Abstract
Over 20 genetic loci with abnormal expansions of short tandem repeats have been associated with human hereditary neurological diseases. Of these, specific trinucleotide repeats located in non-coding and coding regions of individual genes implicated in these disorders are strongly overrepresented. Expansions of CTG, CGG and CAG repeats are linked to, respectively, myotonic dystrophy type 1 (DM1), fragile X-associated tremor/ataxia syndrome (FXTAS), as well as Huntington's disease (HD) and a number of spinocerebellar ataxias (SCAs). Expanded CAG repeats in translated exons trigger the most disorders for which a protein gain-of-function mechanism has been proposed to explain neurodegeneration by polyglutamine-rich (poly-Q) proteins. However, the results of last years showed that RNA composed of mutated CAG repeats can also be toxic and contribute to pathogenesis of polyglutamine disorders through an RNA-mediated gain-of-function mechanism. This mechanism has been best characterized in the non-coding repeat disorder DM1 and is also implicated in several other diseases, such as FXTAS, spinocerebellar ataxia type 8 (SCA8), Huntington's disease-like 2 (HDL2), as well as in myotonic dystrophy type 2 (DM2), spinocerebellar ataxia type 10 (SCA10) and type 31 (SCA31). in this review, we summarize recent findings that emphasize the participation of coding mutant CAG repeat RNA in the pathogenesis of polyglutamine disorders, and we discuss the basis of an RNA gain-of-function model in non-coding diseases such as DM1, FXTAS and SCA8.
Key words: DM1, FXTAS, MBNL sequestration, polyglutamine disorders, RNA gain-of-function, SCA8
Introduction
About half of the human genome is composed of repeated sequences of various types; of these, short tandem repeats, such as trinucleotide repeats, represent a substantial portion.1 CAG, CGG, CCG, CTG and AGG repeats are strongly overrepresented in the human exome as compared with the genome, and these repeats are also present in translated sequences of mRNAs because their frequent length variation does not change the open reading frame.2,3 The characteristic features of short tandem repeats are their genetic instability and their ability to expand4,5 in germline and somatic cells. Short tandem repeats act as trigger in over 20 neurodegenerative and neuromuscular human disorders (reviewed in refs. 6–8) collectively known as triplet repeat expansion diseases (TREDs). These disorders include FXTAS, several SCAs, HD, dentatorubral-pallidoluysian atrophy (DRPLA), Friedreich's ataxia (FRDA) and DM1. Although unstable trinucleotide repeats are the most common repeats that cause neurological disorders, other repeats, such as tetra- and pentanucleotides, may also expand, resulting in DM2 as well as SCA10 and SCA31.9–11
Intragenically located expanded trinucleotide repeats exert their pathogenic effect on transcript and/or protein levels. For repeat mutations present in non-protein coding sequences, an RNA gain-of-function mechanism, which explains how transcripts of expanded alleles exert toxicity, has been proposed (reviewed in refs. 7, 8 and 12). DM1, which is caused by a 3′UTR CTG repeat expansion, is the best characterized example of a disorder in which mutant RNA is thought to induce toxicity. The involvement of other repeat-harboring transcripts in neurodegeneration via an RNA gain-of-function mechanism has been described not only for trinucleotide repeat disorders, such as FXTAS, SCA8 and HDL2,13–15 but also for DM2, SCA10 and SCA31.9,11,16,17
In the RNA gain-of-function mechanism, the expanded repeat of a flawed RNA exerts its toxic function by sequestration and subsequent loss-of-function of RNA binding proteins, such as the muscleblind-like (MBNL) family. Resultant ribonucleoprotein complexes become trapped in the nucleus where they form microscopic bodies and become toxic. Punctate intranuclear foci that label with antibodies against muscleblind 1 (MBNL1) protein are a characteristic feature of cells expressing non-coding expansions of CUG,14,18–20 CGG or CCUG repeats.13,16 More recently, transcripts harboring translated CAG repeat expansions have been observed to form intranuclear MBNL1-positive RNA foci in human HD fibroblasts.21 This result is in agreement with earlier reports describing nuclear titration of muscleblind 1 protein by exogenous CAG repeat expansions expressed in COSM6 cells,22 by SCA3 RNA mutation in Drosophila23 and by untranslated CAG expansion in transgenic Caenorhabditis elegans24 and transgenic mice.25
In cells expressing mutant repeat RNA, nuclear sequestration of host proteins interferes with developmentally regulated alternative splicing.13,14,26–28 In DM1, SCA8 and DM2, altered alternative splicing is partially a consequence of MBNL family protein sequestration by nuclear repeat RNA foci. In FXTAS, wherein mutant CGG repeat RNA titrates, among other proteins also MBNL1, only Sam68 sequestration, which is a strong and early event, interferes with splicing regulation of its pre-mRNA targets.13 This result questions the significance of MBNL1 colocalization with CAG repeat RNA that has been reported by other investigators.21–25 This observation seems to be especially important because MBNL1 protein is likely to play a central role in various aspects of repeat-mediated diseases. Depending on the nucleotide composition of the repeated sequence and the intragenic context of the repeat mutations, MBNL1 can enhance or decrease the deleterious effects of pathogenic RNA and proteins.23,24,29
In the vast majority of diseases triggered by CAG repeat expansions, the causative mutation is present in translated exons that give rise to elongated stretches of polyglutamine in mutant proteins (reviewed in ref. 6). These diseases include several SCAs, HD and others for which a protein gain-of-function mechanism has been proposed. Nonetheless, the growing body of evidence demonstrates that transcripts composed of mutated CAG repeats can also be toxic and participate in the pathogenesis of polyglutamine disorders through an RNA-mediated gain-of-function mechanism.21–25,30–32 This review summarizes several findings that emphasize the participation of coding mutant CAG repeat RNA in the pathogenesis of polyglutamine disorders and presents current knowledge about the RNA gain-of-function mechanism of non-coding diseases.
Mechanisms of Pathogenesisin Microsatellites Diseases
The mechanisms that have been proposed to explain trinucleotide repeat pathogenesis are based on RNA gain-of-function, protein gain-of-function and protein loss-of-function mechanisms. The last mechanism explains neurodegeneration in two disorders, fragile X syndrome (FXS) and FRDA, in which massive expansions of triplet repeats in non-coding sequences lead to transcriptional silencing and the absence of the encoded proteins Fragile X Mental Retardation Protein (FMRP) and frataxin, respectively.33,34 In contrast, in several disorders (SCA1, SCA2, SCA3, SCA6, SCA7, SCA17, HD, DRPLA and spinobulbar muscular atrophy, SBMA) where a protein gain-of-function mechanism has been proposed, coding CAG repeat mutations give rise to polyglutamine-rich proteins that form brain-specific inclusions. These inclusions contain cellular components such as ubiquitin, HSP70, the proteaosome and transcription factors (reviewed in ref. 6). The RNA gain-of-function mechanism initially described in DM1,18,20,35 wherein repeat expansions are found to be toxic only at the RNA level, now also explains some aspects of pathogenesis in other non-coding expansion disorders, including FXTAS, SCA8, SCA31, HDL2 as well as SCA10 and DM2.11,13–17 SCA8 is a unique among these diseases because bidirectional transcription through the repeat region results in an overlap of at least two mechanisms: toxic protein gain-of-function and toxic RNA gain-of-function. Principally, in the RNA gain-of-function model, the mutated gene is only a source of toxic RNA that impacts the expression of other genes, whereas in the protein gain-of-function model, repeat mutations in translated exons can give rise to both toxic RNA and toxic protein. However the way we have been approaching repeat-associated diseases may need some revision because of the latest discovery by Zu et al.36 who described translation initiation at expanded CAG/CTG repeats in the absence of an ATG start codon. As shown, lengthened CAG and CUG repeat transcripts may often express homopolymeric expansion proteins in all frames (i.e., polyGln, polySer and polyAla) raising the possibility that the proteins contribute to the pathogenesis of triplet repeat-associated diseases (Figs. 1 and 2). Regardless, the pathogenesis of all the TREDs has been associated with inclusion bodies (either nuclear or cytoplasmic) composed of toxic protein and/or toxic transcript, and both toxic agents sequester and diminish functional levels of host cell proteins.
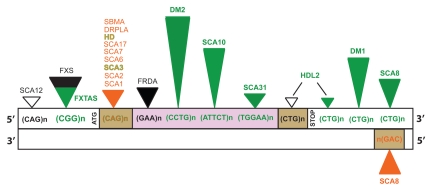
Figure 1.
Localization of simple repetitive sequences and their association with human neurodegenerative disorders. Green triangles represent RNA gain-of-function; black triangles, protein loss-of-function; orange triangles, protein gain-of-function; empty triangles, an unknown mechanism; the height of triangles corresponds roughly to repeat expansion size; brown boxes represent protein coding regions; purple box, intron; white boxes, 5′UTR and 3′UTR.
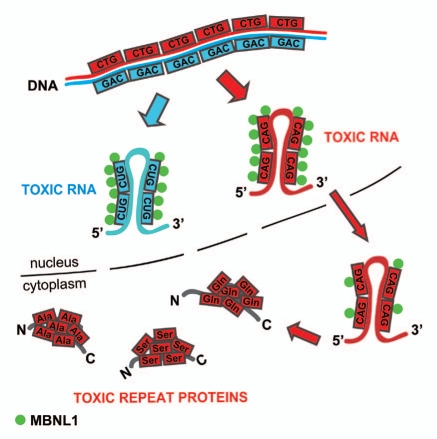
Figure 2.
Nuclear toxicity of expanded CAG repeat RNA. Transcript containing CAG repeat expansions (red) is stopped over in the nucleus where it forms MBNL1-positive ribonucleoprotein aggregates that resemble features of mutant CUG repeat RNA (blue). The effect of toxic gain-of-function of CAG repeats is extended in the cytoplasm where the mutant RNA expresses polyglutamine through canonical translation and may also undergo non-ATG translation into polyserine and polyalanine expansion proteins.
RNA Gain-of-Function Mechanism in DM1
DM1 is a multisystemic, phenotypically variable, adult onset muscular dystrophy caused by tandem CTG repeat expansion in the 3′UTR of the DMPK gene.19,37 Its pathomechanism was the first among neuromuscular disorders to be linked to aberrant functioning of mutant mRNA. The prevailing paradigm is that DM1 is a toxic RNA gain-of-function disease mediated by the expression of mutant (CUG)n expansion,38–40 and its transcription appears to be both necessary and sufficient to cause disease.18,20 Mutant RNA becomes trapped in the nucleus where it forms insoluble foci19,41 that interfere with RNA splicing by altering functional levels of RNA-binding proteins such as the muscleblind-like family and CUG-binding protein 1 (CUGBP1).42–45 These RNA-binding proteins are important splicing factors essential for the maturation of pre-mRNAs, and misregulation of their activity is considered a leading trigger of DM1 pathogenesis. Whereas binding of MBNL1, MBNL2 and MBNL3 proteins to mutant DMPK RNA causes their sequestration and inactivation,29 association of CUGBP1 with the 3′UTR of DMPK mRNA harboring the CUG repeat expansion causes stabilization and higher in vivo concentration of CUGBP1, a protein normally with a short half-life.46 A mechanism that has been proposed to explain upregulation of CUGBP1 involves activation of protein kinase C (PKC) and subsequent PKC-dependent phosphorylation of CUGBP1.47 This mechanism may elucidate how the protein becomes activated even though (in contrast to MBNL1) it neither directly interacts with nor colocalizes with the RNA CUG repeat expansion.
The altered functional activity of MBNL1 and CUGBP1 proteins and the misregulation of developmentally regulated alternative splicing events contribute to the characteristic symptoms of DM1 owing to a lack of essential transcripts. For example, myotonia was proved to be caused by chloride channel (CLCN1) mis-regulation and inappropriate expression of fetal isoforms of the gene in DM1 adult tissues.39 On the other hand, insulin resistance (INSR) and heart conduction defects associated with DM1 have been linked to mis-regulation of, respectively, INSR splicing27 and perturbed regulation of the exclusion of exon 5 in the cardiac TNNT2 (cTNT) pre-mRNA.26 However, thus far, no direct evidence demonstrated that the splicing misregulation of INSR and TNNT2 genes leads to, respectively, insulin resistance and conduction defects in DM1. Several other pre-mRNA transcripts regulated by MBNL1 and CUGBP1 proteins are aberrantly spliced in DM1, such as sarcoplasmic/endoplasmic reticulum Ca2+ ATPase 1 (ATP2A1, also known as SERCA1),48 ryanodine receptor (RyR),48 fast skeletal troponin T (TNNT3),49 and myotubularin-related protein 1 (MTMR1).50 Data that support the involvement of MBNL1 and CUGBP1 splicing factors in DM1 pathogenesis come from mouse models that have recapitulated DM splicing patterns and related skeletal and cardiac muscle abnormalities through either gene knock-out of Mbnl1,49 or transgenic overexpression of CUGBP1.50 From these studies, it appears that identifying additional protein factors and their pre-mRNA targets might help to understand the variety of symptoms associated with DM1. Although MBNL1 co-localization with expanded CUG repeat RNA seems to play a central but complex role in the pathogenesis of DM1, its association with other mutant repeat RNAs in different protein environments might have distinct downstream effects regardless of the presence of MBNL1 in retained nuclear RNA foci.
CGG Repeat RNA Gain-of-Function in FXTAS
The importance of identifying auxiliary proteins that participate in foci formation was demonstrated in FXTAS, wherein MBNL1 has recently been shown to be recruited within CGG repeat RNA aggregates.13 FXTAS is a common inherited neurodegenerative disorder caused by a premutation (55-200 CGG repeat expansions) in the 5′UTR of the fragile X syndrome gene FMR1. FXS, on the other hand, which is associated with the same locus, results from the full mutation (over 200 CGG repeats). As a consequence, FXS pathogenesis involves a protein loss-of-function mechanism whereas in FXTAS, elevated FMR1 mRNA levels induce a toxic RNA gain-of-function mechanism.51–55 Neurodegeneration in FXTAS is essentially linked to the accumulation of ubiquitin-positive intranuclear inclusions in various regions of the brain containing expanded FMR1 mRNA and numerous proteins.56,57
An earlier report modeled FXTAS in Drosophila and found that the expression of expanded CGG RNA (90 repeats) induced neurodegeneration as evidenced by a disrupted eye phenotype and the lethality of neurons.55 The mutant RNA induced formation of nuclear and cytoplasmic inclusions containing ubiquitin and Hsp70, hence mimicking the polyglutamine inclusions observed in poly-Q disorders such as Huntington disease where the inclusions were also found present in both nuclei and cytoplasm.58 Moreover, overexpression of Hsp70 was a positive modifier of RNA-mediated neurodegeneration. However, the most recent experimental evidence suggests that the common toxic RNA gain-of-function mechanism described in DM1 also contributes to the pathogenesis of FXTAS. Intranuclear RNA foci composed of expanded CGG repeats and sequestered RNA-binding splicing factors have been found in cells with endogenous and exogenously expressed CGG expansions. The proteins titrated were possibly recruited within CGG repeat RNA aggregates through protein-protein interactions.13 In FXTAS, the CGG repeat forms dynamic and enlarged aggregates that expand over time. The observation of proteins sequestration, which is typical for DM1, recently led investigators to also assign FXTAS as a spliceopathy.13,59 Interestingly, although MBNL1 is recruited to CGG RNA foci, its function as measured by changes in alternative splicing of regulated pre-mRNAs is not compromised. Conceivably, in this context MBNL1 binding is a late event in foci formation that leaves the activity of this protein in splicing regulation intact; in fact, only the early recruited protein Sam68 showed loss-of-function as detected by aberrant splicing of a set of genes regulated by this protein.
Thus, the toxicity of the FXTAS mutation may involve features of either polyglutamine peptides or repeat RNA. In one neurodegenerative disease, SCA8, these two agents have been reported to be co-expressed.
SCA8 as a Disease of Both RNA and Protein Gain-of-Function Mechanisms
SCA8 is a dominantly inherited, late-onset neurodegenerative disease with CTG•CAG expansions expressed in the brain and cerebellum.60 Experimental data have revealed the existence of bidirectional transcription through the repeat region in the SCA8 locus. The expression of ATXN8 antisense CAG-containing transcript is correlated with its translation into a nearly pure polyglutamine protein, ataxin 8, which is found in intranuclear inclusions in Purkinje cells and in brainstem neurons of both SCA8 patients and BAC-SCA8 mice.61 The concomitant expression of an untranslated sense CTG mutation in the 3′UTR of the ATXN8OS gene leads to the generation of CUG repeat transcript that becomes toxic via a gain-of-function mechanism, similarly to what is observed in DM1. In fact, in mice expressing a BAC transgene with the SCA8 mutation, the accumulation of SCA8 transcript with expanded CUG repeats leads to alternative splicing changes of GABA-A transporter 4 (Gabt4) in the cerebellum as a result of dysregulation of the two key splicing factors Mbnl1 and Cugbp1 in the central nervous system (CNS).14,61 Alternative splicing changes in a number of CNS transcripts including the amyloid precursor protein (APP), microtubule-associated protein tau (MAPT), glutamate receptor NMDAR1 and MBNL1 are also found in human and mouse DM1 tissues.29,62 Although the underlying mechanism for the CNS splicing changes in DM1 and SCA8 is unknown, the results suggest that the two CUG mutant transcripts cause brain defects via an RNA gain-of-function mechanism. Although, the SCA8 mutation is of the same type as DM1, its pathogenic length threshold is shorter than that of DM1.
Attempts to understand the role of ATXN8 polyglutamine protein of antisense CAG transcript in SCA8 disease gave lately rise to very important discovery. The presence of homopolymeric polyalanine and polyserine expansion proteins, in addition to polyglutamine, was shown in cells transfected with SCA8 constructs, in SCA8 mouse model and postmortem brain from SCA8 patients.36 These proteins were expressed simultaneously in the cells and in the absence of an ATG start codon. The non-ATG translation products were shown to form cytoplasmic and nuclear aggregates which are toxic for cells and trigger apoptosis.
Based on expression of mutant CUG and CAG repeat transcripts as well as homopolymeric expansion proteins, SCA8 may be considered a complex gain-of-function disease. However, it is presently unknown which mechanism prevails. A question that remains to be addressed is whether the mutant CAG-harboring transcript resembles features of the CUG RNA gain-of-function mechanism. If so, then does co-expression of both mutant RNAs in SCA8 result in synergistic toxicity considering their individual harmful effects in other TREDs?
CAG Repeat RNA as an Auxiliary Toxic Agent in Polyglutamine Disorders
Numerous human neurodegenerative diseases (Fig. 1) are caused by CAG repeats that form abnormally long tracts in translated exons of various genes, giving rise to elongated stretches of polyglutamine in the resultant proteins (reviewed in ref. 6). The most studied mechanism of pathogenesis of polyglutamine disorders is the gain-of-function mechanism centered on the enhanced ability of poly-Q proteins to form ubiquitin-positive aggregates and large inclusions by attracting host proteins. Initially, a pathogenic effect of these protein bodies was strongly suspected; however, subsequent experimental data questioned their direct pathogenic involvement by providing evidence for a protective role of poly-Q inclusion bodies (IBs). It was shown that in Huntington disease the formation of IBs composed of poly-Q protein expansion is not required for neuronal death, and that less aggregated or possibly monomeric species of mutant huntingtin (Htt) are toxic instead.63,64 Because protein IB formation is found to be protective when mutant Htt is expressed in neurons, could a similar function be attributed to the polyCAG RNA foci that were recently found in human HD cells?
Over the past few years, experimental data from several laboratories have provided evidence that clearly indicates the following: (1) transcripts containing mutant CAG repeats form intranuclear RNA foci, (2) mutant CAG RNA foci sequester MBNL1 protein and (3) transcribed but untranslated RNA containing CAG expansions can induce pathogenic features previously attributed only to mutant poly-Q proteins. These conclusions, derived from work performed in vitro in various cell lines as well as in fly worm and mouse models indicate that CAG repeat RNAs can exert toxicity regardless of the system of expression. In an earlier report by Ho et al.22 a long (CAG)960 repeat RNA was expressed in COSM6 cells and mimicked the foci formation and MBNL1 co-localization observed in these cells expressing (CUG)960. However, despite the presence of sequestered MBNL1 protein in the CAG foci, their toxic effects were milder in comparison with CUG foci, and they did not considerably alter the splicing of the pre-mRNAs for cTNT or INSR. Three later reports that utilized transgenic expression models of SCA3 in Drosophila23 and of mutant and untranslated CAG repeat in C. elegans24 and in mice25 have provided more evidence that mutant CAG RNA contributes to poly-Q-triggered degeneration. The CAG expansions in these systems were shown to form muscleblind protein 1-positive nuclear foci in fly eyes and worm and mouse muscles, but their toxicity in the SCA3 model, as indicated by disrupted eye morphology and induced lethality of neurons, was mitigated by the presence of interruptions altering the CAG repeat sequence. This observation might explain why the long and interrupted CAG repeat RNA used in the study by Ho et al. was not potent enough to induce the alternative splicing aberrations typical for CUG repeats.22 Studies found that in cultured human HD21 and SCA3 fibroblast cells (Mykowska A, et al. unpublished observations), endogenous transcripts harboring uninterrupted CAG expansions are sequestered in nuclei where they form RNA foci that colocalize with MBNL1 protein. Investigations are underway to determine if expressing CAG repeat mutations in human cells and clonal lines alters the splicing of MBNL1-dependent pre-mRNAs. From these studies, it appears that CAG repeat RNA can mimic, to some extent, the pathogenic activity of CUG repeats. Because CAG repeat RNA expression leads to the sequestration of muscleblind 1 protein by nuclear RNA foci, it fulfills a fundamental feature of the RNA gain-of-function model.
Toxic RNA in Other Repeat Disorders—HDL2, DM2, SCA10 and SCA31
HDL2, DM2, SCA31 and SCA10 are examples of non-coding microsatellite expansion disorders whose pathogenesis is associated with mutant RNA and similar basic RNA gain-of-function mechanism.11,15–17 In HDL2, the CTG repeat occurs in variably spliced exons and is either in an untranslated region or encodes a polyleucine or polyalanine protein.15 RNA-dependent pathogenesis of HDL2 includes nuclear retention of the JPH3 transcript with expanded CUG repeats in neurons of the frontal cortex and other brain regions, and MBNL1 co-localization with the CUG RNA foci, which results in cell toxicity.15 However, no splicing aberrations in HDL2 cells have yet been reported.
In addition to SCA31, DM2 and SCA10 are the only human disorders caused by non-trinucleotide microsatellite expansions. Intronic (CCTG)n and (ATTCT)n repeat mutations are properly spliced out of the mutant transcripts but are resistant to degradation.16,65 In DM2 cells, expanded CCUG repeat RNA is the only part of intron 1 from the ZNF9 gene that is retained in the nucleus within MBNL1-positive punctate aggregates. This effect in turn triggers splicing defects similar to those seen in DM1.16 In SCA10, AUUCU repeat expansions aberrantly accumulate in the nucleus and cytoplasm and interact with the RNA-binding protein hnRNP K. This interaction results in loss-of-function of the protein and eventually activates apoptosis in SCA10 cells.
Concluding Remarks
This review presents recent evidence that RNAs containing expanded trinucleotide repeats of various compositions (CUG, CGG and CAG) and different intragenic locations can exert toxicity through a common RNA gain-of-function mechanism.13–15,20–24,55,66 Various investigations have provided strong evidence that mutant CAG repeats exhibit toxic potency at the RNA level and, therefore, ought to be considered an additional factor mediating pathogenesis in polyglutamine disorders (Fig. 2). Collectively, these studies indicate that CAG repeat RNA can mimic, to some extent, the toxic effects induced by CUG repeat RNA.21–25 The picture now emerging is that general hallmarks of RNA toxicity, such as ribonuclear foci formation, protein sequestration and alternative splicing misregulation, are shared between diseases caused by CUG and CAG repeat expansions. However, mutant CUG and CAG repeats that trigger pathogenesis show both similarities and differences in their molecular architecture30–32,67 and likely also in protein binding properties. These same similarities and differences apply to the morphology and protein composition of nuclear foci that they form and to downstream mis-splicing effects.
What we have learned is not only that very long exogenous CAG repeats form nuclear foci that co-localize with the muscle-blind 1 protein but also that much shorter mutant CAG repeats do the same in cells derived from HD and SCA3 patients. We now need to know whether the same mechanism applies to other poly-Q diseases as well. Does the presence of such foci correlate with alternative splicing aberrations, and if so, how are these aberrations related to those observed in cells of DM1 patients? It is likely that the above questions will be answered in the short term. In the long term, we anticipate that various next-generation sequencing, microarray and proteomics platforms will provide copious genome-wide information on dysregulated transcriptomes and proteomes in affected tissues. A global view of transcriptome and proteome dynamics at various stages of disease development will help identify new research avenues and targets that will keep TRED researchers busy for the next decade.
Acknowledgements
Ministry of Science and Higher Education (N N401 097536); European Regional Development Fund within Innovative Economy Programme (POIG.01.03.01-00-098/08).
Abbreviations
- DM
myotonic dystrophy
- FXTAS
fragile X-associated tremor ataxia syndrome
- HD
Huntington's disease
- SCA
spinocerebellar ataxia
- poly-Q
polyglutamine
- HDL2
Huntington disease-like 2
- TREDs
Triplet Repeat Expansion Diseases
- UTR
untranslated region
- MBNL1
muscleblind-like 1
- CUGBP1
CUG-binding protein 1
References
- 1.Jasinska A, Krzyzosiak WJ. Repetitive sequences that shape the human transcriptome. FEBS Lett. 2004;567:136–141. doi: 10.1016/j.febslet.2004.03.109. [DOI] [PubMed] [Google Scholar]
- 2.Jasinska A, Michlewski G, de Mezer M, Sobczak K, Kozlowski P, Napierala M, et al. Structures of trinucleotide repeats in human transcripts and their functional implications. Nucleic Acids Res. 2003;31:5463–5468. doi: 10.1093/nar/gkg767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kozlowski P, de Mezer M, Krzyzosiak WJ. Trinucleotide repeats in human genome and exome. Nucleic Acids Res. 2010;38:4027–4039. doi: 10.1093/nar/gkq127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 5.Lopez Castel A, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat Rev Mol Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 6.Orr HT, Zoghbi HY. Trinucleotide repeat disorders. Annu Rev Neurosci. 2007;30:575–621. doi: 10.1146/annurev.neuro.29.051605.113042. [DOI] [PubMed] [Google Scholar]
- 7.Todd PK, Paulson HL. RNA-mediated neurodegeneration in repeat expansion disorders. Ann Neurol. 2010;67:291–300. doi: 10.1002/ana.21948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamori M, Thornton C. Epigenetic changes and non-coding expanded repeats. Neurobiol Dis. 2010;39:21–27. doi: 10.1016/j.nbd.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, et al. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 10.Matsuura T, Fang P, Lin X, Khajavi M, Tsuji K, Rasmussen A, et al. Somatic and germline instability of the ATTCT repeat in spinocerebellar ataxia type 10. Am J Hum Genet. 2004;74:1216–1224. doi: 10.1086/421526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato N, Amino T, Kobayashi K, Asakawa S, Ishiguro T, Tsunemi T, et al. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet. 2009;85:544–557. doi: 10.1016/j.ajhg.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu Rev Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 13.Sellier C, Rau F, Liu Y, Tassone F, Hukema RK, Gattoni R, et al. Sam68 sequestration and partial loss of function are associated with splicing alterations in FXTAS patients. EMBO J. 2010;29:1248–1261. doi: 10.1038/emboj.2010.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daughters RS, Tuttle DL, Gao W, Ikeda Y, Moseley ML, Ebner TJ, et al. RNA gain-of-function in spinocerebellar ataxia type 8. PLoS Genet. 2009;5:1000600. doi: 10.1371/journal.pgen.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudnicki DD, Holmes SE, Lin MW, Thornton CA, Ross CA, Margolis RL. Huntington's disease—like 2 is associated with CUG repeat-containing RNA foci. Ann Neurol. 2007;61:272–282. doi: 10.1002/ana.21081. [DOI] [PubMed] [Google Scholar]
- 16.Margolis JM, Schoser BG, Moseley ML, Day JW, Ranum LP. DM2 intronic expansions: evidence for CCUG accumulation without flanking sequence or effects on ZNF9 mRNA processing or protein expression. Hum Mol Genet. 2006;15:1808–1815. doi: 10.1093/hmg/ddl103. [DOI] [PubMed] [Google Scholar]
- 17.White MC, Gao R, Xu W, Mandal SM, Lim JG, Hazra TK, et al. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Amack JD, Paguio AP, Mahadevan MS. Cis and trans effects of the myotonic dystrophy (DM) mutation in a cell culture model. Hum Mol Genet. 1999;8:1975–1984. doi: 10.1093/hmg/8.11.1975. [DOI] [PubMed] [Google Scholar]
- 19.Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3′ untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 21.de Mezer M, Wojciechowska M, Napierala M, Sobczak K, Krzyzosiak WJ. Mutant CAG repeats of Huntingtin transcript fold into hairpins, form nuclear foci and are targets for RNA interference. Nucleic Acids Res. 2011;39:3852–3863. doi: 10.1093/nar/gkq1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscle-blind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J Cell Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 23.Li LB, Yu Z, Teng X, Bonini NM. RNA toxicity is a component of ataxin-3 degeneration in Drosophila. Nature. 2008;453:1107–1111. doi: 10.1038/nature06909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang LC, Chen KY, Pan H, Wu CC, Chen PH, Liao YT, et al. Muscleblind participates in RNA toxicity of expanded CAG and CUG repeats in Caenorhabditis elegans. Cell Mol Life Sci. 2010;68:1255–1267. doi: 10.1007/s00018-010-0522-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hsu RJ, Hsiao KM, Lin MJ, Li CY, Wang LC, Chen LK, et al. Long tract of untranslated CAG repeats is deleterious in transgenic mice. PLoS One. 2011;6:16417. doi: 10.1371/journal.pone.0016417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 27.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 28.Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 29.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 30.Michlewski G, Krzyzosiak WJ. Molecular architecture of CAG repeats in human disease related transcripts. J Mol Biol. 2004;340:665–679. doi: 10.1016/j.jmb.2004.05.021. [DOI] [PubMed] [Google Scholar]
- 31.Sobczak K, Krzyzosiak WJ. Imperfect CAG repeats form diverse structures in SCA1 transcripts. J Biol Chem. 2004;279:41563–1572. doi: 10.1074/jbc.M405130200. [DOI] [PubMed] [Google Scholar]
- 32.Sobczak K, Krzyzosiak WJ. CAG repeats containing CAA interruptions form branched hairpin structures in spinocerebellar ataxia type 2 transcripts. J Biol Chem. 2005;280:3898–3910. doi: 10.1074/jbc.M409984200. [DOI] [PubMed] [Google Scholar]
- 33.O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- 34.Campuzano V, Montermini L, Lutz Y, Cova L, Hindelang C, Jiralerspong S, et al. Frataxin is reduced in Friedreich ataxia patients and is associated with mitochondrial membranes. Hum Mol Genet. 1997;6:1771–1780. doi: 10.1093/hmg/6.11.1771. [DOI] [PubMed] [Google Scholar]
- 35.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zu T, Gibbens B, Doty NS, Gomes-Pereira M, Huguet A, Stone MD, et al. Non-ATG-initiated translation directed by microsatellite expansions. Proc Natl Acad Sci USA. 2011;108:260–265. doi: 10.1073/pnas.1013343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- 38.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum Mol Genet. 2006;15:162–169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 39.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, et al. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 44.Timchenko LT, Miller JW, Timchenko NA, DeVore DR, Datar KV, Lin L, et al. Identification of a (CUG) (CUG)n triplet repeat RNA-binding protein and its expression in myotonic dystrophy. Nucleic Acids Res. 1996;24:4407–4414. doi: 10.1093/nar/24.22.4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Timchenko LT, Timchenko NA, Caskey CT, Roberts R. Novel proteins with binding specificity for DNA CTG repeats and RNA CUG repeats: implications for myotonic dystrophy. Hum Mol Genet. 1996;5:115–121. doi: 10.1093/hmg/5.1.115. [DOI] [PubMed] [Google Scholar]
- 46.Timchenko NA, Cai ZJ, Welm AL, Reddy S, Ashizawa T, Timchenko LT. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–7826. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 47.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kimura T, Nakamori M, Lueck JD, Pouliquin P, Aoike F, Fujimura H, et al. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/ endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum Mol Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 49.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 50.Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- 51.Iwahashi CK, Yasui DH, An HJ, Greco CM, Tassone F, Nannen K, et al. Protein composition of the intranuclear inclusions of FXTAS. Brain. 2006;129:256–271. doi: 10.1093/brain/awh650. [DOI] [PubMed] [Google Scholar]
- 52.Berman RF, Willemsen R. Mouse models of fragile x-associated tremor ataxia. J Investig Med. 2009;57:837–841. doi: 10.231/JIM.0b013e3181af59d6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brouwer JR, Huizer K, Severijnen LA, Hukema RK, Berman RF, Oostra BA, et al. CGG-repeat length and neuropathological and molecular correlates in a mouse model for fragile X-associated tremor/ataxia syndrome. J Neurochem. 2008;107:1671–1682. doi: 10.1111/j.1471-4159.2008.05747.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jin P, Duan R, Qurashi A, Qin Y, Tian D, Rosser TC, et al. Pur alpha binds to rCGG repeats and modulates repeat-mediated neurodegeneration in a Drosophila model of fragile X tremor/ataxia syndrome. Neuron. 2007;55:556–564. doi: 10.1016/j.neuron.2007.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jin P, Zarnescu DC, Zhang F, Pearson CE, Lucchesi JC, Moses K, et al. RNA-mediated neurodegeneration caused by the fragile X premutation rCGG repeats in Drosophila. Neuron. 2003;39:739–747. doi: 10.1016/s0896-6273(03)00533-6. [DOI] [PubMed] [Google Scholar]
- 56.Greco CM, Hagerman RJ, Tassone F, Chudley AE, Del Bigio MR, Jacquemont S, et al. Neuronal intranuclear inclusions in a new cerebellar tremor/ataxia syndrome among fragile X carriers. Brain. 2002;125:1760–1771. doi: 10.1093/brain/awf184. [DOI] [PubMed] [Google Scholar]
- 57.Greco CM, Berman RF, Martin RM, Tassone F, Schwartz PH, Chang A, et al. Neuropathology of fragile X-associated tremor/ataxia syndrome (FXTAS) Brain. 2006;129:243–255. doi: 10.1093/brain/awh683. [DOI] [PubMed] [Google Scholar]
- 58.Li H, Li SH, Johnston H, Shelbourne PF, Li XJ. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat Genet. 2000;25:385–389. doi: 10.1038/78054. [DOI] [PubMed] [Google Scholar]
- 59.Sofola OA, Jin P, Qin Y, Duan R, Liu H, de Haro M, et al. RNA-binding proteins hnRNP A2/B1 and CUGBP1 suppress fragile X CGG premutation repeat-induced neurodegeneration in a Drosophila model of FXTAS. Neuron. 2007;55:565–571. doi: 10.1016/j.neuron.2007.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Koob MD, Moseley ML, Schut LJ, Benzow KA, Bird TD, Day JW, et al. An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8) Nat Genet. 1999;21:379–384. doi: 10.1038/7710. [DOI] [PubMed] [Google Scholar]
- 61.Moseley ML, Zu T, Ikeda Y, Gao W, Mosemiller AK, Daughters RS, et al. Bidirectional expression of CUG and CAG expansion transcripts and intranuclear poly-glutamine inclusions in spinocerebellar ataxia type 8. Nat Genet. 2006;38:758–769. doi: 10.1038/ng1827. [DOI] [PubMed] [Google Scholar]
- 62.Dhaenens CM, Schraen-Maschke S, Tran H, Vingtdeux V, Ghanem D, Leroy O, et al. Overexpression of MBNL1 fetal isoforms and modified splicing of Tau in the DM1 brain: two individual consequences of CUG trinucleotide repeats. Exp Neurol. 2008;210:467–478. doi: 10.1016/j.expneurol.2007.11.020. [DOI] [PubMed] [Google Scholar]
- 63.Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- 64.Miller J, Arrasate M, Shaby BA, Mitra S, Masliah E, Finkbeiner S. Quantitative relationships between huntingtin levels, polyglutamine length, inclusion body formation and neuronal death provide novel insight into Huntington's disease molecular pathogenesis. J Neurosci. 30:10541–10550. doi: 10.1523/JNEUROSCI.0146-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wakamiya M, Matsuura T, Liu Y, Schuster GC, Gao R, Xu W, et al. The role of ataxin 10 in the pathogenesis of spinocerebellar ataxia type 10. Neurology. 2006;67:607–613. doi: 10.1212/01.wnl.0000231140.26253.eb. [DOI] [PubMed] [Google Scholar]
- 66.Mankodi A, Teng-Umnuay P, Krym M, Henderson D, Swanson M, Thornton CA. Ribonuclear inclusions in skeletal muscle in myotonic dystrophy types 1 and 2. Ann Neurol. 2003;54:760–768. doi: 10.1002/ana.10763. [DOI] [PubMed] [Google Scholar]
- 67.Napierala M, Krzyzosiak WJ. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J Biol Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]