Abstract
Interleukin-6 (IL-6) is a proinflammatory cytokine that exerts a wide range of cellular, physiological, and pathophysiological responses. Pyrrolidine dithiocarbamate (PDTC) antagonizes the cellular responsiveness to IL-6 through impairment in signal transducer and activator of transcription-3 activation and downstream signaling. To further elucidate the biological properties of PDTC, global gene expression profiling of human HepG2 hepatocellular carcinoma cells was carried out after treatment with PDTC or IL-6 for up to 8 h. Through an unbiased pathway analysis method, gene array analysis showed dramatic and temporal differences in expression changes in response to PDTC versus IL-6. A significant number of genes associated with metabolic pathways, inflammation, translation, and mitochondrial function were changed, with ribosomal protein genes and DNA damage-inducible transcript 4 protein (DDIT4) primarily up-regulated with PDTC but down-regulated with IL-6. Quantitative polymerase chain reaction and Western blot analyses validated the microarray data and showed the reciprocal expression pattern of the mammalian target of rapamycin (mTOR)-negative regulator DDIT4 in response to PDTC versus IL-6. Cell treatment with PDTC resulted in a rapid and sustained activation of Akt and subsequently blocked the IL-6-mediated increase in mTOR complex 1 function through up-regulation in DDIT4 expression. Conversely, down-regulation of DDIT4 with small interfering RNA dampened the capacity of PDTC to block IL-6-dependent mTOR activation. The overall protein biosynthetic capacity of the cells was severely blunted by IL-6 but increased in a rapamycin-independent pathway by PDTC. These results demonstrate a critical effect of PDTC on mTOR complex 1 function and provide evidence that PDTC can reverse IL-6-related signaling via induction of DDIT4.
Introduction
Pyrrolidine dithiocarbamate (PDTC) is a clinically tolerated small thiol compound with antioxidant and anti-inflammatory properties (Chabicovsky et al., 2010). It has been suggested that PDTC prevents dyslipidemia and renal lesions in rats fed a high-fat diet, most likely because of attenuation of proinflammatory gene expression and improvement of metabolic parameters (Ebenezer et al., 2009). Glucose-stimulated insulin secretion in human islets exposed to high glucose is restored after exposure to PDTC (Maedler et al., 2002). Furthermore, PDTC provides neuroprotection in hypoxic-ischemic injury and against liver injury during intestinal ischemia and reperfusion in rats (Nurmi et al., 2006; Tian et al., 2006). Although some studies have suggested that PDTC significantly reduces inflammatory processes through the inhibition of the transcription factor nuclear factor-κB (Schreck et al., 1992; Cuzzocrea et al., 2002; Ebenezer et al., 2009), this has not been observed in all studies (Malm et al., 2007; Huang et al., 2008). In fact, PDTC confers adaptive protection of stressed cells from proinflammatory conditions through activation of the metal-activated transcription factor heat shock factor 1 (HSF1) (Song et al., 2010). Moreover, PDTC is a potent inhibitor of interleukin-6 (IL-6) transcriptional activity, thereby leading to decreased synthesis of type II acute-phase proteins in the human HepG2 hepatocellular carcinoma cell line (He et al., 2006; Xie et al., 2009). Collectively, these studies suggest that PDTC is a potent pharmacological agent with complex biological functions in the context of inflammation and other stressors. There is, however, no comprehensive information on the effects of PDTC on global gene expression profiles and related biological processes in hepatocytes.
The proinflammatory role of IL-6 is initiated by binding to its cell surface receptor with subsequent activation of three canonical signaling pathways (Neurath and Finotto, 2011). IL-6 activates Janus family kinases, which leads to phosphorylation of a latent pool of signal transducer and activator of transcription-3 (STAT3) at Tyr705, promoting its nuclear translocation, DNA binding, and subsequent target gene expression. The phosphorylation of phosphoinositol 3-kinase in response to IL-6 results in Akt activation and, hence, cross-talks with growth factor signaling pathways. In addition, IL-6 activates the pro-oncogenic Ras/Raf/mitogen-activated protein kinase kinase/extracellular signal-regulated kinase 1/2 signaling pathway. Given the notion that inflammation may serve as a precursor to many human diseases (e.g., cancer and type 2 diabetes), it is likely that down-modulation in IL-6 signaling pathway may have therapeutic value against pathological inflammatory conditions.
Our recent work clearly established that treatment of HepG2 cells with PDTC elicits rapid change in the expression of stress-related genes through up-regulation of the HSF1 transcription factor (Song et al., 2010). In that study, it was found that genes encoding molecular chaperones and cochaperones were activated rapidly in response to PDTC (within 1–4 h) and the corresponding transcripts were made soon after stimulation by PDTC. Here, we opted to carry out gene profiling to gain insight into the nature of the genes that were modulated transiently and rapidly (up to 8 h) in response to PDTC versus IL-6, because many of these genes probably will encode transcription factors and coregulators, secreted proteins, enzymes, and other proteins involved in the early regulation of cellular homeostasis. We report that PDTC time-dependently induced significant and qualitative changes in gene expression that were remarkably different to the effect of IL-6 in HepG2 cells. PDTC was found to inhibit the IL-6-dependent increase in mammalian target of rapamycin complex 1 (mTORC1) activity by preventing the reduction in the expression of DDIT4, a negative regulator of the mTOR. The negative regulation of mTORC1 involves the binding of DDIT4 to the regulatory molecule 14-3-3 and the subsequent release of the tumor suppressor tuberous sclerosis complex (TSC) 2, which is then free to form a complex with TSC1 and attenuate mTORC1 activity (DeYoung et al., 2008). mTORC1 plays a key role in the overall protein biosynthetic capacity (Foster and Fingar, 2010), and genes encoding ribosomal proteins and translation factors are often coregulated to efficiently adjust the global rates of protein synthesis of the cell (Mayer and Grummt, 2006; Jastrzebski et al., 2007). Here, additional experiments showed significant differences in the pattern of expression of ribosomal protein genes and global translational activity in response to treatment with PDTC versus IL-6.
Materials and Methods
Cell Lines.
Human HepG2 and PANC-1 cells were purchased from the American Type Culture Collection (Manassas, VA). HepG2 cells were maintained in minimal essential medium (Invitrogen, Carlsbad, CA) supplemented with 4 mM l-glutamine, 1 mM sodium pyruvate, 10% fetal bovine serum (Thermo Fisher Scientific, Waltham, MA), and 1% penicillin-streptomycin, while PANC-1 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 4.5 g · l−1 glucose, 4 mM l-glutamine, 1 mM sodium pyruvate, 1.5 g · l−1 sodium bicarbonate, penicillin-streptomycin, and 10% fetal bovine serum. All cell lines were cultured at 37°C under humidified 5% CO2 in air, and the medium was replenished every 3 days. Cells were subcultured as they reached confluence.
Microarray Experiment.
Serum-starved HepG2 cells were left alone or incubated with either 50 μM PDTC (Sigma-Aldrich, St. Louis, MO) or 20 ng/ml recombinant human IL-6 (R&D Systems, Minneapolis, MN) for 1, 2, 4, and 8 h. Total cellular RNA was extracted using an RNeasy plus mini kit (QIAGEN, Valencia, CA), and its quality was assessed using an Agilent BioAnalyzer (Agilent Technologies, Santa Clara, CA). Microarray analysis was performed using a human Ref-8 v2 Expression Bead Chip containing 22,000 clones (Illumina, San Diego, CA). In brief, a 0.5-μg aliquot of total RNA from each sample was labeled using the Illumina TotalPrep RNA Amplification kit from Ambion (Austin, TX). After a 16-h hybridization of biotin-labeled cRNA to the bead chip, the arrays were washed, blocked, and then hybridized. Biotinylated cRNA was detected with streptavidin-Cy3 and quantitated using Illumina's Bead Station 500GX Genetic Analysis Systems scanner. Image processing and data extraction were performed using Bead Studio v15 (Illumina), and the final results were analyzed using DIANE 6.0, a spreadsheet-based microarray analysis program, which can be found online at http://www.grc.nia.nih.gov/branches/rrb/dna/diane_software.pdf.
Expression Data Analysis.
Z-score transformation was used to normalize the raw microarray signal fluorescent values as described previously (Cheadle et al., 2003). To calculate gene expression changes after cell treatment with PDTC or IL-6 and identify significant gene profile changes between experimental groups, Z scores were converted to Z ratios, which represent fold-like changes for each gene and the false discovery rate. Parameterized significant analysis was completed according to the “significance analysis of microarray” protocol (Tusher et al., 2001) with analysis of variance filtering (p ≤ 0.05). Statistical analysis was based on an increase or decrease of individual genes with Z ratio ≥1.5, P value ≤0.05, and false discovery rate ≤0.30. All microarray data have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) with the accession number GSE14632. Principal components analysis was performed to identify changing patterns within groups. Hierarchical clustering and k-mean clustering methods were used to find out correlations and distinguish patterns among various treatment groups. Ingenuity pathways analysis was used to identify biological networks with the greatest number of differentially expressed genes. For each network or pathway, probability scores were calculated using the right-tailed Fisher's exact test.
Reverse Transcription and Real-Time PCR Analysis.
Total RNA from cultured cells was isolated using the RNeasy Mini Kit (QIAGEN), and first-strand cDNA was synthesized using the Omniscript Reverse Transcript kit (QIAGEN). Real-time PCRs were carried out with the TaqMan Gene Expression Assay system method on an ABI Prism 7300 sequence detection system (Applied Biosystems, Foster City, CA). Primer pairs used for the reactions were from Applied Biosystems (DDIT4, ID Hs00430304_g1; glyceraldehyde-3-phosphate dehydrogenase, ID Hs99999905_m1). Relative quantitation of gene expression was performed using the threshold cycle. The mRNA levels were compared with standard curves (generated using serial RNA dilutions), and differences in mRNA expression were calculated by the 2−ΔΔCT method after normalization to glyceraldehyde-3-phosphate dehydrogenase mRNA. Each analyzed sample was performed in three biological replicates, and at least three reactions were used. Fidelity of the PCR was determined by melting temperature analysis.
Western Blotting.
Unless otherwise indicated, cells were lysed in radioimmunoprecipitation assay buffer supplemented with phosphatase and protease inhibitor cocktails (Song et al., 2010). Insoluble material was removed by centrifugation (10,000g, 20 min at 4°C), and protein concentration in the clarified lysates was determined using the bicinchoninic acid assay (Thermo Fisher Scientific). Proteins were separated by SDS-polyacrylamide gel electrophoresis under reducing conditions and transferred onto polyvinylidene difluoride membranes using iBlot (Invitrogen). After a blocking step, membranes were probed with specific primary antibodies followed by incubation with horseradish peroxidase-conjugated secondary antibody. Visualization of immunoreactive bands was performed by enhanced chemiluminescence and quantified by volume densitometry using ImageJ software (National Institutes of Health, Bethesda, MD) and normalization to β-tubulin or heat shock protein 90α as loading control. The primary antibodies were directed against DDIT4 (rabbit; Proteintech Group, Chicago, IL), STAT3 (rabbit; Santa Cruz Biotechnology Inc., Santa Cruz, CA), heat shock protein 90α (mouse; BD Biosciences, San Jose, CA); phospho-4EBP1 (Thr37/Thr46), phospho-S6K1 (Thr389), phosphoSTAT3 (Tyr705), phosphoAkt (Ser473), phosphoAkt (Thr308), phosphoTSC2 (Thr1462), phosphoGSK3β (Ser9), S6K1, Akt, and β-tubulin (1:1000; rabbit; Cell Signaling Technology, Danvers, MA).
Small Interfering RNA Knockdown Experiments.
The small interfering RNA (siRNA) duplex used in this study was targeted against DDIT4 mRNA (Ambion/Applied Biosystems). The sequence of the DDIT4 siRNA duplex was as follows: (sense) 5′-ACGCAUGAAUGUAAGAGUAtt-3′, (antisense) 5′-UACUCUUACAUUCAUGCGUct-3′. The negative control siRNA was the “AllStars Neg. control siRNA” (QIAGEN) that has no known target gene. HepG2 cells (2–3 × 105 cells/ml) in antibiotics-free minimal essential medium were subjected to reverse transfection using Lipofectamine RNAiMAX reagent (Invitrogen) and 20 to 40 nM siRNA duplex per 35-mm plate according to the supplier's instructions. Specific target gene silencing was confirmed by real-time PCR and immunoblotting 2 days after reverse transfection.
Global 35S-Labeled Protein Translation Assay.
Serum-starved HepG2 cells (seeded on 35-mm plates) were treated with PDTC or IL-6 as indicated. After a series of washes in phosphate-buffered saline, cells were incubated with Met/Cys-free medium for 30 min at 37°C followed by the addition of 100 μCi 35S Protein labeling mix (NEG-072; PerkinElmer Life and Analytical Sciences, Waltham, MA) and further incubation for 15 min. Cells were lysed in radioimmunoprecipitation assay buffer, and equal amounts of proteins were loaded on SDS-polyacrylamide gels, transferred to a polyvinylidene difluoride membrane, and visualized with a PhosphorImager (GE Healthcare, Chalfont St. Giles, Buckinghamshire, UK). The membrane was then reprobed for β-actin by Western blotting.
Statistical Analysis.
Results are expressed as mean ± S.D. Statistical analysis was performed using unpaired Student's t test or one-way analysis of variance followed by Fisher's least significant difference post hoc test as appropriate. A value of P ≤ 0.05 was considered statistically significant. All calculations were carried out using Kaleidagraph v.4.01 (Synergy Software, Reading, PA).
Results
Genomewide Comparison of Expression Profiles between HepG2 Cells Treated with PDTC and IL-6.
Oligonucleotide DNA microarray analysis was performed to identify changes in gene expression after treatment of HepG2 cells with either PDTC or IL-6. Principal component analysis and hierarchical clustering algorithm demonstrated that the global expression patterns of PDTC-treated groups were distinctly different from those of IL-6-treated groups over time (Supplemental Fig. 1, A and B). Of the 22,000 genes and expressed sequence tags that are represented on the chip, a total of 1043, 1159, 1417, and 1601 genes were differentially expressed after PDTC addition for 1, 2, 4, and 8 h, respectively, compared with the control group (P ≤ 0.05). Under these experimental conditions, IL-6 treatment led to 533, 1234, 1048, and 1116 regulated genes after 1, 2, 4, and 8 h, respectively, of which 123, 244, 275, and 329 genes, respectively, were shared with PDTC. A heat map representing an abridged list of “shared” genes showed an opposite direction of change for a number of these genes when IL-6 and PDTC treatment groups were compared (Supplemental Fig. 1C). These include RPLP1 (ribosomal protein, large, P1), IER3 (radiation-inducible immediate-early gene IEX-1), and JUND (jun D) (Supplemental Table 1). On the other hand, several genes showed a similar direction of change after cell treatment with PDTC and IL-6 (Supplemental Table 2).
Analysis of Gene Sets in HepG2 Cells Treated with PDTC and IL-6.
Supervised analyses, such as parameterized analysis of gene set enrichment (Kim and Volsky, 2005), provide insight into regulated signaling pathways or biological processes as well as a list of differentially expressed genes. From the collection of more than 182 gene sets we identified sets of genes that were altered by PDTC and IL-6 over time (P ≤ 0.05; Table 1). There were 104 and 102 gene sets whose expression was altered after 8-h treatment with PDTC and IL-6, respectively, of which 36 gene sets were shared (Table 2; Supplemental Table 3). It is noteworthy that scatter plot analysis of the cumulative Z scores of these shared gene sets showed a reverse relationship between PDTC and IL-6 (Supplemental Fig. 1D; R2 = 0.6835), with nine of the 36 enriched gene sets that were directly associated with metabolic pathways, eight with inflammation and cytokine signaling, five with mitochondrial function, and three with cancer biology. This analysis was repeated for the 1-, 2-, and 4-h time points, of which 4, 21, and 38 sets of genes were shared, respectively (Supplemental Table 4). The temporal expression of gene sets that were the most differentially regulated by PDTC or IL-6 is also depicted (Supplemental Fig. 2A). Therefore, the effects on PDTC on gene expression patterns in HepG2 cells sharply differed from that observed with IL-6.
TABLE 1.
Number of gene sets affected by PDTC and IL-6 in HepG2 cells
All gene sets were significant at P < 0.05.
| Time | PDTC |
IL-6 |
||||
|---|---|---|---|---|---|---|
| Total | Up (%) | Down (%) | Total | Up (%) | Down (%) | |
| h | ||||||
| 1 | 61 | 21 (34.4) | 40 (65.6) | 57 | 46 (80.7) | 11 (19.3) |
| 2 | 66 | 30 (45.5) | 36 (54.5) | 103 | 69 (67.0) | 34 (33.0) |
| 4 | 95 | 44 (46.3) | 51 (53.7) | 104 | 69 (66.3) | 35 (33.7) |
| 8 | 104 | 54 (51.9) | 50 (48.1) | 102 | 73 (71.6) | 29 (28.4) |
TABLE 2.
List of shared gene sets after 8-h treatment with PDTC and IL-6 in HepG2 cells
Size indicates the number of genes in each gene set. A brief description of each gene set is summarized in Supplemental Table 3. All gene sets were significant at P < 0.05.
| Size | Name | PDTC |
IL-6 |
||
|---|---|---|---|---|---|
| Z Score | Rank | Z Score | Rank | ||
| 95 | Breast_cancer_estrogen_signaling | 6.09 | 1 | −2.67 | 96 |
| 105 | LEU_UP | 4.65 | 2 | 2.78 | 37 |
| 138 | GPCRs_class_A_rhodopsin-like | 3.28 | 9 | −3.70 | 99 |
| 28 | nktPathway | 2.67 | 11 | −2.59 | 93 |
| 28 | inflamPathway | 2.67 | 12 | −1.97 | 89 |
| 15 | stemPathway | 2.59 | 13 | −1.89 | 87 |
| 17 | th1th2Pathway | 2.47 | 14 | −2.33 | 91 |
| 21 | cytokinePathway | 1.98 | 16 | −2.64 | 95 |
| 131 | Cell_surface_receptor_linked_signal_transduction | 1.90 | 18 | −3.00 | 98 |
| 23 | GPCRs_class_B_secretin-like | 1.25 | 31 | −1.44 | 85 |
| 10 | il5Pathway | 1.00 | 35 | −1.22 | 84 |
| 8 | asbcellPathway | 0.95 | 36 | −0.77 | 75 |
| 15 | tall1Pathway | 0.88 | 37 | −0.89 | 78 |
| 4 | slrp2Pathway | 0.65 | 49 | −1.05 | 79 |
| 6 | neurotransmittersPathway | 0.61 | 51 | −1.75 | 86 |
| 3 | aifPathway | −1.55 | 59 | 1.83 | 65 |
| 6 | CR_transport | −1.58 | 60 | 1.31 | 71 |
| 15 | MAP00510_N_glycans_biosynthesis | −1.61 | 61 | 3.37 | 23 |
| 7 | KET | −1.75 | 62 | 1.57 | 68 |
| 22 | MAP00500_starch_and_sucrose_metabolism | −2.10 | 66 | 2.93 | 35 |
| 15 | TCA | −2.11 | 67 | 2.05 | 60 |
| 20 | MAP00650_butanoate_metabolism | −2.47 | 72 | 3.93 | 13 |
| 37 | CR_REPAIR | −2.47 | 73 | 2.75 | 38 |
| 18 | MAP00020_citrate_cycle_TCA_cycle | −2.79 | 80 | 3.15 | 28 |
| 3 | torPathway | −2.99 | 83 | 1.22 | 72 |
| 49 | mRNA_splicing | −3.08 | 84 | 4.12 | 10 |
| 28 | Krebs-TCA_cycle | −3.22 | 88 | 2.63 | 42 |
| 19 | Glycogen | −3.38 | 90 | 3.09 | 30 |
| 42 | mRNA_processing | −3.48 | 92 | 3.09 | 31 |
| 7 | MAP00720_reductive_carboxylate_cycle (CO2 fixation) | −4.04 | 96 | 3.25 | 25 |
| 21 | MAP00120_bile_acid_biosynthesis | −4.05 | 97 | 3.55 | 17 |
| 10 | MAP00100_sterol_biosynthesis | −4.11 | 98 | 3.16 | 27 |
| 19 | MAP00640_propanoate_metabolism | −4.50 | 100 | 4.27 | 8 |
| 24 | MAP00280_valine_leucine_and_isoleucine degradation | −5.61 | 102 | 7.44 | 4 |
| 393 | Human_mitoDB_6_2002 | −6.56 | 103 | 3.07 | 32 |
| 404 | Mitochondr | −7.46 | 104 | 3.53 | 20 |
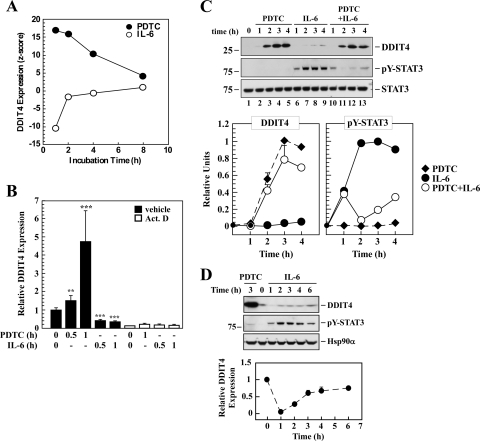
To focus on conditions under which the actions of PDTC toward IL-6 signaling could be best explored, we selected the mTOR complex, a key regulator in the control of protein synthesis, mitochondrial function, and inflammation-mediated tumor development (Lee et al., 2007; Ramanathan and Schreiber, 2009; Foster and Fingar, 2010). The choice of mTOR was borne out by the fact that expression of more than 50 ribosomal protein genes was sharply reduced with IL-6 but up-regulated with PDTC after a 2-h treatment (Table 3; Supplemental Table 5). Moreover, the mTOR-associated GO term “translation” (GO:0006412) was one of the most affected biological processes that were reciprocally affected by PDTC and IL-6 (Supplemental Fig. 2B). Among the changed genes, there was a significant 17-fold increase in DDIT4 mRNA expression at 1 h after PDTC treatment, whereas IL-6 elicited a ∼11-fold reduction compared with untreated cells (Fig. 1A). DDIT4 encodes a protein transcript (DDIT4, also known as REDD1 and Rtp801) that has been identified as a negative regulator of mTOR complex 1 (Foster and Fingar, 2010). Our microarray analysis demonstrated a time-dependent return of DDIT4 expression to almost basal levels 4 to 8 h after the initial stimulation with either PDTC or IL-6 (Fig. 1A).
TABLE 3.
Partial list of ribosomal protein genes whose expression is reciprocally regulated by a 2-h treatment with PDTC and IL-6 in HepG2 cells
All genes were significant at P < 0.05 when compared to untreated controls. A complete list of ribosomal protein genes can be found in Supplemental Table 5.
| Accession | Symbol | Gene Name | PDTC Z Ratio | IL-6 Z Ratio |
|---|---|---|---|---|
| NM_001003.2 | RPLP1 | Ribosomal protein, large, P1 | 9.232 | −5.024 |
| NM_003973.2 | RPL14 | Ribosomal protein L14 | 4.409 | −4.296 |
| NM_000978.2 | RPL23 | Ribosomal protein L23 | 2.684 | −9.910 |
| NM_000988.2 | RPL27 | Ribosomal protein L27 | 1.204 | −7.297 |
| NM_001020.4 | RPS16 | Ribosomal protein S16 | 3.328 | −7.474 |
| NM_001005.3 | RPS3 | Ribosomal protein S3 | 2.543 | −7.059 |
| NM_001031.4 | RPS28 | Ribosomal protein S28 | 2.435 | −6.941 |
| NM_001030009.1 | RPS15A | Ribosomal protein S15a | 1.216 | −8.458 |
Fig. 1.
Effects of PDTC and IL-6 on the regulation of DDIT4 mRNA and protein. A, temporal expression of the DDIT4 gene in HepG2 cells treated with PDTC (●) or IL-6 (○). Data were obtained from the microarray analysis. B, serum-starved HepG2 cells were left alone or pretreated with actinomycin D (Act. D; 1 μg/ml) for 1 h followed by the addition of PDTC (50 μM) or IL-6 (20 ng/ml) for 0.5 and 1 h. Total RNA was extracted and then analyzed by real-time PCR. Data are presented as fold increase relative to the untreated group. Bars represent the average ± S.D. of two independent experiments, each performed in triplicate dishes. **, P < 0.01; ***, P < 0.001 versus untreated controls. C, cells were serum-starved and then treated with either PDTC, IL-6, or a combination for 1 to 4 h. Top, cell lysates were prepared, and Western blot analysis was performed using primary antibodies raised against DDIT4 and total and tyrosine phosphorylated STAT3 (pY-STAT3). STAT3 was included as a loading control. Data are representative of at least three independent experiments. Bottom, graphical representation of DDIT4 and pY-STAT3 blots is shown. D, serum-starved HepG2 cells were incubated with vehicle, PDTC for 3 h, or IL-6 for 1 to 6 h. Top, representative immunoblots of DDIT4, pY-STAT3, and that of the loading control, Hsp90α, are shown. Bottom, graphical representation of DDIT4 is shown.
A number of significantly changed genes and one not significantly changed (HSF1) were selected for quantitative real-time PCR validation of the microarray data. Positive validation was obtained for DDIT4 (Fig. 1B) and BAG3, HSPA1A, DEDD2, and MCL1 (data not shown). Compared with vehicle-treated controls, there was a 4.8- ± 1.6-fold increase in DDIT4 mRNA levels with PDTC for 1 h, but a 2.9- ± 0.3-fold reduction in IL-6-treated HepG2 cells (P < 0.001; Fig. 1B). The role of transcription in the observed up-regulation of DDIT4 expression by PDTC was investigated by preincubating HepG2 cells with actinomycin D. Inhibition of transcription resulted in a near total suppression in both the constitutive and inducible expression of DDIT4 mRNA (Fig. 1B).
PDTC Blocks IL-6 Suppression of DDIT4 and Associated Up-Regulation of mTOR Complex 1.
Western blot analysis indicated an increase in DDIT4 expression by PDTC within 2 h and peaked at 3 to 4 h (Fig. 1C) before returning to basal levels by 6 h (data not shown). The low abundance of DDIT4 protein in vehicle-treated cells made it difficult to ascertain whether DDIT4 stability can be altered by IL-6. However, the use of long-exposure films enabled the detection of DDIT4 in control cells, whose level was rapidly reduced by a 1-h treatment with IL-6 before slowly returning to control levels (Fig. 1D). PDTC is a potent inhibitor of IL-6 signaling cascade whose inactivation is associated with a loss in STAT3 target gene expression (Xie et al., 2009). Here, it is apparent that changes in gene expression in response to PDTC were at the very least STAT3-independent, because no tyrosine phosphorylation of STAT3 was detectable up to 4 h (Fig. 1C, top, lanes 2–5). Treatment of HepG2 cells with IL-6 under serum-free conditions elicited significant tyrosine phosphorylation of STAT3 compared with the vehicle-treated group (Fig. 1C, top, lanes 6–9 versus lane 1). Although IL-6 responsiveness vis-à-vis STAT3 phosphorylation was not affected by PDTC at the 1-h time point, longer treatment with PDTC clearly blocked the IL-6-induced phospho-STAT3 levels at 2 to 4 h (Fig. 1C, top, lanes 10–13). Under these conditions, the ability of PDTC to stimulate DDIT4 protein expression was unchanged by the presence of IL-6 (Fig. 1C, top, lanes 10–13).
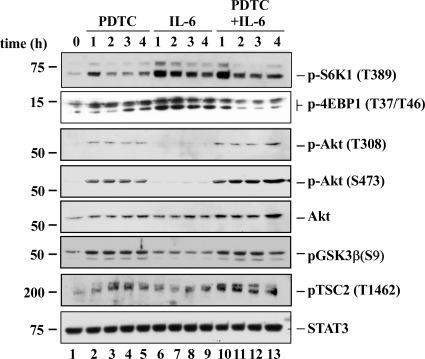
To measure the effect of PDTC and IL-6 on mTORC1 function, we monitored the phosphorylation of the translation regulatory protein 4E-BP1 and the 70-kDa isoform of S6K1, which are frequently used as surrogate markers of mTOR kinase activity (Wullschleger et al., 2006). Here, PDTC induced rapid Thr389 phosphorylation of S6K1 by 1 h, but continued treatment for 2 to 4 h led to S6K1 dephosphorylation (Fig. 2, pS6K1). In contrast, phosphorylation of 4E-BP1 at Thr37/46 was low and remained attenuated throughout PDTC treatment up to 4 h (Fig. 2, p4E-BP1). When cells were treated with IL-6, maximal phosphorylation of S6K1 and 4E-BP1 was achieved by 1 h and then slowly declined thereafter. Although IL-6-mediated phosphorylation of S6K1 was not affected by the coincubation with PDTC at the 1-h time point, longer treatment with PDTC elicited only modest decline in IL-6 responsiveness at 2 to 4 h (Fig. 2, pS6K1, lanes 10–13). In contrast, the presence of PDTC resulted in marked reduction in IL-6-inducible 4E-BP1 phosphorylation at the 2- to 4-h time points, but not at the 1-h time point (Fig. 2, p4E-BP1, lanes 10–13). Taken together, these results indicate that a latency period of 1 h is required before the onset of inhibition of IL-6-dependent mTORC1 signaling by PDTC.
Fig. 2.
PDTC blocked IL-6-induced increase in mTORC1 function. Serum-starved HepG2 cells were treated either with PDTC, IL-6, or a combination for the indicated periods of time. Cell lysates were prepared and Western blot analysis was performed, looking at the phosphorylation of mTORC1 downstream targets, S6K1 (Thr389) and 4EBP1 (Thr37/46), as well as that of Akt (Thr308 and Ser473) and its direct targets, GSK3β (Ser9) and TSC2 (Thr1462). STAT3 was included as a loading control. Similar results were obtained in two other separate experiments.
Selective activation of the phosphatidylinositol 3-kinase/Akt pathway has been found to be key to early mTORC1 activation and downstream regulation of protein synthesis (Foster and Fingar, 2010). Akt activity was assessed by measuring Akt phosphorylation levels on Thr308 and Ser473 and phosphorylation of direct downstream targets, such as GSK3β on Ser9 and TSC2 on Thr1462. Treatment of HepG2 cells with IL-6 did not affect Akt activity; however, exposure to PDTC alone or combined with IL-6 resulted in rapid and sustained phosphorylation of Akt and its downstream targets up to 4 h (Fig. 2). Thus, early activation of mTORC1 signaling by PDTC and IL-6 (1-h time point) is likely to occur through distinct mechanisms.
Then, we analyzed whether the cellular response to PDTC, IL-6, and a combination could be confirmed in the human pancreatic carcinoma cell line PANC-1. The reciprocal regulation of DDIT4 protein levels by PDTC versus IL-6 and the ability of PDTC to suppress IL-6-mediated phosphorylation of STAT3 were similar in both cancer cell types, including the latency period of 1 h before the onset of PDTC inhibition (Supplemental Fig. 3A). Moreover, treatment of PANC-1 cells with PDTC was associated with early activation with the Akt/mTORC1 pathway up to 2 h, followed by a marked reduction in the phosphorylation of Akt and mTORC1 downstream targets, S6K1 and 4E-BP1, by 4 h (Supplemental Fig. 3B). As anticipated, this phosphorylation kinetics was in the opposite direction compared with the expression of the mTORC1-negative regulator DDIT4 (Supplemental Fig. 3A, top). Exposure of PANC-1 cells to IL-6 resulted in very weak activation of the Akt/mTORC1 pathway. Altogether, these results indicated that PDTC could be an effective mTORC1 repressor by up-regulating DDIT4 in two types of cancer cell lines.
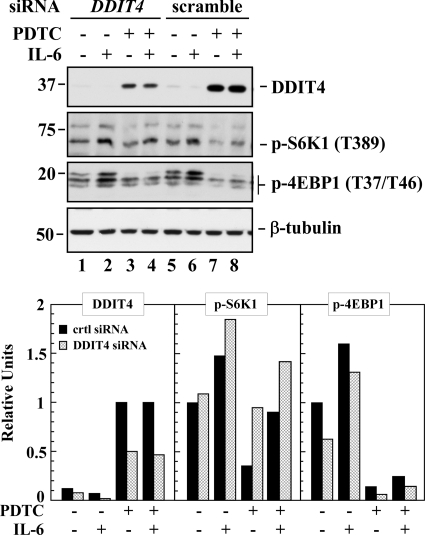
To assess the role of DDIT4 in the down-modulation of mTORC1 by PDTC at the 2- to 4-h time points, HepG2 cells were incubated with the DDIT4 siRNA and nonsilencing siRNA control for 48 h. Compared with the nonsilencing control group, DDIT4 knockdown blocked the inducible expression of DDIT4 protein by 50% in response to a 2-h treatment with PDTC (Fig. 3, top). The levels of β-tubulin (Fig. 3) and STAT3 and Hsp90α (data not shown) were unaffected, indicating a low probability of nonspecific silencing effects. Down-regulation of DDIT4 resulted in the protection against PDTC-mediated decrease in Thr389 phosphorylation of S6K1 while having little or no effect on 4E-BP1 phosphorylation (Fig. 3, top, lanes 3 versus 7). In contrast, the ability of IL-6 to promote S6K1 and 4E-BP1 phosphorylation was unaffected by DDIT4 down-regulation (Fig. 3, top, lanes 2 versus 6). As indicated earlier, treatment with PDTC dramatically attenuated IL-6-induced STAT3 activation and associated mTOR downstream signaling. Here, in cells transfected with the DDIT4 siRNA, PDTC continued to exert its suppressive effects on IL-6-mediated phosphorylation of 4E-BP1 (Fig. 3, top, lanes 4 versus 8).
Fig. 3.
Effect of DDIT4 down-regulation on mTORC1 function. HepG2 cells were transfected with the negative control siRNA (filled bars) or DDIT4 siRNA (hatched bars) for 48 h. Cells were serum-starved and then treated with PDTC, IL-6, or a combination for 2 h. Top, lysates were immunoblotted with the indicated primary antibodies using β-tubulin as a loading control. Bottom, quantitative analysis of the immunoblots is shown and normalized to the PDTC signal (DDIT4) or vehicle-treated group (pS6K1 and p4E-BP1) from cells transfected with the nonsilencing siRNA. Similar results were obtained in a second independent experiment. The migration of molecular mass markers (values in kDa) is shown on the left of the immunoblots.
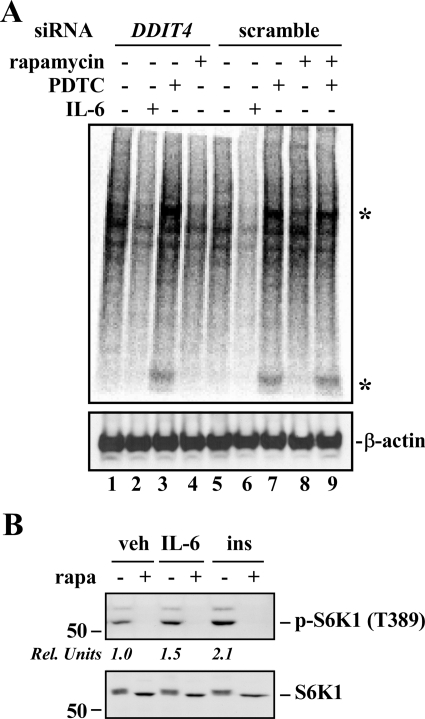
In light of our observation that mTORC1 activity is reciprocally regulated by PDTC and IL-6, we asked whether the expression of DDIT4 was necessary for control of the global protein translation machinery. As shown in Fig. 4A, there was clear increase in the levels of 35S-labeled proteins in HepG2 cells in response to a 2-h treatment with PDTC, even in the presence of the mTORC1 inhibitor rapamycin (Fig. 4A, denoted by*). Down-regulation of DDIT4 with siRNA had no effect on the basal and PDTC-induced increase in protein synthesis. Under these conditions, rapamycin completely inhibited insulin- and IL-6-mediated S6K1 phosphorylation at Thr389 (Fig. 4B). It is noteworthy that IL-6 sharply reduced global protein translation in both control and DDIT4 siRNA-treated cells, consistent with lower ribosome biogenesis and function (see results above).
Fig. 4.
Effects of PDTC and IL-6 on global protein translation. A, transfection with the negative control siRNA or DDIT4 siRNA was performed for 48 h in HepG2 cells, after which cells were serum-starved and then incubated with vehicle, rapamycin (20 nM), PDTC, PDTC + rapamycin, or IL-6 for 2 h. After washing, cells were incubated in Met/Cys-free medium for 30 min followed by [35S]Met/Cys labeling for 15 min. Cell lysates were resolved by SDS-polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membrane, and autoradiography was performed. The membrane was probed with anti-β-actin antibody to confirm equal protein load in each lane. This experiment was repeated twice with comparable results. *, denotes increased translation in response to PDTC. B, serum-starved HepG2 cells were incubated in the absence (−) or presence (+) of rapamycin (20 nM) for 1 h followed by the addition of IL-6 or insulin (100 nM) for 30 min. Cell lysates were immunoblotted with antibodies against phosphorylated (Thr389) and total S6K1. Rel. Units, the ratios of phospho/total S6K1 are shown relative to vehicle-treated controls. Similar results were obtained in a second independent experiment.
Discussion
In the present study, we explored and compared the effects of PDTC with that of the proinflammatory cytokine IL-6 on global gene expression profiling and the overall protein biosynthetic capacity in cultured hepatoma cells. To better understand the actions of PDTC in IL-6 signaling, the combination of IL-6 and PDTC was tested vis-à-vis the regulation of DDIT4 expression and its impact on mTORC1 signaling. The novel findings of this study were: 1) treatment with IL-6 versus PDTC time-dependently induced substantial changes in gene expression patterns in HepG2 cells compared with the untreated control groups; 2) the changes in gene set enrichment were either stimulus-specific or shared by IL-6 and PDTC, with ribosomal protein genes and DDIT4 predominantly up-regulated with PDTC but down-regulated with IL-6; 3) silencing of DDIT4 slowed the adverse effects of PDTC on IL-6-dependent mTORC1 activation; and 4) the overall protein biosynthetic capacity of HepG2 cells was severely blunted by IL-6, but increased in a rapamycin-independent pathway by PDTC.
A variety of cytoprotective pathways were activated in response to PDTC, promoting changes in gene expression that facilitate cell survival and recovery from stress. For example, PDTC activates the transcription factor HSF1 (Song et al., 2010), which mediates the heat shock response, a process that controls the expression of a conserved set of inducible heat shock proteins, their DnaJ cohorts, and a variety of cochaperones. From our microarray data, we identified a large variety of heat shock proteins that were induced by PDTC but not IL-6. A large fraction of genes involved in amino acid metabolism, mitochondrial function, and mRNA processing showed decreased expression with PDTC, whereas the opposite effect was observed with IL-6. In specifically looking for growth arrest and cell cycle-related gene products, it was found that PDTC induces a rapid but transient increase in the levels of the mTORC1-negative regulator, DDIT4, in HepG2 cells. In contrast, IL-6 treatment sharply decreased DDIT4 expression. In addition to HepG2 cells, PDTC elicited a rapid increase in DDIT4 expression in different types of human cancer cells, including PANC-1 (Supplemental Fig. 3A) and A7 melanoma cells, and in freshly isolated human peripheral blood mononuclear cells (S. Song and M. Bernier, unpublished work).
Initially described as a hypoxia-inducible gene through the activation of the transcription factor hypoxia-inducible factor 1α (Shoshani et al., 2002), DDIT4 has since been reported to be a cell stress response gene and the transcriptional target of several transacting factors (Lin et al., 2005). Here, we found that the DDIT4 mRNA levels changed in opposite directions during a 1-h treatment with PDTC and IL-6 in HepG2 cells, consistent with the involvement of immediate-early genes in the regulation of DDIT4 expression. In HL-60 cells, administration of PDTC resulted in the up-regulation of the zinc finger transcription factor early growth response protein-1 (Della Ragione et al., 2002). Although hypoxia-inducible factor 1α and early growth response protein-1 mRNA levels were markedly up-regulated by PDTC in HepG2 cells, these transcription factors did not participate in the induction of DDIT4 during PDTC treatment (S. Song and M. Bernier, unpublished data). Therefore, the identification of the immediate-early genes encoding transcription factors that regulate DDIT4 expression in response to PDTC and IL-6 will require additional studies, which are beyond the scope of this article.
In addition to transcriptional regulation, DDIT4 is subject to post-transcriptional and translational control. The rate of DDIT4 translation is tightly regulated, and DDIT4 mRNA, like the protein, has been shown to have a very short half-life. DDIT4 mRNA translation is inhibited by microRNA binding at the 3′ untranslated region, and ectopic expression of these microRNAs correlates with tumorigenesis through down-modulation of DDIT4 mRNA levels (Pineau et al., 2010; Hwang-Verslues et al., 2011). There is a large body of work detailing one of the mechanisms by which the IL-6/STAT3 pathway promotes tumor cell survival, which includes deregulated expression of microRNAs (Meng et al., 2007). Work is underway to assess the temporal expression profiles of microRNA during PDTC and IL-6 challenge in HepG2 cells and their impact on DDIT4 mRNA regulation.
Akt-mediated TSC2 phosphorylation on Thr1462 is known to relieve the inhibitory effects of the TSC1/TSC2 complex on mTORC1 (Foster and Fingar, 2010). This Akt-dependent phosphorylation of TSC2 could explain the early activation of mTORC1 by PDTC (1-h time point). However, through increased DDIT4 protein translation (2- and 4-h time points), PDTC might then exert a negative feedback mechanism caused by reactivation of the inhibitory tonic activity of TSC2. Because IL-6 activates mTORC1 signaling independently of Akt, multisite phosphorylation of TSC2 by other signaling modules, such as the Ras/extracellular signal-regulated kinase module (Ma et al., 2005), might provide a mechanism that leads to mTORC1 activation by this cytokine.
PDTC has been shown to increase protein S-glutathionylation in HepG2 cells, reaching a maximum by 2 h (Wang et al., 2009). Coincidentally, a 2-h exposure with PDTC is required for the marked reduction in IL-6-mediated phosphorylation of STAT3 and activation of mTORC1. Another study has demonstrated that the ability of PDTC to promote S-glutathionylation of STAT3 renders it a poor substrate for the IL-6 receptor/Janus tyrosine kinase complex (Xie et al., 2009). Therefore, the temporal effects of PDTC on both the DDIT4 expression and cellular redox may ultimately contribute to the refractoriness in IL-6 signaling.
Dysregulation in DDIT4 levels has been recently linked to several human diseases, such as liver cancer (Pineau et al., 2010), and marked increase in tumorigenicity was observed in vivo in DDIT4 knockout mice (Horak et al., 2010). Induction of DDIT4 suppresses mTOR activity and its associated trophic effect on protein synthesis in HeLa cells exposed to oxidative and endoplasmic reticulum stress (Jin et al., 2009). The connection between DDIT4 and the TSC1/TSC2-mTORC1 pathway is supported by the observation that cellular stressors have been shown to block the translational machinery through DDIT4-mediated suppression of mTORC1 activity (Corradetti et al., 2005; Sofer et al., 2005). However, we observed a striking increase in global protein translation in PDTC-treated cells but suppression with IL-6. In the latter case, defect in ribosome biogenesis in IL-6-treated cells (see above) certainly contributes to lower cellular biosynthetic capacity despite mTORC1 activation. Furthermore, mTORC1/cap-dependent translation initiation would have only a minimal role in the contribution of PDTC to the rapamycin-resistant translational control of protein synthesis.
It has been found that mTORC2 associates with ribosomal proteins to regulate mRNA translation (Dormond et al., 2008; Kuehn et al., 2011). In addition to mTORC1-mediated cap-dependent translation, translation initiation can occur in the middle of an mRNA via a nucleotide sequence known as internal ribosome entry site (IRES) (reviewed by Komar and Hatzoglou, 2011). The latter process enables the recruitment of 40S ribosome in the vicinity of the initiation codon with the help of IRES transacting factors. It is noteworthy to mention that IRES-mediated translation has been found to occur for mRNAs encoding proteins involved in either stress protection or apoptosis (Komar and Hatzoglou, 2011). Perhaps the mTORC2/40S ribosome/IRES pathway functions as a rate-limiting factor for global mRNA translational responses depending on the stimulus and cell type. Nevertheless, the mechanism by which PDTC promotes global protein translation has not been investigated further in this study.
Our results suggest that mTORC2 mediates the translational activity of PDTC because PDTC strongly increases the phosphorylation of Akt on Ser473, a recognized effector of mTORC2 (Sarbassov et al., 2005), whose activity can be considered to be upstream of mTORC1. TSC2-Thr1462 has been found to be a direct Akt phosphorylation target site; however, TSC2 phosphorylation on additional sites may also contribute to the stimulation of mTORC2 by PDTC. The activity of mTORC2 responds to PDTC, but how mTORC2 is regulated is unclear. Sin1 (stress-activated protein kinase-interacting protein 1) and rictor maintain mTORC2 integrity and mediates mTORC2 function (Foster and Fingar, 2010). It is possible that PDTC stimulation of mTORC2 might come from enhanced binding of rictor and Sin1 to mTOR. As alluded to earlier, PDTC may also modulate mTORC2 signaling and contribute to global protein translation through phosphorylation and inactivation of TSC2.
Significance and Potential Impact
Elevated circulating levels of IL-6 is one of many factors involved in the pathophysiology of chronic inflammatory diseases and cancer cachexia. It is noteworthy that PDTC attenuates muscle and adipose tissue loss in a cachetic mouse model and reduces IL-6 synthesis from inoculated tumor tissues (Nai et al., 2007). From this and several in vivo studies showing minimal genotoxicity (Chabicovsky et al., 2010), PDTC qualifies as a valuable drug candidate. However, there is much to be learned about whether PDTC confers protection against IL-6-mediated reduction in ribosomal protein biogenesis and subsequent decline in global protein translation. We propose that the anti-inflammatory action of PDTC calls for the inhibition of IL-6-induced mTORC1 activity through a mechanism involving the tumor suppressor DDIT4.
Supplementary Material
Acknowledgments
We thank William H. Wood 3rd from the Gene Expression and Genomics Unit, Research Resources Branch at the National Institute on Aging for invaluable assistance with the cDNA microarray analysis and Sutapa Kole for expert technical assistance.
This research was supported by the Intramural Research Program of the National Institutes of Health, National Institute on Aging.
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/jpet.111.185678.
The online version of this article (available at http://jpet.aspetjournals.org) contains supplemental material.
- PDTC
- pyrrolidine dithiocarbamate
- HSF1
- heat shock factor 1
- IL-6
- interleukin-6
- STAT3
- signal transducer and activator of transcription-3
- mTOR
- mammalian target of rapamycin
- mTORC
- mTOR complex
- TSC
- tuberous sclerosis complex
- IRES
- internal ribosome entry site
- DDIT4
- DNA damage-inducible transcript 4 protein
- PCR
- polymerase chain reaction
- siRNA
- small interfering RNA
- GSK
- glycogen synthase kinase.
Authorship Contributions
Participated in research design: Song, Abdelmohsen, Becker, and Bernier.
Conducted experiments: Song and Abdelmohsen.
Performed data analysis: Song, Abdelmohsen, Zhang, Becker, Gorospe, and Bernier.
Wrote or contributed to the writing of the manuscript: Song, Zhang, and Bernier.
References
- Chabicovsky M, Prieschl-Grassauer E, Seipelt J, Muster T, Szolar OH, Hebar A, Doblhoff-Dier O. (2010) Pre-clinical safety evaluation of pyrrolidine dithiocarbamate. Basic Clin Pharmacol Toxicol 107:758–767 [DOI] [PubMed] [Google Scholar]
- Cheadle C, Vawter MP, Freed WJ, Becker KG. (2003) Analysis of microarray data using Z score transformation. J Mol Diagn 5:73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corradetti MN, Inoki K, Guan KL. (2005) The stress-inducted proteins RTP801 and RTP801L are negative regulators of the mammalian target of rapamycin pathway. J Biol Chem 280:9769–9772 [DOI] [PubMed] [Google Scholar]
- Cuzzocrea S, Chatterjee PK, Mazzon E, Dugo L, Serraino I, Britti D, Mazzullo G, Caputi AP, Thiemermann C. (2002) Pyrrolidine dithiocarbamate attenuates the development of acute and chronic inflammation. Br J Pharmacol 135:496–510 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Della Ragione F, Cucciolla V, Criniti V, Indaco S, Borriello A, Zappia V. (2002) Antioxidants induce different phenotypes by a distinct modulation of signal transduction. FEBS Lett 532:289–294 [DOI] [PubMed] [Google Scholar]
- DeYoung MP, Horak P, Sofer A, Sgroi D, Ellisen LW. (2008) Hypoxia regulates TSC1/2-mTOR signaling and tumor suppression through REDD1-mediated 14-3-3 shuttling. Genes Dev 22:239–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dormond O, Contreras AG, Meijer E, Datta D, Flynn E, Pal S, Briscoe DM. (2008) CD40-induced signaling in human endothelial cells results in mTORC2- and Akt-dependent expression of vascular endothelial growth factor in vitro and in vivo. J Immunol 181:8088–8095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebenezer PJ, Mariappan N, Elks CM, Haque M, Soltani Z, Reisin E, Francis J. (2009) Effects of pyrrolidine dithiocarbamate on high-fat diet-induced metabolic and renal alterations in rats. Life Sci 85:357–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster KG, Fingar DC. (2010) Mammalian target of rapamycin (mTOR): conducting the cellular signaling symphony. J Biol Chem 285:14071–14077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HJ, Zhu TN, Xie Y, Fan J, Kole S, Saxena S, Bernier M. (2006) Pyrrolidine dithiocarbamate inhibits interleukin-6 signaling through impaired STAT3 activation and association with transcriptional coactivators in hepatocytes. J Biol Chem 281:31369–31379 [DOI] [PubMed] [Google Scholar]
- Horak P, Crawford AR, Vadysirisack DD, Nash ZM, DeYoung MP, Sgroi D, Ellisen LW. (2010) Negative feedback control of HIF-1 through REDD1-regulated ROS suppresses tumorigenesis. Proc Natl Acad Sci U S A 107:4675–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Kaminski PM, Edwards JG, Yeh A, Wolin MS, Frishman WH, Gewitz MH, Mathew R. (2008) Pyrrolidine dithiocarbamate restores endothelial cell membrane integrity and attenuates monocrotaline-induced pulmonary artery hypertension. Am J Physiol Lung Cell Mol Physiol 294:L1250–L1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang-Verslues WW, Chang PH, Wei PC, Yang CY, Huang CK, Kuo WH, Shew JY, Chang KJ, Lee EY, Lee WH. (2011) miR-495 is up-regulated by E12/E47 in breast cancer stem cells, and promotes oncogenesis and hypoxia resistance via down-regulation of E-cadherin and REDD1. Oncogene 30:2463–2474 [DOI] [PubMed] [Google Scholar]
- Jastrzebski K, Hannan KM, Tchoubrieva EB, Hannan RD, Pearson RB. (2007) Coordinate regulation of ribosome biogenesis and function by the ribosomal protein S6 kinase, a key mediator of mTOR function. Growth Factors 25:209–226 [DOI] [PubMed] [Google Scholar]
- Jin HO, Seo SK, Woo SH, Kim ES, Lee HC, Yoo DH, An S, Choe TB, Lee SJ, Hong SI, et al. (2009) Activating transcription factor 4 and CCAAT/enhancer-binding protein-β negatively regulate the mammalian target of rapamycin via Redd1 expression in response to oxidative and endoplasmic reticulum stress. Free Radic Biol Med 46:1158–1167 [DOI] [PubMed] [Google Scholar]
- Kim SY, Volsky DJ. (2005) PAGE: parametric analysis of gene set enrichment. BMC Bioinformatics 6:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar AA, Hatzoglou M. (2011) Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10:229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuehn HS, Jung MY, Beaven MA, Metcalfe DD, Gilfillan AM. (2011) Prostaglandin E2 activates and utilizes mTORC2 as a central signaling locus for the regulation of mast cell chemotaxis and mediator release. J Biol Chem 286:391–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DF, Kuo HP, Chen CT, Hsu JM, Chou CK, Wei Y, Sun HL, Li LY, Ping B, Huang WC, et al. (2007) IKKβ suppression of TSC1 links inflammation and tumor angiogenesis via the mTOR pathway. Cell 130:440–455 [DOI] [PubMed] [Google Scholar]
- Lin L, Stringfield TM, Shi X, Chen Y. (2005) Arsenite induces a cell stress-response gene, RTP801, through reactive oxygen species and transcription factors Elk-1 and CCAAT/enhancer-binding protein. Biochem J 392:93–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. (2005) Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell 121:179–193 [DOI] [PubMed] [Google Scholar]
- Maedler K, Sergeev P, Ris F, Oberholzer J, Joller-Jemelka HI, Spinas GA, Kaiser N, Halban PA, Donath MY. (2002) Glucose-induced β cell production of IL-1β contributes to glucotoxicity in human pancreatic islets. J Clin Invest 110:851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malm TM, Iivonen H, Goldsteins G, Keksa-Goldsteine V, Ahtoniemi T, Kanninen K, Salminen A, Auriola S, Van Groen T, Tanila H, et al. (2007) Pyrrolidine dithiocarbamate activates Akt and improves spatial learning in APP/PS1 mice without affecting β-amyloid burden. J Neurosci 27:3712–3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer C, Grummt I. (2006) Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25:6384–6391 [DOI] [PubMed] [Google Scholar]
- Meng F, Henson R, Wehbe-Janek H, Smith H, Ueno Y, Patel T. (2007) The microRNA let-7a modulates interleukin-6-dependent STAT-3 survival signaling in malignant human cholangiocytes. J Biol Chem 282:8256–8264 [DOI] [PubMed] [Google Scholar]
- Nai YJ, Jiang ZW, Wang ZM, Li N, Li JS. (2007) Prevention of cancer cachexia by pyrrolidine dithiocarbamate (PDTC) in colon 26 tumor-bearing mice. JPEN J Parenter Enteral Nutr 31:18–25 [DOI] [PubMed] [Google Scholar]
- Neurath MF, Finotto S. (2011) IL-6 signaling in autoimmunity, chronic inflammation and inflammation-associated cancer. Cytokine Growth Factor Rev 22:83–89 [DOI] [PubMed] [Google Scholar]
- Nurmi A, Goldsteins G, Närväinen J, Pihlaja R, Ahtoniemi T, Gröhn O, Koistinaho J. (2006) Antioxidant pyrrolidine dithiocarbamate activates Akt-GSK signaling and is neuroprotective in neonatal hypoxia-ischemia. Free Radic Biol Med 40:1776–1784 [DOI] [PubMed] [Google Scholar]
- Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A. (2010) miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci U S A 107:264–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramanathan A, Schreiber SL. (2009) Direct control of mitochondrial function by mTOR. Proc Natl Acad Sci U S A 106:22229–22232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307:1098–1101 [DOI] [PubMed] [Google Scholar]
- Schreck R, Meier B, Männel DN, Dröge W, Baeuerle PA. (1992) Dithiocarbamates as potent inhibitors of nuclear factor κB activation in intact cells. J Exp Med 175:1181–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoshani T, Faerman A, Mett I, Zelin E, Tenne T, Gorodin S, Moshel Y, Elbaz S, Budanov A, Chajut A, et al. (2002) Identification of a novel hypoxia-inducible factor 1-responsive gene, RTP801, involved in apoptosis. Mol Cell Biol 22:2283–2293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofer A, Lei K, Johannessen CM, Ellisen LW. (2005) Regulation of mTOR and cell growth in response to energy stress by REDD1. Mol Cell Biol 25:5834–5845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Kole S, Precht P, Pazin MJ, Bernier M. (2010) Activation of heat shock factor 1 plays a role in pyrrolidine dithiocarbamate-mediated expression of the co-chaperone BAG3. Int J Biochem Cell Biol 42:1856–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian XF, Yao JH, Li YH, Gao HF, Wang ZZ, Yang CM, Zheng SS. (2006) Protective effect of pyrrolidine dithiocarbamate on liver injury induced by intestinal ischemia-reperfusion in rats. Hepatobiliary Pancreat Dis Int 5:90–95 [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. (2001) Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A 98:5116–5121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Xie Y, Bernier M, Wainer IW. (2009) Determination of free and protein-bound glutathione in HepG2 cells using capillary electrophoresis with laser-induced fluorescence detection. J Chromatogr A 1216:3533–3537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wullschleger S, Loewith R, Hall MN. (2006) TOR signaling in growth and metabolism. Cell 124:471–484 [DOI] [PubMed] [Google Scholar]
- Xie Y, Kole S, Precht P, Pazin MJ, Bernier M. (2009) S-glutathionylation impairs signal transducer and activator of transcription 3 activation and signaling. Endocrinology 150:1122–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.