Abstract
During the course of our research efforts to understand the kinetics of human aldehyde oxidase as a xenobiotic-clearing enzyme, we investigated the effect of eight different inhibitors on the oxidation of the probe substrate phthalazine. Saturation kinetic parameters for phthalazine oxidation in human liver cytosol were found to be the following: Km = 8.0 ± 0.4 μM and Vmax = 4.3 ± 0.1 nmol · min−1 · mg protein−1. Inhibitory potency of the inhibitors tested ranged from 0.1 to 5 μM. Of the eight different inhibitor compounds tested, seven were observed to inhibit through a mixed mode and one through a strictly competitive mode. A ratio of the Kii and Kis values was used to assess the relative competitiveness of each inhibitor. For the mixed inhibitors, the mode of inhibition varied from mostly uncompetitive to predominantly competitive (Kii/Kis values ranging from 0.1 to 15). The implications for potential drug-drug interactions and inhibition mechanism are discussed. We found two inhibitors, clozapine and chlorpromazine, that have a moderate predicted risk of drug-drug interactions based on the Ki value relative to the inhibitor concentration in human plasma, having a calculated [I]/Ki value of 0.4 and 0.8, respectively.
Introduction
Knowledge of drug-drug interactions (DDIs) is of great importance in the process of drug development. Without prior knowledge of potential DDIs, a drug can fail in clinical trials or after final approval, leading to higher costs of pharmaceuticals and the lack of potentially life-saving therapeutic agents. DDIs can be divided into two major categories. The first, the pharmacodynamic effect, occurs when the presence of one drug alters the response of a second administered drug through either an additive or antagonistic effect. The second, the pharmacokinetic effect, occurs when one drug (the “precipitant” drug) affects the absorption, distribution, metabolism, or excretion of a second drug (the “object” drug). Numerous examples of both classes of interactions have been shown to cause adverse effects in patients; however, instances involving the pharmacokinetic effect are much more common.
Because various cytochrome P450 isoforms are responsible for phase I metabolism of approximately 80% of drugs on the market (Ortiz de Montellano, 2005), the drug interactions ascribed to clearance by these enzymes remain to be the major focus of clinical practice. However, the potential for drug interactions for the remaining 20% of drugs that are metabolized via pathways other than cytochromes P450 still exist. Little is known about the pharmacokinetic interactions of these other xenobiotic-clearing enzymes such as the one of current focus: aldehyde oxidase.
Aldehyde oxidase (AOX) is a member of a group of molybdoflavo enzymes that includes the more extensively studied xanthine oxidase. AOX has been found to play a role in the metabolism of many drugs and xenobiotic compounds. As its name implies, AOX is known for its ability to oxidize aldehydes to carboxylic acids. However, AOX is also involved in the oxidation of nitrogen-containing heterocyclic compounds, as well as iminium ion intermediates (Brandänge and Lindblom, 1979; Whittlesea and Gorrod, 1993).
In a recent review, the authors predicted the growing importance of aldehyde oxidase in drug metabolism (Pryde et al., 2010). This is primarily due to the recent growth in the number of aromatic azaheterocycle moieties that are found in drug leads. These groups are typically installed in lead compounds for reasons including increased metabolic stability to cytochrome P450 oxidation (Chiba et al., 2001; Peng et al., 2010) and enhancement of desired pharmacodynamic properties.
Unlike those with cytochrome P450 (Bibi, 2008), the potential drug interactions in human AOX have not been well characterized. To date only one pharmacokinetic drug interaction due to the inhibition of AOX, between cimetidine and zaleplon, has been discovered (Lake et al., 2002). Predicting drug interactions with AOX is also relatively more difficult than with cytochrome P450 primarily because animal models are poor predictors of human AOX activity owing to the marked interspecies variation of expression levels and number of isoforms (Garattini and Terao, 2011). In a recent study, researchers have made steps to further the understanding of the inhibition of human AOX and the implications on drug interactions (Obach, 2004; Obach et al., 2004).
In 2004, Obach et al. (2004) screened a library of 239 drugs and xenobiotics for potential inhibition of AOX. Of these 239 compounds, 36 inhibited oxidase activity by 80% or greater at a test concentration of 50 μM. The inhibitory potency of these 36 compounds was measured using an IC50 determination assay. A number of different classes of drugs exhibited potent inhibition of AOX. Of particular note was the most potent inhibitor of all compounds that were tested, raloxifene, which has an IC50 value of approximately 3 nM.
A subsequent study by Obach (2004) focused on the mode and mechanism by which AOX was inhibited by raloxifene. For oxidation reactions studied, Obach found that the mode of inhibition was strictly uncompetitive. From this finding, the author concluded that there was a low risk of drug-drug interaction by raloxifene through an AOX-mediated pathway.
In the present study, eight different compounds of widely differing chemical structure were chosen to investigate the mode of inhibition of AOX (Fig. 1). Of these eight, six were previously shown to have relatively high inhibition potency through an IC50 value determination assay. Our primary goal was to elucidate the mode of inhibition for a number of potent inhibitors with varied chemical structure to provide insight into the potential for drug interactions, as well as to provide information regarding the mechanism for inhibition. Second, we wanted to compare the previously measured IC50 values of inhibitors with Ki values to assess the overall effectiveness of an IC50 screening assay for predicting potential drug interactions.
Fig. 1.
Structures of human liver aldehyde oxidase inhibitors.
Materials and Methods
Chemicals Used and Enzyme Source.
Phthalazine, 1-phthalazinone, 2-methyl-4(3H)-quinazolinone, estradiol, ethinyl estradiol, chlorpromazine, clozapine, menadione, domperidone, and 2,6-dichlorophenolindophenol were purchased from Sigma-Aldrich (St. Louis, MO). 6-Chloroquinazolinone was synthesized in our laboratory using methods described previously (Alfaro et al., 2009). Human liver cytosol (HLC), pooled from 10 individual donors, was purchased from BD Biosciences (Woburn, MA).
Incubation Conditions.
Incubations were performed using a modified technique as described previously by Obach et al. (2004). Incubation mixtures consisted of phthalazine (with a final concentration ranging from 1.6 to 100 μM) and inhibitor of varying concentration in 25 mM potassium phosphate buffer, pH 7.4, containing 0.1 mM EDTA. Inhibitor stock solutions were made up in dimethyl sulfoxide (DMSO) and added to the incubation such that the total concentration of DMSO was exactly 1% (v/v). To the incubations without inhibitor, 1% undiluted DMSO was added. Previous experiments demonstrated that this concentration of DMSO had no effect on phthalazine oxidase activity (Obach et al., 2004).
The reaction was initiated at 37°C in a shaking water bath incubator by addition of preincubated HLC at a final concentration of 0.05 mg of total protein/ml in the reaction mixture. The final volume of reaction solution was 800 μl. Reaction vials were shaken in open air for 2.5 min. Then the reactions were quenched with 200 μl of 1 M formic acid, containing a known concentration of 2-methyl-4(3H)-quinazolinone as internal standard. Quenched samples were centrifuged for 10 min at 5000 rpm using an Eppendorf centrifuge 5415D, and the supernatant was collected for analysis.
High-Performance Liquid Chromatography-MS Assay.
Samples were analyzed using an 1100 series high-performance liquid chromatography system (Agilent Technologies, Santa Clara, CA) and an API 4000 tandem mass spectrometry system manufactured by Applied Biosystems/MDS Sciex (Foster City, CA). Chromatography was performed on a Synergi Polar reverse-phase column (30 × 3.0 mm, 4-μm; Phenomenex, Torrance, CA).
Mobile phase A comprised 0.05% formic acid and 0.2% acetic acid in water, and mobile phase B consisted of 90% CH3CN, 9.9% water, and 0.1% formic acid. The column was equilibrated at initial conditions of 95% mobile phase A for 0.3 min. Chromatographic separation was achieved using a linear gradient over the next 2.2 min to 25% mobile phase A. Mobile phase A was then held at 25% over 0.5 min, followed by a linear gradient back to 95% A over 0.5 min. The column was reequilibrated to the initial conditions over the next 1.5 min. The total chromatographic assay time was 5 min/sample. The retention times for internal standard and metabolite were 2.5 and 2.8 min, respectively. An example of a chromatogram for internal standard and metabolite is shown in Supplemental Fig. 1.
The optimized mass spectrometer tune parameters for 1-phthalazinone were as follows: collision gas, 8; curtain gas, 15; ion source gas 1, 4; ion source gas 2, 1; ionspray voltage, 3500; desolvation temperature 300; declustering potential, 50; entrance potential, 3; collision energy, 30; collision cell exit potential, 10.
The analyte (1-phthalazinone) and internal standard [2-methyl-4(3H)-quinazolinone] were detected using multiple reaction monitoring mode by monitoring the m/z transition from 161 to 120 and 147 to 118, respectively. The MS system was operated in positive ion mode. Quantitation of product was achieved by extrapolating from a standard curve ranging from 0 to 3.2 μM metabolite.
Experimental Duplication and Controls.
Two controls were run with each batch of samples. The first control included omission of cytosol from the reaction and the second included omission of phthalazine (substrate).
All Ki determinations were measured in duplicate for each inhibitor using different batches of pooled HLC. In the first Ki measurement, a total of six inhibitor concentrations (including no inhibitor) and six substrate concentrations (1.6, 3.1, 6.3, 25, 50, and 100 μM) were used. In the second (duplicate) Ki measurement done on a different day, a total of four concentrations of inhibitor (including no inhibitor) and four concentrations of substrate (1.6, 6.3, 50, and 100 μM) were used. Fewer concentrations for inhibitor and substrate were analyzed for duplicate assays to expedite data collection and conserve resources and instrument usage.
Each individual reaction vial was measured by liquid chromatography-MS in triplicate. The average value was used for all calculations and plots.
Data Analysis.
Lineweaver-Burk plots and the subsequent replots of both y-intercept versus inhibitor concentration and slope versus inhibitor concentration were made to visualize the kinetic data (Cook and Cleland, 2007) using GraphPad Prism (version 4.03; GraphPad Software Inc., San Diego, CA). Mode of inhibition was determined by visual inspection of Lineweaver-Burk plots and statistical analysis of slope and y-intercept replot data. The slopes of replot linear regression lines were analyzed using an F-test feature that is built into the GraphPad Prism software. The F-test generates a p value that is used to determine statistical significance. p ≤ 0.05 was used as the threshold value for whether the slope for each linear regression line of the replots was significantly nonzero. Graphs with significantly nonzero slopes in the regression line of the replot data for both slope and y-intercept were determined to be mixed-mode type of inhibition. Graphs with a significant nonzero slope in the regression line of the slope replot but not in the y-intercept replot were determined to be strictly competitive. In contrast, graphs with a significant nonzero slope in the regression line of the y-intercept replot but not in the slope replot were determined to be uncompetitive inhibition.
Once the mode of inhibition was selected, the data were fit to the appropriate nonlinear regression model. Equations used for each model are shown below:
Competitive inhibition model:
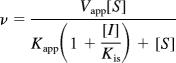 |
Mixed competitive inhibition model:
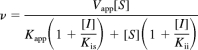 |
where v is the reaction velocity, Vapp is the apparent Vmax, [S] is the substrate concentration, Kapp is the apparent Michaelis-Menten constant, [I] is the concentration of inhibitor, Kis is the dissociation constant for the enzyme-inhibitor complex, and Kii is the dissociation constant for enzyme-substrate-inhibitor complex.
Results
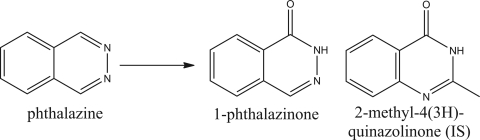
The AOX substrate chosen for this study was the aromatic azaheterocycle compound phthalazine. The oxidation reaction monitored in kinetic assays is shown in Fig. 2. Phthalazine has been shown previously to be a substrate with a relatively high reaction rate and a high specificity for AOX (Panoutsopoulos and Beedham, 2004). In addition, both phthalazine and its metabolite are commercially available compounds so quantitation of metabolite and substrate availability was not an issue.
Fig. 2.
Oxidation reaction of phthalazine to 1-phthalazinone. The structure of the internal standard (IS) used for mass spectrometry assay is provided on the right.
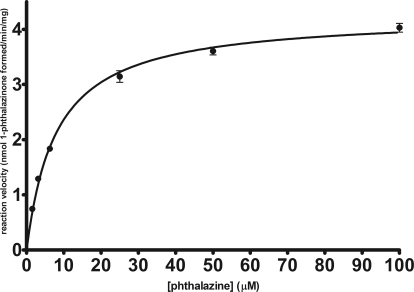
Multiple concentrations of phthalazine (substrate) and inhibitor were incubated in the presence of pooled HLC from 10 individual donors. After a period of 2.5 min, the reaction was terminated with a formic acid solution containing a known concentration of internal standard, 2-methyl-4(3H)-quinazolinone. Kinetic data were measured using a liquid chromatography-tandem mass spectrometry assay that monitored the formation of metabolite (1-phthalazinone). Figure 3 shows the substrate saturation curve for phthalazine oxidation. Saturation kinetic parameters were determined by nonlinear regression analysis using the Michaelis-Menten kinetic model as follows: Km = 8.0 ± 0.4 μM and Vmax = 4.3 ± 0.1 nmol · min−1 · mg protein−1.
Fig. 3.
Saturation kinetics plot for oxidation of phthalazine. Each point represents an average ± S.E. for a minimum of eight different experiments.
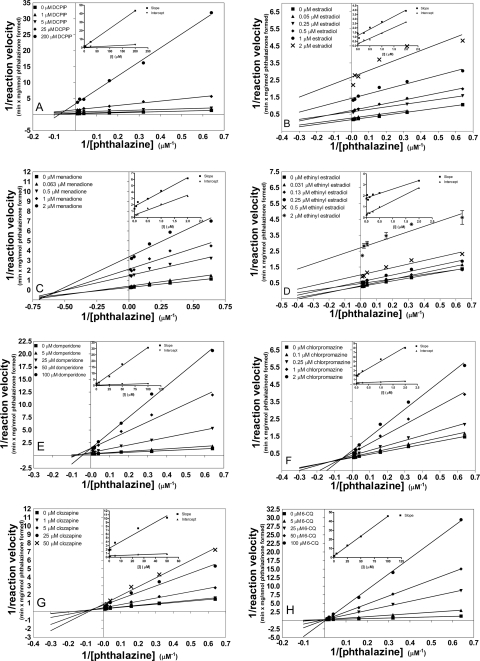
We then determined the mode of inhibition by visual inspection of Lineweaver-Burk (LB) plots as well as analysis of the statistical significance in both slope and intercept replots. The details of statistical analysis can be found under Materials and Methods. The LB plots and replots for all eight inhibitors are shown in Fig. 4, and the LB plots for duplicate measurements can be found in Supplemental Fig. 2. All Ki values were determined by nonlinear regression analysis using a global fit to the appropriate kinetic model.
Fig. 4.
Lineweaver-Burk plots and slope/intercept replots of phthalazine oxidation inhibition by eight different inhibitors: DCPIP (A), estradiol (B), menadione (C), ethinyl estradiol (D), domperidone (E), chlorpromazine (F), clozapine (G), and 6-chloroquinazolinone (6-CQ) (H). Each point reflects the average of triplicate determinations.
Of the eight compounds tested, seven showed a mixed-mode type of inhibition, exhibiting some character of both uncompetitive and competitive inhibition. Table 1 provides a full account of the Kis and Kii values obtained for the compounds tested. Kis is a measure of the affinity the inhibitor has for the free enzyme, whereas Kii is the affinity the inhibitor has for the enzyme-substrate complex. In addition, a Kii/Kis value was calculated for each compound as a metric of the degree of inhibition competitiveness. A larger Kii/Kis value indicates a mode of inhibition that is relatively more competitive, whereas a lower Kii/Kis value reflects a more uncompetitive type of inhibition. In other words, as the mode of inhibition becomes completely competitive, Kii → ∞ and, therefore, Kii/Kis approaches ∞.
TABLE 1.
Kis and Kii values for AOX
Each value reflects an average ± S.E. for two determinations.
| Inhibitor | Kis | Kii | Kii/Kis | IC50a |
|---|---|---|---|---|
| μM | ||||
| DCPIP | 19 ± 1.8 | 2.4 ± 0.18 | 0.13 | N.D. |
| β-Estradiol | 0.9 ± 0 | 0.13 ± 0 | 0.14 | 0.29 ± 0.07 |
| Menadione | 0.75 ± 0.18 | 0.12 ± 0 | 0.16 | 0.20 ± 0.04 |
| Ethinyl estradiol | 1.1 ± 0.32 | 0.23 ± 0.011 | 0.21 | 0.57 ± 0.15 |
| Domperidone | 5.3 ± 2.6 | 14 ± 2.8 | 2.6 | 3.0 ± 1.4 |
| Chlorpromazine | 0.62 ± 0.23 | 3.3 ± 0.53 | 5.3 | 0.57 ± 0.15 |
| Clozapine | 3.9 ± 0.35 | 60 ± 1.8 | 15 | 4.4 ± 1.8 |
| 6-Chloroquinazolinone | 5.4 ± 0.92 | N.D. | ||
N.D., not detected.
Measured in a previous study by Obach et al. (2004).
Inhibitors showed a wide variation in potency, but, more importantly, also varied in degree of competitiveness. In Table 1, inhibitors are ranked in order from least competitive to most competitive. 2,6-Dichlorophenolindophenol (DCPIP) inhibited phthalazine oxidation with moderate micromolar potency and was the most uncompetitive of the compounds tested. Similar to the findings previously reported by Obach (2004) for raloxifene inhibition, the β-estradiol compounds of similar structure inhibited phthalazine oxidation predominantly through an uncompetitive mode. However, unlike raloxifene, which was found to be exclusively uncompetitive, we observed a mixture of both competitive and uncompetitive inhibition. Menadione was also predominantly uncompetitive, but both Kis and Kii had a high degree of potency (nanomolar range). Domperidone inhibited mostly through a competitive mode, however, only with moderate micromolar potency. Chlorpromazine was mostly competitive with a Kis value in the nanomolar range. Of the compounds tested, only one compound, 6-chloroquinazolinone, was found to be exclusively competitive. This result was not surprising, because 6-chloroquinazolinone is a known substrate of AOX (Alfaro et al., 2009).
Discussion
In a previous report, researchers determined that the selective estrogen receptor modulator raloxifene inhibited AOX through an exclusively uncompetitive mechanism (Obach, 2004). In general, uncompetitive inhibitors are not considered to be significant candidates for drug interactions because the effect of such an inhibitor is only realized at very high substrate concentrations (near saturating conditions). In vivo, the substrate concentrations are typically very low relative to the Km and, thus, an uncompetitive inhibitor would have very little effect on the rate of metabolism. However, because we have demonstrated that the mode of inhibition for these inhibitors is mixed (i.e., containing both competitive and uncompetitive characteristics), the effect of the inhibitor is predicted to be significant even at substrate concentrations around the Km. Therefore, at relatively low substrate concentrations, those typically found in vivo, the potential for drug-drug interactions is much greater than was previously suspected.
Predicting the magnitude of drug interactions in vivo using in vitro data is a difficult challenge. One coarse method for such a prediction uses the ratio of [I]/Ki as a metric, where [I] is equal to the concentration of inhibitor in vivo. A value of 1 or greater is considered high risk for drug interactions, and a value between 1 and 0.1 is considered moderate risk. Any value less than 0.1 is considered a low risk of drug interactions (Tucker et al., 2001) and generally indicates that further in vivo study is not required.
One of the key challenges with use of this method to predict in vivo DDIs is selecting the most appropriate value for [I]. Previous attempts to define the value for [I] that best approximates the concentration of inhibitor at the enzyme site have used peripheral vein concentration, hepatic portal vein concentration, average plasma concentration, maximum plasma concentration, hepatic input concentration, and hepatocyte concentration (Blanchard et al., 2004; Ito et al., 2004; Bachmann, 2006). For each of these values, both bound and unbound fractions can be used. To date, different choices for [I] have had varying amounts of success, and there seems to be no clear consensus on which value for [I] is best (Bachmann, 2006). For the sake of simplicity, we chose to use the maximal plasma concentration for [I] for this discussion. This value is easily accessible in the literature and provides a reasonable, albeit relatively less conservative, estimate for the concentration of inhibitor at the enzyme site.
Of the compounds tested, we looked at four (ethinyl estradiol, β-estradiol, clozapine, and chlorpromazine) for drug interaction potential using the above criteria. These compounds were selected for discussion because of their clinical importance in man: ethinyl estradiol, clozapine, and chlorpromazine are commonly administered drugs, and β-estradiol is an endogenous hormone.
Peak drug concentrations and [I]/Kis values for the four clinically relevant inhibitors are shown in Table 2. Despite the Kii values for some inhibitors being much more potent, Kis values were used in the calculation for all inhibitors to best approximate the potential drug interactions in vivo. For reasons discussed earlier, use of a Kii value for this calculation may lead to an erroneous prediction of potential drug interactions.
TABLE 2.
Human peak plasma concentrations ([I]) and Kis values for AOX inhibitors
[I]/Kis represents an estimate of the likelihood of drug interactions in vivo.
| Inhibitor | Kis | [I]/Kis | |
|---|---|---|---|
| nM | |||
| Ethinyl estradiol | 0.44a | 1100 | 0.0004 |
| β-Estradiol | 12b | 900 | 0.01 |
| Clozapine | 1700c | 3900 | 0.4 |
| Chlorpromazine | 470d | 620 | 0.8 |
For normal women given a 30-μg dose (Westhoff et al., 2010).
In pregnant women (Piwowarska et al., 2010).
For patients given a 150-mg dose twice a day for 7 days (Brunton et al., 2006).
For patients given 100 mg/day for 30 days (Dahl and Strandjord, 1977).
The results display an enormous range of [I]/Kis values that are largely dependent on the concentration of drug found in vivo. Even in pregnant women, when β-estradiol levels are elevated, the concentration in vivo is very low relative to the Kis. Likewise, ethinyl estradiol is typically taken at very low doses in modern cocktails used as contraceptives. As such, the plasma concentration never reaches a value close to the Kis. The likelihood of drug interactions by the estrogenic compounds was found to be almost zero. However, the other two drugs we looked at are typically given to patients at much higher doses. Previous research has shown that the plasma concentrations of clozapine and chlorpromazine can reach as high as 1.7 and 0.5 μM, respectively (Dahl and Strandjord, 1977; Brunton et al., 2006). For these drugs, the [I]/Kis values are both between 0.1 and 1, indicating a moderate risk of drug interaction (Tucker et al., 2001). Further study is required in vivo to determine whether a potential for drug interaction exists. For example, coadministration of either chlorpromazine or clozapine with the sedative, zaleplon (a known substrate for AOX1), could possibly lead to an undesired intensification of the sedative effect of the drug.
It should be noted, however, that the examples of potential object drugs that may be involved in drug interactions are few. The previously mentioned substrate zaleplon, a sedative drug used primarily for the treatment of insomnia, is a nonbenzodiazepine hypnotic that is cleared primarily by AOX (Lake et al., 2002). In addition, famciclovir, an antiviral prodrug used primarily to treat herpes virus infection is activated by AOX to its active 6-oxo form, penciclovir (Clarke et al., 1995; Rashidi et al., 1997). Ziprasidone, which is used to treat the symptoms of schizophrenia, is metabolized by AOX (Beedham et al., 2003), as is the anticancer agent methotrexate (Jordan et al., 1999). These clinically used substrates are primarily but not exclusively metabolized by AOX with some portion of metabolism being through a pathway mediated by cytochrome P450 enzymes. Thus, the likelihood of DDI due to inhibition of AOX is reduced. There is currently a lack of a highly selective substrate on the market that is metabolized via a pathway that is exclusively mediated by AOX. Therefore, a clinical study of DDIs caused by the inhibition of AOX may not currently be achievable. Still, despite relatively few substrates presently on the market, the importance of AOX as a drug-clearing enzyme is predicted to increase markedly in the coming years primarily due to the growth in the number of nitrogen-containing heterocycles as drug candidates (Pryde et al., 2010). Perhaps a clinical substrate that is exclusively metabolized by AOX will be available in the near future.
The inhibition data described herein have implications not only for DDIs but also for the inhibition mechanism. Mixed-mode inhibition implies that the inhibitor has an affinity for both the “free” enzyme and the enzyme-substrate (E-S) complex. A lower Kii value relative to the Kis value means that the inhibitor has a higher affinity for the E-S complex than for the free enzyme. The traditional model for mixed inhibition involves one inhibitor binding site that is removed from the active site. This site may have its conformation altered by the presence or absence of substrate at the active site, such that the affinity for inhibitor is different for free enzyme and the E-S complex, which would explain the variation in degree of competitiveness for inhibitors found in the results of this study.
Researchers have previously found evidence of multiple egress sites for electron transfer (Rajagopalan and Handler, 1964). One possible explanation for inhibition is binding of the inhibitor to the electron transfer site, which prevents the electron transfer from reduced form of enzyme to molecular oxygen. This is supported by the fact that DCPIP, a known electron acceptor for AOX, was found to inhibit largely uncompetitively.
As shown in Table 1, the Ki values we measured were quite similar to the IC50 values determined by Obach et al. (2004). This result demonstrates that an IC50 screening assay of HLC is effective for determining potent inhibitors of AOX in HLC from a large library of drugs. However, as was shown in the case of raloxifene and other estrogenic compounds, taking into account the IC50 value alone without any information regarding the mode of inhibition may lead to an overprediction of drug interactions. We have shown that many inhibitors of AOX act primarily through an uncompetitive means such that any chance of drug interaction is slight. Therefore, because of the variation in competitiveness of various inhibitors, it appears that an effective strategy for determining whether or not the potential for drug interactions in vitro exists would be to first screen compounds for the IC50 and subsequently determine the mode of inhibition by measuring Ki for the most potent inhibitors screened.
In conclusion, the inhibition of oxidation of phthalazine by human aldehyde oxidase was characterized for eight different inhibitor compounds of largely varying structure. Of these eight, seven were demonstrated to inhibit the enzyme though a mixed mode. Potential drug-drug interactions for potent pharmacologically relevant inhibitors were discussed. Further study in vivo is warranted to assess the prediction of a moderate risk of drug interactions with clozapine and chlorpromazine.
Supplementary Material
Acknowledgments
Special thanks are given to Upendra Dahal for helpful discussion concerning mass spectrometry assays.
This work was supported by the National Institutes of Health National Institute of General Medical Sciences [Grant GM84546].
Article, publication date, and citation information can be found at http://jpet.aspetjournals.org.
doi:10.1124/dmd.111.041806.
The online version of this article (available at http://dmd.aspetjournals.org) contains supplemental material.
- DDI
- drug-drug interaction
- AOX
- aldehyde oxidase
- HLC
- human liver cytosol
- DMSO
- dimethyl sulfoxide
- MS
- mass spectrometry
- LB
- Lineweaver-Burk
- DCPIP
- 2,6-dichlorophenolindophenol
- E-S
- enzyme-substrate.
Authorship Contributions
Participated in research design: Barr and Jones.
Conducted experiments: Barr.
Performed data analysis: Barr.
Wrote or contributed to the writing of the manuscript: Barr and Jones.
References
- Alfaro JF, Joswig-Jones CA, Ouyang W, Nichols J, Crouch GJ, Jones JP. (2009) Purification and mechanism of human aldehyde oxidase expressed in Escherichia coli. Drug Metab Dispos 37:2393–2398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann KA. (2006) Inhibition constants, inhibitor concentrations and the prediction of inhibitory drug drug interactions: pitfalls, progress and promise. Curr Drug Metab 7:1–14 [DOI] [PubMed] [Google Scholar]
- Beedham C, Miceli JJ, Obach RS. (2003) Ziprasidone metabolism, aldehyde oxidase, and clinical implications. J Clin Psychopharmacol 23:229–232 [DOI] [PubMed] [Google Scholar]
- Bibi Z. (2008) Role of cytochrome P450 in drug interactions. Nutr Metab (Lond) 5:27. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Blanchard N, Richert L, Coassolo P, Lavé T. (2004) Qualitative and quantitative assessment of drug-drug interaction potential in man, based on Ki, IC50 and inhibitor concentration. Curr Drug Metab 5:147–156 [DOI] [PubMed] [Google Scholar]
- Brandänge S, Lindblom L. (1979) The enzyme “aldehyde oxidase” is an iminium oxidase. Reaction with nicotine Δ1′(5′) iminium ion. Biochem Biophys Res Commun 91:991–996 [DOI] [PubMed] [Google Scholar]
- Brunton LL, Gilman A, Goodman LS. (2006) Goodman & Gilman's the Pharmacological Basis of Therapeutics, 12 ed, McGraw-Hill, New York [Google Scholar]
- Chiba M, Jin L, Neway W, Vacca JP, Tata JR, Chapman K, Lin JH. (2001) P450 interaction with HIV protease inhibitors: relationship between metabolic stability, inhibitory potency, and P450 binding spectra. Drug Metab Dispos 29:1–3 [PubMed] [Google Scholar]
- Clarke SE, Harrell AW, Chenery RJ. (1995) Role of aldehyde oxidase in the in vitro conversion of famciclovir to penciclovir in human liver. Drug Metab Dispos 23:251–254 [PubMed] [Google Scholar]
- Cook PB, Cleland WW. (2007) Enzyme Kinetics and Mechanism, Garland Science, New York [Google Scholar]
- Dahl SG, Strandjord RE. (1977) Pharmacokinetics of chlorpromazine after single and chronic dosage. Clin Pharmacol Ther 21:437. [DOI] [PubMed] [Google Scholar]
- Garattini E, Terao M. (2011) Increasing recognition of the importance of aldehyde oxidase in drug development and discovery. Drug Metab Rev 43:374–386 [DOI] [PubMed] [Google Scholar]
- Ito K, Brown HS, Houston JB. (2004) Database analyses for the prediction of in vivo drug-drug interactions from in vitro data. Br J Clin Pharmacol 57:473–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan CG, Rashidi MR, Laljee H, Clarke SE, Brown JE, Beedham C. (1999) Aldehyde oxidase-catalysed oxidation of methotrexate in the liver of guinea-pig, rabbit and man. J Pharm Pharmacol 51:411–418 [DOI] [PubMed] [Google Scholar]
- Lake BG, Ball SE, Kao J, Renwick AB, Price RJ, Scatina JA. (2002) Metabolism of zaleplon by human liver: evidence for involvement of aldehyde oxidase. Xenobiotica 32:835–847 [DOI] [PubMed] [Google Scholar]
- Obach RS. (2004) Potent inhibition of human liver aldehyde oxidase by raloxifene. Drug Metab Dispos 32:89–97 [DOI] [PubMed] [Google Scholar]
- Obach RS, Huynh P, Allen MC, Beedham C. (2004) Human liver aldehyde oxidase: inhibition by 239 drugs. J Clin Pharmacol 44:7–19 [DOI] [PubMed] [Google Scholar]
- Ortiz de Montellano PR. (2005) Cytochrome P450: Structure, Mechanism, and Biochemistry, 3rd ed, Plenum, New York [Google Scholar]
- Panoutsopoulos GI, Beedham C. (2004) Enzymatic oxidation of phthalazine with guinea pig liver aldehyde oxidase and liver slices: inhibition by isovanillin. Acta Biochim Pol 51:943–951 [PubMed] [Google Scholar]
- Peng CC, Pearson JT, Rock DA, Joswig-Jones CA, Jones JP. (2010) The effects of type II binding on metabolic stability and binding affinity in cytochrome P450 CYP3A4. Arch Biochem Biophys 497:68–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piwowarska J, Radowicki S, Pachecka J. (2010) Simultaneous determination of eight estrogens and their metabolites in serum using liquid chromatography with electrochemical detection. Talanta 81:275–280 [DOI] [PubMed] [Google Scholar]
- Pryde DC, Dalvie D, Hu Q, Jones P, Obach RS, Tran TD. (2010) Aldehyde oxidase: an enzyme of emerging importance in drug discovery. J Med Chem 53:8441–8460 [DOI] [PubMed] [Google Scholar]
- Rajagopalan KV, Handler P. (1964) Hepatic aldehyde oxidase. 3. The substrate-binding site.J Biol Chem 239:2027–2035 [PubMed] [Google Scholar]
- Rashidi MR, Smith JA, Clarke SE, Beedham C. (1997) In vitro oxidation of famciclovir and 6-deoxypenciclovir by aldehyde oxidase from human, guinea pig, rabbit, and rat liver. Drug Metab Dispos 25:805–813 [PubMed] [Google Scholar]
- Tucker GT, Houston JB, Huang SM. (2001) Optimizing drug development: strategies to assess drug metabolism/transporter interaction potential—toward a consensus. Pharm Res 18:1071–1080 [DOI] [PubMed] [Google Scholar]
- Westhoff CL, Torgal AH, Mayeda ER, Pike MC, Stanczyk FZ. (2010) Pharmacokinetics of a combined oral contraceptive in obese and normal-weight women. Contraception 81:474–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittlesea CM, Gorrod W. (1993) The enzymology of the in-vitro oxidation of prolintane to oxoprolintane. J Clin Pharm Ther 18:357–364 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.