Abstract
The role of epigenetic alterations in various human chronic diseases has gained increasing attention and has resulted in a paradigm shift in our understanding of disease susceptibility. In the field of cancer research, e.g., genetic abnormalities/mutations historically were viewed as primary underlying causes; however, epigenetic mechanisms that alter gene expression without affecting DNA sequence are now recognized as being of equal or greater importance for oncogenesis. Methylation of DNA, modification of histones, and interfering microRNA (miRNA) collectively represent a cadre of epigenetic elements dysregulated in cancer. Targeting the epigenome with compounds that modulate DNA methylation, histone marks, and miRNA profiles represents an evolving strategy for cancer chemoprevention, and these approaches are starting to show promise in human clinical trials. Essential micronutrients such as folate, vitamin B-12, selenium, and zinc as well as the dietary phytochemicals sulforaphane, tea polyphenols, curcumin, and allyl sulfur compounds are among a growing list of agents that affect epigenetic events as novel mechanisms of chemoprevention. To illustrate these concepts, the current review highlights the interactions among nutrients, epigenetics, and prostate cancer susceptibility. In particular, we focus on epigenetic dysregulation and the impact of specific nutrients and food components on DNA methylation and histone modifications that can alter gene expression and influence prostate cancer progression.
Prostate cancer is the uncontrolled growth of abnormal cells originating from the prostate, a small gland in the male reproductive system. Hallmarks of this disease include the following: 1) genomic instability; 2) the capacity of abnormal cells to resist cell death, evade growth suppression, and induce proliferative signals; 3) chronic inflammation resulting in tumor promotion; and 4) angiogenesis, invasion, and metastasis to distant organs (1). These hallmarks are often driven, at the most fundamental level, by dysregulation of gene expression. The classic view of cancer etiology is that genetic alterations, arising from exposure to exogenous genotoxic agents and endogenous oxidants, damage DNA and induce mutations, resulting in nonfunctional proteins that underlie disease progression. Historically, genetic abnormalities and mutations were cited as primary causative factors; however, epigenetic mechanisms are now recognized as playing an equal or perhaps greater role in cancer development (2).
Epigenetics is the study of changes in gene expression that occur independent of alterations in nucleotide sequence (3). Importantly, dietary factors can modulate epigenetics and influence disease susceptibility. The role of epigenetics in chronic disease development has gained increasing attention and has resulted in a new understanding of the etiology and susceptibility to several chronic diseases. Epigenetic mechanisms implicated in prostate cancer include gene silencing via DNA promoter methylation, histone modifications, and changes in miRNA profiles (4, 5). In the latter case, ~50 miRNA6 have been reported to be aberrantly expressed in prostate cancer, only a few of which have been experimentally verified as direct contributors to the disease [for review, see Catto et al. (6)]. A comprehensive review of dietary miRNA modulators has recently been published (7). Thus, the present review focuses on DNA methylation and histone modifications in prostate cancer and the dietary agents that target these mechanisms, possibly leading to prostate cancer prevention.
Epigenetic alterations during prostate cancer
DNA methylation.
The majority of methylation marks on DNA are found on CpG dinucleotides, regions in which a cytosine is followed by a guanine residue in the 5′ to 3′ direction. The genome contains both CpG-rich and CpG-poor regions. CpG-rich “islands” within promoter regions are highly susceptible to methylation. In general, methylation of CpG islands results in gene silencing. The addition of methylation marks is catalyzed by DNMT enzymes. The DNMT family has several members, including DNMT1, a maintenance enzyme that alters DNA methylation during cell replication, and DNMT3a and DNMT3b, which are responsible for de novo methylation. Although DNMT3a and DNMT3b have been largely associated with de novo methylation during processes such as embryogenesis, recent research suggests that these enzymes are also important in the maintenance of DNA methylation patterns. After replication, DNMT3a and DNMT3b complete the methylation process and correct errors left by DNMT1 (8). DNA methylation is an important event in several biological processes such as X chromosome inactivation and silencing of germ line-specific genes and repetitive elements. There is a wide body of evidence that suggests that DNA methylation patterns are tissue specific (9, 10). During cancer progression, there appears to be dysregulation of DNA methylation patterns. For example, cancer cells are characterized by global hypomethylation of DNA and site-specific hypermethylation of specific genes involved in cancer progression [reviewed in (11, 12)].
In prostate cancer cells, disease progression has been associated with both DNA hypo- and hyper-methylation. Global cytosine hypomethylation has been associated with metastatic prostate cancer, chromosome instability, and disease progression (13–15). Repetitive DNA regions, like LINE1, are also hypomethylated in a high proportion of prostate cancer patients (16). There have been a handful of specific genes that are hypomethylated in prostate cancer; however, the majority of genes are characterized by site-specific hypermethylation, including over 50 genes that are aberrantly hypermethylated [for review, see (12)]. The latter include genes involved in hormonal response [androgen receptor (AR)], cell cycle control (cyclinD2 and cyclin-dependent kinase inhibitor 2a), signal transduction (RASSF1A, Runx3), tumor invasion (E-cadherin, TIMP metallopeptidase inhibitor 3), DNA damage repair (O6-methylguanine DNA methyltransferase), detoxification (glutathione S-transferase M1 and P1), and apoptosis (Bcl2) (12, 17). Importantly, many of these methylation events are also found in early high-grade prostatic intraepithelial neoplasia lesions (18, 19), suggesting that aberrant DNA methylation changes occur early during carcinogenesis.
Histone modifications.
Chromatin remodeling and histone modification also play an important role in regulating gene expression during prostate cancer development. In the nucleus, histone proteins interact with each other and with DNA to form a structure termed the nucleosome (Fig. 1A). The nucleosome contains 2 copies of 4 core histones (H2A, H2B, H3, and H4). The amino acid tails of these histones are susceptible to various reversible, covalent, post-translational modifications, such as acetylation, methylation, phosphorylation, ADP-ribosylation, ubiquitination, sumoylation, and biotinylation (20). In prostate cancer, the observed histone alterations are commonly associated with changes in several histone–modifying enzymes such as HDAC, histone methyltransferases, and histone demethylases (12). Histone methylation occurs on either arginine or lysine residues. Arginines can be mono- or di-methylated by the family of protein arginine methyltransferases and results in transcriptional activation (21). Lysine residues can be mono-, di-, or tri-methylated by a diverse set of histone methylation transferases belonging to the SET domain protein family (22). Depending on the residue and number of methyl groups added, histone lysine methylation can result in either transcriptional activation or repression (Table 1). For example, H3K9 can be either acetylated or methylated. H3K9ac is an activating mark, whereas, in general, H3K9me, H3K9me2, and H3K9me3 are repressing marks. However, H3K9me3 can also result in gene activation and appears to be dependent on the balance between H3K9 acetylation and methylation (23). Interestingly, histone methylation marks were originally thought to be irreversible. However, histone demethylases have also been recently characterized and play an important role in demethylating lysines. Methylated arginines are not directly demethylated. Instead, peptidylarginine deaminase 4 converts these residues to citrulline. Upregulation of Enhancer of zeste homolog 2, a histone methyltransferase that catalyzes the trimethylation of H3K27 (24), is associated with repression of tumor suppressor genes, high proliferation rates, and increased tumor aggressiveness in prostate cancer (25). Overexpression of lysine-specific demethylase (LSD1) is also associated with increases in cell proliferation, tumor aggressiveness, and hormone refractory prostate cancer. The molecular mechanism by which this occurs is complicated. LSD1 removes mono- or di-methyl groups from H3K4. H3K4 methylation is an activating mark; hence, demethylation results in repression. In contrast, LSD1 also demethylates H3K9, a repressive mark important in AR-mediated transcription, resulting in gene activation (26). Aberrant histone methylation has also been implicated in the regulation of gene expression during epithelial to mesenchymal transition in the prostate (27).
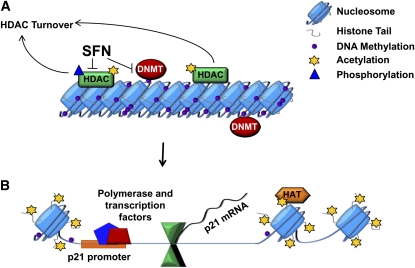
Figure 1.
Histone acetylation and DNA methylation: epigenetic mechanisms that affect gene expression. (A) Closed chromatin structure and transcriptional silencing. Hypoacetylation due to HDAC activity combined with localized promoter hypermethylation, regulated by DNMT. (B) Open chromatin structure and gene activation. Dietary compounds, such as SFN inhibit HDAC and DNMT, facilitating reexpression of silenced tumor suppressors in cancers, such as p21. Open chromatin is associated with increased acetylation of histone tails by HAT and the recruitment of transcription factors and their coactivator complexes. DNMT, DNA methyltransferase; HAT, histone acetyltransferase; HDAC, histone deacetylase; SFN, sulforaphane.
Table 1.
Examples of histone modifications resulting in transcription activation or repression
| Histone | Amino acid residue | ||
| Acetylation | H2A | K5, K9, K13 | Activating |
| H2B | K5, K12, K15, K20 | Activating | |
| H3 | K9, K14, K18, K23, K56 | Activating | |
| H4 | K5, K8, K13, K16 | Activating | |
| Methylation | H3 | K4, K36, K79 | Activating |
| K9,1 K27 | Repressing | ||
| R17, R23 | Activating | ||
| H4 | R3 | Activating | |
| K20 | Repressing | ||
| Phosphorylation | H3 | T3, S10, S28 | Activating |
| Y41 | Activating | ||
| Ubiquitination | H2B | K120 | Activating |
| Sumoylation | H4 | K5, K8, K12, K16, K20 | Repressing |
| Biotinylation | H4 | K8, K12 | Repressing |
H3K9me3 can in some cases be an activating mark (23).
A considerable amount of research has focused on dysregulation of histone acetylation during cancer. When DNA tightly interacts with histones through their protein tails, a closed DNA structure is formed, which is transcriptionally repressed [Fig. 1A; reviewed in (11)]. This is usually accompanied by a high level of methylation of the DNA at cytosine residues, which is regulated by DNMT. This repressed chromatin structure is also generally associated with the hypoacetylation of histone tails, catalyzed by HDAC (Fig. 1A). Acetylation of histones by HAT and removal of DNA methylation causes the DNA structure to become more accessible to transcription factors and their coactivator complexes, which bind DNA and initiate gene transcription (Fig. 1B). In cancer cells, the balance between HAT and HDAC activities is disrupted, often associated with higher levels of HDAC proteins and epigenetic silencing of tumor suppressor genes [reviewed in (11)]. HDAC can be divided into 4 classes based on their structure and sequence homology: class I consists of HDAC 1, 2, 3, and 8; class II includes HDAC 4, 5, 6, 7, 9, and 10; class III enzymes are HDAC originally found in yeast and include Sir2-related proteins; and Class IV comprises the sole member HDAC11.
Prostate cancer cells also exhibit aberrant histone and nonhistone protein acetylation patterns. It has been shown that HDAC activity increases in metastatic prostate cancer cells compared with prostate hyperplasia (28). A 4-fold increase in HDAC1 protein in PC3 prostate cancer cells results in an 1.5-fold increase in cell proliferation and an overall decrease in cell differentiation as determined by a marked decline in cytokeratin18 staining and increased cytokeratine staining (29). Increased expression of HDAC may be of particular importance in the progression to androgen independence, because accumulation of HDAC1 protein as determined by immunohistochemistry or increased nuclear localization of HDAC4 coincides with loss of androgen sensitivity (30). Lysine acetylation also appears to play an important role in AR regulation. Many AR coactivators and repressors alter transcriptional activity via modulation of acetylation (31). AR activity is downregulated by HDAC1, HDAC2, HDAC3, HDAC6, and Sirt1. It has also been shown that Sirt1, the predominant class III NAD+-dependent HDAC, is overexpressed in both human and mouse prostate cancers (32). In human patient samples, global decreases in histone acetylation state corresponded with increased grade of cancer and risk of prostate cancer recurrence (33). Importantly, inhibitors of HDAC, including suberoylanilide hydroxamic acid, valproic acid, depsipeptide, and sodium butyrate, are effective against prostate cancer cell lines and xenograft models (34, 35).
Dietary modulators of DNA methylation
Global hypomethylation and site-specific hypermethylation of cancer-related genes occur in many tumors, including prostate cancer. Hypomethylation can result in genome instability, reactivation of transposons, and upregulation/activation of proto-oncogenes. In contrast, hypermethylation during cancer development can silence genes involved in cancer protection; targets include genes involved in DNA repair, detoxification, apoptosis, cell cycle control, and other tumor suppressor genes (36, 37). Strategies to alter aberrant methylation patterns are of considerable interest. Dietary strategies that influence DNA methylation fall into 3 broad categories: 1) methyl donors that directly contribute to the methyl pool and are substrates involved in DNA methylation; 2) agents that indirectly affect the methyl pool by modulating the activity of enzymes that regulate the methyl pool; or 3) compounds that act as DNMT inhibitors (Fig. 2). An overview of dietary compounds that modulate DNA methylation in prostate cancer in each of these categories is outlined below and summarized in Figure 2.
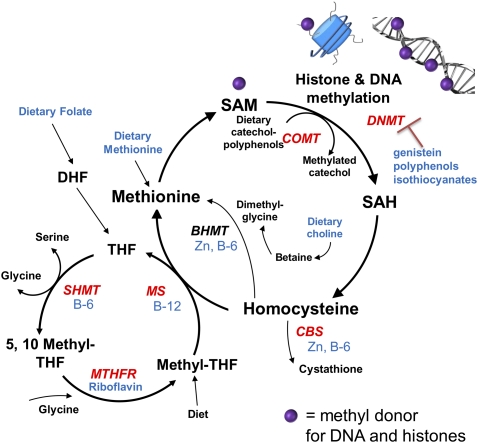
Figure 2.
Overview of mechanisms by which diet and dietary factors influence DNA and histone methylation. DNA methylation is influenced by the methyl pool involving SAM, which acts as a substrate for DNMT that transfer one-carbon units to DNA. SAM also serves as donor for histone methylation. Dietary factors that can regulate DNA and histone methylation include the following: 1) vitamins: vitamin B-6, vitamin B-12, and riboflavin; 2) minerals: zinc and selenium; 3) phytochemicals: genistein, tea polyphenols, and isothiocyanates; and 4) other dietary factors: methionine and choline. BHMT, betaine-homocysteine methyltransferase; CBS, cystathionine β-synthase; COMT, catechol-O-methyltransferase; DHF, dihydrofolate; MS, methionine synthase; MTHFR, 5, 10-methylenetetrahydrofolate reductase; SAH, S-adenosyl homocysteine; SAM, S-adenosyl methionine; SHMT, serine hydroxymethyltransferase; THF, tetrahydrofolate.
Methyl donors and one-carbon metabolism nutrients (folate, vitamins B-12 and B-6, choline, betaine, methionine).
Nutrients involved in one-carbon metabolism play a critical role in maintaining DNA methylation (Fig. 2). Folate aids in the transfer of one-carbon units and is involved in multiple cellular processes, including specific amino acid synthesis, DNA synthesis, and DNA methylation. The metabolism of folate is a complex process. Folate from dietary sources is reduced to dihydrofolate and THF. THF can then go through several different metabolic pathways, including formation of 5-methyl-THF or 10-formyl THF, and these intermediates act as cofactors for various enzymes. Of relevance to DNA methylation, folate plays an important role in the synthesis of methionine as a cofactor in the enzyme methionine synthase. 5-methyl-THF transfers the methyl group to homocysteine to produce methionine, which plays a critical role as a precursor to SAM, the universal methylation donor. The methylation reaction catalyzed by DNMT requires SAM as the methyl group donor, as does the methylation of histone tails. Thus, factors that limit SAM supply could have an important impact on DNA methylation and histone marks. Several other one-carbon metabolism nutrients also contribute to the methionine pool and DNA methylation in the cell by acting as either direct methyl donors or cofactors in enzymes contributing to the methyl pool. For example, methionine levels can be modulated directly through consumption of methionine in protein-rich diets. Choline is another important methyl donor contributing to the methionine pool. Vitamin B-12, as methyl-malonyl-cobalamin, is also an essential cofactor for methionine synthase. Vitamin B-6 is a cofactor for several enzymes involved in homocysteine regeneration, methionine synthesis, and metabolism of folate. A lack or excess of any of these nutrients affects one-carbon metabolism, alters the availability of SAM in the methionine cycle, and has the potential to perturb DNA and histone methylation patterns (Fig. 2). Diets deficient in methionine, choline, vitamin B-12, or folate induce global hypomethylation and site-specific hypermethylation and have been linked to increased cancer development (38–43). Interestingly, as methyl donors are slowly depleted in the diet, alterations in histone methylation (to be discussed in next section) precede alterations in DNA methylation (44).
Epidemiological and human intervention studies utilizing one carbon related nutrients in prostate cancer have yielded inconsistent results. In several trials, serum folate, vitamins B-6 and B-12, and homocysteine levels have shown no association with prostate cancer (45, 46). Studies examining polymorphisms in folate metabolism genes also do not appear to support significant contributions to prostate cancer risk (47, 48). Moreover, some reports have shown an increase in prostate cancer risk with increasing folate (49). In a recent meta-analysis, circulating vitamin B-12 and folate were positively associated with prostate cancer risk (50). A positive association between folate and other cancers, such as colon, has also been observed (51, 52). It has been postulated that dose, timing of folate administration, and stage of cancer may play an important role in the preventive compared to promoting effects of folate (53, 54). Because folate and other one-carbon nutrients function in multiple pathways controlling DNA integrity and cell proliferation, their relative contributions to aberrant DNA and histone methylation patterns in prostate cancer remain unclear.
Soy isoflavones and phytoestrogens.
Epidemiological studies suggest that the consumption of soy foods is associated with lower risk of prostate cancer (55). The plasma and serum phytoestrogen concentrations in Japanese men are at least 10-fold higher than their Caucasian counterparts in the UK (56, 57). Soy isoflavones, classified as phytoestrogens, act as both estrogen agonists and antagonists by differentially binding to estrogen receptor α or β (58, 59) and/or altering enzymes involved in hormone metabolism (56, 58, 59). Such observations led to hypotheses that soy or components in soy exert anticancer effects. Most have attributed these effects to soy isoflavones, especially genistein (60). The anticarcinogenic effects of genistein are summarized in several reviews (61, 62). More recently, soy isoflavones such as genistein and daidzein have been shown to modulate DNMT activity and promoter methylation. In both androgen-dependent and -independent prostate cancer cell lines, genistein (20–50 μmol/L) dose dependently inhibited DNMT activity (63–66). The IC50 of genistein was ∼67 μmol/L. Other isoflavones such as daidzein also inhibited DNMT but to a lesser extent (67). Kinetic studies suggest that genistein inhibits DNA methyltransferase activity in both a competitive and noncompetitive manner depending on the substrate (68), unlike EGCG, which appears to function as a competitive inhibitor of DNA methyltransferase (discussed in the next section). DNMT inhibition with genistein leads to hypomethylation and reactivation of silenced genes in prostate cancer, such as GSTp1 and tumor suppressor proteins such as B-cell translocation gene 3 (BTG3), Ras association domain family 1 (RASSF1a), and p16INK4a (67, 69, 70). However, feeding studies with genistein increased DNA methylation in vivo. For example, Dolinoy et al. (71) demonstrated that maternal consumption of genistein in mice resulted in hypermethylation in Agouti offspring, resulting in gene silencing, a shift in coat color, and decreased obesity. In studies using differential methylation arrays, mice fed a genistein-rich diet for 4 wk had increased CpG methylation in the prostate (72). Although the effects of soy isoflavones on DNA methylation in prostate cancer patients have not been evaluated, Qin et al. (73) examined mammary intraductal specimens from healthy premenopausal women for hypermethylated genes associated with breast cancer. Isoflavone supplementation and genistein levels were correlated with hypermethylation of RARβ2 and cyclinD2 in these tissues (73). Mechanistic studies are warranted on target sites for hyper- or hypomethylation and additional in vivo studies are necessary to better understand the impact of soy on DNA methylation, gene expression, and cancer outcomes.
Tea polyphenols.
Green tea (Camellia sinensis) is one of the most widely consumed beverages, especially in Asia. The bioactive components of green tea include catechins, EGCG, epigallocatechin, epicatechin gallate, and epicatechin, and theaflavins. Catechins are present at high levels in green tea, accounting for 30–40% of its dry weight (74). Green tea polyphenols, especially EGCG, exert anticarcinogenic effects at multiple stages of carcinogenesis. In vitro, EGCG is a strong antioxidant and an inhibitor of carcinogen activation. EGCG also induces cell cycle arrest and apoptosis in cultured cancer cells, but which of the various mechanisms predominates in vivo remains unclear, including during the attenuation of tumor promotion (75). The actions of EGCG in prostate cancer prevention and treatment were summarized by Stuart et al. (76).
Experimental studies show that EGCG is an effective DNMT inhibitor in vitro [for review, see (68)]. EGCG can be methylated by COMT using SAM as a substrate (Fig. 2). This pathway is largely responsible for the biotransformation and inactivation of catechol molecules. Demethylation of SAM results in an increase in S-adenosyl homocysteine levels, which as noted above are inhibitors of DNMT. COMT and DMNT belong to the same superfamily of SAM-dependent methyltransferases and share common core structures. EGCG can act as a mixed-type (competitive and noncompetitive) inhibitor for COMT (77). Molecular docking studies have revealed that EGCG also interacts with the active pocket of DNMT1, acting as a competitive inhibitor (67). In vitro, research has shown that EGCG causes a concentration- and time-dependent reversal of hypermethylation of tumor suppressor genes in several cancer types (78, 79). In prostate cancer cell lines, EGCG treatment also reactivates genes silenced by methylation (80).
However, the effects of tea polyphenols on prostate cancer progression and DNA methylation in vivo are less conclusive. In both animal models and humans, supplementation with green tea polyphenols has shown promise as chemopreventive agents for prostate cancer. In human participants with high-grade prostatic intraepithelial neoplasia lesions, oral supplementation with green tea extracts for 1 y decreased the prevalence of prostate cancer (81). In prostate cancer patients consuming 6 cups of green tea per day for 3–6 wk prior to prostatectomy, significant increases in EGCG were found in the prostate tissue, with 48% of total EGCG in methylated form (82). These studies provide proof of concept that tea catechins are susceptible to methylation via COMT and potentially modulate the methyl pool. Using the TRAMP model, an oral infusion of green tea catechins resulted in a decrease in prostate tumors (83,84). However, not all studies using TRAMP mice have observed protection with tea polyphenol supplementation. Further work suggested that tea polyphenols had limited effectiveness in advanced disease (85); thus, supplementation might be necessary at early stages of prostate cancer. However, Morey Kinney et al. (86) provided both wild-type and TRAMP mice with 0.3% green tea polyphenols in drinking water, starting at 4 wk of age, and no change was detected in prostate cancer incidence at 12 and 24 wk. In addition, no alternations in markers of DNA methylation were observed. 5-methyl-deoxycytidine levels, methylation of the B1 repetitive element, and methylation of the Mage-a8 gene did not differ from control mice. Genome-wide DNA methylation profiling using the HpaII tiny fragment enrichment by ligation-mediated PCR assay also revealed no significant hypomethylating effect of green tea polyphenols (86). Discrepancies between these studies may be attributed to different mouse strains, doses, and/or formulations of green tea polyphenols, but additional in vivo research is warranted before clear conclusions can be made on the role of catechins as DNMT inhibitors in prostate cancer.
ITC.
ITC are found in cruciferous vegetables such as broccoli, Brussels sprouts, cauliflower, and cabbage. SFN is an ITC derived from cruciferous vegetables and is especially high in broccoli and broccoli sprouts (87), where it exists as the glucosinolate precursor, glucoraphanin. When the plant is consumed, plant myrosinases or hydrolases present in gut bacteria convert glucoraphanin to SFN (88). Epidemiological studies have shown an inverse association between cruciferous vegetable intake and cancer risk in many tissues, including the prostate. Interestingly, a recent analysis of the EPIC-Heidelberg cohort study showed a significant decrease in the risk of prostate cancer specifically with increasing glucosinolate intake (89).
SFN is an effective chemoprotective agent in carcinogen-induced animal models (87, 90, 91) as well as in xenograft and genetic models of prostate cancer (92, 93). Recent studies suggest that SFN may target aberrant gene hypermethylation by modulating DNMT expression. SFN decreased expression of DNMT1 and DNMT3a in breast cancer cells (94). Similarly, in LnCap prostate cancer cells, SFN treatment downregulated DNMT1 and 3b but not DNMT3a (95). These results suggest that the responses of individual DNMT to SFN treatment may differ according to cancer cell type. Our recent study indicated that SFN may downregulate DNMT, resulting in demethylation of the cyclin D2 promoter and enhanced transcription of cyclin D2 mRNA, suggesting a novel mechanism behind SFN’s growth inhibitory effects on prostate cancer cells (95). Other ITC have shown an ability to alter DNA methylation patterns. For example, PEITC caused demethylation and reactivation of GSTp1, a commonly hypermethylated gene in LnCap prostate cancer cells (96).
Minerals (selenium and zinc).
Several trace elements can indirectly affect one-carbon metabolism and methyl donor availability. Selenium has been proposed to react with homocysteine to form selenohomocysteine. This reaction may limit homocysteine in the methionine cycle. Indeed, rats supplemented with SM show decreases in global hypomethylation and site-specific increases in methylation of p53 (97, 98). Although there has been considerable interest in selenium as a prostate cancer chemoprevention agent (see next section), the modulatory effects of selenium on methylation events in the prostate remain to be clarified.
Zinc also acts as key cofactor in several enzymes involved in the methionine/transsulfuration pathway, a key pathway for generating methyl donation equivalents such as SAM and betaine (Fig. 2). Betaine-homocysteine methyltransferase and methionine synthase are also zinc-dependent enzymes (99). Serine hydroxymethyltransferase, a key enzyme in folate metabolism that helps transfer methyl units from serine into the methionine cycle, is regulated by zinc-dependent transcription factors, including metal-regulatory transcription factor 1. This strongly suggests that zinc likely has an important function in maintaining methylation status in the cell. Thus, zinc deficiency may cause a methyl deficiency similar to other methyl donors like folate, resulting in abnormal gene expression and developmental defects. In rats, zinc deficiency decreased turnover of SAM and depressed DNA and histone methylation in the liver (100). In Agouti mice, supplementation with methyl donors (choline, betaine, folate, vitamin B-12, methionine) and zinc epigenetically regulated the expression of Agouti in the offspring (101).
Similar to selenium, there is also considerable interest in zinc in prostate cancer development. The prostate contains the highest concentration of zinc of any soft tissue and secretes high amounts of zinc in the prostatic fluid. Zinc concentrations in malignant prostate tissues are ∼10–25% of that in healthy prostates (102), suggesting that high zinc concentrations may be essential for the maintenance of prostate health. Several mechanisms by which zinc may protect prostate cells from malignancies have been proposed, including zinc as an inhibitor to mitochondrial-aconitase activity, apoptogenic effects, and as a protector for DNA integrity [for review, see (103)]. The possible interplay between zinc and DNA methylation may be a novel mechanism by which zinc deficiency increases the risk of prostate cancer.
Dietary modulators of histone modifications
The use of Class I and Class II HDAC inhibitors in cancer chemoprevention and therapy has gained considerable interest (104). Modulators of histone methylation and other histone marks have not been extensively investigated in prostate cancer. 3-deazaneplanocin-A, a pharmacological inhibitor for histone methylation, has been studied in various cancers (105) and has recently been shown to limit tumor growth in prostate cancer cells both in vitro and in mouse xenografts (106). To date, specific dietary modulators of histone methylation have not been clearly identified. In contrast, considerable work has been done with targeting histone acetylation in prostate cancer, both utilizing pharmacological and dietary approaches. Several clinical trials are currently ongoing aimed at establishing the chemotherapeutic efficacy of HDAC inhibitors, based on evidence that cancer cells undergo cell cycle arrest, differentiation, and apoptosis in vitro and that tumor volume and/or tumor number may be reduced by such agents in preclinical animal models. HDAC inhibitors have been shown to increase global acetylation as well as acetylation associated with specific gene promoters (107). Although the equilibrium is shifted toward greater histone acetylation after treatment with HDAC inhibitors, the expression of only a relatively small number of genes is altered in an upward or downward direction (107). Importantly, only neoplastically transformed cells appear to respond to increased acetylation by undergoing differentiation, cell cycle arrest, or apoptosis; normal cells, despite the increased acetylation, do not respond in this manner to HDAC inhibitors (108). An overview of several diet-based histone modulators follows.
Dietary fiber (butyrate).
Butyrate is one of the earliest and smallest known HDAC inhibitors [reviewed in (109)], containing a simple 3 carbon spacer attached to a carboxylic acid group. Most of the chemoprevention research with butyrate has focused on colorectal cancer, because the colonocytes are the primary site of production. Butyrate decreases HDAC activity and triggers cell cycle arrest or apoptosis, likely as a competitive HDAC inhibitor (110). The effects of butyrate in prostate cancer are less well studied, especially given that high concentrations of butyrate (mmol/L range) are necessary to exert HDAC inhibitory and proapoptotic effects (111, 112). In prostate cancer cells, sodium butyrate (>1 mmol/L) decreased both HDAC2 and HDAC3 expression. In addition, sodium butyrate had effects on the AR. The transcriptional activities of AR and other members of the nuclear receptor superfamily are modulated by coregulatory proteins. In particular, the corepressor SMRT forms a complex with NCOR and with HDAC3 regulate AR expression (113). Recent research suggests that the addition of sodium butyrate to androgen-responsive prostate cancer cells may decrease AR expression by modulating the SMRT corepressor interaction (114).
Garlic compounds.
Epidemiological studies suggest that increased dietary intake of Allium vegetables, such as garlic, is associated with protection against prostate and other cancers (115). The protective effects of garlic have been attributed to several organosulfur compounds that are released when these foods are crushed and/or chewed, such as diallyl sulfide, DADS, and DATS. Some of these compounds have reported HDAC inhibitory activity. For example, in colon cancer cells, DADS causes histone acetylation both in vitro and in vivo (116). Preclinical studies in vitro and in animal models also support the notion that administration of these garlic-derived organosulfur compounds results in prostate cancer chemoprevention. DATS treatment also potently induces cell cycle arrest, apoptosis, and repression of the AR in prostate cancer both in vitro and in vivo (117–120). Increases in histone acetylation and apoptosis with DAD treatment is found in prostate cancer cells (121). Like DADS, S-allylmercaptocysteine, and several other garlic compounds (116, 121), DATS can be metabolized to AM, a competitive HDAC inhibitor. The effects of AM on prostate cancer cells are not known, but treatment of human colon cancer cells with AM induced rapid histone acetylation along with HDAC inhibition (122). As a consequence, increased association of acetylated histones and Sp3 transcription factor binding to the promoter element of p21Waf1 occurred, thereby increasing both p21 mRNA and protein expression, triggering cell cycle arrest (122).
SFN and other ITC.
Multiple mechanisms have been postulated for the chemopreventive effects of SFN. The majority of studies focused on SFN as a potent Phase 2 enzyme inducer, with additional evidence for cell cycle arrest and apoptosis in vitro. More recently it has been found that SFN metabolites inhibit HDAC activity in vitro, the greatest inhibition involving SFN-NAC and SFN-Cys metabolites [reviewed in (123)]. Molecular modeling in the active site of an HDAC enzyme provided evidence for SFN-Cys as a possible competitive inhibitor (124). In BPH1, PC3, and LnCap prostate cancer cells, SFN inhibited HDAC activity with a concomitant increase in global histone acetylation, increased acetylated histone H4 interactions with the p21 and Bax promoter, induction of p21 and Bax mRNA and protein levels, and increased cell cycle arrest (125).
In addition to competitive enzyme inhibition, recent research has shown that SFN lowers the expression of specific HDAC proteins. Thus, treatment of transformed prostate cells with 15 μmol/L SFN caused a selective decrease in both HDAC3 (class I) and HDAC6 (class II) proteins (126). Loss of HDAC6 has important implications in modulating the acetylation of nonhistone proteins such as tubulin and HSP90, which have roles in controlling apoptosis/autophagy pathways and AR stability, respectively. Loss of HDAC3 may be related to turnover/degradation pathways triggered by disruption of the HDAC3/SMRT corepressor complex and nuclear/cytoplasmic trafficking (127). Interestingly, similar to pharmacological HDAC inhibitors, we have found that 15 μmol/L SFN causes potent HDAC inhibition and G2/M arrest in LnCaP and PC3 cancer cells but has no effect on normal prostate epithelial cells (126). These data support the hypothesis that HDAC inhibition may be an important mechanism of chemoprevention for SFN and similar pharmacological HDAC inhibitors, producing cytotoxic effects specific to cancer cells and not normal cells. SFN may also coordinate its effects by altering other histone marks, such as histone phosphorylation and histone methylation. As reported in the earlier section, SFN modulates DNA methylation patterns; thus, the epigenetic effects of SFN may not be limited to HDAC inhibition. Possible coordination between these other histone marks and/or DNA methylation represents an important area for future research.
Other ITC and compounds derived from cruciferous vegetables also have reported effects on prostate cancer cells via histone modifications and HDAC inhibition. Structurally related ITC such as sulforaphene, erucin, and phenylbutyl isothiocyanate had comparable HDAC inhibitory activities in vitro (128). Phenhexyl isothiocyanate, BITC, and PEITC have shown antiproliferative effects in prostate cancer cells as well as chemopreventive properties in prostate cancer models in vivo (129–134). Phenhexyl isothiocyanate administered to androgen-dependent LnCap cells resulted in decreased HDAC activity and HDAC1 and HDAC2 expression and increased acetylated histone and p21 expression that was concomitant with G1 cell cycle arrest and apoptosis (133). Similar to SFN, PEITC appears to have dual effects on DNA methylation and HDAC inhibition (96). Concurrent with demethylation effects, PEITC (2–5 μmol/L) inhibited HDAC activity and increased acetylated histone status. At the doses tested, PEITC was more effective toward promoter demethylation and HDAC inhibition than prototypical DNMT and HDAC inhibitors, 5-aza and trichostatin A (TSA) (96). Although BITC and HDAC inhibition has not been examined in prostate cancer cells, in pancreatic cells, BITC treatment resulted in HDAC inhibition and decreased HDAC1 and HDAC3 levels. Importantly, the antiproliferative effects of BITC were abrogated by HDAC1 or HDAC3 overexpression, suggesting that HDAC inhibition was a critical mechanism leading to BITC-induced cell cycle arrest and apoptosis (135).
Indoles.
I3C is another phytochemical derived from cruciferous vegetables. In the acidic environment of the stomach, I3C undergoes extensive and rapid self-condensation and produces many oligomeric products. A major condensation product is DIM, formed from the dimerization of 2 molecules of I3C. DIM was shown to be protective against prostate cancer both in vitro and in vivo (136, 137). Interestingly, DIM alters HDAC expression in human colon cancer cells. Specifically, treatment with 0–60 μmol/L DIM decreased the levels of HDAC1, HDAC2, HDAC3, and HDAC8 proteins and there was a decline in the expression of survivin and Cyclin B1 (138, 139). DIM also produced a decline in the expression of class I HDAC in mice bearing colon cancer xenografts (139). Change in HDAC expression was associated with increased Bax, p21, and p27 levels (classic HDAC target genes) and increased cell cycle arrest and apoptosis (139). Class II HDAC were not affected by DIM and the selective degradation of class I HDAC was shown to be mediated by the proteasome (139). A study of similar mechanisms in prostate cancer cells appears to be warranted for DIM and related dietary indoles.
Selenium and its metabolites.
Based on both epidemiological and preclinical studies, there has been considerable interest in the mineral selenium as a prostate cancer chemoprevention agent. Many studies have focused on the role of selenium in selenoproteins, such as glutathione peroxidase and thioredoxin reductase, and their involvement in antioxidant and redox function in the cell [reviewed in (140)]. The biological activity of selenium is highly dependent on the chemical form (i.e. inorganic or organoselenium compound) and metabolic conversion in the cell. Depending on the form of selenium used and/or metabolic transformation in the cell, the molecular targets, including epigenetic targets, can differ markedly. In vitro, inorganic selenite caused decreases in HDAC activity, increases in acetylated histones, and reactivation of GSTp1. At the same time, selenite decreased DNMT1 and methylated H3K9 expression associated with the GSTp1 promoter (141).
More recently, α-keto acid metabolites of organoselenium compounds have also been identified as novel HDAC inhibitors in prostate cancer cells. From the diet, the major organoselenium compounds are MSC and SM. These forms can either be metabolized by β/γ lyase elimination reactions forming methylselenol or undergo transamination/oxidation reactions forming α-keto acids. Methylselenol has been implicated as the putative “active” metabolite responsible for modulating cell signaling and redox pathways leading to apoptosis [for review, see (142)]. However, α-keto acids such as MSP and KMSB may exert epigenetic effects. Specifically, MSP formed from the transamination of MSC and KMSB generated from the transamination of SM both inhibited HDAC activity, increased acetylated histones and p21 promoter activity, and triggered cell cycle arrest and apoptosis in prostate and colon cancer cells at concentrations as low as 2 μmol/L (143, 144). Further research revealed that whereas both transamination reactions occur in liver, in prostate cancer cells, MSC is effectively converted to MSP, whereas SM is poorly metabolized to KMSB.
To date, large-scale, prospective, randomized, double-blinded, placebo-controlled prostate cancer trials utilizing selenium, such as the SELECT trial, have yielded disappointing results (145, 146). In the SELECT trial, healthy men were randomized to supplementation of SM (200 μg/d) alone or in combination with vitamin E (400 IU/d) and prostate cancer incidence was monitored. SM supplementation had no effect on prostate cancer and caused a nonsignificant increase in the incidence of diabetes (146). As a result, the SELECT trail was halted in 2008. Some researchers have postulated that the inability of prostate cancer cells to metabolize SM to the α-keto acid and the lack of HDAC inhibition could be a contributing factor to poor responses in this trial (147). As a corollary, MSC may have been a better candidate due to its conversion to the HDAC inhibitor MSP. Further research is needed on form, dose, tissue-specific metabolism, and HDAC inhibition by mechanistically relevant organoselenium compounds in prostate carcinogenesis.
Curcumin.
The polyphenol curcumin is a natural phytochemical derived from the spice turmeric. Evidence suggests that curcumin suppresses multiple stages of carcinogenesis: initiation, promotion, and progression. The antiinflammatory actions and inhibition of NF-κB have been most widely studied as mechanisms accounting for the efficacy of curcumin [for review, see (148)]. Curcumin exhibits chemopreventive potential in a wide variety of cancers in both preclinical and clinical trials [reviewed in (149)], although one confounding factor may be in vivo uptake and bioavailability following typical oral/dietary intake. Recent research suggests that curcumin modulates global histone and NF-κB acetylation and alters proinflammatory processes as a HAT inhibitor (150). In PC3 prostate cancer cells, curcumin promoted proteasome-dependent degradation of the HAT p300/CREB binding protein (CBP) (151). In addition to HAT inhibition, there are also several reports of possible HDAC inhibitory effects of curcumin. In lymphoma cells, curcumin treatment decreases HDAC1, HDAC3, and HDAC8 protein expression (152). However, in monocytes/macrophages, curcumin treatment inhibited inflammation and overcame glucocorticoid resistance by upregulating HDAC2 activity in lung (153). In other sites, including prostate, HDAC and HAT responses to curcumin are not well understood; however, HAT compared to HDAC effects may depend on tissue type and related pathophysiological conditions, such as oxidative stress and chronic inflammation.
Tea polyphenols.
The majority of epigenetic studies with tea polyphenols have focused on EGCG as a DNMT inhibitor; however, recent research also suggests that catechins modulate histone marks. Similar to DNA methylation, EGCG appears to be the most potent histone modifier among the tea catechins (154). Addition of green tea polyphenols to prostate cancer cells decreased HDAC1 and HDAC3 expression and increased acetylated histones H3 (K9/18) and H4 (80). It is not clear if the effect of green tea polyphenols on histone acetylation is an independent event or if it is related to the cross talk between DNA methylation and chromatin remodeling to control gene silencing. Interestingly, DNMT1 also appears to direct histone modifications by recruiting HDAC (155). Methylation of CpG sequences by DNMT1 binds specific MBD proteins such as MeCP2 and MBD2. This MBD binding complex recruits a complex of transcriptional repressors, including HDAC, which results in chromatin-associated gene silencing (156, 157). Green tea polyphenol also decreased the expression of MBD1, MBC4, and MeCP2 and decreased the association of MBD2 with the GSTp1 promoter (80). These studies clearly demonstrate a dual potential of green tea polyphenols to alter both DNA methylation and histone modifications and the associated players involved in chromatin remodeling. Cell-specific effects of tea catechins and their metabolites on HAT and HDAC have not been thoroughly examined in prostate or other tissues.
Sirtuin modulators.
Compared to Class I and II HDAC inhibitors, studies examining the modulation of sirtuins in prostate cancer are much more limited. The sirtuins are Class III HDAC that utilize NAD as a cofactor (158). Seven homologs (Sirt1–7) have been identified in the human genome and predominantly target nonhistone proteins for deacetylation. Each Sirt has a conserved structure, but their targets and cellular localization differ. Sirt1 is the best characterized sirtuin in mammals and has a wide variety of targets in both the nucleus and cytoplasm. Of particular relevance to cancer, Sirt1 appears to play an important role in DNA repair, recombination, and cell survival. Some examples of Sirt1 targets include AR, p53, RB protein, forkhead transcription factors, Bax, Ku70 repair protein, and p300 (159).
More recently, Sirt1 was shown to be overexpressed in prostate cancer cells compared to normal controls (160). Sirt1 inhibition with nicotinamide and RNAi strategies led to significant growth inhibition and cell death in the cancer cells, with no effects in normal cells, suggesting that Sirt1 may have oncogenic activity. In addition to Sirt1, overexpression of nicotinamide phosphoribosyltransferase, the rate-limiting enzyme for regenerating NAD, also has been reported in prostate cancer (161). However, there is considerable controversy as to whether Sirt1 is actually an oncogenic or tumor suppressor protein. In contrast to work in vitro, studies in vivo suggest that Sirt1 acts a tumor suppressor. Deletion of the Sirt1 gene in mice resulted in prostatic intraepithelial neoplasia, and genome-wide expression analysis of these lesions revealed that loss of Sirt1 in the prostate resulted in an inhibition of autophagy pathways (162).
Dietary strategies that target Sirt1 are extremely limited at present. Resveratrol, a putative activator of sirtuins, has been studied in prostate and appears to have antiproliferative effects (163, 164). However, Sirt1 inhibitors have also shown an ability to limit prostate cancer development. Kikuno et al. (165) showed that genistein treatment caused demethylation and acetylation of H3K9. At the same time, reductions in Sirt1 activity were detected and associated with acetylation of H3K9 at p53 and forkhead transcription factor 3a promoters. It is likely that the impact on Sirt1 depends on factors such as the level of expression and timing of the disease. For example, endogenous Sirt1 may be essential for maintaining prostate gland early during development, whereas aberrant overexpression of Sirt1, with age or during later stages of carcinogenesis, may lead to enhanced proliferation and tumor development. In this scenario, knockdown of endogenous levels of Sirt1 early in life may be cancer promoting, whereas Sirt1 inhibition after cancer has developed may be protective. Additional studies are necessary both in vitro and in vivo to address these issues and identify key Sirt1 targets involved in prostate cancer development.
Conclusions
Great strides have been made in recent years in advancing our understanding of diet-gene interactions and their influence on cancer development. In the burgeoning field of epigenetics, an increased understanding of the mechanisms that regulate gene expression has produced a paradigm shift in how we consider cancer etiology and the relevant molecular biomarkers for cancer prevention. In contrast to genetic alterations, epigenetic marks are potentially reversible, making them promising targets for both cancer prevention and therapy. Researchers have identified new and novel epigenetic mechanisms for many candidate dietary chemoprevention agents. However, control of gene expression often requires the cooperation, interaction, and interplay among multiple epigenetic mechanisms. For example, the relationship between DNA methylation and chromatin remodeling suggests significant cross-talk among distinct epigenetic pathways that control gene silencing/unsilencing. Indeed, the combination of pharmacological DNMT inhibitors and HDAC inhibitors has been explored as a potential antitumor therapy (166, 167). However, both HDAC and DNMT inhibitor drugs have potential hazards and side effects, including cardiotoxicity in some clinical scenarios (168, 169). Interestingly, several miRNA are known to be regulated by DNA methylation status in cells. In a recent study, up to one-third of dysregulated miRNA loci showed consistent patterns of DNA methylation and H3K9 acetylation (170), highlighting the complex interplay among multiple epigenetic regulators. Dovetailing this work with mechanistic studies of dietary epigenetic modulators deserves greater attention. In particular, understanding the relevant doses, timing, and possibly synergy and/or antagonism of various epigenetic marks to coordinate biological effects is of utmost importance.
To complicate matters, there is also significant interplay between genetic and epigenetic targets. For example, the traditional dogma for chemoprotection by SFN considered reactive thiol groups in Keap1 as key targets, promoting Nrf2 dissociation, nuclear trafficking, and activation of ARE-driven gene expression. Recent studies, however, have found that Nrf2 itself is under epigenetic regulation. Yu et al. (171) demonstrated that Nrf2 expression was suppressed in prostate tumors derived from TRAMP mice. Moreover, silencing of Nrf2 was attributed to both promoter hypermethylation and histone modifications. Pharmacological DNMT inhibitors and/or HDAC inhibitors restored the expression and activity of Nrf2 (171). Thus, dietary agents such as SFN may coordinate Nrf2 activation at the epigenetic/chromatin level in addition to effects on Keap1. As we move forward, the lines between diet-genome and diet-epigenome interactions are becoming increasingly blurred. Individual susceptibility to disease and efficacy of specific nutrients and phytochemicals may be determined by a combination of both genetic and epigenetic control mechanisms. No doubt, these will be important issues in the future development of better optimized dietary chemoprevention strategies.
Understanding the precise actions of dietary bioactive nutrients in cancer prevention is a complicated and difficult task. Many such compounds exhibit pleiotropic effects, which can be viewed as either beneficial or deleterious. Advocates consider the benefit of targeting multiple players along the pathway to malignancy, whereas skeptics typically cite potential “off-target” concerns. For many nutrients, the relative priority of genetic compared to epigenetic mechanisms remains poorly defined. Importantly, additional research on dose, metabolism, timing, and tissue specificity is sorely needed. It is possible that doses that optimally target epigenetics may not target other known genetic targets and vice versa. Establishing dose responses, including possible toxicities, will be extremely important prior to making recommendations. Nonetheless, targeting the epigenome with modifiers of miRNA, DNA methylation, and histone marks provides an attractive avenue for future research, with considerable promise for cancer clinical trials. Identification of dietary epigenetic modulators and their clinical application, either alone or in combination, may enhance the efficacy of ongoing anticancer therapies/prevention strategies while reducing the unwanted side effects.
Acknowledgments
All authors have read and approved the final manuscript.
Footnotes
Supported by NIH grants CA90890, CA65525, CA122906, CA122959, and CA80176 and by National Institute of Environmental Health Sciences (NIEHS) Center grant P30 ES00210.
Author disclosures: E. Ho, L. M. Beaver, D. E. Williams, and R. H. Dashwood, no conflicts of interest.
Abbreviations used: AM, allyl mercaptan; AR, androgen receptor; BITC, benzethyl isothiocyanate; DADS, diallyl sulfide, diallyl disulfide; DATS, diallyl trisulfide; DIM, 3,3′-diindolylmethane; DNMT, DNA methyltransferase; EGCG, epigallocatechin-3-gallate; HAT, histone acetyltransferase; HDAC, histone deacetylase; I3C, indole-3-carbinol; ITC, isothiocyanate; KMSB, α-keto-methylselenobutyrate; miRNA, microRNA; MSC, methylselenocysteine; MSP, methylselenopyruvate; PEITC, phenethyl isothiocyanate; SAM, S-adenosyl methionine; SELECT, Selenium and Vitamin E Cancer Prevention Trial; SFN, sulforaphane; SM, selenomethionine; TRAMP, Transgenic Adenocarcinoma of Mouse Prostate; THF, tetrahydrofolate.
Literature Cited
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74 [DOI] [PubMed] [Google Scholar]
- 2.Boumber Y, Issa JP. Epigenetics in cancer: what's the future? Oncology. 2011;25:220–6, 228 [PubMed] [Google Scholar]
- 3.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–40 [DOI] [PubMed] [Google Scholar]
- 4.Abbas A, Gupta S. The role of histone deacetylases in prostate cancer. Epigenetics. 2008;3:300–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobosy JR, Roberts JLW, Fu VX, Jarrard DF. The expanding role of epigenetics in the development, diagnosis and treatment of prostate cancer and benign prostatic hyperplasia. J Urol. 2007;177:822–31 [DOI] [PubMed] [Google Scholar]
- 6.Catto JW, Alcaraz A, Bjartell AS, De Vere White R, Evans CP, Fussel S, Hamdy FC, Kallioniemi O, Mengual L, Schlomm T, et al. MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur Urol. 2011;59:671–81 [DOI] [PubMed] [Google Scholar]
- 7.Parasramka MA, Ho E, Williams DE, Dashwood RH. MicroRNAs, diet, and cancer: new mechanistic insights on the epigenetic actions of phytochemicals. Mol Carcinog. Epub 2011 Jul 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nat Rev Genet. 2009;10:805–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ollikainen M, Smith KR, Joo EJ, Ng HK, Andronikos R, Novakovic B, Abdul Aziz NK, Carlin JB, Morley R, Saffery R, et al. DNA methylation analysis of multiple tissues from newborn twins reveals both genetic and intrauterine components to variation in the human neonatal epigenome. Hum Mol Genet. 2010;19:4176–88 [DOI] [PubMed] [Google Scholar]
- 10.Schneider E, Pliushch G, El Hajj N, Galetzka D, Puhl A, Schorsch M, Frauenknecht K, Riepert T, Tresch A, Muller AM, et al. Spatial, temporal and interindividual epigenetic variation of functionally important DNA methylation patterns. Nucleic Acids Res. 2010;38:3880–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perry AS, Watson RW, Lawler M, Hollywood D. The epigenome as a therapeutic target in prostate cancer. Nat Rev Urol. 2010;7:668–80 [DOI] [PubMed] [Google Scholar]
- 12.Jeronimo C, Bastian PJ, Bjartell A, Carbone GM, Catto JW, Clark SJ, Henrique R, Nelson WG, Shariat SF. Epigenetics in prostate cancer: biologic and clinical relevance. Eur Urol. 2011;60:753–66 [DOI] [PubMed] [Google Scholar]
- 13.Bedford MT, van Helden PD. Hypomethylation of DNA in pathological conditions of the human prostate. Cancer Res. 1987;47:5274–6 [PubMed] [Google Scholar]
- 14.Santourlidis S, Warskulat U, Florl AR, Maas S, Pulte T, Fischer J, Muller W, Schulz WA. Hypermethylation of the tumor necrosis factor receptor superfamily 6 (APT1, Fas, CD95/Apo-1) gene promoter at rel/nuclear factor kappaB sites in prostatic carcinoma. Mol Carcinog. 2001;32:36–43 [DOI] [PubMed] [Google Scholar]
- 15.Schulz WA, Elo JP, Florl AR, Pennanen S, Santourlidis S, Engers R, Buchardt M, Seifert HH, Visakorpi T. Genomewide DNA hypomethylation is associated with alterations on chromosome 8 in prostate carcinoma. Genes Chromosomes Cancer. 2002;35:58–65 [DOI] [PubMed] [Google Scholar]
- 16.Santourlidis S, Florl A, Ackermann R, Wirtz HC, Schulz WA. High frequency of alterations in DNA methylation in adenocarcinoma of the prostate. Prostate. 1999;39:166–74 [DOI] [PubMed] [Google Scholar]
- 17.Li LC, Okino ST, Dahiya R. DNA methylation in prostate cancer. Biochim Biophys Acta. 2004;1704:87–102 [DOI] [PubMed] [Google Scholar]
- 18.Jerónimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, Oliveira J, Teixeira MR, Lopes C, Sidransky D. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8 [DOI] [PubMed] [Google Scholar]
- 19.Yang B, Sun H, Lin W, Hou W, Li H, Zhang L, Li F, Gu Y, Song Y, Li Q, et al. Evaluation of global DNA hypomethylation in human prostate cancer and prostatic intraepithelial neoplasm tissues by immunohistochemistry. Urol Oncol. Epub 2011 Jun 23 [DOI] [PubMed] [Google Scholar]
- 20.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705 [DOI] [PubMed] [Google Scholar]
- 21.Pal S, Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. J Cell Physiol. 2007;213:306–15 [DOI] [PubMed] [Google Scholar]
- 22.Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol Cell. 2005;19:381–91 [DOI] [PubMed] [Google Scholar]
- 24.Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science. 2002;298:1039–43 [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi Y, Absher DM, Gulzar ZG, Young SR, McKenney JK, Peehl DM, Brooks JD, Myers RM, Sherlock G. DNA methylation profiling reveals novel biomarkers and important roles for DNA methyltransferases in prostate cancer. Genome Res. 2011;21:1017–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Metzger E, Wissmann M, Yin N, Muller JM, Schneider R, Peters AH, Gunther T, Buettner R, Schule R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–9 [DOI] [PubMed] [Google Scholar]
- 27.Ke XS, Qu Y, Cheng Y, Li WC, Rotter V, Oyan AM, Kalland KH. Global profiling of histone and DNA methylation reveals epigenetic-based regulation of gene expression during epithelial to mesenchymal transition in prostate cells. BMC Genomics. 2010;11:669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patra SK, Patra A, Dahiya R. Histone deacetylase and DNA methyltransferase in human prostate cancer. Biochem Biophys Res Commun. 2001;287:705–13 [DOI] [PubMed] [Google Scholar]
- 29.Halkidou K, Gaughan L, Cook S, Leung HY, Neal DE, Robson CN. Upregulation and nuclear recruitment of HDAC1 in hormone refractory prostate cancer. Prostate. 2004;59:177–89 [DOI] [PubMed] [Google Scholar]
- 30.Halkidou K, Cook S, Leung HY, Neal DE, Robson CN. Nuclear accumulation of histone deacetylase 4 (HDAC4) coincides with the loss of androgen sensitivity in hormone refractory cancer of the prostate. Eur Urol. 2004;45:382–9, author reply 389 [DOI] [PubMed] [Google Scholar]
- 31.Korkmaz CG, Fronsdal K, Zhang Y, Lorenzo PI, Saatcioglu F. Potentiation of androgen receptor transcriptional activity by inhibition of histone deacetylation–rescue of transcriptionally compromised mutants. J Endocrinol. 2004;182:377–89 [DOI] [PubMed] [Google Scholar]
- 32.Huffman DM, Grizzle WE, Bamman MM, Kim JS, Eltoum IA, Elgavish A, Nagy TR. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–8 [DOI] [PubMed] [Google Scholar]
- 33.Seligson DB, Horvath S, Shi T, Yu H, Tze S, Grunstein M, Kurdistani SK. Global histone modification patterns predict risk of prostate cancer recurrence. Nature. 2005;435:1262–6 [DOI] [PubMed] [Google Scholar]
- 34.Fr#x00F8nsdal K, Saatcioglu F. Histone deacetylase inhibitors differentially mediate apoptosis in prostate cancer cells. Prostate. 2005;62:299–306 [DOI] [PubMed] [Google Scholar]
- 35.Butler LM, Agus DB, Scher HI, Higgins B, Rose A, Cordon-Cardo C, Thaler HT, Rifkind RA, Marks PA, Richon VM. Suberoylanilide hydroxamic acid, an inhibitor of histone deacetylase, suppresses the growth of prostate cancer cells in vitro and in vivo. Cancer Res. 2000;60:5165–70 [PubMed] [Google Scholar]
- 36.Li LC, Carroll PR, Dahiya R. Epigenetic changes in prostate cancer: implication for diagnosis and treatment. J Natl Cancer Inst. 2005;97:103–15 [DOI] [PubMed] [Google Scholar]
- 37.Esteller M. Aberrant DNA methylation as a cancer-inducing mechanism. Annu Rev Pharmacol Toxicol. 2005;45:629–56 [DOI] [PubMed] [Google Scholar]
- 38.James SJ, Pogribny IP, Pogribna M, Miller BJ, Jernigan S, Melnyk S. Mechanisms of DNA damage, DNA hypomethylation, and tumor progression in the folate/methyl-deficient rat model of hepatocarcinogenesis. J Nutr. 2003;133:S3740–7 [DOI] [PubMed] [Google Scholar]
- 39.Pogribny IP, James SJ, Jernigan S, Pogribna M. Genomic hypomethylation is specific for preneoplastic liver in folate/methyl deficient rats and does not occur in non-target tissues. Mutat Res. 2004;548:53–9 [DOI] [PubMed] [Google Scholar]
- 40.Pogribny IP, Ross SA, Wise C, Pogribna M, Jones EA, Tryndyak VP, James SJ, Dragan YP, Poirier LA. Irreversible global DNA hypomethylation as a key step in hepatocarcinogenesis induced by dietary methyl deficiency. Mutat Res. 2006;593:80–7 [DOI] [PubMed] [Google Scholar]
- 41.Poirier LA. The effects of diet, genetics and chemicals on toxicity and aberrant DNA methylation: an introduction. J Nutr. 2002;132:S2336–9 [DOI] [PubMed] [Google Scholar]
- 42.Chandar N, Lombardi B, Locker J. c-myc gene amplification during hepatocarcinogenesis by a choline-devoid diet. Proc Natl Acad Sci USA. 1989;86:2703–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhave MR, Wilson MJ, Poirier LA. c-H-ras and c-K-ras gene hypomethylation in the livers and hepatomas of rats fed methyl-deficient, amino acid-defined diets. Carcinogenesis. 1988;9:343–8 [DOI] [PubMed] [Google Scholar]
- 44.Pogribny IP, Tryndyak VP, Muskhelishvili L, Rusyn I, Ross SA. Methyl deficiency, alterations in global histone modifications, and carcinogenesis. J Nutr. 2007;137:S216–22 [DOI] [PubMed] [Google Scholar]
- 45.Beilby J, Ambrosini GL, Rossi E, de Klerk NH, Musk AW. Serum levels of folate, lycopene, beta-carotene, retinol and vitamin E and prostate cancer risk. Eur J Clin Nutr. 2010;64:1235–8 [DOI] [PubMed] [Google Scholar]
- 46.Weinstein SJ, Hartman TJ, Stolzenberg-Solomon R, Pietinen P, Barrett MJ, Taylor PR, Virtamo J, Albanes D. Null association between prostate cancer and serum folate, vitamin B(6), vitamin B(12), and homocysteine. Cancer Epidemiol Biomarkers Prev. 2003;12:1271–2 [PubMed] [Google Scholar]
- 47.Collin SM, Metcalfe C, Zuccolo L, Lewis SJ, Chen L, Cox A, Davis M, Lane JA, Donovan J, Smith GD, et al. Association of folate-pathway gene polymorphisms with the risk of prostate cancer: a population-based nested case-control study, systematic review, and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2009;18:2528–39 [DOI] [PubMed] [Google Scholar]
- 48.Eussen SJ, Vollset SE, Igland J, Meyer K, Fredriksen A, Ueland PM, Jenab M, Slimani N, Boffetta P, Overvad K, et al. Plasma folate, related genetic variants, and colorectal cancer risk in EPIC. Cancer Epidemiol Biomarkers Prev. 2010;19:1328–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomaszewski JJ, Cummings JL, Parwani AV, Dhir R, Mason JB, Nelson JB, Bacich DJ, O'Keefe DS. Increased cancer cell proliferation in prostate cancer patients with high levels of serum folate. Prostate. 2011;71:1287–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Collin SM, Metcalfe C, Refsum H, Lewis SJ, Zuccolo L, Smith GD, Chen L, Harris R, Davis M, Marsden G, et al. Circulating folate, vitamin B12, homocysteine, vitamin B12 transport proteins, and risk of prostate cancer: a case-control study, systematic review, and meta-analysis. Cancer Epidemiol Biomarkers Prev. 2010;19:1632–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neuhouser ML, Nijhout HF, Gregory JF, III, Reed MC, James SJ, Liu A, Shane B, Ulrich CM. Mathematical modeling predicts the effect of folate deficiency and excess on cancer related biomarkers. Cancer Epidemiol Biomarkers Prev. 2011;20:1912–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith AD, Kim YI, Refsum H. Is folic acid good for everyone? Am J Clin Nutr. 2008;87:517–33 [DOI] [PubMed] [Google Scholar]
- 53.Mason JB. Folate, cancer risk, and the Greek god, Proteus: a tale of two chameleons. Nutr Rev. 2009;67:206–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YI. Folate and colorectal cancer: an evidence-based critical review. Mol Nutr Food Res. 2007;51:267–92 [DOI] [PubMed] [Google Scholar]
- 55.Yan L, Spitznagel EL. Meta-analysis of soy food and risk of prostate cancer in men. Int J Cancer. 2005;117:667–9 [DOI] [PubMed] [Google Scholar]
- 56.Adlercreutz H, Bannwart C, Wahala K, Makela T, Brunow G, Hase T, Arosemena PJ, Kellis JT, Jr, Vickery LE. Inhibition of human aromatase by mammalian lignans and isoflavonoid phytoestrogens. J Steroid Biochem Mol Biol. 1993;44:147–53 [DOI] [PubMed] [Google Scholar]
- 57.Morton MS, Arisaka O, Miyake N, Morgan LD, Evans BA. Phytoestrogen concentrations in serum from Japanese men and women over forty years of age. J Nutr. 2002;132:3168–71 [DOI] [PubMed] [Google Scholar]
- 58.Lee H, Wang HW, Su HY, Hao NJ. The structure-activity relationships of flavonoids as inhibitors of cytochrome P-450 enzymes in rat liver microsomes and the mutagenicity of 2-amino-3-methyl-imidazo[4,5-f]quinoline. Mutagenesis. 1994;9:101–6 [DOI] [PubMed] [Google Scholar]
- 59.Mäkelä S, Poutanen M, Kostian ML, Lehtimaki N, Strauss L, Santti R, Vihko R. Inhibition of 17beta-hydroxysteroid oxidoreductase by flavonoids in breast and prostate cancer cells. Proc Soc Exp Biol Med. 1998;217:310–6 [DOI] [PubMed] [Google Scholar]
- 60.Park OJ, Surh YJ. Chemopreventive potential of epigallocatechin gallate and genistein: evidence from epidemiological and laboratory studies. Toxicol Lett. 2004;150:43–56 [DOI] [PubMed] [Google Scholar]
- 61.Magee PJ, Rowland IR. Phyto-oestrogens, their mechanism of action: current evidence for a role in breast and prostate cancer. Br J Nutr. 2004;91:513–31 [DOI] [PubMed] [Google Scholar]
- 62.Sarkar FH, Li Y. Soy isoflavones and cancer prevention. Cancer Invest. 2003;21:744–57 [DOI] [PubMed] [Google Scholar]
- 63.Evans BA, Griffiths K, Morton MS. Inhibition of 5 alpha-reductase in genital skin fibroblasts and prostate tissue by dietary lignans and isoflavonoids. J Endocrinol. 1995;147:295–302 [DOI] [PubMed] [Google Scholar]
- 64.Fritz WA, Wang J, Eltoum IE, Lamartiniere CA. Dietary genistein down-regulates androgen and estrogen receptor expression in the rat prostate. Mol Cell Endocrinol. 2002;186:89–99 [DOI] [PubMed] [Google Scholar]
- 65.Hamilton-Reeves JM, Rebello SA, Thomas W, Slaton JW, Kurzer MS. Isoflavone-rich soy protein isolate suppresses androgen receptor expression without altering estrogen receptor-beta expression or serum hormonal profiles in men at high risk of prostate cancer. J Nutr. 2007;137:1769–75 [DOI] [PubMed] [Google Scholar]
- 66.Handayani R, Rice L, Cui Y, Medrano TA, Samedi VG, Baker HV, Szabo NJ, Shiverick KT. Soy isoflavones alter expression of genes associated with cancer progression, including interleukin-8, in androgen-independent PC-3 human prostate cancer cells. J Nutr. 2006;136:75–82 [DOI] [PubMed] [Google Scholar]
- 67.Fang MZ, Chen D, Sun Y, Jin Z, Christman JK, Yang CS. Reversal of hypermethylation and reactivation of p16INK4a, RARbeta, and MGMT genes by genistein and other isoflavones from soy. Clin Cancer Res. 2005;11:7033–41 [DOI] [PubMed] [Google Scholar]
- 68.Fang M, Chen D, Yang CS. Dietary polyphenols may affect DNA methylation. J Nutr. 2007;137:S223–8 [DOI] [PubMed] [Google Scholar]
- 69.Majid S, Dar AA, Shahryari V, Hirata H, Ahmad A, Saini S, Tanaka Y, Dahiya AV, Dahiya R. Genistein reverses hypermethylation and induces active histone modifications in tumor suppressor gene B-Cell translocation gene 3 in prostate cancer. Cancer. 2010;116:66–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vardi A, Bosviel R, Rabiau N, Adjakly M, Satih S, Dechelotte P, Boiteux JP, Fontana L, Bignon YJ, Guy L, et al. Soy phytoestrogens modify DNA methylation of GSTP1, RASSF1A, EPH2 and BRCA1 promoter in prostate cancer cells. In Vivo. 2010;24:393–400 [PubMed] [Google Scholar]
- 71.Dolinoy DC, Weidman JR, Waterland RA, Jirtle RL. Maternal genistein alters coat color and protects Avy mouse offspring from obesity by modifying the fetal epigenome. Environ Health Perspect. 2006;114:567–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Day JK, Bauer AM, DesBordes C, Zhuang Y, Kim BE, Newton LG, Nehra V, Forsee KM, MacDonald RS, Besch-Williford C, et al. Genistein alters methylation patterns in mice. J Nutr. 2002;132:S2419–23 [DOI] [PubMed] [Google Scholar]
- 73.Qin W, Zhu W, Shi H, Hewett JE, Ruhlen RL, MacDonald RS, Rottinghaus GE, Chen YC, Sauter ER. Soy isoflavones have an antiestrogenic effect and alter mammary promoter hypermethylation in healthy premenopausal women. Nutr Cancer. 2009;61:238–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Graham HN. Green tea composition, consumption, and polyphenol chemistry. Prev Med. 1992;21:334–50 [DOI] [PubMed] [Google Scholar]
- 75.Na HK, Surh YJ. Intracellular signaling network as a prime chemopreventive target of (-)-epigallocatechin gallate. Mol Nutr Food Res. 2006;50:152–9 [DOI] [PubMed] [Google Scholar]
- 76.Stuart EC, Scandlyn MJ, Rosengren RJ. Role of epigallocatechin gallate (EGCG) in the treatment of breast and prostate cancer. Life Sci. 2006;79:2329–36 [DOI] [PubMed] [Google Scholar]
- 77.Chen D, Wang CY, Lambert JD, Ai N, Welsh WJ, Yang CS. Inhibition of human liver catechol-O-methyltransferase by tea catechins and their metabolites: structure-activity relationship and molecular-modeling studies. Biochem Pharmacol. 2005;69:1523–31 [DOI] [PubMed] [Google Scholar]
- 78.Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, Welsh W, Yang CS. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003;63:7563–70 [PubMed] [Google Scholar]
- 79.Berletch JB, Liu C, Love WK, Andrews LG, Katiyar SK, Tollefsbol TO. Epigenetic and genetic mechanisms contribute to telomerase inhibition by EGCG. J Cell Biochem. 2008;103:509–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Pandey M, Shukla S, Gupta S. Promoter demethylation and chromatin remodeling by green tea polyphenols leads to re-expression of GSTP1 in human prostate cancer cells. Int J Cancer. 2010;126:2520–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bettuzzi S, Brausi M, Rizzi F, Castagnetti G, Peracchia G, Corti A. Chemoprevention of human prostate cancer by oral administration of green tea catechins in volunteers with high-grade prostate intraepithelial neoplasia: a preliminary report from a one-year proof-of-principle study. Cancer Res. 2006;66:1234–40 [DOI] [PubMed] [Google Scholar]
- 82.Wang P, Aronson WJ, Huang M, Zhang Y, Lee RP, Heber D, Henning SM. Green tea polyphenols and metabolites in prostatectomy tissue: implications for cancer prevention. Cancer Prev Res (Phila). 2010;3:985–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Caporali A, Davalli P, Astancolle S, D'Arca D, Brausi M, Bettuzzi S, Corti A. The chemopreventive action of catechins in the TRAMP mouse model of prostate carcinogenesis is accompanied by clusterin over-expression. Carcinogenesis. 2004;25:2217–24 [DOI] [PubMed] [Google Scholar]
- 84.Gupta S, Hastak K, Ahmad N, Lewin JS, Mukhtar H. Inhibition of prostate carcinogenesis in TRAMP mice by oral infusion of green tea polyphenols. Proc Natl Acad Sci USA. 2001;98:10350–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Adhami VM, Siddiqui IA, Sarfaraz S, Khwaja SI, Hafeez BB, Ahmad N, Mukhtar H. Effective prostate cancer chemopreventive intervention with green tea polyphenols in the TRAMP model depends on the stage of the disease. Clin Cancer Res. 2009;15:1947–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morey Kinney SR, Zhang W, Pascual M, Greally JM, Gillard BM, Karasik E, Foster BA, Karpf AR. Lack of evidence for green tea polyphenols as DNA methylation inhibitors in murine prostate. Cancer Prev Res (Phila). 2009;2:1065–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fahey JW, Zhang Y, Talalay P. Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc Natl Acad Sci USA. 1997;94:10367–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Higdon JV, Delage B, Williams DE, Dashwood RH. Cruciferous vegetables and human cancer risk: epidemiologic evidence and mechanistic basis. Pharmacol Res. 2007;55:224–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Steinbrecher A, Nimptsch K, Husing A, Rohrmann S, Linseisen J. Dietary glucosinolate intake and risk of prostate cancer in the EPIC-Heidelberg cohort study. Int J Cancer. 2009;125:2179–86 [DOI] [PubMed] [Google Scholar]
- 90.Chung FL, Conaway CC, Rao CV, Reddy BS. Chemoprevention of colonic aberrant crypt foci in Fischer rats by sulforaphane and phenethyl isothiocyanate. Carcinogenesis. 2000;21:2287–91 [DOI] [PubMed] [Google Scholar]
- 91.Zhang Y, Kensler TW, Cho CG, Posner GH, Talalay P. Anticarcinogenic activities of sulforaphane and structurally related synthetic norbornyl isothiocyanates. Proc Natl Acad Sci USA. 1994;91:3147–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Singh AV, Xiao D, Lew KL, Dhir R, Singh SV. Sulforaphane induces caspase-mediated apoptosis in cultured PC-3 human prostate cancer cells and retards growth of PC-3 xenografts in vivo. Carcinogenesis. 2004;25:83–90 [DOI] [PubMed] [Google Scholar]
- 93.Singh SV, Warin R, Xiao D, Powolny AA, Stan SD, Arlotti JA, Zeng Y, Hahm ER, Marynowski SW, Bommareddy A, et al. Sulforaphane inhibits prostate carcinogenesis and pulmonary metastasis in TRAMP mice in association with increased cytotoxicity of natural killer cells. Cancer Res. 2009;69:2117–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Meeran SM, Patel SN, Tollefsbol TO. Sulforaphane causes epigenetic repression of hTERT expression in human breast cancer cell lines. PLoS ONE. 2010;5:e11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hsu A, Wong CP, Zhen Y, Williams DE, Dashwood RH, Ho E. Promoter de-methylation of cyclin D2 by sulforaphane in prostate cancer cells. Clin Epigenetics. 2011; in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wang LG, Beklemisheva A, Liu XM, Ferrari AC, Feng J, Chiao JW. Dual action on promoter demethylation and chromatin by an isothiocyanate restored GSTP1 silenced in prostate cancer. Mol Carcinog. 2007;46:24–31 [DOI] [PubMed] [Google Scholar]
- 97.Davis CD, Uthus EO. Dietary folate and selenium affect dimethylhydrazine-induced aberrant crypt formation, global DNA methylation and one-carbon metabolism in rats. J Nutr. 2003;133:2907–14 [DOI] [PubMed] [Google Scholar]
- 98.Zeng H, Yan L, Cheng WH, Uthus EO. Dietary selenomethionine increases exon-specific DNA methylation of the p53 gene in rat liver and colon mucosa. J Nutr. 2011;141:1464–8 [DOI] [PubMed] [Google Scholar]
- 99.Evans JC, Huddler DP, Jiracek J, Castro C, Millian NS, Garrow TA, Ludwig ML. Betaine-homocysteine methyltransferase: zinc in a distorted barrel. Structure. 2002;10:1159–71 [DOI] [PubMed] [Google Scholar]
- 100.Wallwork JC, Duerre JA. Effect of zinc deficiency on methionine metabolism, methylation reactions and protein synthesis in isolated perfused rat liver. J Nutr. 1985;115:252–62 [DOI] [PubMed] [Google Scholar]
- 101.Wolff GL, Kodell RL, Moore SR, Cooney CA. Maternal epigenetics and methyl supplements affect agouti gene expression in Avy/a mice. FASEB J. 1998;12:949–57 [PubMed] [Google Scholar]
- 102.Costello LC, Franklin RB. The clinical relevance of the metabolism of prostate cancer; zinc and tumor suppression: connecting the dots. Mol Cancer. 2006;5:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ho E, Song Y. Zinc and prostatic cancer. Curr Opin Clin Nutr Metab Care. 2009;12:640–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Dashwood RH, Ho E. Dietary histone deacetylase inhibitors: from cells to mice to man. Semin Cancer Biol. 2007;17:363–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Miranda TB, Cortez CC, Yoo CB, Liang G, Abe M, Kelly TK, Marquez VE, Jones PA. DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther. 2009;8:1579–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Crea F, Hurt EM, Mathews LA, Cabarcas SM, Sun L, Marquez VE, Danesi R, Farrar WL. Pharmacologic disruption of Polycomb Repressive Complex 2 inhibits tumorigenicity and tumor progression in prostate cancer. Mol Cancer. 2011;10:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mitsiades CS, Mitsiades NS, McMullan CJ, Poulaki V, Shringarpure R, Hideshima T, Akiyama M, Chauhan D, Munshi N, Gu X, et al. Transcriptional signature of histone deacetylase inhibition in multiple myeloma: biological and clinical implications. Proc Natl Acad Sci USA. 2004;101:540–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Brinkmann H, Dahler AL, Popa C, Serewko MM, Parsons PG, Gabrielli BG, Burgess AJ, Saunders NA. Histone hyperacetylation induced by histone deacetylase inhibitors is not sufficient to cause growth inhibition in human dermal fibroblasts. J Biol Chem. 2001;276:22491–9 [DOI] [PubMed] [Google Scholar]
- 109.Davie JR. Inhibition of histone deacetylase activity by butyrate. J Nutr. 2003;133:S2485–93 [DOI] [PubMed] [Google Scholar]
- 110.Sekhavat A, Sun JM, Davie JR. Competitive inhibition of histone deacetylase activity by trichostatin A and butyrate. Biochem Cell Biol. 2007;85:751–8 [DOI] [PubMed] [Google Scholar]
- 111.Hinnebusch BF, Meng S, Wu JT, Archer SY, Hodin RA. The effects of short-chain fatty acids on human colon cancer cell phenotype are associated with histone hyperacetylation. J Nutr. 2002;132:1012–7 [DOI] [PubMed] [Google Scholar]
- 112.Waldecker M, Kautenburger T, Daumann H, Busch C, Schrenk D. Inhibition of histone-deacetylase activity by short-chain fatty acids and some polyphenol metabolites formed in the colon. J Nutr Biochem. 2008;19:587–93 [DOI] [PubMed] [Google Scholar]
- 113.Trtková K, Paskova L, Matijescukova N, Kolar Z. Formation of AR-SMRT binding in prostate cancer cells treated with natural histone deacetylase inhibitor. Cancer Biomark. 2010;7:79–90 [DOI] [PubMed] [Google Scholar]
- 114.Trtkova K, Paskova L, Matijescukova N, Strnad M, Kolar Z. Binding of AR to SMRT/N-CoR complex and its co-operation with PSA promoter in prostate cancer cells treated with natural histone deacetylase inhibitor NaB. Neoplasma. 2010;57:406–14 [DOI] [PubMed] [Google Scholar]
- 115.Powolny AA, Singh SV. Multitargeted prevention and therapy of cancer by diallyl trisulfide and related Allium vegetable-derived organosulfur compounds. Cancer Lett. 2008;269:305–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Druesne N, Pagniez A, Mayeur C, Thomas M, Cherbuy C, Duee PH, Martel P, Chaumontet C. Diallyl disulfide (DADS) increases histone acetylation and p21(waf1/cip1) expression in human colon tumor cell lines. Carcinogenesis. 2004;25:1227–36 [DOI] [PubMed] [Google Scholar]
- 117.Herman-Antosiewicz A, Kim YA, Kim SH, Xiao D, Singh SV. Diallyl trisulfide-induced G2/M phase cell cycle arrest in DU145 cells is associated with delayed nuclear translocation of cyclin-dependent kinase 1. Pharm Res. 2010;27:1072–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Kim SH, Bommareddy A, Singh SV. Garlic constituent diallyl trisulfide suppresses x–linked inhibitor of apoptosis protein in prostate cancer cells in culture and in vivo. Cancer Prev Res (Phila). 2011;4:897–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Stan SD, Singh SV. Transcriptional repression and inhibition of nuclear translocation of androgen receptor by diallyl trisulfide in human prostate cancer cells. Clin Cancer Res. 2009;15:4895–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Xiao D, Zeng Y, Singh SV. Diallyl trisulfide-induced apoptosis in human cancer cells is linked to checkpoint kinase 1-mediated mitotic arrest. Mol Carcinog. 2009;48:1018–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Nian H, Delage B, Ho E, Dashwood RH. Modulation of histone deacetylase activity by dietary isothiocyanates and allyl sulfides: studies with sulforaphane and garlic organosulfur compounds. Environ Mol Mutagen. 2009;50:213–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nian H, Delage B, Pinto JT, Dashwood RH. Allyl mercaptan, a garlic-derived organosulfur compound, inhibits histone deacetylase and enhances Sp3 binding on the P21WAF1 promoter. Carcinogenesis. 2008;29:1816–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Rajendran P, Williams DE, Ho E, Dashwood RH. Metabolism as a key to histone deacetylase inhibition. Crit Rev Biochem Mol Biol. 2011;46:181–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Myzak MC, Karplus PA, Chung FL, Dashwood RH. A novel mechanism of chemoprotection by sulforaphane: inhibition of histone deacetylase. Cancer Res. 2004;64:5767–74 [DOI] [PubMed] [Google Scholar]
- 125.Myzak MC, Hardin K, Wang R, Dashwood RH, Ho E. Sulforaphane inhibits histone deacetylase activity in BPH-1, LnCaP and PC-3 prostate epithelial cells. Carcinogenesis. 2006;27:811–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Clarke JD, Hsu A, Yu Z, Dashwood RH, Ho E. Differential effects of sulforaphane on histone deacetylases, cell cycle arrest and apoptosis in normal prostate cells versus hyperplastic and cancerous prostate cells. Mol Nutr Food Res. 2011;55:999–1009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rajendran P, Delage B, Dashwood WM, Yu TW, Wuth B, Williams DE, Ho E, Dashwood RH. Histone deacetylase turnover and recovery in sulforaphane-treated colon cancer cells: competing actions of 14–3-3 and Pin1 in HDAC3/SMRT corepressor complex dissociation/reassembly. Mol Cancer. 2011;10:68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Myzak MC, Ho E, Dashwood RH. Dietary agents as histone deacetylase inhibitors. Mol Carcinog. 2006;45:443–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Hu J, Straub J, Xiao D, Singh SV, Yang HS, Sonenberg N, Vatsyayan J. Phenethyl isothiocyanate, a cancer chemopreventive constituent of cruciferous vegetables, inhibits cap-dependent translation by regulating the level and phosphorylation of 4E–BP1. Cancer Res. 2007;67:3569–73 [DOI] [PubMed] [Google Scholar]
- 130.Wang LG, Liu XM, Chiao JW. Repression of androgen receptor in prostate cancer cells by phenethyl isothiocyanate. Carcinogenesis. 2006;27:2124–32 [DOI] [PubMed] [Google Scholar]
- 131.Xiao D, Lew KL, Zeng Y, Xiao H, Marynowski SW, Dhir R, Singh SV. Phenethyl isothiocyanate-induced apoptosis in PC-3 human prostate cancer cells is mediated by reactive oxygen species-dependent disruption of the mitochondrial membrane potential. Carcinogenesis. 2006;27:2223–34 [DOI] [PubMed] [Google Scholar]
- 132.Xiao D, Zeng Y, Choi S, Lew KL, Nelson JB, Singh SV. Caspase-dependent apoptosis induction by phenethyl isothiocyanate, a cruciferous vegetable-derived cancer chemopreventive agent, is mediated by Bak and Bax. Clin Cancer Res. 2005;11:2670–9 [DOI] [PubMed] [Google Scholar]
- 133.Beklemisheva AA, Fang Y, Feng J, Ma X, Dai W, Chiao JW. Epigenetic mechanism of growth inhibition induced by phenylhexyl isothiocyanate in prostate cancer cells. Anticancer Res. 2006;26:1225–30 [PubMed] [Google Scholar]
- 134.Liu KC, Huang YT, Wu PP, Ji BC, Yang JS, Yang JL, Chiu TH, Chueh FS, Chung JG. The roles of AIF and Endo G in the apoptotic effects of benzyl isothiocyanate on DU 145 human prostate cancer cells via the mitochondrial signaling pathway. Int J Oncol. 2011;38:787–96 [DOI] [PubMed] [Google Scholar]
- 135.Batra S, Sahu RP, Kandala PK, Srivastava SK. Benzyl isothiocyanate-mediated inhibition of histone deacetylase leads to NF-kappaB turnoff in human pancreatic carcinoma cells. Mol Cancer Ther. 2010;9:1596–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Aggarwal BB, Ichikawa H. Molecular targets and anticancer potential of indole-3-carbinol and its derivatives. Cell Cycle. 2005;4:1201–15 [DOI] [PubMed] [Google Scholar]
- 137.Sarkar FH, Li Y. Indole-3-carbinol and prostate cancer. J Nutr. 2004;134:S3493–8 [DOI] [PubMed] [Google Scholar]
- 138.Bhatnagar N, Li X, Chen Y, Zhou X, Garrett SH, Guo B. 3,3′-diindolylmethane enhances the efficacy of butyrate in colon cancer prevention through down-regulation of survivin. Cancer Prev Res (Phila). 2009;2:581–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li Y, Li X, Guo B. Chemopreventive agent 3,3′-diindolylmethane selectively induces proteasomal degradation of class I histone deacetylases. Cancer Res. 2010;70:646–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Dennert G, Zwahlen M, Brinkman M, Vinceti M, Zeegers MP, Horneber M. Selenium for preventing cancer. Cochrane Database Syst Rev. 2011;5:CD005195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Xiang N, Zhao R, Song G, Zhong W. Selenite reactivates silenced genes by modifying DNA methylation and histones in prostate cancer cells. Carcinogenesis. 2008;29:2175–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Zeng H, Wu M, Botnen JH. Methylselenol, a selenium metabolite, induces cell cycle arrest in G1 phase and apoptosis via the extracellular-regulated kinase 1/2 pathway and other cancer signaling genes. J Nutr. 2009;139:1613–8 [DOI] [PubMed] [Google Scholar]
- 143.Lee JI, Nian H, Cooper AJ, Sinha R, Dai J, Bisson WH, Dashwood RH, Pinto JT. Alpha-keto acid metabolites of naturally occurring organoselenium compounds as inhibitors of histone deacetylase in human prostate cancer cells. Cancer Prev Res (Phila). 2009;2:683–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Nian H, Bisson WH, Dashwood WM, Pinto JT, Dashwood RH. Alpha-keto acid metabolites of organoselenium compounds inhibit histone deacetylase activity in human colon cancer cells. Carcinogenesis. 2009;30:1416–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dunn BK, Richmond ES, Minasian LM, Ryan AM, Ford LG. A nutrient approach to prostate cancer prevention: The Selenium and Vitamin E Cancer Prevention Trial (SELECT). Nutr Cancer. 2010;62:896–918 [DOI] [PubMed] [Google Scholar]
- 146.Lippman SM, Klein EA, Goodman PJ, Lucia MS, Thompson IM, Ford LG, Parnes HL, Minasian LM, Gaziano JM, Hartline JA, et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the Selenium and Vitamin E Cancer Prevention Trial (SELECT). JAMA. 2009;301:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Pinto JT, Lee JI, Sinha R, MacEwan ME, Cooper AJ. Chemopreventive mechanisms of alpha-keto acid metabolites of naturally occurring organoselenium compounds. Amino Acids. 2011;41:29–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Aggarwal BB. Prostate cancer and curcumin: add spice to your life. Cancer Biol Ther. 2008;7:1436–40 [DOI] [PubMed] [Google Scholar]
- 149.Goel A, Aggarwal BB. Curcumin, the golden spice from Indian saffron, is a chemosensitizer and radiosensitizer for tumors and chemoprotector and radioprotector for normal organs. Nutr Cancer. 2010;62:919–30 [DOI] [PubMed] [Google Scholar]
- 150.Kang J, Chen J, Shi Y, Jia J, Zhang Y. Curcumin-induced histone hypoacetylation: the role of reactive oxygen species. Biochem Pharmacol. 2005;69:1205–13 [DOI] [PubMed] [Google Scholar]
- 151.Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, Neckers L. Curcumin is an inhibitor of p300 histone acetylatransferase. Med Chem. 2006;2:169–74 [DOI] [PubMed] [Google Scholar]
- 152.Liu HL, Chen Y, Cui GH, Zhou JF. Curcumin, a potent anti-tumor reagent, is a novel histone deacetylase inhibitor regulating B-NHL cell line Raji proliferation. Acta Pharmacol Sin. 2005;26:603–9 [DOI] [PubMed] [Google Scholar]
- 153.Meja KK, Rajendrasozhan S, Adenuga D, Biswas SK, Sundar IK, Spooner G, Marwick JA, Chakravarty P, Fletcher D, Whittaker P, et al. Curcumin restores corticosteroid function in monocytes exposed to oxidants by maintaining HDAC2. Am J Respir Cell Mol Biol. 2008;39:312–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Link A, Balaguer F, Goel A. Cancer chemoprevention by dietary polyphenols: promising role for epigenetics. Biochem Pharmacol. 2010;80:1771–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Dobosy JR, Selker EU. Emerging connections between DNA methylation and histone acetylation. Cell Mol Life Sci. 2001;58:721–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ng HH, Zhang Y, Hendrich B, Johnson CA, Turner BM, Erdjument-Bromage H, Tempst P, Reinberg D, Bird A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61 [DOI] [PubMed] [Google Scholar]
- 157.Fuks F, Burgers WA, Brehm A, Hughes-Davies L, Kouzarides T. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91 [DOI] [PubMed] [Google Scholar]
- 158.Fabrizio P, Gattazzo C, Battistella L, Wei M, Cheng C, McGrew K, Longo VD. Sir2 blocks extreme life-span extension. Cell. 2005;123:655–67 [DOI] [PubMed] [Google Scholar]
- 159.Lim CS. Human SIRT1: a potential biomarker for tumorigenesis? Cell Biol Int. 2007;31:636–7 [DOI] [PubMed] [Google Scholar]
- 160.Jung-Hynes B, Nihal M, Zhong W, Ahmad N. Role of sirtuin histone deacetylase SIRT1 in prostate cancer. A target for prostate cancer management via its inhibition? J Biol Chem. 2009;284:3823–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang B, Hasan MK, Alvarado E, Yuan H, Wu H, Chen WY. NAMPT overexpression in prostate cancer and its contribution to tumor cell survival and stress response. Oncogene. 2011;30:907–21 [DOI] [PubMed] [Google Scholar]
- 162.Powell MJ, Casimiro MC, Cordon-Cardo C, He X, Yeow WS, Wang C, McCue PA, McBurney MW, Pestell RG. Disruption of a Sirt1-dependent autophagy checkpoint in the prostate results in prostatic intraepithelial neoplasia lesion formation. Cancer Res. 2011;71:964–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Seeni A, Takahashi S, Takeshita K, Tang M, Sugiura S, Sato SY, Shirai T. Suppression of prostate cancer growth by resveratrol in the transgenic rat for adenocarcinoma of prostate (TRAP) model. Asian Pac J Cancer Prev. 2008;9:7–14 [PubMed] [Google Scholar]
- 164.Wang TT, Hudson TS, Wang TC, Remsberg CM, Davies NM, Takahashi Y, Kim YS, Seifried H, Vinyard BT, Perkins SN, et al. Differential effects of resveratrol on androgen-responsive LNCaP human prostate cancer cells in vitro and in vivo. Carcinogenesis. 2008;29:2001–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Kikuno N, Shiina H, Urakami S, Kawamoto K, Hirata H, Tanaka Y, Majid S, Igawa M, Dahiya R. Genistein mediated histone acetylation and demethylation activates tumor suppressor genes in prostate cancer cells. Int J Cancer. 2008;123:552–60 [DOI] [PubMed] [Google Scholar]
- 166.Gore SD, Baylin S, Sugar E, Carraway H, Miller CB, Carducci M, Grever M, Galm O, Dauses T, Karp JE, et al. Combined DNA methyltransferase and histone deacetylase inhibition in the treatment of myeloid neoplasms. Cancer Res. 2006;66:6361–9 [DOI] [PubMed] [Google Scholar]
- 167.Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet. 1999;21:103–7 [DOI] [PubMed] [Google Scholar]
- 168.Ghoshal K, Bai S. DNA methyltransferases as targets for cancer therapy. Drugs Today (Barc). 2007;43:395–422 [DOI] [PubMed] [Google Scholar]
- 169.Jüttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hulf T, Sibbritt T, Wiklund ED, Bert S, Strbenac D, Statham AL, Robinson MD, Clark SJ. Discovery pipeline for epigenetically deregulated miRNAs in cancer: integration of primary miRNA transcription. BMC Genomics. 2011;12:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Yu S, Khor TO, Cheung KL, Li W, Wu TY, Huang Y, Foster BA, Kan YW, Kong AN. Nrf2 expression is regulated by epigenetic mechanisms in prostate cancer of TRAMP mice. PLoS ONE. 2010;5:e8579. [DOI] [PMC free article] [PubMed] [Google Scholar]