Abstract
Dietary transitions in human history have been suggested to play important roles in the evolution of mankind. Genetic variations caused by adaptation to diet during human evolution could have important health consequences in current society. The advance of sequencing technologies and the rapid accumulation of genome information provide an unprecedented opportunity to comprehensively characterize genetic variations in human populations and unravel the genetic basis of human evolution. Series of selection detection methods, based on various theoretical models and exploiting different aspects of selection signatures, have been developed. Their applications at the species and population levels have respectively led to the identification of human specific selection events that distinguish human from nonhuman primates and local adaptation events that contribute to human diversity. Scrutiny of candidate genes has revealed paradigms of adaptations to specific nutritional components and genome-wide selection scans have verified the prevalence of diet-related selection events and provided many more candidates awaiting further investigation. Understanding the role of diet in human evolution is fundamental for the development of evidence-based, genome-informed nutritional practices in the era of personal genomics.
Introduction
Food represents one of the most important environmental factors for humans. Genetic adaptations to the diet consumed throughout human history have sculpted the human genome and influenced a variety of human traits. Local adaptations to regionally specific dietary components might have been one critical shaping force of the human genome, driving population differentiation and laying the genetic basis for human diversity. Genomic adaptations to environmental factors through increasing frequency of advantageous mutations in the human population usually take hundreds of generations (thousands of years), whereas societal, cultural, and dietary transformations in the human society are ever-accelerating. Maladaptations of the lagging genome to rapid dietary shifts may underlie a wide range of so-called civilization diseases, such as diabetes, obesity, cardiovascular diseases, and cancers. Extensive efforts have been invested into the examination of genomic adaptations to diet with the hope of elucidating the genetic basis of complex disorders and tailoring strategies of disease management through personalized medicine and nutrition (1–3).
The objective of this review is to briefly overview the shaping effects of food on the human genome and to discuss insights learned from evolutionary research into genome-informed clinical and nutritional practices. Along the way, we will summarize the main dietary transitions in human history, the mode of natural selection, approaches utilized to detect selection signals, and knowledge gained about the genetic basis of dietary adaptation. Comprehensive reviews could be found elsewhere of evidence for adaptations to diet (2, 3), or more generally of human demographic history and evolutionary adaptations (4–6). Specialized reviews of approaches for detecting selection and their specific advantages and limitations are also available elsewhere (7–9).
Major dietary transitions during human evolution
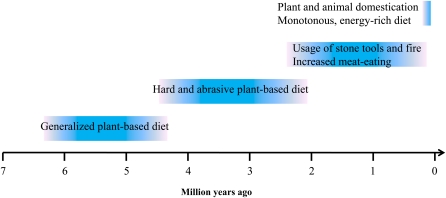
There might be 3 major dietary transitions in human history (Fig. 1). Upon the split of humans and chimpanzees ∼4.6–6.2 mya3 (10), the early hominid (Ardipithecus ramidus) was suggested to reside in a predominantly wooded habit and exploit a generalized plant-based diet (11, 12). Around 4 mya, Australopithecus, the predecessor of Homo, began to transfer to more open landscapes and consume harder and more abrasive foods, such as large seeds with tough shells and underground storage organs. This dietary pattern was supported by the adaptively enlarged teeth and thickened enamel (11). Homo arose ∼2.5 mya and began to use stone tools, leading to an increased consumption of animal source foods through scavenging and hunting (11, 13). Another important technological innovation in this period, around 1.6 mya onward, was the controlled use of fire (14), which also contributed to this dietary shift. This dual dietary strategy, providing essential nutrients from animal source foods and energy mainly from plant source foods, made possible the evolution of humans as a large, active, and highly social primate with an unraveled complex brain (15).
FIGURE 1.
Major dietary transitions along human history. The dietary pattern of each period is depicted with a box with blurry boundaries representing the transitional processes.
Around 200 kya anatomically modern humans (Homo sapiens) originated in Africa and expanded to other parts of the world (16, 17). The Neolithic Revolution, which was characterized by the domestication of plants and animals, occurred ∼10 kya and stimulated another dramatic dietary transformation in human history. Varying environmental resources on different continents led to domestications of local species and thus adoptions of regionally specific dietary patterns. In spite of the pronounced explosion of food production, only a few wild species were domesticated and have since then been heavily relied on (18). Some agricultural societies might draw 90% of their energy input from domesticated plants (19). Overall, this dietary transformation resulted in loss of nutritional diversity, excess of caloric availability, and sedentary lifestyle, which might contribute to many of the health challenges in modern society (20–22).
Adaptations to dietary transitions during human evolution have left specific signatures in our genome. Identifying these signatures with the hope of unraveling human evolutionary history and improving human health has been an active research direction in the fields of evolutionary genomics, population genetics, and molecular evolution.
Genetic variation and its functional significance
Genetic variation underlies the tremendous range of phenotypic diversity and disease susceptibility in human populations. Any 2 individuals are estimated to be different in 1–3% of their genomes (23). Each person is estimated to carry ∼250–300 genes with loss-of-function variants, 50–100 of which are implicated in inherited disorders (24). With the rapid advance of sequencing technologies, especially the advent of next and next-next generation sequencing platforms, extensive efforts have been invested into unraveling human genetic variations and understanding their general features, forces shaping their pattern, and, most importantly, their clinical consequence. Genome-wide association studies have been successful in linking some genetic variations to complex traits and common diseases. In spite of confronting difficulty in explaining all genetic impacts, it still holds the promise of elucidating the genetic architecture of human health, providing interesting hypotheses and pointing out possible directions for medical research (25).
Genetic variations include SNPs, SV, and chromosomal abnormality. SNP is the simplest, most common, and most well-studied variation. It is estimated that there are in total 10–15 million common SNPs with a frequency > 1% in human genomes and 3 million are estimated to be present in each individual (24, 26–28). SV, according to the molecular mechanisms of their origin, could be classified into deletion, insertion, reversion, translocation, duplication, and their complicated combinations. Some SV result in different copy numbers of sequence unit and are called copy number variations. The importance of SV has been underestimated until the recent development of sequencing technologies, which makes its characterization possible. It is estimated that copy number variations cover 12–30% of the human genome (29, 30). A big proportion of genetic variations do not have fitness consequence, meaning that their existences will not make their carriers more sick or healthier. Only functional variations that influence the fitness of their carriers are subject to natural selection. Functional variations in coding regions may directly change protein structure or its function and those in noncoding regions may interfere with the regulation of gene expression. Usually, coding SNPs are classified into synonymous SNPs and nonsynonymous SNPs. Synonymous SNPs do not change the peptide sequence, because 2 codons encode the same amino acids. Nonsynonymous SNPs may change the amino acid (missense SNPs) or introduce a stop codon (nonsense SNPs). Nonsynonymous SNPs are more likely than synonymous ones to have phenotypic consequences.
The pattern of genetic variation in human populations is the product of interactions among demographic history (e.g. effective population size, population structure, and migration), gene-specific factors (mutation and recombination rate), selection pressure, and random processes (also called genetic drift) (31). Mutation rate determines how many variations are introduced into the genome. Under simple demographic models without selection, the mutation rate equals the evolutionary rate. Recombination rate is another shaping force of variation pattern, especially when coupled with selection and demographic history. The recombination rate is the main parameter determining the degree of linkage disequilibrium between adjacent variants and the coupling of their evolutionary fate. Regions identified as targets of selection show a drastically reduced level of recombination, probably due to the fact that a low recombination rate decelerates the deterioration of selection signatures (8, 32). For the same reason, regions of high recombination rate attenuate the sweeping effect of selection and tend to have high variability, though there is also a hypothesis that recombination could be mutagenic (33). Demographic history is an important determinant of variation patterns in different populations (34). Demographic events, such as population bottleneck and population growth, can create variation patterns that mimic those of selection. Thus, it is always important to take into account the effect of demographic history when interpreting potential selection signals (31, 35). Selection occurs only on functional variations and thus creates dramatically different patterns of evolution and population dynamics between functional and nonfunctional variants, which usually are regarded as important indicators of selection (9).
Different modes of natural selection
Natural selection can be broadly defined as the process in which beneficial heritable traits increase in frequency and unfavorable ones decrease. The reflection of this process at the molecular level is the spread of advantageous alleles and the purification of deleterious ones in the population. Natural selection happens in a variety of modes. According to the trajectory of frequency shift for the allele under study, selection can be classified into 3 categories: positive selection, negative selection, and balancing selection. Positive selection refers to the process whereby a beneficial allele spreads in the population and ultimately reaches fixation. In contrast, negative selection is the process by which a deleterious allele decreases in frequency and ultimately becomes extinct. Balancing selection occurs under 2 scenarios. In one scenario, the presence of multiple conflicting selective pressures, instead of a single selective pressure or multiple selective pressures in a single direction, renders the targeted allele both beneficial and deleterious. Ultimately, the frequency of the targeted allele will fluctuate around a specific level. The other scenario is called heterozygous advantage under which the heterozygote is favored and both alleles are maintained in the population at the equilibrium frequency.
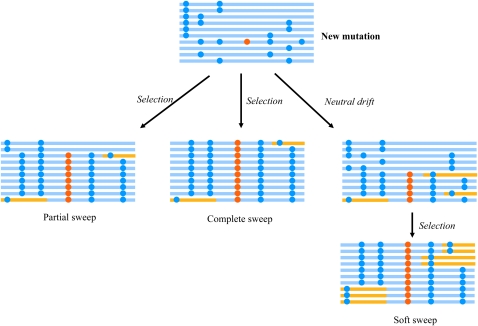
Positive selection is of special interest, because it underlies regional adaptation and the development of novel traits. Positive selection alone has a wide range of modes, leaving distinct signatures along the selected genomic region (36). Two extensively studied models of positive selection are complete sweep and partial sweep (Fig. 2), both of which emphasize the onset of selection on a newly mutated allele, driving it to rapidly increase in frequency. Complete sweep describes the scenario in which the allele reaches fixation in the population, whereas partial sweep refers to the situation that the allele only reaches high frequency but not fixation. Under both scenarios, the mutation of the advantageous allele happens in a specific genomic background (haplotype) and is linked with specific alleles of nearby variable sites. The strong selection on the favored allele rapidly drags the haplotype to high frequency or fixation in such a short time that mutation and recombination do not drastically degrade the selected haplotype, leaving specific selection signatures, including long-range haplotype with high frequency, skewed SFS (excess of rare variants or excess of high-frequency derived alleles), reduced variation level in the selected region, etc. (7, 9).
FIGURE 2.
Models of hard sweep (including partial sweep and complete sweep) and soft sweep. Each horizontal line represents a haplotype defined by different combinations of alleles of adjacent variable sites. Blue lines are the haplotype present before selection, and the new haplotype generated from recombination are depicted with an additional color (yellow). The derived allele (mutation) of a variable site is illustrated as a dot. The orange dot represents the beneficial mutation under selection. Hard sweep, both partial and complete, emphasizes the onset of selection on a new advantageous mutation. Haplotypes, old or new, carrying the selected mutation will rapidly increase in frequency to high frequency (partial sweep) or fixation (complete sweep). The time to reach high frequency or fixation is so short that not many recombination events occur. Soft sweep occurs on a standing variation rather than a new mutation. Before the onset of selection, the mutation is neutral and its frequency fluctuates randomly. In this period of neutral drift, recombination creates new haplotypes carrying the mutation. Once environmental change renders the mutation beneficial, all haplotypes carrying this mutation will rapidly spread in the population. Although the selected mutation also reaches high frequency or fixation, the haplotype homogeneity in the surrounding regions is degraded. Part of the figure was adapted from (8).
In contrast with complete sweep and partial sweep, both of which are also called hard sweep, soft sweep depicts another situation under which selection operates on standing variations or simultaneously on multiple alleles of a locus (Fig. 2). Standing variations refer to variations that have been present and evolved neutrally before the action of selection when environmental shifts render them adaptive. The initial neutral drifting phase allows mutation and recombination to erode the homogeneous genetic background upon which the new mutations arise. When multiple alleles of a locus are similarly adaptive, all of them will be selected, but none of them will be able to reach fixation. In this situation, the selected alleles have limited effects on the linked sites (37).
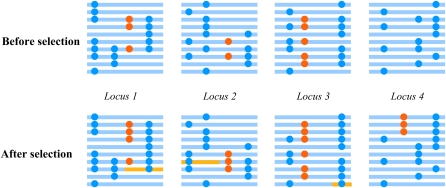
Selective sweeps, both hard and soft, emphasize the drastic frequency change before and after selection. However, adaptation could also happen through subtle allele frequency shifts of many loci, which was recently proposed as a model of polygenic adaptation (Fig. 3) (36–38). For complex traits controlled by multiple genes of small effect, a new optimum phenotype could be reached by simultaneous subtle allele frequency shifts across many loci without dramatic allele frequency changes or fixation events. The relative importance of hard sweep, soft sweep, and polygenic adaptation to human evolution is still difficult to estimate, because most studies have focused on hard sweep and it was only possible in the past decade to perform genome-wide assessment of natural selection events. Moreover, both soft sweep and polygenic adaptation received attention only recently. Theoretical modeling and empirical genome-wide examination are required to understand their relative importance.
FIGURE 3.
Polygenic adaptation model. Multiple loci (4 in this case) are responsible for a single trait. Most loci (loci 1, 2, 3) harbor mutations, later becoming beneficial, while there are also loci (locus 4) gaining new advantageous mutations after the onset of selection. The selected mutations are present at different frequencies in the population before selection as a result of random drift. Upon environmental change, selection acts on all these beneficial mutations and elevates their frequency. However, each mutation only needs to have a slight increase in frequency.
Neutrality tests for detecting natural selection
A series of neutrality tests has been developed to identify selection signatures that are distinct from those of neutral evolution. Different tests exploit different aspects of selection signatures, have specific underlying assumptions (about selection mode, strength, scale, etc.), and have varying power for different selective modes. Human-specific adaptation events after the split of humans with other primates can be detected by comparative genomic analysis between human and nonhuman primates, and regional adaptation events along with human global migration can be analyzed using human population genomic data from populations of different habitats. For each neutrality test, it is essential to assess the significance of the signal, i.e., to differentiate a true selection signal from those by neutral processes. For this purpose, neutral signals are usually gained from simulations with equivalent demographic models without selection or from empirical distributions of functionally neutral loci. Due to the complexity of human demographic history, it is usually difficult to develop an accurate, realistic model. With the availability of genome-wide variation data, however, using empirical neutral distribution is more practical and convenient. Neutrality tests have been applied for specific genes and for genome-wide selection scans. The candidate gene approach is hypothesis driven and usually provides detailed evolutionary stories, whereas genome-wide scans are unbiased, which can reveal a complete picture of adaptation events and generate hypotheses for further candidate gene approach. A large number of loci under selection have been revealed with the application of these methods.
Comparative analysis.
Without selection, the evolutionary rate of functional mutations (e.g. nonsynonymous mutations, regulatory mutations) should be the same as that of nonfunctional ones (e.g. most synonymous mutations). However, if there were repeated selective events on multiple functional mutations in a gene that drove these mutations to fixation, we would find an elevated evolutionary rate of functional mutations compared with that of neutral ones (dN/dS test) (39–41). In the absence of selection, the polymorphism levels within species and sequence divergence between species should be positively correlated. Similarly, the ratio of nonsynonymous:synonymous changes between species should be equal to that within species. Departures from these neutral expectations are signals of natural selection and could be tested by HKA and MK tests, respectively (42, 43). A creative comparison of the evolutionary rate at the promoter regions with that at neutral intron regions yielded a surprising discovery that nutrition-related genes have experienced positive selection at the regulatory regions (41). It is worthwhile to point out that classical examples of metabolic adaptation, such as lactose digestion (44) and starch metabolism (45), experienced their selections on gene regulation and adapted to varying food sources by changing gene expression levels. Selection on regulatory regions may have played an important role in the evolution of metabolic genes, though its relative importance compared to selection on coding regions during human evolution is still difficult to assess.
SFS-based tests.
After a complete selection sweep, the variation levels along the selected regions were reduced, because most variations were wiped out due to fixation of the selected alleles. Although new mutations began to accumulate in this region, they were present only in low frequency given limited time after the selection event. Thus, the SFS skewed with an excess of low-frequency polymorphisms. Several tests, such as Tajima’s D test (46) and Fu and Li’s tests (47), have been developed for this type of signature left by complete sweep. Under the scenario of partial sweep, the rapid increase of a beneficial allele to high frequency drags alleles of nearby loci to similarly high frequencies (a phenomenon called hitchhiking). If the derived alleles of linked loci happened to be in linkage with the favored allele, the hitchhiking effect will result in an excess of high-frequency derived alleles, which would otherwise remain at low frequency given their young age. This type of skewed SFS can be detected with Fay and Wu’s H test (48). Because complete resequencing data are needed to capture the accurate SFS with extremely rare variations, SFS-based tests are usually applied in candidate gene studies. For instance, CYP3A4 and CYP3A5 genes have a marked excess of rare variants in Asian and European populations as revealed by negative Tajima’s D. CYP3A4 gene also has an excess of high-frequency derived alleles in both populations (49). Taken together with other evidence, these 2 genes are suggested to be adaptive to climate-related selection, contributing to the current population disparity in the prevalence of salt-sensitive hypertension (49).
Haplotype-based tests.
When a new advantageous mutation arose, it did so in a specific haplotype background. Positive selection on the beneficial variant dragged the haplotype to high frequency in such a short time that mutation and recombination did not substantially break down the haplotype. Thus, a long-range haplotype with an unusually high frequency in a population is a signature of positive selection (50, 51). To control for regional heterogeneity of recombination rate, several derived tests were developed to compare the haplotype of interest with other haplotypes in the same region (50, 52). Although these tests are powerful for partial sweeps, they lose power for complete sweeps or when the selected allele approaches fixation, because there are few alternative alleles to serve as control. A comparison between populations may solve this problem, because when the selected allele reaches fixation in one population, it remains polymorphic in the others (53). A genome-wide selection scan using haplotype-based tests in 3 main continental populations (African, European, and Asian) found genes with exceptionally long common haplotypes in one or more populations. Examples include LCT (a gene responsible for lactose digestion) in Europeans, GBA (a gene associated glycogen-storage disorder) in East Asians, NKX2–2 (a gene associated with carbohydrate metabolism and blood sugar regulation) in Europeans, and CYP genes (involving in detoxification of foreign compounds) in Asians and Europeans (52).
Population differentiation and environmental correlation.
The global environmental disparity creates an adaptive landscape of drastic heterogeneity. The presence or absence of certain environmental factors confers selective pressure only in some populations but not others. Besides dichotomous environmental factors, continuous ones with geographical clines confer a gradient of selective strength, leading to gradual adaptive frequency changes. Furthermore, similar selective pressure in different populations, either geographically distant or close, might result in parallel adaptive frequency shifts. These spatial patterns of variation as a result of adaptation to varying environmental variables can be detected by population differentiation or environmental correlation methods. Population differentiation measures the difference of allele frequency between populations. Large population differentiation usually indicates the presence of natural selection and local adaptation. Wright’s fixation index and its derivatives have been used for this purpose (54, 55). ADH1B in alcohol metabolism (56), HFE in iron metabolism (57), LCT in lactose digestion (58), among many other metabolic genes, have variations with an unusually high level of population differentiation.
A direct correlation between allele frequency and environmental variables is also a signature of adaptation to various degrees of selective pressure. This method is called environmental correlation. One advantage of this method is that it is applied with the knowledge of selective pressure, whereas all other approaches described above only examine the sequence information. Another advantage is that it is powerful even for subtle allele frequency shifts, making it especially useful for selection on standing variations or polygenic adaptation model. One confounding factor in environmental correlation is population structure, because geographically close populations tend to share similar frequency whereas those of distant populations are different. Although there are parallel allele frequency shifts in distant populations due to similar selective pressure, these changes might not be apparent unless the regional averages are controlled for (37, 59). A Bayesian linear model was recently developed to assess the evidence of allele frequency and environmental variable correlation while controlling for the covariance of allele frequencies between populations as a result of population structure (60). Two genome-wide selection scans with this new method utilized 61 global populations and assessed the correlations between ∼650,000 SNPs and 20 environmental factors. They discovered strong signals of adaptation to polar eco-region, foraging subsistence, diets rich in roots and tubers, and several climate factors (e.g. temperature, precipitation, and solar radiation) (61, 62). Genes involved in metabolism were especially found to play an important role in adaptation to these environmental factors (61, 62). For instance, pathways of starch and sucrose metabolism, and folate biosynthesis are enriched with strong signals of adaptation to a diet rich in roots and tubers, which are poor dietary sources for folic acids (61). Intriguingly, genes underlying complex diseases, including common metabolic disorders (e.g. type 2 diabetes, obesity, hypertension, and dyslipidemia), show a significant overlap with these adaptive signals (61–63).
Applications of these statistical methods in candidate gene investigations and/or genome-wide selection scans have revealed a large number of genes, functional groups, and pathways under selection during human evolution. Well-studied cases of adaptation to food or diet-related practices, which are discussed in detail hereafter, include regulatory variants of the lactase gene adaptive to milk consumption, copy number variation of amylase gene adaptive to starchy food, enhanced ADH1B activity that prevents alcohol overconsumption, increased sensitivity of bitter taste receptor gene TAS2R16 to avoid ingestion of plant toxins, etc. Beside individual genes, certain pathways or functional groups are also enriched with genes or polymorphisms under selection. Genome-wide scans for recent selection signatures found that selective signals are enriched in starch/sucrose metabolism and folate biosynthesis pathway (61), carbohydrate, steroid, phosphate metabolism and vitamin/cofactor transport (52), protein and DNA metabolism (51), etc. Ancient selection signatures were found to be enriched in the promoter regions of nutrition-related genes (41), nucleic acid metabolism (64), amino acid metabolism (40), etc. It is advisable to bear in mind that different approaches of selection detection utilize different types of data, exploit different aspects of selection signatures, and detect selective events at varying levels and timescale. Between-species comparisons (dN/dS test, MK test, HKA test) examine selective events older than 1 mya at the early stage of human evolution, whereas population genomics analyses (haplotype-based tests, frequency spectrum-based tests, population differentiation, and environmental correlation) will detect more recent adaptation events, usually more recent than 10 kya (4, 8, 9).
To illustrate the action of natural selection and approaches to identify its signatures, we will discuss several well-studied cases of adaptation of metabolism and perception to dietary components and/or diet-related practices. We need to point out that our discussion here is intended to be representative but surely not exhaustive and we apologize for missing some interesting studies in this area.
Specific examples for diet-related adaptation
LP.
Lactase, an enzyme expressed in intestine, is required for the digestion of lactose in milk. Most mammals, including humans, lose the ability to digest lactose after weaning due to reduced lactase expression. The persistence of the ability to digest milk lactose into adulthood is called LP, which has a genetic basis and is inherited as a dominant trait. The prevalence of LP is high among northern Europeans (>90% in Swedes and Danes), decreases across southern Europe and the Middle East (∼50% in Spanish, French, and pastoralist Arab populations), and is low in the African and Asian agriculturalists (∼5–20% in West African and ∼1% in Chinese). Notably, LP is also prevalent in African pastoralist groups, like the Tutsi (∼90%) and Fulani (∼50%) (44). Interestingly, LP has a different genetic basis in these populations. Two SNPs, C/T-13910 and G/A-22018 (rs4988235 and rs182549) identified in the cis-regulatory elements of the gene encoding lactase (LCT), are highly associated with LP in European populations and C/T-13910 is proposed to be the causal regulatory site (65). However, the SNP C/T-13910 is present only with low frequency in a few West African pastoralist populations and absent in others. Recent studies in Eastern African populations identified 3 new SNPs to be significantly associated with LP and enhance transcription of the LCT in vitro (44). A more recent study in Middle Easterners also revealed a population-specific haplotype responsible for LP (66). Evolutionary analyses (population divergence, long-range haplotype, etc.) detected significant selection signatures on these functional variants and age estimates of mutations (∼8000–9000 y ago in Europeans, ∼2700–6800 y ago in East Africans, ∼4000 y ago in Saudi Arabians) are consistent with the timeframe of animal domestication in each region, indicating convergent adaptations in different regions to similar selective pressures of milk consumption (44, 58, 66).
Starch digestion.
The gene AMY1, encoding salivary amylase for starch digestion, shows extensive variations in copy number among individuals and between human populations. The copy number of the gene is positively correlated with its protein expression. Populations consuming high-starch diets, such as agricultural populations of European Americans, Japanese, and Hadza hunter-gatherers, have a higher copy number of AMY1 than low-starch populations, like hunter-gatherers in the rainforests and near the Arctic Circle. Comparative analyses with other primates suggest that the additional copy of AMY1 was gained in the human lineage. The low amount of nucleotide divergence among different gene copies might be a result of recent origin that fell within the timeframe of modern human origins (∼200,000 y ago). Taken together, the copy number variations of AMY1 among different populations are suggested to be a result of regional adaptation to diets with varying starch contents (45).
Alcohol metabolism.
Ethanol is oxidized to acetaldehyde by the enzyme alcohol dehydrogenase (ADH), and acetaldehyde is subsequently oxidized to acetic acid by aldehyde dehydrogenase (ALDH). The metabolism of ethanol varies widely among individuals due to genetic variations in these 2 genes. One of the best-studied polymorphisms influencing ethanol metabolism is ADH1B Arg47His (rs1229984). The derived allele ADH1B*47His changes the pKa of alcohol dehydrogenase from 8.5 to 10.0, leading to a 40- to 100-fold increase in Km and Vmax, respectively, of alcohol metabolism (67). The global distribution of ADH1B*47His reveals that this allele reaches high frequencies only in western and eastern Asia but is nearly absent in other regions (68). Population differentiation tests and long-range haplotype tests both provide evidence of positive selection in East Asian populations (56, 67, 69). Interestingly, molecular dating suggests that the emergence of this allele (7000∼10,000 y ago) coincides with the origin of rice domestication in East Asia and that there is a strong correlation between ADH1B*47His frequency and the age of rice domestication (67). Thus, it was proposed that the rise of ADH1B*47His frequency was an adaptation to rice domestication and the subsequent production and consumption of fermented food and/or beverages. ADH1B*47His enables the rapid accumulation of acetaldehyde after alcohol consumption. Acetaldehyde is toxic and can cause a flushing reaction, which is proposed to be protective from alcohol overconsumption (67). Consistently, ADH1B*47His has long been shown to be protective against alcoholism (70, 71).
Perception and bitter taste.
Our perceptions of food, including vision, olfaction, and taste shape our dietary preference and influence our health. Humans overall have remarkably reduced sensory capabilities compared with other mammals. In accordance with the phenotypic reduction, genes related with olfaction, taste pheromone, and hearing have experienced relaxation of selective constraint and loss of function during human evolution (72–76). However, in contrast to this general trend, some perception-related genes are still targets of natural selection. The long-wavelength opsin responsible for red color vision was suggested to be under positive selection during human evolution (77). The gain of full trichromatic color vision was proposed to contribute to the deterioration of genes related to olfaction and pheromone, because the reliance on visual signals to communicate social and reproductive status might reduce the importance of chemical signals (73, 78). Moreover, bitter-taste receptor gene TAS2R38 has signatures of balancing selection (79), whereas TAS2R16 is suggested to be target of positive selection (80).
Taste is of specific interest, because it is the major determinant of food selection. Our perception of bitter taste is mediated by a highly variable family of seven-transmembrane G protein-coupled receptor genes (TAS2Rs), which experienced family expansion by tandem gene duplication during mammalian evolution (81, 82). There are 38 putatively functional TAS2R genes and 5 pseudogenes with disrupted open reading frames, organized in 3 clusters and located in chromosomes 5, 7, and 12 (83). Population and comparative genetic analyses of 25 TAS2R genes among humans and other primates suggest that this group of genes has undergone relaxation of selective constraints and been in the process of loss of function through pseudogenization (75, 84). Evidence supporting relaxed selective constraints can be drawn from equal evolutionary rates of nonsynonymous to synonymous polymorphisms within the human species, equal rates of nonsynonymous to synonymous substitutions between humans and other mammals, and the presence of nonsense mutation at medium frequency and pseudogenes (75, 84). It was suggested that the dietary shift at the early stage of human evolution with increased intake of animal-based food reduced the ingestion of plant toxins and the controlled use of fire helped detoxify poisonous compounds, so the importance of bitter-tasting was reduced and related genes were free to deteriorate (75).
In spite of the general trend of relaxed selection, one bitter taste receptor gene, TAS2R16, shows signatures of adaptation. TAS2R16 has an excess of high frequency-derived alleles as measured by Fay and Wu’s (80) H statistics in worldwide populations. The derived allele at one of its nonsynonymous mutations, K172N (rs846664), has been shown to increase sensitivity to toxic β-glucopyranosides, which are ubiquitous in nature and synthesized by over 2500 plants and insects as a mean of protection against predators. Molecular dating revealed that this allele arose 78,700–791,000 y ago in the Middle Pleistocene and before the expansion of early humans out of Africa. Thus, it was suggested that K172N drove the positive selection of TAS2R16 at the early stage of human evolution, because it protected its carriers from consuming cyanogenic plant toxins (80). Another bitter taste receptor gene known to be under selection is TAS2R38, which is responsible for the classic phenotype of phenylthiocarbamide tasting. TAS2R38 has 2 dominating intermediate-frequency haplotypes corresponding to PTC-taster and -nontaster phenotypes. These 2 haplotypes have similar frequencies in African, European, and Asian populations and have little population differentiation (low Wright’s fixation index) among them, indicating a similar evolutionary history. SFS-based tests (Tajima’s D, Fu and Li’s D and F) revealed an excess of intermediate-frequency variants, suggesting a role of balancing selection. However, the selective pressures are yet to be found (79, 85).
Insight into nutritional practices from evolutionary research
Evolutionary analyses of genetic adaptations to food and dietary-related behaviors not only contribute to our knowledge of human evolution but also increase our understanding of the genetic bases of complex traits or diseases and thus facilitate clinical and nutritional practices. Many genetic loci adaptive to specific food sources or dietary practices are also involved in pathological processes. For instance, the ancestral allele of K172N in TAS2R16 contributes to a higher risk of alcohol dependence because of its decreased sensitivity to bitter-taste stimuli, including alcoholic beverages (86, 87). Similarly, the nontaster genotype of TAS2R38 reduces bitter sensitivity and correlates with higher alcohol consumption (87, 88). Furthermore, the understanding of the human nature of bitterness avoidance as an adaptive mechanism in the evolutionary past emphasizes the importance of comprehensive examination of food properties and rational dietary choice, because not all bitter substances are poisonous and harmful. For instance, plant-derived phytonutrients (phenols, flavonoids, isoflavones, terpenes, glucosinolates, etc.) are usually bitter and aversive to consumers, but they have chemoprotective effects against cancer and cardiovascular diseases. The bitter taste has been a barrier for the consumption of phytonutrients and renders them removed from products in food industry. The conflicting demands of taste and health call for innovative solutions to incorporate bitter but beneficial components into our diet (89).
More generally, evolutionary research provides an evolutionary perspective of complex diseases and nutritional practices. The thrifty gene hypothesis is one of the leading evolutionary theories explaining the explosion of metabolic syndromes, including type 2 diabetes and obesity, in modern society. This hypothesis proposed the existence of thrifty genes that promote more efficient food utilization, fat deposition, and rapid weight gain during times of food abundance and confer to their carriers a higher chance of survival during times of famine. These genes were adaptive to the ancient hunter-gatherer lifestyle with frequent cycles of feast and famine but are maladaptive in modern society with its excess of energy availability, which can result in the prevalence of metabolic disorders (20). Another similar theory, the carnivore connection hypothesis, proposes that insulin resistance was probably advantageous to accommodate low glucose intake during the early human evolution when the Ice Ages dominated and restricted the ancient human diet to be low in carbohydrate and high in protein. However, the carbohydrate-rich diet after the advent of agriculture and the more recent high glycemic index diet after the Industrial Revolution rendered insulin resistance deleterious, contributing to the prevalence of diabetes (21, 22). In contrast to the dramatic societal, cultural, and dietary transformations during the past 10,000 y, our genome remains almost the same as that of our Paleolithic ancestors. Whereas our genome was shaped by millions of years of selection to be adaptive to the dietary pattern in the Paleolithic era, it is maladaptive to our modern dietary habits. This maladaption of the genome to modern diet may be the underlying evolutionary cause of “civilization diseases.” Based on this theory, a dietary regimen called the Paleolithic diet has been proposed to shift our diet to resemble that of our Paleolithic ancestors to meet the limitation of our genome and to prevent us from complex diseases (90, 91). Although these theories about complex diseases and nutritional practices are still controversial (92), they provide valuable evolutionary perspectives and guidance for further investigations.
Future directions
The hunt for diet-adaptive genes is still at an early age. The rapid advance in sequencing technologies and the plummet of sequencing cost will stimulate an explosion of genomic information in the foreseeable future. Genome sequences from indigenous groups will be especially valuable in this endeavor because of their genetic homogeneity and characteristic dietary patterns. For instance, some indigenous populations, such as the Inuit and the Maasai, traditionally consuming an animal-based diet, will be great subjects for studying adaptation to a high-fat/low-carbohydrate diet. The increase of genomic sequences, coupled with an accumulation of dietary and phenotypic information, will provide an unprecedented opportunity to detect even small frequency shifts in response to environmental factors, including dietary components (59). The increase of population genomics data in nonhuman primates will also facilitate the elucidation of adaptation events at the early stage of human evolution. Another field of specific interest is the study of human gut microbiome. Investigation into the symbiotic relationship between gut microbiome and the human host will not only shed light on human dietary history but also uncover their coadaptations to the human diet (93, 94).
Research of human evolutionary adaptations to dietary changes has tremendous implications for human health, especially during the era of personal genomics. Incorporating this type of knowledge with genome-wide association study and functional investigations will provide a comprehensive understanding of the evolutionary and molecular causes of complex metabolic diseases. Individualized nutritional practices will be subsequently made possible to tailor optimal strategies for health management by taking into account personal genomic information.
Acknowledgments
We are grateful to Ryan A. Coots and Liuqi Gu for their valuable comments. Both authors have read and approved the final manuscript.
Footnotes
Supported by startup funds from Cornell University, International Life Sciences Institute Future Leader Award in Nutrition, National Science Foundation grant DEB-0949556, and NIH grant 1R01AI085286 awarded to Z.G.
Author disclosures: K. Ye and Z. Gu, no conflicts of interest.
Abbreviations used: kya, thousand years ago; LP, lactase persistence; mya, million years ago; SFS, site frequency spectrum; SNP, single nucleotide polymorphism; SV, structure variation.
Literature Cited
- 1.Feero WG, Guttmacher AE, Collins FS. Genomic medicine: an updated primer. N Engl J Med. 2010;362:2001–11 [DOI] [PubMed] [Google Scholar]
- 2.Luca F, Perry GH, Di Rienzo A. Evolutionary adaptations to dietary changes. Annu Rev Nutr. 2010;30:291–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Babbitt CC, Warner LR, Fedrigo O, Wall CE, Wray GA. Genomic signatures of diet-related shifts during human origins. Proc Biol Sci. 2011;278:961–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kelley JL, Swanson WJ. Positive selection in the human genome: from genome scans to biological significance. Annu Rev Genomics Hum Genet. 2008;9:143–60 [DOI] [PubMed] [Google Scholar]
- 5.Shi H, Su B. Molecular adaptation of modern human populations. Int J Evol Biol. 2011;484769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akey JM. Constructing genomic maps of positive selection in humans: where do we go from here? Genome Res. 2009;19:711–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nielsen R. Molecular signatures of natural selection. Annu Rev Genet. 2005;39:197–218 [DOI] [PubMed] [Google Scholar]
- 8.Nielsen R, Hellmann I, Hubisz M, Bustamante C, Clark AG. Recent and ongoing selection in the human genome. Nat Rev Genet. 2007;8:857–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oleksyk TK, Smith MW, O'Brien SJ. Genome-wide scans for footprints of natural selection. Philos Trans R Soc Lond B Biol Sci. 2010;365:185–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen FC, Li WH. Genomic divergences between humans and other hominoids and the effective population size of the common ancestor of humans and chimpanzees. Am J Hum Genet. 2001;68:444–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.White TD, Asfaw B, Beyene Y, Haile-Selassie Y, Lovejoy CO, Suwa G, WoldeGabriel G. Ardipithecus ramidus and the paleobiology of early hominids. Science. 2009;326:75–86 [PubMed] [Google Scholar]
- 12.Hernández Fernández M, Vrba ES. Plio-Pleistocene climatic change in the Turkana Basin (East Africa): evidence from large mammal faunas. J Hum Evol. 2006;50:595–626 [DOI] [PubMed] [Google Scholar]
- 13.de Heinzelin J, Clark JD, White T, Hart W, Renne P, WoldeGabriel G, Beyene Y, Vrba E. Environment and behavior of 2.5-million-year-old Bouri hominids. Science. 1999;284:625–9 [DOI] [PubMed] [Google Scholar]
- 14.Roebroeks W, Villa P. On the earliest evidence for habitual use of fire in Europe. Proc Natl Acad Sci USA. 2011;108:5209–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Milton K. The critical role played by animal source foods in human (Homo) evolution. J Nutr. 2003;133:S3886–92 [DOI] [PubMed] [Google Scholar]
- 16.White TD, Asfaw B, DeGusta D, Gilbert H, Richards GD, Suwa G, Howell FC. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–7 [DOI] [PubMed] [Google Scholar]
- 17.Liu H, Prugnolle F, Manica A, Balloux F. A geographically explicit genetic model of worldwide human-settlement history. Am J Hum Genet. 2006;79:230–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Diamond J. Evolution, consequences and future of plant and animal domestication. Nature. 2002;418:700–7 [DOI] [PubMed] [Google Scholar]
- 19.Fairweather-Tait SJ. Human nutrition and food research: opportunities and challenges in the post-genomic era. Philos Trans R Soc Lond B Biol Sci. 2003;358:1709–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Neel JV. Diabetes mellitus: a "thrifty" genotype rendered detrimental by "progress"? Am J Hum Genet. 1962;14:353–62 [PMC free article] [PubMed] [Google Scholar]
- 21.Miller JC, Colagiuri S. The carnivore connection: dietary carbohydrate in the evolution of NIDDM. Diabetologia. 1994;37:1280–6 [DOI] [PubMed] [Google Scholar]
- 22.Colagiuri S, Brand MJ. The ‘carnivore connection’: evolutionary aspects of insulin resistance. Eur J Clin Nutr. 2002;56 Suppl 1:S30–5 [DOI] [PubMed] [Google Scholar]
- 23.Venter JC. Multiple personal genomes await. Nature. 2010;464:676–7 [DOI] [PubMed] [Google Scholar]
- 24.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manolio TA, Brooks LD, Collins FSA. HapMap harvest of insights into the genetics of common disease. J Clin Invest. 2008;118:1590–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eichler EE, Nickerson DA, Altshuler D, Bowcock AM, Brooks LD, Carter NP, Church DM, Felsenfeld A, Guyer M, Lee C, et al. Completing the map of human genetic variation. Nature. 2007;447:161–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J, Wang W, Li R, Li Y, Tian G, Goodman L, Fan W, Zhang J, Li J, Zhang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang F, Gu W, Hurles ME, Lupski JR. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet. 2009;10:451–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tishkoff SA, Verrelli BC. Patterns of human genetic diversity: implications for human evolutionary history and disease. Annu Rev Genomics Hum Genet. 2003;4:293–340 [DOI] [PubMed] [Google Scholar]
- 32.Keinan A, Reich D. Human population differentiation is strongly correlated with local recombination rate. PLoS Genet. 2010;6:e1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lercher MJ, Hurst LD. Human SNP variability and mutation rate are higher in regions of high recombination. Trends Genet. 2002;18:337–40 [DOI] [PubMed] [Google Scholar]
- 34.Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR. Whole-genome patterns of common DNA variation in three human populations. Science. 2005;307:1072–9 [DOI] [PubMed] [Google Scholar]
- 35.Nielsen R, Hubisz MJ, Hellmann I, Torgerson D, Andres AM, Albrechtsen A, Gutenkunst R, Adams MD, Cargill M, Boyko A, et al. Darwinian and demographic forces affecting human protein coding genes. Genome Res. 2009;19:838–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pritchard JK, Pickrell JK, Coop G. The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr Biol. 2010;20:R208–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hancock AM, Alkorta-Aranburu G, Witonsky DB, Di Rienzo A. Adaptations to new environments in humans: the role of subtle allele frequency shifts. Philos Trans R Soc Lond B Biol Sci. 2010;365:2459–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pritchard JK, Di Rienzo A. Adaptation: not by sweeps alone. Nat Rev Genet. 2010;11:665–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen R, Bustamante C, Clark AG, Glanowski S, Sackton TB, Hubisz MJ, Fledel-Alon A, Tanenbaum DM, Civello D, White TJ, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:e170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clark AG, Glanowski S, Nielsen R, Thomas PD, Kejariwal A, Todd MA, Tanenbaum DM, Civello D, Lu F, Murphy B, et al. Inferring nonneutral evolution from human-chimp-mouse orthologous gene trios. Science. 2003;302:1960–3 [DOI] [PubMed] [Google Scholar]
- 41.Haygood R, Fedrigo O, Hanson B, Yokoyama KD, Wray GA. Promoter regions of many neural- and nutrition-related genes have experienced positive selection during human evolution. Nat Genet. 2007;39:1140–4 [DOI] [PubMed] [Google Scholar]
- 42.Hudson RR, Kreitman M, Aguade M. A test of neutral molecular evolution based on nucleotide data. Genetics. 1987;116:153–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–4 [DOI] [PubMed] [Google Scholar]
- 44.Tishkoff SA, Reed FA, Ranciaro A, Voight BF, Babbitt CC, Silverman JS, Powell K, Mortensen HM, Hirbo JB, Osman M, et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nat Genet. 2007;39:31–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Perry GH, Dominy NJ, Claw KG, Lee AS, Fiegler H, Redon R, Werner J, Villanea FA, Mountain JL, Misra R, et al. Diet and the evolution of human amylase gene copy number variation. Nat Genet. 2007;39:1256–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu YX, Li WH. Statistical tests of neutrality of mutations. Genetics. 1993;133:693–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fay JC, Wu CI. Hitchhiking under positive Darwinian selection. Genetics. 2000;155:1405–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thompson EE, Kuttab-Boulos H, Witonsky D, Yang L, Roe BA, Di Rienzo A. CYP3A variation and the evolution of salt-sensitivity variants. Am J Hum Genet. 2004;75:1059–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabeti PC, Reich DE, Higgins JM, Levine HZ, Richter DJ, Schaffner SF, Gabriel SB, Platko JV, Patterson NJ, McDonald GJ, et al. Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–7 [DOI] [PubMed] [Google Scholar]
- 51.Wang ET, Kodama G, Baldi P, Moyzis RK. Global landscape of recent inferred Darwinian selection for Homo sapiens. Proc Natl Acad Sci USA. 2006;103:135–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sabeti PC, Varilly P, Fry B, Lohmueller J, Hostetter E, Cotsapas C, Xie X, Byrne EH, McCarroll SA, Gaudet R, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weir BS. Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–70 [DOI] [PubMed] [Google Scholar]
- 55.Weir BS, Hill WG. Estimating F-statistics. Annu Rev Genet. 2002;36:721–50 [DOI] [PubMed] [Google Scholar]
- 56.Han Y, Gu S, Oota H, Osier MV, Pakstis AJ, Speed WC, Kidd JR, Kidd KK. Evidence of positive selection on a class I ADH locus. Am J Hum Genet. 2007;80:441–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Toomajian C, Kreitman M. Sequence variation and haplotype structure at the human HFE locus. Genetics. 2002;161:1609–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bersaglieri T, Sabeti PC, Patterson N, Vanderploeg T, Schaffner SF, Drake JA, Rhodes M, Reich DE, Hirschhorn JN. Genetic signatures of strong recent positive selection at the lactase gene. Am J Hum Genet. 2004;74:1111–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Novembre J, Di Rienzo A. Spatial patterns of variation due to natural selection in humans. Nat Rev Genet. 2009;10:745–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Coop G, Witonsky D, Di Rienzo A, Pritchard JK. Using environmental correlations to identify loci underlying local adaptation. Genetics. 2010;185:1411–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hancock AM, Witonsky DB, Ehler E, Alkorta-Aranburu G, Beall C, Gebremedhin A, Sukernik R, Utermann G, Pritchard J, Coop G, et al. Human adaptations to diet, subsistence, and ecoregion are due to subtle shifts in allele frequency. Proc Natl Acad Sci USA. 2010;107 Suppl 2:8924–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hancock AM, Witonsky DB, Alkorta-Aranburu G, Beall CM, Gebremedhin A, Sukernik R, Utermann G, Pritchard JK, Coop G, Di Rienzo A. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 2011;7:e1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hancock AM, Witonsky DB, Gordon AS, Eshel G, Pritchard JK, Coop G, Di Rienzo A. Adaptations to climate in candidate genes for common metabolic disorders. PLoS Genet. 2008;4:e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bustamante CD, Fledel-Alon A, Williamson S, Nielsen R, Hubisz MT, Glanowski S, Tanenbaum DM, White TJ, Sninsky JJ, Hernandez RD, et al. Natural selection on protein-coding genes in the human genome. Nature. 2005;437:1153–7 [DOI] [PubMed] [Google Scholar]
- 65.Enattah NS, Sahi T, Savilahti E, Terwilliger JD, Peltonen L, Jarvela I. Identification of a variant associated with adult-type hypolactasia. Nat Genet. 2002;30:233–7 [DOI] [PubMed] [Google Scholar]
- 66.Enattah NS, Jensen TG, Nielsen M, Lewinski R, Kuokkanen M, Rasinpera H, El-Shanti H, Seo JK, Alifrangis M, Khalil IF, et al. Independent introduction of two lactase-persistence alleles into human populations reflects different history of adaptation to milk culture. Am J Hum Genet. 2008;82:57–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Peng Y, Shi H, Qi XB, Xiao CJ, Zhong H, Ma RL, Su B. The ADH1B Arg47His polymorphism in east Asian populations and expansion of rice domestication in history. BMC Evol Biol. 2010;10:15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Li H, Mukherjee N, Soundararajan U, Tarnok Z, Barta C, Khaliq S, Mohyuddin A, Kajuna SL, Mehdi SQ, Kidd JR, et al. Geographically separate increases in the frequency of the derived ADH1B*47His allele in eastern and western Asia. Am J Hum Genet. 2007;81:842–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, Gu S, Cai X, Speed WC, Pakstis AJ, Golub EI, Kidd JR, Kidd KK. Ethnic related selection for an ADH Class I variant within East Asia. PLoS One. 2008;3:e1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thomasson HR, Edenberg HJ, Crabb DW, Mai XL, Jerome RE, Li TK, Wang SP, Lin YT, Lu RB, Yin SJ. Alcohol and aldehyde dehydrogenase genotypes and alcoholism in Chinese men. Am J Hum Genet. 1991;48:677–81 [PMC free article] [PubMed] [Google Scholar]
- 71.Chen CC, Lu RB, Chen YC, Wang MF, Chang YC, Li TK, Yin SJ. Interaction between the functional polymorphisms of the alcohol-metabolism genes in protection against alcoholism. Am J Hum Genet. 1999;65:795–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gilad Y, Man O, Paabo S, Lancet D. Human specific loss of olfactory receptor genes. Proc Natl Acad Sci USA. 2003;100:3324–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gilad Y, Przeworski M, Lancet D. Loss of olfactory receptor genes coincides with the acquisition of full trichromatic vision in primates. PLoS Biol. 2004;2:E5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang J, Webb DM. Evolutionary deterioration of the vomeronasal pheromone transduction pathway in catarrhine primates. Proc Natl Acad Sci USA. 2003;100:8337–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X, Thomas SD, Zhang J. Relaxation of selective constraint and loss of function in the evolution of human bitter taste receptor genes. Hum Mol Genet. 2004;13:2671–8 [DOI] [PubMed] [Google Scholar]
- 76.Nance WE, Kearsey MJ. Relevance of connexin deafness (DFNB1) to human evolution. Am J Hum Genet. 2004;74:1081–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Verrelli BC, Lewis CJ, Stone AC, Perry GH. Different selective pressures shape the molecular evolution of color vision in chimpanzee and human populations. Mol Biol Evol. 2008;25:2735–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liman ER, Innan H. Relaxed selective pressure on an essential component of pheromone transduction in primate evolution. Proc Natl Acad Sci USA. 2003;100:3328–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wooding S, Kim UK, Bamshad MJ, Larsen J, Jorde LB, Drayna D. Natural selection and molecular evolution in PTC, a bitter-taste receptor gene. Am J Hum Genet. 2004;74:637–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Soranzo N, Bufe B, Sabeti PC, Wilson JF, Weale ME, Marguerie R, Meyerhof W, Goldstein DB. Positive selection on a high-sensitivity allele of the human bitter-taste receptor TAS2R16. Curr Biol. 2005;15:1257–65 [DOI] [PubMed] [Google Scholar]
- 81.Shi P, Zhang J, Yang H, Zhang YP. Adaptive diversification of bitter taste receptor genes in Mammalian evolution. Mol Biol Evol. 2003;20:805–14 [DOI] [PubMed] [Google Scholar]
- 82.Conte C, Ebeling M, Marcuz A, Nef P, Andres-Barquin PJ. Evolutionary relationships of the Tas2r receptor gene families in mouse and human. Physiol Genomics. 2003;14:73–82 [DOI] [PubMed] [Google Scholar]
- 83.Bachmanov AA, Beauchamp GK. Taste receptor genes. Annu Rev Nutr. 2007;27:389–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Fischer A, Gilad Y, Man O, Paabo S. Evolution of bitter taste receptors in humans and apes. Mol Biol Evol. 2005;22:432–6 [DOI] [PubMed] [Google Scholar]
- 85.Wooding S, Bufe B, Grassi C, Howard MT, Stone AC, Vazquez M, Dunn DM, Meyerhof W, Weiss RB, Bamshad MJ. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature. 2006;440:930–4 [DOI] [PubMed] [Google Scholar]
- 86.Hinrichs AL, Wang JC, Bufe B, Kwon JM, Budde J, Allen R, Bertelsen S, Evans W, Dick D, Rice J, et al. Functional variant in a bitter-taste receptor (hTAS2R16) influences risk of alcohol dependence. Am J Hum Genet. 2006;78:103–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang JC, Hinrichs AL, Bertelsen S, Stock H, Budde JP, Dick DM, Bucholz KK, Rice J, Saccone N, Edenberg HJ, et al. Functional variants in TAS2R38 and TAS2R16 influence alcohol consumption in high-risk families of African-American origin. Alcohol Clin Exp Res. 2007;31:209–15 [DOI] [PubMed] [Google Scholar]
- 88.Duffy VB, Davidson AC, Kidd JR, Kidd KK, Speed WC, Pakstis AJ, Reed DR, Snyder DJ, Bartoshuk LM. Bitter receptor gene (TAS2R38), 6-n-propylthiouracil (PROP) bitterness and alcohol intake. Alcohol Clin Exp Res. 2004;28:1629–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Drewnowski A, Gomez-Carneros C. Bitter taste, phytonutrients, and the consumer: a review. Am J Clin Nutr. 2000;72:1424–35 [DOI] [PubMed] [Google Scholar]
- 90.Eaton SB, Konner M. Paleolithic nutrition. A consideration of its nature and current implications. N Engl J Med. 1985;312:283–9 [DOI] [PubMed] [Google Scholar]
- 91.Eaton SB, Eaton SR, Konner MJ. Paleolithic nutrition revisited: a twelve-year retrospective on its nature and implications. Eur J Clin Nutr. 1997;51:207–16 [DOI] [PubMed] [Google Scholar]
- 92.Speakman JR. Thrifty genes for obesity, an attractive but flawed idea, and an alternative perspective: the ‘drifty gene’ hypothesis. Int J Obes (Lond). 2008;32:1611–7 [DOI] [PubMed] [Google Scholar]
- 93.Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–12 [DOI] [PubMed] [Google Scholar]
- 94.Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124:837–48 [DOI] [PubMed] [Google Scholar]