Abstract
The emergence of genome-wide analysis to interrogate cellular DNA, RNA, and protein content has revolutionized the study of the control network that mediates cellular homeostasis. Nutrigenomics addresses the effect of nutrients on gene expression, which provides a basis for understanding the biological activity of dietary components. Translation of mRNAs represents the last step of genetic flow and primarily defines the proteome. Translational regulation is thus critical for gene expression, in particular, under nutrient excess or deficiency. Until recently, it was unclear how the global effects of translational control are influenced by nutrient signaling. An emerging concept of translational reprogramming addresses how to maintain the expression of specific proteins during pathophysiological conditions by translation of selective mRNAs. Here we describe recent advances in our understanding of translational control, nutrient signaling, and their dysregulation in aging and cancer. The mechanistic understanding of translational regulation in response to different nutrient conditions may help identify potential dietary and therapeutic targets to improve human health.
Introduction
The past decade witnessed stunning progress in the molecular biology techniques. The elucidation of the human genome and the explosion of next-generation sequencing technologies are fueling a revolution in a variety of sciences including nutrition (1, 2). We have come to appreciate the dynamic state of genomics, including DNA modifications, RNA quantitative and qualitative changes, and proteome landscapes in a diverse array of species. Nutrigenomics is research focusing on identifying and understanding the molecular-level interactions between nutrients and other dietary bioactive molecules with the genome and the functional consequences in gene expression (3). Nutrigenomics encompasses the fields of genomics, epigenomics, posttranslational modifications, proteomics, and metabolomics. The excitement about nutrigenomics comes from a growing awareness of the potential for modifications of food or diet to support health and reduce the risk of diet-related diseases (4). By understanding how nutrients interact with the genome and influence gene expression, better dietary regimens may be formulated and novel therapeutic approaches may be designed for human diseases such as diabetes, cancer, and neurodegenerative disorders.
A fundamental question in nutrigenomics is how cells respond to the availability of nutrients and adapt to nutrient deficiencies by changing the flow of genomic information. After transcription, genomic information in the nucleotide sequences begins a long journey toward translation into the amino acids of a protein. Proteins constitute vital components of life, and protein synthesis represents one of the most fundamental biochemical processes. Many recent studies using comparative genomic and proteomic profiling of cells have documented a lack of correlation between the mRNA and protein levels of numerous genes (5). This indicates that posttranscriptional regulation events, including mRNA degradation, translational control, and protein turnover, are more important than is often assumed. It is becoming increasingly evident that the regulation of translation provides the cell with the plasticity to respond to rapid changes in the environment (6). Given the considerable time lag associated with the synthesis, processing, and exporting of de novo synthesized mRNA, the use of existing mRNAs by a controlled translation mechanism allows for an immediate and rapid response to changes in physiological conditions (7). Similar to transcriptional regulation, translational control also exhibits specificity because certain mRNAs can override the general repression of protein synthesis. Defining the precise mechanisms by which subsets of mRNAs are differentially regulated under a variety of conditions is fundamental to our understanding of posttranscriptional control of gene expression. Our laboratory has long-standing interest in protein synthesis and its regulation by nutrient signaling. In this review, we focus on the functional interpretation of nutrigenomics from the perspective of translational control and discuss the implications in human disease.
Current status of knowledge
Translational control
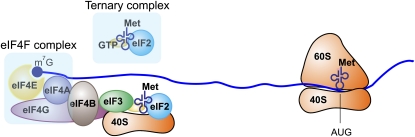
To better illustrate the translational control mechanism, it is necessary to briefly revisit what we have learned regarding mRNA translation in eukaryotes. mRNA translation can be divided into three stages: initiation, elongation, and termination. The initiation stage, which ends in the assembly of the elongation-competent 80S ribosomes at the initiation codon, is considered to be the rate-limiting step. The initiation is a complex multistep process involving a large number of protein factors required for protein synthesis (Fig. 1). First, the methionine-loaded initiator tRNA binds to the GTP-coupled eukaryotic initiation factor (eIF)5 2, yielding the ternary complex, which combines with the 40S small ribosomal subunit and other factors to form a 43S preinitiation complex (PIC) (8). PIC then binds to the mRNA through the eIF4F complex assembled at the 5′ terminus of the mRNA. eIF4E, one element of the eIF4F complex, recognizes the cap structure and then recruits additional components, including eIF4G and eIF4A. eIF4G acts as a scaffold protein mediating the interaction between the eIF4F complex and the PIC, whereas eIF4A functions as a helicase to unwind secondary structures in the 5′ untranslated region (UTR) and facilitate 43S PIC scanning (9–11). Next, the loaded 43S PIC scans along the 5′UTR in a 5′ → 3′ direction until it encounters an initiation codon, at which the 60S subunit joins and protein synthesis begins (12–14). Despite the enormous progress in the past decade in dissecting molecular mechanisms of eukaryotic translation initiation, several key questions remain to be fully addressed. For example, what are the determinants of start codon selection? Which factors regulate 43S PIC binding? What additional regulators are required for subunit joining?
Figure 1.
Translation initiation in eukaryotic cells. The ternary complex (eIF2-GTP-Met-tRNAi) and eIF4 complex are highlighted because they are the major targets for translational regulation. The recognition of m7G cap structure at the 5′ end of the mRNA is mediated by the cap-binding protein eIF4E, which is part of eIF4F. In addition to eIF4E, eIF4F complex consists of eIF4A (an RNA helicase) and eIF4G (a scaffold protein). The 40S subunit, which is associated with the ternary complex, is recruited to the eIF4F complex by eIF3, forming the preinitiation complex (PIC). PIC scans the mRNA in the 5′ → 3′ direction until it locates an initiation codon (most often AUG) in an optimal sequence context, where it is joined by the 60S subunit to form an 80S initiation complex. eIF, eukaryotic initiation factor.
Translation consumes a lion’s share of energy, and it is not surprising that multiple translational regulation mechanisms exist in cells to affect protein synthesis in a global or mRNA-specific manner. Two key targets have been well characterized to manipulate global translation, including the cap recognition by the eIF4F complex assembly and the control of tertiary complex formation by eIF2. A family of proteins called eIF4E-binding proteins (4E-BPs) share a domain similar to that within eIF4G that mediates the interaction with eIF4E. Thus, 4E-BPs bind to eIF4E, competing with the eIF4G association and then blocking the cap-dependent initiation. Under several stimuli such as nutrients and growth factors, signaling pathways trigger the phosphorylation of 4E-BPs and then the release from the bound eIF4E, allowing the assembly of eIF4F and the activation of translation (15). Interestingly, genome-wide translation profiling analysis uncovered a different mRNA specificity of 4E-BPs and the collaboration with the 3′UTR binding proteins to fine-tune translation (16). Moreover, 4E-BPs have been shown to act in a hierarchical network of protein translational control during stem cell proliferation and differentiation (17–19).
Another well-established mechanism controlling the global translation is the modulation of the ternary complex. GTP, coupled with eIF2 in the ternary complex, undergoes hydrolysis during initiation codon recognition. The resulting GDP must be substituted by another GTP for the recycling of the ternary complex, which is activated by eIF2B, a guanine nucleotide exchange factor. eIF2 consists of three subunits: α, β, and γ. One of the key mechanisms of translational control during stress is the phosphorylation of eIF2α subunit on Ser51, converting eIF2-GDP into a competitive inhibitor of eIF2B and decreasing the ternary complex assembly (20). There are four different eIF2α kinases in mammals activated by different stresses: protein kinase R (PKR, double-stranded RNA in virus infection), PKR-like ER kinase (PERK, unfolded proteins in endoplasmic reticulum), heme regulated inhibitor (HRI, heme deprivation), and general control nonderepressible protein 2 (GCN2, amino acid starvation). Although this integrated stress response involves a downregulation of general protein synthesis, the translation of a subset of mRNAs can be increased (21, 22). Most of these mRNAs encode proteins critical for stress response and cell survival.
There are two major forms of translational control. In one, regulation is global and modulates the rates of protein synthesis, thereby contributing to the overall regulation of cell growth and metabolism. In the second form, a specific mRNA or subsets of mRNAs is regulated. The mRNA-specific regulation of protein synthesis is primarily due to the existence of mRNA sequence features (cis elements) and the interaction with corresponding factors (trans-acting factors). Most cis elements reside in the 5′UTR and 3′UTR and modulate the activities of translational factors through RNA binding proteins. Cells adopt several distinct mechanisms to manipulate mRNA-specific translation. Examples include steric hindrance of the 43S complex recruitment (e.g., ferritin mRNAs and iron regulatory proteins), interfering with the eIF4F complex (e.g., maternal mRNAs and cytoplasmic polyadenylation element binding protein), and cap-independent inhibition of early initiation (e.g., msl-2 mRNA and Sex-lethal protein) (23). One particularly interesting translational regulation involves the SECIS element in the 3′UTR of mRNA, which allows the incorporation of selenocysteine encoded by a UGA stop codon (24). This unique mechanism gives rise to different proteins from the same transcript. Another well-studied mRNA-specific regulation is the microRNA (miRNA)-mediated repression of target mRNAs. miRNAs are short oligonucleotides (∼22) that are major regulators of gene expression at the posttranscription level. The functional specificity is determined by the imperfect complementarity between miRNAs and their target mRNA sites, which are usually located within the 3′UTR. A large number of studies, both in vivo and in vitro, demonstrate that miRNAs either inhibit translation, destabilize the mRNA, or both, depending on many factors. A recent genome-wide comparison of mRNA level and translation efficiency in response to miRNA suppression showed that the mRNA instability rather than the translation repression is the predominant effect of miRNA on gene expression (25, 26). Nevertheless, it is obvious that miRNAs could suppress target mRNA translation at multiple stages, including initiation repression, post-initiation inhibition, and the increase in deadenylation (27).
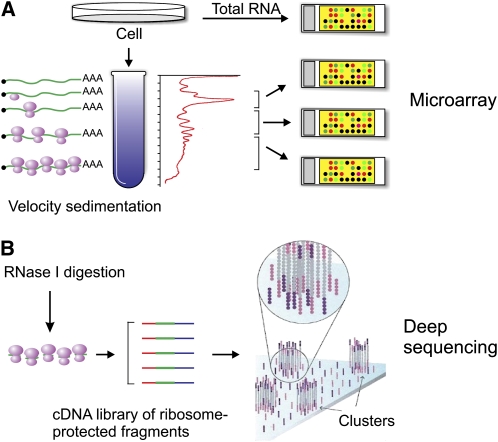
As we gain better insights into the mechanisms of translation, it is clear that the use of emerging technologies will lead to a more complete understanding of this paramount cellular process. A reliable measure of the translation of cellular mRNA is the degree of its association with ribosomes. Actively translated mRNAs are typically bound by several ribosomes (polysomes) and can be separated from individual 40S and 60S ribosomal subunits and 80S monosomes by centrifugation through a linear sucrose gradient. In combination with microarray analysis, a global view of the translation status of the entire transcriptome can be observed (Fig. 2A). This polysome microarray approach has been widely used to monitor the differences in ribosomal association of mRNAs with changes in growth conditions or genetic background (28–32). A shortcoming of this approach is the lack of information about the ribosome positions or distribution along mRNAs. To circumvent this limitation, a recent method called genome-wide ribosome footprinting has been developed that involves isolation and sequencing of RNA fragments that are protected by ribosomes (33) (Fig. 2B). The genome-wide ribosome footprinting provides a wealth of information about both the position and density of ribosomes on mRNAs, allowing the monitoring of translation efficiency as well as the dissection of the regulatory aspects of mRNA translation under different conditions.
Figure 2.
Genome-wide analysis of mRNA translation. (A) Combination of density gradient centrifugation and microarray analysis. Ribosome fractions are separated by ultracentrifugation through linear sucrose gradients. RNA is then isolated from light and heavy ribosome fractions for subsequent microarray analysis. This establishes the polysome profile for the mRNA, where actively translated mRNA molecules are distributed in the heavy ribosome fractions and translationally inactive messages are found in the lighter fractions. (B) Combination of density gradient centrifugation and deep sequencing of ribosome-protected mRNA fragments. Typically, cellular lysates are treated with RNase I to digest mRNA not protected by ribosomes. The ribosome protected fragments (∼30 nucleotides) are isolated and converted to cDNA library suitable for deep sequencing. The next-generation sequencing technology allows single-molecule sequencing in parallel, giving rise to a large volume of data sets. This establishes genome-wide footprinting to quantify ribosome positions on the entire transcriptome.
Nutrient signaling pathway
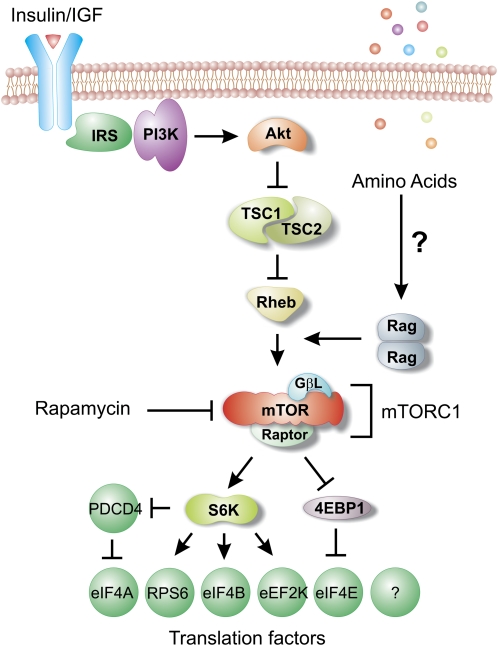
Coupling of the availability of nutrients and growth factors to cellular growth is essential for all organisms. Cells have evolved to establish a sophisticated sensor and signal transduction system to detect nutrient availability and adjust the cellular processes to meet their bioenergetics needs. A key signaling pathway that regulates growth and metabolism is the mammalian target of rapamycin complex 1 (mTORC1) (34). mTORC1 is an evolutionarily conserved serine/threonine kinase that senses signals from extracellular stimuli, amino acid availability, and the energy status of the cell. In a typical signaling cascade, the binding of insulin or insulin-like growth factors to their receptors triggers the phosphoinositide 3-kinase (PI3K) pathways, leading to the phosphorylation and activation of Akt (Fig. 3). Activated Akt then phosphorylates and suppresses the tuberous sclerosis complex (TSC) 2, which is in complex with TSC1 and acts as a negative regulator of mTORC1 signaling (35, 36). TSC1/TSC2 functions as the GTPase activating protein of Ras homolog enriched in brain (Rheb) (37, 38). Rheb coupled with GTP activates the kinase activity of mTORC1, whereas the GDP form of Rheb resulting from TSC1/TSC2 loses the stimulating function (39). Therefore, mTORC1 integrates environmental cues via the PI3K-Akt pathway to achieve the metabolic balance between growth factors and cellular growth.
Figure 3.
The mammalian target of rapamycin complex 1 (mTORC1) signaling pathway. Upstream signaling inputs such as phosphoinositide 3-kinase (PI3K)-Akt positively regulates mTORC1 by inhibiting the negative regulator tuberous sclerosis complex (TSC)1/TSC2. mTORC1 also senses amino acid sufficiency via Rag proteins, which serve to control the subcellular localization of mTORC1 independent of TSC. Two downstream targets of mTORC1, S6 kinase (S6K) and eIF4E-binding protein 1 (4EBP1), regulates a number of translation factors, thereby controlling mRNA translation at both initiation and elongation stages. eIF, eukaryotic initiation factor; GβL, G-protein β-subunit-like (also known as mLST8, mammalian lethal with SEC13 protein 8); IGF, insulin-like growth factor; IRS, insulin receptor substrate; mTOR, mammalian target of rapamycin; PDCD4, programmed cell death protein 4; Rheb, Ras homologue enriched in brain.
Amino acids are the building blocks of polypeptides, so it is critical for cells to adjust the translation level according to their availability. mTORC1 senses amino acid sufficiency and controls the protein synthesis rate accordingly. The transporters on the cytoplasm and the recycled amino acids through autophagy primarily determine the intracellular level of essential amino acids (40). The sensor system of amino acids is not fully characterized, but some of the downstream mediators have been identified, including the mitogen-activated protein kinase kinase kinase kinase 3 (41) and the PI3K catalytic subunit type 3 (42). The most important regulator linking amino acids to mTORC1 is the Rag family of small GTPases (43, 44). These GTPases are heterodimers of either RagA or RagB with RagC or RagD. Without the activation by amino acids, RagA/B binds with GDP, whereas RagC/D binds with GTP. These inactivated Rag GTPases are attached to the membrane of late lysosome through three small regulators: p14, mitogen-activated protein kinase scaffold protein 1, and p18 (45). In the presence of amino acids, Rag GTPases switch to the active form, which consists of the GTP-coupled RagA/B and GDP-bound RagC/D. Activated Rag GTPases then recruit mTORC1 to the lysosome membrane, which subsequently interacts with its activator of the small GTPase Rheb (39).
In response to a wide range of upstream inputs, mTORC1 regulates cell growth by maintaining the appropriate balance between anabolic processes, such as macromolecular synthesis and nutrient storage, and catabolic processes, such as autophagy and the use of energy stores. Among multiple downstream targets of mTORC1, the two best characterized are 4E-BPs and ribosomal protein S6 kinase (RPS6K) (46). The 4E-BPs, when nonphosphorylated, sequester the eIF4E mRNA cap-binding proteins, prevent the assembly of the eIF4F complex at the 5′ cap structure, and thus block cap-dependent mRNA translation (47). The activation of RPS6K by mTORC1-mediated phosphorylation promotes mRNA translation through several substrates, including eukaryotic translation initiation factor 4B(eIF4B), programmed cell death protein 4(PDCD4), eukaryotic elongation factor-2 kinase(eEF2K). Phosphorylated eIF4B further promotes the helicase activity of eIF4A, which unwinds the secondary structure in the 5′UTR and facilitates the scanning process of 43S PIC (48). PDCD4 acts as a negative regulator of eIF4A. RPS6K-mediated PDCD4 phosphorylation triggers its degradation, thereby relieving its repression of eIF4A activity. S6 kinase (S6K) 1 also affects the elongation stage of mRNA translation by phosphorylating eEF2K. Nonphosphorylated eEF2K negatively regulates eukaryotic elongation factor-2 (eEF2), and therefore S6K1 activates eEF2 by releasing the inhibition. Collectively, mTORC1 influences the global protein synthesis rate through both initiation and elongation steps.
mTORC1 not only regulates the activity of the translational machinery as a whole, but also specifically controls the translation of a subset of mRNAs that are thought to promote cell growth and proliferation. For example, we observed differential regulation of cap-dependent and cap-independent mRNA translation by mTORC1. As a consequence, hyperactive mTORC1 activity suppresses the translation of Hsp70, an important stress responsive protein (49). In addition, activated mTORC1 preferentially stimulates the translation of a subset mRNAs with a 5′ tract of oligopyrimidine (TOP) sequences. The 5′TOP mRNAs mostly encode ribosomal proteins and translation factors, which increase the available translational machinery and thus global protein synthesis. This preferential regulation of TOP mRNAs is supported by the translation profiling analysis in TSC1/TSC2-depleted cells and serum stimulative responses (50). Despite years of study, the underlying mechanism through which mTORC1 controls TOP mRNA translation remains unclear.
Amino acid response pathway
Cells respond to the stress of amino acid deprivation through multiple systems. In addition to the mTORC1 pathway, another well-characterized pathway capable of sensing amino acid deficiency is the amino acid response (AAR) pathway (51). When an essential amino acid is inadequate due to either limited extracellular supply or the block of intracellular recycling, unchanged tRNA species accumulate and trigger GCN2 kinase. Subsequently, activated GCN2 kinase phosphorylates eIF2α at Ser51. Phosphorylated eIF2α reduces the formation of the ternary complex and then attenuates global protein synthesis (52). Interestingly, transcriptome study in yeast discovered the integration of the AAR and TOR pathways in nutrient-sensing and starvation responses (53).
Despite the decrease in global protein synthesis under stresses such as starvation, a subset of mRNAs acquire increased translation. One of the best studied examples is transcription factor GCN4 in yeast and its mammalian counterpart, activating transcription factor 4 (ATF4) (54, 55). The ATF4 transcript contains one very short upstream open reading frame (uORF) 1 and another longer uORF 2 overlapping with the main coding sequence (CDS). This special configuration of uORF length and position modulates the expression of ATF4 through a reinitiation mechanism. After the translation of uORF1, ribosomes dissociate into subunits at the stop codon, leaving the 40S subunits to resume scanning. These 40S subunits acquire ternary complex during the scanning and reinitiate at downstream open reading frames. The scanning time and distance before reinitiation largely depend on the abundance of ternary complexes. Under normal conditions with high ternary complex levels, most ribosomes that resume scanning reinitiate at uORF2 in a relatively short amount of time, resulting in low basal levels of ATF4. However, the decreased ternary complex level under starvation prolongs the reinitiation and bypasses uORF2, leading to increased expression of ATF4. The increased level of ATF4 then stimulates the transcription of target genes involved in stress resistance. Notably, a recent study using deep sequencing–based ribosome profiling analysis reported a prevailing non-AUG translation in the 5′UTR under starvation (33). The physiological significance of this nonclassic mode of translation events remains to be identified.
Aging
Aging is a complex biological process characterized by progressive functional and structural deterioration of multiple organ systems that eventually leads to death. Aging is also an important factor for most of the common diseases, including type 2 diabetes, cardiovascular disease, cancer, and neurodegeneration. During the past decade, it became clear that the rate of aging, like many other processes in biology, is subject to regulation. Multiple signaling pathways have been linked to the aging process, including highly conserved insulin and insulin-like growth factor 1, TOR, and sirtuin signaling pathways. These signaling pathways are coordinated with nutrient levels, energy status, DNA damage, mitochondria function, as well as protein homeostasis in controlling growth and aging. Here we focus on the role of the nutrient signaling pathway and protein synthesis in aging. Readers are directed to several recent excellent reviews of other aspects of aging regulation (56–59).
Studies conducted during the past 70 y in rodents have shown that their life span is extended by calorie restriction (60). Similar effects have been observed in a wide range of organisms, including protozoans, yeast, nematodes, and canines (61). Preliminary results from studies in monkeys indicate that a similar phenomenon occurs in primates (62). Calorie-restricted organisms not only live longer but also have increased resistance to disease and seem to physically age slower. For example, calorie restriction postpones many signs of aging, including skin changes, obesity, decline in motor function, and loss of learning and memory. In addition, calorie-restricted rodents are less prone to a large number of diseases of aging, including nephropathy, immune dysfunction, heart disease, and neurodegeneration (62). The exact mechanism by which calorie restriction has such a dramatic effect on primary aging is not yet known. Many interrelated and overlapping factors have been proposed to play a role.
Many conditions that shift cells from states of nutrient use and growth to states of cell maintenance extend the life span. The TOR pathway has warranted increased attention from the aging-research community due to its apparently conserved influence on the life span of a number of organisms. Decreased TOR signaling (using RNAi or a hypomorphic TOR mutant) has been shown to extend the life span of the nematode Caenorhabditis elegans (63). Likewise, overexpression of a dominant-negative allele of TOR or inhibitors of TOR (Tsc1 and Tsc2) extends the life span of Drosophila (64). Deletion of the Saccharomyces cerevisiae TOR1 gene was shown to increase the replicative life span (65). A recent high-throughput screen for gene deletions that extends the chronological life span yielded a number of genes involved in nutrient sensing and influenced in part by the TOR pathway (66). Remarkably, administration of a specific TOR inhibitor rapamycin in adult mice was sufficient to cause a increase in their life span (67). Further supporting the mechanistic connection between TOR signaling and longevity, modulating TOR downstream targets S6K and 4E-BP1 also affects the life span in a wide range of species. Data from all three aging models support the notion that TOR deficiency extends the life span.
The most prominent consequence of mTORC1 inactivation is the attenuation of global protein synthesis. Protein synthesis is a key regulated cellular process that links nutrients to organism growth. Classic studies in diverse organisms, including humans, have demonstrated that aging is accompanied by marked alterations in both general and specific protein synthesis (68). These early observations established a link between the aging process and the regulation of protein synthesis. Three recent papers reported that decreasing protein synthesis in C. elegans can increase life span (69). Pan et al. (70) reported that inhibition of various genes in the translation initiation complex including eIF4G and S6K results in life span extension. In a study by Hansen et al. (71), it was found that decreasing the levels of ribosomal proteins, S6K, or translation initiation factors increases the life span. Syntichaki et al. (72) reported that loss of a specific eIF4E isoform that functions in somatic tissues decreases global protein synthesis, protects against oxidative stress, and extends the life span in C. elegans. Interestingly, overexpression of eIF4E was recently shown to increase cellular senescence in mice as measured by β-galactosidase staining, implicating its potential involvement in mammalian aging (73). The regulation of cap-dependent translation may therefore be a conserved response to nutrient limitation in different species (74).
It is suggested that the general reduction of protein synthesis due to the decreased frequency of mRNA translation also lowers the cellular load of erroneously synthesized polypeptides that the constitutive protein homeostatic apparatus (proteases and chaperones proteins) normally eliminates. This situation results in “spare" proteolytic and chaperone function that can then deal with those proteins modified posttranslationally (e.g., by oxidation and/or glycation), which are thought to contribute to the senescent phenotype (68). This increased availability of proteolytic and chaperone functions may thereby contribute to the observed increase in organism stress resistance and life span. Interestingly, translational profiling analysis revealed the preferential expression of stress resistance transcripts under the repression of nutrient signaling. As a consequence, these stress resistance proteins promote the cell survival and increase the life span (75). A recent report on Drosophila indicated that the upregulation of 4E-BP in dietary restriction activates the translation of mitochondrial genes, leading to improved mitochondria activity and increased life span (76). In addition, studies on worms showed that eIF4G is downregulated during starvation, which results in the suppression of genes for growth but enhanced stress response genes at the translation level. These stress proteins are required for increased life span when eIF4G is inhibited (77). Altogether, those recent genome-wide studies of translation profiling under dietary restriction and stress conditions highlight the significance of translational control in aging.
Cancer
Most cancers are caused by dysregulation of signaling pathways that control cell growth and proliferation. As the central sensor of nutrients and controller of growth, mTORC1 signaling promotes tumor development by constitutively stimulating the synthesis of macromolecules. Consistent with this notion, mutations in the negative regulators in mTORC1 signaling such as the TSC1/TSC2 complex, liver kinase B1 (LKB1), and phosphatase and tensin homolog (PTEN) lead to tumorigenesis (78). Conversely, treatment of mTORC1 inhibitors represses tumorigenesis (79). In many cancers, the capacity of translational machinery is increased to meet excessive cell growth and proliferation. Indeed, mutations of translation factors have been shown to be involved in tumor development and transformation (80–82). 4E-BPs are the most well-known negative regulators of translation in response to nutrient signaling, which act as key controllers of cell proliferation but not cell size (83). Inactivation of 4E-BPs results in the hyperactivity of eIF4E and malignant transformation (84). Translational profiling of hyperactive eIF4E revealed that it promotes the preferential expression of a subset of mRNAs encoding protumorigenic proteins participating in the cell cycle, antiapoptosis, growth and proliferation (85–88). In contrast, inhibition of eIF4E by either constitutively active 4E-BP1 or small interfering RNA led to the repression of tumor growth derived from PI3K signaling mutations (89, 90). In addition to eIF4E, other initiation factors such as eIF6 and eukaryotic elongation factor -1 A1 have also been shown to promote tumorigenesis and transformation (91, 92).
In addition to mTORC1 and downstream targets, the dysregulation of the eIF2 pathway is frequently observed in cancer cells (93). Overexpression of nonphosphorylatable eIF2α or dominant-negative eIF2α kinase lead to the constitutive activation of eIF2α, upregulation of translation initiation, and the malignant transformation of cells in culture (94, 95). In brief, nutrient signaling–mediated translational control of both global protein synthesis and specific mRNAs is critical for cell proliferation and tumorigenesis (96, 97).
CONCLUSIONS
Translational control in eukaryotic cells is critical for gene expression during nutrient deprivation and stress, development and differentiation, nervous system functions, aging, and disease. In eukaryotes, two major pathways, TOR and AAR, sense both the sufficiency and deficiency of amino acids. The process of translation requires substantial cellular resources. Thus, it is critical for cells to modulate global protein synthesis to save energy expenditure. The biological implications of selective translation are clearly important. This translation reprogramming constitutes cellular stress response to maintain metabolic homeostasis. Consequently, translational dysregulation causes a series of human diseases, including aging, cancer, and neurodegenerative disorders.
By definition, nutrigenomics applies high-throughput genomic tools in nutrition research. However, the development of high-throughput methods to capture the nutrition-relevant translatome changes lags far behind the trancriptome and proteome tools. Recent advances in microarray and next-generation sequencing techniques allow us to monitor the translational efficiency in a genome-wide manner. The progress largely fills the gap between transcriptome and proteome in the regulation of gene expression, enabling the characterization of genome-wide translational signatures under both health and disease conditions. These large-scale tools also assist the mechanistic understanding of the pathology of diet-relevant diseases at the translation level. Ultimately, applications of newly developed high-throughput tools in nutrigenomics will allow the development of early diagnostic methods and effective therapeutic strategies for diet-related diseases.
Acknowledgments
We apologize for the many studies that were not discussed because of limited space. We thank Crystal Conn and Alex Coots at Qian lab for the critical reading of this review paper. Both authors read and approved the final manuscript.
Footnotes
Supported by Ellison Medical Foundation (AG-NS-0605-09), National Institutes of Health (1 DP2 OD006449-01), and Department of Defense (TS100078) (to S.-B.Q).
Author disclosures: B. Liu and S.-B. Qian, no conflict of interest.
Abbreviations used: AAR, amino acid response; ATF4, activating transcription factor 4; eEF2K, eukaryotic elongation factor-2 kinase; eIF, eukaryotic initiation factor; 4E-BP, eIF4E-binding protein; GCN, general control nonderepressible protein; miRNA, microRNA; mTORC1, mammalian target of rapamycin complex 1; PDCD4, programmed cell death protein 4; PIC, preinitiation complex; PI3K, phosphoinositide 3-kinase; Rheb, Ras homologue enriched in brain; RPSK, ribosomal protein S6 kinase; S6K, S6 kinase; TOP, tract of oligopyrimidine; TSC, tuberous sclerosis complex; uORF, upstream open reading frame; UTR, untranslated region.
Literature Cited
- 1.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–97 [DOI] [PubMed] [Google Scholar]
- 2.Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–45 [DOI] [PubMed] [Google Scholar]
- 3.Grayson M. Nutrigenomics. Nature. 2010;468:S1. [DOI] [PubMed] [Google Scholar]
- 4.Fenech M, El-Sohemy A, Cahill L, Ferguson LR, French TA, Tai ES, Milner J, Koh WP, Xie L, et al. Nutrigenetics and nutrigenomics: viewpoints on the current status and applications in nutrition research and practice. J Nutrigenet Nutrigenomics. 2011;4:69–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ebner OA, Selbach M. Whole cell proteome regulation by microRNAs captured in a pulsed SILAC mass spectrometry approach. Methods Mol Biol. 2011;725:315–31 [DOI] [PubMed] [Google Scholar]
- 6.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spriggs KA, Bushell M, Willis AE. Translational regulation of gene expression during conditions of cell stress. Mol Cell. 2010;40:228–37 [DOI] [PubMed] [Google Scholar]
- 8.Pisarev AV, Hellen CU, Pestova TV. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gross JD, Moerke NJ, von der Haar T, Lugovskoy AA, Sachs AB, McCarthy JE, Wagner G. Ribosome loading onto the mRNA cap is driven by conformational coupling between eIF4G and eIF4E. Cell. 2003;115:739–50 [DOI] [PubMed] [Google Scholar]
- 10.Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc Natl Acad Sci U S A. 2008;105:9564–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pestova TV, Lomakin IB, Lee JH, Choi SK, Dever TE, Hellen CU. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–5 [DOI] [PubMed] [Google Scholar]
- 15.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–7 [DOI] [PubMed] [Google Scholar]
- 16.Cridge AG, Castelli LM, Smirnova JB, Selley JN, Rowe W, Hubbard SJ, McCarthy JE, Ashe MP, Grant CM, Pavitt GD. Identifying eIF4E-binding protein translationally-controlled transcripts reveals links to mRNAs bound by specific PUF proteins. Nucleic Acids Res. 2010;38:8039–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sampath P, Pritchard DK, Pabon L, Reinecke H, Schwartz SM, Morris DR, Murry CE. A hierarchical network controls protein translation during murine embryonic stem cell self-renewal and differentiation. Cell Stem Cell. 2008;2:448–60 [DOI] [PubMed] [Google Scholar]
- 18.Grech G, Blazquez-Domingo M, Kolbus A, Bakker WJ, Mullner EW, Beug H, von Lindern M. Igbp1 is part of a positive feedback loop in stem cell factor-dependent, selective mRNA translation initiation inhibiting erythroid differentiation. Blood. 2008;112:2750–60 [DOI] [PubMed] [Google Scholar]
- 19.Chen J, Melton C, Suh N, Oh JS, Horner K, Xie F, Sette C, Blelloch R, Conti M. Genome-wide analysis of translation reveals a critical role for deleted in azoospermia-like (Dazl) at the oocyte-to-zygote transition. Genes Dev. 2011;25:755–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sonenberg N, Hershey JWB, Mathews M. Translational control of gene expression. 2nd ed Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2000 [Google Scholar]
- 21.Thomas JD, Johannes GJ. Identification of mRNAs that continue to associate with polysomes during hypoxia. RNA. 2007;13:1116–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shenton D, Smirnova JB, Selley JN, Carroll K, Hubbard SJ, Pavitt GD, Ashe MP, Grant CM. Global translational responses to oxidative stress impact upon multiple levels of protein synthesis. J Biol Chem. 2006;281:29011–21 [DOI] [PubMed] [Google Scholar]
- 23.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat Rev Mol Cell Biol. 2004;5:827–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donovan J, Copeland PR. Threading the needle: getting selenocysteine into proteins. Antioxid Redox Signal. 2010;12:881–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hendrickson DG, Hogan DJ, McCullough HL, Myers JW, Herschlag D, Ferrell JE, Brown PO. Concordant regulation of translation and mRNA abundance for hundreds of targets of a human microRNA. PLoS Biol. 2009;7:e1000238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mata J, Marguerat S, Bahler J. Post-transcriptional control of gene expression: a genome-wide perspective. Trends Biochem Sci. 2005;30:506–14 [DOI] [PubMed] [Google Scholar]
- 29.Pradet-Balade B, Boulme F, Beug H, Mullner EW, Garcia-Sanz JA. Translation control: bridging the gap between genomics and proteomics? Trends Biochem Sci. 2001;26:225–9 [DOI] [PubMed] [Google Scholar]
- 30.Kudo K, Xi Y, Wang Y, Song B, Chu E, Ju J, Russo JJ. Translational control analysis by translationally active RNA capture/microarray analysis (TrIP-Chip). Nucleic Acids Res. 2010;38:e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lackner DH, Beilharz TH, Marguerat S, Mata J, Watt S, Schubert F, Preiss T, Bahler J. A network of multiple regulatory layers shapes gene expression in fission yeast. Mol Cell. 2007;26:145–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, et al. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648–57 [DOI] [PubMed] [Google Scholar]
- 36.Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658–65 [DOI] [PubMed] [Google Scholar]
- 37.Inoki K, Li Y, Xu T, Guan KL. Rheb GTPase is a direct target of TSC2 GAP activity and regulates mTOR signaling. Genes Dev. 2003;17:1829–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Gao X, Saucedo LJ, Ru B, Edgar BA, Pan D. Rheb is a direct target of the tuberous sclerosis tumour suppressor proteins. Nat Cell Biol. 2003;5:578–81 [DOI] [PubMed] [Google Scholar]
- 39.Saucedo LJ, Gao X, Chiarelli DA, Li L, Pan D, Edgar BA. Rheb promotes cell growth as a component of the insulin/TOR signalling network. Nat Cell Biol. 2003;5:566–71 [DOI] [PubMed] [Google Scholar]
- 40.Nicklin P, Bergman P, Zhang B, Triantafellow E, Wang H, Nyfeler B, Yang H, Hild M, Kung C, et al. Bidirectional transport of amino acids regulates mTOR and autophagy. Cell. 2009;136:521–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan L, Mieulet V, Burgess D, Findlay GM, Sully K, Procter J, Goris J, Janssens V, Morrice NA, Lamb RF. PP2A T61 epsilon is an inhibitor of MAP4K3 in nutrient signaling to mTOR. Mol Cell. 2010;37:633–42 [DOI] [PubMed] [Google Scholar]
- 42.Gulati P, Gaspers LD, Dann SG, Joaquin M, Nobukuni T, Natt F, Kozma SC, Thomas AP, Thomas G. Amino acids activate mTOR complex 1 via Ca2+/CaM signaling to hVps34. Cell Metab. 2008;7:456–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim E, Goraksha-Hicks P, Li L, Neufeld TP, Guan KL. Regulation of TORC1 by Rag GTPases in nutrient response. Nat Cell Biol. 2008;10:935–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–18 [DOI] [PubMed] [Google Scholar]
- 47.Haghighat A, Mader S, Pause A, Sonenberg N. Repression of cap-dependent translation by 4E-binding protein 1: competition with p220 for binding to eukaryotic initiation factor-4E. EMBO J. 1995;14:5701–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Raught B, Peiretti F, Gingras AC, Livingstone M, Shahbazian D, Mayeur GL, Polakiewicz RD, Sonenberg N, Hershey JW. Phosphorylation of eucaryotic translation initiation factor 4B Ser422 is modulated by S6 kinases. EMBO J. 2004;23:1761–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sun J, Conn CS, Han Y, Yeung V, Qian SB. PI3K-mTORC1 attenuates stress response by inhibiting cap-independent Hsp70 translation. J Biol Chem. 2011;286:6791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bilanges B, Argonza-Barrett R, Kolesnichenko M, Skinner C, Nair M, Chen M, Stokoe D. Tuberous sclerosis complex proteins 1 and 2 control serum-dependent translation in a TOP-dependent and -independent manner. Mol Cell Biol. 2007;27:5746–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kilberg MS, Pan YX, Chen H, Leung-Pineda V. Nutritional control of gene expression: how mammalian cells respond to amino acid limitation. Annu Rev Nutr. 2005;25:59–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang P, McGrath BC, Reinert J, Olsen DS, Lei L, Gill S, Wek SA, Vattem KM, Wek RC, et al. The GCN2 eIF2alpha kinase is required for adaptation to amino acid deprivation in mice. Mol Cell Biol. 2002;22:6681–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Staschke KA, Dey S, Zaborske JM, Palam LR, McClintick JN, Pan T, Edenberg HJ, Wek RC. Integration of general amino acid control and target of rapamycin (TOR) regulatory pathways in nitrogen assimilation in yeast. J Biol Chem. 2010;285:16893–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Blais JD, Filipenko V, Bi M, Harding HP, Ron D, Koumenis C, Wouters BG, Bell JC. Activating transcription factor 4 is translationally regulated by hypoxic stress. Mol Cell Biol. 2004;24:7469–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vattem KM, Wek RC. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc Natl Acad Sci U S A. 2004;101:11269–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bishop NA, Lu T, Yankner BA. Neural mechanisms of ageing and cognitive decline. Nature. 2010;464:529–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sahin E, Depinho RA. Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature. 2010;464:520–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaeberlein M. Lessons on longevity from budding yeast. Nature. 2010;464:513–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kenyon CJ. The genetics of ageing. Nature. 2010;464:504–12 [DOI] [PubMed] [Google Scholar]
- 60.McCay CM, Crowell MF, Maynard LA. The effect of retarded growth upon the length of life span and upon the ultimate body size. 1935. Nutrition. 1989;5:155–71, discussion 72 [PubMed] [Google Scholar]
- 61.Guarente L, Picard F. Calorie restriction--the SIR2 connection. Cell. 2005;120:473–82 [DOI] [PubMed] [Google Scholar]
- 62.Fontana L. The scientific basis of caloric restriction leading to longer life. Curr Opin Gastroenterol. 2009;25:144–50 [DOI] [PubMed] [Google Scholar]
- 63.Vellai T, Takacs-Vellai K, Zhang Y, Kovacs AL, Orosz L, Muller F. Genetics: influence of TOR kinase on lifespan in C. elegans. Nature. 2003;426:620. [DOI] [PubMed] [Google Scholar]
- 64.Kapahi P, Zid BM, Harper T, Koslover D, Sapin V, Benzer S. Regulation of lifespan in Drosophila by modulation of genes in the TOR signaling pathway. Curr Biol. 2004;14:885–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaeberlein M, Powers RW, 3rd, Steffen KK, Westman EA, Hu D, Dang N, Kerr EO, Kirkland KT, Fields S, Kennedy BK. Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science. 2005;310:1193–6 [DOI] [PubMed] [Google Scholar]
- 66.Powers RW, 3rd, Kaeberlein M, Caldwell SD, Kennedy BK, Fields S. Extension of chronological life span in yeast by decreased TOR pathway signaling. Genes Dev. 2006;20:174–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Harrison DE, Strong R, Sharp ZD, Nelson JF, Astle CM, Flurkey K, Nadon NL, Wilkinson JE, Frenkel K, et al. Rapamycin fed late in life extends lifespan in genetically heterogeneous mice. Nature. 2009;460:392–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hipkiss AR. Accumulation of altered proteins and ageing: causes and effects. Exp Gerontol. 2006;41:464–73 [DOI] [PubMed] [Google Scholar]
- 69.Hipkiss AR. On why decreasing protein synthesis can increase lifespan. Mech Ageing Dev. 2007;128:412–4 [DOI] [PubMed] [Google Scholar]
- 70.Pan KZ, Palter JE, Rogers AN, Olsen A, Chen D, Lithgow GJ, Kapahi P. Inhibition of mRNA translation extends lifespan in Caenorhabditis elegans. Aging Cell. 2007;6:111–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hansen M, Taubert S, Crawford D, Libina N, Lee SJ, Kenyon C. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell. 2007;6:95–110 [DOI] [PubMed] [Google Scholar]
- 72.Syntichaki P, Troulinaki K, Tavernarakis N. eIF4E function in somatic cells modulates ageing in Caenorhabditis elegans. Nature. 2007;445:922–6 [DOI] [PubMed] [Google Scholar]
- 73.Ruggero D, Montanaro L, Ma L, Xu W, Londei P, Cordon-Cardo C, Pandolfi PP. The translation factor eIF-4E promotes tumor formation and cooperates with c-Myc in lymphomagenesis. Nat Med. 2004;10:484–6 [DOI] [PubMed] [Google Scholar]
- 74.Steffen KK, MacKay VL, Kerr EO, Tsuchiya M, Hu D, Fox LA, Dang N, Johnston ED, Oakes JA, et al. Yeast life span extension by depletion of 60s ribosomal subunits is mediated by Gcn4. Cell. 2008;133:292–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.McColl G, Rogers AN, Alavez S, Hubbard AE, Melov S, Link CD, Bush AI, Kapahi P, Lithgow GJ. Insulin-like signaling determines survival during stress via posttranscriptional mechanisms in C. elegans. Cell Metab. 2010;12:260–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zid BM, Rogers AN, Katewa SD, Vargas MA, Kolipinski MC, Lu TA, Benzer S, Kapahi P. 4E-BP extends lifespan upon dietary restriction by enhancing mitochondrial activity in Drosophila. Cell. 2009;139:149–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rogers AN, Chen D, McColl G, Czerwieniec G, Felkey K, Gibson BW, Hubbard A, Melov S, Lithgow GJ, Kapahi P. Life span extension via eIF4G inhibition is mediated by posttranscriptional remodeling of stress response gene expression in C. elegans. Cell Metab. 2011;14:55–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Janes MR, Limon JJ, So L, Chen J, Lim RJ, Chavez MA, Vu C, Lilly MB, Mallya S, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 2010;16:205–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Scheper GC, van der Knaap MS, Proud CG. Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet. 2007;8:711–23 [DOI] [PubMed] [Google Scholar]
- 82.Dürig J, Nuckel H, Huttmann A, Kruse E, Holter T, Halfmeyer K, Fuhrer A, Rudolph R, Kalhori N, et al. Expression of ribosomal and translation-associated genes is correlated with a favorable clinical course in chronic lymphocytic leukemia. Blood. 2003;101:2748–55 [DOI] [PubMed] [Google Scholar]
- 83.Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, et al. mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science. 2010;328:1172–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Petroulakis E, Parsyan A, Dowling RJ, LeBacquer O, Martineau Y, Bidinosti M, Larsson O, Alain T, Rong L, et al. p53-dependent translational control of senescence and transformation via 4E-BPs. Cancer Cell. 2009;16:439–46 [DOI] [PubMed] [Google Scholar]
- 85.Larsson O, Perlman DM, Fan D, Reilly CS, Peterson M, Dahlgren C, Liang Z, Li S, Polunovsky VA, et al. Apoptosis resistance downstream of eIF4E: posttranscriptional activation of an anti-apoptotic transcript carrying a consensus hairpin structure. Nucleic Acids Res. 2006;34:4375–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hsieh AC, Costa M, Zollo O, Davis C, Feldman ME, Testa JR, Meyuhas O, Shokat KM, Ruggero D. Genetic dissection of the oncogenic mTOR pathway reveals druggable addiction to translational control via 4EBP-eIF4E. Cancer Cell. 2010;17:249–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wendel HG, De Stanchina E, Fridman JS, Malina A, Ray S, Kogan S, Cordon-Cardo C, Pelletier J, Lowe SW. Survival signalling by Akt and eIF4E in oncogenesis and cancer therapy. Nature. 2004;428:332–7 [DOI] [PubMed] [Google Scholar]
- 88.Wendel HG, Silva RL, Malina A, Mills JR, Zhu H, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Teruya-Feldstein J, et al. Dissecting eIF4E action in tumorigenesis. Genes Dev. 2007;21:3232–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.She QB, Halilovic E, Ye Q, Zhen W, Shirasawa S, Sasazuki T, Solit DB, Rosen N. 4E-BP1 is a key effector of the oncogenic activation of the AKT and ERK signaling pathways that integrates their function in tumors. Cancer Cell. 2010;18:39–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Graff JR, Konicek BW, Vincent TM, Lynch RL, Monteith D, Weir SN, Schwier P, Capen A, Goode RL, et al. Therapeutic suppression of translation initiation factor eIF4E expression reduces tumor growth without toxicity. J Clin Invest. 2007;117:2638–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gandin V, Miluzio A, Barbieri AM, Beugnet A, Kiyokawa H, Marchisio PC, Biffo S. Eukaryotic initiation factor 6 is rate-limiting in translation, growth and transformation. Nature. 2008;455:684–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hussey GS, Chaudhury A, Dawson AE, Lindner DJ, Knudsen CR, Wilce MC, Merrick WC, Howe PH. Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 2011;41:419–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Rosenwald IB. Upregulated expression of the genes encoding translation initiation factors eIF-4E and eIF-2alpha in transformed cells. Cancer Lett. 1996;102:113–23 [DOI] [PubMed] [Google Scholar]
- 94.Blais JD, Addison CL, Edge R, Falls T, Zhao H, Wary K, Koumenis C, Harding HP, Ron D, et al. Perk-dependent translational regulation promotes tumor cell adaptation and angiogenesis in response to hypoxic stress. Mol Cell Biol. 2006;26:9517–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Donzé O, Jagus R, Koromilas AE, Hershey JW, Sonenberg N. Abrogation of translation initiation factor eIF-2 phosphorylation causes malignant transformation of NIH 3T3 cells. EMBO J. 1995;14:3828–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Calkhoven CF, Muller C, Leutz A. Translational control of gene expression and disease. Trends Mol Med. 2002;8:577–83 [DOI] [PubMed] [Google Scholar]
- 97.Rajasekhar VK, Viale A, Socci ND, Wiedmann M, Hu X, Holland EC. Oncogenic Ras and Akt signaling contribute to glioblastoma formation by differential recruitment of existing mRNAs to polysomes. Mol Cell. 2003;12:889–901 [DOI] [PubMed] [Google Scholar]