as a series of overlapping peptides synthesized on a solid support and overlaying with a recombinant version of the second protein. Antibodies to the second protein are then used to detect the presence of the protein on the array. In this way, not only are you achieving verification of the protein-protein interaction, but the procedure will reveal the binding site of the overlayed protein on the immobilized protein (peptides) (Fig. 1A). Another clear advantage of peptide arrays is that they can be used to map protein interactions to specific amino acids. Once the binding site of a protein is defined to a region of a protein, alanine substitution (sometimes called alanine walking) can be used to determine the critical amino acid in the interaction. To do this, a new array is generated based on sequences of the parent (first scan) sequence. In this array, peptide 1 is the parent peptide, peptide 2 will be almost identical to the parent apart from the first amino acid which is changed to an alanine. The subsequent peptide has the second amino acid changed to an alanine while changing the first amino acid back to correspond to the parent peptide and so on, until an array is generated where each peptide differs from the parent peptide by one amino acid. Overlaying with a recombinant version of the second protein together with antibodies to the second protein detects the presence of the protein on the array (Fig. 1B). In this way, not only are you achieving verification of the protein-protein interaction, but the procedure will reveal the exact binding site of the overlayed protein on the immobilized protein (peptides). In combination with an Odyssey® infrared imager (LI-COR Biosciences) competition between two proteins for binding to the same immobilized protein (peptide) can be investigated. This is based on using secondary antibodies conjugated with dyes that are detected at different wavelengths (color detection) (Fig. 1C).
Figure 1.
Using peptide arrays to study protein-protein interactions. (A) When 2 interacting proteins have been identified, the interaction site of one of the proteins on the other can be identified if one of the proteins is available in a purified recombinant form. To do this, protein 1 is synthesized on a cellulose support using SPOT synthesis. The protein is made in short overlapping peptides (in this figure, a simulated protein sequence is shown which is then synthesized as a set of overlapping 10-mer peptides which are shifted to the right by 2 amino acids). When overlayed with recombinant protein 2, the binding site of the overlayed protein on the immobilized protein (peptides) will be revealed. (B) To refine the binding site and identify the specific amino acids in Protein 1 required for the interaction of protein 2, an alanine substitution peptide array is synthesized. To do this, a parent peptide (or several) is chosen from the first screen (A), in this case; PQRSTVWYAC from protein 1. New immobilized peptides are synthesized but in this case, each peptide differs from the parent peptide by 1 amino acid (each amino acid is subsequently changed to an alanine, alanines are changed to aspartic acid). The array is overlayed with protein 2 as described above, and the critical amino acids required for the interaction are revealed (ST from the example shown). (C) When 2 proteins are suspected to compete for binding to another protein, both competing proteins can be overlayed in equal concentration on an immobilized protein (peptide). One protein can be conjugated to a green dye, one protein can be conjugated to a red dye. In combination with an Odyssey® infrared imager (LI-COR Biosciences) competition between two proteins for binding to the same immobilized protein (peptide) can be investigated. ‘Green spots’ indicate a binding preference for protein 1, ‘red spots’ indicate a binding preference for protein 2 and ‘yellow spots’ indicate that the two proteins compete for binding to the immobilized peptide sequence.
There are numerous aspects of the process that need to be considered before employing the method and careful consideration must be given to peptide design and to the screening/reporter assay to be used. In most cases, assay signals are detected directly on the array using standard antibody based detection or by fluorescence scanning. The main disadvantage of the SPOT process is the question surrounding the purity of the peptides. As the peptides are synthesized on a solid support, they do not undergo purification and so the purity and yield cannot be accurately predicted. However reports indicate that the quality is as high as standard solid phase techniques (reviewed in ref. 100).
Fluorescence microscopy.
Microscopy has become an essential confirmatory tool for investigating protein-protein interactions and many different types of microscopy are employed including; Widefield, confocal, 2-photon and Total Internal Reflection Fluorescence (TIRF) microscopy (reviewed in ref. 100–104). The principle of most microscopes is an adjustment on the basic epifluorescence (widefield) microscope: reflected light fluorescence. Light is emitted from a light source from above (or below in inverted microscopes) through the objective lens. The light is reflected by a dichroic mirror onto the sample and the energy is absorbed by fluorescent molecules (fluorophores). Once the fluorophore absorbs the photon of light, it causes an electron to transition to a higher orbital producing a molecule in an excited state.101 In order to return to the ground state, the electron must lose the excess energy. It can accomplish this by radiative transmission—emitting fluorescent light at a longer wavelength than it absorbed.101 The dichroic mirror is extremely important for the principle of fluorescent microscopy. The mirror reflects the light used in the excitation beam down onto the sample while allowing the emitted light to pass through the mirror and into the objective lens or camera due to its longer wavelength.105
Fluorophores used for fluorescence microscopy for investigating protein-protein interactions can be used indirectly (as a tag on a secondary antibody) or directly (as a tag on a primary antibody, or by using fluorescent protein tags, e.g., GFP). Green fluorescent protein (GFP) is a naturally occurring fluorophore produced by the jellyfish Aequorea victoria.106,107 which has had such a wide reaching impact on the world of biological research, particularly in the study of protein-protein interactions. The success of GFP is due to nature of the protein: either alone or expressed in a genetic fusion with another protein, a visible fluorescence is observed without the need for a cofactor; only oxygen (O2) is required.108,109 Various mutants of GFP have been made over the years to generate blue fluorescent protein (BFP),110 cyan fluorescent protein (CFP) 110 and yellow fluorescent protein (YFP).111 Extensive mutagenesis studies were performed to find a ‘red’ GFP but were unsuccessful,112 however, in 1999, a red fluorescent protein (RFP) was isolated from a non-bioluminescent reef coral (class Anthozoa, genus Discosoma).113 The use of these fluorescent proteins as a tag for a protein of interest is a major advantage. Once the tagged proteins are transfected into cells, the cellular localization of the protein can be monitored by fluorescence microscopy without the need to undertake further processing of the sample.108 This makes them equally useful in both fixed and live samples109 and very useful for colocalization studies, Fluorescence (or Förster) Resonance Energy Transfer (FRET) and Bioluminescence Resonance Energy Transfer (BRET).
Colocalization of proteins in a sample is indicated by an overlap of two colors in one pixel (or one 3-dimensional voxel) (e.g., red and green signals overlap to form yellow). Care should be taken with the choice of multiple fluorophores to ensure the chosen ones are excited by different wavelength lasers in order to decrease crosstalk or ‘bleed-through’ between colors.114 Resolution of the image captured is extremely important for colocalization studies: resolution decreases as voxel size increases resulting in separate structures merging to appear as one.115 This can greatly affect colocalization studies and lead to overestimation of the level of protein-protein interactions occurring or result in false localization of interactions being reported. Basic epifluorescence (widefield) technology can produce images with high background and low resolution resulting in erroneous reports of colocalization in the samples. For this reason confocal microscopy is used for studies of protein-protein interactions. There are three types of confocal microscopy: laser scanning (LSM),105 Nipkow disk116 and slit-scanning114 and the practical guidelines for fluorescence microscopy are extensively reviewed in references 101, 105, 117 and 118. Although the use of LSM has brought advantages to the field of fluorescent microscopy such as increased resolution and decreased background fluorescence compared with widefield, there are also several disadvantages including photobleaching and phototoxicity due to the length of time taken to scan samples.119
Confocal microscopy can be used to confirm interactions of two or more proteins in a complex. Specific primary antibodies can be used to target proteins of interest and fluorescent tagged secondary antibodies (e.g., Alexa 405, Cy3, DyLight 488) can be used to detect the localization of the protein of interest in the sample. Localization of two or more proteins can be determined if the primary antibodies are of different species and the secondary antibodies have different emission spectra. Overlap of emitted fluorescence signal in one pixel of the image suggests colocalization of the proteins. It is important that quantification of colocalization is performed to confirm interactions with appropriate statistical analysis.120 There are several colocalization coefficients available for analysis with Pearson's correlation coefficient and Manders' overlap coefficient being the most popular. Pearson's correlation coefficient is a standard measure of pattern recognition and can be used to measure the strength of the linear relationship between two fluorescence images.120,121 It compares the similarity of shapes and ignores signal intensity giving a value between −1 and 1, where −1 suggests an entirely negative correlation.121 Manders' overlap coeffiecient is derived from the Pearson's correlation coefficient and indicates the overlap of two signals and hence the true degree of colocalization.121 This coefficient gives a value between 0 and 1, with a value of 1 being complete colocalization and is very useful where one antigen displays stronger fluorescence than the other.121 Adler et al. undertook a critical review of methods used to quantify colocalization comparing four different coefficients to quantify colocalization in selected images. This group found that Pearson's is better than Manders' and suggested that Manders' is not suitable for colocalization studies. Many different types of software are available for quantification of degrees of colocalization and many aspects of image acquisition need to be considered before quantitation is performed (reviewed in ref. 121). Confocal microscopy can be used to confirm protein-protein interactions in both tissues and cells and has been used as a verification system for protein-protein interactions between Hepatitis C virus non-structural proteins that were identified by GST-pulldown assays and Y2H.123 The use of the confocal microscope to view 3D reconstruction of a sample is possible through collection of Z-stacks during image acquisition. The top and bottom of a cell or tissue sample can be set and the microscope will take optical slices at a set interval. These slices are then reconstructed into a 3D image by the microscope software.105 Colocalization analysis can be performed on the 3D image or on the individual slices to determine level of protein-protein interactions in different areas of the cell.
2-photon microscopy is commonly used for imaging tissue samples124,125 and has been used for studying the microbial ecology of biofilm systems.126 It overcomes a problem associated with the laser scanning confocal microscope: the excitation of the specimen both above and below the focal-plane.105 The principle involves using two low-energy photons to excite a fluorophore instead of the one high-energy photon used in confocal microscopy.127 Two-photon microscopy has many advantages such as less photobleaching, decreased autofluorescence and high 3D resolution.128,129 A major advantage is the ability to image deeper into a tissue sample than with confocal (with a depth of ∼400 µm vs. ∼100 µm).105,130
The optical resolution of a microscope can limit scientists trying to determine co-localization in a sample, with limits of ∼200 nm in the xy plane and 600 nm in the z-axis.114 The confocal microscope can be used for studying protein-protein interactions in systems that aim to overcome this limitation such as FRET and BRET. FRET131,132 involves the use of two fluorophores with overlapping emission/absorption spectra which when separated by less than 10 nm, transfer energy from a donor to an acceptor molecule and fluoresce. This technique can be used to investigate binding interactions between two sets of molecules by labeling the proteins with fluorophores. A commonly used pair are both mutants of GFP (mentioned previously): CFP (as donor) and YFP (as acceptor). RFP can also be used in conjunction with GFP in FRET applications.133 FRET has proved useful in the investigation of host-pathogen interactions (reviewed in ref. 134). The use of three chromophore FRET to analyze multiprotein interactions has also been described.135 BRET is similar to FRET but instead of having two fluorescent tags, one of the tags is a bioluminescent donor enzyme, luciferase.136 The luciferase generates blue light by catalyzing an oxidative degradation interaction and can subsequently act as a donor for a fluorophore such as GFP or YFP.137 As with FRET, the signal will only occur if the proteins are in close proximity and is widely used as a verification system for the detection of protein-protein interactions.138,139 BRET was utilized by Xu et al. to show an interaction of the cyanobacterium circadian clock protein kaiB with itself. Utilization of YFP-tagged kaiB and luciferase-tagged kaiB allowed this group to show formation of a kaiB homodimer.140
All the types of microscopy mentioned in this review still do not have the resolving power of the electron microscope which is capable of resolving down to the level of <1 nm, with a recent report showing resolution of <50 pm.141 Reports of fluorescent technologies capable of resolution of ∼10 nm have emerged and are being described as super-resolution microscopy procedures (reviewed in refs. 142–145). There are exciting possibilities for the use of these technologies in the study of protein-protein interactions in the future.
Future Perspectives
For researchers beginning a protein interaction screen, the first step should be to consult the large resources of information available in the public domain. However, the researcher must tread carefully through this data because unfortunately, where many new interactions are reported, few are actually confirmed. Methods that confirm and characterize specific protein-protein interactions are delivering key pieces of information about how proteins interact. It is essential that this information is integrated better with the data coming from large scale genome wide screening methods.
We have discussed how X-ray crystallography is playing a very important role in deciphering how proteins interact and advances in the speed at which X-ray crystallography can be performed will be very welcome. Advances in structural biological approaches such as electron cryo-microscopy techniques146 and high-speed X-ray diffraction experiments147 are providing very detailed information about protein binding interfaces. Research is firmly focused on visualizing the specific protein location inside intact cells. For example, cryo-electron microscopy performed in bacterial cells is helping to identify structures and localize specific proteins inside the cell.148 New tools are continuously emerging to better improve how researchers examine how multi protein complexes are assembled. Most of these techniques are based on new modifications and combinations of older techniques. Pelletier et al. have developed a method to analyze protein-protein interactions in Gram-negative bacteria based on expression of affinity-tagged “bait” proteins from a medium copy-number plasmid.149 This method has the capacity to incorporate a wide variety of affinity, fluorescent or other tags and is based on the success of affinity tagging which is discussed in detail in this review. Three-chromophore FRET has also emerged as a method used to study the temporal organization and regulation of signaling events in living cells. This system has harnessed the potential of affinity tagging and fluorescence microscopy and is based on the mutually dependent energy transfer between three different fluorescent labels on three different proteins.135 These tags can be placed on different proteins or can be incorporated into chimeric proteins. Another combination of affinity tagging and fluorescence microscopy has seen the emergence of Biomolecular Fluorescence Complementation (BiFC) as a tool used to investigate and verify protein-protein interactions.150 In this system, two halves of a fluorescent tag is placed on two separate proteins that are predicted to interact. These proteins do not fluoresce unless they are in close proximity (suggesting interaction) which promotes the association of the two halves of the fluorescent protein to restore fluorescence.
The key challenge now is to determine the spatial and temporal regulation of protein complex assembly because many screening methods only capture a ‘moment in time’ inside the cell and do not reveal information about how different environmental cues are integrated during a signaling event. High throughput screens that reflect the dynamic assembly of protein complexes and address hierarchy of binding during complex assembly while trapping more of the transient protein-protein interactions will be very valuable.
Acknowledgments
We gratefully acknowledge the financial support of the Health Research Board of Ireland (Grant number HRA/2009/188) and the Irish Cancer Society.
Abbreviations
- M
molar
- µM
micro molar
- Y2H
yeast-two-hybrid
- MS
mass spectrometry
- TAP
tandem affinity purification
- GST
glutathione S-transferase
- GFP
green fluorescent protein
References
- 1.Berggård T, Linse S, James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7:2833–2842. doi: 10.1002/pmic.200700131. [DOI] [PubMed] [Google Scholar]
- 2.Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, et al. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631–636. doi: 10.1038/nature04532. [DOI] [PubMed] [Google Scholar]
- 3.Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, et al. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. [DOI] [PubMed] [Google Scholar]
- 4.Perkins JR, Diboun I, Dessailly BH, Lees JG, Orengo C. Transient protein-protein interactions: structural, functional and network properties. Structure. 2010;18:1233–1243. doi: 10.1016/j.str.2010.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Li X, Keskin O, Ma B, Nussinov R, Liang J. Protein-protein interactions: hot spots and structurally conserved residues often locate in complemented pockets that pre-organized in the unbound states: implications for docking. J Mol Biol. 2004;344:781–795. doi: 10.1016/j.jmb.2004.09.051. [DOI] [PubMed] [Google Scholar]
- 6.Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, et al. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or “interologs”. Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hamadeh A, Roberts MA, August E, McSharry PE, Maini PK, Armitage JP, et al. Feedback control architecture and the bacterial chemotaxis network. PLOS Comput Biol. 2011;7:1001130. doi: 10.1371/journal.pcbi.1001130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neumann S, Hansen CH, Wingreen NS, Sourjik V. Differences in signalling by directly and indirectly binding ligands in bacterial chemotaxis. EMBO J. 2010;29:3484–3495. doi: 10.1038/emboj.2010.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sourjik V, Armitage JP. Spatial organization in bacterial chemotaxis. EMBO J. 2010;29:2724–2733. doi: 10.1038/emboj.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kentner D, Sourjik V. Spatial organization of the bacterial chemotaxis system. Curr Opin Microbiol. 2006;9:619–624. doi: 10.1016/j.mib.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 11.Galperin MY, Koonin EV. Who's your neighbor? New computational approaches for functional genomics. Nat Biotechnol. 2000;18:609–613. doi: 10.1038/76443. [DOI] [PubMed] [Google Scholar]
- 12.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLOS Comput Biol. 2007;3:42. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valencia A, Pazos F. Computational methods for the prediction of protein interactions. Curr Opin Struct Biol. 2002;12:368–373. doi: 10.1016/S0959-440X(02)00333-0. [DOI] [PubMed] [Google Scholar]
- 14.Rajasekaran S. Computational techniques for motif search. Front Biosci. 2009;14:5052–5065. doi: 10.2741/3586. [DOI] [PubMed] [Google Scholar]
- 15.Bader GD, Donaldson I, Wolting C, Ouellette BF, Pawson T, Hogue CW. BIND—The Biomolecular Interaction Network Database. Nucleic Acids Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bader GD, Hogue CW. BIND—a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16:465–477. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- 17.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:449–451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: the database of interacting proteins. Nucleic Acids Res. 2000;28:289–291. doi: 10.1093/nar/28.1.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nooren IM, Thornton JM. Diversity of protein-protein interactions. EMBO J. 2003;22:3486–3492. doi: 10.1093/emboj/cdg359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nooren IM, Thornton JM. Structural characterisation and functional significance of transient protein-protein interactions. J Mol Biol. 2003;325:991–1018. doi: 10.1016/S0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 21.Shoemaker BA, Panchenko AR. Deciphering protein-protein interactions. Part II. Computational methods to predict protein and domain interaction partners. PLOS Comput Biol. 2007;3:43. doi: 10.1371/journal.pcbi.0030043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tuncbag N, Kar G, Keskin O, Gursoy A, Nussinov R. A survey of available tools and web servers for analysis of protein-protein interactions and interfaces. Brief Bioinform. 2009;10:217–232. doi: 10.1093/bib/bbp001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tuncbag N, Gursoy A, Keskin O. Prediction of protein-protein interactions: unifying evolution and structure at protein interfaces. Phys Biol. 2011;8:35006. doi: 10.1088/1478-3975/8/3/035006. [DOI] [PubMed] [Google Scholar]
- 24.Tuncbag N, Keskin O, Gursoy A. HotPoint: hot spot prediction server for protein interfaces. Nucleic Acids Res. 2010;38:402–406. doi: 10.1093/nar/gkq323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tuncbag N, Gursoy A, Keskin O. Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics. 2009;25:1513–1520. doi: 10.1093/bioinformatics/btp240. [DOI] [PubMed] [Google Scholar]
- 26.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhardwaj N, Lu H. Correlation between gene expression profiles and protein-protein interactions within and across genomes. Bioinformatics. 2005;21:2730–2738. doi: 10.1093/bioinformatics/bti398. [DOI] [PubMed] [Google Scholar]
- 28.Ge H, Liu Z, Church GM, Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet. 2001;29:482–486. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- 29.Jansen R, Greenbaum D, Gerstein M. Relating whole-genome expression data with protein-protein interactions. Genome Res. 2002;12:37–46. doi: 10.1101/gr.205602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 31.Berrade L, Garcia AE, Camarero JA. Protein microarrays: novel developments and applications. Pharm Res. 2011;28:1480–1499. doi: 10.1007/s11095-010-0325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abramovitz M, Leyland-Jones B. A systems approach to clinical oncology: focus on breast cancer. Proteome Sci. 2006;4:5. doi: 10.1186/1477-5956-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arenkov P, Kukhtin A, Gemmell A, Voloshchuk S, Chupeeva V, Mirzabekov A. Protein microchips: use for immunoassay and enzymatic reactions. Anal Biochem. 2000;278:123–131. doi: 10.1006/abio.1999.4363. [DOI] [PubMed] [Google Scholar]
- 34.Lee KB, Park SJ, Mirkin CA, Smith JC, Mrksich M. Protein nanoarrays generated by dip-pen nanolithography. Science. 2002;295:1702–1705. doi: 10.1126/science.1067172. [DOI] [PubMed] [Google Scholar]
- 35.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 36.Luckert K, Gotschel F, Sorger PK, Hecht A, Joos TO, Potz O. Snapshots of protein dynamics and posttranslational modifications in one experiment—beta-catenin and its functions. Mol Cell Proteomics. 2011;10:110–7377. doi: 10.1074/mcp.M110.007377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jeong JS, Rho HS, Zhu H. A functional protein microarray approach to characterizing posttranslational modifications on lysine residues. Methods Mol Biol. 2011;723:213–223. doi: 10.1007/978-1-61779-043-0_14. [DOI] [PubMed] [Google Scholar]
- 38.Le Meur N, Gentleman R. Modeling synthetic lethality. Genome Biol. 2008;9:135. doi: 10.1186/gb-2008-9-9-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pagano JM, Clingman CC, Ryder SP. Quantitative approaches to monitor protein-nucleic acid interactions using fluorescent probes. RNA. 2011;17:14–20. doi: 10.1261/rna.2428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park SH, Raines RT. Fluorescence gel retardation assay to detect protein-protein interactions. Methods Mol Biol. 2004;261:155–160. doi: 10.1385/1-59259-762-9:155. [DOI] [PubMed] [Google Scholar]
- 41.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 42.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal structure of the eukaryotic 40S ribosomal subunit in complex with initiation factor 1. Science. 2011;331:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 43.Kobe B, Guncar G, Buchholz R, Huber T, Maco B, Cowieson N, et al. Crystallography and protein-protein interactions: biological interfaces and crystal contacts. Biochem Soc Trans. 2008;36:1438–1441. doi: 10.1042/BST0361438. [DOI] [PubMed] [Google Scholar]
- 44.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 45.Fields S, Sternglanz R. The two-hybrid system: an assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-U. [DOI] [PubMed] [Google Scholar]
- 46.Warbrick E. Two's company, three's a crowd: the yeast two hybrid system for mapping molecular interactions. Structure. 1997;5:13–17. doi: 10.1016/S0969-2126(97)00162-7. [DOI] [PubMed] [Google Scholar]
- 47.Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 48.Brückner A, Polge C, Lentze N, Auerbach D, Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;10:2763–2788. doi: 10.3390/ijms10062763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ratushny V, Golemis E. Resolving the network of cell signaling pathways using the evolving yeast two-hybrid system. Biotechniques. 2008;44:655–662. doi: 10.2144/000112797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Koegl M, Uetz P. Improving yeast two-hybrid screening systems. Brief Funct Genomic Proteomic. 2007;6:302–312. doi: 10.1093/bfgp/elm035. [DOI] [PubMed] [Google Scholar]
- 51.Drees BL. Progress and variations in two-hybrid and three-hybrid technologies. Curr Opin Chem Biol. 1999;3:64–70. doi: 10.1016/S1367-5931(99)80012-X. [DOI] [PubMed] [Google Scholar]
- 52.Bak G, Hwang SW, Ko Y, Lee J, Kim Y, Kim K, et al. On-off controllable RNA hybrid expression vector for yeast three-hybrid system. BMB Rep. 2010;43:110–114. doi: 10.5483/BMBRep.2010.43.2.110. [DOI] [PubMed] [Google Scholar]
- 53.Wurster SE, Maher LJ., 3rd Selections that optimize RNA display in the yeast three-hybrid system. RNA. 2010;16:253–258. doi: 10.1261/rna.1880410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wurster SE, Bida JP, Her YF, Maher LJ., 3rd Characterization of anti-NFkappaB RNA aptamer-binding specificity in vitro and in the yeast three-hybrid system. Nucleic Acids Res. 2009;37:6214–6224. doi: 10.1093/nar/gkp670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jaeger S, Eriani G, Martin F. Results and prospects of the yeast three-hybrid system. FEBS Lett. 2004;556:7–12. doi: 10.1016/S0014-5793(03)01434-0. [DOI] [PubMed] [Google Scholar]
- 56.Alcaide-German ML, Vara-Vega A, Garcia-Fernandez LF, Landazuri MO, del Peso L. A yeast three-hybrid system that reconstitutes mammalian hypoxia inducible factor regulatory machinery. BMC Cell Biol. 2008;9:18. doi: 10.1186/1471-2121-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fiebitz A, Nyarsik L, Haendler B, Hu YH, Wagner F, Thamm S, et al. High-throughput mammalian two-hybrid screening for protein-protein interactions using transfected cell arrays. BMC Genomics. 2008;9:68. doi: 10.1186/1471-2164-9-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo M, Xia Z, Ma H. Functional phosphosite screening for targeted protein-protein interactions by combining phosphoproteomics strategies and mammalian two-hybrid assays. Mol Biosyst. 2011;7:1838–1841. doi: 10.1039/c1mb05053b. [DOI] [PubMed] [Google Scholar]
- 59.Fiebitz A, Vanhecke D. High-throughput mammalian two-hybrid screening for protein-protein interactions using transfected cell arrays (CAPPIA) Methods Mol Biol. 2011;723:165–183. doi: 10.1007/978-1-61779-043-0_11. [DOI] [PubMed] [Google Scholar]
- 60.Aloy P, Bottcher B, Ceulemans H, Leutwein C, Mellwig C, Fischer S, et al. Structure-based assembly of protein complexes in yeast. Science. 2004;303:2026–2029. doi: 10.1126/science.1092645. [DOI] [PubMed] [Google Scholar]
- 61.Aloy P, Russell RB. Ten thousand interactions for the molecular biologist. Nat Biotechnol. 2004;22:1317–1321. doi: 10.1038/nbt1018. [DOI] [PubMed] [Google Scholar]
- 62.Aloy P, Russell RB. Taking the mystery out of biological networks. EMBO Rep. 2004;5:349–350. doi: 10.1038/sj.embor.7400129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lu L, Arakaki AK, Lu H, Skolnick J. Multimeric threading-based prediction of protein-protein interactions on a genomic scale: application to the Saccharomyces cerevisiae proteome. Genome Res. 2003;13:1146–1154. doi: 10.1101/gr.1145203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Russell RB, Alber F, Aloy P, Davis FP, Korkin D, Pichaud M, et al. A structural perspective on protein-protein interactions. Curr Opin Struct Biol. 2004;14:313–324. doi: 10.1016/j.sbi.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 65.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, et al. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 66.Bickle MB, Dusserre E, Moncorge O, Bottin H, Colas P. Selection and characterization of large collections of peptide aptamers through optimized yeast two-hybrid procedures. Nat Protoc. 2006;1:1066–1091. doi: 10.1038/nprot.2006.32. [DOI] [PubMed] [Google Scholar]
- 67.Bolger GB, Baillie GS, Li X, Lynch MJ, Herzyk P, Mohamed A, et al. Scanning peptide array analyses identify overlapping binding sites for the signalling scaffold proteins, beta-arrestin and RACK1, in cAMP-specific phosphodiesterase PDE4D5. Biochem J. 2006;398:23–36. doi: 10.1042/BJ20060423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kittanakom S, Chuk M, Wong V, Snyder J, Edmonds D, Lydakis A, et al. Analysis of membrane protein complexes using the split-ubiquitin membrane yeast two-hybrid (MYTH) system. Methods Mol Biol. 2009;548:247–271. doi: 10.1007/978-1-59745-540-4_14. [DOI] [PubMed] [Google Scholar]
- 69.Adams CC, Gross DS. The yeast heat shock response is induced by conversion of cells to spheroplasts and by potent transcriptional inhibitors. J Bacteriol. 1991;173:7429–7435. doi: 10.1128/jb.173.23.7429-7435.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buckley DA, Cheng A, Kiely PA, Tremblay ML, O'Connor R. Regulation of insulin-like growth factor type I (IGF-I) receptor kinase activity by protein tyrosine phosphatase 1B (PTP-1B) and enhanced IGF-I-mediated suppression of apoptosis and motility in PTP-1B-deficient fibroblasts. Mol Cell Biol. 2002;22:1998–2010. doi: 10.1128/MCB.22.7.1998-2010.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hansen LH, Knudsen S, Sorensen SJ. The effect of the lacY gene on the induction of IPTG inducible promoters, studied in Escherichia coli and Pseudomonas fluorescens. Curr Microbiol. 1998;36:341–347. doi: 10.1007/s002849900320. [DOI] [PubMed] [Google Scholar]
- 72.Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci USA. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kumar R, Singh J. A truncated derivative of nmt 1 promoter exhibits temperature-dependent induction of gene expression in Schizosaccharomyces pombe. Yeast. 2006;23:55–65. doi: 10.1002/yea.1343. [DOI] [PubMed] [Google Scholar]
- 74.Wu C. Heat shock transcription factors: structure and regulation. Annu Rev Cell Dev Biol. 1995;11:441–469. doi: 10.1146/annurev.cb.11.110195.002301. [DOI] [PubMed] [Google Scholar]
- 75.Einhauer A, Jungbauer A. The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J Biochem Biophys Methods. 2001;49:455–465. doi: 10.1016/S0165-022X(01)00213-5. [DOI] [PubMed] [Google Scholar]
- 76.Einhauer A, Jungbauer A. Affinity of the monoclonal antibody M1 directed against the FLAG peptide. J Chromatogr A. 2001;921:25–30. doi: 10.1016/S0021-9673(01)00831-7. [DOI] [PubMed] [Google Scholar]
- 77.Brizzard BL, Chubet RG, Vizard DL. Immunoaffinity purification of FLAG epitope-tagged bacterial alkaline phosphatase using a novel monoclonal antibody and peptide elution. Biotechniques. 1994;16:730–735. [PubMed] [Google Scholar]
- 78.Chiang CM, Roeder RG. Expression and purification of general transcription factors by FLAG epitope-tagging and peptide elution. Pept Res. 1993;6:62–64. [PubMed] [Google Scholar]
- 79.Prendergast FG, Mann KG. Chemical and physical properties of aequorin and the green fluorescent protein isolated from Aequorea forskalea. Biochemistry. 1978;17:3448–3453. doi: 10.1021/bi00610a004. [DOI] [PubMed] [Google Scholar]
- 80.Ward TH, Lippincott-Schwartz J. The uses of green fluorescent protein in mammalian cells. Methods Biochem Anal. 2006;47:305–337. doi: 10.1002/0471739499.ch14. [DOI] [PubMed] [Google Scholar]
- 81.Haseloff J, Siemering KR. The uses of green fluorescent protein in plants. Methods Biochem Anal. 2006;47:259–284. [PubMed] [Google Scholar]
- 82.Hazelrigg T, Mansfield JH. Green fluorescent protein applications in Drosophila. Methods Biochem Anal. 2006;47:227–257. [PubMed] [Google Scholar]
- 83.Hitchcock AL, Kahana JA, Silver PA. The uses of green fluorescent protein in yeasts. Methods Biochem Anal. 2006;47:179–201. doi: 10.1002/0471739499.ch9. [DOI] [PubMed] [Google Scholar]
- 84.Valdivia RH, Cormack BP, Falkow S. The uses of green fluorescent protein in prokaryotes. Methods Biochem Anal. 2006;47:163–178. [PubMed] [Google Scholar]
- 85.Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol. 2010;11:427–439. doi: 10.1038/nrm2900. [DOI] [PubMed] [Google Scholar]
- 86.ten Have S, Boulon S, Ahmad Y, Lamond AI. Mass spectrometry-based immuno-precipitation proteomics—the user's guide. Proteomics. 2011;11:1153–1159. doi: 10.1002/pmic.201000548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Y. Commonly used tag combinations for tandem affinity purification. Biotechnol Appl Biochem. 2010;55:73–83. doi: 10.1042/BA20090273. [DOI] [PubMed] [Google Scholar]
- 88.Puig O, Caspary F, Rigaut G, Rutz B, Bouveret E, Bragado-Nilsson E, et al. The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods. 2001;24:218–229. doi: 10.1006/meth.2001.1183. [DOI] [PubMed] [Google Scholar]
- 89.Völkel P, Le Faou P, Angrand PO. Interaction proteomics: characterization of protein complexes using tandem affinity purification-mass spectrometry. Biochem Soc Trans. 2010;38:883–887. doi: 10.1042/BST0380883. [DOI] [PubMed] [Google Scholar]
- 90.Xu X, Song Y, Li Y, Chang J, Zhang H, An L. The tandem affinity purification method: an efficient system for protein complex purification and protein interaction identification. Protein Expr Purif. 2010;72:149–156. doi: 10.1016/j.pep.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 91.Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, et al. A physical and functional map of the human TNFalpha/NFkappaB signal transduction pathway. Nat Cell Biol. 2004;6:97–105. doi: 10.1038/ncb1086. [DOI] [PubMed] [Google Scholar]
- 92.Matsumoto M, Oyamada K, Takahashi H, Sato T, Hatakeyama S, Nakayama KI. Large-scale proteomic analysis of tyrosine-phosphorylation induced by T-cell receptor or B-cell receptor activation reveals new signaling pathways. Proteomics. 2009;9:3549–3563. doi: 10.1002/pmic.200900011. [DOI] [PubMed] [Google Scholar]
- 93.Nittis T, Guittat L, LeDuc RD, Dao B, Duxin JP, Rohrs H, et al. Revealing novel telomere proteins using in vivo cross-linking, tandem affinity purification and label-free quantitative LC-FTICR-MS. Mol Cell Proteomics. 2010;9:1144–1156. doi: 10.1074/mcp.M900490-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Melcher K. New chemical crosslinking methods for the identification of transient protein-protein interactions with multiprotein complexes. Curr Protein Pept Sci. 2004;5:287–296. doi: 10.2174/1389203043379701. [DOI] [PubMed] [Google Scholar]
- 95.Robinette D, Neamati N, Tomer KB, Borchers CH. Photoaffinity labeling combined with mass spectrometric approaches as a tool for structural proteomics. Expert Rev Proteomics. 2006;3:399–408. doi: 10.1586/14789450.3.4.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Sinz A. Investigation of protein-protein interactions in living cells by chemical crosslinking and mass spectrometry. Anal Bioanal Chem. 2010;397:3433–3440. doi: 10.1007/s00216-009-3405-5. [DOI] [PubMed] [Google Scholar]
- 97.Elion EA. Detection of protein-protein interactions by coprecipitation. Curr Protoc Neurosci. 2006;5:25. doi: 10.1002/0471142301.ns0525s35. [DOI] [PubMed] [Google Scholar]
- 98.Frank R, Heikens W, Heisterberg-Moutsis G, Blocker H. A new general approach for the simultaneous chemical synthesis of large numbers of oligonucleotides: segmental solid supports. Nucleic Acids Res. 1983;11:4365–4377. doi: 10.1093/nar/11.13.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Frank R. The SPOT-synthesis technique. Synthetic peptide arrays on membrane supports—principles and applications. J Immunol Methods. 2002;267:13–26. doi: 10.1016/S0022-1759(02)00137-0. [DOI] [PubMed] [Google Scholar]
- 100.Katz C, Levy-Beladev L, Rotem-Bamberger S, Rito T, Rudiger SG, Friedler A. Studying protein-protein interactions using peptide arrays. Chem Soc Rev. 2011;40:2131–2145. doi: 10.1039/c0cs00029a. [DOI] [PubMed] [Google Scholar]
- 101.Lichtman JW, Conchello JA. Fluorescence microscopy. Nat Methods. 2005;2:910–919. doi: 10.1038/nmeth817. [DOI] [PubMed] [Google Scholar]
- 102.Paddock SW. Principles and practices of laser scanning confocal microscopy. Mol Biotechnol. 2000;16:127–149. doi: 10.1385/MB:16:2:127. [DOI] [PubMed] [Google Scholar]
- 103.So PT, Dong CY, Masters BR, Berland KM. Two-photon excitation fluorescence microscopy. Annu Rev Biomed Eng. 2000;2:399–429. doi: 10.1146/annurev.bioeng.2.1.399. [DOI] [PubMed] [Google Scholar]
- 104.Mattheyses AL, Simon SM, Rappoport JZ. Imaging with total internal reflection fluorescence microscopy for the cell biologist. J Cell Sci. 2010;123:3621–3628. doi: 10.1242/jcs.056218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Semwogerere D, Weeks ER. Confocal Microscopy. In: Gary E, Wnek GLB, editors. Encyclopedia of Biomaterials and Biomedical Engineering. Informa Healthcare; 2008. pp. 705–714. [Google Scholar]
- 106.Cody CW, Prasher DC, Westler WM, Prendergast FG, Ward WW. Chemical structure of the hexapeptide chromophore of the Aequorea green-fluorescent protein. Biochemistry. 1993;32:1212–1218. doi: 10.1021/bi00056a003. [DOI] [PubMed] [Google Scholar]
- 107.Prasher DC, Eckenrode VK, Ward WW, Prendergast FG, Cormier MJ. Primary structure of the Aequorea victoria green-fluorescent protein. Gene. 1992;111:229–233. doi: 10.1016/0378-1119(92)90691-H. [DOI] [PubMed] [Google Scholar]
- 108.Chiesa A, Rapizzi E, Tosello V, Pinton P, de Virgilio M, Fogarty KE, et al. Recombinant aequorin and green fluorescent protein as valuable tools in the study of cell signalling. Biochem J. 2001;355:1–12. doi: 10.1042/0264-6021:3550001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science. 2006;312:217–224. doi: 10.1126/science.1124618. [DOI] [PubMed] [Google Scholar]
- 110.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/S0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 111.Ormö M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 112.Delagrave S, Hawtin RE, Silva CM, Yang MM, Youvan DC. Red-shifted excitation mutants of the green fluorescent protein. Biotechnology (NY) 1995;13:151–154. doi: 10.1038/nbt0295-151. [DOI] [PubMed] [Google Scholar]
- 113.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, et al. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 114.Smith CL. Basic confocal microscopy. Curr Protoc Mol Biol. 2008;14:1. doi: 10.1002/0471142727.mb1411s81. [DOI] [PubMed] [Google Scholar]
- 115.Scriven DR, Lynch RM, Moore ED. Image acquisition for colocalization using optical microscopy. Am J Physiol Cell Physiol. 2008;294:1119–1122. doi: 10.1152/ajpcell.00133.2008. [DOI] [PubMed] [Google Scholar]
- 116.Xiao GQ, Corle TR, Kino GS. Real-time confocal scanning optical microscope. Appl Phys Lett. 1988;53:716–718. doi: 10.1063/1.99814. [DOI] [PubMed] [Google Scholar]
- 117.Henthorn DC, Jaxa-Chamiec AA, Meldrum EA. GAL4-based yeast three-hybrid system for the identification of small molecule-target protein interactions. Biochem Pharmacol. 2002;63:1619–1628. doi: 10.1016/S0006-2952(02)00884-5. [DOI] [PubMed] [Google Scholar]
- 118.North AJ. Seeing is believing? A beginners' guide to practical pitfalls in image acquisition. J Cell Biol. 2006;172:9–18. doi: 10.1083/jcb.200507103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Holmes TJ, Cheng PC. Basic Principles of Imaging. In: Yu HCPCLPCKFJ, editor. Multi-Modality Microscopy. Singapore: World Scientific Publishing Co., Pte., Ltd.; 2006. pp. 1–74. [Google Scholar]
- 120.Barlow AL, Macleod A, Noppen S, Sanderson J, Guerin CJ. Colocalization analysis in fluorescence micrographs: verification of a more accurate calculation of pearson's correlation coefficient. Microsc Microanal. 2010;16:710–724. doi: 10.1017/S143192761009389X. [DOI] [PubMed] [Google Scholar]
- 121.Zinchuk V, Zinchuk O, Okada T. Quantitative colocalization analysis of multicolor confocal immunofluorescence microscopy images: pushing pixels to explore biological phenomena. Acta Histochem Cytochem. 2007;40:101–111. doi: 10.1267/ahc.07002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Adler J, Parmryd I. Quantifying colocalization by correlation: the Pearson correlation coefficient is superior to the Mander's overlap coefficient. Cytometry A. 2010;77:733–742. doi: 10.1002/cyto.a.20896. [DOI] [PubMed] [Google Scholar]
- 123.Dimitrova M, Imbert I, Kieny MP, Schuster C. Protein-protein interactions between hepatitis C virus nonstructural proteins. J Virol. 2003;77:5401–5414. doi: 10.1128/JVI.77.9.5401-14.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Helmchen F, Denk W. Deep tissue two-photon microscopy. Nat Methods. 2005;2:932–940. doi: 10.1038/nmeth818. [DOI] [PubMed] [Google Scholar]
- 125.Konjufca V, Miller MJ. Two-photon microscopy of host-pathogen interactions: acquiring a dynamic picture of infection in vivo. Cell Microbiol. 2009;11:551–559. doi: 10.1111/j.1462-5822.2009.01289.x. [DOI] [PubMed] [Google Scholar]
- 126.Neu TR, Woelfl S, Lawrence JR. Three-dimensional differentiation of photo-autotrophic biofilm constituents by multi-channel laser scanning microscopy (single-photon and two-photon excitation) J Microbiol Methods. 2004;56:161–172. doi: 10.1016/j.mimet.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 127.Denk W, Strickler JH, Webb WW. Two-photon laser scanning fluorescence microscopy. Science. 1990;248:73–76. doi: 10.1126/science.2321027. [DOI] [PubMed] [Google Scholar]
- 128.Xu C, Zipfel W, Shear JB, Williams RM, Webb WW. Multiphoton fluorescence excitation: new spectral windows for biological nonlinear microscopy. Proc Natl Acad Sci USA. 1996;93:10763–10768. doi: 10.1073/pnas.93.20.10763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Zipfel WR, Williams RM, Webb WW. Nonlinear magic: multiphoton microscopy in the biosciences. Nat Biotechnol. 2003;21:1369–1377. doi: 10.1038/nbt899. [DOI] [PubMed] [Google Scholar]
- 130.Diaspro A, Sheppard C. Two-photon microscopy: basic principles and architectures. In: Diaspro A, editor. Confocal and Two-Photon Microscopy Foundations, Applications and Advances. New York: Wiley-Liss Inc.; 2002. pp. 39–73. [Google Scholar]
- 131.Piston DW, Kremers GJ. Fluorescent protein FRET: the good, the bad and the ugly. Trends Biochem Sci. 2007;32:407–414. doi: 10.1016/j.tibs.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 132.Sahoo H, Schwille P. FRET and FCS—friends or foes? ChemPhysChem. 2011;12:532–541. doi: 10.1002/cphc.201000776. [DOI] [PubMed] [Google Scholar]
- 133.Mizuno H, Sawano A, Eli P, Hama H, Miyawaki A. Red fluorescent protein from Discosoma as a fusion tag and a partner for fluorescence resonance energy transfer. Biochemistry. 2001;40:2502–2510. doi: 10.1021/bi002263b. [DOI] [PubMed] [Google Scholar]
- 134.Hayward RD, Goguen JD, Leong JM. No better time to FRET: shedding light on host pathogen interactions. J Biol. 2010;9:12. doi: 10.1186/jbiol225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Galperin E, Verkhusha VV, Sorkin A. Three-chromophore FRET microscopy to analyze multiprotein interactions in living cells. Nat Methods. 2004;1:209–217. doi: 10.1038/nmeth720. [DOI] [PubMed] [Google Scholar]
- 136.Boute N, Jockers R, Issad T. The use of resonance energy transfer in high-throughput screening: BRET versus FRET. Trends Pharmacol Sci. 2002;23:351–354. doi: 10.1016/S0165-6147(02)02062-X. [DOI] [PubMed] [Google Scholar]
- 137.Bevan N, Rees S. Pharmaceutical applications of GFP and RCFP. In: Martin Chalfie SK, editor. Green fluorescent protein: properties, applications and protocols. John Wiley and Sons; 2006. pp. 361–390. [Google Scholar]
- 138.Pfleger KD, Eidne KA. Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET) Nat Methods. 2006;3:165–174. doi: 10.1038/nmeth841. [DOI] [PubMed] [Google Scholar]
- 139.Pfleger KD, Seeber RM, Eidne KA. Bioluminescence resonance energy transfer (BRET) for the real-time detection of protein-protein interactions. Nat Protoc. 2006;1:337–345. doi: 10.1038/nprot.2006.52. [DOI] [PubMed] [Google Scholar]
- 140.Xu Y, Piston DW, Johnson CH. A bioluminescence resonance energy transfer (BRET) system: application to interacting circadian clock proteins. Proc Natl Acad Sci USA. 1999;96:151–156. doi: 10.1073/pnas.96.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Erni R, Rossell MD, Kisielowski C, Dahmen U. Atomic-resolution imaging with a sub-50-pm electron probe. Phys Rev Lett. 2009;102:096101. doi: 10.1103/PhysRevLett.102.096101. [DOI] [PubMed] [Google Scholar]
- 142.Hell SW. Far-field optical nanoscopy. Science. 2007;316:1153–1158. doi: 10.1126/science.1137395. [DOI] [PubMed] [Google Scholar]
- 143.Patterson G, Davidson M, Manley S, Lippincott-Schwartz J. Superresolution imaging using single-molecule localization. Annu Rev Phys Chem. 2010;61:345–367. doi: 10.1146/annurev.physchem.012809.103444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lippincott-Schwartz J, Manley S. Putting super-resolution fluorescence microscopy to work. Nat Methods. 2009;6:21–23. doi: 10.1038/nmeth.f.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Schermelleh L, Heintzmann R, Leonhardt H. A guide to super-resolution fluorescence microscopy. J Cell Biol. 2010;190:165–175. doi: 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fujii T, Iwane AH, Yanagida T, Namba K. Direct visualization of secondary structures of F-actin by electron cryomicroscopy. Nature. 2010;467:724–728. doi: 10.1038/nature09372. [DOI] [PubMed] [Google Scholar]
- 147.Luger P. Fast electron density methods in the life sciences—a routine application in the future? Org Biomol Chem. 2007;5:2529–2540. doi: 10.1039/b706235d. [DOI] [PubMed] [Google Scholar]
- 148.Pilhofer M, Ladinsky MS, McDowall AW, Jensen GJ. Bacterial TEM: new insights from cryo-microscopy. Methods Cell Biol. 2010;96:21–45. doi: 10.1016/S0091-679X(10)96002-0. [DOI] [PubMed] [Google Scholar]
- 149.Pelletier DA, Hurst GB, Foote LJ, Lankford PK, McKeown CK, Lu TY, et al. A general system for studying protein-protein interactions in Gram-negative bacteria. J Proteome Res. 2008;7:3319–3328. doi: 10.1021/pr8001832. [DOI] [PubMed] [Google Scholar]
- 150.Kerppola TK. Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat Protoc. 2006;1:1278–1286. doi: 10.1038/nprot.2006.201. [DOI] [PMC free article] [PubMed] [Google Scholar]