Abstract
Herpes simplex encephalitis (HSE) is the most common sporadic viral encephalitis of childhood. Autosomal recessive (AR) UNC-93B and TLR3 deficiencies and autosomal dominant (AD) TLR3 and TRAF3 deficiencies underlie HSE in some children. We report here unrelated HSE children with AR or AD TRIF deficiency. The AR form of the disease was found to be due to a homozygous nonsense mutation that resulted in a complete absence of the TRIF protein. Both the TLR3- and the TRIF-dependent TLR4 signaling pathways were abolished. The AD form of disease was found to be due to a heterozygous missense mutation, resulting in a dysfunctional protein. In this form of the disease, the TLR3 signaling pathway was impaired, whereas the TRIF-dependent TLR4 pathway was unaffected. Both patients, however, showed reduced capacity to respond to stimulation of the DExD/H-box helicases pathway. To date, the TRIF-deficient patients with HSE described herein have suffered from no other infections. Moreover, as observed in patients with other genetic etiologies of HSE, clinical penetrance was found to be incomplete, as some HSV-1–infected TRIF-deficient relatives have not developed HSE. Our results provide what we believe to be the first description of human TRIF deficiency and a new genetic etiology for HSE. They suggest that the TRIF-dependent TLR4 and DExD/H-box helicase pathways are largely redundant in host defense. They further demonstrate the importance of TRIF for the TLR3-dependent production of antiviral IFNs in the CNS during primary infection with HSV-1 in childhood.
Introduction
Herpes simplex encephalitis (HSE) is a rare and potentially fatal manifestation of herpes simplex virus-1 (HSV-1) infection, with an incidence of about 1 in 250,000 individuals per year (1). With the introduction of acyclovir from the 1980s onward, HSE mortality rates, which used to be as high as 70%, have declined significantly (2, 3), although most patients, affected children in particular, continue to suffer life-long neurological sequelae (4–6). The incidence of HSE peaks between the ages of 6 months and 3 years, a period during which the vast majority of cases are a consequence of primary infection with HSV-1 (7–9). The pathogenesis of HSE, first described in 1941, remained elusive until the demonstration of an underlying role in this devastating disease, in at least some children, of autosomal recessive (AR) UNC-93B deficiency in 2006, autosomal dominant (AD) TLR3 deficiency in 2007, and, more recently, AD TNF receptor-associated factor 3 (TRAF3) and AR TLR3 deficiencies (10–13). Fibroblasts from patients with UNC-93B, TLR3, and TRAF3 deficiencies do not respond to stimulation with TLR3 agonists or infection with HSV-1 or vesicular stomatitis virus (VSV). HSE, together with other infectious diseases, was also reported in 2 children with mutations in STAT-1 and NEMO (10–15). These genetic deficiencies thus highlighted the importance of the TLR3-dependent production of IFN-α/β and IFN-λ after infection of the CNS with HSV-1 (6, 16, 17). In fibroblasts from patients with UNC-93B, TLR3, and TRAF3 deficiency (10–12) and in iPS-derived CNS cells (M. Lafaille, unpublished observations), impaired IFN production has been shown to result in enhanced viral replication and higher levels of cell death.
However, most cases of childhood HSE remain unexplained. We hypothesize that HSE is a genetically heterogeneous disease, involving a collection of single-gene inborn errors of immunity to HSV-1 in the CNS during the course of primary infection (18). Specifically, we hypothesize that mutations in genes controlling the TLR3 pathway may predispose children to HSE. Human TLR3-mediated immune responses are initiated by dsRNA intermediates in vivo or via their synthetic analog polyinosinic-polycytidylic acid [poly(I:C)] in vitro, leading to the induction of IFN-β via the NF-κB, IRF3, and AP-1 pathways (19). A principal candidate gene for HSE encodes the Toll/IL-1R (TIR) domain-containing adaptor inducing IFN-β (TRIF) protein, also known as TIR domain-containing adaptor molecule 1 (TICAM-1), due to its role as the sole adaptor of TLR3 (20–23). However, this molecule also serves as an adaptor for the MyD88-independent pathway downstream from TLR4 (24–26), raising the possibility that TRIF mutations may confer a distinct phenotype. A recent report has also shown TRIF to be involved in the detection of cytosolic dsRNA via the DExD/H-box helicase complex DDX1-DDX21-DHX36 (27). After TLR3 activation, TRIF is thought to act as a molecular platform for subsequent signaling events, recruiting TRAF3, TANK-binding kinase 1 (TBK1), NF-κB–activating kinase-associated protein 1, receptor-interacting protein 1 (RIP1), and IFN regulatory factor 3 (IRF3), in particular (28, 29). Mice lacking TRIF do not respond to poly(I:C), display impaired LPS-induced inflammatory cytokine production, and show increased susceptibility to mouse CMV and vaccinia virus infections (25, 26). Given the key role of TRIF in the TLR3 pathway demonstrated in mice, our previous demonstration of the role of the TLR3-IFN pathway in preventing the spread of HSV-1 to the CNS, and despite the potential involvement of human TRIF in TLR4 and helicase responses, we focused our candidate gene approach on TRIF by sequencing the TRIF gene in a cohort of children with HSE.
Results
Homozygous TRIF nonsense mutation in patient 1.
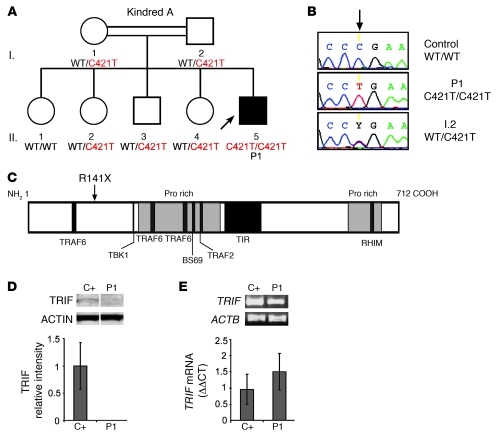
A patient (P1) born to consanguineous Saudi parents presented HSE at the age of 2 years (Figure 1A). This patient is now 3.5 years old and has had no other unusually severe infectious disease. No mutations were found in the coding regions of UNC93B1 and TRAF3, consistent with the normal PBMC responses to TLR3, TLR7, TLR8, and TLR9 agonists (leukocyte responses to TLR3 agonists have been shown to be TLR3 independent) observed in this patient (11, 13) (Supplemental Figure 1, A and B; supplemental material available online with this article; doi: 10.1172/JCI59259DS1). No mutations were found in the coding regions of TLR3. However, both genomic DNA (gDNA) and cDNA from the leukocytes and fibroblasts of P1 displayed a homozygous nonsense mutation in TRIF at nucleotide position 421 (c.421C>T), resulting in a premature termination codon replacing an arginine residue at amino acid position 141 (R141X) (Figure 1, B and C). P1 is the proband and the only member of this family homozygous for this mutation. There are no other reports of premature termination mutations in TRIF, and this mutation was found neither in the NCBI or Ensembl databases nor in the 1,234 unrelated healthy controls sequenced, including 1,050 individuals from the Centre d’Etude du Polymorphisme Humain–Human Genome Diversity (CEPH-HGD) panel and 182 Saudi Arabian controls (a total of 2,464 chromosomes). The premature termination codon occurs at position 141, resulting in no detectable protein in SV40 fibroblasts and EBV-transformed B cells (EBV-B cells) from the patient, as shown by Western blotting (Figure 1D and Supplemental Figure 2A). TRIF mRNA levels in fibroblasts and EBV-B cells from P1 were similar to those in controls, as shown by quantitative real-time PCR (Q-PCR) and full-length TRIF RT-PCR, suggesting that there is little or no nonsense-mediated mRNA decay (Figure 1E and Supplemental Figure 2B). These data suggest that TRIF R141X is a null allele, causing complete TRIF deficiency in P1.
Figure 1. AR TRIF deficiency in P1.
(A) Family pedigree of kindred A with allele segregation of the mutation. The HSE patient is shaded in black. Roman numerals (left margin) indicate generations. An arrow indicates the proband. (B) Automated sequencing profiles for the TRIF C421>T mutation in gDNA isolated from leukocytes from a healthy unrelated control; the patient, P1; and the father, I.2. The arrow indicates the position of the mutation. (C) A schematic representation of the TRIF protein (1–712 amino acids) indicating the amino acid position, R141X, affected by the C421>T mutation. Proline-rich domains (pro rich) are shaded in gray; functional domains are shaded in black (TRAF6 binding, TIR, RIP homotypic interacting motif [RHIM]). (D) TRIF protein expression by immunoblot analysis of SV40 fibroblast cell lysates from a healthy control (C+) and P1. Samples were migrated on the same blot. TRIF expression levels were quantified by densitometry results normalized with respect to ACTIN levels and expressed as relative intensity of TRIF. This is a representative blot from 3 independent experiments (mean ± SEM). (E) RT-PCR of full-length TRIF cDNA is shown with ACTB cDNA as an internal control. TRIF cDNA levels were assessed by real-time PCR in control fibroblasts (C+) and P1. Data are represented as relative fold change (ΔΔCt units), where GUS was used for normalization. An average of 3 independent experiments is represented (mean ± SEM).
Heterozygous TRIF missense mutation in P2.
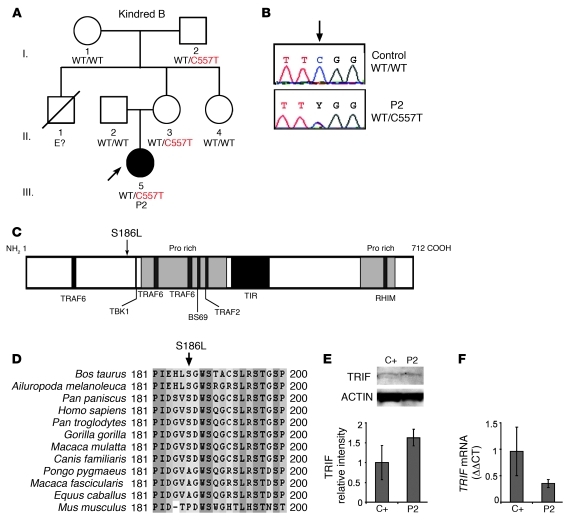
A girl (P2) of mixed European descent (French, Portuguese, and Swiss) presented HSE at the age of 21 months (Figure 2A). The patient is now 18 years old and has had no other unusually severe infectious disease. Mutations in all known HSE-causing genes were excluded, not only for the coding regions of UNC93B1 and TRAF3, consistent with the normal responses to TLR3, TLR7, TLR8, and TLR9 agonists observed (Supplemental Figure 1, C and D), but also for the coding regions of TLR3. However, both gDNA) and cDNA from the leukocytes and fibroblasts of P2 displayed a heterozygous nucleotide substitution in TRIF at position 557 (c.557C>T), resulting in the substitution of a leucine for a serine residue at amino acid position 186 (S186L) (Figure 2, B and C). P2 is the proband and the only member of this family to have developed HSE. However, the mother and maternal grandfather of P2 also carry the S186L mutation and have HSV-1–specific serum antibodies. This mutation was not found in the NCBI and Ensembl databases or in the 1,050 unrelated healthy controls from the CEPH-HGD panel sequenced, including 289 Europeans (a total of 2,100 chromosomes). Serine 186 is conserved in 8 out of the 11 animal species with TRIF proteins sharing over 50% homology to the human protein (Figure 1D). Leucine has never been found in position 186 in any species, although 2 nonhuman primates and the horse carry an alanine residue at this position, whereas the mouse has a proline residue. The S186L mutation affects the N-terminal region of the protein (Figure 1C). Previous studies have shown that the N-terminal region of TRIF plays an important role, as an N-terminal deletion mutant (including S186) displayed a specific loss of IRF3 activation and IFN-β promoter induction (21, 25). TRIF mRNA and protein levels in the fibroblasts and EBV-B cells of P2 were found to be similar to those in controls, suggesting that the transcription and expression of the gene were not affected by this or any other undetected mutation (Figure 1, E and F, and Supplemental Figure 2, A and B). These data suggest that TRIF S186L is a rare allele and that the missense mutation may cause an AD form of TRIF deficiency, conferring a predisposition to HSE with incomplete clinical penetrance (similar to both UNC-93B and TLR3 deficiencies) (10, 11).
Figure 2. AD TRIF deficiency in P2.
(A) Family pedigree of kindred B with allele segregation of the mutation. The HSE patient is shaded in black. Roman numerals (left margin) indicate generations. An arrow indicates the proband, and “E?” indicates an undetermined genotype. (B) Automated sequencing profiles for the TRIF C557>T mutation in gDNA isolated from leukocytes from a healthy unrelated control and in the patient, P2. The arrow indicates the position of the mutation. (C) A representation of the TRIF protein (1–712 amino acids) indicating the amino acid position, S186L, affected by the C557>T mutation. Proline-rich domains are shaded in gray; functional domains are shaded in black (TRAF6 binding, TIR, and RHIM). (D) Multiple alignment of TRIF amino acid sequence surrounding the mutation S186L. (E) TRIF protein expression by immunoblot analysis of SV40 fibroblast cell lysates from a healthy control (C+) and P2. TRIF expression levels were quantified by densitometry results normalized with respect to ACTIN levels and expressed as relative intensity of TRIF. This is a representative blot from 3 independent experiments (mean ± SEM). (F) TRIF cDNA levels were assessed by real-time PCR in control fibroblasts and P2. Data are represented as relative fold change (ΔΔCt units), where GUS was used for normalization. An average of 3 independent experiments is represented (mean ± SEM).
Impaired cellular responses to TLR3 agonists.
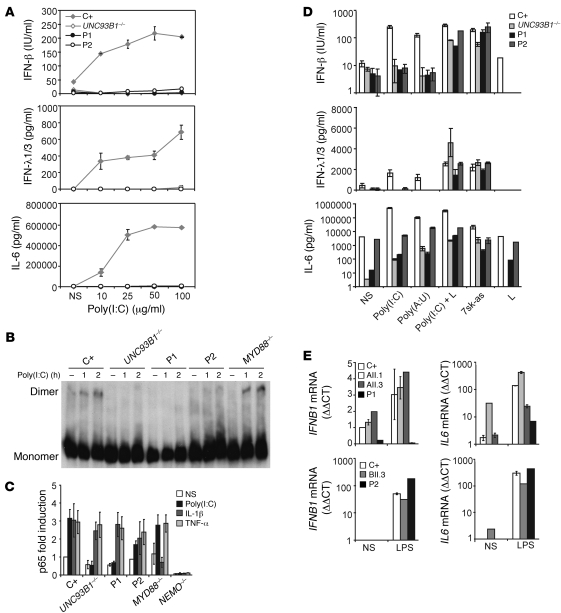
Dermal fibroblasts from P1 and P2 were used to investigate the AR R141X and AD S186L TRIF mutations. After 24 hours of stimulation with poly(I:C), the fibroblasts of P1 and P2 displayed impaired production of IFN-β, IFN-λ1/3 (IL-29/IL-28B), and IL-6 similar to that observed in cells from UNC-93B–deficient patients and contrasting with the situation in cells from a healthy control (P < 0.05 for all comparisons) (Figure 3A). Cells from another TRIF heterozygote, the mother of P2, also showed impaired production of IFN-β, IFN-λ1/3, and IL-6 compared with that of a control (P < 0.05 for all comparisons) after 24 hours of stimulation with various doses of poly(I:C) (Supplemental Figure 3). Consistent with the lack of IFN-β induction in P1, no IRF3 dimers were found in P1 or in cells from a patient with complete UNC-93B deficiency (negative control). In P2, IRF3 dimerization in response to poly(I:C) was impaired but not abolished. By contrast, cells from a patient with MyD88 deficiency and cells from a healthy individual (positive control) were able to form dimers as early as 1 hour after stimulation (Figure 3B). Nuclear translocation of the p65 subunit of NF-κB after poly(I:C) stimulation was also abolished in P1 cells (P < 0.05) and reduced in P2 cells (P < 0.05) compared with that in a control (Figure 3C). However, NF-κB activation in P1 and P2 cells was normal after stimulation with IL-1β and TNF-α. As expected, there was no p65 translocation upon IL-1β stimulation in MyD88-deficient cells or in NEMO-deficient cells. We then tested the responses of P1 and P2 to a specific noncommercial TLR3 agonist, polyadenylic-polyuridylic acid [poly(A:U)], poly(I:C) delivered intracellularly with lipofectamine, and the RIG-I–specific ligand, 7sk-as (30). There was no response to the TLR3 agonist poly(A:U) in P1 and P2 cells, whereas IFN-β, IFN-λ1/3, and IL-6 were induced in cells from P1 and P2 after transfection with poly(I:C) or 7sk-as (Figure 3D). Interestingly, both patients induced lower levels of IFN-β and IL-6 in response to transfected poly(I:C) compared with those of a control (P < 0.05 for all comparisons). Intracellular poly(I:C) is known to activate cytosolic dsRNA receptors, such as RIG-I and MDA5, which use the adaptor VISA to induce IRF3 and IFN-β (31, 32). Recently, however, TRIF was also shown to participate in cytosolic dsRNA receptors pathways (27). These findings demonstrate that the recognition of extracellular dsRNA was impaired in P1 and P2 cells, consistent with the established role of TRIF in the TLR3 pathway. The response to intracellular dsRNA in the cytosol was modestly affected in both patients, in line with recent reports of TRIF’s involvement in cytosolic pathways. In addition, we carried out genome-wide transcriptional analysis of the TLR3 pathway in fibroblasts of a control, P1, an AR TLR3-deficient patient, and an AR MyD88-deficient patient after 4 hours of poly(I:C) or IL-1β stimulation. In control fibroblasts, there were 506 transcripts found to be differentially regulated upon poly(I:C) (Supplemental Figure 4A). Unlike control cells or MyD88-deficient cells, P1 cells as well as the TLR3-deficient cells failed to respond to poly(I:C) (Supplemental Figure 4A), whereas their responses were normal upon IL-1β stimulation, unlike MyD88-deficient cells (Supplemental Figure 4B). Investigation of the functional pathways induced by poly(I:C) lead to the identification of genes that were induced in control cells but not in P1. These genes included IFN-regulated genes (IFIH1, IFIT1, IFIT2, and IFIT3), proinflammatory cytokines (IFNB1, IL12A, IL15, IL29, TNFSF13B, TNFAIP6, and TNFSF10), chemokines (CXCL11, CCL3, CCL3L1, CCL3L3, CCL8, CXCL9, CCL5, and CXCR7), and cell death/apoptosis genes (RIPK2, BCL10, CASP7, BIRC2, and BIRC3) (Supplemental Figure 4C). These experiments suggest that the R141X mutation results in a null allele causing AR complete TRIF deficiency, whereas the S186L TRIF allele is dysfunctional and causes AD dominant partial TRIF deficiency.
Figure 3. Response to TLR3 and TLR4 ligands.
(A) SV40 fibroblasts were stimulated with increasing doses of poly(I:C) for 24 hours, and production of cytokines was assessed. C+ is a healthy control; UNC93B1–/– served as a negative control. Values (mean ± SEM) were calculated from 3 independent experiments. (B) Native gel Western blot showing IRF3 dimers from total fibroblast cell lysates after stimulation with poly(I:C) for 1 or 2 hours. C+ and MYD88–/– cells were used as positive controls to poly(I:C) stimulation, and UNC93B1–/– was used as a negative control. (C) Nuclear protein extracts from SV40 fibroblasts stimulated for 30 minutes with IL-1β or TNF-α and 120 minutes with poly(I:C) were tested for the presence of the p65 subunit of NF-κB by ELISA. NEMO–/– cells were used as a negative control for all stimuli; MyD88–/– was used as a negative control for IL-1β; and UNC93B1–/– was used for poly(I:C) stimulation. Values (mean ± SEM) were calculated from 3 independent experiments. NS, nonstimulated. (D) SV40 fibroblasts were stimulated with a TLR3-specific ligand, poly(A:U), transfected poly(I:C), RIG-I–specific ligand (7sk-as), or lipofectamine alone (L) for 24 hours and tested for cytokine production. Values (mean ± SEM) were calculated from 3 independent experiments. (E) PBMCs from healthy controls, various family members from kindred A (AII.1 and AII.3) and B (BII.3), and P1 and P2 were stimulated with LPS for 2 hours. Induction of IFNB1 and IL6 mRNA was assessed by real-time PCR. Values are expressed as relative fold change using the ΔΔCt method, where GUS was used for normalization; values (mean ± SEM) from 3 independent experiments were calculated.
Cellular response to LPS.
TRIF is known to interact with TLR4 in mice and humans (22) and has been shown to control IFN induction by LPS in the mouse model (24–26). We therefore assessed IFNB1 mRNA induction in response to LPS stimulation in PBMCs from the patients. Compared with those from with a healthy control or his WT or heterozygous siblings, PBMCs from P1 did not induce the production of IFNB1 mRNA (P < 0.05, P < 0.05, and P < 0.05, respectively), whereas IL-6 was induced, albeit to a lower extent than in the control (P < 0.05), probably via the intact TLR4-MYD88–dependent pathway in P1 (Figure 3E). PBMCs from P2 and her mother, both of whom are heterozygous for the S186L mutation, were able to induce IFNB1 and IL6 mRNAs after LPS stimulation (Figure 3E), suggesting that the S186L TRIF mutation does not affect the TLR4-dependent induction of antiviral IFNs. The induction of IFNB1 mRNA was variable among controls and subjects, but P1 was the only patient for whom no induction was observed. Consistent with this observation, results from a genome-wide transcriptional analysis of the TLR4 pathway in PBMCs of a control, P1, and an AR MyD88-deficient patient showed impaired responses to LPS in P1 (Supplemental Figure 5A), although P1 responded normally to the TLR7/8 ligand R848 unlike the MyD88-deficient cells (Supplemental Figure 5B). As expected, MyD88-defcient cells did not respond normally to LPS, and a different pattern of LPS dysregulation was observed in the MyD88-deficient cells when compared with that in cells from P1, demonstrating the distinct pathways involved in the TLR4-MyD88 dependent pathway (Supplemental Figure 5, A and C). We found that the functional pathways regulated by LPS in control PBMCs consisted of genes involved in the production of various cytokines (IFNB1, IL23A, IL19, IL1A, and IL1F9), chemokine-related genes (CCR1, CCL3L1, CCL3L3, CCL14, CCL20, and CCL23), and IFN-regulated genes (IFIT3 and IRF8) (Supplemental Figure 5C). The nonsense mutation in P1 is consistent with a null mutation affecting all aspects of TRIF-specific signaling, including TLR4-TRIF responses. By contrast, the collective data for the kindred carrying a missense TRIF allele point to a specific defect in the TLR3 signaling pathway, suggesting that the TRIF missense allele is dysfunctional and dominant for TLR3 but not TLR4 responses and for the induction of antiviral IFNs in particular.
Impaired cellular responses to viruses.
We then assessed the responses of fibroblasts to viral infection. P1 and P2 cells showed impaired production of IFN-β, IFN-λ1/3, and IL-6 upon infection with VSV and HSV-1, like UNC-93B–deficient cells and unlike control cells (P < 0.05 for all comparisons) (Figure 4, A and D). Cell viability after infection with VSV and HSV-1 was tested in the presence and absence of exogenous IFN-α (Figure 4, B and E). In control cells, there was a small decrease or no decrease in the number of living cells upon infection with VSV or HSV-1, respectively. However, cells from P1 and P2 both had significantly lower percentages of surviving cells as compared with those of control cells, at MOIs of 10–100 with VSV (P < 0.05 for all comparisons) and upon HSV-1 infection (P < 0.05 for all comparisons). However, the addition of exogenous IFN-α resulted in very little cell death in cells from either P1 or P2, in contrast with what was observed for STAT1-deficient cells, which were unable to respond normally to IFN-α. This is consistent with the higher VSV viral titers in P1 and P2 compared with those in a healthy control (P < 0.05 for both comparisons), which were reduced to control levels after pretreatment with IFN-α (Figure 4C). Similarly, both P1 and P2 showed higher levels of HSV-1 replication compared with those of a control, which was also reduced with the addition of IFN-α (Figure 4F). Thus, the susceptibility of P1 and P2 to viruses probably results from a lack of protective endogenous IFN-α/β and/or IFN-λ production during viral infection. These data suggest that both P1 and P2 harbor a specific defect in the TLR3-dependent induction of IFN-β and IFN-λ, in response to HSV-1 and VSV, resulting in enhanced viral growth and cell death, at least in fibroblasts. This phenotype is consistent with those reported for both UNC-93B and TLR3 deficiencies and suggests that the R141X and S186L TRIF alleles are deleterious, with complete cellular penetrance but incomplete clinical penetrance (10, 11).
Figure 4. Response to viral infections.
(A) IFN-β, IFN-λ1/3, and IL-6 production in SV40 fibroblasts 24 hours after infection with VSV. Values (mean ± SEM) were calculated from 3 independent experiments. (B) Cell viability of SV40 fibroblasts after 24 hours of infection with VSV in the absence (left) or presence (right) of recombinant IFN-α. The percentage of surviving cells was assessed using resazurin. C+ is a healthy control; UNC93B1–/– and STAT1–/– cells were used as negative controls. Values (mean ± SEM) were calculated from 3 independent experiments. (C) VSV replication in SV40 fibroblasts was estimated at various time points after infection using an MOI of 10. Cells were pretreated with IFN-α or media alone. VSV titer estimation was carried out on Vero cells. Values (mean ± SEM) were calculated from 3 independent experiments. (D) IFN-β, IFN-λ1/3, and IL-6 production after infection with 1 MOI of HSV-1 in SV40 fibroblasts after 24 hours. Values (mean ± SEM) were calculated from 3 independent experiments. (E) Cell viability of SV40 fibroblasts was assessed using resazurin after 24 hours of HSV-1 infection. C+ is a healthy control; UNC93B1–/– and STAT1–/– cells were used as negative controls. Cells were pretreated with media (left) or recombinant IFN-α (right). Values (mean ± SEM) were calculated from 3 independent experiments. (F) Replication of HSV-1 GFP was assessed in SV40 fibroblasts after 24 hours of infection. Cells were pretreated with media (left) or recombinant IFN-α (right). Values (mean ± SEM) were calculated from 3 independent experiments.
Functional characterization and complementation of the R141X mutation.
We further investigated the impact of the R141X nonsense mutation by transiently transfecting 293HEK-TLR3 cells, which are responsive to poly(I:C) stimulation, with WT, R141X, or ΔNC TRIF constructs. The ΔNC TRIF construct encodes a truncated dominant-negative TRIF, consisting of only the 162 amino acids of the TIR domain (20). As expected, the R141X TRIF plasmid did not induce expression from the IFN-β or NF-κB promoters, unlike the WT plasmid, as shown by luciferase assays (Figure 5A). The level of induction was similar to that of cells transfected with empty plasmids, consistent with a null allele, and contrasting with the dominant-negative effect observed with the ΔNC plasmid. When the cells were transfected with equal amounts of WT plasmid and R141X or ΔNC, lower levels of luciferase activity were observed only in WT/ΔNC cells, suggesting that the R141X allele does not exert a dominant-negative effect. By contrast to what was observed with cells from P1, analysis of the proteins produced by transfected cells revealed the presence of an approximately 15-kDa truncated R141X protein (Figure 5B). The induction of the IFN-β promoter in response to LPS is strictly TRIF dependent. We therefore transiently transfected LPS-responsive 293HEK-TLR4-MD2-CD14 cells with the TRIF plasmids (Figure 5C). The R141X plasmid, unlike the WT plasmid, did not induce luciferase gene expression from the IFN-β plasmid, and cotransfection with the WT plasmid did not decrease luciferase induction, consistent with a null effect. However, the R141X and mock plasmids were able to induce NF-κB after LPS stimulation in these cells, as the induction of the NF-κB promoter is primarily MyD88 dependent, consistent with the known role of TRIF in TLR4 signaling. Transient transfection of control fibroblasts or fibroblasts from the patient with WT TRIF cDNA led to rapid apoptosis (data not shown), as previously shown in other systems (33, 34). Complementation of the patient’s fibroblast phenotype was therefore achieved by retroviral transduction. A doxycycline-regulated TRIF expression vector was used for the retroviral transduction of fibroblasts from P1, such that TRIF expression was repressed in the presence of doxycycline, making it possible to obtain stably transduced clones (Supplemental Figure 6A). The transduction of cells from P1 with the WT TRIF plasmid (P1+WT) restored poly(I:C)-induced IFN-β, IFN-λ, and IL-6 production, whereas cells transduced with the R141X plasmid (P1+R141X) remained unresponsive (Figure 5D). Viral replication after HSV-1 infection was also rescued in P1+WT cells but not in P1+R141X cells (Figure 5E). Thus, the R141X mutation in P1 is responsible for TRIF deficiency and the abolition of TLR3 signaling as well as susceptibility to HSV-1.
Figure 5. Molecular characterization of the R141X AR TRIF mutation.
(A) 293 HEK-TLR3 cells were transfected with 10 ng empty vector, WT, R141X, or ΔNC TRIF along with IFN-β–Luc/NF-κB–Luc and RL-TK vectors to assess induction upon overexpression of TRIF. ΔNC TRIF serves as a dominant-negative construct of TRIF, containg only the TIR domain. Transfected cells were left unstimulated or stimulated with 50 μg/ml poly(I:C) for 4 hours. Firefly luciferase values were normalized using renilla values. Total transfected DNA was held constant by adding empty vector. Values (mean ± SEM) were calculated from 3 independent experiments. (B) Immunoblot analysis of protein lysates from 293HEK-TLR3–transfected cells, using an antibody recognizing the N-terminal of TRIF. CFP was cotransfected as a control for transfection efficiency. Samples were migrated on the same blot. (C) 293HEK-TLR4-MD2-CD14 cells were transfected with 10 ng TRIF vectors, including WT, R141X, and ΔNC TRIF, and the induction of IFN-β–Luc/NF-κB–Luc was assessed by luciferase. Values (mean ± SEM) were calculated from 3 independent experiments. (D) Control (C+) SV40 fibroblasts and P1’s fibroblasts retrovirally transduced with empty vector, WT TRIF, or R141X TRIF were stimulated with 25 μg/ml poly(I:C) for 24 hours, and production of cytokines was assessed by ELISA. Values (mean ± SEM) were calculated from 3 independent experiments. (E) HSV-1 GFP replication was assessed after 24 hours of infection in control SV40 fibroblasts (C+) and SV40 fibroblasts from P1 retrovirally transduced with empty vector, WT, or R141X TRIF. Cells were pretreated with media (left) or recombinant IFN-α (right).Values (mean ± SEM) were calculated from 3 independent experiments.
Functional characterization of the S186L mutation in 293-TLR3 cells.
The molecular mechanism by which the S186L TRIF mutation exerts its deleterious effects was then investigated. We transfected 293-TLR3 cells with expression plasmids encoding WT or S186L TRIF. Reporter assays were performed with the IFN-β and NF-κB–luciferase constructs. In WT and S186L TRIF-transfected cells, marked induction of IFN-β and NF-κB luciferase was observed in the absence of poly(I:C) stimulation, consistent with previous reports in which TRIF overexpression led to TLR3-independent activation of IFN-β and NF-κB (Figure 6A, Supplemental Figure 7A, and refs. 21, 25). We assessed the ability of these mutations to interact with TLR3 upon poly(I:C) stimulation by transfecting 293-TLR3 cells with the mutant plasmids and the IFN-β luciferase construct and then stimulating them with poly(I:C) (Supplemental Figure 7B). Transfection with the S186L allele resulted in the induction of IFN-β upon poly(I:C) stimulation, as observed for the WT TRIF plasmid. After activation by TLR3, TRIF forms homo-oligomers in the cytosol that are responsible for all downstream signaling events (29). These homo-oligomers may be visualized on confocal microscopy as speckle-like structures. Activated TRIF was observed in HeLa cells after poly(I:C) stimulation in cells transfected with the WT or S186L plasmid, suggesting that mutant TRIF was able to interact with TLR3 and homo-oligomerize, like WT TRIF (Supplemental Figure 7C and ref. 29). Data for transient transfection with the S186L allele revealed no difference between the functions of this mutant form and the WT TRIF. The effect of this mutation was undetectable in transient transfection experiments, even if carried out with as little as 10 pg plasmid (data not shown).
Figure 6. Functional characterization of the S186L AD TRIF mutation.
(A) 293HEK-TLR3 cells were transfected with empty vector, WT, or S186L TRIF plasmids along with IFN-β–Luc/NF-κB–Luc and RL-TK vectors to assess IFN-β and NF-κB promoter induction upon overexpression of TRIF. Values (mean ± SEM) were calculated from at least 3 independent experiments. (B) SV40 fibroblasts from control (C+) and P1’s fibroblasts retrovirally transduced with either empty vector, WT TRIF, or S186L TRIF were stimulated with 25 μg/ml poly(I:C) for 24 hours, and the production of cytokines was assessed by ELISA. Values (mean ± SEM) were calculated from at least 3 independent experiments. (C) SV40 fibroblasts from control (C+) and P2’s fibroblasts retrovirally transduced with empty vector, WT TRIF, or S186L TRIF were stimulated with 25 μg/ml poly(I:C) for 24 hours, and the production of cytokines was assessed by ELISA. Values (mean ± SEM) were calculated from at least 3 independent experiments. (D) Replication of HSV-1 GFP was assessed after 24 hours of infection in control SV40 fibroblasts (C+); SV40 fibroblasts from P1 retrovirally transduced with empty vector, WT, or S186L TRIF; and (E) SV40 fibroblasts from P2 retrovirally transduced with empty vector, WT, or S186L. Cells were pretreated with media (left) or recombinant IFN-α (right). Values (mean ± SEM) were calculated from at least 3 independent experiments.
Investigating TRIF overexpression in transient transfection.
We then addressed the issue of the dominance of the S186L allele. However, as the hypomorphic mutation alone demonstrated normal TRIF function, cotransfection of WT and S186L alleles also revealed normal activation comparable to WT TRIF in 293-TLR3 cells (data not shown). We hypothesize that TRIF levels were already saturated in transient transfection experiments and were therefore not sensitive enough to reveal subtle differences. One of the problems inherent to studies of TRIF overexpression is that the protein is constitutively active and induces apoptosis (20, 33, 34), which does not occur in cells producing small amounts of endogenous TRIF. Indeed, TRIF is normally produced in very small amounts in cells, only transducing and recruiting downstream signaling molecules upon stimulation by TLR3 (29, 35). Thus, the amounts of TRIF protein produced in conditions of overexpression do not reflect physiologically relevant functions of TRIF, as they cause constitutive activation and apoptosis. The transient transfection system has been successfully used by many groups to study the function of TRIF in conditions of overexpression (20, 21, 28, 29, 33, 36). However, a review of the literature showed that no study has ever demonstrated a hypomorphic effect of a point mutation in the context of this transfection system; only null effects have been characterized. These findings support the notion that subtle effects cannot be studied accurately in the context of overexpression.
Functional characterization of the S186L allele in retroviral transduced fibroblasts.
We then made use of the fibroblasts available from the patient with AR TRIF deficiency (P1) for retroviral transfection experiments with the S186L allele. Retroviral transduction yielded much lower TRIF levels, although these levels were nonetheless higher than endogenous levels (Supplemental Figure 6). The lack of constitutive activation in retrovirally transduced cells (Figure 5D and Figure 6, B and C) and the low levels of apoptosis suggest that TRIF levels in this system were similar to endogenous levels. They may, therefore, provide ideal conditions for the detection of subtle mutations. The S186L-transduced cells (P1+S186L cells) resulted in significantly lower levels of cytokine production in response to poly(I:C) stimulation compared with P1+WT cells (P < 0.05 for all comparisons), suggesting that the S186L allele is hypomorphic (Figure 6B and Supplemental Figure 6A). Furthermore, complementation of fibroblasts from P2, which express endogenous TRIF, was achieved with the WT TRIF allele (P2+WT), whereas the S186L allele (P2+S186L) showed impaired responses in terms of the production of IFN-β, IFN-λ, and IL-6 compared with P2+WT cells (P < 0.05 for all comparisons) (Figure 6C and Supplemental Figure 6B). Moreover, viral replication after HSV-1 infection in P1+WT cells was comparable to that of a control, whereas P1+S186L cells had significantly higher viral titers compared with those of a control or with P1+WT cells (P < 0.05 for both comparisons). There was lower replication in P1+S186L cells than in cells from P1 retrovirally transduced with empty vector (P1+Empty cells) (P < 0.05), consistent with a hypomorphic effect (Figure 6D). Increased HSV-1 replication observed in P2+Empty cells was also complemented in P2+WT cells (P < 0.05) (Figure 6E). P2+S186L cells showed a trend toward higher HSV-1 titers compared with that of P2+WT cells, although the difference was not statistically significant, which can be attributed to the hypomorphic nature of the S186L allele. Overall, these data suggest that the S186L mutation is hypomorphic, affecting both the IRF3/IFN-β and NF-κB/IL-6 pathways and controlling susceptibility to HSV-1 infection.
A heterozygous nondeleterious TRIF missense mutation in P3.
We identified a third distinct heterozygous mutation in TRIF in a boy of Iranian origin (P3), who was born to nonconsanguineous parents and who suffered from HSE at 4.5 years of age (Supplemental Figure 8A). The patient is now 7 years old and has not suffered from other unusual or severe infections. Mutations in the coding regions of UNC93B1, TLR3, and TRAF3 were excluded. Further sequencing of leukocyte gDNA from P3 lead to the discovery of a heterozygous missense mutation in TRIF at nucleotide position 1875 (c.1875C>T), resulting in a leucine at amino acid residue 625 in place of a proline in the proline-rich domain of the C terminus of TRIF (P625L) (Supplemental Figure 8, B and C). This mutation was inherited from P3’s father who is seronegative for anti–HSV-1 antibodies; hence, P3 remains the only family member affected by HSE. This mutation was not found in the NCBI database, the 1,050 unrelated healthy controls from the CEPH-HGD panel, or in an additional 109 healthy Iranian controls tested, confirming that c.1875C>T is a not a polymorphism. Proline at position 625 was conserved in 7 out of the 11 animal species in which TRIF shares over 50% homology with human TRIF (Supplemental Figure 8D). TRIF protein and mRNA expression were normal in EBV-B cells from P3 (Supplemental Figure 8E and data not shown). This mutation occurred in the proline-rich C-terminal region, implicated in homo-oligomerization, RIP1 binding, NF-κB activation, and apoptosis-inducing functions of TRIF (29, 33). However, despite a poly(I:C) nonresponsive fibroblast phenotype (Supplemental Figure 8F), we did not observe complementation of the phenotype by retroviral transduction of the WT TRIF allele in P3’s cells (Supplemental Figure 8H). Moreover, P3’s mutation P625L transduced into P1’s cells did not differ from WT TRIF–transduced cells in its ability to induce cytokines after poly(I:C) stimulation (Supplemental Figure 8F and Supplemental Figure 6A). Hence, although not found in healthy controls, we were unable to show that this particular mutation is deleterious with the assays available. We hypothesize that P3 carries a pathogenic mutation affecting another gene in the TLR3 signaling pathway. These data neatly illustrate the importance of careful functional characterization of rare mutant alleles, as shown for P2, to ascribe disease responsibility.
Discussion
We report here the first 2 kindreds with AR and AD TRIF deficiencies conferring a predisposition to childhood HSE to our knowledge. The loss-of-expression, loss-of-function, nonsense R141X allele results in the abolition of TLR3-mediated signaling and TRIF-dependent TLR4 responses in the cells of P1. Most family members of P1 are heterozygous carriers of this mutation but have not suffered from HSE or any other infection otherwise. Despite diminished TRIF protein in cells from heterozygous individuals, their leukocytes responded to LPS by the induction of IFN-β transcripts, suggesting that TRIF function was maintained. Moreover, the R141X allele was not dominant negative in 293-TLR3 transfection assays. We did not have access to heterozygous fibroblasts for further testing of the TLR3 pathway, but these results suggest that this defect is purely recessive and that there is no cellular or clinical haploinsufficiency at the TRIF locus. The TLR4-TRIF pathway was the only TIR pathway for which no inborn defect had been detected, as it is not defective in patients with UNC-93B, TLR3, MYD88, and IRAK4 deficiencies (10, 11, 13, 37–40). The amounts of IFN-α/β produced in response to the human TRIF-dependent TLR4 pathway are modest, and the role of this pathway in host defense has remained elusive (41, 42). The identification of complete TRIF deficiency in P1, who did not suffer from any infection other than HSE, opportunistic or otherwise, suggests that the human TLR4-TRIF pathway is largely redundant in host defense. Moreover, our study suggests that the recently described role of TRIF in DExD/H-box helicases-dependent cytosolic pathways is also probably redundant for protective immunity (27).
We also identified a patient, P2, with a heterozygous missense mutation in the N-terminal region of TRIF (S186L), defining AD TRIF deficiency as a genetic etiology of HSE. Cells from P2 displayed impaired TLR3-mediated signaling but the maintenance of TRIF-dependent TLR4 responses. The mutant allele showed complete penetrance at the cellular level, because fibroblasts from the 2 TRIF heterozygotes tested, P2 and her mother, showed impaired TLR3 responses. The N-terminal region of TRIF, including the TBK1-binding domain, is indispensable for the IRF3 pathway, consistent with the cellular phenotype of P2, in which IFN-β production and IRF3 dimerization were more affected than the production of IL-6 and NF-κB nuclear translocation after poly(I:C) stimulation (21, 25, 36). P2 fibroblasts were rescued by the WT — but not the mutant TRIF — allele, indicating that the allele is deleterious. The S186L allele appeared functional when used for the transient transfection of 293-TLR3 cells, whereas it was dysfunctional when introduced by retroviral transduction into TRIF-deficient fibroblasts from P1, suggesting that it is hypomorphic but not a complete loss-of-function allele. Data for the kindred with AR complete TRIF deficiency suggest that there is no haploinsufficiency at the TRIF locus, pointing toward the notion of the S186L mutation being dominant negative. However, it was difficult to determine experimentally whether the S186L allele was actually dominant negative, as overexpressed TRIF is constitutively active and a potent inducer of apoptosis, preventing fine analysis of the TLR3 pathway. Nonetheless, the available data suggest that the hypomorphic S186L allele of TRIF is probably dominant negative and responsible for AD TRIF deficiency and HSE.
The clinical penetrance of AR TRIF deficiency appears, so far, to be complete, as only one patient with this deficiency has been identified and has had HSE. Similarly, the only known patients with AR TLR3 or AD TRAF3 deficiency also show complete clinical penetrance (12, 13). In contrast, the penetrance of AD TRIF deficiency is incomplete, as only 1 out of the 3 S186L TRIF heterozygotes developed HSE upon infection with HSV-1. This is consistent with AR UNC-93B and AD TLR3 deficiencies, which also show incomplete clinical penetrance (10, 11). Whole-exome sequencing carried out in P2 failed to identify overt deleterious mutations that may explain the variable penetrance (data not shown). In patients with mutations in any of these 4 genes, HSE probably resulted from the impairment of antiviral IFN production in the CNS, leading to higher levels of viral replication and the death of resident cells. The fibroblastic and clinical phenotypes of AR and AD TRIF deficiencies are strikingly similar to those of UNC-93B deficiency (10), TLR3 deficiency (11, 13), and TRAF3 deficiency (12), consistent with the known role of TRIF as the sole adaptor for TLR3. This identification of a fourth gene underlying susceptibility to HSE in 2 kindreds is consistent with our model for the genetic architecture of infectious diseases (18, 43). We proposed that life-threatening pediatric infectious diseases can result from the inheritance of rare single gene mutations, highly deleterious at the cellular level but of variable clinical penetrance (18, 44). Although unlikely, the lack of infectious phenotype or the selective susceptibility to HSE observed in known patients with inborn errors of TLR3 immunity might result from compensatory variations at other loci. The TLR3 pathway is vital for protection against HSE, at least in some children, which may additionally ensure that chronic latency of HSV-1 heightens immunity against other microbes, as shown in the mouse model (45). In any case, HSE provides proof of principle that a collection of single-gene variations displaying incomplete penetrance at the clinical level and affecting different but immunologically related genes may account for severe, sporadic, and common infectious diseases of childhood.
Methods
Patients
We investigated 3 unrelated patients who suffered from HSE during childhood. P1 is a Saudi Arabian boy of consanguineous parents (first cousins). There is no family history of encephalitis, and there was no remarkable infectious history until P1 developed viral meningitis of unknown origin at 1.5 years of age. At the age of 2, he developed HSE, presenting with irritability, fever, and partial seizure. HSE was confirmed by a positive PCR for HSV-1 in the CSF. An MRI scan showed atrophy of the left temporal lobe, and an EEC revealed epileptogenic activity. He suffers from neurological sequelae, mainly consisting of delayed speech. He also has recurrent herpetic stomatitis. He has not yet been exposed to other herpes viruses, determined by negative serology for CMV and VZV.
P2 is a French, Swiss, and Portuguese girl, of nonconsanguineous parents. There is no family history of encephalitis. The mother suffered from meningitis of unknown origin at the age of 9, from which she fully recovered. The mother and maternal grandmother have had herpes labialis. The mother, the maternal grandfather, and P2 are all seropositive for HSV-1. Of note, P2 had a rectovaginal fistula surgical intervention at birth. There was no remarkable infectious history until P2 suffered from serologically determined HSE at 21 months, presenting with high fever. HSE was confirmed by a positive PCR for HSV-1 in the CSF. Serum analysis detected recent HSV-1 seroconversion. Treatment with acyclovir was administered 4 days after symptoms of fever, after which the patient was hospitalized for treatment. There was no relapse, and she has never suffered from herpes labialis. She suffers from neurological sequelae, mainly consisting of blindness and epilepsy. Following HSE, she has been exposed to other herpes viruses, as shown by positive serology for VZV and EBV with no complications. She has been vaccinated with MMR with no adverse effects.
P3 is an Iranian boy of nonconsanguineous parents. P3 presented with fever, vomiting, and convulsions, with right temporal involvement as shown by MRI. HSE diagnosis was confirmed by an HSV-1–positive PCR in the CSF. Treatment was initiated with acyclovir with no significant improvement. Subsequent treatment with foscarnet for 21 days was successful. At day 23, symptoms of low consciousness and choreatic movement were observed along with a positive PCR for HSV-1. Treatment with cidofovir was administered for 5 days, and he has fully recovered. P3 has not yet been exposed to other herpes viruses, such as VZV and EBV, as determined by negative serology. Of note, the father who carries the mutation is seronegative for serum antibodies to HSV, whereas the mother is seropositive. There is no remarkable medical history in the family.
Cell lines
Skin biopsy specimens from the patients as well as family members, the UNC-93B–deficient patient, TLR3-deficient patient, the MyD88-deficient patient, the STAT1-deficient patient, and healthy controls were obtained for the establishment of primary fibroblasts. Primary fibroblasts were cultured in DMEM supplemented with 10% FBS, 10 U/ml penicillin, 10 μg/ml streptomycin, and 62.5 ng/ml amphotericin B (Invitrogen). Fibroblasts were immortalized with the SV40 large T antigen, as previously described (46). NEMO-deficient immortalized fibroblasts were derived from a fetus with incontinentia pigmenti (47). All cell lines were incubated at 37°C, under an atmosphere containing 5% CO2. PBMCs from patients and healthy controls were harvested from fresh blood by Ficoll-Hypaque density gradient centrifugation (GE Healthcare). PBMCs were cultured in RPMI supplemented with 10% FBS (Invitrogen) or 1% human sera.
Activation by TLR ligands
A synthetic analog of dsRNA, poly(I:C) (TLR3 agonist, at 25 μg/ml), and LPS (LPS Salmonella minnesota, TLR-4 agonist, at 10 to 100 ng/ml) were purchased from Invivogen; resiquimod hydrochloride (R-848; TLR-7, and TLR-8 agonist, at 3 μg/ml) was purchased from GLSynthesis Inc.; unmethylated CpG DNA CpG-A (D19) (TLR-9 agonist, at 5 μg/ml) was provided by Robert Coffman and Frank Barrat (both from Dynavax Technologies, Berkeley, California); and IL-1β (at 20 ng/ml) and TNF-α (at 20 ng/ml) were obtained from R&D Systems. All agonists and reagents were endotoxin free. In all stimulations of PBMCs with TLR agonists other than LPS, the cells were incubated with 10 μg/ml polymyxin B (Sigma-Aldrich), at 37°C, for 30 minutes before activation. TLR agonists were used to stimulate PBMCs for 48 hours, at a concentration of 2 × 106 cells per ml in RPMI 1640 supplemented with 10% FBS or 1% human serum. The SV40-fibroblast cell lines were activated in 24-well plates, at a density of 105 cells per well, for 24 hours with poly(I:C) (25 μg/ml), TNF-α (20 ng/ml), IL-1β (20 ng/ml), IPH 3102 or poly(A:U) (50 μg/ml) TLR3-specific ligand (Innate Pharma), and 7sk-as (provided by Caetano Reis e Sousa, London Research Institute, London, United Kingdom).
Viral infections for cytokine determination
PBMCs isolated by Ficoll-Paque density gradient centrifugation were stimulated with various intact viruses for 24 hours, at a density of 2 × 106 cells per ml in RPMI 1640 supplemented with 10% FBS or, for some experiments, 2% FBS. The following viruses were used: the dsDNA virus HSV-1 (KOS-1 strain, MOI = 1) and the ss(–)RNA virus VSV (Indiana strain, MOI = 1). The SV40-fibroblast cell lines were activated in 24-well plates, at a density of 105 cells per well, for 24 hours with HSV-1 or VSV. Cell supernatants were recovered and tested for cytokine production by ELISA.
Cytokine determinations
The production of IFN-α, IFN-β, IFN-λ1/3, and IL-6 was assessed by ELISA, according to the kit manufacturer’s instructions, respectively: Human IFN-α Module Set (AbCys SA), Human IFN-β ELISA Kit (TFB, Fujirebio Inc.), Duo Set Human IL29/IL28B (IFN-λ1/3) (R&D Systems), and Human IL-6 (Sanquin).
Q-PCR
Total RNA was extracted from cells either unstimulated or stimulated with TLR agonists. All RNA samples were treated with RNase-free DNase (Qiagen) after isolation with the RNeasy Extraction Kit (Qiagen). RNA was reverse transcribed directly, with the High Capacity RNA-to-cDNA Master Mix (Applied Biosystems). Q-PCR was performed with Applied Biosystems Assays-on-Demand probe/primer combinations specific for IFN-β, IL-6, TRIF, and β-glucuronidase (GUS), which was used for normalization. Results are expressed according to the ΔΔCt method, as described by the manufacturer.
Western blots
Total cell extracts were prepared from 293-NULL, 293-TLR3, 293-TLR4-MD2-CD14, SV40 fibroblasts, and EBV-B cells. Equal amounts of protein from each sample were separated on SDS-PAGE gels and blotted onto iBLOT gel transfer stacks (Invitrogen), which were then probed with antibodies and developed with peroxidase-conjugated secondary antibodies and ECL Western blotting substrate (GE Healthcare). TRIF antibodies binding to the N-terminal region of human TRIF (Alexis Biochemicals) were used for Western blots. Anti-CFP (Abcam), anti-ACTIN, anti-GAPDH (Santa Cruz Biotechnology Inc.), and anti-HA antibodies (Sigma-Aldrich) were also used.
Signal transduction studies
Nuclear cell extracts were prepared from SV40-fibroblasts after incubation with or without poly(I:C), TNF-α, and IL-1β. TransAM NF-κB p65 ELISA (Active Motif) was performed with an NF-κB p65-specific DNA probe and 10 μg nuclear extract. The Bio-Rad protein assay was used to determine protein concentrations. For the detection of IRF3 dimerization, whole-cell extracts were prepared from SV40-fibroblasts with or without poly(I:C) treatment. The IRF3 monomers and dimers were separated by native PAGE in the presence of 1% sodium deoxycholate (DOC) (Sigma-Aldrich). Total cell extracts (25 μg of protein) were diluted 1:2 in nondenaturing sample buffer (125 mM Tris-HCl, pH 6.8, 30% glycerol, and 2% DOC) and separated by electrophoresis in a 7.5% polyacrylamide gel, in 25 mM Tris and 192 mM glycine, pH 8.4, with 1% DOC present in the cathode chamber only. The gel was blotted onto a membrane, which was then probed with the anti-IRF3 antibody (FL-425, Santa Cruz Biotechnology Inc.).
Transient transfections and reporter assays
The mutant alleles were generated with the QuikChange II Site-Directed Mutagenesis Kit from Qiagen for introduction of the C421T, C557T, and C1875T mutations into the pEF-Bos-TRIF plasmid C-terminally tagged with HA (provided by Misako Matsumoto). The pcDNA3-CFP vector (Addgene) was used to control for transfection efficiency. The ΔNC TRIF construct was a gift from Osamu Takeuchi (Osaka University, Osaka, Japan). pGL4.32[luc2P/NF-κB-RE/Hygro] NF-κB luciferase and pRL-TK Renilla luciferase plasmids were purchased from Promega Corporation. The IFN-β promoter-luciferase construct was provided by Christopher Basler (Mount Sinai School of Medicine, New York, New York, USA). 293-NULL, 293-hTLR3, or 293-hTLR4A-MD2-CD14 cells (Invivogen) were plated in 24-well plates and transfected with the expression vectors in the presence of lipofectamine LTX reagent, according to the manufacturer’s instructions (Invitrogen). The total amount of DNA was kept constant with empty vector (1 μg/well). Apoptosis was inhibited by adding 20 μM z-VAD-fmk (Sigma-Aldrich) to each well. Twenty hours after transfection, the cells were incubated for 4 hours more, with or without 50 μg/ml poly(I:C). Cell lysates were obtained and analyzed with the Dual-Luciferase Reporter Assay System (Promega Corporation), according to the manufacturer’s instructions. Firefly luciferase values were normalized against Renilla luciferase activity and expressed as fold change with respect to cells transfected with the empty vector.
Confocal microscopy
HeLa cells were transfected with the expression plasmid for WT TICAM-1 (WT) or the S186L mutant (0.2 ng); 24 hours after transfection, cells were stimulated with buffer alone or with 10 μg/ml poly(I:C) for 30 minutes. Cells were fixed in 4% paraformaldehyde for 30 minutes and permeabilized with 0.2% Triton X-100 in PBS for 10 minutes. Fixed cells were blocked in 1% bovine serum albumin in PBS and labeled by incubation with anti-HA pAb for 60 minutes at room temperature. Alexa Fluor 594–conjugated secondary antibodies (1:400) were used to visualize the primary antibody. Nuclei were stained with DAPI (2 μg/ml) in PBS for 10 minutes, before being mounted on glass slides in PBS supplemented with 2.3% 1,4-diazabicyclo[2.2.2]octane and 50% glycerol. Cells were visualized at ×63 magnification, with an LSM510 META microscope (Zeiss).
Retroviral transduction of SV40 fibroblasts
The doxycycline-inducible expression plasmid (pLINX-Flag) and pLINX-Flag-TRIF were provided by Sang Hoon Rhee (UCLA, Los Angeles, California, USA). The C421T, C557T, and C1875T mutations were introduced into the pLINX-Flag-TRIF plasmid by site-directed mutagenesis, as described above. 293T cells (ATCC no. CRL-11268) were grown in DMEM (Gibco BRL/Invitrogen) supplemented with 10% heat-inactivated FBS, 50 IU/ml penicillin, and 50 μg/ml streptomycin (Gibco BRL/Invitrogen) at 37°C in a humidified atmosphere containing 5% CO2. Retroviral vectors pseudotyped with the vesicular stomatitis G protein (VSV-G) were generated as previously described (48) by calcium phosphate transfection into 293T cells of a packaging construct, pMNold gag-pol; a plasmid producing the VSV-G envelope (pMD.G); and each of the pLINX vectors. Culture medium was collected at 24, 48, and 72 hours, pooled, 0.45-μ filtered, and concentrated approximately 1,000 fold by ultracentrifugation. Fibroblasts (1.5 × 105 cells per well) were seeded into a 12-well dish and infected on the following day, with 8 to 16 μl of each retroviral vector for 4 hours. Twenty-four hours later, the medium was replaced with DMEM containing 1 μg/ml doxycycline (Clonetech) to prevent expression of the TRIF cDNA insert. Forty-eight hours later, DMEM containing doxycycline and 1 mg/ml G418 (Invivogen) was added to the cells. Selected cells were amplified. The stably transduced cells were then plated in 24-well plates without doxycycline for 24 hours and stimulated with 25 μg/ml poly(I:C) for an additional 24 hours. The cells were lysed after 48 hours of culture in the absence of doxycycline.
Viral replication
For viral titration of VSV, 105 SV40-fibroblasts were plated in each well of a 24-well plate and infected at a MOI of 10 in DMEM supplemented with 2% FBS. The Stat-1–deficient patient has been described elsewhere (46). After 30 minutes, cells were washed and incubated in 500 μl of medium. All cells and supernatants were harvested and frozen at the time points indicated in the figures. Viral titers were determined by calculating the 50% end point (TCID50), as described by Reed and Muench, after the inoculation of Vero cell cultures in 96-well plates. For HSV-1 replication, HSV-1 GFP (strain KOS) (49), at MOIs ranging from 0.1–10, was used to infect the 104 SV40 fibroblasts plated in 96-well plates. The GFP fluorescence of the samples was quantified after 24 hours. For assays of cell protection upon viral stimulation, cells were treated with IFN-α2b (Schering-Plough, 1 × 105 IU/ml) 18 hours before infection and during infection.
Cell viability assay
The viability of SV40-fibroblasts was assessed by resazurin oxidoreduction (TOX-8) (Sigma-Aldrich). Cells were plated in triplicate in 96-well flat-bottomed plates (2 × 104 cells/well) in DMEM supplemented with 2% FBS. Cells were infected with VSV or HSV-1 for 24 hours at the indicated MOI. Resazurin dye solution was then added, and the samples were incubated for an additional 2 hours at 37°C. Fluorescence was then measured at a wavelength of 590/560 nm. Background fluorescence, calculated for dye and complete medium alone (in the absence of cells), was then subtracted from the values for all the other samples; 100% viability corresponds to the fluorescence of uninfected cells. For assays of cell protection upon viral stimulation, cells were treated with IFN-α2b (Schering-Plough, 1 × 105 IU/ml) for 18 hours before infection and during infection.
Genome-wide transcriptional profile studies in fibroblasts and PBMCs
Microarray.
SV40 fibroblasts obtained from patients or control subjects were stimulated with 25 μg/ml poly(I:C) or 20 ng/ml IL-1β or left unstimulated for 4 hours. Previously frozen PBMCs from patients or control subjects were left overnight in 10% FBS RPMI and then stimulated with 10 ng/ml LPS or 3 μg/ml R848 or left unstimulated for 2 hours. Total RNA was isolated (RNeasy Kit, Qiagen), and RNA integrity was assessed on an Agilent 2100 Bioanalyzer (Agilent). Biotinylated cRNA targets were prepared from 100 to 250 ng total RNA, using the Illumina TotalPrep RNA Amplification Kit (Ambion). The labelled cRNA (750 ng) were then incubated for 16 hours to HT-12 v4 BeadArrays (48,323 probes). Beadchip arrays were then washed, stained, and scanned on an Illumina HiScanSQ according to the manufacturer’s instructions.
Data preprocessing.
For the analysis of SV40 fibroblasts and PBMCs, after background subtraction, the raw signal values extracted with Illumina Beadstudio (version 2) were scaled using quantile normalization. Minimum intensity was set to 10, and all the intensity values were log2 transformed. Only the probes called present in at least 1 sample (P < 0.01) were retained for downstream analysis (n = 23,281 and n = 22,881 for fibroblasts and PBMCs, respectively).
Data analysis.
Transcripts differentially regulated upon stimulation were defined based on a minimum 2-fold change (upregulation or downregulation) and a minimum absolute raw intensity difference of 100 with respect to the respective unstimulated sample. Heat maps were generated using R (version 2.12.2). Raw data from this study are available in the Gene Expression Omnibus repository (accession no. GSE32390; http://www.ncbi.nlm.nih.gov/gds).
DNA samples
Control gDNA samples were obtained from the CEPH for the 1,052 individuals from the CEPH-HGD panel. An additional 182 genomic samples were obtained from Prince Naif Center for Immunology Research, King Khalid University Hospital (KKUH), Riyadh, Saudi Arabia, for a panel of control Saudi Arabian children aged between 6 and 8 years old. This study was approved by the College of Medicine Research Council (CMRC) Ethical Committee of the KKUH. Informed written consent was obtained from the patients’ parents or guardians. The male-to-female ratio was approximately 2:1. In order to validate the P625L variant in a control Iranian population, we used 109 gDNA samples collected from healthy blood donors referred to the Medical Laboratories of the Arad General Hospital in Tehran. All individuals were from both sexes, from Tehran, and represent different ethnic groups, which are considered to be representative of the general Iranian population. DNA was extracted from whole blood. All data and the collection of samples were approved by the Iranian National Ethics Committee.
Statistics
Unless otherwise specified, mean values ± SEM were calculated for all results. The Student’s t test (2 tailed) was used to determine significance, where P < 0.05 was found to be statistically significant.
Study approval
The experiments described here were conducted in France in accordance with local regulations approved by the CMRC Ethical Committee of the KKUH, the IRB of Necker Enfants Malades Hospital, and the IRB of Mofid Children Hospital.
Supplementary Material
Acknowledgments
We thank the members of the Laboratory of Human Genetics of Infectious Diseases, in particular Mélanie Migaud, Isabelle Melki, Martine Courat, Tony Leclerc, and Yelena Nemirovskaya; M. Oritz Lombardo for helpful discussions and technical assistance; Osamu Takeuchi for the dominant-negative TRIF vector; Sang Hoon Rhee for the doxycycline-inducible TRIF plasmid; Christopher Basler for the IFNB-luciferase plasmid; and Marianne Leruez for viral serology. We thank the children and their families for participating in this study. V. Sancho-Shimizu was supported by the Marie Curie Intra-European Fellowship 2008-2010. J.-L. Casanova was an international scholar of the Howard Hughes Medical Institute from 2005 to 2008. The Laboratory of Human Genetics of Infectious Diseases is supported by grants from the Agence national de la Recherche (ANR-08-MNP-014), The Rockefeller University Center for Clinical and Translational Science (grant no. 5UL1RR024143-03), and The Rockefeller University.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2011;121(12):4889–4902. doi:10.1172/JCI59259.
References
- 1.Whitley RJ. Herpes simplex virus in children. Curr Treat Options Neurol. 2002;4(3):231–237. doi: 10.1007/s11940-002-0040-2. [DOI] [PubMed] [Google Scholar]
- 2.Whitley RJ, et al. Vidarabine versus acyclovir therapy in herpes simplex encephalitis. N Engl J Med. 1986;314(3):144–149. doi: 10.1056/NEJM198601163140303. [DOI] [PubMed] [Google Scholar]
- 3.Whitley RJ, Lakeman F. Herpes simplex virus infections of the central nervous system: therapeutic and diagnostic considerations. Clin Infect Dis. 1995;20(2):414–420. doi: 10.1093/clinids/20.2.414. [DOI] [PubMed] [Google Scholar]
- 4.McGrath N, Anderson NE, Croxson MC, Powell KF. Herpes simplex encephalitis treated with acyclovir: diagnosis and long term outcome. J Neurol Neurosurg Psychiatry. 1997;63(3):321–326. doi: 10.1136/jnnp.63.3.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon B, Selnes OA, Hart J, Jr, Hanley DF, Whitley RJ. Long-term cognitive sequelae of acyclovir–treated herpes simplex encephalitis. Arch Neurol. 1990;47(6):646–647. doi: 10.1001/archneur.1990.00530060054017. [DOI] [PubMed] [Google Scholar]
- 6.Sancho-Shimizu V, et al. Genetic susceptibility to herpes simplex virus 1 encephalitis in mice and humans. Curr Opin Allergy Clin Immunol. 2007;7(6):495–505. doi: 10.1097/ACI.0b013e3282f151d2. [DOI] [PubMed] [Google Scholar]
- 7.Whitley RJ, Gnann JW. Viral encephalitis: familiar infections and emerging pathogens. Lancet. 2002;359(9305):507–513. doi: 10.1016/S0140-6736(02)07681-X. [DOI] [PubMed] [Google Scholar]
- 8.Abel L, et al. Age-dependent Mendelian predisposition to herpes simplex virus type 1 encephalitis in childhood. J Pediatr. 2010;157(4):623–629. doi: 10.1016/j.jpeds.2010.04.020. [DOI] [PubMed] [Google Scholar]
- 9.De Tiege X, Rozenberg F, Heron B. The spectrum of herpes simplex encephalitis in children. Eur J Paediatr Neurol. 2008;12(2):72–81. doi: 10.1016/j.ejpn.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 10.Casrouge A, et al. Herpes simplex virus encephalitis in human UNC-93B deficiency. Science. 2006;314(5797):308–312. doi: 10.1126/science.1128346. [DOI] [PubMed] [Google Scholar]
- 11.Zhang SY, et al. TLR3 deficiency in patients with herpes simplex encephalitis. Science. 2007;317(5844):1522–1527. doi: 10.1126/science.1139522. [DOI] [PubMed] [Google Scholar]
- 12.Perez de Diego R, et al. Human TRAF3 adaptor molecule deficiency leads to impaired Toll-like receptor 3 response and susceptibility to herpes simplex encephalitis. Immunity. 2010;33(3):400–411. doi: 10.1016/j.immuni.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Guo Y, et al. Herpes simplex virus encephalitis in a patient with complete TLR3 deficiency: TLR3 is otherwise redundant in protective immunity. J Exp Med. 2011;208(10):2083–2098. doi: 10.1084/jem.20101568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dupuis S, et al. Impaired response to interferon-alpha/beta and lethal viral disease in human STAT1 deficiency. Nat Genet. 2003;33(3):388–391. doi: 10.1038/ng1097. [DOI] [PubMed] [Google Scholar]
- 15.Audry M, et al. NEMO is a key component of NF-κB- and IRF-3–dependent TLR3-mediated immunity to herpes simplex virus. J Allergy Clin Immunol. 2011;128(3):610–617. doi: 10.1016/j.jaci.2011.04.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouanguy E, et al. Human primary immunodeficiencies of type I interferons. Biochimie. 2007;89(6–7):878–883. doi: 10.1016/j.biochi.2007.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Zhang SY, et al. Human Toll-like receptor–dependent induction of interferons in protective immunity to viruses. Immunol Rev. 2007;220:225–236. doi: 10.1111/j.1600-065X.2007.00564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Alcaïs A, Quintana-Murci L, Thaler DS, Schurr E, Abel L, Casanova JL. Life-threatening infectious diseases of childhood: single-gene inborn errors of immunity? Ann N Y Acad Sci. 2010;1214:18–33. doi: 10.1111/j.1749-6632.2010.05834.x. [DOI] [PubMed] [Google Scholar]
- 19.Matsumoto M, Kikkawa S, Kohase M, Miyake K, Seya T. Establishment of a monoclonal antibody against human Toll-like receptor 3 that blocks double-stranded RNA–mediated signaling. Biochem Biophys Res Commun. 2002;293(5):1364–1369. doi: 10.1016/S0006-291X(02)00380-7. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto M, et al. Cutting edge: a novel Toll/IL-1 receptor domain–containing adapter that preferentially activates the IFN-beta promoter in the Toll-like receptor signaling. J Immunol. 2002;169(12):6668–6672. doi: 10.4049/jimmunol.169.12.6668. [DOI] [PubMed] [Google Scholar]
- 21.Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3–mediated interferon-beta induction. Nat Immunol. 2003;4(2):161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald KA, et al. LPS-TLR4 signaling to IRF-3/7 and NF-kappaB involves the toll adapters TRAM and TRIF. J Exp Med. 2003;198(7):1043–1055. doi: 10.1084/jem.20031023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oshiumi H, et al. TIR-containing adapter molecule (TICAM)-2, a bridging adapter recruiting to toll-like receptor 4 TICAM-1 that induces interferon-beta. J Biol Chem. 2003;278(50):49751–49762. doi: 10.1074/jbc.M305820200. [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto M, et al. TRAM is specifically involved in the Toll-like receptor 4–mediated MyD88-independent signaling pathway. Nat Immunol. 2003;4(11):1144–1150. doi: 10.1038/ni986. [DOI] [PubMed] [Google Scholar]
- 25.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301(5633):640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 26.Hoebe K, et al. Identification of Lps2 as a key transducer of MyD88-independent TIR signalling. Nature. 2003;424(6950):743–748. doi: 10.1038/nature01889. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Z, et al. DDX1, DDX21, and DHX36 helicases form a complex with the adaptor molecule TRIF to sense dsRNA in dendritic cells. Immunity. 2011;34(6):866–878. doi: 10.1016/j.immuni.2011.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang Z, Mak TW, Sen G, Li X. Toll-like receptor 3–mediated activation of NF-kappaB and IRF3 diverges at Toll–IL-1 receptor domain-containing adapter inducing IFN-beta. Proc Natl Acad Sci U S A. 2004;101(10):3533–3538. doi: 10.1073/pnas.0308496101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Funami K, Sasai M, Oshiumi H, Seya T, Matsumoto M. Homo-oligomerization is essential for Toll/interleukin-1 receptor domain–containing adaptor molecule-1–mediated NF-kappaB and interferon regulatory factor–3 activation. J Biol Chem. 2008;283(26):18283–18291. doi: 10.1074/jbc.M801013200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pichlmair A, et al. RIG-I–mediated antiviral responses to single-stranded RNA bearing 5′-phosphates. Science. 2006;314(5801):997–1001. doi: 10.1126/science.1132998. [DOI] [PubMed] [Google Scholar]
- 31.Meylan E, Tschopp J. Toll-like receptors and RNA helicases: two parallel ways to trigger antiviral responses. Mol Cell. 2006;22(5):561–569. doi: 10.1016/j.molcel.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 32.Yoneyama M, Fujita T. Function of RIG-I–like receptors in antiviral innate immunity. J Biol Chem. 2007;282(21):15315–15318. doi: 10.1074/jbc.R700007200. [DOI] [PubMed] [Google Scholar]
- 33.Kaiser WJ, Offermann MK. Apoptosis induced by the toll–like receptor adaptor TRIF is dependent on its receptor interacting protein homotypic interaction motif. J Immunol. 2005;174(8):4942–4952. doi: 10.4049/jimmunol.174.8.4942. [DOI] [PubMed] [Google Scholar]
- 34.Han KJ, et al. Mechanisms of the TRIF–induced interferon–stimulated response element and NF-kappaB activation and apoptosis pathways. J Biol Chem. 2004;279(15):15652–15661. doi: 10.1074/jbc.M311629200. [DOI] [PubMed] [Google Scholar]
- 35.Funami K, et al. Spatiotemporal mobilization of Toll/IL-1 receptor domain-containing adaptor molecule–1 in response to dsRNA. J Immunol. 2007;179(10):6867–6872. doi: 10.4049/jimmunol.179.10.6867. [DOI] [PubMed] [Google Scholar]
- 36.Tatematsu M, et al. A molecular mechanism for Toll–IL-1 receptor domain-containing adaptor molecule–1–mediated IRF-3 activation. J Biol Chem. 2010;285(26):20128–20136. doi: 10.1074/jbc.M109.099101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Bernuth H, et al. Pyogenic bacterial infections in humans with MyD88 deficiency. Science. 2008;321(5889):691–696. doi: 10.1126/science.1158298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Picard C, et al. Pyogenic bacterial infections in humans with IRAK-4 deficiency. Science. 2003;299(5615):2076–2079. doi: 10.1126/science.1081902. [DOI] [PubMed] [Google Scholar]
- 39.Ku CL, et al. IRAK4 and NEMO mutations in otherwise healthy children with recurrent invasive pneumococcal disease. J Med Genet. 2007;44(1):16–23. doi: 10.1136/jmg.2006.044446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Casanova JL, Abel L, Quintana-Murci L. Human TLRs and IL-1Rs in host defense: natural insights from evolutionary, epidemiological, and clinical genetics. Annu Rev Immunol. 2011;29:447–491. doi: 10.1146/annurev-immunol-030409-101335. [DOI] [PubMed] [Google Scholar]
- 41.Yang K, et al. Human TLR-7–, -8–, and -9–mediated induction of IFN-alpha/beta and -lambda is IRAK-4 dependent and redundant for protective immunity to viruses. Immunity. 2005;23(5):465–478. doi: 10.1016/j.immuni.2005.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ku CL, et al. Inherited disorders of human Toll-like receptor signaling: immunological implications. Immunol Rev. 2005;203:10–20. doi: 10.1111/j.0105-2896.2005.00235.x. [DOI] [PubMed] [Google Scholar]
- 43.Alcais A, Abel L, Casanova JL. Human genetics of infectious diseases: between proof of principle and paradigm. J Clin Invest. 2009;119(9):2506–2514. doi: 10.1172/JCI38111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casanova JL, Abel L. Primary immunodeficiencies: a field in its infancy. Science. 2007;317(5838):617–619. doi: 10.1126/science.1142963. [DOI] [PubMed] [Google Scholar]
- 45.Barton ES, et al. Herpesvirus latency confers symbiotic protection from bacterial infection. Nature. 2007;447(7142):326–329. doi: 10.1038/nature05762. [DOI] [PubMed] [Google Scholar]
- 46.Chapgier A, et al. Human complete Stat-1 deficiency is associated with defective type I and II IFN responses in vitro but immunity to some low virulence viruses in vivo. J Immunol. 2006;176(8):5078–5083. doi: 10.4049/jimmunol.176.8.5078. [DOI] [PubMed] [Google Scholar]
- 47.Smahi A, et al. Genomic rearrangement in NEMO impairs NF-kappaB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature. 2000;405(6785):466–472. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- 48.Barde I, et al. Efficient control of gene expression in the hematopoietic system using a single Tet-on inducible lentiviral vector. Mol Ther. 2006;13(2):382–390. doi: 10.1016/j.ymthe.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 49.Desai P, Person S. Incorporation of the green fluorescent protein into the herpes simplex virus type 1 capsid. J Virol. 1998;72(9):7563–7568. doi: 10.1128/jvi.72.9.7563-7568.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.