In plants grown under field conditions, seed dormancy varies considerably with the time of year the seed is set, and this variation is important for the coordination of plant life history and seed quality. This work elucidates the mechanisms through which environmental temperature during seed set affects the germination behavior of seeds when released from the mother plant.
Abstract
Summer annuals overwinter as seeds in the soil seed bank. This is facilitated by a cold-induced increase in dormancy during seed maturation followed by a switch to a state during seed imbibition in which cold instead promotes germination. Here, we show that the seed maturation transcriptome in Arabidopsis thaliana is highly temperature sensitive and reveal that low temperature during seed maturation induces several genes associated with dormancy, including DELAY OF GERMINATION1 (DOG1), and influences gibberellin and abscisic acid levels in mature seeds. Mutants lacking DOG1, or with altered gibberellin or abscisic acid synthesis or signaling, in turn show reduced ability to enter the deeply dormant states in response to low seed maturation temperatures. In addition, we find that DOG1 promotes gibberellin catabolism during maturation. We show that C-REPEAT BINDING FACTORS (CBFs) are necessary for regulation of dormancy and of GA2OX6 and DOG1 expression caused by low temperatures. However, the temperature sensitivity of CBF transcription is markedly reduced in seeds and is absent in imbibed seeds. Our data demonstrate that inhibition of CBF expression is likely a critical feature allowing cold to promote rather than inhibit germination and support a model in which CBFs act in parallel to a low-temperature signaling pathway in the regulation of dormancy.
INTRODUCTION
In temperate regions, plants coordinate their lifecycle with the passing of the seasons. Central to this process is the ability of plants to process and integrate environmental information, with temperature and photoperiod the most important seasonal cues. Understanding the regulation of the timing of phenological events has become an important goal across biology, especially given the sensitivity of both plant and invertebrate phenology to climate change.
Arabidopsis thaliana accessions can be split broadly into summer and winter annual accessions, with the latter requiring a prolonged vernalization period for flowering by virtue of the expression of high levels of FLOWERING LOCUS C (FLC) gene expression (Michaels and Amasino, 1999; Sheldon et al., 1999). These accessions are characterized by germination during late summer or autumn, vegetative overwintering, and flowering during spring or early summer. By contrast, summer annual accessions lack genes such as FRIGIDA (Johanson et al., 2000) that are necessary for high FLC expression and flower in the year of germination. The seeds of summer annuals overwinter in the soil seed bank and germinate in response to spring cues, which in Arabidopsis remain only partly understood. During seed maturation, the level of dormancy is highly dependent on the prevailing environmental conditions with low temperatures and to a lesser extent short photoperiods, inducing high levels of dormancy and modifying the cold responsiveness of germination (Munir et al., 2001; Schmuths et al., 2006). Genetic influences on the induction of strong primary dormancy by low seed maturation temperatures have been uncovered, with roles for both phytochrome and FLC having been proposed (Donohue et al., 2008; Chiang et al., 2009).
The level of seed dormancy is set during seed maturation, and the phytohormone abscisic acid (ABA) is believed to be a central player. Mutants deficient in ABA synthesis or signaling in general show reduced dormancy, often accompanied by defects in the seed maturation program, such as reduced reserve accumulation and desiccation tolerance (Nambara et al., 1994). In seeds, the action of ABA is antagonized by that of gibberellin (GA), and numerous studies have shown that an intricate web of cross-regulation between ABA and GA levels lies at the heart of the control of seed germination (Seo et al., 2006; Piskurewicz et al., 2008, 2009). Environmental signals that influence dormancy or germination have been shown to result in the transcriptional regulation of GA and ABA metabolism in the imbibed seed. In particular, light and temperature have been shown to influence GA levels through the transcriptional regulation of bioactive GA synthesis through GA3 oxidase (GA3ox) and GA inactivation through GA2 oxidase (GA2ox; Yamaguchi et al., 1998; Yamauchi et al., 2004; Oh et al., 2006). In lettuce (Lactuca sativa), temperature regulation of ABA synthesis through 9-cis-epoxycarotenoid dioxygenase 4 (NCED4) is necessary for thermoinhibition (Argyris et al., 2011), whereas ABA catabolism through CYTOCHROME P450 707A2 (CYP707A2) is a target of after-ripening (Millar et al., 2006). Using forward genetic screens and natural variation studies, several other loci have been identified with important roles in dormancy regulation, most notably REDUCED DORMANCY4 (RDO4; Liu et al., 2007) and DELAY OF GERMINATION1 (DOG1), the major quantitative trait loci underlying the strong dormancy of many wild Arabidopsis accessions (Bentsink et al., 2006). However, it is not yet clear which, if any, of these pathways are important in the induction of high levels of dormancy by low temperatures and through what mechanism the temperature regulation occurs.
During the cooler seasons, plants have evolved a suite of mechanisms that facilitate their survival of adverse conditions. The best characterized of these is the process of cold acclimation, in which the central players are a small group of AP2-domain transcription factors known as C-REPEAT BINDING FACTORS (CBFs; Stockinger et al., 1997). CBF transcript levels increase quickly in response to falling temperatures and are maximally sensitive 8 h after dusk. Overexpression of CBFs confers freezing tolerance in the absence of cold acclimation due to the increased expression of a suite of genes involved in metabolic and physiological changes that aid resistance to freezing temperatures (Jaglo-Ottosen et al., 1998; Gilmour et al., 2000; Vogel et al., 2005). One notable feature of both low temperatures and CBF overexpression is that both cause marked growth retardation, and this has been shown to be through the promotion of GA catabolism by at least two CBF-regulated isoforms of GA2 oxidase, GA2ox3 and GA2ox6, and the subsequent accumulation of DELLA proteins (Achard et al., 2008).
In this study, we use a transcriptomic comparison of dry Arabidopsis seeds set under warm and cool seed maturation temperatures to identify low-temperature-regulated gene sets. Strikingly, both DOG1 and GA2ox show a marked cold induction during seed maturation, and subsequent experiments showed dog1 and GA signaling mutants are deficient in their ability to enter highly dormant states. We show that CBFs are required for dormancy but surprisingly are not temperature regulated in seeds. Our data suggest that a mechanism for the suppression of the cold induction of CBFs is an essential component of temperature responses in seeds and that CBFs have a temperature-independent role in the induction of strongly dormant states.
RESULTS
Identification of Temperature-Dependent Transcripts in Maturing Seeds
Previous work has shown that lowering seed maturation temperatures induces high levels of dormancy in rapid cycling Arabidopsis ecotypes (Schmuths et al., 2006; Donohue et al., 2008; Chiang et al., 2009) as well as other species (Fenner, 1991; Gu et al., 2006). We confirmed that this was indeed a dormancy phenomenon by stimulating high levels of germination in seeds matured at low temperatures by applying multiple dormancy breaking treatments or by removing coat-imposed dormancy by nicking the seed coat (see Supplemental Figure 1A online) and by showing that embryo and seed coat morphology was normal in seeds developed at 10°C (see Supplemental Figures 1B and 1C online). To begin to understand the molecular basis of this signaling pathway, we developed a system in which plants were grown to flowering under our standard laboratory conditions (see Methods) and then switched to either warm or cool temperatures from first flowering until the end of seed maturation. Reduction of the seed maturation temperature caused an incremental increase in seed dormancy, until at lower temperatures, even 2 weeks of cold stratification was insufficient to promote high levels of germination (Figure 1A). Seeds set in this way were then used as a basis for a transcriptomic analysis, and replicate batches of mature dry seeds set at either 20 or 10°C were compared to understand the consequences of variation in the seed maturation temperature in terms of the seed transcriptome. Dry seeds were chosen for the transcriptome analysis to maximize the chances of a like-for-like comparison because the delay in developmental timing caused by low seed maturation temperatures complicated the selection of comparable states during development. This is consistent with a recent analysis of dry seed transcriptomes for comparison of the mechanisms of action of major dormancy-controlling quantitative trait loci (Bentsink et al., 2010). Data from Affymetrix Ath1 chips were analyzed by significance analysis of microarrays (Tusher et al., 2001) to identify stringent lists of differentially regulated genes. We used 10°C as the lowest likely wild seed maturation temperature and an extreme temperature to maximize our chances of robustly detecting differentially expressed transcripts using microarrays; however, follow-up experiments were performed at a range of temperatures (12 to 17°C) to sample the range of likely behaviors of seed set under wild conditions. Using such a range avoids placing undue emphasis on behavior observed only at one particular temperature. A temperature reduction from 20 to 10°C during seed maturation resulted in the altered expression of a large number of transcripts in dry seeds, with over 275 genes upregulated at 10°C compared with 20°C and 681 genes downregulated, even after a relatively stringent cutoff of a 1% false discovery rate and at least a threefold change in mean expression (Figure 1B). To compare this to the situation in vegetative tissues, we used a data set in which seedlings at 12 and 22°C were analyzed (Nascarrays 147; see Methods). Interestingly, the number of differentially up- and downregulated genes in seeds is approximately an order of magnitude higher than in seedlings, suggesting that the transcriptome of seeds is comparatively hypersensitive to temperature. There was also very little overlap between the two gene sets: for instance, HSP70, a gene previously shown to be highly temperature sensitive and used to identify temperature-signaling components (Kumar and Wigge, 2010), was strongly regulated in the seedling data set but not in seeds. Similarly, there were temperature-regulated transcripts in seeds that did not appear in the seedling data set, such as DOG1 (see Supplemental Data Set 1 online). This suggests that there may be differences in the temperature signal transduction pathways between the two tissues.
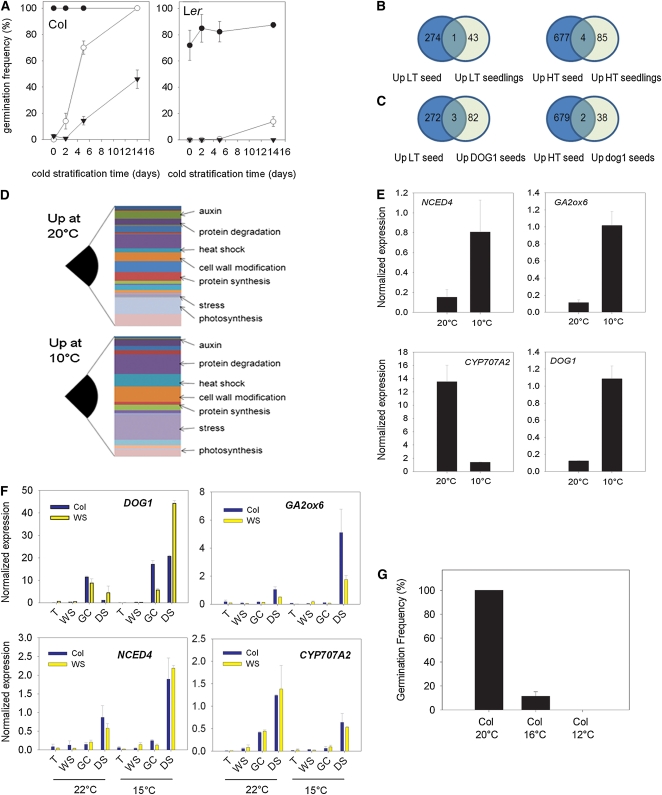
Figure 1.
Transcriptional Changes in Dry Seeds Matured at 20°C versus 10°C.
(A) Germination behavior of freshly harvested wild-type Col-0 and Ler seeds matured at 10°C (closed triangles), 15°C (open circles), and 20°C (closed circles) and the response to cold stratification. Data represent mean and se (n = 5).
(B) Numbers of transcripts significantly different between the 10°C (LT) seeds and the 20°C (HT) seeds and a comparison with a similar experiment performed on 10-d-old seedlings. Numbers for transcripts significantly different (P < 0.01) with a magnitude change of at least threefold are shown.
(C) Comparison of our data with that from DOG1 and dog1 loci (Bentsink et al., 2010). There is little overlap between the two data sets.
(D) Analysis of differentially regulated genes by TAGGIT (Carrera et al., 2007). For description of overrepresented gene ontologies, see main text.
(E) Confirmation of microarray gene expression changes by real-time RT-PCR. Low seed maturation temperatures induce GA2ox6, DOG1, and NCED4 expression, whereas warm seed maturation temperatures induce high CYP707A2 expression in dry seeds. Data represent the mean and se of two biological replicate samples for each temperature.
(F) Temperature-regulated gene expression during seed development and maturation. Analysis on Col and Ws backgrounds confirms the microarray results, which are from Ler. Data represent the mean and se of two biological replicates per time point. T, torpedo stage; WS, walking stick stage; GC, green cotyledon stage; DS, dry seed at maturity.
(G) High seed maturation temperatures remove the light requirement for germination. Seeds matured at three temperatures were incubated in the dark at 4°C for 50 d and germination scored. Data represents the mean and se of five independent seed batches
Next, we scanned the temperature-regulated gene lists for genes with known roles in dormancy or germination control (Table 1). Strikingly, we found that DOG1 expression was highly upregulated in response to low temperatures, as was GA2ox6, a gene previously shown to be important for the control of germination by light quality. In terms of ABA metabolism, we found that NCED4 expression is significantly upregulated, whereas CYP707A2 (Kushiro et al., 2004) is one of the genes most strongly downregulated by low seed maturation temperature (confirming the analysis of Chiang et al., 2009). Thus, we could identify gene expression associated with both dormancy and hormone balance that may underlie the dormancy changes caused by low seed maturation temperatures. RDO4 expression was also significantly different between the two treatments, but in this case, the low dormant seeds from the higher temperature showed higher RDO4 expression, inconsistent with the hypothesis that the temperature regulation of RDO4 transcripts is important in germination control. Because the effect of the DOG1 locus on the dry seed transcriptome has been previously reported (Bentsink et al., 2010), we compared temperature- and DOG1-dependent gene expression in dry seeds (Figure 1C). Again, very little overlap was discovered between the two gene sets, suggesting that either very few DOG1-dependent genes are important in dormancy regulation or that the important effects of DOG1 are not represented in the dry seed transcriptome. Two phytochrome isoforms, phyB and phyE, were both downregulated by low temperature. Both of these are important for dormancy and germination (Heschel et al., 2007), and this suggests that more dormant seeds might desensitize their germination to light via the inhibition of phytochrome transcription. Genes involved in ABA signaling in general showed an increase in expression at the warmer temperature, although these changes were in general only twofold or less and are expected to be much less important than the large difference in CYP707A2 expression and the magnitude of the effect of ABA on their protein levels. Finally, to identify groups of genes regulated by seed maturation temperatures, we used the TAGGIT ontology (Carrera et al., 2007) to compare gene expression between the two data sets (Figure 1D). In the dormant low-temperature-set seeds, cell wall modification, heat shock, and protein degradation were the most prominent categories, whereas in the low dormant seeds, translation, photosynthesis, seed storage proteins, and ethylene were better represented. Broadly speaking, this result is in line with previous studies that have compared seed dormancy states (Finch-Savage et al., 2007), which also identified increased gene expression associated with translation and photosynthesis as indicative of low dormant states. We confirmed the differential expression in dry seeds set at 10 or 20°C by real-time PCR using independent samples (Figure 1E).
Table 1.
Genes with Known Roles in the Regulation of Dormancy or Germination and the Effect of Temperature during Seed Maturation on Transcript Levels in Dry Seeds
| Gene Name | Locus | Average Expression at 20°C | Average Expression at 10°C | Fold Change 10°C/20°C | Q Value (%) |
| Upregulated | |||||
| DOG1 | AT5g45830 | 506.6 ± 305.5 | 6,809.8 ± 925.7 | 13.4423 | 0 |
| GA2ox6 | AT1g02400 | 167.4 ± 24.9 | 816.5 ± 277.1 | 4.8781 | 0.3293 |
| NCED4 | AT4g19170 | 1,412.1 ± 3,637 | 3,546.0 ± 638.6 | 2.5111 | 0.3878 |
| Unchanged | |||||
| GA2ox2 | AT1g30040 | 138.1 ± 85.5 | 319.1 ± 113.2 | 2.3094 | 4.3481 |
| GAI | AT1g14920 | 715.1 ± 312.8 | 1,343.1 ± 68.8 | 1.8782 | 5.1642 |
| RGL3 | AT5g17490 | 1,504.2 ± 391.7 | 2,584.3 ± 68.8 | 1.7181 | 5.1642 |
| ABI2 | AT5g57050 | 1,672.9 ± 184.5 | 2,648.4 ± 88.6 | 1.5832 | 4.3481 |
| RGA | AT2g01570 | 154.9 ± 50.8 | 242.3 ± 44.0 | 1.5650 | 12.9371 |
| RGL2 | AT3g03450 | 212.0 ± 49.7 | 207.4 ± 45.4 | 0.9780 | 20.9340 |
| ABA2 | AT1g52340 | 194.4 ± 10.9 | 176.5 ± 44.8 | 0.9079 | 20.9340 |
| SLY1 | AT4g24210 | 711.0 ± 282.8 | 625.9 ± 81.7 | 0.8803 | 20.9340 |
| FLC | At5g10140 | 1,897.3 ± 300.9 | 1,537.28 ± 306.9 | 0.8102 | 11.050 |
| SPT | AT4g36930 | 100.7 ± 24.6 | 78.1 ± 37.3 | 0.7757 | 11.050 |
| GA3 | AT5g25900 | 3,645.7 ± 864.6 | 2,479.0 ± 206.2 | 0.6799 | 4.3481 |
| GA2 | AT1g79460 | 139.3 ± 68.7 | 80.4 ± 15.3 | 0.5774 | 5.1642 |
| ABI5 | AT2g36270 | 10,066.3 ± 1,560.8 | 5,467.0 ± 1,419.2 | 0.5431 | 1.1283 |
| Downregulated | |||||
| ABI1 | AT4g26080 | 2,087.1 ± 147.2 | 1,044.9 ± 205.0 | 0.5007 | 0.6089 |
| ABI8 | AT3g08550 | 267.4 ± 48.8 | 116.1 ± 53.1 | 0.4342 | 0.9796 |
| ABI3 | AT3g24650 | 2,083.2 ± 87.1 | 859.7 ± 215.6 | 0.4127 | 0.6089 |
| PHYB | AT2g18790 | 1,336.2 ± 170.6 | 423.3 ± 191.1 | 0.3168 | 0.6089 |
| ABA1 | AT5g67030 | 7,696.9 ± 1,126.6 | 2,526.3 ± 792.6 | 0.3144 | 0.6089 |
| PHYE | AT4g18130 | 4,028.0 ± 213.9 | 1,083.3 ± 430.3 | 0.2690 | 0.6089 |
| RDO4/HUB1 | AT2g44950 | 821.3 ± 182.8 | 189.4 ± 60.0 | 0.2305 | 0.6089 |
| NIA2 | AT1g37130 | 238.7 ± 142.0 | 55.3 ± 26.5 | 0.1506 | 0.6089 |
| CYP707A2 | AT2g29090 | 2,603.9 ± 1,033.6 | 263.0 ± 14.8 | 0.1010 | 0.6089 |
Based on a standard 1% false discovery rate, significantly up- and downregulated genes are shown at the top and bottom of the table, respectively. In between, selected genes with no significant change in expression are listed. Values represent mean and sd of the three replicate arrays. The Q value indicates the expected frequency of false positives present in a list of differentially expressed genes containing that probe set, and values of 1% or below are considered significant.
The level of dormancy is set during seed maturation, but our gene expression data were restricted to the analysis of dry seeds. Therefore, we analyzed the expression of genes by real-time PCR at four stages of seed maturation, which were identified by embryo morphology due to differences in growth rates in seeds maturing at different temperatures. RNA was collected from torpedo stage, walking stick, green cotyledon, and dry seeds at two maturation temperatures. We also wanted to understand whether the gene expression changes we observed in Landsberg erecta (Ler) were robust across ecotypes and so chose to analyze both Columbia (Col) and Wassilewskija (Ws) (Figure 1F). DOG1 expression was interesting in that it increased markedly at the green cotyledon stage at 20°C, only to decline again by maturity. At 10°C, DOG1 expression levels also increased at a similar stage but remained elevated in mature seeds. In the case of NCED4, CYP707A2, and GA2ox6, the highest expression was observed at maturity, and temperature affected steady state levels in a similar manner to that shown on the microarrays. As this experiment was performed in the Col and Ws backgrounds, this also shows that these effects of temperature are conserved across ecotypes.
Light is an important germination-inducing signal, and our data suggested that seeds matured at lower temperatures may exhibit altered phytochrome levels and, therefore, light sensitivity upon imbibition. In nature, primary dormant seeds must remain ungerminated in darkness to enter the soil seed bank, where low temperature drives the transition of imbibed seeds into secondary dormancy (Finch-Savage et al., 2007). To test this, we performed experiments testing the ability of seeds matured at three temperatures to resist germination in the dark during extended chilling (Figure 1G). Remarkably, seeds matured at 20°C were unable to remain dormant in the dark during chilling, suggesting that these seeds would not be able to overwinter in the soil seed bank under wild conditions. Seeds matured at lower temperatures (16 or 12°C) could resist germination during dark chilling, showing that reducing the seed maturation temperature produced seeds that could in theory enter the seed bank and remain dormant. Thus, at least for Col-0, seeds matured at warm temperatures would be unlikely to persist long in the seed bank and may be unable to enter secondary dormancy. This suggests that increased phytochrome levels may lead to a lack of light requirement in warm-matured seeds.
Genetic Regulation of Strong Dormancy Induced by Cool Seed Maturation Conditions
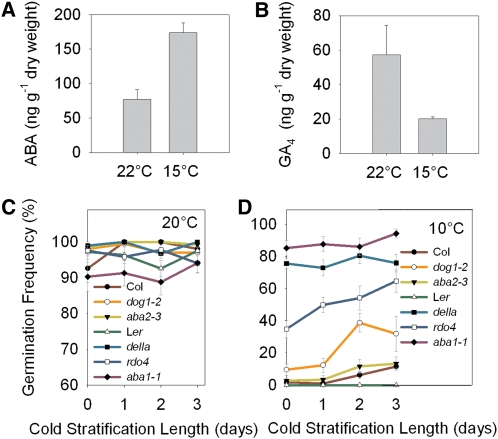
Very little is known about the genetic pathways required for dormancy caused by low seed maturation temperatures, and we wished to understand which, if any, of the genes and hormones implicated by the transcriptomic study are necessary for the strongly dormant state. First, we compared levels of bioactive GAs and ABA in dry seeds set under low or high temperatures (Figures 2A and 2B). Low temperature caused an increase in ABA levels in dry seeds, coupled with a decrease in GA4 levels, when compared with seeds set under warm temperatures, indicating that temperature affects the poise of hormone balance but without disruption of the ABA/GA cross-regulation (i.e., the negative correlation between ABA and GA levels still holds).
Figure 2.
Genetic Regulation of Strong Dormancy Induced by Low Seed Maturation Temperature.
(A) and (B) Low seed maturation temperatures increase ABA levels and decrease GA4 levels, the principle active GA in Arabidopsis seeds. Dry seeds of Ler ecotype were set at the indicated temperature during seed maturation and hormone levels measured at maturity. Data represents the mean and se of three biological replicates per treatment.
(C) The germination frequency for seeds of the indicated wild type and mutant lines set at 20°C are shown after various lengths of cold stratification. All lines show low dormancy when set at 20°C.
(D) Primary dormancy and response to cold of freshly harvested Arabidopsis mutants previously shown to have low dormancy; seeds were matured at 10°C. Our data show that ABA, GA, DOG1, and RDO4 are all required for low-temperature-induced dormancy. Germination data represent the mean and se of at least five replicate seed batches per genotype per treatment.
Most genetic examination of low dormancy mutants has taken place in rapid cycling ecotypes using seeds set in glasshouses or standard laboratory conditions so that even wild-type seeds under the same conditions have only low levels of dormancy. Hence, it is still largely unclear how important these loci are when seeds have higher levels of dormancy, for instance, similar to those of ecotype Cape Verde Island. To understand further the mechanism through which high dormancy levels are achieved in response to low seed maturation temperatures, we analyzed the role of ABA and GA by examining the ability of ABA biosynthetic and DELLA mutants to enter into the strongly dormant states caused by low temperature as well as that of other mutants shown to have reduced dormancy, including dog1 (Figures 2C and 2D). When seed was set at 20°C wild-type seeds showed little or no dormancy, so that the decrease in dormancy of the mutants in the assay was hard to quantify (Figure 2C). Surprisingly, dog1 mutants set at 10°C were dormant at harvest, with germination frequencies only slightly higher than the wild type, suggesting that low temperature promotes dormancy by a DOG1-independent mechanism. However, after short periods of cold stratification, the lower dormancy of dog1 mutants was revealed, confirming that DOG1 is essential for dormancy induced by cool conditions. We also found that quadruple DELLA loss-of-function mutants were severely compromised in low-temperature induced dormancy, showing the central role of high DELLA levels in the maintenance of dormant states. We also found that ABA was absolutely required for the strongly dormant state, as aba1 mutants showed no dormancy even when matured at low temperatures (Figure 2D). The aba2-3 mutant was able to induce dormancy at low temperatures (Figure 2D), perhaps indicating that this allele is not a null or that ABA is less important in the Col background. To confirm the role of ABA in the Col background, we subjected additional mutants to a dormancy screen with seed set at low temperature (see Supplemental Figure 2 online). We found that the aba1-6 and aba3-1 mutants showed little or no dormancy when set at low temperature (see Supplemental Figure 2 online). Because low temperature during seed maturation strongly reduces CYP707A2 expression (Table 1) and increases ABA levels in dry seeds (Figure 2) and because ABA-deficient lines cannot enter the strongly dormant states induced by low maturation temperatures, these results show that high ABA levels are central to the increased dormancy induced by low seed maturation temperatures. Taken together, our results show that DOG1 and the regulation of the balance of GA and ABA levels are both important for the induction of strong dormancy by low temperatures and that there may therefore be multiple mechanisms through which temperature affects dormancy levels.
CBFs Are Required for Dormancy in Arabidopsis but Are Not Cold Regulated in Seeds
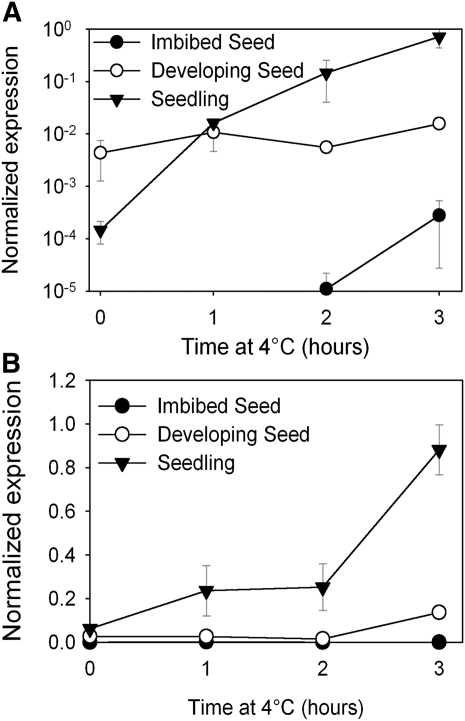
In vegetative tissues, cold sensing takes place through at least two known mechanisms: the transcriptional regulation of CBFs and the chromatin reorganization of the FLC locus (Bastow et al., 2004). FLC has previously been described to have a role in dormancy regulation but only if imbibed seeds are also incubated at low temperatures (Chiang et al., 2009). Given that low seed maturation temperature leads to high dormancy at warm imbibition temperatures (Figure 1A), and at warmer temperatures FLC was shown to have little or no role (Chiang et al., 2009), we investigated whether CBF-dependent pathways have a role in dormancy induction by low maturation temperatures. Investigation of available microarray data using the EFP browser (Bassel et al., 2008) also suggested that CBFs might be expressed during seed development. To analyze the temperature regulation of CBF transcription in seeds, we performed time-course experiments in which seeds or seedlings were transferred from 22 to 4°C and the expression of CBF1 recorded by RT-PCR. We chose CBF1 because the low availability of material from developing seeds necessitated an RT-PCR approach, and our analysis of previously published CBF primers (Franklin and Whitelam, 2007) revealed that their specificity for any one isoform was questionable, and only for CBF1 could we reliably develop a specific Taqman assay (see Methods). These experiments revealed that in maturing seeds, CBF1 expression is higher in developing seeds than in seedlings in the absence of cold (Figure 3A) but unexpectedly that the ability of cold to increase transcript abundance to high levels was attenuated compared with vegetative tissues. In imbibed seeds, our ability to detect CBF1 expression was questionable, and cold had no significant effect on the transcript abundance. The inactivity of CBFs in imbibed seeds was confirmed by the observation that CBF-induced COR15b transcription was also unresponsive to cold in imbibed seeds and only minimally responsive in maturing seeds (Figure 3B). Together, these data suggest that a mechanism exists to suppress CBF expression in imbibed seeds and that this is already partially active during seed maturation. Furthermore, because the key property of CBFs that places them at the center of cold signaling in leaves is their transcriptional response to low temperatures and given that this response is largely absent in seeds, it seemed unlikely that they are directly involved in temperature signal transduction in seed tissues. Because CBFs act to reduce GA levels in response to cold in vegetative tissues, we hypothesized that this repression of CBFs in imbibed seeds may be necessary to permit the promotion of germination by cold stratification (which requires an increase in GA levels; Yamauchi et al., 2004).
Figure 3.
CBF Transcript Levels Are Not Cold Inducible in Seeds.
The transcript response of CBF1 (A) and the CBF target COR15b (B) in response to sudden cold shock of 4°C in seedlings, imbibed seeds, or during seed maturation, measured by real-time RT-PCR. CBF1 expression levels are induced by cold over 4 levels of magnitude in seedlings but not in seeds. A similar response for COR15b suggests that our analysis of CBF1 is representative of all three CBFs. Data represent the mean and se of two biological replicates per treatment.
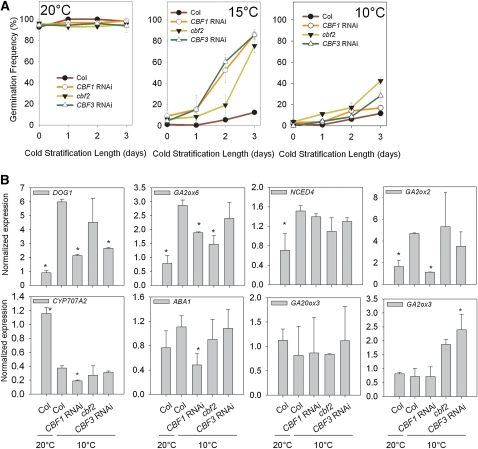
To investigate whether CBFs have a role in the regulation of dormancy during seed development, we subjected loss- and gain-of-function lines to variation in seed maturation temperatures and analyzed the effect on dormancy. We focused on analyzing dormancy breakage by cold because during autumn or winter, cold is the more likely dormancy breaking signal (after-ripening requires the warm temperatures and low soil moisture normally associated with summer) and because a priori there appeared a possibility that CBFs could play a role in both the induction and breakage of dormancy by low temperatures. We first analyzed the ability of CBF loss-of-function lines to enter into dormancy by maturing seeds at 20, 15, or 10°C (Figure 4A). At 20°C, none of the lines showed any obvious dormancy, consistent with observations that Col ecotype seeds show little or no dormancy under standard laboratory conditions. At lower temperatures a strong primary dormancy was induced, but each of the CBF-deficient lines showed an increased germination phenotype, consistent with a lower level of dormancy induced in these lines. However, at 10°C when the strongest dormancy was induced, high levels of dormancy were also present in lines lacking one CBF gene. In our view, these data shows that CBFs are required for the induction of normal levels of seed dormancy but also suggest that CBF-independent processes also play a role, especially at lower temperatures. However, we cannot completely rule out that redundant function obscures the analysis.
Figure 4.
CBFs Are Required for Primary Seed Dormancy and for the Normal Cold-Responsive Gene Expression Required for Dormancy.
(A) The primary dormancy and response to cold stratification of seeds of loss-of-function lines for CBFs 1, 2, and 3, set at either 10, 15, or 20°C. Data represent the mean and se of five independent seed lots for each genotype.
(B) RT-PCR analysis of low-temperature-induced gene expression in wild-type Col-0 seeds and cbf loss-of-function lines. Data represent the mean and se of three biological replicates per genotype. Significant differences from the wild type (t test, P < 0.01) are indicated by asterisks.
Given that we identified cold-induced gene expression with a role in the induction or maintenance of high dormancy states, we analyzed cbf lines for alterations in this gene expression program (Figure 4B). These data again confirmed the microarray analysis that GA2ox6, DOG1, and NCED4 were all increased in expression at 10°C, whereas CYP707A2 was increased at 20°C. In general, only GA2ox6 and DOG1 expression was lower in CBF loss-of-function lines, whereas expression of NCED4 and CYP707A2 was not affected. The role of CBF1 in the regulation of GA2ox6 expression has been observed previously in vegetative tissues (Achard et al., 2008). We could also confirm that the vegetative CBF target, GA2ox3 (Achard et al., 2008), is not temperature regulated in seeds, as seen in the microarray analysis. We analyzed our low seed maturation temperature-induced gene set for the presence of the CBF binding low temperature response element in their promoters (see Supplemental Data Set 1 online). Interestingly, of the genes tested above, only DOG1 has a putative CBF binding site in the promoter region, suggesting that GA2ox6 is not a direct target of CBFs. Together, our data support the hypothesis that DOG1 and GA2ox6 are coregulated in maturing seeds and that CBFs are required for wild-type expression levels.
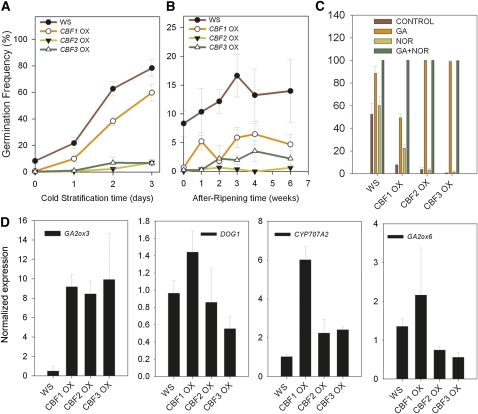
To further our analysis of CBF function during seed maturation, lines overexpressing the three CBFs were analyzed for dormancy (Stockinger et al., 1997; Gilmour et al., 2000; Figure 5). These experiments revealed that each CBF, when overexpressed, could confer an increase in seed dormancy that persisted even after cold stratification or after-ripening (Figures 5A and 5B), although in our hands the parent ecotype Ws after-ripens very poorly. The increased dormancy of 35S:CBF lines was rescued by the addition of exogenous GA but not by inhibition of ABA biosynthesis, suggesting that low GA levels in the imbibed seed may be responsible for this phenotype (Figure 5C). However, we cannot rule out that GA is simply overcoming the effect of higher ABA levels in the dry seed. We could not analyze the phenotype of CBF-overexpressing seeds matured at low temperatures because under these conditions the lines developed very slowly after flowering and made few, if any, seeds. However, at the warmer temperature, we were able to analyze gene expression in mature seeds. Surprisingly, overexpression of CBFs did not lead to an increase in either GA2ox6 or DOG1 gene expression (Figure 5D). Instead, we found that each line strongly overexpressed GA2ox3, as has been observed previously in vegetative tissues (Achard et al., 2008). Thus, the CBF loss- and gain-of-function appear to affect dormancy through two distinct processes (discussed in detail below).
Figure 5.
CBF Overexpression Results in the Inhibition of Germination and Increased GA2ox3 Expression in Seeds Set at 20°C.
(A) CBF overexpression leads to strong primary dormancy, which in the case of CBF2OX and CBF3OX responds poorly to cold stratification.
(B) The increased dormancy of CBFOX lines is not removed by after-ripening (note that Ws after-ripens very poorly in our hands).
(C) The application of exogenous 100 μM GA restores full germination to CBFOX lines, but application of the ABA biosynthetic inhibitor norflurazon does not. All germination data represent the mean and se or five replicate seed batches per treatment.
(D) Gene expression changes in dry seeds caused by CBF overexpression using seeds set at 20°C. Data represent the mean and se of three biological replicates per genotype.
Toward a Pathway of Temperature Signal Transduction during Seed Maturation
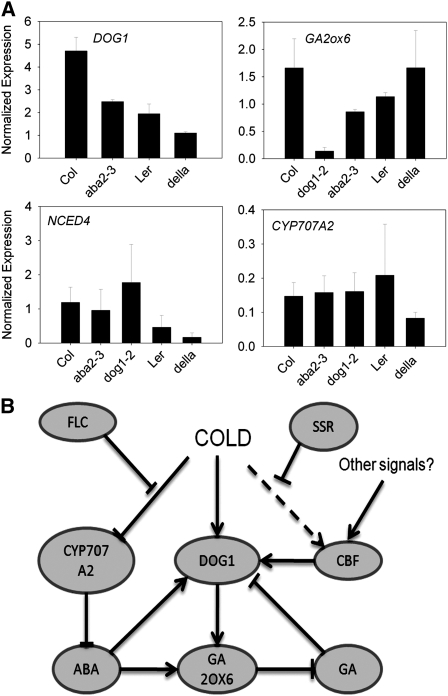
We have shown that temperature and the CBFs independently affect the transcripts of important dormancy-regulating genes during seed maturation. To better understand the roles of DOG1, GA, and ABA in the regulatory network that leads to high levels of dormancy in seeds set at low temperature, we analyzed gene expression in dog1, aba3, and della mutant seeds set at low temperatures (Figure 6A). Strikingly, dog1 mutants showed a 10-fold decrease in GA2ox6 expression compared with the wild type, showing that one role of DOG1 is the promotion of GA catabolism. In return, aba2 and della mutants showed a 50% reduction in DOG1 transcript levels, showing that GA and ABA have some influence on DOG1. We found that dog1, aba2, and della mutants all show roughly wild-type levels of CYP707A2 and NCED4 (Figure 6A), showing that the temperature regulation of ABA metabolism is largely independent of both DOG1 and DELLAs.
Figure 6.
Pathways Mediating the Effect of Seed Maturation Temperature on Dormancy.
(A) Expression of DOG1, GA2ox6, NCED4, and CYP707A2 in dog1, aba2, and della mutant dry seeds matured at 17°C. Values represent the mean and se of two biological replicates. Germination data for the above seed lots are shown in Supplemental Figure 3 online.
(B) A proposed pathway mediating the regulation of dormancy by cold during seed maturation. Cold induction of CBF transcription is inhibited by an unknown seed-specific Repressor of CBF expression (SSR), although expression in the absence of cold means CBFs are still required for normal dormancy. Cold induces high levels of dormancy through more than one mechanism, including the elevation of DOG1 transcription and the action of DOG1 in the promotion of GA catabolism. ABA also promotes GA2ox6 expression during seed maturation (Seo et al., 2006). As dog1 and della mutants show little misregulation of ABA-related gene expression, we propose that this constitutes a second pathway, perhaps relying on FLC, which is required for normal CYP707A2 expression (Chiang et al., 2009).
DISCUSSION
The Role of CBFs in the Configuration of the Low-Temperature-Responsive Transcriptome and Seed Dormancy
Our phenotypic analyses clearly define a role for CBFs in the regulation of dormancy and the gene expression required for seed dormancy in rapid cycling ecotypes. Loss of function leads to lower levels of DOG1 and GA2ox6 in dry seeds, and DOG1, GA, and ABA levels are clearly central to the temperature response of dormancy (Figure 2). However, somewhat surprisingly, we could detect very little elevation in CBF levels in maturing seeds when low temperatures are applied, in strong contrast with the established paradigm in vegetative tissues. In addition, we do not observe large numbers of CBF targets, the COR genes, being expressed in response to low temperatures in seeds (see Supplemental Data Set 1 online) nor does temperature regulation of transcription in seeds overlap significantly with that in vegetative tissues (Figure 1). Because the CBFs themselves are not cold regulated at the transcript level in seeds, in our view this supports a model in which CBFs in seeds are not directly involved in the temperature signal transduction pathway during seed maturation. One further possibility is that the temperature regulation of CBFs in maternal tissues could be important for dormancy. We could not test for a maternal function because the loss-of-function phenotypes did vary somewhat in our hands, although the data presented are representative of a large number of experiments. However, we consider this unlikely because CBF transcription is not temperature responsive at normal red/far-red ratios in the temperature range used for this study (Franklin and Whitelam, 2007). It would be interesting to test whether CBFs have a role in the regulation of dormancy by light quality (as opposed to germination; Heschel et al., 2007).
However, in the absence of cold, CBF levels are generally higher in developing seeds than in seedlings, a conclusion also supported by available microarray data. Thus, CBFs may have a general role during seed development, independent of a cold signaling pathway. This view is supported by phenotypic analysis of loss-of-function lines of each CBF, which show that under conditions when significant levels of dormancy are induced, CBFs are required for wild-type dormancy levels (Figure 4A). The fact that CBF loss-of-function lines continue to show progressively higher levels of dormancy as the seed maturation temperature is reduced is further evidence that this dormancy is induced by a CBF-independent mechanism. Analysis of cbf triple mutants would be necessary to confirm this unequivocally. Yet, CBFs appear to be necessary for the normal levels of cold-regulated gene expression in seeds, suggesting that this second pathway and the CBFs share some common elements.
CBF overexpression leads to decreased germination that is completely restored by the application of exogenous GA during imbibition. Our analysis shows that this is most likely attributable to the strong upregulation of GA3ox3, the isoform most strongly regulated by CBF overexpression and in seedlings (Achard et al., 2008; GA2ox6 is also affected in this study, but to a much lesser extent). Strikingly, GA2ox3 is not upregulated by low-temperature treatments in wild-type seeds nor misregulated by CBF loss of function during seed maturation, despite its importance for growth inhibition by low temperatures in vegetative tissues. Thus, CBF gain and loss of function, although superficially leading to opposite phenotypes, affect germination through distinct mechanisms. In CBF overexpressors, it is possible that some affects of overexpression are obscured by the downstream consequences of high GA2ox3 levels and the affect this has on GA levels and known feedback mechanisms. GA applied during imbibition is enough to provoke high germination in 35S:CBF seeds, suggesting that 35S:CBF may act to lower GA levels in the imbibed seed.
One of our most striking observations is that in imbibed seeds, CBFs are virtually undetectable, and their transcripts are completely unresponsive to low-temperature pulses. In addition, low temperature in imbibed seeds does not induce the expression of the CBF target COR15b (Figure 3). Given that increased CBF levels lowers GA content through the promotion of GA2ox levels, the positive effect of cold on the germination of dormant seeds would not be predicted to occur in the presence of strong cold-regulated CBF expression, as this requires an increase in GA levels (Ogawa et al., 2003; Yamauchi et al., 2004). Hence, the strong repression of CBFs is likely to be an essential component of gene regulatory networks in imbibed seeds that permit cold to promote, rather than inhibit, germination. The partial suppression of the cold induction of CBF transcripts during seed maturation may be the beginning of the expression of this mechanism. It is noteworthy that in some species cold continues to be an inhibitor of germination and establishment, particularly in crops from warm environments, such as soybean (Glycine max) and maize (Zea mays), and it will be interesting to understand the behavior of CBF-regulated pathways in the seeds of these species.
GAs Are Central to the Induction of Strong Dormancy Induced by Low Temperatures
In the past, it has been contentious whether GA has a role in dormancy regulation or simply a role in the promotion of germination in seed with no dormancy, as suggested by Bewley (1997). The latter hypothesis appeared to be supported by data showing that during germination, GA biosynthesis is strongly upregulated and that GA synthesis and degradation are under the close control of light and phytochromes, photoreceptors principally characterized as regulators of germination (Yamaguchi et al., 1998). By contrast, we showed that under standard glasshouse conditions, della mutants failed to enter dormancy even in the absence of GA synthesis (Penfield et al., 2006). Furthermore, phytochromes clearly have a role in regulating dormancy, in addition to germination (Donohue et al., 2008). Here, we show that reduced GA levels are associated with the strongly dormant states promoted by environmental conditions that mimic the coming of winter and that della loss-of-function mutants are among the most highly compromised in their ability to enter dormant states. We also note that the GA-insensitive mutant sleepy1 is able to block germination of the nondormant aba insensitive1 mutant in multiple genetic backgrounds (Steber et al., 1998; Ariizumi and Steber, 2007). Because both GA and ABA levels in mature seeds, as well as gene expression affecting the metabolism of both hormones, are affected by low seed maturation temperatures, we suggest that GA and ABA are equally important regulators of dormancy in Arabidopsis.
A Model for the Regulation of Dormancy by the Seed Maturation Environment
Temperature is the key signal used by Arabidopsis to sense the proximity to winter and induce high levels of dormancy, whereas photoperiod is less important (Munir et al., 2001). In the mother plant, FLC has been shown to play a role in dormancy and may be part of a dormancy-regulating temperature signal transduction pathway, regulating CYP707A2 expression (Chiang et al., 2009). However, the FLC genotype affects dormancy only if the seeds are subsequently incubated at 10°C, suggesting that there must be other pathways influencing this trait. The temperature of 10°C itself has a dormancy-breaking effect on Arabidopsis seeds (Chiang et al., 2009; Penfield et al., 2010), suggesting that FLC is important only when close to the germination threshold. However, differences in the severity of ABA-deficient lines between Col and Ler may be due to a known polymorphism at the FLC locus (Michaels et al., 2003).
Unlike previous analyses using standard conditions (Bentsink et al., 2006), in seeds set at low temperatures, DOG1 is not critically required for primary dormancy (Figure 2), showing the importance of DOG1-independent pathways, at least in the Col-0 background. Our analysis shows that the maintenance of DOG1 expression through to maturity is an essential target of low temperature and also that DOG1 is required for high GA2ox6 transcript levels in dry seeds (Figure 6B). The consequence of this is to lower GA levels in the seed. Our data also show that hormone balance feeds back to affect DOG1 transcripts, as loss of either ABA or DELLAs leads to a 50% decline in DOG1 levels in dry seeds. The fact that loss of DELLAs and ABA both result in a similar halving of DOG1 expression suggests that they act through a common intermediary. For instance, ABA may affect DOG1 expression indirectly through the regulation of GA2ox6 (Seo et al., 2006). Further work will be needed to define the relationship of DOG1 action to GA and ABA action in seeds and to investigate whether the cold regulation of these transcripts is also a feature of dormancy cycling in soil seed banks. However, because dog1 mutants retain primary dormancy, a second DOG1-independent pathway must act in parallel. One possibility is that this is the maternal effect pathway influenced by FLC (Chiang et al., 2009).
Loss of CBFs in seeds appears to affect primarily DOG1 and GA2ox6, so the most parsimonious model is one whereby CBFs principally act to regulate DOG1 expression alone (as DOG1 is necessary for GA2ox6 expression). Although CBFs show little transcriptional regulation by temperature in our experiments, their temperature-independent expression still allows them to affect DOG1 levels. We cannot rule out that CBFs are important for the response to further environmental signals that may influence dormancy during seed maturation.
Our data also show that lower seed maturation temperatures cause a reduction in phytochrome B and E transcripts in dry seeds. This is accompanied by the induction of a light requirement for germination completion. Our experiments in the laboratory predict that seeds of rapid cycling ecotypes could not enter the soil seed bank if they are matured at warm temperatures (Figure 1G) because they germinate easily in the dark during treatments that induce secondary dormancy. It has previously been shown that loss of phytochrome confers unresponsiveness to cold stratification in seeds matured at 15°C (Donohue et al., 2008). Taken together, this suggests that lowering phytochrome levels is important for the decreased germination in response to chilling and for nongermination in the absence of light. Therefore, temperature regulation of phytochromes is likely a key mechanism for increasing the probability of a transition to secondary dormancy and persistence in the seed bank.
METHODS
Plant Material
CBF-overexpressing lines were a gift from Michael Thomashow and have been described (Jaglo-Ottosen et al., 1998; Gilmour et al., 2000). They are in the Ws background. CBF loss-of-function lines were obtained from Julio Salinas and have been described previously (Novillo et al., 2007; Col background). dog1-2 (Col; Bentsink et al., 2006) and rdo4 mutants (Ler) were a gift from Marten Koornneef and Wim Soppe, and the gai-t6 rga-t2 rgl2-1 rgl1-1 quadruple DELLA loss-of-function lines were obtained from Nicholas Harberd (Ler background; Achard et al., 2006). aba1-1 (N21), aba1-6 (N3772), aba2-3 (N158), aba3-1 (N157), and abi4-1 (N8104) were ordered from the Nottingham Arabidopsis Stock Centre (NASC).
Dormancy and Germination Assays
For seed generation, plants were germinated on agar plates and transferred to John Innes seed compost (Levington) in P40 trays. Plants were grown to flowering at 20°C under standard long days using fluorescent white light at ~70 to 100 μM m−2 s−1 until first flowering (defined as anthesis of the first flowers), at which point they were transferred to a second growth cabinet running the same conditions, but with the indicated seed maturation temperature. Plants were left to set seed until dehiscence began, and seed was then harvested and dried for germination analysis. Poorly filled seeds were excluded from germination trials using a 250-μm sieve (Fisher Scientific). In general, freshly harvested seeds were sown within 24 h of harvest, on 0.9% water agar plates, in replicate seed batches from different parents as biological repeats. Each experiment was repeated at least three times, and representative data sets are shown. Germination was scored as radicle emergence after 7 d of incubation at 22°C in a 12-h photoperiod in a Sanyo MLR growth chamber. Cold stratification was achieved by preincubation of plates at 4°C in the dark for the indicated time periods using a Sanyo MIR-154 incubator. GA3 (Sigma Aldrich) was added at 100 μM and norflurazon (Greyhound Chromatography) at 50 μM where indicated.
Phytohormone Assays
GAs and ABA were quantified from replicate batches of 100 mg of dry seeds flash frozen in liquid nitrogen when freshly harvested and stored at −80°C until analysis. Hormone determination was performed by ultraperformance liquid chromatography–mass spectrometry analysis of citrate-buffered acetone extracts as described previously (Dave et al., 2011).
Microarray Analysis
Seeds of the Ler genotype were set at either 20 or 10°C as described for the germination assays and stored at −80°C until analysis. RNA was extracted from dry seeds as described previously (Penfield et al., 2005) and labeled using the Affymetrix one-cycle labeling kit (Affymetrix) using the manufacturer’s protocol, before hybridization to the Affymetrix Ath1 chip. For each treatment, three replicate seed batches each derived from a unique parent plant were used to create the biological replicate probes for the experiment. Raw data were normalized by MAS5 (www.Affymetrix.com) to a target signal of 500 before analysis by significance analysis of microarrays (Tusher et al., 2001) to identify statistically supported up- and downregulated gene sets, using a freely available Excel macro. Key findings were replicated in an independent experiment with independent samples by real-time RT-PCR. TAGGIT analysis was performed with an Excel macro (Carrera et al., 2007), a gift from Mike Holdsworth. Microarray data are deposited at NASC (NASCARRAYS-594) and Gene Expression Omnibus with series number GSE28747. NASC-ARRAYS-147 was used for the comparison shown in Figure 1B. A list of DOG1-regulated genes in dry seeds used in the comparison shown in Figure 1C was downloaded from the supplemental data files accompanying Bentsink et al. (2010). The presence of the DRE motif (CCGAC) in the promoters of the genes with maturation temperature-dependent expression was analyzed using Athena (O’Connor et al., 2005).
Real-Time RT-PCR
RNA was extracted from ~10 mg of dry, developing, or imbibed seeds and cDNA synthesized as previously described (Penfield et al., 2005). cDNA was synthesized from 2 μg total RNA and diluted 1:30 with distilled water before use for real-time RT-PCR with SYBR Green (or Taqman for CBF1) detection using an ABI Prism 7300 thermocycler (Applied Biosystems). The Taqman detection system was used to analyze CBF1 expression to ensure specificity, avoiding amplification of CBF2 and CBF3. Further assays used SYBR-green detection. Transcript levels were detected in two biological replicates for each sample using a standard curve derived from one reference sample with an arbitrary value set to one. Transcripts were normalized to mean expression of both ACTIN2 and AT3G06240 for Figures 1E, 1F, 4B, 5D, and 6A and TUBULIN 9 and AT3G06240 for Figure 3A. Analysis of publicly available Affymetrix array data shows that AT3G06240 is expressed stably throughout the transition from torpedo stage developing seeds to seedlings. PCR primers used can be found in Supplemental Table 1 online.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: DOG1, At5g45830; CBF1, At4g25490; CBF2, At4g25470; CBF3, At4g25480; CYP707A2, At2g29090; GA2ox6, At1g02400; NCED4, At4g19170; and COR15b, At2g42530.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Low Temperature during Seed Maturation Inhibits Germination by Inducing a Strongly Dormant State.
Supplemental Figure 2. Testing the Lack of Requirement for ABA for Dormancy in the Columbia Background.
Supplemental Figure 3. The Dormancy and Germination Characteristics of the Seed Lots Used for the Real-Time RT-PCR Data Presented in Figure 6A.
Supplemental Table 1. Primers Used for Real-Time RT-PCR Assays.
Supplemental Data Set 1. Differential Gene Expression in Mature Seeds in Response to Temperature Variation during Seed Development and Maturation.
Acknowledgments
We thank Micheal Thomashow, Julio Salinas, and Wim Soppe for sharing plant material. We also thank Michael Holdsworth for providing the TAGGIT Excel macro. Sarah Kendall was supported by a Biotechnology and Biological Sciences Research Council quota studentship and Steven Penfield by a Royal Society Fellowship.
AUTHOR CONTRIBUTIONS
S.L.K. performed the experimental work, except where attributed to other authors, and cowrote the manuscript. A.H. performed the extraction and data analysis for Figure 2A supervised and using experimental methods designed by I.A.G. P.M. contributed the data for Supplemental Figure 2 online. C.W. performed by microarray hybridizations and initial data analysis. S.P. performed experimental work for Figures 1A to 1C, designed experiments, and cowrote the manuscript.
References
- Achard P., Cheng H., De Grauwe L., Decat J., Schoutteten H., Moritz T., Van Der Straeten D., Peng J., Harberd N.P. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311: 91–94 [DOI] [PubMed] [Google Scholar]
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Argyris J., Truco M.J., Ochoa O., McHale L., Dahal P., Van Deynze A., Michelmore R.W., Bradford K.J. (2011). A gene encoding an abscisic acid biosynthetic enzyme (LsNCED4) collocates with the high temperature germination locus Htg6.1 in lettuce (Lactuca sp.). Theor. Appl. Genet. 122: 95–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ariizumi T., Steber C.M. (2007). Seed germination of GA-insensitive sleepy1 mutants does not require RGL2 protein disappearance in Arabidopsis. Plant Cell 19: 791–804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel G.W., Fung P., Chow T.F., Foong J.A., Provart N.J., Cutler S.R. (2008). Elucidating the germination transcriptional program using small molecules. Plant Physiol. 147: 143–155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Bentsink L., et al. (2010). Natural variation for seed dormancy in Arabidopsis is regulated by additive genetic and molecular pathways. Proc. Natl. Acad. Sci. USA 107: 4264–4269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentsink L., Jowett J., Hanhart C.J., Koornneef M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. USA 103: 17042–17047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J.D. (1997). Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera E., Holman T., Medhurst A., Peer W., Schmuths H., Footitt S., Theodoulou F.L., Holdsworth M.J. (2007). Gene expression profiling reveals defined functions of the ATP-binding cassette transporter COMATOSE late in phase II of germination. Plant Physiol. 143: 1669–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang G.C., Barua D., Kramer E.M., Amasino R.M., Donohue K. (2009). Major flowering time gene, flowering locus C, regulates seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 106: 11661–11666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A., Hernández M.L., He Z., Andriotis V.M., Vaistij F.E., Larson T.R., Graham I.A. (2011). 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23: 583–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K., Heschel M.S., Butler C.M., Barua D., Sharrock R.A., Whitelam G.C., Chiang G.C. (2008). Diversification of phytochrome contributions to germination as a function of seed-maturation environment. New Phytol. 177: 367–379 [DOI] [PubMed] [Google Scholar]
- Fenner M. (1991). The effects of the parent environment on seed germinability. Seed Sci. Res. 1: 75–84 [Google Scholar]
- Finch-Savage W.E., Cadman C.S., Toorop P.E., Lynn J.R., Hilhorst H.W. (2007). Seed dormancy release in Arabidopsis Cvi by dry after-ripening, low temperature, nitrate and light shows common quantitative patterns of gene expression directed by environmentally specific sensing. Plant J. 51: 60–78 [DOI] [PubMed] [Google Scholar]
- Franklin K.A., Whitelam G.C. (2007). Light-quality regulation of freezing tolerance in Arabidopsis thaliana. Nat. Genet. 39: 1410–1413 [DOI] [PubMed] [Google Scholar]
- Gilmour S.J., Sebolt A.M., Salazar M.P., Everard J.D., Thomashow M.F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124: 1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu X.Y., Kianian S.F., Foley M.E. (2006). Dormancy genes from weedy rice respond divergently to seed development environments. Genetics 172: 1199–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heschel M.S., Selby J., Butler C., Whitelam G.C., Sharrock R.A., Donohue K. (2007). A new role for phytochromes in temperature-dependent germination. New Phytol. 174: 735–741 [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen K.R., Gilmour S.J., Zarka D.G., Schabenberger O., Thomashow M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280: 104–106 [DOI] [PubMed] [Google Scholar]
- Johanson U., West J., Lister C., Michaels S., Amasino R., Dean C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290: 344–347 [DOI] [PubMed] [Google Scholar]
- Kumar S.V., Wigge P.A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cell 140: 136–147 [DOI] [PubMed] [Google Scholar]
- Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., Hirai N., Koshiba T., Kamiya Y., Nambara E. (2004). The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: Key enzymes in ABA catabolism. EMBO J. 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Koornneef M., Soppe W.J. (2007). The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell 19: 391–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., Amasino R.M. (1999). FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels S.D., He Y., Scortecci K.C., Amasino R.M. (2003). Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100: 10102–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar A.A., Jacobsen J.V., Ross J.J., Helliwell C.A., Poole A.T., Scofield G., Reid J.B., Gubler F. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: The role of ABA 8′-hydroxylase. Plant J. 45: 942–954 [DOI] [PubMed] [Google Scholar]
- Munir J., Dorn L.A., Donohue K., Schmitt J. (2001). The effect of maternal photoperiod on seasonal dormancy in Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 88: 1240–1249 [PubMed] [Google Scholar]
- Nambara E., Keith K., McCourt P., Naito S. (1994). Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant Cell Physiol. 35: 509–513 [PubMed] [Google Scholar]
- Novillo F., Medina J., Salinas J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Natl. Acad. Sci. USA 104: 21002–21007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor T.R., Dyreson C., Wyrick J.J. (2005). Athena: A resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics 21: 4411–4413 [DOI] [PubMed] [Google Scholar]
- Ogawa M., Hanada A., Yamauchi Y., Kuwahara A., Kamiya Y., Yamaguchi S. (2003). Gibberellin biosynthesis and response during Arabidopsis seed germination. Plant Cell 15: 1591–1604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E., Yamaguchi S., Kamiya Y., Bae G., Chung W.I., Choi G. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47: 124–139 [DOI] [PubMed] [Google Scholar]
- Penfield S., Gilday A.D., Halliday K.J., Graham I.A. (2006). DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Curr. Biol. 16: 2366–2370 [DOI] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Halliday K.J. (2010). A role for an alternative splice variant of PIF6 in the control of Arabidopsis primary seed dormancy. Plant Mol. Biol. 73: 89–95 [DOI] [PubMed] [Google Scholar]
- Penfield S., Josse E.M., Kannangara R., Gilday A.D., Halliday K.J., Graham I.A. (2005). Cold and light control seed germination through the bHLH transcription factor SPATULA. Curr. Biol. 15: 1998–2006 [DOI] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20: 2729–2745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Turecková V., Lacombe E., Lopez-Molina L. (2009). Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J. 28: 2259–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmuths H., Bachmann K., Weber W.E., Horres R., Hoffmann M.H. (2006). Effects of preconditioning and temperature during germination of 73 natural accessions of Arabidopsis thaliana. Ann. Bot. (Lond.) 97: 623–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo M., et al. (2006). Regulation of hormone metabolism in Arabidopsis seeds: Phytochrome regulation of abscisic acid metabolism and abscisic acid regulation of gibberellin metabolism. Plant J. 48: 354–366 [DOI] [PubMed] [Google Scholar]
- Sheldon C.C., Burn J.E., Perez P.P., Metzger J., Edwards J.A., Peacock W.J., Dennis E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber C.M., Cooney S.E., McCourt P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149: 509–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger E.J., Gilmour S.J., Thomashow M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94: 1035–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher V.G., Tibshirani R., Chu G. (2001). Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. USA 98: 5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J.T., Zarka D.G., Van Buskirk H.A., Fowler S.G., Thomashow M.F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41: 195–211 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Smith M.W., Brown R.G., Kamiya Y., Sun T. (1998). Phytochrome regulation and differential expression of gibberellin 3beta-hydroxylase genes in germinating Arabidopsis seeds. Plant Cell 10: 2115–2126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y., Ogawa M., Kuwahara A., Hanada A., Kamiya Y., Yamaguchi S. (2004). Activation of gibberellin biosynthesis and response pathways by low temperature during imbibition of Arabidopsis thaliana seeds. Plant Cell 16: 367–378 [DOI] [PMC free article] [PubMed] [Google Scholar]