Abstract
“Germ granules” are cytoplasmic, nonmembrane-bound organelles unique to germline. Germ granules share components with the P bodies and stress granules of somatic cells, but also contain proteins and RNAs uniquely required for germ cell development. In this review, we focus on recent advances in our understanding of germ granule assembly, dynamics, and function. One hypothesis is that germ granules operate as hubs for the posttranscriptional control of gene expression, a function at the core of the germ cell differentiation program.
Germ granules are RNA-rich structures unique to the cytoplasm of germ cells. They contain factors required for germ cell development and may operate as hubs for posttranscriptional regulation of gene expression.
“Germ granules” is a general term used to refer to the RNA-rich cytoplasmic bodies of germ cells. By electron microscopy, germ granules appear as compact granulo-fibrillar aggregates with no surrounding membrane (Fig. 1A,D) (Eddy 1975; Guraya 1979). Germ granules contain proteins and RNAs required for germ cell development, and may carry out posttranscriptional regulation specific to germ cells. In some organisms (i.e., Drosophila, Xenopus, Caenorhabditis elegans, and zebrafish), germ granules are present continuously throughout development, with the exception of mature sperm. In those organisms, germ granules are transmitted from oocyte to embryo as part of the germ plasm, a specialized cytoplasm that segregates with the germ lineage and is sufficient to specify germ cell fate. In other organisms, notably mammals, germ granules are not detected in oocytes or early embryos, but are formed de novo in primordial germ cells shortly after their specification. The current view is that, while not all organisms require germ granules to specify germ cell fate, all organisms depend on germ granules for germ cell function. Molecular studies have revealed intriguing similarities between germ granules and RNA granules present in somatic cells. In this review, we focus on recent advances in our understanding of germ granule composition, variety, dynamics, and function.
Figure 1.
Electron micrographs of the major germ granule classes. (A) Perinuclear P granule in a meiotic germ cell of C. elegans. Distinct subdomains are visible: the crest (white arrow) and base (arrowhead) overlying a cluster of nuclear pores (black arrows). Scale bar: 500 nm. (Panel A is adapted from Sheth et al. [2010] and reproduced, with permission, from The Company of Biologists © 2010.) (B) Balbiani body in a mouse oocyte. Black arrow points to mitochondria clustered around Golgi membranes (BB). N, nucleus. Scale bar: 5 um. (Panel B is adapted from Pepling et al. [2007] and reproduced, with permission, from National Academy of Sciences, USA © 2007.) (C) Sponge bodies in the Drosophila nurse cell. Sponge bodies (asterisks) intermingled with mitochondria (m) and ER cisternae (arrows) near an intercellular bridge (black arrowheads) connecting the nurse cell (nc) to the oocyte (o). ld, lipid droplet. Scale bar: 500 nm. (Panel C adapted from Jaglarz et al. [2011] and reproduced, with permission, from Springer © 2011.) (D) Perinuclear chromatoid body in a mouse stage I round spermatid. Scale bar: 500 nm. (Panel D is adapted from Vasileva et al. [2009] and reproduced here, with permission, from Elsevier © 2009.) (E) Intermitochondrial cement in a mouse spermatocyte. IMC (arrowheads) forms in the spaces between clustered mitochondria. Scale bar: 1 um. (Panel E is adapted from Chuma et al. [2006] and reproduced, with permission, from National Academy of Sciences, USA © 2006.) (F) Polar granules (pg) in the cortical cytoplasm of a Drosophila embryo. Polysomes (arrows) extend from the surface of the granule. Scale bar: 100 nm. (Panel F is adapted from Amikura et al. [2001] and reproduced, with permission, from the Company of Biologists 2001.)
TYPES OF GERM GRANULES
Cytoplasmic aggregates that stained with RNA dyes were first detected in the germ cells of insects in 1890 (Ritter 1890), and have since been observed by cytochemistry or electron microscopy in over 80 species across the animal kingdom from rotifers to mammals (Eddy 1975). Today, antibodies against the Vasa family of RNA helicases are commonly used to identify germ granules. Many other conserved components of germ granules have also been identified (Tables 1 and 2). Some, like Vasa family members, are present in most germ granules, others are found only in specific stages. The morphology and localization of germ granules also changes during development. Here we describe the major classes of germ granules found in immature and differentiating germ cells (nuage), gametes (sponge, balbiani, and chromatoid bodies), and embryos (germ plasm) (Fig. 1) (Eddy 1975; Guraya 1979; also see de Sousa Lopes and Roelen 2010).
Table 1.
Conserved components of germ granules—listed are components localizing to germ granules in two or more species
| Function | Organism: Protein name and germ granule type | References | |
|---|---|---|---|
| Proteins | |||
| Vasa and related DEAD box RNA helicases | RNP remodeling | Drosophila: Vasa in nuage and polar granules; C. elegans: GLH-1-4 in P granules; Xenopus: XVLG1, nuage in oogonia, perinuclear in PGCs; Zebrafish: Vasa, perinuclear in PGCs; Mouse: MVH in chromatoid body; Humans: Vasa, perinuclear in fetal oocytes | Hay et al. 1988; Liang et al. 1994 Gruidl et al. 1996a Ikenishi et al. 1996; Bilinski et al. 2004 Knaut et al. 2000 Toyooka et al. 2000 Castrillon et al. 2000 |
| Argonaute family | Small RNA (miRNA/piRNA) regulation | Drosophila: Aubergine in nuage and polar granules, PIWI in polar granules, Ago3 in nuage; Mouse: Miwi, Mili, Ago2, Ago3 in chromatoid body; Zebrafish: Ziwi, Zili in germ granules, perinuclear in PGC and germ cells; C. elegans: PRG-1, CSR-1 in P granules | Harris and Macdonald 2001; Megosh et al. 2006; Nishida et al. 2007; Li et al. 2009 Kotaja et al. 2006a,b Houwing et al. 2007, 2008; Batista et al. 2008; Claycomb et al. 2009 |
| Dicer | miRNA production | Mouse: Dicer in chromatoid body; C. elegans: DRH-3, DCR-1 in P granules | Kotaja et al. 2006a Claycomb et al. 2009 Beshore et al. 2011 |
| Maelstrom (HMG box) | Chromatin and piRNA regulation | Drosophila: Maelstrom in nuage; Mouse: Maelstrom in chromatoid body | Findley et al. 2003 Costa et al. 2006 |
| Tudor domain proteins (methyl-binding modules) | Recruitment of methylated proteins to germ granules; recruitment of mitochondrial ribosomal RNA to polar granules in Drosophila | Drosophila: Tudor in nuage and polar granules, Krimp, Tejas, and PAPI in nuage; Mouse: Mtr (TDRD1-7;9) in intermitochondrial cement and chromatoid body, Tdrd4/Rnf17 in Rnf17-granule | Bardsley et al. 1993 Arkov et al. 2006; Lim and Kai 2007; Patil and Kai 2010; Liu et al. 2011 Chuma et al. 2003; Pan et al. 2005; Hosokawa et al. 2007; Vasileva et al. 2009 Aravin et al. 2009; Shoji et al. 2009 |
| WD repeat proteins | Cofactors of protein arginine methyltransferase; targeting of methylation? | Drosophila: Valois (homolog of MEP50) in polar granules and nuage; Mouse: WDR77 in chromatoid bodies | Anne and Mechler 2005 Vagin et al. 2009b |
| Sm proteins | Splicing in the spliceosome, germ granule localization and germ cell fate in C. elegans | C. elegans: Sm in P granules; Xenopus: Sm in nuage in oocytes; Mouse: Sm in chromatoid body; Drosophila: SmB and SmD3 in polar granules | Barbee et al. 2002 Bilinski et al. 2004 Moussa et al. 1994; Chuma et al. 2003 Anne 2010; Gonsalvez et al. 2010 |
| Dcp1 | Decapping of mRNAs in P bodies, localization of Oskar RNA to the germ plasm in Drosophila | Drosophila: dDcp1 in sponge bodies; Mouse: Dcp1a in chromatoid body, and perinuclear spermatocyte nuage | Lin et al. 2006 Kotaja et al. 2006a Aravin et al. 2009 |
| Dcp2 | Decapping of mRNAs in P bodies | Drosophila: dDcp2 in sponge bodies; C. elegans: DCP-2 in P granules | Lin et al. 2006 Lall et al. 2005 |
| Dhh1p/Rck (DEAD box RNA helicase) | Repression of translation (also in P bodies) | Drosophila: Me31b in sponge bodies, and polar granules; C. elegans: CGH-1 in P granules | Nakamura et al. 2001; Thomson et al. 2008 Navarro et al. 2001 |
| Scd6p/Rap55 (Sm-like domain) | Repression of translation (also in P bodies) | Drosophila: Trailer Hitch in Balbiani body (early oocyte), nuage and sponge bodies (oogenesis); C. elegans: CAR-1 in P granules; mouse: Trailer hitch in Balbiani body (early oocytes), perinuclear granule (spermatocytes) | Wilhelm et al. 2005 Liu et al. 2011 Audhya et al. 2005; Boag et al. 2005; Squirrell et al. 2006 Pepling et al. 2007 |
| Xrn1 | Exonuclease degrading mRNA | Drosophila: PCM in nuage; Mouse: Xrn1 in perinuclear spermatocyte nuage | Lim et al. 2009 Aravin et al. 2009 |
| Nanos | Repression of translation | Zebrafish: in perinucelar germ granules of PGCs; C. elegans: in P granules of embryos | Koprunner et al. 2001 Subramaniam and Seydoux 1999 |
| eIF4F complex | Translational regulation by components or binding partners of eIF4F | Drosophila: eIF4A polar granules; eIF4E-binding protein Cup in Babliani body and sponge bodies (oogenesis); C. elegans: IFE-1(eIF4E) in P granules | Cox and Spradling 2003; Wilhelm et al. 2003; Nakamura et al. 2004; Thomson et al. 2008 Amiri et al. 2001 |
| RNAs | |||
| Nanos (mRNA) | Repression of translation | Drosophila: Nanos RNA enriched in germ plasm; C. elegans: nos-2 RNA enriched in P granules; Xenopus: Xcat2 RNA in balbiani body; Zebrafish: nos1 RNA enriched in germ plasm; Pea Aphid: nanos RNA in nuage-like structure in oocytes | Forrest and Gavis 2003 Subramaniam and Seydoux 1999 Mosquera et al. 1993; Bilinski et al. 2004 Koprunner et al. 2001 Chang et al. 2006 |
| Mitochondrial large and small ribosomal RNAs (noncoding RNAs) | Translation of germ cell less (gcl) in Drosophila | Drosophila: on surface of polar granules of syncytial embryo; Xenopus: in germinal granules; planaria: on surface of chromatoid bodies | Kashikawa et al. 1999 Kashikawa et al. 2001 Sato et al. 2001 |
| piRNA | Repression of transposons | Zebrafish: oocyte germ granules; mouse: chromatoid body | Houwing et al. 2007 Melkar et al. 2010 |
Table 2.
Components of the mouse chromatoid body
| Component | Function | References |
|---|---|---|
| MVH | Dead box RNA helicase that is required for spermatogenesis | Chuma et al. 2003; Kotaja et al. 2006a,b |
| MIWI | Argonaute/PIWI family RNA binding protein that is required for spermatogenesis. MIWI and MILI repress transposone element mediated by piRNA | Deng and Lin 2002; Grivna et al. 2006; Kotaja et al. 2006a,b |
| RanBPM | RanGTP binding protein that is involved in microtubule nucleation. It associates with MVH | Shibata et al. 2004 |
| Kif17b | Kinesin motor protein that is involved in the transport of the coactivator protein ACT and mRNAs | Kotaja et al. 2006a |
| Tdrd1 | Tudor domain containing protein that is required for spermatogenesis. It interacts with MILI | Hosokawa et al. 2007; Kojima et al. 2009; Wang et al. 2009 |
| Tdrd6 | Tudor domain containing protein that is required for spermatogenesis and CB formation | Vasileva et al. 2009 |
| Tdrd7 | Tudor domain containing protein | Hosokawa et al. 2007 |
| p48 and p52 | Germ cell specific RNA binding proteins | Oko et al. 1996 |
| Ago2 and Ago3 | Main catalytic engine of RISC | Kotaja et al 2006b |
| Dicer | Cytoplasmic RNase that produces siRNA and miRNA from their precursors | Kotaja et al. 2006b |
| Dcp1a | Decapping enzyme that remove 5'cap of mRNA | van Dijk et al. 2002 |
| GW182 | P-body localized RNA binding protein that is required for RNAi | Kotaja et al. 2006b |
| mRNA | Translation template | Kotaja et al. 2006b |
| MiRNA | Small RNA that represses complement target mRNA expression | Kotaja et al. 2006b |
| SnRNP | Essential component of the spliceosomal complex that function in pre-mRNA processing | Paniagua et al. 1985; Moussa et al. 1994; Biggiogera et al. 1990 |
| GRTH | Dead box RNA helicase that is required for spermatogenesis. Deficient of GRTH shows impaired CB formation | Tsai-Morris et al. 2004 |
| GW182 | Essential component for si/miRNA induced gene silencing | Kotaja et al. 2006b |
| H2B | Chromatin formation | Haraguchi et al. 2005 |
| H4 | Chromatin formation | Werner and Werner 1995 |
| Acetylated H3 | Chromatin regulation | Haraguchi et al. 2005 |
| Acetylated H4 | Chromatin regulation | Haraguchi et al. 2005 |
| Actin | Cytoskeletal protein | Walt and Armbruster 1984; Haraguchi et al. 2005 |
| Vimentin | Intermediate filament | Haraguchi et al. 2005 |
| Ubiquitin | Targeting signal for several pathways | Haraguchi et al. 2005 |
| E2 | Ubiquitin conjugating enzyme | Haraguchi et al. 2005 |
| p52 | 26S proteasome subunit | Haraguchi et al. 2005 |
| pA700 | Proteasome activator | Haraguchi et al. 2005 |
| COX1 | Cytochrome C oxidase subunit 1 | Haraguchi et al. 2005 |
| HSP70 | protein chaperon | Haraguchi et al. 2005 |
| F1α | ATP synthase subunit α | Haraguchi et al. 2005 |
| F1β | ATP synthase subunit β | Haraguchi et al. 2005 |
| LDH | Dehydrogenation of 2-hydroxybutylate | Haraguchi et al. 2005 |
| Enolase | Conversion from 2-phosphogylcerate to phosphoenoylpyruvate | Haraguchi et al. 2005 |
| PHGPx | Reduces lipid hydroperoxides in membranes | Haraguchi et al. 2005 |
| Cytochrome C | Electron transfer component | Hess et al. 1993 |
| Ca2+, Mg2+ | Andonov and Chaldakov 1989; Rouelle-Rossier et al. 1993 | |
| Histocompatibility antigen | Immune system | Head and Kresge 1985 |
| LAMP1 and LAM2 | Lysosome localized glycoproteins | Haraguchi et al. 2005 |
| Acid phosphatase | Lysosome localized phosphatase | Anton 1983 |
| Cathepsins B,D, H, and L | Protease | Haraguchi et al. 2005 |
| LAP | Leucin aminopeptidase | Haraguchi et al. 2005 |
| DNase | hydrolysis of DNA | Haraguchi et al. 2005 |
| RNase | hydrolysis of RNA | Haraguchi et al. 2005 |
| NADPase | hydrolysis of NADP | Tang et al. 1982; Thorne-Tjomsland et al. 1988 |
| CMPase | hydrolysis of monophosphate polysaccharides | Tang et al. 1982; Thorne-Tjomsland et al. 1988 |
| Krimer and Esponda 1980 |
Many more components have been identified for the individual model organisms, and may later prove conserved. For component lists reflecting component diversity in popular model organisms see: mouse chromatoid body Table 2 in this manuscript; Drosophila sponge bodies (Snee and Macdonald 2009); C. elegans P granules (Updike and Strome 2009b).
Perinuclear Granules
During most of germline development, germ granules appear as rounded fibrillar aggregates (or nuage, from the French for “cloud”) that cluster around nuclei. This arrangement has been documented in the primordial germ cells (PGCs) of Drosophila (Mahowald 1968, 1971), C. elegans (Strome and Wood 1982, 1983), Xenopus (Ikenishi et al. 1996), and zebrafish (Knaut et al. 2000). In mice, small granules surrounded by fibrillar matrix and mitochondria become apparent around the nuclei of PGCs two days after their formation at 9–9.5 days of gestation (Spiegelman and Bennett 1973; Clark and Eddy 1975). Perinuclear granules persist after the PGCs become incorporated into the somatic gonad. Electron micrographic studies of C. elegans germ cells in meiotic prophase have shown that germ granules overlie dense clusters of nuclear pores, corresponding to the main sites of mRNA export from the nucleus (Fig. 1A) (Strome and Wood 1982; Pitt et al. 2000; Sheth et al. 2010). Close association of the nuage with nuclear pores has also been reported in ultrastructural studies of Xenopus (Czolowska 1969), zebrafish (Knaut et al. 2000; reviewed in Kloc et al. 2004), and mouse (reviewed in Chuma et al. 2009).
Balbiani Body
In differentiating gametes, germ granules often adopt unique morphologies. The Balbiani body, or mitochondrial cloud, is a transient structure, containing mitochondria, Golgi, endoplasmic reticulum (ER) and RNA, that forms in the young (previtellogenic) oocytes of insects and vertebrates (Cox and Spradling 2003; Kloc et al. 2004; Wilk et al. 2005; Pepling et al. 2007). In Drosophila, ER membranes associated with the Balbiani body are enriched for the ER resident protein reticulon, suggesting that these membranes represent a specialized subset of smooth ER (Roper 2007). The Balbiani body initially forms near the nucleus and spreads to the cytoplasm during oocyte growth. In Drosophila and Xenopus, the aggregates eventually localize to a subregion of the oocyte cortex, in which they incorporate with other germ granule material to form the germ plasm that will be transmitted to the germline of the next generation (see below). It has been proposed that the Balbiani body selects the best mitochondria for incorporation in the germline (Cox and Spradling 2003). In Xenopus, mitochondria in the germ plasm have lower respiratory activity than mitochondria destined for the soma (Kogo et al. 2011). Lower respiration is hypothesized to decrease oxidative stress and mutational damage. A Drosophila mutant that disrupts mitochondrial transport and association with the Balbiani body, however, had no apparent effect on fertility (Cox and Spradling 2006).
Balbiani bodies, consisting of ER and mitochondria interspersed with nuage surrounding Golgi stacks, have also been reported in mouse oocytes, which do not form germ plasm (Pepling et al. 2007). The mouse Balbiani bodies are present in the cyst and primordial follicle stages, and disappear during follicular growth (Fig. 1B) (Pepling et al. 2007). Their function is not yet known.
Sponge Bodies
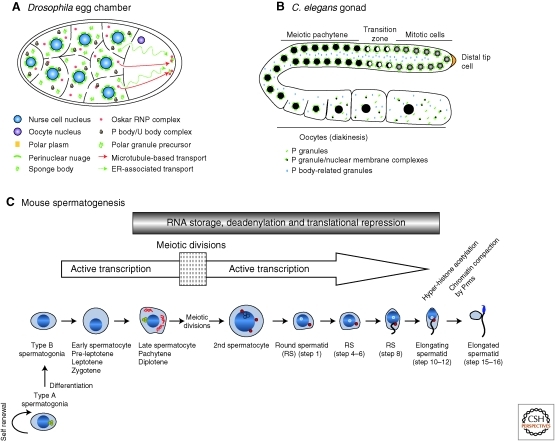
During Drosophila oogenesis, each growing oocyte is connected to 15 nurse cells through intercellular bridges or “ring canals” (Fig. 2A). In addition to perinuclear nuage, nurse cells and the oocyte contain sponge bodies, which are stacks of endoplasmic reticulum (ER) cisternae embedded in an electron-dense matrix scattered through the cytoplasm (Fig. 1C) (Wilsch-Brauninger et al. 1997). Sponge bodies appear to derive from fragments of perinuclear nuage that associate with ER membranes, detach from the nucleus, and intermingle in the cytoplasm with other P-body-like granules (see below) (Wilsch-Brauninger et al. 1997; Jaglarz et al. 2011). Sponge bodies accumulate in the oocyte, apparently by movement along ER membranes that extend from the nurse cells through the ring canals into the oocyte (Jaglarz et al. 2011). Sponge bodies movement has been hypothesized to transport perinuclear nuage material from the nurse cells to the germ (pole) plasm of the oocyte (Nakamura et al. 2001). The composition of sponge bodies, however, is dynamic and differs between nurse cells and oocytes, suggesting that sponge bodies are not simply a mobile version of nuage, but represent a range of heterogeneous RNP assemblies (Snee and Macdonald 2004, 2009).
Figure 2.
Germ granules during gametogenesis. (A) Transport of germ granule material in a Drosophila egg chamber. Various germ granule RNPs intermingle in the nurse cells’ cytoplasm. Green dashed lines represent migration of sponge bodies, red lines indicate the oskar RNP transport pathway. (Panel A adapted from Jaglarz et al. [2011] and reproduced, with permission, from Springer © 2011.) (B) Germ granules in the germline of an adult C. elegans hermaphrodite. Germ cells progress along an “assembly line” of development from mitotic stem cells at the distal end to the oocytes at the proximal end. Germ cells share a common cytoplasm, and are individualized late in oogenesis. P granules (green) are docked at the germ cell nuclei until diplotene, and transition to the cytoplasm along with nuclear pore complexes (green/black). P body-related granules (blue) accumulate in the shared cytoplasm (rachis) and in oocytes. (C) Mammalian spermatogenesis. Intermitochondrial cement (yellow dots) is present in type A spermatogonia, and disappears in type B spermatogonia until meiosis. The chromatoid body (CB; red filaments then becoming dots) appears in late pachytene cells as thick cytoplasmic fibers. After the first meiotic division, the CB starts condensing as one or two round foci. Except during meiotic divisions, transcription is active until the elongating spermatid stage, and many transcripts are translationally silenced and located in the CB. In elongated spermatids, transcripts are released from translational arrest.
Membrane-rich structures reminiscent of sponge bodies have also been reported in C. elegans oocytes (Fig. 2B). During the oocyte growth phase, the perinuclear germ granules detach from the nuclear envelope, apparently taking with them membrane fragments containing clusters of nuclear pores (Pitt et al. 2000). Stressed and aging oocytes assemble even larger cytoplasmic RNP complexes, whose formation has been correlated with dramatic nuclear membrane blebbing (Jud et al. 2008; Patterson et al. 2011). Drosophila sponge bodies also change morphology under different environmental conditions (Snee and McDonald 2009), suggesting a role for these membranous structures in protecting oocytes from stress.
Germ Granules in Germ Plasm
In mature oocytes of animals with “germ plasm,” materials transported in the Balbiani and sponge bodies coalesce in a subregion of the oocyte cortex at a position that predicts the site of PGC formation, as seen in Xenopus (Czolowska 1969), zebrafish (Knaut et al. 2000), and Drosophila (Illmensee and Mahowald 1974). Immediately after fertilization, in Drosophila, germ granules appear in small clusters surrounded by ribosomes (Fig. 1F) (Amikura et al. 2001; Mahowald 2001) near membranes of the endoplasmic reticulum (Thomson et al. 2008). During germ (pole) cell formation, the germ granules are transported on microtubules toward the nuclei of the future pole cells (Lerit and Gavis 2011). In zebrafish embryos, the germ plasm forms on the distal ends of the first and second cleavage furrows, in a complex process involving microtubules, microfilaments, and two distinct RNA segregation pathways (Knaut et al. 2000; Theusch et al. 2006). By the 32-cell stage, the germ plasm aggregates are found in four cells that will give rise to the germline (Knaut et al. 2000).
In C. elegans, germ granules detach from nuclei during oogenesis and remain uniformly distributed in the cytoplasm through fertilization. In the zygote, after embryonic polarity is specified, the germ granules become enriched in the posterior cytoplasm and, as a result, are inherited primarily by the posterior daughter, the germline blastomere P1. This process is repeated for another three asymmetric divisions separating soma from germline. By the time of the birth of the germline founder cells, the germ granules have returned to their perinuclear position (Strome and Wood 1982).
Chromatoid Body
During mouse spermatogenesis (Fig. 2C), germ granules undergo a number of morphological changes (Eddy 1975). Perinuclear nuage and cytoplasmic granules associated with mitochondria (“intermitochondrial cement”) are found in early male germ cells (Fig. 1E) (Chuma et al. 2006; reviewed in Chuma et al. 2009). During meiosis, spermatocytes assemble a unique type of germ granule called the chromatoid body (Fig. 1D) (Kotaja and Sassone-Corsi 2007). The chromatoid body arises from intermitochondrial fibers that disperse during the meiotic divisions and consolidate postmeiosis into a single sphere apposed against the nucleus. The site of chromatoid body/nucleus contact is rich in nuclear pores (Fawcett et al. 1970) and labeling of the nucleus with tritiated uridine also labels the chromatoid body, suggesting that RNA synthesized in the nucleus is transferred to the chromatoid body (Soderstrom and Parvinen 1976). Remarkably, the chromatoid body is motile, moving both around the nucleus and into neighboring cells through cytosolic bridges. This movement has been proposed to mediate sharing of RNAs between the haploid spermatids (Ventela et al. 2003). The chromatoid body relocalizes to the base of the flagellum in elongating spermatids (Fig. 2C), and disassembles later during spermiogenesis (Parvinen 2005). No germ granules have been reported in the mature sperm of any organism.
Chromatoid-like bodies have also been described in the neoblasts of planaria; the totipotent stem cells that give rise to both somatic and germ cells (Coward 1974; Hori 1997). Chromatoid bodies are lost when neoblast progeny differentiate into somatic lineages.
P Bodies
Two main classes of cytoplasmic RNA granules have been described in somatic cells: processing bodies and stress granules (Balagopal and Parker 2009). Processing bodies (P bodies) and stress granules share components implicated in RNA storage, turnover, and translational regulation. Each granule type also possesses unique features: P bodies contain components of the mRNA decay machinery, and stress granules contain TIA proteins, poly-A binding protein, and stalled translation initiation complexes (Anderson and Kedersha 2009). Several P body and stress granule components have been detected in germ granules (Tables 1 and 2). This overlap has generated some speculation as to whether germ granules might correspond to P bodies and/or stress granules. Although functional parallels are likely, a direct correspondence remains unclear. So far, stalled initiation complexes have not been reported in germ granules, suggesting that germ granules may not be directly equivalent to stress granules. In contrast, most components of the RNA decay machinery (DCP1, DCP2, CGH-1/Me31B/Rck, CAR-1/Tral/RAP55, and PCM/Xrn1) have been detected in the perinuclear nuage, sponge bodies, germ plasm, and/or chromatoid bodies, suggesting a strong connection to P bodies (Nakamura et al. 2001; Navarro et al. 2001; Audhya et al. 2005; Boag et al. 2005; Lall et al. 2005; Wilhelm et al. 2005; Kotaja et al. 2006a; Lin et al. 2006; Squirrell et al. 2006; Kotaja and Sassone-Corsi 2007; Pepling et al. 2007; Thomson et al. 2008; Aravin et al. 2009; Lim et al. 2009; Yabuta et al. 2011). Careful examination of the distribution of P body proteins in germ cells has revealed, however, that these factors also accumulate in foci that do not contain core germ granule proteins. For example, mature mouse oocytes, which lack VASA-positive granules, contain foci positive for the decapping enzyme Dcp1a (Swetloff et al. 2009). Mouse spermatocytes also contain canonical P bodies distinct from the germ granules (Aravin et al. 2009; Shoji et al. 2009). In C. elegans, several types of P-body related granules have been observed in pachytene germ cells and in maturing oocytes (Navarro et al. 2001; Audhya et al. 2005; Boag et al. 2005; Noble et al. 2008). In embryos, granules containing the decapping activator PATR-1 and other P body proteins, but lacking the germ granule component PGL-1, dock around the germ granules (Audhya et al. 2005; Lall et al. 2005; Gallo et al. 2008). Unlike germ granules, the PATR-1 granules are partitioned symmetrically during mitosis and recruit additional P body proteins after segregation into somatic blastomeres (Gallo et al. 2008). Similarly, in the sponge bodies of Drosophila nurse cells, structures resembling the P body/U body complexes of somatic cells (Liu and Gall 2007) can be distinguished from nuage (Lim et al. 2009; Jaglarz et al. 2011). Together, these observations suggest that germ cells contain bona fide P-bodies, or P body-related granules, which interact and may even exchange components with germ granules, but are maintained as distinct RNP assemblies. Whether germ granules represent specialized P-bodies, P-body/stress granule hybrids, or a distinct granule type unique to the germline, remains to be determined.
DYNAMICS
In electron micrographs, germ granules appear as dense, fibrillar structures that in some cases appear to have distinct subcompartments (Fig. 1A,D) (Fawcett et al. 1970; Sheth et al. 2010). In live recordings, perinuclear germ granules in Drosophila nurse cells (nuage) and in C. elegans meiotic cells (P granules) have been observed to maintain their shape and relative position for up to 40 minutes, the maximum recording time (Snee and Macdonald 2004; Sheth et al. 2010). Despite this apparent stability, experiments using fluorescence-recovery after photobleaching (FRAP) have revealed that individual components are in fact highly dynamic. GFP::Vasa is replaced in the Drosophila nuage with a half-time of 60 sec (Snee and Macdonald 2004). Similarly, GFP::PGL-1 diffuses within seconds inside the perinuclear granules of C. elegans (Brangwynne et al. 2009), and whole perinuclear clusters recover from photobleaching with a half-time of 20 sec (Sheth et al. 2010). Whether germ granules also contain stable components that form static scaffolds is not known.
Under some circumstances, whole granules can rapidly change size or position. During the asymmetric cleavages of the C. elegans embryo, germ granules in cytoplasm destined for the germline grow and fuse into larger granules, whereas germ granules in cytoplasm destined for the soma shrink in size until no longer visible (Brangwynne et al. 2009). These dynamics have been compared to those of liquid droplets undergoing condensation and dissolution (Brangwynne et al. 2009). In zygotes, a subset of germ granules have also been observed to be move directionally over several microns (Hird et al. 1996). This movement is microtubule-independent (Hill and Strome 1990) and may be mediated by cytoplasmic flows that develop in response to actomyosin contractions in the zygote’s cortex (Marston and Goldstein 2006).
Microtubules have been implicated in germ granules dynamics in Drosophila and zebrafish. During Drosophila oogenesis, transport of oskar RNP particles from nurse cell to the posterior pole occurs along microtubules in kinesin-dependent manner (reviewed in Becalska and Gavis 2009). In Drosophila embryos, translocation of VASA-containing granules from the cortex to nuclei occurs along astral microtubules in a process that requires dynein (Lerit and Gavis 2011). In zebrafish, microtubules promote germ granule aggregation in early embryos (Theusch et al. 2006), and have been implicated in germ granule partitioning during mitosis in primordial germ cells (Strasser et al. 2008). Disruption of microtubules or dynein causes the germ granules to coalesce into larger granules, suggesting a role in regulating both granule dynamics and size (Strasser et al. 2008).
In mouse spermatids, the chromatoid body is highly mobile, tracking around the nucleus and shuttling from cell to cell via cytoplasmic bridges (Fawcett et al. 1970; Ventela et al. 2003). Microtubule-depolymerizing drugs disrupt these movements and cause the chromatoid body to disperse (Ventela et al. 2003). The testis-specific kinesin KIF17b is a possible candidate for mediating chromatoid body movement. KIF17b was first identified as a nucleus-cytoplasm shuttling protein that transports ACT (activator of CREM in testis), a transcriptional coregulator of postmeiotic transcription (Fimia et al. 1999). KIF17b also contributes to the transport of TB-RBP, Testis-brain RNA-binding protein, which associates with CREM regulated mRNAs (Chennathukuzhi et al. 2003a,b). Importantly, KIF17b is also found in the chromatoid body (Kotaja et al. 2006b), where it may participate in RNA transport and/or mediate chromatoid body movement.
ASSEMBLY OF GERM GRANULES
Unlike other cellular organelles, germ granules are not bound by membranes, yet maintain distinct boundaries and shapes. What molecular interactions underlie germ granule form and integrity? Although no single assembly pathway accounts for the wide range of germ granule morphologies, several have been documented.
Nucleation by Seed Proteins
Genetic analyses in Drosophila have led to a hierarchical model for the organization of germ plasm: a single protein, Oskar, is necessary and sufficient to initiate a cascade of interactions that assemble germ plasm at the site of Oskar localization (Mahowald 2001). Oskar exists in two forms: a short form required for germ granule assembly (Markussen et al. 1995), and a long form required to anchor the granules to the cell cortex (Vanzo and Ephrussi 2002). Short Oskar initiates granule formation likely through direct interactions with Vasa, which in turn recruits Tudor (Breitwieser et al. 1996; Anne 2010), Valois, and Piwi-family Argonautes (Kirino et al. 2009; Nishida et al. 2009). Germ granule mRNAs (i.e., nanos, germ-cell-less, and polar granule component) are synthesized in the nurse cells and transferred to the oocyte during nurse cell cytoplasm “dumping” (reviewed in Becalska and Gavis 2009). Localization to the germ plasm depends on sequences in the 3′UTR and an actin-dependent, cortex-anchoring mechanism initiated by Oskar (Becalska and Gavis 2009; Rangan et al. 2009). How the long form of Oskar anchors the germ plasm to the cortex is not fully understood, but it involves recruitment of endosomal proteins and stimulation of endocytosis, which eventually reorganizes F-actin in the oocyte cortex (Tanaka and Nakamura 2008).
Oskar homologs have not been found outside of insects, but proteins with similar germ plasm “seed” functions have been identified in zebrafish and C. elegans. In zebrafish, the Balbiani body component Bucky Ball is necessary and sufficient to induce germ plasm aggregation (Bontems et al. 2009). Bucky ball is conserved among vertebrates but does not possess recognizable functional domains; its mode of action remains unknown (Bontems et al. 2009). In C. elegans, the RGG-domain proteins PGL-1 and PGL-3 are required for germ granule integrity in embryos, and can form cytoplasmic granules when ectopically expressed in tissue culture cells or in intestinal cells (Hanazawa et al. 2011; Updike et al. 2011). Ectopic PGL granules recruit RNA and other germ granule components. Granule formation depends on a domain in the PGL proteins that mediates self-association, whereas recruitment of RNA and RNA-binding proteins requires the RGG domain (Hanazawa et al. 2011). PGL-1 and PGL-3 also form transient granules in the somatic blastomeres of early embryos, but in those cells, granule formation depends on the autophagy machinery, which ultimately targets the PGL granules for degradation (Zhang et al. 2009).
Nucleation by Organelles
Germ granules often form near mitochondria, raising the possibility that nucleating factors also exist on the membranes of these organelles. Two recent reports implicate such mechanism in generation of the intermitochondrial cement (IMC) in mouse spermatocytes. Mutants lacking the mitochondrial phospholipase D MitoPLD do not form IMC and mislocalize IMC components (Huang et al. 2011; Watanabe et al. 2011). MitoPLD generates the signaling lipid phosphatidic acid. Remarkably, a deficiency in the phosphatidic acid-metabolizing enzyme, Lipin 1 causes hyper-aggregation of electron-dense nuage around mitochondria (Huang et al. 2011). Phosphatidic acid may therefore function as the organizing signal for IMC.
Binding of Tudor Domains to Methylated Arginines
The Tudor domain is a small (∼55 amino acid) protein–protein interaction module that recognizes methylated arginines or lysines. Tudor domains have been found in several proteins localized to perinuclear nuage, germ plasm, and/or chromatoid bodies (see Table 1 for references). The founding member of the family, Drosophila Tudor, has 11 Tudor domains, but Tudor domains can also be found in single copy or combined with other conserved motifs, such as in SpnE/TDRD9 (reviewed in Arkov and Ramos 2010).
The Tudor domain forms an aromatic cage that interacts directly with the methylated amino acid (reviewed in Arkov and Ramos 2010; Vourekas et al. 2010). Crystallographic studies of Tudor proteins complexed with methyl-arginine-containing peptides show that the Tudor module is embedded in a larger 170 amino-acid domain that contacts the peptide at other sites (Liu et al. 2010; Vourekas et al. 2010). These additional interactions may help Tudor-containing proteins discriminate among potential partners.
Piwi-family Argonautes, Vasa helicases and Sm proteins in Drosophila, Xenopus, and mouse all contain methylated arginines and thus are potential targets for Tudor domain-containing proteins (Kuramochi-Miyagawa et al. 2004; Gonsalvez et al. 2006; Thomson et al. 2008; Chen et al. 2009; Kirino et al. 2009; Kojima et al. 2009; Nishida et al. 2009; Reuter et al. 2009; Shoji et al. 2009; Vagin et al. 2009a,b; Wang et al. 2009; Kirino et al. 2010a,b; Patil and Kai 2010; Liu et al. 2011). Symmetrical dimethylation of arginines (sDMA) is performed in flies and mice by the methyltransferase Capsuleen/PRMT and its cofactor Valois/MEP50/WDR77 (Gonsalvez et al. 2006; Anne et al. 2007). In Drosophila, sDMA modifications of the Argonaute Aubergine are recognized by Tudor, and are necessary for Aubergine-Tudor interaction and germ granule assembly (Anne et al. 2007; Kirino et al. 2009; Nishida et al. 2009). sDMA modifications of the spliceosomal Sm proteins SmB and SmD3 also depend on Capsuleen/Valois, and are required for their localization to germ granules; however, the relevant Tudor-domain partners of these proteins have not been identified (Anne 2010; Gonsalvez et al. 2010). In the mouse, several Tudor domain proteins interact with the sDMA-modified Piwi family Argonautes and are required for spermatogenesis (reviewed in Arkov and Ramos 2010; Vourekas et al. 2010). Lack of interactions causes both the Tudor domain proteins and Piwi-family Argonautes to become mislocalized, suggesting a mutual dependency for localization.
Low Specificity Interactions
Tudor-sDMA interactions have the potential to create high affinity/specificity complexes that are stable and long-lived. A different type of interaction, however, may be needed to bring these complexes into higher-order dynamic structures. Studies in C. elegans has led to the proposal that germ granules show liquid-like behaviors (Brangwynne et al. 2009). In response to pressure, germ granules in the nuage of meiotic germ cells appeared to “drip” off nuclei and fuse into large drops; germ granules also “condense” and “dissolve” during the asymmetric divisions of the embryo (Brangwynne et al. 2009). Liquid behaviors are thought to be mediated by weak, fast-rearranging molecular interactions, as could arise from low-affinity protein–RNA interactions. Consistent with a role for RNA in maintaining germ granule integrity, depletion of the VegT and Xlsirts RNAs disrupts the organization of germ granules in the vegetal cortex of Xenopus oocytes (Kloc and Etkin 1994; Kloc et al. 2005, 2007). Inhibition of transcription causes loss of PGL-1 and GLH-2 from the C. elegans nuage (Sheth et al. 2010) and loss of the electron-dense core of the mouse chromatoid body (Soderstrom 1977). Mutations in genes implicated in the biogenesis of small RNAs (ego-1, csr-1, drh-3) cause a dramatic increase in the size and number of germ granules in meiotic germ cells (Updike and Strome 2009a).
Other studies in C. elegans have implicated weak hydrophobic interactions involving phenylalanine-glycine (FG)-repeats. FG-repeats are naturally unfolded and flexible, and thus well suited for dynamic binding to multiple partners, as in the liquid droplet model above. FG-repeats are found in nucleoporins that mediate active transport across the nuclear pore and prevent passive diffusion of molecules greater than > 30 kD (Elad et al. 2009; Terry and Wente 2009). Perinuclear germ granules exclude dextrans larger than 40 kDa and are sensitive to treatments predicted to disrupt FG-repeat interactions (Updike et al. 2011). Consistent with a role for FG-repeats, three C. elegans VASA family members, GLH-1, 2, and 4, contain FG repeats (Kuznicki et al. 2000), and the FG-containing nucleoporin Nup98 and mRNA export factor DDX-19 colocalize with germ granules (Sheth et al. 2010; Voronina and Seydoux 2010). RNAi screens have also identified several nucleoporins required for germ granule integrity in embryos (Updike and Strome 2009a; Voronina and Seydoux 2010) and for the assembly of sponge body-like aggregates in stressed oocytes (Patterson et al. 2011).
Recently, FG-repeat proteins have been recognized as a subtype of prions (Ader et al. 2010). Remarkably, overexpression in mouse neuroblastoma N2a cells of the cytosolic form of the prion PrP causes the formation of a large RNP aggregate near the cell nucleus, resembling a chromatoid body (Beaudoin et al. 2009). The “PrP body” associates with mitochondria, and is able to recruit mRNAs, FG-repeat nucleoporins, Dicer, Dcp1a, DDX6, and SmB proteins reminiscent of germ granules (Beaudoin et al. 2009; Roucou 2009). Nucleation of stress granules in somatic cells also depends on a prion-domain containing protein, TIA-1 (Gilks et al. 2004). An intriguing possibility is that prion-like templating properties give germ granules not only their unique structural characteristics, but also the ability to propagate from germ cell to germ cell, and from generation to generation in animals with germ plasm.
HYPOTHESES FOR FUNCTION OF GERM GRANULES
Germ Cell Fate Specification and Differentiation
The first proposed function for germ granules was to support the specification of the germline in embryos where germ granules are inherited maternally as a part of the germ plasm. In Xenopus and Drosophila embryos, irradiation of a specific region of the cytoplasm (germ plasm) results in embryos that lack germ cells (Geigy 1931; Bounoure 1937; Smith 1966). In Drosophila, transplantation of germ plasm to an ectopic site is sufficient to reprogram cells to become germ cells (Illmensee and Mahowald 1974). In fact, ectopic localization of the germ plasm component Oskar is sufficient to induce ectopic germ granules (see above) and form ectopic germ cells (Ephrussi and Lehmann 1992). Similarly, overexpression of the Balbiani body protein Bucky ball in zebrafish embryos induces extra germ cells (Bontems et al. 2009). These experiments suggest that germ plasm, and perhaps the granule granules therein, contain the factors that specify germ cell fate.
In Drosophila, mothers with mutations in vasa generate embryos that lack germ cells (Schupbach and Wieschaus 1986; Lasko and Ashburner 1990; Styhler et al. 1998). In Xenopus embryos, morpholino treatments that reduce Vasa protein levels decrease germ cell numbers (Ikenishi and Tanaka 1997; Ikenishi et al. 2006). Genetic analyses have identified several other germ granule proteins like Vasa that are required for germ cell specification, but none (besides Oskar) is sufficient to induce germ cell fate on its own (Rongo and Lehmann 1996; Harris and Macdonald 2001; Megosh et al. 2006; Anne 2010; Gonsalvez et al. 2010). Germ granules in germ plasm associate with ribosomes, raising the possibility that translation of mRNAs stored in the germ granules is the critical factor in germ cell fate specification (Mahowald 2001). Consistent with this hypothesis, mRNAs enriched in germ granules encode proteins (i.e., Nanos and Pgc) that are required for germ cell development (see below). Again, however, no single RNA was found to be sufficient to induce germ cell fate.
Recent data has challenged the simple model that germ granules are sufficient to specify germ cell fate. Mutants that mis-express multiple germ granule proteins in larvae have been identified in Drosophila and C. elegans (Unhavaithaya et al. 2002; Wang et al. 2005; Janic et al. 2010; Petrella et al. 2011). These mutants show somatic defects, but do not develop extra germ cells. Mutants that mispartition germ granules in embryos also do not make extra germ cells (Strome et al. 1995; Gallo et al. 2010). In fact, a C. elegans mutant, in which germ granules disassemble at each mitosis and components segregate symmetrically, is still viable and fertile (Gallo et al. 2010). These observations suggest that germ granules are not essential organizers of germ plasm, and that other factors maintain the distinction between soma and germline in embryos. Consistent with this view, germ granules are not present at the time of germ cell specification in mammals.
In contrast, an essential role for germ granules during germ cell differentiation is well supported by genetic analyses in several animals. For example, loss of Nanos causes PGCs to fail to reach the somatic gonad and eventually die in Drosophila, C. elegans, and zebrafish embryos (Kobayashi et al. 1996; Subramaniam and Seydoux 1999; Koprunner et al. 2001). Nanos is also required for PGC viability in mice (Tsuda et al. 2003; Suzuki et al. 2007), for maintenance and regeneration of the germline in planaria (Wang et al. 2007), and for the in vitro differentiation of human germ cells from embryonic stem cells (Julaton and Reijo Pera 2011). Members of the Vasa family of RNA helicases and the PIWI family of Argonautes have also been implicated in several aspects of germ cell proliferation and differentiation in Drosophila, C. elegans, zebrafish, and mouse (Lin and Spradling 1997; Cox et al. 1998; Kuznicki et al. 2000; Tanaka et al. 2000; Deng and Lin 2002; Kuramochi-Miyagawa et al. 2004; Carmell et al. 2007; Houwing et al. 2007; Batista et al. 2008; Houwing et al. 2008; Spike et al. 2008; Li et al. 2009a; Unhavaithaya et al. 2009). Phenotypes include failure to maintain germline stem cells, germ cell death before meiosis onset, reduced germ cell proliferation, and meiotic defects. The challenge now is to identify the molecular mechanisms that underlie this wide range of phenotypes. In this respect, much progress has been made recently in flies and mice, in which meiotic failure has been traced back to specific defects in chromatin and transposon regulation.
Production of piRNAs
Transposition must be under tight regulation in germ cells to prevent spreading of deleterious DNA rearrangements to gametes and the next generation. Recent studies in several organisms have shown that the PIWI subfamily of Argonautes blocks transposition by producing small RNAs, called piRNAs, against transposon sequences (Vagin et al. 2006; Aravin et al. 2007; Houwing et al. 2007; reviewed in Thomson and Lin 2009). The PIWI Argonautes function in a two-step “ping-pong” amplification to generate sense and antisense piRNAs from piRNA precursor loci and expressed transposon sequences (Aravin et al. 2006, 2007; Grivna et al. 2006; Brennecke et al. 2007; reviewed in Senti and Brennecke 2010).
In Drosophila ovaries, the Argonautes Aubergine and Ago3 colocalize with Vasa and other factors required for piRNA production in the perinuclear nuage of nurse cells (reviewed in Senti and Brennecke 2010). In vasa mutant ovaries, piRNA-pathway proteins do not concentrate in the nuage and transposon mRNAs are up-regulated (Snee and Macdonald 2004; Vagin et al. 2004). Genetic and biochemical analyses suggest that Tudor-domain proteins Tudor, Tejas, and PAPI are important for recruitment of Aub and Ago3 to the nuage (Nishida et al. 2009; Patil and Kai 2010; Liu et al. 2011). Additionally, Aubergine/Tudor and Ago3/PAPI complexes interact with P-body proteins Dhh1/Me31B and Rap55/Trailer Hitch implicating the mRNA degradation machinery in transposon silencing (Thomson et al. 2008; Liu et al. 2011). Consistent with this view, the P-body proteins CCR4/TWIN, DCP1, SKI3, and Rap55/TRAL are all required for transposon silencing (Lim et al. 2009; Liu et al. 2011).
piRNA pathway proteins are also found in the germ plasm of Drosophila oocytes and embryos, raising the possibility that piRNAs are transmitted maternally with the germ plasm to the next generation. Consistent with this possibility, piRNAs targeting I-elements are found in the daughters of mothers with I-elements, but not in the daughters of fathers with I-elements. The latter, but not the former, are sterile (hybrid dysgenesis) because of I-element activation (Brennecke et al. 2008).
As in Drosophila, zebrafish Piwi family Argonautes Zili and Ziwi localize to nuage and are required for piRNA biogenesis and repression of transposons in germ cells (Houwing et al. 2007, 2008). In both species, as well as in mouse (see also below), piRNAs are modified at their 3′ end by 2′-hydroxyl methylation (Vagin et al. 2006; Horwich et al. 2007; Houwing et al. 2007; Kirino and Mourelatos 2007b; Ohara et al. 2007). The enzyme responsible for this modification is Hen1 (Horwich et al. 2007), which interacts with Piwi proteins (Horwich et al. 2007; Kirino and Mourelatos 2007a; Saito et al. 2007). Hen1 is a component of nuage in zebrafish, and functions in Drosophila and zebrafish to stabilize piRNAs and maximize transposon repression (Horwich et al. 2007; Kamminga et al. 2010).
In the mouse, piRNAs are most abundant in spermatocytes in which they associate with Piwi family Argonautes (Aravin et al. 2007, 2008) and accumulate in the chromatoid body (Meikar et al. 2010). The Argonautes MILI and MIWI2 and Tudor proteins TDRD1, TDRD5, and TDRD9 are all required for piRNA production, methylation of transposon promoters, and repression of transposon transcription (Aravin et al. 2007; Carmell et al. 2007; Reuter et al. 2009; Shoji et al. 2009; Vagin et al. 2009b; Yabuta et al. 2011). piRNA production and transposon silencing also require Vasa (MVH), and the germ granule assembly factors MAEL, GASZ, and MitoPLD (Soper et al. 2008; Ma et al. 2009; Kuramochi-Miyagawa et al. 2010; Watanabe et al. 2011). Spermatocytes lacking MitoPLD or MVH do not form intermitochondrial cement, the germ granule precursor to the chromatoid body. In contrast, intermitochondrial cement is made in MILI-deficient spermatocytes, which lack most piRNAs, indicating that piRNAs are not required for IMC formation. Unlike in Drosophila, the piRNA pathway in mice functions primarily in the male germline: mutations in this pathway block spermatogenesis, but do not affect oogenesis. Repression of transposons is mediated by a different group of small RNAs in oocytes: the Dicer-dependent endo-siRNAs formed from double-stranded RNA precursors (Murchison et al. 2007; Tam et al. 2008; Watanabe et al. 2008).
A spermatogenesis-specific function for Piwi-family Argonautes is also observed in C. elegans. Piwi-family member PRG-1 is a component of germ granules that binds to 21U-RNAs, analogous to piRNAs of other species (Batista et al. 2008). A subclass of 21U-RNAs targets the transposon Tc3. prg-1 mutants lack 21U-RNAs and have higher Tc3 RNA levels (Batista et al. 2008) and Tc3 transposition (Das et al. 2008). Transposon regulation in C. elegans also depends on a greatly expanded subgroup of worm-specific argonautes (WAGOs) that produce a different class of small RNAs, termed 22G-RNAs (Batista et al. 2008; Das et al. 2008; Gu et al. 2009). PRG-1 and WAGOs are hypothesized to function in a two-step mechanism, in which primary 21U-RNAs trigger secondary 22G-RNA production required for efficient silencing of transposons and pseudogenes (Batista et al. 2008; Das et al. 2008).
Mutations in the piRNA pathway lead to both transposon activation and meiotic defects, suggesting that the two may be linked (reviewed in Ollinger et al. 2010). Transposition could cause meiotic defects by inducing DNA damage, interfering with homolog recognition, and/or cytotoxicity of retrotransposon-encoded proteins. Another possibility, however, is that the piRNA pathway regulates transposons and meiosis in parallel.
Mitosis and Chromatin Organization
The 22G-RNAs of C. elegans target not only repetitive elements, but also unique sequences, including many annotated genes (Gu et al. 2009). 22G-RNAs targeting unique genes associate preferentially with the Argonaute CSR-1 (Claycomb et al. 2009). CSR-1 is a component of the nuage in which it functions with the Dicer-related helicase DRH-3, the RNA-dependent RNA polymerase EGO-1, and the Tudor domain protein EKL-1 to generate 22G-RNAs. Surprisingly, mutations in csr-1 do not affect the expression of most genes targeted by 22G-RNAs, but cause dramatic defects in chromosome organization during meiosis and mitosis (Claycomb et al. 2009). In embryos, CSR-1 and its partners associate with mitotic chromosomes and are required for the proper distribution of kinetochore proteins (Claycomb et al. 2009). These observation suggest that CSR-1 could function in the delineation of centromeres, as has been shown for Ago1 in Schizosaccharomyces pombe (Verdel et al. 2004).
A role for nuage components in chromosome dynamics has also been reported in Drosophila (Pek and Kai 2011a). vasa mutant germ cells display lagging chromosomes at anaphase that have reduced levels of type I condensins. Vasa coimmunoprecipitates with type I condensins and localizes to perichromosomal foci during mitosis. aubergine and spnE mutants lack Vasa-positive perichromosomal foci and show mitotic defects similar to those seen in the vasa mutants (Pek and Kai 2011b). These findings suggest that small RNAs produced in the nuage also regulate chromatin compaction during mitosis (Pek and Kai 2011b). Interestingly, in the sea urchin, Vasa is also required for chromosome segregation but this function does not appear to be specific to the germline: Vasa associates with mitotic spindles in all early embryonic blastomeres (Yajima and Wessel 2011). These observations suggest that Vasa may have an ancestral role in mitosis, distinct from its role in germ cell development.
Regulation of mRNAs
As germ granules are enriched in RNA-binding proteins and developmentally regulated mRNAs, they have been hypothesized to function as hubs of posttranscriptional regulation; specifically to regulate mRNA localization, levels, and translational activity (Kotaja and Sassone-Corsi 2007; Rangan et al. 2009).
mRNA Localization
Germ granules have been implicated in RNA localization during oogenesis in several organisms (for details see reviews by Kloc et al. 2004; Zhou and King 2004; Becalska and Gavis 2009). Localized mRNAs associate with Balbiani and sponge bodies in the oocytes of Drosophila, Xenopus, and zebrafish (Ephrussi et al. 1991; Kim-Ha et al. 1991; Wang and Lehmann 1991; Kloc et al. 2002; Cox and Spradling 2003; Wilk et al. 2005; Kosaka et al. 2007). The Balbiani body protein Bucky Ball is required for the Balbiani body aggregation and localization of several mRNAs to the vegetal cortex of zebrafish eggs (Dosch et al. 2004; Marlow and Mullins 2008; Bontems et al. 2009). Several RNA-binding proteins associated with sponge bodies are required for the directed transport of oskar to the posterior pole, including Exuperantia, Yps, Rap55/Trailer Hitch, and the hnRNP Hrp48 (Wilhelm et al. 2000, 2005; Yano et al. 2004). nanos localization to the germ plasm, in contrast, appears to depend on the trapping of a minority of mRNA molecules in the germ (polar) granules (Wang et al. 1994; Rongo et al. 1997). The Argonaute Aubergine is a candidate tether for nanos in the polar granules (Becalska et al. 2011). Aub is in a complex with nanos RNA, and aub/+ embryos show defects in nanos RNA localization but no defect in Oskar localization. Remarkably Aubergine’s function in nanos localization appears to be independent of its role in the piRNA pathway (Becalska et al. 2011).
mRNA Translation
Germ granules have been implicated in both the repression and activation of translation. Granules implicated in RNA transport such as in Balbiani and sponge bodies typically contain mRNAs that are translationally repressed and become activated after release from the granules or after transition to another granule type.
Several mechanisms of translation repression have been described for germ granule associated mRNAs. In Drosophila nurse cells, repressed mRNAs are often found in sponge bodies (Nakamura et al. 2001). Repression of oskar mRNA in sponge bodies depends on the sequence-specific translational repressor Bruno (Kim-Ha et al. 1995). Bruno binds to Bruno-response elements in the oskar 3′UTR and promotes the formation of large (up to 80S) oskar RNA-containing complexes that exclude ribosomes (Chekulaeva et al. 2006). Bruno additionally represses oskar translation through interactions with Cup. Cup is a component of sponge bodies that forms a complex with the translation initiation factor eIF4E and the DEAD-box helicase Dhh1/Me31B (Nakamura et al. 2001, 2004; Wilhelm et al. 2003). eIF4E-binding proteins (4E-BPs) such as Cup are thought to modulate translational initiation by blocking the interaction between eIF4E and eIF4G in a reversible manner (Gingras et al. 1998). Mutations in Cup that disrupt binding to eIF4E cause premature translation of oskar mRNA (Nakamura et al. 2004). A similar mechanism has been proposed for the C. elegans germ granule component SPN-2 (Li et al. 2009b). SPN-2 is a 4E-BP protein that is recruited to the mei-1 mRNA by interaction with the sequence-specific RNA-binding proteins OMA-1 and OMA-2 (Li et al. 2009b).
During C. elegans oogenesis, translationally-repressed mRNAs are targeted to P-body like granules (grP bodies), that also contain the sequence-specific RNA-binding protein and translational repressor PUF-5 (Noble et al. 2008). These granules become larger in arrested oocytes and may correspond to the membrane-associated, sponge body-like structures characterized by electron microscopy (Jud et al. 2008; Patterson et al. 2011). Loss of the P-body protein CAR1/Tral/RAP55 disrupts grP bodies, and causes ectopic translation of glp-1 mRNA in oocytes, as is observed upon loss of PUF-5 and related PUFs (Noble et al. 2008).
Unlike the repressive sponge bodies, germ granules in the Drosophila germ (pole) plasm have been implicated in translational activation. For example, nanos mRNA in the cytoplasm is maintained translationally repressed by mechanisms that block translation both before and after initiation (Andrews et al. 2011; Jeske et al. 2011). Only mRNA localized with the germ (polar) granules is translated. Electron microscopy studies have shown that poly-ribosome chains extend from the surface of germ granules (Mahowald 1968, 1971; Amikura et al. 2001), suggesting that the granules could deliver mRNAs directly to the translational machinery. Translational activation of mRNAs in the germ plasm occurs in discrete temporal patterns dictated by specific sequences in the 3'UTR (Rangan et al. 2009), and may involve disassembly of silencing complexes. Translation of nanos in the germ plasm requires Oskar, which binds directly to the repressor Smaug and is sufficient in vitro to prevent assembly of a Smaug-dependent silencing complex on the nanos 3′UTR (Ephrussi and Lehmann 1992; Zaessinger et al. 2006; Jeske et al. 2011). Another key player is likely to be Vasa. Vasa binds to the initiation factor eIF5B and this interaction is required for germ cell formation in embryos (Carrera et al. 2000; Johnstone and Lasko 2004). Several germ plasm mRNAs bind to Vasa (Liu et al. 2009). Translation of at least one of them, mei-p26, is dependent on Vasa, and requires the Vasa-eIF5B interaction (Liu et al. 2009).
In Xenopus and mouse, translational activation has been correlated with exit from germ granules. In Xenopus, Xcat2 mRNA is maintained in germ granules in the Balbiani body and in the germ plasm through oogenesis and early embryogenesis (Kloc et al. 2002). Release of Xcat2 from germ granules correlates with its translational derepression (Kloc et al. 2002). Similarly, in mouse spermatids, translation of Brd2 and GCNF correlates with their exit from the chromatoid body (Nguyen Chi et al. 2009). When in the chromatoid body, the mRNAs cofractionate with Vasa (MVH) in ribonucleoprotein complexes away from polysomes (Grivna et al. 2006; Nguyen Chi et al. 2009). In stage V and older spermatids, mRNAs exit the chromatoid body, associate with polysomes, and become translated (Nguyen Chi et al. 2009). Remarkably, at that time, MVH also becomes associated with polysomes, consistent with a possible role in translational activation (Nguyen Chi et al. 2009).
mRNA Stability
Germ granules have also been implicated in the regulation of mRNA levels, by promoting either mRNA storage or mRNA turnover at specific developmental stages. In the mouse, genes required for spermiogenesis are transcribed several days before the corresponding proteins are needed (Kleene 1993). The untranslated transcripts are stored stably without degradation for up to a week. Some of these transcripts localize to chromatoid body, suggesting that this organelle provides a stable storage site for mRNAs (Soderstrom and Parvinen 1976; Kotaja et al. 2006a; Nguyen Chi et al. 2009). A role for germ granules in mRNA storage has also been proposed in C. elegans, based on the function of the DEAD-box RNA helicase Dhh1/CGH-1 (Boag et al. 2008; Noble et al. 2008). In somatic cells, Dhh1/CGH-1 functions with PATR-1 to promote mRNA decapping and degradation in P bodies. In contrast, in germ cells, Dhh1/CGH-1 functions independently from PATR-1, and associates with ∼300 mRNAs that are stably maintained during oogenesis and passed on to the embryo (Boag et al. 2008). Loss of Dhh1/CGH-1 causes destabilization of many of these mRNAs and sterility (Boag et al. 2008; Noble et al. 2008).
Other germ granule proteins in contrast have been implicated in mRNA turnover. The C. elegans Argonaute family members ALG-3 and ALG-4 generate 26G-RNAs that are required for meiosis and spermiogenesis, especially at elevated temperatures (Conine et al. 2010). Most 26G-RNAs target mRNAs are expressed during spermatogenesis, and 27% of stringently defined targets are up-regulated by twofold or more in alg-3/4 mutant worms (Conine et al. 2010). Similarly, the RNA-dependent RNA polymerase EGO-1 is required to produce a population of small triphosphorylated antisense RNAs that target hundreds of germline transcripts expressed at different stages of germline development (Maniar and Fire 2011). In ego-1 mutants, the small RNAs are not made and their target mRNAs are moderately overexpressed (Maniar and Fire 2011). One possibility is that alg-3/4 and ego-1 provide temporal fine-tuning of gene expression during germline development by targeting specific mRNAs for degradation.
CONCLUDING REMARKS
Germ granules are remarkable not only for their morphological diversity but also for the many functions they perform during germ cell development. This complexity can be dizzying, but the studies summarized here suggest two important principles and questions for future investigations.
First, many germ granule proteins have been shown to play fundamental roles in RNA regulation. Control of gene expression at the RNA level is common in germ cells, and may be essential to preserve the plasticity of the germline genome (Seydoux and Braun 2006). In fact, Vasa, PIWI, and Nanos homologs have been proposed to form a “conserved multipotency program” used not only by germ cells, but also by multipotent progenitors that retain the ability to generate both somatic and germline tissues (Juliano et al. 2010). An important challenge will be to determine whether specific steps of RNA regulation occur within germ granules, or whether germ granules function primarily as sorting stations where RNAs and regulators meet up before transitioning to the cytoplasm.
Second, unlike other organelles, germ granules are maintained in the cytoplasm without the help of surrounding membranes. The molecular interactions that give germ granules their unique combination of macro-level stability and micro-level dynamics are only beginning to be elucidated. An understanding of the unique microenvironment of germ granules is likely to yield fundamental new insights into the physical principles that organize the flow of genetic information in cells.
ACKNOWLEDGMENTS
We gratefully acknowledge support from NIH grants 5F32GM080923 (E.V.), HD37047 (G.S.), GM081634 (P.S-C.), the Howard Hughes Medical Institute (G.S. and E.V.), and the California Institute for Regenerative Medicine (I.N.).
Footnotes
Editors: Paolo Sassone-Corsi, Margaret T. Fuller, and Robert Braun
Additional Perspectives on Germ Cells available at www.cshperspectives.org
REFERENCES
- Ader C, Frey S, Maas W, Schmidt HB, Gorlich D, Baldus M 2010. Amyloid-like interactions within nucleoporin FG hydrogels. Proc Natl Acad Sci 107: 6281–6285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amikura R, Kashikawa M, Nakamura A, Kobayashi S 2001. Presence of mitochondria-type ribosomes outside mitochondria in germ plasm of Drosophila embryos. Proc Natl Acad Sci 98: 9133–9138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amiri A, Keiper BD, Kawasaki I, Fan Y, Kohara Y, Rhoads RE, Strome S 2001. An isoform of eIF4E is a component of germ granules and is required for spermatogenesis in C. elegans. Development 128: 3899–3912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson P, Kedersha N 2009. RNA granules: Post-transcriptional and epigenetic modulators of gene expression. Nat Rev Mol Cell Biol 10: 430–436 [DOI] [PubMed] [Google Scholar]
- Andonov MD, Chaldakov GN 1989. Morphological evidence for calcium storage in the chromatoid body of rat spermatids. Experientia 45: 377–378 [DOI] [PubMed] [Google Scholar]
- Andrews S, Snowflack DR, Clark IE, Gavis ER 2011. Multiple mechanisms collaborate to repress nanos translation in the Drosophila ovary and embryo. RNA 17: 967–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne J 2010. Targeting and anchoring Tudor in the pole plasm of the Drosophila oocyte. PLoS One 5: e14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anne J, Mechler BM 2005. Valois, a component of the nuage and pole plasm, is involved in assembly of these structures, and binds to Tudor and the methyltransferase Capsuleen. Development 132: 2167–2177 [DOI] [PubMed] [Google Scholar]
- Anne J, Ollo R, Ephrussi A, Mechler BM 2007. Arginine methyltransferase Capsuleen is essential for methylation of spliceosomal Sm proteins and germ cell formation in Drosophila. Development 134: 137–146 [DOI] [PubMed] [Google Scholar]
- Anton E 1983. Association of Golgi vesicles containing acid phosphatase with the chromatoid body of rat spermatids. Experientia 39: 393–394 [DOI] [PubMed] [Google Scholar]
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442: 203–207 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316: 744–747 [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell 31: 785–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, van der Heijden GW, Castaneda J, Vagin VV, Hannon GJ, Bortvin A 2009. Cytoplasmic compartmentalization of the fetal piRNA pathway in mice. PLoS Genet 5: e1000764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov AL, Ramos A 2010. Building RNA-protein granules: Insight from the germline. Trends Cell Biol 20: 482–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arkov AL, Wand JY, Ramos A, Lehmann R 2006. The role of Tudor domains in germline development and polar granule architecture. Development 133: 4053–4062 [DOI] [PubMed] [Google Scholar]
- Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR III, Desai A, Oegema K 2005. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol 171: 267–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balagopal V, Parker R 2009. Polysomes, P bodies and stress granules: States and fates of eukaryotic mRNAs. Curr Opin Cell Biol 21: 403–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbee SA, Lublin AL, Evans TC 2002. A novel function for the Sm proteins in germ granule localization during C. elegans embryogenesis. Curr Biol 12: 1502–1506 [DOI] [PubMed] [Google Scholar]
- Bardsley A, McDonald K, Boswell RE 1993. Distribution of tudor protein in the Drosophila embryo suggests separation of functions based on site of localization. Development 119: 207–219 [DOI] [PubMed] [Google Scholar]
- Batista PJ, Ruby JG, Claycomb JM, Chiang R, Fahlgren N, Kasschau KD, Chaves DA, Gu W, Vasale JJ, Duan S, et al. 2008. PRG-1 and 21U-RNAs interact to form the piRNA complex required for fertility in C. elegans. Mol Cell 31: 67–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaudoin S, Vanderperre B, Grenier C, Tremblay I, Leduc F, Roucou X 2009. A large ribonucleoprotein particle induced by cytoplasmic PrP shares striking similarities with the chromatoid body, an RNA granule predicted to function in posttranscriptional gene regulation. Biochim Biophys Acta 1793: 335–345 [DOI] [PubMed] [Google Scholar]
- Becalska AN, Gavis ER 2009. Lighting up mRNA localization in Drosophila oogenesis. Development 136: 2493–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becalska AN, Kim YR, Belletier NG, Lerit DA, Sinsimer KS, Gavis ER 2011. Aubergine is a component of a nanos mRNA localization complex. Dev Biol 349: 46–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beshore EL, McEwen TJ, Jud MC, Marshall JK, Schisa JA, Bennett KL 2011. C. elegans Dicer interacts with the P-granule component GLH-1 and both regulate germline RNPs. Dev Biol 350: 370–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggiogera M, Fakan S, Leser G, Martin TE, Gordon J 1990. Immunoelectron microscopical visualization of ribonucleoproteins in the chromatoid body of mouse spermatids. Mol Reprod Dev 26: 150–158 [DOI] [PubMed] [Google Scholar]
- Bilinski SM, Jaglarz MK, Szymanska B, Etkin LD, Kloc M 2004. Sm proteins, the constituents of the spliceosome, are components of nuage and mitochondrial cement in Xenopus oocytes. Exp Cell Res 299: 171–178 [DOI] [PubMed] [Google Scholar]
- Boag PR, Nakamura A, Blackwell TK 2005. A conserved RNA-protein complex component involved in physiological germline apoptosis regulation in C. elegans. Development 132: 4975–4986 [DOI] [PubMed] [Google Scholar]
- Boag PR, Atalay A, Robida S, Reinke V, Blackwell TK 2008. Protection of specific maternal messenger RNAs by the P body protein CGH-1 (Dhh1/RCK) during Caenorhabditis elegans oogenesis. J Cell Biol 182: 543–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bontems F, Stein A, Marlow F, Lyautey J, Gupta T, Mullins MC, Dosch R 2009. Bucky ball organizes germ plasm assembly in zebrafish. Curr Biol 19: 414–422 [DOI] [PubMed] [Google Scholar]
- Bounoure L 1937. Le sort de la lignee germinale chez la Grenouille rousse apres l’action des rayons ultraviolets sur le pole inferieur de l’oeuf. C r hebd Acad Sci 204: 1837 [Google Scholar]
- Brangwynne CP, Eckmann CR, Courson DS, Rybarska A, Hoege C, Gharakhani J, Julicher F, Hyman AA 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science 324: 1729–1732 [DOI] [PubMed] [Google Scholar]
- Breitwieser W, Markussen FH, Horstmann H, Ephrussi A 1996. Oskar protein interaction with Vasa represents an essential step in polar granule assembly. Genes Dev 10: 2179–2188 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128: 1089–1103 [DOI] [PubMed] [Google Scholar]
- Brennecke J, Malone CD, Aravin AA, Sachidanandam R, Stark A, Hannon GJ 2008. An epigenetic role for maternally inherited piRNAs in transposon silencing. Science 322: 1387–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Girard A, van de Kant HJ, Bourc’his D, Bestor TH, de Rooij DG, Hannon GJ 2007. MIWI2 is essential for spermatogenesis and repression of transposons in the mouse male germline. Dev Cell 12: 503–514 [DOI] [PubMed] [Google Scholar]
- Carrera P, Johnstone O, Nakamura A, Casanova J, Jackle H, Lasko P 2000. VASA mediates translation through interaction with a Drosophila yIF2 homolog. Mol Cell 5: 181–187 [DOI] [PubMed] [Google Scholar]
- Castrillon DH, Quade BJ, Wang TY, Quigley C, Crum CP 2000. The human VASA gene is specifically expressed in the germ cell lineage. Proc Natl Acad Sci 97: 9585–9590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Lee WC, Cook CE, Lin GW, Chang T 2006. Germ-plasm specification and germline development in the parthenogenetic pea aphid Acyrthosiphon pisum: Vasa and Nanos as markers. Int J Dev Biol 50: 413–421 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M, Hentze MW, Ephrussi A 2006. Bruno acts as a dual repressor of oskar translation, promoting mRNA oligomerization and formation of silencing particles. Cell 124: 521–533 [DOI] [PubMed] [Google Scholar]
- Chen C, Jin J, James DA, Adams-Cioaba MA, Park JG, Guo Y, Tenaglia E, Xu C, Gish G, Min J, et al. 2009. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc Natl Acad Sci 106: 20336–20341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennathukuzhi V, Morales CR, El-Alfy M, Hecht NB 2003a. The kinesin KIF17b and RNA-binding protein TB-RBP transport specific cAMP-responsive element modulator-regulated mRNAs in male germ cells. Proc Natl Acad Sci 100: 15566–15571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chennathukuzhi V, Stein JM, Abel T, Donlon S, Yang S, Miller JP, Allman DM, Simmons RA, Hecht NB 2003b. Mice deficient for testis-brain RNA-binding protein exhibit a coordinate loss of TRAX, reduced fertility, altered gene expression in the brain, and behavioral changes. Mol Cell Biol 23: 6419–6434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hiyoshi M, Yamamoto A, Hosokawa M, Takamune K, Nakatsuji N 2003. Mouse Tudor Repeat-1 (MTR-1) is a novel component of chromatoid bodies/nuages in male germ cells and forms a complex with snRNPs. Mech Dev 120: 979–990 [DOI] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Kitamura K, Kasai S, Fujioka M, Hiyoshi M, Takamune K, Noce T, Nakatsuji N 2006. Tdrd1/Mtr-1, a tudor-related gene, is essential for male germ-cell differentiation and nuage/germinal granule formation in mice. Proc Natl Acad Sci 103: 15894–15899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuma S, Hosokawa M, Tanaka T, Nakatsuji N 2009. Ultrastructural characterization of spermatogenesis and its evolutionary conservation in the germline: Germinal granules in mammals. Mol Cell Endocrinol 306: 17–23 [DOI] [PubMed] [Google Scholar]
- Clark JM, Eddy EM 1975. Fine structural observations on the origin and associations of primordial germ cells of the mouse. Dev Biol 47: 136–155 [DOI] [PubMed] [Google Scholar]
- Claycomb JM, Batista PJ, Pang KM, Gu W, Vasale JJ, van Wolfswinkel JC, Chaves DA, Shirayama M, Mitani S, Ketting RF, et al. 2009. The Argonaute CSR-1 and its 22G-RNA cofactors are required for holocentric chromosome segregation. Cell 139: 123–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conine CC, Batista PJ, Gu W, Claycomb JM, Chaves DA, Shirayama M, Mello CC 2010. Argonautes ALG-3 and ALG-4 are required for spermatogenesis-specific 26G-RNAs and thermotolerant sperm in Caenorhabditis elegans. Proc Natl Acad Sci 107: 3588–3593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa Y, Speed RM, Gautier P, Semple CA, Maratou K, Turner JM, Cooke HJ 2006. Mouse MAELSTROM: The link between meiotic silencing of unsynapsed chromatin and microRNA pathway? Hum Mol Genet 15: 2324–2334 [DOI] [PubMed] [Google Scholar]
- Coward SJ 1974. Chromatoid bodies in somatic cells of the planarian: Observations on their behavior during mitosis. Anat Rec 180: 533–545 [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC 2003. A Balbiani body and the fusome mediate mitochondrial inheritance during Drosophila oogenesis. Development 130: 1579–1590 [DOI] [PubMed] [Google Scholar]
- Cox RT, Spradling AC 2006. Milton controls the early acquisition of mitochondria by Drosophila oocytes. Development 133: 3371–3377 [DOI] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H 1998. A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czolowska R 1969. Observations on the origin of the “germinal cytoplasm” in Xenopus laevis. J Embryol Exp Morphol 22: 229–251 [PubMed] [Google Scholar]
- Das PP, Bagijn MP, Goldstein LD, Woolford JR, Lehrbach NJ, Sapetschnig A, Buhecha HR, Gilchrist MJ, Howe KL, Stark R, et al. 2008. Piwi and piRNAs act upstream of an endogenous siRNA pathway to suppress Tc3 transposon mobility in the Caenorhabditis elegans germline. Mol Cell 31: 79–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Lin H 2002. miwi, a murine homolog of piwi, encodes a cytoplasmic protein essential for spermatogenesis. Dev Cell 2: 819–830 [DOI] [PubMed] [Google Scholar]
- de Sousa Lopes SM, Roelen BA 2010. An overview on the diversity of cellular organelles during the germ cell cycle. Histol Histopathol 25: 267–276 [DOI] [PubMed] [Google Scholar]
- Dosch R, Wagner DS, Mintzer KA, Runke G, Wiemelt AP, Mullins MC 2004. Maternal control of vertebrate development before the midblastula transition: Mutants from the zebrafish I. Dev Cell 6: 771–780 [DOI] [PubMed] [Google Scholar]
- Eddy EM 1975. Germ plasm and the differentiation of the germ cell line. Int Rev Cytol 43: 229–280 [DOI] [PubMed] [Google Scholar]
- Elad N, Maimon T, Frenkiel-Krispin D, Lim RY, Medalia O 2009. Structural analysis of the nuclear pore complex by integrated approaches. Curr Opin Struct Biol 19: 226–232 [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Lehmann R 1992. Induction of germ cell formation by oskar. Nature 358: 387–392 [DOI] [PubMed] [Google Scholar]
- Ephrussi A, Dickinson LK, Lehmann R 1991. Oskar organizes the germ plasm and directs localization of the posterior determinant nanos. Cell 66: 37–50 [DOI] [PubMed] [Google Scholar]
- Fawcett DW, Eddy EM, Phillips DM 1970. Observations on the fine structure and relationships of the chromatoid body in mammalian spermatogenesis. Biol Reprod 2: 129–153 [DOI] [PubMed] [Google Scholar]
- Fimia GM, De Cesare D, Sassone-Corsi P 1999. CBP-independent activation of CREM and CREB by the LIM-only protein ACT. Nature 398: 165–169 [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H 2003. Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130: 859–871 [DOI] [PubMed] [Google Scholar]
- Forrest KM, Gavis ER 2003. Live imaging of endogenous RNA reveals a diffusion and entrapment mechanism for nanos mRNA localization in Drosophila. Curr Biol 13: 1159–1168 [DOI] [PubMed] [Google Scholar]
- Gallo CM, Munro E, Rasoloson D, Merritt C, Seydoux G 2008. Processing bodies and germ granules are distinct RNA granules that interact in C. elegans embryos. Dev Biol 323: 76–87 [DOI] [PubMed] [Google Scholar]
- Gallo CM, Wang JT, Motegi F, Seydoux G 2010. Cytoplasmic partitioning of P granule components is not required to specify the germline in C. elegans. Science 330: 1685–1689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geigy R 1931. Action de l’ultraviolet sur le pole germinal dans l’oeuf de Drosophila melanogaster (Castration et mutabilite). Rev Suisse Zool 38: 187–288 [Google Scholar]
- Gilks N, Kedersha N, Ayodele M, Shen L, Stoecklin G, Dember LM, Anderson P 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol Biol Cell 15: 5383–5398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AC, Kennedy SG, O’Leary MA, Sonenberg N, Hay N 1998. 4E-BP1, a repressor of mRNA translation, is phosphorylated and inactivated by the Akt(PKB) signaling pathway. Genes Dev 12: 502–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Tian L, Matera AG 2006. The Sm-protein methyltransferase, dart5, is essential for germ-cell specification and maintenance. Curr Biol 16: 1077–1089 [DOI] [PubMed] [Google Scholar]
- Gonsalvez GB, Rajendra TK, Wen Y, Praveen K, Matera AG 2010. Sm proteins specify germ cell fate by facilitating oskar mRNA localization. Development 137: 2341–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivna ST, Pyhtila B, Lin H 2006. MIWI associates with translational machinery and PIWI-interacting RNAs (piRNAs) in regulating spermatogenesis. Proc Natl Acad Sci 103: 13415–13420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruidl ME, Smith PA, Kuznicki KA, McCrone JS, Kirchner J, Roussell DL, Strome S, Bennett KL 1996. Multiple potential germ-line helicases are components of the germ-line-specific P granules of Caenorhabditis elegans. Proc Natl Acad Sci 93: 13837–13842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Shirayama M, Conte D Jr, Vasale J, Batista PJ, Claycomb JM, Moresco JJ, Youngman EM, Keys J, Stoltz MJ, et al. 2009. Distinct argonaute-mediated 22G-RNA pathways direct genome surveillance in the C. elegans germline. Mol Cell 36: 231–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guraya SS 1979. Recent advances in the morphology, cytochemistry, and function of Balbiani’s vitelline body in animal oocytes. Int Rev Cytol 59: 249–321 [DOI] [PubMed] [Google Scholar]
- Hanazawa M, Yonetani M, Sugimoto A 2011. PGL proteins self associate and bind RNPs to mediate germ granule assembly in C. elegans. J Cell Biol 192: 929–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haraguchi CM, Mabuchi T, Hirata S, Shoda T, Hoshi K, Akasaki K, Yokota S 2005. Chromatoid bodies: Aggresome-like characteristics and degradation sites for organelles of spermiogenic cells. J Histochem Cytochem 53: 455–465 [DOI] [PubMed] [Google Scholar]
- Harris AN, Macdonald PM 2001. Aubergine encodes a Drosophila polar granule component required for pole cell formation and related to eIF2C. Development 128: 2823–2832 [DOI] [PubMed] [Google Scholar]
- Hay B, Jan LY, Jan YN 1988. A protein component of Drosophila polar granules is encoded by vasa and has extensive sequence similarity to ATP-dependent helicases. Cell 55: 577–587 [DOI] [PubMed] [Google Scholar]
- Head JR, Kresge CK 1985. Reaction of the chromatoid body with a monoclonal antibody to a rat histocompatibility antigen. Biol Reprod 33: 1001–1008 [DOI] [PubMed] [Google Scholar]
- Hess RA, Miller LA, Kirby JD, Margoliash E, Goldberg E 1993. Immunoelectron microscopic localization of testicular and somatic cytochromes c in the seminiferous epithelium of the rat. Biol Reprod 48: 1299–1308 [DOI] [PubMed] [Google Scholar]
- Hill DP, Strome S 1990. Brief cytochalasin-induced disruption of microfilaments during a critical interval in 1-cell C. elegans embryos alters the partitioning of developmental instructions to the 2-cell embryo. Development 108: 159–172 [DOI] [PubMed] [Google Scholar]
- Hird SN, Paulsen JE, Strome S 1996. Segregation of germ granules in living Caenorhabditis elegans embryos: Cell-type-specific mechanisms for cytoplasmic localisation. Development 122: 1303–1312 [DOI] [PubMed] [Google Scholar]
- Hori I 1997. Cytological approach to morphogenesis in the planarian blastema. II. The effect of neuropeptides. J Submicrosc Cytol Pathol 29: 91–97 [PubMed] [Google Scholar]
- Horwich MD, Li C, Matranga C, Vagin V, Farley G, Wang P, Zamore PD 2007. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr Biol 17: 1265–1272 [DOI] [PubMed] [Google Scholar]
- Hosokawa M, Shoji M, Kitamura K, Tanaka T, Noce T, Chuma S, Nakatsuji N 2007. Tudor-related proteins TDRD1/MTR-1, TDRD6 and TDRD7/TRAP: Domain composition, intracellular localization, and function in male germ cells in mice. Dev Biol 301: 38–52 [DOI] [PubMed] [Google Scholar]
- Houwing S, Kamminga LM, Berezikov E, Cronembold D, Girard A, van den Elst H, Filippov DV, Blaser H, Raz E, Moens CB, et al. 2007. A role for Piwi and piRNAs in germ cell maintenance and transposon silencing in Zebrafish. Cell 129: 69–82 [DOI] [PubMed] [Google Scholar]
- Houwing S, Berezikov E, Ketting RF 2008. Zili is required for germ cell differentiation and meiosis in zebrafish. EMBO J 27: 2702–2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Gao Q, Peng X, Choi SY, Sarma K, Ren H, Morris AJ, Frohman MA 2011. piRNA-associated germline nuage formation and spermatogenesis require MitoPLD profusogenic mitochondrial-surface lipid signaling. Dev Cell 20: 376–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikenishi K, Tanaka TS 1997. Involvement of the protein of Xenopus vasa homolog (Xenopus vasa-like gene 1, XVLG1) in the differentiation of primordial germ cells. Dev Growth Differ 39: 625–633 [DOI] [PubMed] [Google Scholar]
- Ikenishi K, Tanaka TS, Komiya T 1996. Spatio-temporal distribution of the protein of Xenopus vasa homologue (Xenopus vasa-like gene 1, XVLG1) in embryos. Dev Growth Differ 38: 527–535 [DOI] [PubMed] [Google Scholar]
- Ikenishi K, Nishiumi F, Komiya T 2006. The Xdsg protein in presumptive primordial germ cells (pPGCs) is essential to their differentiation into PGCs in Xenopus. Dev Biol 297: 483–492 [DOI] [PubMed] [Google Scholar]
- Illmensee K, Mahowald AP 1974. Transplantation of posterior polar plasm in Drosophila. Induction of germ cells at the anterior pole of the egg. Proc Natl Acad Sci 71: 1016–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaglarz MK, Kloc M, Jankowska W, Szymanska B, Bilinski SM 2011. Nuage morphogenesis becomes more complex: Two translocation pathways and two forms of nuage coexist in Drosophila germline syncytia. Cell Tissue Res 344: 169–181 [DOI] [PubMed] [Google Scholar]
- Janic A, Mendizabal L, Llamazares S, Rossell D, Gonzalez C 2010. Ectopic expression of germline genes drives malignant brain tumor growth in Drosophila. Science 330: 1824–1827 [DOI] [PubMed] [Google Scholar]
- Jeske M, Moritz B, Anders A, Wahle E 2011. Smaug assembles an ATP-dependent stable complex repressing nanos mRNA translation at multiple levels. EMBO J 30: 90–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnstone O, Lasko P 2004. Interaction with eIF5B is essential for Vasa function during development. Development 131: 4167–4178 [DOI] [PubMed] [Google Scholar]
- Jud MC, Czerwinski MJ, Wood MP, Young RA, Gallo CM, Bickel JS, Petty EL, Mason JM, Little BA, Padilla PA, et al. 2008. Large P body-like RNPs form in C. elegans oocytes in response to arrested ovulation, heat shock, osmotic stress, and anoxia and are regulated by the major sperm protein pathway. Dev Biol 318: 38–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julaton VT, Reijo Pera RA 2011. NANOS3 function in human germ cell development. Hum Mol Genet 20: 2238–2250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juliano CE, Swartz SZ, Wessel GM 2010. A conserved germline multipotency program. Development 137: 4113–4126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamminga LM, Luteijn MJ, den Broeder MJ, Redl S, Kaaij LJ, Roovers EF, Ladurner P, Berezikov E, Ketting RF 2010. Hen1 is required for oocyte development and piRNA stability in zebrafish. EMBO J 29: 3688–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashikawa M, Amikura R, Nakamura A, Kobayashi S 1999. Mitochondrial small ribosomal RNA is present on polar granules in early cleavage embryos of Drosophila melanogaster. Dev Growth Differ 41: 495–502 [DOI] [PubMed] [Google Scholar]
- Kashikawa M, Amikura R, Kobayashi S 2001. Mitochondrial small ribosomal RNA is a component of germinal granules in Xenopus embryos. Mech Dev 101: 71–77 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Smith JL, Macdonald PM 1991. oskar mRNA is localized to the posterior pole of the Drosophila oocyte. Cell 66: 23–35 [DOI] [PubMed] [Google Scholar]
- Kim-Ha J, Kerr K, Macdonald PM 1995. Translational regulation of oskar mRNA by bruno, an ovarian RNA-binding protein, is essential. Cell 81: 403–412 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z 2007a. The mouse homolog of HEN1 is a potential methylase for Piwi-interacting RNAs. RNA 13: 1397–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Mourelatos Z 2007b. Mouse Piwi-interacting RNAs are 2′-O-methylated at their 3′ termini. Nat Struct Mol Biol 14: 347–348 [DOI] [PubMed] [Google Scholar]
- Kirino Y, Kim N, de Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z 2009. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat Cell Biol 11: 652–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Kim N, de Lima Alves F, Rappsilber J, Klein PS, Jongens TA, Mourelatos Z 2010a. Arginine methylation of vasa protein is conserved across phyla. J Biol Chem 285: 8148–8154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino Y, Vourekas A, Sayed N, de Lima Alves F, Thomson T, Lasko P, Rappsilber J, Jongens TA, Mourelatos Z 2010b. Arginine methylation of Aubergine mediates Tudor binding and germ plasm localization. RNA 16: 70–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene KC 1993. Multiple controls over the efficiency of translation of the mRNAs encoding transition proteins, protamines, and the mitochondrial capsule selenoprotein in late spermatids in mice. Dev Biol 159: 720–731 [DOI] [PubMed] [Google Scholar]
- Kloc M, Etkin LD 1994. Delocalization of Vg1 mRNA from the vegetal cortex in Xenopus oocytes after destruction of Xlsirt RNA. Science 265: 1101–1103 [DOI] [PubMed] [Google Scholar]
- Kloc M, Dougherty MT, Bilinski S, Chan AP, Brey E, King ML, Patrick CW Jr, Etkin LD 2002. Three-dimensional ultrastructural analysis of RNA distribution within germinal granules of Xenopus. Dev Biol 241: 79–93 [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Etkin LD 2004. The Balbiani body and germ cell determinants: 150 years later. Curr Top Dev Biol 59: 1–36 [DOI] [PubMed] [Google Scholar]
- Kloc M, Wilk K, Vargas D, Shirato Y, Bilinski S, Etkin LD 2005. Potential structural role of non-coding and coding RNAs in the organization of the cytoskeleton at the vegetal cortex of Xenopus oocytes. Development 132: 3445–3457 [DOI] [PubMed] [Google Scholar]
- Kloc M, Bilinski S, Dougherty MT 2007. Organization of cytokeratin cytoskeleton and germ plasm in the vegetal cortex of Xenopus laevis oocytes depends on coding and non-coding RNAs: Three-dimensional and ultrastructural analysis. Exp Cell Res 313: 1639–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaut H, Pelegri F, Bohmann K, Schwarz H, Nusslein-Volhard C 2000. Zebrafish vasa RNA but not its protein is a component of the germ plasm and segregates asymmetrically before germline specification. J Cell Biol 149: 875–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi S, Yamada M, Asaoka M, Kitamura T 1996. Essential role of the posterior morphogen nanos for germline development in Drosophila. Nature 380: 708–711 [DOI] [PubMed] [Google Scholar]
- Kogo N, Tazaki A, Kashino Y, Morichika K, Orii H, Mochii M, Watanabe K 2011. Germ-line mitochondria exhibit suppressed respiratory activity to support their accurate transmission to the next generation. Dev Biol 349: 462–469 [DOI] [PubMed] [Google Scholar]
- Kojima K, Kuramochi-Miyagawa S, Chuma S, Tanaka T, Nakatsuji N, Kimura T, Nakano T 2009. Associations between PIWI proteins and TDRD1/MTR-1 are critical for integrated subcellular localization in murine male germ cells. Genes Cells 14: 1155–1165 [DOI] [PubMed] [Google Scholar]
- Koprunner M, Thisse C, Thisse B, Raz E 2001. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev 15: 2877–2885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosaka K, Kawakami K, Sakamoto H, Inoue K 2007. Spatiotemporal localization of germ plasm RNAs during zebrafish oogenesis. Mech Dev 124: 279–289 [DOI] [PubMed] [Google Scholar]
- Kotaja N, Sassone-Corsi P 2007. The chromatoid body: A germ-cell-specific RNA-processing centre. Nat Rev Mol Cell Biol 8: 85–90 [DOI] [PubMed] [Google Scholar]
- Kotaja N, Bhattacharyya SN, Jaskiewicz L, Kimmins S, Parvinen M, Filipowicz W, Sassone-Corsi P 2006a. The chromatoid body of male germ cells: Similarity with processing bodies and presence of Dicer and microRNA pathway components. Proc Natl Acad Sci 103: 2647–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotaja N, Lin H, Parvinen M, Sassone-Corsi P 2006b. Interplay of PIWI/Argonaute protein MIWI and kinesin KIF17b in chromatoid bodies of male germ cells. J Cell Sci 119: 2819–2825 [DOI] [PubMed] [Google Scholar]
- Krimer DB, Esponda P 1980. Presence of polysaccharides and proteins in the chromatoid body of mouse spermatids. Cell Biol Int Rep 4: 265–270 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Kimura T, Ijiri TW, Isobe T, Asada N, Fujita Y, Ikawa M, Iwai N, Okabe M, Deng W, et al. 2004. Mili, a mammalian member of piwi family gene, is essential for spermatogenesis. Development 131: 839–849 [DOI] [PubMed] [Google Scholar]
- Kuramochi-Miyagawa S, Watanabe T, Gotoh K, Takamatsu K, Chuma S, Kojima-Kita K, Shiromoto Y, Asada N, Toyoda A, Fujiyama A, et al. 2010. MVH in piRNA processing and gene silencing of retrotransposons. Genes Dev 24: 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznicki KA, Smith PA, Leung-Chiu WM, Estevez AO, Scott HC, Bennett KL 2000. Combinatorial RNA interference indicates GLH-4 can compensate for GLH-1; these two P granule components are critical for fertility in C. elegans. Development 127: 2907–2916 [DOI] [PubMed] [Google Scholar]
- Lall S, Piano F, Davis RE 2005. Caenorhabditis elegans decapping proteins: Localization and functional analysis of Dcp1, Dcp2, and DcpS during embryogenesis. Mol Biol Cell 16: 5880–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasko PF, Ashburner M 1990. Posterior localization of vasa protein correlates with, but is not sufficient for, pole cell development. Genes Dev 4: 905–921 [DOI] [PubMed] [Google Scholar]
- Lerit DA, Gavis ER 2011. Transport of germ plasm on astral microtubules directs germ cell development in Drosophila. Curr Biol 21: 439–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Vagin VV, Lee S, Xu J, Ma S, Xi H, Seitz H, Horwich MD, Syrzycka M, Honda BM, et al. 2009a. Collapse of germline piRNAs in the absence of Argonaute3 reveals somatic piRNAs in flies. Cell 137: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, DeBella LR, Guven-Ozkan T, Lin R, Rose LS 2009b. An eIF4E-binding protein regulates katanin protein levels in C. elegans embryos. J Cell Biol 187: 33–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim AK, Kai T 2007. Unique germ-line organelle, nuage, functions to repress selfish genetic elements in Drosophila melanogaster. Proc Natl Acad Sci 104: 6714–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang L, Diehl-Jones W, Lasko P 1994. Localization of vasa protein to the Drosophila pole plasm is independent of its RNA-binding and helicase activities. Development 120: 1201–1211 [DOI] [PubMed] [Google Scholar]
- Lim AK, Tao L, Kai T 2009. piRNAs mediate posttranscriptional retroelement silencing and localization to pi-bodies in the Drosophila germline. J Cell Biol 186: 333–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Spradling AC 1997. A novel group of pumilio mutations affects the asymmetric division of germline stem cells in the Drosophila ovary. Development 124: 2463–2476 [DOI] [PubMed] [Google Scholar]
- Lin MD, Fan SJ, Hsu WS, Chou TB 2006. Drosophila decapping protein 1, dDcp1, is a component of the oskar mRNP complex and directs its posterior localization in the oocyte. Dev Cell 10: 601–613 [DOI] [PubMed] [Google Scholar]
- Liu JL, Gall JG 2007. U bodies are cytoplasmic structures that contain uridine-rich small nuclear ribonucleoproteins and associate with P bodies. Proc Natl Acad Sci 104: 11655–11659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N, Han H, Lasko P 2009. Vasa promotes Drosophila germline stem cell differentiation by activating mei-P26 translation by directly interacting with a (U)-rich motif in its 3′ UTR. Genes Dev 23: 2742–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Wang JY, Huang Y, Li Z, Gong W, Lehmann R, Xu RM 2010. Structural basis for methylarginine-dependent recognition of Aubergine by Tudor. Genes Dev 24: 1876–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Qi H, Wang J, Lin H 2011. PAPI, a novel TUDOR-domain protein, complexes with AGO3, ME31B and TRAL in the nuage to silence transposition. Development 138: 1863–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Buchold GM, Greenbaum MP, Roy A, Burns KH, Zhu H, Han DY, Harris RA, Coarfa C, Gunaratne PH, et al. 2009. GASZ is essential for male meiosis and suppression of retrotransposon expression in the male germline. PLoS Genet 5: e1000635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahowald AP 1968. Polar granules of Drosophila. II. Ultrastructural changes during early embryogenesis. J Exp Zool 167: 237–261 [DOI] [PubMed] [Google Scholar]
- Mahowald AP 1971. Polar granules of Drosophila. 3. The continuity of polar granules during the life cycle of Drosophila. J Exp Zool 176: 329–343 [DOI] [PubMed] [Google Scholar]
- Mahowald AP 2001. Assembly of the Drosophila germ plasm. Int Rev Cytol 203: 187–213 [DOI] [PubMed] [Google Scholar]
- Maniar JM, Fire AZ 2011. EGO-1, a C. elegans RdRP, modulates gene expression via production of mRNA-templated short antisense RNAs. Curr Biol 21: 449–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markussen FH, Michon AM, Breitwieser W, Ephrussi A 1995. Translational control of oskar generates short OSK, the isoform that induces pole plasma assembly. Development 121: 3723–3732 [DOI] [PubMed] [Google Scholar]
- Marlow FL, Mullins MC 2008. Bucky ball functions in Balbiani body assembly and animal-vegetal polarity in the oocyte and follicle cell layer in zebrafish. Dev Biol 321: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marston DJ, Goldstein B 2006. Symmetry breaking in C. elegans: Another gift from the sperm. Dev Cell 11: 273–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megosh HB, Cox DN, Campbell C, Lin H 2006. The role of PIWI and the miRNA machinery in Drosophila germline determination. Curr Biol 16: 1884–1894 [DOI] [PubMed] [Google Scholar]
- Meikar O, Da Ros M, Liljenback H, Toppari J, Kotaja N 2010. Accumulation of piRNAs in the chromatoid bodies purified by a novel isolation protocol. Exp Cell Res 316: 1567–1575 [DOI] [PubMed] [Google Scholar]
- Mosquera L, Forristall C, Zhou Y, King ML 1993. A mRNA localized to the vegetal cortex of Xenopus oocytes encodes a protein with a nanos-like zinc finger domain. Development 117: 377–386 [DOI] [PubMed] [Google Scholar]
- Moussa F, Oko R, Hermo L 1994. The immunolocalization of small nuclear ribonucleoprotein particles in testicular cells during the cycle of the seminiferous epithelium of the adult rat. Cell Tissue Res 278: 363–378 [DOI] [PubMed] [Google Scholar]
- Murchison EP, Stein P, Xuan Z, Pan H, Zhang MQ, Schultz RM, Hannon GJ 2007. Critical roles for Dicer in the female germline. Genes Dev 21: 682–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Amikura R, Hanyu K, Kobayashi S 2001. Me31B silences translation of oocyte-localizing RNAs through the formation of cytoplasmic RNP complex during Drosophila oogenesis. Development 128: 3233–3242 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Sato K, Hanyu-Nakamura K 2004. Drosophila cup is an eIF4E binding protein that associates with Bruno and regulates oskar mRNA translation in oogenesis. Dev Cell 6: 69–78 [DOI] [PubMed] [Google Scholar]
- Navarro RE, Shim EY, Kohara Y, Singson A, Blackwell TK 2001. cgh-1, a conserved predicted RNA helicase required for gametogenesis and protection from physiological germline apoptosis in C elegans. Development 128: 3221–3232 [DOI] [PubMed] [Google Scholar]
- Nguyen Chi M, Chalmel F, Agius E, Vanzo N, Khabar KS, Jegou B, Morello D 2009. Temporally regulated traffic of HuR and its associated ARE-containing mRNAs from the chromatoid body to polysomes during mouse spermatogenesis. PLoS One 4: e4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Saito K, Mori T, Kawamura Y, Nagami-Okada T, Inagaki S, Siomi H, Siomi MC 2007. Gene silencing mechanisms mediated by Aubergine piRNA complexes in Drosophila male gonad. RNA 13: 1911–1922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida KM, Okada TN, Kawamura T, Mituyama T, Kawamura Y, Inagaki S, Huang H, Chen D, Kodama T, Siomi H, et al. 2009. Functional involvement of Tudor and dPRMT5 in the piRNA processing pathway in Drosophila germlines. EMBO J 28: 3820–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SL, Allen BL, Goh LK, Nordick K, Evans TC 2008. Maternal mRNAs are regulated by diverse P body-related mRNP granules during early Caenorhabditis elegans development. J Cell Biol 182: 559–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohara T, Sakaguchi Y, Suzuki T, Ueda H, Miyauchi K, Suzuki T 2007. The 3′ termini of mouse Piwi-interacting RNAs are 2′-O-methylated. Nat Struct Mol Biol 14: 349–350 [DOI] [PubMed] [Google Scholar]
- Oko R, Korley R, Murray MT, Hecht NB, Hermo L 1996. Germ cell-specific DNA and RNA binding proteins p48/52 are expressed at specific stages of male germ cell development and are present in the chromatoid body. Mol Reprod Dev 44: 1–13 [DOI] [PubMed] [Google Scholar]
- Ollinger R, Reichmann J, Adams IR 2010. Meiosis and retrotransposon silencing during germ cell development in mice. Differentiation 79: 147–158 [DOI] [PubMed] [Google Scholar]
- Pan J, Goodheart M, Chuma S, Nakatsuji N, Page DC, Wang PJ 2005. RNF17, a component of the mammalian germ cell nuage, is essential for spermiogenesis. Development 132: 4029–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paniagua R, Nistal M, Amat P, Rodriguez MC 1985. Presence of ribonucleoproteins and basic proteins in the nuage and intermitochondrial bars of human spermatogonia. J Anat 143: 201–206 [PMC free article] [PubMed] [Google Scholar]
- Parvinen M 2005. The chromatoid body in spermatogenesis. Int J Androl 28: 189–201 [DOI] [PubMed] [Google Scholar]
- Patil VS, Kai T 2010. Repression of retroelements in Drosophila germline via piRNA pathway by the Tudor domain protein Tejas. Curr Biol 8: 724–730 [DOI] [PubMed] [Google Scholar]
- Patterson JR, Wood MP, Schisa JA 2011. Assembly of RNP granules in stressed and aging oocytes requires nucleoporins and is coordinated with nuclear membrane blebbing. Dev Biol 15: 173–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, Kai T 2011a. Non-coding RNAs enter mitosis: Functions, conservation and implications. Cell Div 6: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pek JW, Kai T 2011b. A role for vasa in regulating mitotic chromosome condensation in Drosophila. Curr Biol 21: 39–44 [DOI] [PubMed] [Google Scholar]
- Pepling ME, Wilhelm JE, O’Hara AL, Gephardt GW, Spradling AC 2007. Mouse oocytes within germ cell cysts and primordial follicles contain a Balbiani body. Proc Natl Acad Sci 104: 187–192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrella LN, Wang W, Spike CA, Rechtsteiner A, Reinke V, Strome S 2011. synMuv B proteins antagonize germline fate in the intestine and ensure C. elegans survival. Development 138: 1069–1079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitt JN, Schisa JA, Priess JR 2000. P granules in the germ cells of Caenorhabditis elegans adults are associated with clusters of nuclear pores and contain RNA. Dev Biol 219: 315–333 [DOI] [PubMed] [Google Scholar]
- Rangan P, DeGennaro M, Jaime-Bustamante K, Coux RX, Martinho RG, Lehmann R 2009. Temporal and spatial control of germ-plasm RNAs. Curr Biol 19: 72–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS 2009. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat Struct Mol Biol 16: 639–646 [DOI] [PubMed] [Google Scholar]
- Ritter R 1890. Die Entwicklung der Geschlechtsorgane und des Darmes bei Chiromomus. Zeit furr Wiss Zool Bd 50: 408–427 [Google Scholar]
- Rongo C, Lehmann R 1996. Regulated synthesis, transport and assembly of the Drosophila germ plasm. Trends Genet 12: 102–109 [DOI] [PubMed] [Google Scholar]
- Rongo C, Broihier HT, Moore L, Van Doren M, Forbes A, Lehmann R 1997. Germ plasm assembly and germ cell migration in Drosophila. Cold Spring Harb Symp Quant Biol 62: 1–11 [PubMed] [Google Scholar]
- Roper K 2007. Rtnl1 is enriched in a specialized germline ER that associates with ribonucleoprotein granule components. J Cell Sci 120: 1081–1092 [DOI] [PubMed] [Google Scholar]
- Roucou X 2009. Prion protein and RNA: A view from the cytoplasm. Front Biosci 14: 5157–5164 [DOI] [PubMed] [Google Scholar]
- Rouelle-Rossier VB, Biggiogera M, Fakan S 1993. Ultrastructural detection of calcium and magnesium in the chromatoid body of mouse spermatids by electron spectroscopic imaging and electron energy loss spectroscopy. J Histochem Cytochem 41: 1155–1162 [DOI] [PubMed] [Google Scholar]
- Saito K, Sakaguchi Y, Suzuki T, Suzuki T, Siomi H, Siomi MC 2007. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev 21: 1603–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato K, Sugita T, Kobayashi K, Fujita K, Fujii T, Matsumoto Y, Mikami T, Nishizuka N, Nishizuka S, Shojima K, et al. 2001. Localization of mitochondrial ribosomal RNA on the chromatoid bodies of marine planarian polyclad embryos. Dev Growth Differ 43: 107–114 [DOI] [PubMed] [Google Scholar]
- Schupbach T, Wieschaus E 1986. Germline autonomy of maternal-effect mutations altering the embryonic body pattern of Drosophila. Dev Biol 113: 443–448 [DOI] [PubMed] [Google Scholar]
- Senti KA, Brennecke J 2010. The piRNA pathway: A fly’s perspective on the guardian of the genome. Trends Genet 26: 499–509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seydoux G, Braun RE 2006. Pathway to totipotency: Lessons from germ cells. Cell 127: 891–904 [DOI] [PubMed] [Google Scholar]
- Sheth U, Pitt J, Dennis S, Priess JR 2010. Perinuclear P granules are the principal sites of mRNA export in adult C. elegans germ cells. Development 137: 1305–1314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata N, Tsunekawa N, Okamoto-Ito S, Akasu R, Tokumasu A, Noce T 2004. Mouse RanBPM is a partner gene to a germline specific RNA helicase, mouse vasa homolog protein. Mol Reprod Dev 67: 1–7 [DOI] [PubMed] [Google Scholar]
- Shoji M, Tanaka T, Hosokawa M, Reuter M, Stark A, Kato Y, Kondoh G, Okawa K, Chujo T, Suzuki T, et al. 2009. The TDRD9-MIWI2 complex is essential for piRNA-mediated retrotransposon silencing in the mouse male germline. Dev Cell 17: 775–787 [DOI] [PubMed] [Google Scholar]
- Smith LD 1966. The role of a “germinal plasm” in the formation of primordial germ cells in Rana pipiens. Dev Biol 14: 330–347 [DOI] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM 2004. Live imaging of nuage and polar granules: Evidence against a precursor-product relationship and a novel role for Oskar in stabilization of polar granule components. J Cell Sci 117: 2109–2120 [DOI] [PubMed] [Google Scholar]
- Snee MJ, Macdonald PM 2009. Dynamic organization and plasticity of sponge bodies. Dev Dyn 238: 918–930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soderstrom KO 1977. Effect of actinomycin D on the structure of the chromatoid body in the rat spermatids. Cell Tissue Res 184: 411–421 [DOI] [PubMed] [Google Scholar]
- Soderstrom KO, Parvinen M 1976. Incorporation of (3H)uridine by the chromatoid body during rat spermatogenesis. J Cell Biol 70: 239–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soper SF, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A 2008. Mouse maelstrom, a component of nuage, is essential for spermatogenesis and transposon repression in meiosis. Dev Cell 15: 285–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman M, Bennett D 1973. A light- and electron-microscopic study of primordial germ cells in the early mouse embryo. J Embryol Exp Morphol 30: 97–118 [PubMed] [Google Scholar]
- Spike C, Meyer N, Racen E, Orsborn A, Kirchner J, Kuznicki K, Yee C, Bennett K, Strome S 2008. Genetic analysis of the Caenorhabditis elegans GLH family of P-granule proteins. Genetics 178: 1973–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, Lyons GE, Anderson P, White JG 2006. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell 17: 336–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser MJ, Mackenzie NC, Dumstrei K, Nakkrasae LI, Stebler J, Raz E 2008. Control over the morphology and segregation of Zebrafish germ cell granules during embryonic development. BMC Dev Biol 8: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB 1982. Immunofluorescence visualization of germ-line-specific cytoplasmic granules in embryos, larvae, and adults of Caenorhabditis elegans. Proc Natl Acad Sci 79: 1558–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB 1983. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell 35: 15–25 [DOI] [PubMed] [Google Scholar]
- Strome S, Martin P, Schierenberg E, Paulsen J 1995. Transformation of the germ line into muscle in mes-1 mutant embryos of C. elegans. Development 121: 2961–2972 [DOI] [PubMed] [Google Scholar]
- Styhler S, Nakamura A, Swan A, Suter B, Lasko P 1998. vasa is required for GURKEN accumulation in the oocyte, and is involved in oocyte differentiation and germline cyst development. Development 125: 1569–1578 [DOI] [PubMed] [Google Scholar]
- Subramaniam K, Seydoux G 1999. nos-1 and nos-2, two genes related to Drosophila nanos, regulate primordial germ cell development and survival in Caenorhabditis elegans. Development 126: 4861–4871 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Tsuda M, Saga Y 2007. Functional redundancy among Nanos proteins and a distinct role of Nanos2 during male germ cell development. Development 134: 77–83 [DOI] [PubMed] [Google Scholar]
- Swetloff A, Conne B, Huarte J, Pitetti JL, Nef S, Vassalli JD 2009. Dcp1-bodies in mouse oocytes. Mol Biol Cell 20: 4951–4961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam OH, Aravin AA, Stein P, Girard A, Murchison EP, Cheloufi S, Hodges E, Anger M, Sachidanandam R, Schultz RM, et al. 2008. Pseudogene-derived small interfering RNAs regulate gene expression in mouse oocytes. Nature 453: 534–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Nakamura A 2008. The endocytic pathway acts downstream of Oskar in Drosophila germ plasm assembly. Development 135: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T 2000. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev 14: 841–853 [PMC free article] [PubMed] [Google Scholar]
- Tang XM, Lalli MF, Clermont Y 1982. A cytochemical study of the Golgi apparatus of the spermatid during spermiogenesis in the rat. Am J Anat 163: 283–294 [DOI] [PubMed] [Google Scholar]
- Terry LJ, Wente SR 2009. Flexible gates: Dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell 8: 1814–1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theusch EV, Brown KJ, Pelegri F 2006. Separate pathways of RNA recruitment lead to the compartmentalization of the zebrafish germ plasm. Dev Biol 292: 129–141 [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H 2009. The biogenesis and function of PIWI proteins and piRNAs: Progress and prospect. Annu Rev Cell Dev Biol 25: 355–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson T, Liu N, Arkov A, Lehmann R, Lasko P 2008. Isolation of new polar granule components in Drosophila reveals P body and ER associated proteins. Mech Dev 125: 865–873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorne-Tjomsland G, Clermont Y, Hermo L 1988. Contribution of the Golgi apparatus components to the formation of the acrosomic system and chromatoid body in rat spermatids. Anat Rec 221: 591–598 [DOI] [PubMed] [Google Scholar]
- Toyooka Y, Tsunekawa N, Takahashi Y, Matsui Y, Satoh M, Noce T 2000. Expression and intracellular localization of mouse Vasa-homologue protein during germ cell development. Mech Dev 93: 139–149 [DOI] [PubMed] [Google Scholar]
- Tsai-Morris CH, Sheng Y, Lee E, Lei KJ, Dufau ML 2004. Gonadotropin-regulated testicular RNA helicase (GRTH/Ddx25) is essential for spermatid development and completion of spermatogenesis. Proc Natl Acad Sci 101: 6373–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuda M, Sasaoka Y, Kiso M, Abe K, Haraguchi S, Kobayashi S, Saga Y 2003. Conserved role of nanos proteins in germ cell development. Science 301: 1239–1241 [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Shin TH, Miliaras N, Lee J, Oyama T, Mello CC 2002. MEP-1 and a homolog of the NURD complex component Mi-2 act together to maintain germline-soma distinctions in C. elegans. Cell 111: 991–1002 [DOI] [PubMed] [Google Scholar]
- Unhavaithaya Y, Hao Y, Beyret E, Yin H, Kuramochi-Miyagawa S, Nakano T, Lin H 2009. MILI, a PIWI-interacting RNA-binding protein, is required for germ line stem cell self-renewal and appears to positively regulate translation. J Biol Chem 284: 6507–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Strome S 2009a. A genomewide rnai screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics 183: 1397–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Strome S 2009b. P granule assembly in C. elegans germ cells. J Androl 31: 53–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Updike DL, Hachey SJ, Kreher J, Strome S 2011. P granules extend the nuclear pore complex environment in the C. elegans germ line. J Cell Biol 192: 939–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Klenov MS, Kalmykova AI, Stolyarenko AD, Kotelnikov RN, Gvozdev VA 2004. The RNA interference proteins and vasa locus are involved in the silencing of retrotransposons in the female germline of Drosophila melanogaster. RNA Biol 1: 54–58 [PubMed] [Google Scholar]
- Vagin VV, Sigova A, Li C, Seitz H, Gvozdev V, Zamore PD 2006. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 313: 320–324 [DOI] [PubMed] [Google Scholar]
- Vagin VV, Hannon GJ, Aravin AA 2009a. Arginine methylation as a molecular signature of the Piwi small RNA pathway. Cell Cycle 8: 4003–4004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA 2009b. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev 23: 1749–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk E, Cougot N, Meyer S, Babajko S, Wahle E, Séraphin B 2002. Human Dcp2: A catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J 21: 6915–6924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanzo NF, Ephrussi A 2002. Oskar anchoring restricts pole plasm formation to the posterior of the Drosophila oocyte. Development 129: 3705–3714 [DOI] [PubMed] [Google Scholar]
- Vasileva A, Tiedau D, Firooznia A, Müller-Teichert T, Jessberger R 2009. Tdrd6 is required for spermiogenesis, chromatoid body architecture, and regulation of miRNA expression. Curr Biol 19: 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventela S, Toppari J, Parvinen M 2003. Intercellular organelle traffic through cytoplasmic bridges in early spermatids of the rat: Mechanisms of haploid gene product sharing. Mol Biol Cell 14: 2768–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verdel A, Jia S, Gerber S, Sugiyama T, Gygi S, Grewal SI, Moazed D 2004. RNAi-mediated targeting of heterochromatin by the RITS complex. Science 303: 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronina E, Seydoux G 2010. The C. elegans homolog of nucleoporin Nup98 is required for the integrity and function of germline P granules. Development 137: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Kirino Y, Mourelatos Z 2010. Elective affinities: A Tudor-Aubergine tale of germline partnership. Genes Dev 24: 1963–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walt H, Armbruster BL 1984. Actin and RNA are components of the chromatoid bodies in spermatids of the rat. Cell Tissue Res 236: 487–490 [DOI] [PubMed] [Google Scholar]
- Wang C, Lehmann R 1991. Nanos is the localized posterior determinant in Drosophila. Cell 66: 637–647 [DOI] [PubMed] [Google Scholar]
- Wang C, Dickinson LK, Lehmann R 1994. Genetics of nanos localization in Drosophila. Dev Dyn 199: 103–115 [DOI] [PubMed] [Google Scholar]
- Wang D, Kennedy S, Conte D Jr, Kim JK, Gabel HW, Kamath RS, Mello CC, Ruvkun G 2005. Somatic misexpression of germline P granules and enhanced RNA interference in retinoblastoma pathway mutants. Nature 436: 593–597 [DOI] [PubMed] [Google Scholar]
- Wang Y, Zayas RM, Guo T, Newmark PA 2007. Nanos function is essential for development and regeneration of planarian germ cells. Proc Natl Acad Sci 104: 5901–5906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Saxe JP, Tanaka T, Chuma S, Lin H 2009. Mili interacts with tudor domain-containing protein 1 in regulating spermatogenesis. Curr Biol 19: 640–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T, Totoki Y, Toyoda A, Kaneda M, Kuramochi-Miyagawa S, Obata Y, Chiba H, Kohara Y, Kono T, Nakano T, et al. 2008. Endogenous siRNAs from naturally formed dsRNAs regulate transcripts in mouse oocytes. Nature 453: 539–543 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Chuma S, Yamamoto Y, Kuramochi-Miyagawa S, Totoki Y, Toyoda A, Hoki Y, Fujiyama A, Shibata T, Sado T, et al. 2011. MITOPLD is a mitochondrial protein essential for nuage formation and piRNA biogenesis in the mouse germline. Dev Cell 20: 364–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner G, Werner K 1995. Immunocytochemical localization of histone H4 in the chromatoid body of rat spermatids. J Submicrosc Cytol Pathol 27: 325–330 [PubMed] [Google Scholar]
- Wilhelm JE, Mansfield J, Hom-Booher N, Wang S, Turck CW, Hazelrigg T, Vale RD 2000. Isolation of a ribonucleoprotein complex involved in mRNA localization in Drosophila oocytes. J Cell Biol 148: 427–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Hilton M, Amos Q, Henzel WJ 2003. Cup is an eIF4E binding protein required for both the translational repression of oskar and the recruitment of Barentsz. J Cell Biol 163: 1197–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm JE, Buszczak M, Sayles S 2005. Efficient protein trafficking requires trailer hitch, a component of a ribonucleoprotein complex localized to the ER in Drosophila. Dev Cell 9: 675–685 [DOI] [PubMed] [Google Scholar]
- Wilk K, Bilinski S, Dougherty MT, Kloc M 2005. Delivery of germinal granules and localized RNAs via the messenger transport organizer pathway to the vegetal cortex of Xenopus oocytes occurs through directional expansion of the mitochondrial cloud. Int J Dev Biol 49: 17–21 [DOI] [PubMed] [Google Scholar]
- Wilsch-Brauninger M, Schwarz H, Nusslein-Volhard C 1997. A sponge-like structure involved in the association and transport of maternal products during Drosophila oogenesis. J Cell Biol 139: 817–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabuta Y, Ohta H, Abe T, Kurimoto K, Chuma S, Saitou M 2011. TDRD5 is required for retrotransposon silencing, chromatoid body assembly, and spermiogenesis in mice. J Cell Biol 192: 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yajima M, Wessel GM 2011. The DEAD-box RNA helicase Vasa functions in embryonic mitotic progression in the sea urchin. Development 138: 2217–2222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Lopez de Quinto S, Matsui Y, Shevchenko A, Shevchenko A, Ephrussi A 2004. Hrp48, a Drosophila hnRNPA/B homolog, binds and regulates translation of oskar mRNA. Dev Cell 6: 637–648 [DOI] [PubMed] [Google Scholar]
- Zaessinger S, Busseau I, Simonelig M 2006. Oskar allows nanos mRNA translation in Drosophila embryos by preventing its deadenylation by Smaug/CCR4. Development 133: 4573–4583 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Yan L, Zhou Z, Yang P, Tian E, Zhang K, Zhao Y, Li Z, Song B, Han J, et al. 2009. SEPA-1 mediates the specific recognition and degradation of P granule components by autophagy in C. elegans. Cell 136: 308–321 [DOI] [PubMed] [Google Scholar]
- Zhou Y, King ML 2004. Sending RNAs into the future: RNA localization and germ cell fate. IUBMB Life 56: 19–27 [DOI] [PubMed] [Google Scholar]