The authors identified a bacterial monogalactosyldiacylglycerol synthase enzyme from Chlorobaculum tepidum. Using complementation analysis of a galactolipid synthase mutant of Arabidopsis, they revealed crucial roles of galactolipids for plant and bacterial photosynthesis. In addition, they investigated and discuss evolutionary paths of galactolipid biosynthesis in phototrophs.
Abstract
Monogalactosyldiacylglycerol (MGDG), which is conserved in almost all photosynthetic organisms, is the most abundant natural polar lipid on Earth. In plants, MGDG is highly accumulated in the chloroplast membranes and is an important bulk constituent of thylakoid membranes. However, precise functions of MGDG in photosynthesis have not been well understood. Here, we report a novel MGDG synthase from the green sulfur bacterium Chlorobaculum tepidum. This enzyme, MgdA, catalyzes MGDG synthesis using UDP-Gal as a substrate. The gene encoding MgdA was essential for this bacterium; only heterozygous mgdA mutants could be isolated. An mgdA knockdown mutation affected in vivo assembly of bacteriochlorophyll c aggregates, suggesting the involvement of MGDG in the construction of the light-harvesting complex called chlorosome. These results indicate that MGDG biosynthesis has been independently established in each photosynthetic organism to perform photosynthesis under different environmental conditions. We complemented an Arabidopsis thaliana MGDG synthase mutant by heterologous expression of MgdA. The complemented plants showed almost normal levels of MGDG, although they also had abnormal morphological phenotypes, including reduced chlorophyll content, no apical dominance in shoot growth, atypical flower development, and infertility. These observations provide new insights regarding the importance of regulated MGDG synthesis in the physiology of higher plants.
INTRODUCTION
Life on Earth mostly relies on energy from the sun that is converted to biochemical energy by photosynthesis. The coordinated control of the photochemical reaction is thus critically important for all living organisms. To date, six bacterial phyla with species capable of chlorophyll-based photosynthesis are known. They are Cyanobacteria, Proteobacteria (purple bacteria), Chloroflexi (anoxygenic filamentous bacteria), Chlorobi (green sulfur bacteria), Firmicutes (heliobacteria), and Acidobacteria (Bryant and Frigaard, 2006; Bryant et al., 2007). Photosynthetic bacteria that belong to Proteobacteria and Chloroflexi use the photosystem II (PSII) type reaction center, and those of Chlorobi, Firmicutes, and Acidobacteria use the photosystem I (PSI) type reaction center. Cyanobacteria use both types of reaction centers, which allows them to perform oxygenic photosynthesis. It is widely accepted that chloroplasts in plants and algae originated from an ancient cyanobacterium that was introduced into a proto-plant cell by endosymbiosis.
Although only two types of reaction centers are used by the phototrophs, each species has evolved a wide variety of light-harvesting antennae that absorb photons and transfer excitation energy to the reaction centers (Green, 2003). The variation in antenna systems may reflect an adaptation to different environmental light conditions and the different colors and intensities of sunlight. Among the light-harvesting systems, chlorosomes, found in Chlorobi, Chloroflexi, and Acidobacteria, are the largest and the most efficient antenna systems in nature (Blankenship et al., 1995; Blankenship and Matsuura, 2003; Frigaard and Bryant, 2006; Bryant et al., 2007). Chlorosomes are constructed from carotenoids, quinones, and hundreds of thousands of bacteriochlorophyll (BChl) molecules inside the monolayer membrane vesicle, which is extremely stable and can perform efficient and rapid energy transfer into the reaction center. These properties allow the bacteria to perform photosynthesis under extraordinarily low light conditions. In recent years, chlorosome architecture, including its protein and lipid composition as well as the organization of BChls and the membrane orientation in the antenna systems, has been well characterized (Jochum et al., 2008; Pedersen et al., 2008; Sørensen et al., 2008; Ganapathy et al., 2009; Tronrud et al., 2009; Wen et al., 2009; Saga et al., 2010; Mizoguchi et al., 2011; Yoshitomi et al., 2011). Although these studies have successfully documented the roles and properties of chlorosomes, how this unique antenna system is generated in native cells remains poorly understood (Hohmann-Marriott and Blankenship, 2007; Pedersen et al., 2010). A key factor for chlorosome biogenesis is suggested based on its lipid composition. Specifically, unlike the cytoplasmic bilayer membrane, which is mostly composed of phospholipids, chlorosome membrane accumulates galactoglycerolipids, such as monogalactosyldiacylglycerol (MGDG) and rhamnosylgalactosyldiacylglycerol; these represent ~60% of total chlorosome polar lipids (Sørensen et al., 2008). Thus, control of galactolipid biosynthesis and its accumulation in chlorosome membranes is likely to be a key factor for chlorosome biogenesis.
Gal-containing glycerolipids also play important roles in photosynthesis in cyanobacteria and chloroplasts. Specifically, galactolipids, such as MGDG and digalactosyldiacylglycerol (DGDG), have been firmly established as the predominant lipid components of their photosynthetic membranes (Block et al., 1983; Dörmann and Benning, 2002). Their abundance is particularly high in thylakoid membranes, in which MGDG and DGDG represent ~50 and ~20% of the polar lipids, respectively. Crystal structures have revealed that MGDG molecules are found in both PSI and PSII reaction centers (Jordan et al., 2001; Loll et al., 2005, 2007; Guskov et al., 2009; Umena et al., 2011), suggesting that they not only are bulk constituents of thylakoid membranes but also are an integral component of photosystem complexes. The Arabidopsis thaliana mgd1-2 mutant, which lacks a major MGDG synthase, has disrupted photosynthetic membranes as well as complete impairment of photosynthetic activity (Kobayashi et al., 2007). This mutant also arrests during embryogenesis, indicating that proper MGDG synthesis is important not only for chloroplast biogenesis but also for normal plant development (Kobayashi et al., 2007). In fact, MGDG synthase activity is strictly regulated at the posttranslational level through photosynthesis-dependent oxidation/reduction of a disulfide bond as well as binding/dissociation of phosphatidic acid (Yamaryo et al., 2006; Dubots et al., 2010). However, the importance of the posttranslational control of MGDG synthesis is still unknown in planta. Identification and comparative analysis of alternative MGDG synthases will be useful for elucidating the impact of the control of galactolipid biosynthesis for normal plant development.
Although galactolipids are conserved in many phototrophs, their biosynthetic pathways and enzymes seem to differ among photosynthetic organisms. For example, in chloroplasts, MGDG synthase uses UDP-Gal and sn-1,2-diacylglycerol (DAG) as substrates and transfers the galactosyl moiety of UDP-Gal to DAG to produce MGDG (Shimojima et al., 1997; Benning and Ohta, 2005). DGDG is then synthesized from MGDG by DGDG synthase using UDP-Gal as a substrate (Dörmann et al., 1995; Dörmann and Benning, 2002). On the other hand, cyanobacteria use UDP-Glc for the first step of galactolipid synthesis; the glucosyl moiety of UDP-Glc is transferred to DAG and monoglucosyldiacylglycerol (MGlcDG) is synthesized as an intermediate. The glucosyl head group, MGlcDG, is then isomerized to the galactosyl moiety by an unidentified epimerase to produce MGDG (Sato and Murata, 1982; Awai et al., 2006). Thus, chloroplasts and cyanobacteria use very different enzymes to produce MGDG, although cyanobacteria are thought to be the endosymbiotic ancestors of chloroplasts.
Genomic sequence data from photosynthetic bacteria from different phyla are currently available. Based on these data, homologous genes for plant-type MGDG synthase were identified in phototrophic species from Chloroflexi but not in those from other phyla (Hölzl et al., 2005). Orthologous genes for MGlcDG synthase of cyanobacteria were not found in other species (Awai et al., 2006). Some glycosyltransferases with MGDG or MGlcDG synthase activity were identified in the nonphototrophic bacteria Acholeplasma laidlawii (Berg et al., 2001), Bacillus subtilis (Jorasch et al., 1998), Staphylococcus aureus (Jorasch et al., 2000), Deinococcus radiodurans, Thermotoga maritime, Agrobacterium tumefaciens, and Mesorhizobium loti (Hölzl et al., 2005); however, no homologous gene for the enzymes is found in all classes of photosynthetic bacteria. Thus, there may be unidentified types of MGDG (or MGlcDG) synthases conserved in phototrophic bacteria. Given that galactolipids are important for photosynthesis in different ways in different organisms, as described above, the identification of new enzymes for MGDG biosynthesis is critical for gaining more insight into the function of galactolipids in photosynthesis.
In this study, we aimed to identify a gene responsible for galactolipid synthesis in the chlorosome-containing photosynthetic bacterium Chlorobaculum tepidum (formerly called Chlorobium tepidum). This bacterium has been extensively characterized as a model for the Chlorobi group of green sulfur bacteria because of its ease of cultivation and natural transformability as well as the availability of its genomic sequence data (Frigaard and Bryant, 2001; Eisen et al., 2002; Frigaard et al., 2003). Furthermore, the chlorosome of C. tepidum is well characterized, and protein and lipid components of the antenna system have been well studied (Frigaard et al., 2004b; Sørensen et al., 2008). Using a recently developed computer-aided gene discovery system, we identified a gene encoding MGDG synthase of C. tepidum. Mutational analysis indicated that this enzyme, MgdA, is essential for this bacterium. We also performed a complementation analysis using an Arabidopsis MGDG synthase mutant by MgdA and found that it could only partially complement the mutation. These results give us new insights regarding the importance of the regulated synthesis of MGDG in chloroplasts for higher plant development as well as evolutionary paths of galactolipid biosynthesis.
RESULTS
Identification of Enzymes for MGDG Biosynthesis from C. tepidum
To identify candidate genes encoding enzymes involved in galactolipid biosynthesis in C. tepidum, we first performed a BLAST search analysis of the C. tepidum genome using an Arabidopsis MGDG synthase (MGD1) as the query sequence (Jarvis et al., 2000). A C. tepidum open reading frame (ORF), CT0034, showed the highest identity (~15%) to the Arabidopsis MGDG synthase. However, on the genome sequence database, CT0034 was annotated as UDP-N-acetylglucosamine-N-acetylmuramyl-(pentapeptide) pyrophosphoryl-undecaprenol-N-acetylglucosamine transferase (MurG), which is involved in the biosynthesis of bacterial peptidoglycan (Mengin-Lecreulx et al., 1990). To date, glycosyltransferases have been categorized into 78 families based on deduced amino acid sequences, and the bacterial MurG and plant-type MGDG synthases are classified into the same glycosyltransferase family (GT28), although they have distinct functions (Coutinho et al., 2003). In fact, the amino acid sequence of CT0034 shared more identical residues with those of other bacterial MurG (~35%) than with those of plant MGDG synthases (~15%), suggesting that CT0034 is a MurG ortholog in C. tepidum. We also performed a BLAST search analysis with a cyanobacterial MGlcDG synthase and found that an ORF, CT1987, showed the highest identity (~20%) to the enzyme. However, CT1987 is a chlorobactene glucosyltransferase (named CruC) involved in carotenoid modification in this bacterium (Maresca and Bryant, 2006). These observations suggested that C. tepidum does not have any orthologs for either plant-type MGDG synthases or cyanobacterial MGlcDG synthases. The results also suggested that unidentified enzymes involved in galactolipid biosynthesis in C. tepidum are not conserved in cyanobacteria and plants.
Then, we used a computer-aided gene discovery program, Correlation Coefficient Calculation Tool (CCCT) (Ito et al., 2008), to isolate candidate ORFs for galactolipid biosynthesis in C. tepidum. CCCT is based on a comparative analysis of whole gene sets between different organisms, which allows for the identification of genes involved in a particular function in a certain class of organisms. In this analysis, we made three assumptions: (1) all green sulfur bacteria tested, including C. tepidum, should have a specific enzyme(s) responsible for MGDG synthesis; (2) the enzyme(s) should not be found in cyanobacteria, red algae, green algae, diatoms, and higher plants as well as nonphotosynthetic organisms tested; and (3) the enzyme(s) should have motifs for glycosyltransferase. The program was used with gene sets from 85 species (including nine green sulfur bacteria) whose genome sequences have been determined (see Supplemental Table 1 online). ORFs from the C. tepidum genome were used as a query sequence. In the output data, all ORFs of C. tepidum were ranked according to their correlation coefficients, such that ORFs specifically conserved in the nine species of green sulfur bacteria were expected to show higher values. The top six ORFs in the output data were CT1558 (hypothetical membrane spanning protein), CT0208 (hypothetical protein), CT0344 (β-glycosyltransferase-like protein), CT1314 (trehalose-6-phosphate synthase), CT0480 (hypothetical protein), and CT2205 (hypothetical membrane protein); orthologs of these genes were found only in nine green sulfur bacteria but not in the other 76 examined organisms. Given that putative MGDG synthase(s) may have a motif for glycosyltransferase, CT0344, annotated as β-glycosyltransferase-like protein, was chosen as a good candidate for a MGDG synthase conserved in green sulfur bacteria.
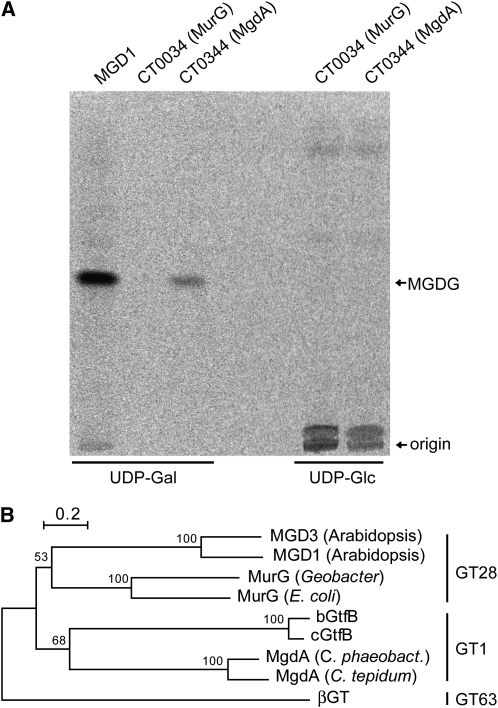
We next tested whether CT0344 has MGDG (or MGlcDG) synthase activity. For this analysis, CT0344 was expressed in Escherichia coli, and its activity was measured using DAG and radiolabeled UDP-Gal (or UDP-Glc) as substrates. We also tested the activity of cucumber (Cucumis sativus) MGDG synthase (MGD1) and a MurG ortholog of C. tepidum (CT0034) as positive and negative controls, respectively. Expressed enzymes that have MGDG (or MGlcDG) synthase activity in the presence of UDP-Gal (or UDP-Glc) should produce radiolabeled spots on thin layer chromatography (TLC), which separates the synthesized lipids. It is of note that the solvent used for TLC analysis can differentiate between anomeric and epimeric glycosyldiacylglycerols (for detail, see Methods). Crude cell extracts of E. coli expressing CT0344 synthesized a radiolabeled lipid from UDP-Gal but not from UDP-Glc (Figure 1A). By contrast, crude extracts of CT0034-expressing E. coli showed no visible incorporation of radioactivity in analyzed lipids, whereas the positive control, cucumber MGDG synthase, was able to use UDP-Gal for MGDG synthesis (Figure 1A). These results clearly show that CT0344 has MGDG synthase activity using DAG and UDP-Gal as substrates. Thus, we referred to CT0344 as MgdA (MGDG synthase A).
Figure 1.
Enzymatic and Phylogenetic Analysis of MgdA.
(A) Genes for C. tepidum CT0344 (MgdA) and CT0034 (MurG) as well as cucumber MGDG synthase (MGD1) were expressed in E. coli, and the activity of crude extracts was measured using radiolabeled UDP-Gal or UDP-Glc. The reaction mixtures were spotted on a TLC plate (indicated as origin) and then separated. MgdA and the positive control MGD1 enzymes synthesize MGDG with UDP-Gal as a substrate.
(B) A phylogenetic tree based on an amino acid sequence alignment of enzymes from GT28, GT1, and GT63 families. The bootstrap values obtained by 1000 replications are given.
We performed BLAST search analyses of mgdA against currently available nucleotide sequence databases. As expected, orthologous genes for mgdA were specifically conserved in green sulfur bacteria in the phylum Chlorobi but not in other photosynthetic organisms. However, genes showing a significant similarity to mgdA were found in some species of the genus Vibrionaceae, which includes Vibrio cholerae and Vibrio vulnificus; these bacterial species cause cholera and food poisoning, respectively (Oliver and Kaper, 2001). In a database for glycosyltransferases (CAZY; http://www.cazy.org), MgdA homologs are classified into the GT1 family. This family is closely related to the GT28 family, in which bacterial MurG and plant MGDG synthases are clustered. Both GT1 and GT28 families were suggested to have branched off from the GT63 family, and the three families form the same clan, termed Clan II (Coutinho et al., 2003). We constructed a phylogenetic tree based on an amino acid sequence alignment of several GT28, GT63, and GT1 proteins (Figure 1B), which clearly showed that MgdA-like proteins are clustered within the GT1 family.
We characterized some enzymatic properties of MgdA. Plant MGDG synthases are activated by phosphatidic acid; Vmax was increased fivefold when the phosphatidic acid concentration in the reaction mixture was changed from 0.15 to 1.5 mol % (Dubots et al., 2010). Km for UDP-Gal of the enzyme is also influenced by phosphatidic acid concentration. This activation is believed to be important for controlling the coupling of phospholipid and galactolipid synthesis in native plant cells (Dubots et al., 2010). We investigated whether a similar activation mechanism is observed for MgdA. The activity of MgdA in the membrane fractions of mgdA-expressing E. coli is not significantly influenced by phosphatidic acid. Specifically, Vmax values (± se) for the specific activity of MgdA in the absence and presence of phosphatidic acid (1.5 mol %) are 7.04 ± 0.26 μmol min–1 mg–1 and 6.55 ± 0.38 μmol min–1 mg–1, respectively. However, we cannot exclude the possibility that endogenous phosphatidic acid in E. coli influences the MgdA activity. To check this possibility, we purified His-tagged MgdA (see Supplemental Figure 1 online) and measured its activity. Vmax values (± se) of the purified MgdA in the presence of low (0.15 mol %) and high (1.5 mol %) concentration of phosphatidic acid are not significantly different; they are 53.1 ± 3.4 nmol min–1 ng–1 and 49.1 ± 3.2 nmol min–1 ng–1, respectively. Km values for UDP-Gal in the presence of 0.15 and 1.5 mol % phosphatidic acid are also not significantly different; they are 18.3 ± 6.6 μM and 15.7 ± 3.2 μM, respectively. The substrate selectivity of MgdA in the membrane fractions of mgdA-expressing E. coli was also investigated. Two DAG molecular species, sn-1,2-dioleoylglycerol (18:1/18:1) and sn-1-oleoyl-sn-2-palmitoyl-glycerol (18:1/16:0), were used to compare the MGDG synthase activity of MgdA. Vmax values (± se) for 18:1/18:1 and 18:1/16:0 DAG did not significantly differ at 7.04 ± 0.26 μmol min–1 mg–1 and 7.51 ± 1.84 μmol min–1 mg–1, respectively.
Functional Analysis of MgdA in C. tepidum
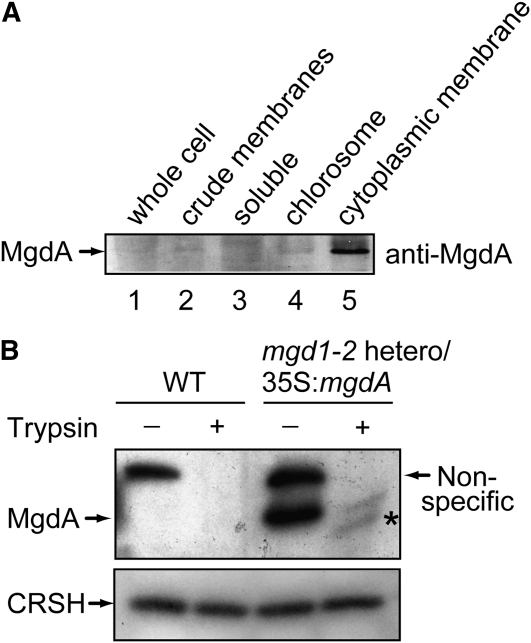
To understand MgdA functions in C. tepidum, we first analyzed the localization of MgdA by immunoblot analysis (Figure 2A). For this purpose, various protein fractions were collected during preparation of chlorosomes and cytoplasmic membranes. Specifically, after ultracentrifugation of the whole-cell extract (lane 1), the precipitate (crude membranes; lane 2) and soluble fraction (lane 3) were collected. The membrane fraction was further separated by Suc density ultracentrifugation, and chlorosome (lane 4) and cytoplasmic membrane (lane 5) fractions were obtained. Enrichment of the chlorosome or cytoplasmic membranes in the fractions was checked by monitoring the absorption spectra (for detail, see Methods). Proteins in each fraction were separated by SDS-PAGE, and MgdA was detected with an MgdA-specific antibody. MgdA was detected in the cytoplasmic membrane fraction (lane 5) but not much observed in the soluble and chlorosome fractions (Figure 2A, lanes 3 and 4). These results indicated that MgdA is localized in cytoplasmic membranes but not in chlorosome membranes.
Figure 2.
Localization of MgdA Examined by Immunoblot Analysis.
(A) Crude cell extracts of C. tepidum were fractionated by centrifugation. Immunoblot analysis was performed with the MgdA antibody.
(B) Chloroplasts isolated from the wild type (WT) and the mgd1-2 heterozygous mutant harboring the 35S:mgdA construct were incubated for 10 min with or without trypsin. After SDS-PAGE, immunoblot analysis was performed with the MgdA and the CRSH antibodies. A fragment appearing after trypsin treatment is marked by an asterisk.
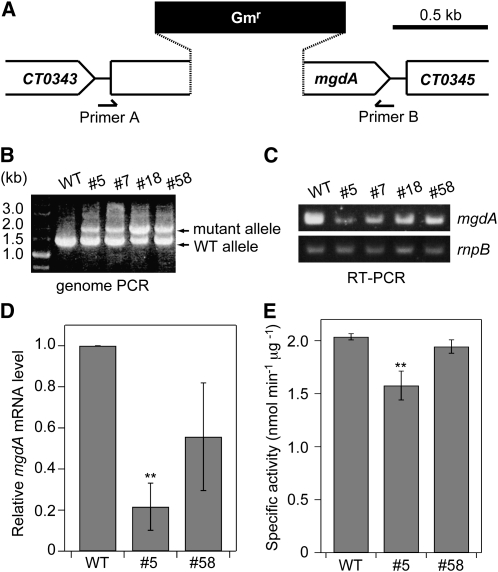
To investigate the biological function of MgdA in C. tepidum, we next attempted to construct a knockout mutant of mgdA from C. tepidum. For this purpose, a gentamicin resistance cassette was inserted into the cloned DNA fragment of mgdA, and the plasmid construct was mixed with C. tepidum wild-type cells to allow homologous recombination between the mutagenized DNA and the chromosomal gene mgdA (Figure 3A). Colonies showing gentamicin resistance were chosen and were applied several times for single-colony isolation. Then, insertion of the gentamicin resistance cassette into chromosomal mgdA was confirmed by PCR analysis. The primers used in the experiment were expected to amplify 1.5- and 2.0-kb DNA fragments with the wild-type mgdA and the mutagenized mgdA, respectively. No mgdA null mutants could be obtained; the mutant locus could not be fully segregated in the gentamicin-resistant cells (Figure 3B). The genotype was stable for further generations under different growth conditions (e.g., low light, high light, low temperature, and exogenous addition of MGDG), indicating that mgdA is an essential gene for C. tepidum. It is of note that we previously obtained a C. tepidum mutant lacking a nonessential gene encoding geranylgeranyl reductase by this method with the same gentamicin resistance cassette (Harada et al., 2008).
Figure 3.
Construction of an mgdA Mutant of C. tepidum.
(A) Schematic description of gene manipulation for insertion of mgdA mutation. Primers A and B used for genome PCR analysis are shown.
(B) PCR analysis for checking the genotypes of the mgdA locus in four gentamicin-resistant strains (#5, #7, #18, and #58). WT, wild type.
(C) RT-PCR analysis of mgdA in the four heterozygous mutant strains. The gene rnpB was used as an internal control.
(D) Relative mgdA mRNA levels of the wild type and #5 and #58 mutants as determined by quantitative real-time PCR analysis. The wild-type level was set to 1.0. The rnpB was used as an internal standard. The values are the means ± sd of three biological replications. **P < 0.05, t test (compared with the wild type).
(E) MGDG synthase activity of C. tepidum wild type and #5 and #58 strains. The activity was measured with crude extracts of each strain. The values are the means ± sd of three biological replications. **P < 0.05, t test (compared with the wild type).
Although no null mutant of mgdA could be segregated, one could assume that the amount of mgdA mRNA in the gentamicin-resistant cells would be reduced compared with wild-type cells because of lower copy numbers of the chromosomal wild-type mgdA gene. We checked this possibility by RT-PCR analysis (Figure 3C). Heterozygous mutants showed a marked reduction in the amount of mgdA mRNA, although the levels were variable in each mutant. We checked the actual levels of mgdA mRNA in two of the mutants by quantitative real-time PCR analysis (Figure 3D) that indicated that mgdA mRNA levels in #5 and #58 mutants were ~20 and ~50%, respectively, compared with that in the wild type. We measured the specific MGDG synthase activity in the wild type and the mutants (Figure 3E). MGDG synthase activity of crude extracts of #5 mutant was reduced to ~80% of those of the wild type, indicating that it is actually a knockdown mutant of mgdA. On the other hand, MGDG synthase activity in #58 mutant is not significantly different from that in the wild type.
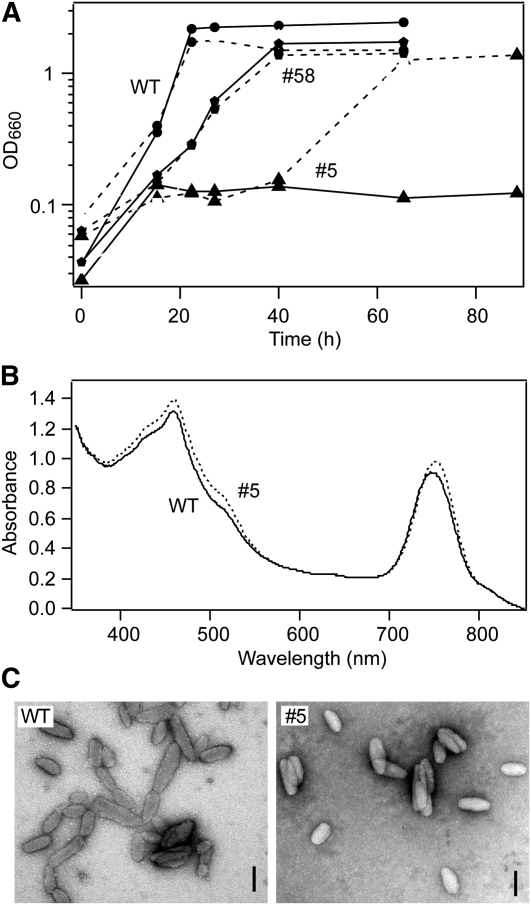
We performed phenotypic analyses of the knockdown mutants. The growth profiles of the wild type and mutants were measured (Figure 4A). During the mutagenesis experiment in which cells were grown at a lower temperature (40°C) because of the presence of an antibiotic, we found that some mutants showed reduced growth rate; they took a longer time to form colonies than did the wild type. This phenotype was more obvious under optimal growth conditions (47°C), such that the growth of strain #5 was almost completely inhibited in the absence of MGDG (Figure 4A; triangles, solid line). The growth of another mutant (#58) was not much different from that observed for the wild type (Figure 4A, pentagons, solid line). It is of note that the mgdA mRNA level as well as MGDG synthase activity in #5 is lower than those in the wild type and #58 mutant (Figures 3C and 3D), suggesting that the growth phenotypes are due to the knockdown mutation of mgdA. The growth impairment of strain #5 was restored by addition of exogenous MGDG, although the growth rate was still slower than that of the wild type (Figure 4A, triangles, dotted line). However, addition of exogenous MGDG did not influence the growth of #58 mutant (Figure 4A, pentagons, dotted line), suggesting that incorporation of MGDG is the rate-limiting step for the growth of strain #5. These results indicated that MgdA-dependent MGDG synthesis is important for photosynthetic growth of C. tepidum. Because the knockdown mutant #5 could not grow at the optimal conditions, we grew the mutant and wild-type cells at a lower temperature (40°C) for further analysis.
Figure 4.
Phenotypes of mgdA Knockdown Mutants.
(A) Growth profiles of the wild type (WT) (circles) and the mgdA knockdown mutants (#5, triangles; #58, pentagons) in the absence (solid lines) or presence (dotted lines) of 100 mM MGDG. Antibiotics were not present in the medium.
(B) Absorption spectra of the C. tepidum wild type (solid line) and the mgdA knockdown mutant (#5) (dotted line).
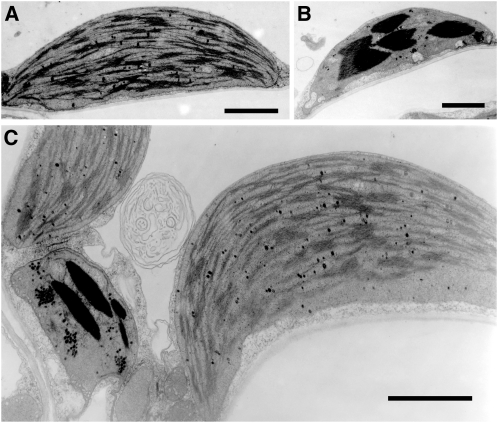
(C) Transmission electron micrographs of negatively stained chlorosomes isolated from the wild type and the #5 mutant. Bars = 100 nm.
We next checked in vivo absorption properties of chlorosomes in mutant strain #5 (Figure 4B). The position of the absorbance maximum of the Qy band of BChl c aggregates (~750 nm) was slightly, but significantly (P < 0.05, t test), shifted to a longer wavelength (by ~5 nm) in strain #5 (λmax = 754.9 ± 0.0 nm; n = 3) compared with that observed in the wild type (λmax = 750.0 ± 0.5 nm; n = 3), indicating that the structure of the BChl c aggregates in the mutant chlorosomes is altered. We also analyzed chlorosome structures of #5 mutant by transmission electron microscopy (Figure 4C). Chlorosomes isolated from #5 mutant showed size and shape differences compared with those from the wild type. The measured dimensions of chlorosomes from the wild type and the #5 mutant were as follows (sample size 50 and 43, respectively): length, 141 ± 40 nm (wild type) and 120 ± 26 nm (#5) (P < 0.05, t test); width, 49 ± 10 nm (wild type) and 53 ± 7 nm (#5) (P < 0.01, t test); and length-to-width ratio, 2.8 ± 0.6 (wild type) and 2.1 ± 0.4 (#5) (P < 0.001, t test). These observations indicated that MgdA has a role in chlorosome biogenesis.
Complementation Analysis of Arabidopsis MGDG Synthase Mutant by MgdA
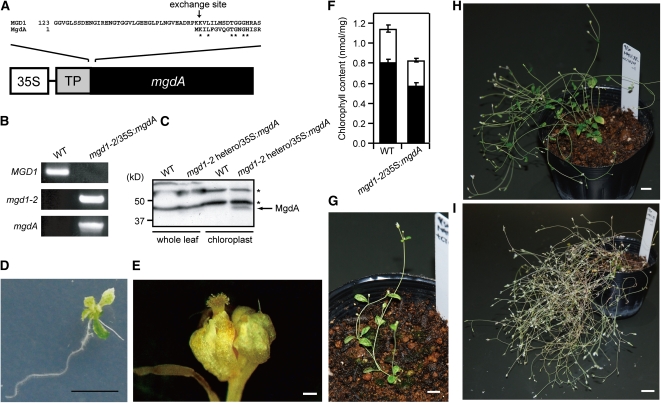
We next expressed mgdA in an Arabidopsis MGDG synthase mutant (mgd1-2) (Kobayashi et al., 2007). The purposes of this experiment were (1) to directly show the enzymatic activity of MgdA in vivo and (2) to determine the impact of replacement of the plant MGDG synthase with bacterial MgdA. Arabidopsis has three MGDG synthases, MGD1, MGD2, and MGD3, all of which are localized in chloroplasts (Benning and Ohta, 2005). Among them, MGD1 is the most important enzyme for MGDG biosynthesis such that its null mutant (mgd1-2) shows a significant reduction in MGDG content, whereas a MGD2 and MGD3 double mutant still retains almost normal levels of MGDG synthesis (Kobayashi et al., 2007, 2009). Figure 5A shows a schematic description of the DNA construct used to produce potentially complementing plants. The chloroplast transit peptide of MGD1 was fused with C. tepidum MgdA at its N terminus. The putative cleavage site of the MGD1 transit peptide is Leu-67 (Yamaryo et al., 2003). The recombinant protein was expressed under the control of the cauliflower mosaic virus 35S promoter. Because the mgd1-2 mutation is lethal at the seedling stage (Kobayashi et al., 2007), the mgdA-expressing construct was initially introduced into the heterozygous mgd1-2 mutant via Agrobacterium-mediated transformation.
Figure 5.
Phenotypes of mgd1-2/35S:mgdA Complementing Plants.
(A) Schematic representation of the DNA construct for producing the recombinant plants. The exchange site between MGD1 transit peptide and MgdA is indicated by an arrow.
(B) PCR analysis of genomic DNA from a wild-type and mgd1-2/35S:mgdA transgenic plant. Amplified bands were specific for MGD1, mgd1-2, and the mgdA locus.
(C) Immunoblot analysis with MgdA antibody. Whole-leaf extracts and chloroplast-enriched fractions were obtained from the wild type and the mgd1-2 heterozygous mutant harboring the 35S:mgdA construct. Asterisks indicate nonspecific bands.
(D) Seedling stage 21 d of a mgd1-2/35S:mgdA transgenic plant. Bar = 1 cm.
(E) A flower bud of the mgd1-2/35S:mgdA transgenic plant. Bar = 1 mm.
(F) Chlorophyll content of wild-type and mgd1-2/35S:mgdA transgenic plants. White and black bars are for chlorophyll b and chlorophyll a, respectively. The values are the means ± sd of three biological replicates.
(G) A 35-d-old mgd1-2/35S:mgdA transgenic plant. Bar = 1 cm.
(H) A 56-d-old mgd1-2/35S:mgdA transgenic plant. Bar = 1 cm.
(I) A 99-d-old mgd1-2/35S:mgdA transgenic plant. Bar = 2 cm.
A heterozygous mgd1-2 mutant showing clear MgdA expression was selected based on the results of protein gel blot analysis with an MgdA-specific antibody (Figure 5C). The molecular weight shown in the analysis (~45 kD) matched well with the value calculated from the deduced amino acid sequence when the chloroplast transit peptide of MGD1 was excluded (47.8 kD), suggesting that the expressed MgdA was successfully targeted to chloroplasts. This idea was further supported by the protein gel blot analysis, in that MgdA accumulated in the chloroplast fractions (Figure 5C). It was previously shown that MGD1 is localized on the outside of the inner envelope membrane in native chloroplasts (Xu et al., 2005). This was evidenced by the protease protection assay with intact chloroplasts that indicated that trypsin, a protease that cannot penetrate the inner envelope membrane, partially digests MGD1 at the certain concentration of the protease (Xu et al., 2005). To determine the exact localization of the targeted MgdA in chloroplasts, we performed the trypsin protection assay with chloroplasts isolated from the heterozygous mgd1-2 mutant harboring the 35S:mgdA construct. For this analysis, CRSH (Ca2+-activated RelA/SpoT Homolog) of Arabidopsis (Masuda et al., 2008) was used as a control. This enzyme is expected to be trypsin resistant, since it is localized within the chloroplast stroma (Masuda et al., 2008). As shown in Figure 2B, MgdA is cleaved by trypsin, although CRSH was not degraded at the trypsin concentration. These results suggest that MgdA expressed in Arabidopsis localizes on the outside of the chloroplast inner envelope membrane as MGD1 does.
A homozygous mgd1-2 mutant containing the mgdA-expressing construct was then obtained by self-pollination of the heterozygous mgd1-2 mutant harboring the 35S:mgdA construct. The homozygous mgd1-2 mutation and the presence of mgdA in the complementing line were confirmed by genome PCR analysis (Figure 5B). Protein gel blot analysis indicated that the level of MgdA in the homozygous mgd1-2 mutant was almost the same compared with that in the parent heterozygous mgd1-2 mutant harboring the 35S:mgdA construct (see Supplemental Figure 2 online), indicating that localization and stability of MgdA are not much affected by the presence or absence of MGD1. Given that 35S promoter–dependent expression of the Arabidopsis MGD1 cDNA fully complements the mgd1-2 mutation (Kobayashi et al., 2007), phenotypes observed in the complementing plants should be due to the functional replacement of MGD1 by MgdA.
The mgd1-2 mutant has abnormal dwarf and albino phenotypes at the seedling stage when its growth is arrested (Kobayashi et al., 2007). By contrast, seedlings of the complementing plants were greening at this stage, although a slightly dwarf phenotype was still observed (Figure 5D). After transferring the seedlings to soil, they maintained growth and produced flowers (Figure 5G). However, the floral organs (Figure 5E) were not fertile; no seeds were obtained from the transgenic plants even when they were artificially pollinated. The flowers are about half in their size compared with those of the wild type, and atypical development of anthers and the pistils were observed (see Supplemental Figure 3 online). The unusual flowers were crossed with wild-type anthers (or wild-type pistils); however, we could not obtain F1 seeds. The complementing plants also showed other unusual phenotypes, including no apical dominance in shoot growth (Figures 5H and 5I), reduced Chl content (~80% of wild-type level; Figure 5F), and increased longevity (approximately twofold compared with the wild type). It is of note that mgdA expression in the wild-type background, which was obtained from self-pollinated parent plants heterozygous for the mgd1-2 mutation, did not show any abnormal phenotypes (see Supplemental Figure 4A online). Furthermore, another independently isolated mgd1-2/35S:mgdA line shows similar unusual shoot growth (see Supplemental Figure 4B online). Thus, the abnormal phenotypes shown in the complementing plants resulted from functional differences between MGD1 and MgdA but not from overexpression of MgdA or the insertion of the 35S:mgdA construct.
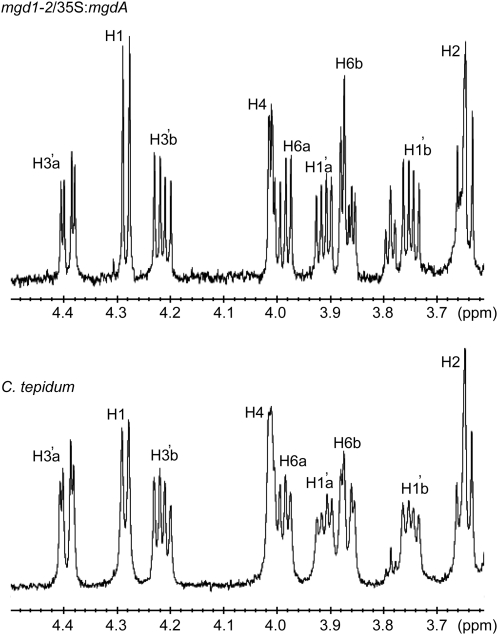
We next checked the properties of synthesized lipids in the complementing plants. There are two possible linkages (α- or β-configuration) for Gal conjugation of MGDG. MGDG in wild-type Arabidopsis is in the β-configuration for Gal conjugation (Xu et al., 2003). To obtain the linkage information of MGDG in the complementing plants, we acquired a 600-MHz 1H NMR spectrum for the MGDG purified from the complementing line. Figure 6 (top) shows the expanded NMR spectrum for the carbohydrate region (3.6 to 4.5 ppm). Anomeric proton resonances at 4.28 ppm (H1) showed relatively large coupling constants (J1,2 = 7.5 Hz), indicating the β-configuration for the sugar residue. Thus, the Gal configuration of MGDG synthesized in the complementing plant should be in the β-configuration as in wild-type plants. The same resonances were detected in MGDG isolated from C. tepidum wild-type cells (Figure 6, bottom). These results indicated that MgdA synthesizes β-linked MGDG in C. tepidum as well as in Arabidopsis.
Figure 6.
MgdA Synthesizes β-Linked MGDG.
1H NMR spectra of MGDG isolated from the mgd1-2/35S:mgdA transgenic plant (top panel) and C. tepidum wild-type cells (bottom panel) in CDCl3. Peak assignments are shown in Methods.
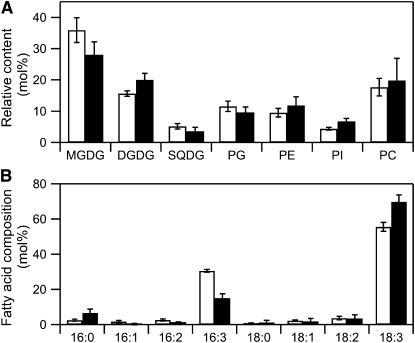
We also measured the lipid composition of the complementing plant. Lipid composition is severely affected by the mgd1-2 mutation such that the amount of MGDG in the mgd1-2 mutant is reduced to ~2% of that in the wild type (Kobayashi et al., 2007). Because MGDG synthesized by MGD1 is also used for DGDG synthesis as a substrate (Benning and Ohta, 2005), DGDG content was also dramatically reduced in the mutant (to ~2% of that in the wild type). Likewise, the amounts of other lipids were also significantly affected by the loss of function of MGDG (Kobayashi et al., 2007). Nearly the same lipid composition was observed in the complementing plants as in the wild type (Figure 7A), indicating that MgdA can restore the loss of function of MGD1 for the biosynthesis of bulk MGDG in chloroplasts. However, there was a small difference between the wild type and the complementing plants in their lipid composition. Specifically, the complementing plants contained 15% less MGDG than did the wild type. A reduced abundance in MGDG in the complementing plants was accompanied by an increased abundance in other major lipids, DGDG, phosphatidylethanolamine, and phosphatidylinositol. On the other hand, the abundance of sulfoquinovosyldiacylglycerol, phosphatidylglycerol, and phosphatidylcholine was not altered.
Figure 7.
Lipid Analyses of mgd1-2/35S:mgdA Transgenic and Wild-Type Plants.
(A) Comparison of polar glycerolipids in wild-type (white bars) and mgd1-2/35S:mgdA (black bars) plants. Values are the mean ± se from three independent measurements. PC, phosphatidylcholine; PE, phosphatidylethanolamine; PG, phosphatidylglycerol; PI, phosphatidylinositol; SQDG, sulfoquinovosyldiacylglycerol.
(B) Fatty acid composition of MGDG isolated from wild-type (white bars) and mgd1-2/35S:mgdA (black bars) plants. Values are the mean ± se from three independent measurements.
We also measured the fatty acid composition of MGDG isolated from the wild type and the complementing plants. The DAG used in MGDG synthesis is derived from two different biosynthetic pathways, the so-called prokaryotic and eukaryotic pathways (Benning and Ohta, 2005). MGDG from the prokaryotic pathway contains 16:3 at the sn-2 position, whereas MGDG from the eukaryotic pathway contains 18-carbon fatty acids at the sn-2 position; both forms contain mostly 18-carbon fatty acids at the sn-1 position. MGDG from the complementing plant contained ~50% less 16:3 (a characteristic constituent of the prokaryotic pathway) and 20% more 18:3 (the principal fatty acid in MGDG derived from the eukaryotic pathway) than did MGDG from the wild type (Figure 7B).
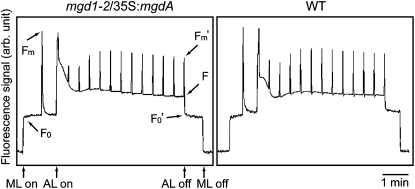
The mgd1-2 mutant cannot grow photoautotrophically because of impaired photosynthetic activities (Kobayashi et al., 2007). To analyze the properties of photosynthetic activity in the complementing line, we performed pulse amplitude modulation (PAM) fluorescence analysis on wild-type and complementing plants. Figure 8 shows representative traces for the PAM analysis. The fluorescence kinetics of the complementing line were almost identical to those of the wild type. In fact, there were no differences in the calculated values for the wild type and the complementing line for their maximal quantum yield of PSII (Fv/Fm), fraction of open PSII (qL), and nonphotochemical quenching. Thus, the photosynthetic activities were almost completely recovered in the complementing plant, although abnormal phenotypes were observed in its growth (Figure 5).
Figure 8.
PAM Analysis of mgd1-2/35S:mgdA Transgenic and Wild-Type Plants.
Saturating pulses were applied at intervals. AL, actinic light; arb. unit, arbitrary unit; ML, measuring light; WT, wild type.
To investigate the effects of the functional replacement of MGD1 with MgdA for thylakoid membrane biogenesis, plastid morphology in the complementing plants was analyzed by transmission electron microscopy. MGD1 is critical for plastid biogenesis; all plastids in the mgd1-2 mutant contain internal membrane structures that are either absent or severely undeveloped with abnormal shapes (Kobayashi et al., 2007). In the complementing plants, however, normal-like mature chloroplasts were always observed (Figure 9A), indicating that MgdA essentially restores the loss of function of MGD1 for chloroplast biogenesis. There were also abnormal plastids in the complementing plants that were rather small and contained electron-dense structures (Figure 9B). It is of note that both normal and abnormal plastids coexisted in the same cell (Figure 9C). These results indicated that MgdA-dependent MGDG synthesis had some effects on chloroplast biogenesis, although the origin of the unusual structures in the abnormal plastids was unclear. We also analyzed ultrastructure of plastids in roots (see Supplemental Figure 5 online) and shoot meristematic tissues (see Supplemental Figure 6 online) of the complementing plant. However, we did not see unusual plastids in the tissues.
Figure 9.
Ultrastructure of Plastids in Mature Leaves of the mgd1-2/35S:mgdA Transgenic Plants.
(A) An usual plastid found in the mgd1-2/35S:mgdA line. Bar = 500 nm.
(B) An unusual plastid found in the mgd1-2/35S:mgdA line. Bar = 500 nm.
(C) Colocalization of usual and usual plastids in a single cell. Bar = 500 nm.
DISCUSSION
In this study, we identified a novel MGDG synthase, MgdA, from the green sulfur bacterium C. tepidum through a computer-aided gene discovery system. MgdA is specifically conserved in green sulfur bacteria but not in other photosynthetic organisms, including cyanobacteria and plants. This indicates that although MGDG is conserved in many phototrophs, its biosynthetic enzymes have been independently established in each organism for adaptation to different environmental niches.
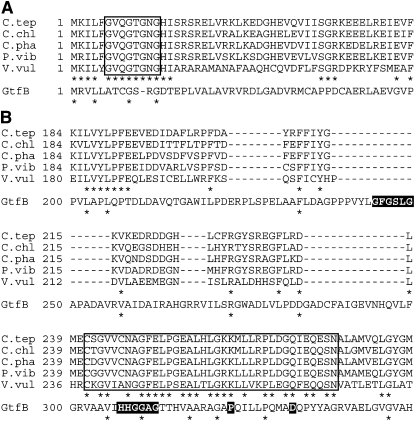
The phylogenetic analysis indicated that MgdA belongs to glycosyltransferase family GT1 (Figure 1B). The GT1 family contains a variety of enzymes with different enzymatic activities in different organisms (archaea, bacteria, virus, animal, plant, and fungus). Among them, GtfB, a UDP-glucosyltransferase that functions in an antibiotic (chloroeremomycin) glycosylation reaction, may be best characterized; the crystal structure of GtfB has been determined (Mulichak et al., 2001). The structure is comprised of two distinct N- and C-terminal domains, which are similar in topology and size. Both domains contain a Rossmann-like β/α/β domain, and each of these is suggested to harbor an acceptor or donor binding site. Although ligands are not in the crystal, it was suggested that a Gly fingerprint sequence immediately following the first β-strand of the C-terminal domain (G244-X-G246-XX-G249; Figure 10B) is important for association with the UDP-phosphate moiety for donor (UDP-Glc) binding. At the corresponding position on the GtfB N-terminal domain, a vestigial Gly-rich sequence was found (G9-XX-G12) as a nonfunctional motif (Figure 10A). However, an amino acid sequence alignment of MgdA homologs and GtfB indicated that the Gly fingerprint motif of the GtfB C-terminal domain, which potentially functions as a donor binding site, is not conserved in MgdA homologs (Figure 10B). By contrast, a typical Gly fingerprint sequence was found in the N-terminal β1 strand region of the MgdA sequence (G6-XX-G9-X-G11-X-G13; Figure 10A). These observations suggest that in the case of MgdA, a site for donor (UDP-Gal) binding is in the N-terminal domain. As for the acceptor binding site, we found a highly conserved region in the C-terminal domain (Cys-241 to Asn-277) of MgdA (Figure 10B). This region is actually the proposed donor binding site of GT1 family enzymes (Mulichak et al., 2001). The donor binding site for MgdA appears to be in its N-terminal domain, whereas the C-terminal highly conserved region of MgdA is a potential site for acceptor (DAG) binding, as opposed to other GT1 family enzymes in which the C-terminal region is involved in donor binding. In this region, the Gly-rich sequence (HHGGAGT) as well as the conserved Pro (Pro-323) and Asp (Asp-332) of GtfB are suggested to be involved in the binding of UDP-phosphate moieties (Mulichak et al., 2001). However, none of these sequences are conserved in MgdA, supporting the idea that the C-terminal region is not the donor binding site but is the acceptor binding site in MgdA.
Figure 10.
Partial Amino Acid Sequence Alignments of MgdA Homologs.
(A) The N-terminal region. The sequence of GtfB (chloroeremomycin gtfB; CAA11775.1) was also aligned as a reference. C.tep, C. tepidum (NP_661248.1); C.chl, C. chlorochromatii (ABB28672.1); C.pha, C. phaeobacteroides (YP_911154.1); P.vib, Prosthecochloris vibrioformis (ABP36524.1); V.vul, Vibrio vulnificus (AAO09163.1). The possible substrate binding site of MgdA is boxed. The conserved amino acids are indicated by asterisks.
(B) The C-terminal region. Gly-rich and Gly fingerprint sequences as well as conserved Pro and Asp residues of GtfB, which are suggested to be involved in substrate binding, are indicated by white letters. The possible substrate binding site of MgdA is boxed. The conserved amino acids are indicated by asterisks.
An analysis of the secondary structure of MgdA by a computer program (SOSUI; http://bp.nuap.nagoya-u.ac.jp/sosui) suggested that it is a nonmembrane spanning protein. However, protein gel blot analysis indicated that MgdA is localized to the cytoplasmic membrane (Figure 2A). Because one of the substrates (DAG) exists in membranes, it seems likely that MgdA attaches on the surface of the cytoplasmic membrane and catalyzes MGDG synthesis from DAG and UDP-Gal. The localization of MgdA also implies the mechanism of chlorosome biogenesis. Because MGDG is a major lipid of the chlorosome membrane system, its biosynthesis should be important for assembly of the unique light-harvesting complex. Our results indicated that MGDG synthesis is achieved on the cytoplasmic, but not the chlorosome, membrane. This observation supports a proposed chlorosome biogenesis model in which glycosyl lipids that have a high affinity for BChl pigments accumulate within the cytoplasmic membrane, and the chlorosome membrane is derived from the cytoplasmic leaflet of the cytoplasmic membrane by a mechanism similar to the biogenesis of the lipid body in eukaryotes (Hohmann-Marriott and Blankenship, 2007). Perhaps MgdA-dependent MGDG synthesis in cytoplasmic membrane is tightly coupled with chlorosome biogenesis. A knockdown mutation in mgdA resulted in some effects on the assembly of chlorosome pigments (Figure 4B), which supports this hypothesis. However, mgdA is conserved only in species of the phylum Chlorobi, although chlorosomes have been found in other photosynthetic bacteria from different phyla (Chloroflexi and Acidobacteria) (Blankenship and Matsuura, 2003; Bryant et al., 2007). This indicates that MgdA-dependent MGDG synthesis itself is not required for chlorosome assembly.
In this study, we achieved complementation of an Arabidopsis MGD1 mutant (mgd1-2) by C. tepidum MgdA that harbors the chloroplast transit peptide of MGD1 (Figure 5A). Protein gel blot analysis indicated that the expressed MgdA could be targeted to chloroplasts with cleavage of the transit peptide (Figure 5C). The targeted MgdA is localized on the outside of the chloroplast inner envelope membrane as examined by the trypsin protection assay (Figure 2B). This localization is same as that observed for the native MGD1 (Xu et al., 2005). MGDG synthases of higher plants are classified into two types, Type A and Type B (Benning and Ohta, 2005). Type A MGDG synthases, including Arabidopsis MGD1, are localized on the inner envelope membrane; on the other hand, Type B MGDG synthases do not have the transit peptide and are localized on the chloroplast outer envelope membrane. Because the MGD1 transit peptide may be cleaved at Leu-67 (Yamaryo et al., 2003), the targeted MgdA still contains MGD1-derived peptide (Leu-67 to Lys-142) at the N terminus. This suggests that the remaining N-terminal region is responsible for translocating MgdA to the outside of the inner envelope membrane. This idea is supported by the amino acid sequence alignment of several MGDG synthases (see Supplemental Figure 7 online) that shows that the Type A enzymes, but not Type B enzymes, show some similarities on the N-terminal region.
The complementing plants we obtained had almost normal levels of lipid constituents (Figure 7A), although the mgd1-2 mutant differs greatly in its lipid composition as compared with the wild type (Kobayashi et al., 2007), indicating that MgdA could complement the function of MGD1 for bulk MGDG synthesis. Given that MgdA uses DAG and UDP-Gal for MGDG synthesis (similar to MGD1; Figure 1A), it is not surprising that the substrates were successfully supplied for MgdA in the complementing line. However, MGDG content in the transgenic line was lower than that in the wild type (~85% of the wild type; Figure 7A). As expected, the MGDG deficiency was accompanied by an increase in the relative abundance of another major plastidic lipid, DGDG. In addition, nonplastidial lipids (phosphatidylethanolamine and phosphatidylinositol) in the mutant were also increased. This result suggests that the chloroplasts in the complementing plants make a smaller contribution to the total lipid pool than do those in wild-type plants. This may imply the chloroplasts in the complementing line are sometimes immature with small and unusual shapes, as was observed (Figure 9).
MGDG synthesized in the complementing plants contained ~50% less 16:3 and ~20% more 18:3 than did MGDG from wild-type plants (Figure 7B). Because 16:3 is a characteristic feature of MGDG derived from the prokaryotic pathway (Benning and Ohta, 2005), these data suggest that MgdA operates in the eukaryotic pathway more than in the prokaryotic pathway. In vitro analysis of the enzymatic activity of MgdA showed no selectivity for DAG from the prokaryotic (18:1/16:0) and eukaryotic (18:1/18:1) pathways (see Results) as observed in Arabidopsis MGD1 (Awai et al., 2001), which suggests that this may not be due to the different substrate specificities of the enzymes. Another possibility to explain the different composition of fatty acid content is to consider the longevity of the complementing plant (approximately twofold longer compared with the wild type). The fatty acid composition of MGDG is stage specific, such that MGDG in 5-d-old Arabidopsis shows ~50% less 16:3 and ~20% more 18:3 than does MGDG from 24-d-old Arabidopsis, although the exact reason is unclear (Awai et al., 2001). These observations suggest that the two MGDG synthetic pathways are regulated in growth stage–specific manners. MGDG synthesis in the complementing plants retained the properties of young wild-type seedlings with respect to substrate provision for MGDG synthesis, even during mature stages of growth.
Two types of chloroplasts were found in the complementing plants, as examined by electron microscopy (Figure 9). One shows normal structures, in which typical stacking thylakoid membranes were observed. The photosynthetic activity in these chloroplasts may be the same as that in wild-type chloroplasts because photosynthetic parameters of the complementing plants do not differ from those in the wild type (Figure 8). However, another type of chloroplast was found in the complementing plant; these chloroplasts are small and contain unusual structures. Formation of the unusual chloroplasts may result in the reduction of chlorophyll content in the complementing line (Figure 5F). It is of note that both normal and abnormal chloroplasts existed in the same cell (Figure 9C), indicating that these unusual structures were not due to different expression levels of MgdA. Perhaps MGD1 activity is tightly controlled at the posttranslational level in each chloroplast, which is important for complete chloroplast biogenesis. The stability of MGD1 is also suggestive to be important for normal chloroplast biogenesis. In fact, MGD1 activity is controlled by redox-dependent oxidation/reduction of disulfide bonds as well as the dissociation/association of phosphatidic acid (Yamaryo et al., 2006; Dubots et al., 2010). Cys residues are not conserved between MGD1 and MgdA, and MgdA activity was not affected by phosphatidic acid, suggesting that such regulatory mechanisms are important, but not essential, for normal chloroplast biogenesis. It was recently shown that chloroplast development is coupled with the activity of other cellular components, such as cytoskeleton and peroxisomes (Albrecht et al., 2010). One possibility is that such interaction is impaired in the abnormal chloroplasts. The complementing plants also showed unusual phenotypes in their growth, including no apical dominance in shoot growth (Figures 5H and 5I) and atypical flower development (Figure 5E; see Supplemental Figure 3 online), although unusual plastids could not be found in non-green tissues, such as roots and shoot meristematic tissues (see Supplemental Figures 5 and 6 online). These observations suggest that regulated MGDG synthesis is also important for normal plant development. Although some mechanisms that control MGD1 activity are firmly established (Yamaryo et al., 2006; Dubots et al., 2010), the physiological meaning of posttranslational regulation is still unclear. The complementing plants obtained in this study should be useful for further investigation of the importance of regulated MGDG synthesis.
In summary, the identification of a novel MGDG synthase from green sulfur bacteria provides a better understanding for mechanisms of chlorosome assembly in photosynthetic bacteria as well as of chloroplast biogenesis in higher plants. Further characterization of the enzyme should be important for gaining more insights into the significance of the mechanistic regulation of MGDG synthesis in the physiology of photosynthetic organisms.
METHODS
Construction of Bacterial Strains and Growth Conditions
All primers used in this study are listed in Supplemental Table 2 online. Chlorobaculum tepidum strain WT2321 (Wahlund and Madigan, 1995) was used as a parent wild-type strain. All mutant and wild-type strains of C. tepidum were grown anaerobically as described (Frigaard et al., 2004a). For growth curve measurements, MGDG (Larodan Fine Chemicals) dissolved in 99.5% ethanol was added to the medium. The same amount of ethanol without MGDG was added to the control medium. mgdA mutagenesis was performed as follows. The DNA fragment for mgdA was amplified by PCR using CT0344-F-NdeI and CT0344-R-EcoRI primers. The amplified fragment was digested with NdeI and EcoRI and cloned into pET28(a) vector (Novagen). The internal region of the cloned mgdA (amino acids 110 to 355) was replaced by a gentamicin resistance cassette, and the resulting plasmid was transferred into C. tepidum cells (Frigaard et al., 2004a). Possible mgdA mutants were selected based on gentamicin resistance, and the homologous recombination between mutagenized mgdA and chromosomal mgdA was verified by PCR using CT0344-F and CT0344-R primers. mgdA mRNA in the mutants was analyzed by RT-PCR. For this purpose, the wild type and mutants were grown until mid-log phase, and total mRNA was isolated using the SV Total RNA isolation system (Promega). Using the isolated RNA, cDNA was synthesized by the TaKaRa RNA PCR kit, version 3.0 (TaKaRa). Then, RT-PCR for mgdA mRNA was performed with CT0344-F-RTPCR and CT0344-R-RTPCR primers. For a control, rnpB mRNA was also analyzed by RT-PCR using rnpB-F-RTPCR and rnpB-R-RTPCR primers. Unsaturated RT-PCR products were analyzed by 1.2% agarose gel electrophoresis.
Quantitative Real-Time PCR
Total RNA was isolated from C. tepidum cells grown mid-log phase as described above. Quantitative real-time PCR was performed using the total RNA as templates, and CT0344-F-RTPCR and CT0344-R-RTPCR primers with PrimeScript RT reagent kit (TaKaRa) on a Thermal Cycler Dice real-time system (TaKaRa). The level of rnpB mRNA was used as an internal standard. The real-time PCR experiment was repeated using three independent biological replicates.
Construction of Transgenic Plants and Growth Conditions
All plants used were the Columbia ecotype of Arabidopsis thaliana. Surface-sterilized seeds were plated on 0.8% (w/v) agar-solidified Murashige and Skoog medium and were grown at 22°C with white light (25 μmol photons m–2 s–1). The complementing plant of mgd1-2 mutant with C. tepidum mgdA was constructed as described below. The DNA construct for the recombinant gene containing MGD1 transit peptide and MgdA (with no tag) was produced by a double-PCR strategy. Specifically, a coding region for the transit peptide of MGD1 was amplified from the MGD1 cDNA clone with ATTB-MGD1-F and MGD1-R-CT0344. The coding region for C. tepidum mgdA was also amplified using CT0344-F-MGD1 and CT0344-R-ATTB primers. The first PCR fragments for the MGD1 transit peptide and MgdA were mixed, and then the second PCR was performed with ATTB-F and ATTB-R primers. The second PCR product was cloned into pDONR/ZeO vector by use of the Gateway system (Invitrogen). After checking for the correct sequence of the inserted DNA, the fragment was cloned into pGWB2 vector (Nakagawa et al., 2007). The resulting construct (Figure 5A) was introduced into heterozygous mgd1-2 mutants via an Agrobacterium tumefaciens–mediated transformation method. Genotypes of mgd1-2 and MGD1 in the complementing plants (mgd1-2/35S:mgdA) were checked by genomic PCR with primers reported previously (Kobayashi et al., 2007). Primers for genotyping of mgdA were ATTB-F and MGD1-R-CT0344.
Activity Measurements
The coding regions for C. tepidum murG (CT0034) and mgdA (CT0344) were amplified by PCR using two primer pairs, CT0034-F-NdeI/CT0034-R-EcoRI and CT0344-F2-NdeI/CT0344-R-EcoRI, respectively. The PCR products digested with NdeI and EcoRI were cloned separately into pET28(a) vector (Novagen), and the resulting plasmids were named pETCT0034 (for murG) and pETCT0344 (for mgdA). The plasmid constructs were used to transform Escherichia coli strain BL21(DE3) (Merck). The E. coli expression strain for cucumber MGD1 was reported previously (Shimojima et al., 1997). The expression of the proteins was induced for 3 h at 37°C with 1 mM isopropyl 1-thio-d-galactopyranoside (TaKaRa). The expressed MgdA was purified from soluble fraction using His-bind resin (Merck) according to the manufacturer’s instructions. Cell-free extracts of the induced cells or purified MgdA were mixed with DAG (1 mg mL–1; dispersed in 0.01% Tween 20) in Buffer A, which contains 0.1 M MOPS/NaOH, pH 7.8, 5 mM MgCl2, and 5 mM DTT, and incubated for 5 min at 30°C. Then, the reaction was started by addition of 0.4 mM UDP-[14C]Gal or -[14C]Glc (~300 Bq/nmol). After incubation for 1 h at 30°C, the reaction was stopped by vortexing with 5 volumes of ethyl acetate. The organic phase was washed with 0.45% (w/v) NaCl, and the upper organic layer was used for TLC analysis with a solvent system of chloroform/hexane/tetrahydrofuran/isopropanol/water (25:50:0.5:40:1, v/v/v/v/v) (Hölzl et al., 2005). Radioactive spots were detected using an imaging plate (Fuji Film). For analyzing substrate selectivity of MgdA, a cell-free extract from the E. coli strain that expresses MgdA was further subjected to ultracentrifugation at 60,000 rpm (~150,000g) using the TA100.3 rotor (Beckman) for 30 min at 4°C. The membrane fraction (precipitate) was suspended in an aliquot of Buffer A and used for activity measurements. After stopping the reaction, the radiolabeled products in each organic phase were quantified with a LS6500 scintillation counter (Beckman). The kinetic parameters for catalysis by MgdA were determined by monitoring the initial rate of MGDG production using the following equation:
where ν is the initial rate, [S] is the substrate (UDP-Gal) concentration, Vmax is the initial rate achieved as [S] approaches infinity, and Km is the apparent value of [S] giving Vmax/2. Obtained data shown in Supplemental Figure 8 online were fitted, and apparent Km values (± se) were calculated using the SIGMAPLOT program (Systat Software). For MGDG synthase activity measurement of C. tepidum, cells were grown until mid-log phase at 40°C, suspended in Buffer A, and broken by sonication. Obtained cell-free extracts were used for the measurement, and the activity was quantified using a scintillation counter as described above. The protein concentration was measured by the Bradford assay kit (Bio-Rad) using BSA as a standard.
Immunoblotting and Chlorophyll Determination
MgdA antibody was prepared as follows: His-tagged MgdA was overexpressed in E. coli strain BL21(DE3)/pETCT0344 (see above) at 37°C for 3 h. The expressed protein was purified from inclusion bodies using His-bind resin (Merck). A rabbit was immunized with ~0.5 mg of the purified His-tagged MgdA, and the antiserum obtained was used for protein gel blot analysis. The anti-CRSH antibody was described previously (Masuda et al., 2008). Samples for protein gel blotting were obtained as follows. Rosette leaves were homogenized in 20 mM phosphate buffer, pH 8.0, containing 50 mM NaCl and 0.4 M Suc, and the homogenate was used as a whole-leaf extract. The homogenate was filtered through Miracloth (Calbiochem) and then centrifuged at 3000g for 10 min. The precipitates were used as the chloroplast-enriched fraction for protein gel blot analysis. For trypsin protection assay, intact chloroplasts were further isolated as described (Douce and Joyard, 1982), suspended in homogenized buffer containing 50 mg L−1 trypsin and 1 mM CaCl2, and incubated for 10 min on ice. Chloroplasts were reisolated by centrifuged at 3000g for 10 min and then subjected to SDS-PAGE. For samples of C. tepidum, cells grown until mid-log phase were harvested and suspended in 20 mM Tris-HCl buffer, pH 7.8, containing 2 M NaSCN. The whole-cell extract, precipitates, and soluble, chlorosome and cytoplasmic membrane fractions were obtained according the methods described (Gerola and Olson, 1986; Wang et al., 1995). Absorption spectra of the isolated chlorosome and cytoplasmic membrane fractions are shown in Supplemental Figure 9 online. The purity of the isolated chlorosome and cytoplasmic membrane fractions could be estimated by the presence and/or absence of BChl c (λmax = ~750 nm) and BChl a (λmax = ~820 nm) in each fraction, since most parts of BChl c and BChl a molecules exist in chlorosomes and cytoplasmic membrans, respectively (Frigaard et al., 2003). The spectrum of the chlorosome fraction shows no detectable BChl a absorbance, indicating that chlorosomes were enriched in the fraction. The relative absorbance ratio of BChl a to BChl c in the cytoplasmic membrane fraction was ~10-fold higher than that in the native cells (Figure 4B), indicating that the cytoplasmic membranes were highly enriched in the fraction. To remove the large amount of pigments, the fractions were mixed with methanol (>80%, v/v). Following centrifugation at 27,500g for 30 min, the precipitates were resuspended in 8 M urea. An equal amount of proteins (5 to 15 μg) was separated on an SDS-PAGE and electroblotted onto a polyvinylidene difluoride membrane. Bands immunoreactive against anti-MgdA and anti-CRSH were detected using the ECL system (GE Healthcare). Chlorophyll content was determined as described (Arnon, 1949) with three biological replications.
Lipid Analysis
Total lipids were extracted from 12-d-old wild-type and 36-d-old mgdA-complemented mgd1-2 plants as described (Bligh and Dyer, 1959). Lipids were separated by two-dimensional TLC using the following solvent systems: chloroform/methanol/7 M ammonia (15:10:1, v/v/v) for the first dimension and chloroform/methanol/acetic acid/water (170:20:17:3, v/v/v/v) for the second dimension. Fatty acid detection and measurement of membrane lipid content were performed as described (Kobayashi et al., 2006). MGDG separated by TLC was further subjected to NMR analysis. 1H NMR spectra were recorded at 600 MHz (Bruker AV-600). Internal tetramethylsilane (δ = 0 ppm) in CDCl3 was used as a standard. Chemical shifts were expressed in parts per million with respect to the standard. The assignments are based on COSY spectra, which are as follows. For mgd1-2/35S:mgdA; 1H NMR (CDCl3): d 4.39 (dd, J2',3′a = 3.5 Hz, J3′a,3′b = 12.1 Hz, 1H, H-3′a), 4.28 (d, J1,2 = 7.5 Hz, 1H, H-1), 4.22 (dd, J2',3′b = 6.5 Hz, J3′a,3′b = 12.1 Hz, 1H, H-3′b), 4.01 (br, 1H, H-4), 3.99 (dd, J5,6a = 5.9 Hz, J6a,6b = 12.2 Hz, 1H, H-6a), 3.92 (dd, J1'a,2' = 5.5 Hz, J1'a,1'b = 11.2 Hz, 1H, H-1’a), 3.87 (dd, J5,6b = 3.8 Hz, J6a,6b = 12.2 Hz, 1H, H-6b), 3.75 (dd, J1'b,2' = 6.3 Hz, J1'a,1'b = 11.2 Hz, 1H, H-1’b), 3.65 (dd, J1,2 = 7.5 Hz, J2,3 = 9.0 Hz, 1H, H-2), 3.55-3.60 (m, 2H, H-3, H-5). For C. tepidum; 1H NMR (CDCl3): d 4.40 (dd, J2',3′a = 3.3, J3′a,3′b = 12.0 Hz, 1H, H-3′a), 4.29 (d, J1,2 = 7.5 Hz, 1H, H-1), 4.22 (dd, J2',3′b = 6.4 Hz, J3′a,3′b = 12.0 Hz, 1H, H-3′b), 4.01 (br, 1H, H-4), 3.99 (dd, J5,6a = 5.7 Hz, J6a,6b = 11.8 Hz, 1H, H-6a), 3.91 (dd, J1'a,2' = 5.3 Hz, J1'a,1'b = 11.5 Hz, 1H, H-1’a), 3.87 (dd, J5,6b = 3.4 Hz, J6a,6b = 11.8 Hz, 1H, H-6b), 3.75 (dd, J1'b,2' = 6.6 Hz, J1'a,1'b = 11.5 Hz, 1H, H-1’b), 3.65 (dd, J1,2 = 7.5 Hz, J2,3 = 9.3 Hz, 1H, H-2), 3.52-3.60 (m, 2H, H-3, H-5).
Fluorescence and Microscopy Analyses
PAM fluorescence analysis was performed with MINI PAM (Heinz Walz). Modulated measuring light of 650 nm was used at a setting of 6 with gain 8. Actinic light with a setting 4 (Fact 0.50) was used to drive photosynthesis. Pulses (0.8 s) of white light at a setting of 4 at 25-s intervals were applied to obtain maximal fluorescence.
Transmission electron microscopy of 30-d-old mgdA-complemented mgd1-2 plants was performed (Kobayashi et al., 2007). Briefly, leaf segments were fixed with 4% (v/v) paraformaldehyde and 1% (v/v) glutaraldehyde in 50 mM cacodylate buffer, pH 7.4. The ultrathin sections were analyzed by a transmission electron microscope (model H-7600; Hitachi High-Technologies). For electron microscopy analysis of chlorosomes, chlorosomes isolated from the wild-type and #5 mutant were adsorbed on a Formvar-coated copper grid and negatively stained with 2% (w/v) uranyl acetate (Bryant et al., 2002).
Phylogenetic Analysis
The amino acid sequence alignment was first produced with PineAlign (Plewniak et al., 2003) and then manually adjusted to optimize the alignment. The final alignment is shown in Supplemental Figure 10 and Supplemental Data Set 1 online. The phylogenetic tree was constructed by MEGA4 (Tamura et al., 2007). All gaps in the sequence alignment were omitted in a complete deletion manner, and the construction of the tree was performed by neighbor-joining methods with the substitution model for Poisson rates.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database under the following accession numbers: Arabidopsis MGDG synthase (AAF65066.1), cucumber MGDG synthase (U62622), and a MurG ortholog of C. tepidum (NP_660940.1). Accession numbers of the amino acid sequences used for phylogenetic analysis are as follows: bGtfB (balhimycin gtfB, CAA76552.1), cGtfB (chloroeremomycin gtfB, CAA11775.1), C. phaeobacteroides MgdA (YP_911154.1), C. tepidum MgdA (NP_661248.1), Arabidopsis MGD1 (NP_849482.1), T4 phage βGT (NP_049658.1), Geobacter metallireducens MurG (YP_383380.1), E. coli MurG (AP_000753.1), and Arabidopsis MGD3 (NP_001118301.1).
Supplemental Data
The following materials are available in the online version of the article.
Supplemental Figure 1. Purification of His-Tagged MgdA Expressed in E. coli.
Supplemental Figure 2. Immunoblot Analysis of the mgd1-2/35S:mgdA Line.
Supplemental Figure 3. Flowers of the mgd1-2/35S:mgdA Line.
Supplemental Figure 4. Phenotype of Wild Type/35S:mgdA and mgd1-2/35S:mgdA (Line 2).
Supplemental Figure 5. Ultrastructure of Plastids in Roots.
Supplemental Figure 6. Ultrastructure of Plastids in Shoot Apical Meristematic Tissues.
Supplemental Figure 7. A Partial Amino Acid Sequence Alignment of MGDG Synthases of Higher Plants.
Supplemental Figure 8. Graphical Representation of the Data Used to Determine Km Values of MgdA.
Supplemental Figure 9. Absorption Spectra of Isolated Chlorosome and Cytoplasmic Membrane Fractions.
Supplemental Figure 10. The Amino Acid Sequence Alignment Used for Phylogenetic Tree Construction.
Supplemental Table 1. Organisms Whose Genome Sequences Were Used for CCCT Analysis.
Supplemental Table 2. Primers Used in This Study.
Supplemental Data Set 1. Text File of the Sequences and Alignment Used for the Phylogenetic Analysis Shown in Figure 1B.
Acknowledgments
We thank Shukun Ren and Keiko Yamamichi for excellent technical assistance. This work was supported in part by a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan to S.M. (23770038) and M.N. (21590321).
AUTHOR CONTRIBUTIONS
S.M. and J.H. designed the research. S.M., J.H., M.Y., Y.Y., M.S., K.M., H.T., H.M., M.M., T.H., and M.K. performed research. All authors analyzed data, and S.M. wrote the article.
References
- Albrecht V., Simková K., Carrie C., Delannoy E., Giraud E., Whelan J., Small I.D., Apel K., Badger M.R., Pogson B.J. (2010). The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22: 3423–3438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnon D.I. (1949). Cooper enzymes in isolated chloroplasts. Polyphenol oxidase in Beta vulgaris. Plant Physiol. 24: 1–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awai K., Kakimoto T., Awai C., Kaneko T., Nakamura Y., Takamiya K., Wada H., Ohta H. (2006). Comparative genomic analysis revealed a gene for monoglucosyldiacylglycerol synthase, an enzyme for photosynthetic membrane lipid synthesis in cyanobacteria. Plant Physiol. 141: 1120–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Awai K., Maréchal E., Block M.A., Brun D., Masuda T., Shimada H., Takamiya K., Ohta H., Joyard J. (2001). Two types of MGDG synthase genes, found widely in both 16:3 and 18:3 plants, differentially mediate galactolipid syntheses in photosynthetic and nonphotosynthetic tissues in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98: 10960–10965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C., Ohta H. (2005). Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J. Biol. Chem. 280: 2397–2400 [DOI] [PubMed] [Google Scholar]
- Berg S., Edman M., Li L., Wikström M., Wieslander A. (2001). Sequence properties of the 1,2-diacylglycerol 3-glucosyltransferase from Acholeplasma laidlawii membranes. Recognition of a large group of lipid glycosyltransferases in eubacteria and archaea. J. Biol. Chem. 276: 22056–22063 [DOI] [PubMed] [Google Scholar]
- Blankenship R.E., Matsuura K. (2003). Antenna complexes from green photosynthetic bacteria. Light-Harvesting Antennas in Photosynthesis, Green B.R., Parson W.W., (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 195–217 [Google Scholar]
- Blankenship R.E., Olson J.M., Miller M. (1995). Antenna complexes from green photosynthetic bacteria. Anoxygenic Photosynthetic Bacteria, Blankenship R.E., Madigan M.T., Bauer C.E., (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp 399–435 [Google Scholar]
- Bligh E.G., Dyer W.J. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37: 911–917 [DOI] [PubMed] [Google Scholar]
- Block M.A., Dorne A.J., Joyard J., Douce R. (1983). Preparation and characterization of membrane fractions enriched in outer and inner envelope membranes from spinach chloroplasts. II. Biochemical characterization. J. Biol. Chem. 258: 13281–13286 [PubMed] [Google Scholar]
- Bryant D.A., Costas A.M., Maresca J.A., Chew A.G., Klatt C.G., Bateson M.M., Tallon L.J., Hostetler J., Nelson W.C., Heidelberg J.F., Ward D.M. (2007). Candidatus Chloracidobacterium thermophilum: An aerobic phototrophic Acidobacterium. Science 317: 523–526 [DOI] [PubMed] [Google Scholar]
- Bryant D.A., Frigaard N.U. (2006). Prokaryotic photosynthesis and phototrophy illuminated. Trends Microbiol. 14: 488–496 [DOI] [PubMed] [Google Scholar]
- Bryant D.A., Vassilieva E.V., Frigaard N.U., Li H. (2002). Selective protein extraction from Chlorobium tepidum chlorosomes using detergents. Evidence that CsmA forms multimers and binds bacteriochlorophyll a. Biochemistry 41: 14403–14411 [DOI] [PubMed] [Google Scholar]
- Coutinho P.M., Deleury E., Davies G.J., Henrissat B. (2003). An evolving hierarchical family classification for glycosyltransferases. J. Mol. Biol. 328: 307–317 [DOI] [PubMed] [Google Scholar]
- Dörmann P., Benning C. (2002). Galactolipids rule in seed plants. Trends Plant Sci. 7: 112–118 [DOI] [PubMed] [Google Scholar]
- Dörmann P., Hoffmann-Benning S., Balbo I., Benning C. (1995). Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7: 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douce R., Joyard J. (1982). Methods in Chloroplast Molecular Biology. (Grenoble, France: Elsevier Biomedical Press) [Google Scholar]
- Dubots E., Audry M., Yamaryo Y., Bastien O., Ohta H., Breton C., Maréchal E., Block M.A. (2010). Activation of the chloroplast monogalactosyldiacylglycerol synthase MGD1 by phosphatidic acid and phosphatidylglycerol. J. Biol. Chem. 285: 6003–6011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen J.A., et al. (2002). The complete genome sequence of Chlorobium tepidum TLS, a photosynthetic, anaerobic, green-sulfur bacterium. Proc. Natl. Acad. Sci. USA 99: 9509–9514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N.U., Bryant D.A. (2001). Chromosomal gene inactivation in the green sulfur bacterium Chlorobium tepidum by natural transformation. Appl. Environ. Microbiol. 67: 2538–2544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N.U., Bryant D.A. (2006). Chlorosomes: Antenna organelles in photosynthetic green bacteria. Microbiology Monographs, Shively J.M., (Berlin: Springer; ), pp. 79–114 [Google Scholar]
- Frigaard N.U., Li H., Milks K.J., Bryant D.A. (2004b). Nine mutants of Chlorobium tepidum each unable to synthesize a different chlorosome protein still assemble functional chlorosomes. J. Bacteriol. 186: 646–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigaard N.U., Sakuragi Y., Bryant D.A. (2004a). Gene inactivation in the cyanobacterium Synechococcus sp. PCC 7002 and the green sulfur bacterium Chlorobium tepidum using in vitro-made DNA constructs and natural transformation. Methods Mol. Biol. 274: 325–340 [DOI] [PubMed] [Google Scholar]
- Frigaard N.U., Chew A.G., Li H., Maresca J.A., Bryant D.A. (2003). Chlorobium tepidum: Insights into the structure, physiology, and metabolism of a green sulfur bacterium derived from the complete genome sequence. Photosynth. Res. 78: 93–117 [DOI] [PubMed] [Google Scholar]
- Ganapathy S., Oostergetel G.T., Wawrzyniak P.K., Reus M., Gomez Maqueo Chew A., Buda F., Boekema E.J., Bryant D.A., Holzwarth A.R., de Groot H.J. (2009). Alternating syn-anti bacteriochlorophylls form concentric helical nanotubes in chlorosomes. Proc. Natl. Acad. Sci. USA 106: 8525–8530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerola P.D., Olson J.M. (1986). A new bacteriochlorophyll a-protein complex associated with chlorosomes of green sulfur bacteria. Biochim. Biophys. Acta 848: 69–76 [DOI] [PubMed] [Google Scholar]
- Green B.R. (2003). The evolution of light-harvesting antennas. Light-Harvesting Antennas in Photosynthesis, Green B.R., Parson W.W., (Dordrecht, The Netherlands: Kluwer Academic Publishers; ), pp. 129–268 [Google Scholar]
- Guskov A., Kern J., Gabdulkhakov A., Broser M., Zouni A., Saenger W. (2009). Cyanobacterial photosystem II at 2.9-A resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16: 334–342 [DOI] [PubMed] [Google Scholar]
- Harada J., Miyago S., Mizoguchi T., Azai C., Inoue K., Tamiaki H., Oh-oka H. (2008). Accumulation of chlorophyllous pigments esterified with the geranylgeranyl group and photosynthetic competence in the CT2256-deleted mutant of the green sulfur bacterium Chlorobium tepidum. Photochem. Photobiol. Sci. 7: 1179–1187 [DOI] [PubMed] [Google Scholar]
- Hohmann-Marriott M.F., Blankenship R.E. (2007). Hypothesis on chlorosome biogenesis in green photosynthetic bacteria. FEBS Lett. 581: 800–803 [DOI] [PubMed] [Google Scholar]
- Hölzl G., Zähringer U., Warnecke D., Heinz E. (2005). Glycoengineering of cyanobacterial thylakoid membranes for future studies on the role of glycolipids in photosynthesis. Plant Cell Physiol. 46: 1766–1778 [DOI] [PubMed] [Google Scholar]
- Ito H., Yokono M., Tanaka R., Tanaka A. (2008). Identification of a novel vinyl reductase gene essential for the biosynthesis of monovinyl chlorophyll in Synechocystis sp. PCC6803. J. Biol. Chem. 283: 9002–9011 [DOI] [PubMed] [Google Scholar]
- Jarvis P., Dörmann P., Peto C.A., Lutes J., Benning C., Chory J. (2000). Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc. Natl. Acad. Sci. USA 97: 8175–8179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jochum T., Reddy C.M., Eichhöfer A., Buth G., Szmytkowski J., Kalt H., Moss D., Balaban T.S. (2008). The supramolecular organization of self-assembling chlorosomal bacteriochlorophyll c, d, or e mimics. Proc. Natl. Acad. Sci. USA 105: 12736–12741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorasch P., Warnecke D.C., Lindner B., Zahringer U., Heinz E. (2000). Novel processive and nonprocessive glycosyltransferases from Staphylococcus aureus and Arabidopsis thaliana synthesize glycoglycerolipids, glycophospholipids, glycosphingolipids and glycosylsterols. Eur. J. Biochem. 267: 3770–3783 [DOI] [PubMed] [Google Scholar]
- Jorasch P., Wolter F.P., Zähringer U., Heinz E. (1998). A UDP glucosyltransferase from Bacillus subtilis successively transfers up to four glucose residues to 1,2-diacylglycerol: expression of ypfP in Escherichia coli and structural analysis of its reaction products. Mol. Microbiol. 29: 419–430 [DOI] [PubMed] [Google Scholar]
- Jordan P., Fromme P., Witt H.T., Klukas O., Saenger W., Krauss N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Awai K., Nakamura M., Nagatani A., Masuda T., Ohta H. (2009). Type-B monogalactosyldiacylglycerol synthases are involved in phosphate starvation-induced lipid remodeling, and are crucial for low-phosphate adaptation. Plant J. 57: 322–331 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Masuda T., Takamiya K., Ohta H. (2006). Membrane lipid alteration during phosphate starvation is regulated by phosphate signaling and auxin/cytokinin cross-talk. Plant J. 47: 238–248 [DOI] [PubMed] [Google Scholar]
- Kobayashi K., Kondo M., Fukuda H., Nishimura M., Ohta H. (2007). Galactolipid synthesis in chloroplast inner envelope is essential for proper thylakoid biogenesis, photosynthesis, and embryogenesis. Proc. Natl. Acad. Sci. USA 104: 17216–17221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2005). Towards complete cofactor arrangement in the 3.0 A resolution structure of photosystem II. Nature 438: 1040–1044 [DOI] [PubMed] [Google Scholar]
- Loll B., Kern J., Saenger W., Zouni A., Biesiadka J. (2007). Lipids in photosystem II: Interactions with protein and cofactors. Biochim. Biophys. Acta 1767: 509–519 [DOI] [PubMed] [Google Scholar]
- Maresca J.A., Bryant D.A. (2006). Two genes encoding new carotenoid-modifying enzymes in the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 188: 6217–6223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S., Mizusawa K., Narisawa T., Tozawa Y., Ohta H., Takamiya K. (2008). The bacterial stringent response, conserved in chloroplasts, controls plant fertilization. Plant Cell Physiol. 49: 135–141 [DOI] [PubMed] [Google Scholar]
- Mengin-Lecreulx D., Texier L., van Heijenoort J. (1990). Nucleotide sequence of the cell-envelope murG gene of Escherichia coli. Nucleic Acids Res. 18: 2810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi T., Yoshitomi T., Harada J., Tamiaki H. (2011). Temperature- and time-dependent changes in the structure and composition of glycolipids during the growth of the green sulfur photosynthetic bacterium Chlorobaculum tepidum. Biochemistry 50: 4504–4512 [DOI] [PubMed] [Google Scholar]
- Mulichak A.M., Losey H.C., Walsh C.T., Garavito R.M. (2001). Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics. Structure 9: 547–557 [DOI] [PubMed] [Google Scholar]
- Nakagawa T., Kurose T., Hino T., Tanaka K., Kawamukai M., Niwa Y., Toyooka K., Matsuoka K., Jinbo T., Kimura T. (2007). Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J. Biosci. Bioeng. 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Oliver J.D., Kaper J. (2001). Vibrio species. Food Microbiology: Fundamentals and Frontiers, Doyle M.P., Beuchat L.R., Montville T.J., (Washington, DC: ASM Press; ), pp. 263–300 [Google Scholar]
- Pedersen M.O., Linnanto J., Frigaard N.U., Nielsen N.C., Miller M. (2010). A model of the protein-pigment baseplate complex in chlorosomes of photosynthetic green bacteria. Photosynth. Res. 104: 233–243 [DOI] [PubMed] [Google Scholar]
- Pedersen M.O., Underhaug J., Dittmer J., Miller M., Nielsen N.C. (2008). The three-dimensional structure of CsmA: A small antenna protein from the green sulfur bacterium Chlorobium tepidum. FEBS Lett. 582: 2869–2874 [DOI] [PubMed] [Google Scholar]
- Plewniak F., et al. (2003). PipeAlign: A new toolkit for protein family analysis. Nucleic Acids Res. 31: 3829–3832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saga Y., Shibata Y., Tamiaki H. (2010). Spectral properties of single light-harvesting complexes in bacterial photosynthesis. J. Photochem. Photobiol. C. 11: 15–24 [Google Scholar]
- Sato N., Murata N. (1982). Lipid biosynthesis in the blue-green alga, Anabaena variabilis I. Lipid classes. Biochim. Biophys. Acta 710: 271–278 [Google Scholar]
- Shimojima M., Ohta H., Iwamatsu A., Masuda T., Shioi Y., Takamiya K. (1997). Cloning of the gene for monogalactosyldiacylglycerol synthase and its evolutionary origin. Proc. Natl. Acad. Sci. USA 94: 333–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen P.G., Cox R.P., Miller M. (2008). Chlorosome lipids from Chlorobium tepidum: Characterization and quantification of polar lipids and wax esters. Photosynth. Res. 95: 191–196 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tronrud D.E., Wen J., Gay L., Blankenship R.E. (2009). The structural basis for the difference in absorbance spectra for the FMO antenna protein from various green sulfur bacteria. Photosynth. Res. 100: 79–87 [DOI] [PubMed] [Google Scholar]
- Umena Y., Kawakami K., Shen J.R., Kamiya N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473: 55–60 [DOI] [PubMed] [Google Scholar]
- Wahlund T.M., Madigan M.T. (1995). Genetic transfer by conjugation in the thermophilic green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 177: 2583–2588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Y., Marx G., Umetsu M., Kobayashi M., Mimuro M., Nozawa T. (1995). Morphology and spectroscopy of chlorosomes from Chlorobium tepidum by alcohol treatments. Biochim. Biophys. Acta 1232: 187–1967488640 [Google Scholar]
- Wen J., Zhang H., Gross M.L., Blankenship R.E. (2009). Membrane orientation of the FMO antenna protein from Chlorobaculum tepidum as determined by mass spectrometry-based footprinting. Proc. Natl. Acad. Sci. USA 106: 6134–6139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Fan J., Froehlich J.E., Awai K., Benning C. (2005). Mutation of the TGD1 chloroplast envelope protein affects phosphatidate metabolism in Arabidopsis. Plant Cell 17: 3094–3110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Fan J., Riekhof W., Froehlich J.E., Benning C. (2003). A permease-like protein involved in ER to thylakoid lipid transfer in Arabidopsis. EMBO J. 22: 2370–2379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaryo Y., Kanai D., Awai K., Shimojima M., Masuda T., Shimada H., Takamiya K., Ohta H. (2003). Light and cytokinin play a co-operative role in MGDG synthesis in greening cucumber cotyledons. Plant Cell Physiol. 44: 844–855 [DOI] [PubMed] [Google Scholar]
- Yamaryo Y., Motohashi K., Takamiya K., Hisabori T., Ohta H. (2006). In vitro reconstitution of monogalactosyldiacylglycerol (MGDG) synthase regulation by thioredoxin. FEBS Lett. 580: 4086–4090 [DOI] [PubMed] [Google Scholar]
- Yoshitomi T., Mizoguchi T., Kunieda M., Tamiaki H. (2011). Characterization of glycolipids in light-harvesting chlorosomes from the green photosynthetic bacterium Chlorobium tepidum. Bull. Chem. Soc. Jpn. 84: 395–402 [Google Scholar]