Nsk1 is a novel fission yeast protein that binds the nucleolus during interphase and the nucleoplasm during early mitosis. After anaphase and following dephosphorylation by Clp1, Nsk1 binds the kinetochore–spindle pole junction and maintains accurate chromosome segregation by promoting the association of kinetochores to spindle poles during anaphase B.
Abstract
Type 1 phosphatase (PP1) antagonizes Aurora B kinase to stabilize kinetochore–microtubule attachments and to silence the spindle checkpoint. We screened for factors that exacerbate the growth defect of Δdis2 cells, which lack one of two catalytic subunits of PP1 in fission yeast, and identified Nsk1, a novel protein required for accurate chromosome segregation. During interphase, Nsk1 resides in the nucleolus but spreads throughout the nucleoplasm as cells enter mitosis. Following dephosphorylation by Clp1 (Cdc14-like) phosphatase and at least one other phosphatase, Nsk1 localizes to the interface between kinetochores and the inner face of the spindle pole body during anaphase. In the absence of Nsk1, some kinetochores become detached from spindle poles during anaphase B. If this occurs late in anaphase B, then the sister chromatids of unclustered kinetochores segregate to the correct daughter cell. These unclustered kinetochores are efficiently captured, retrieved, bioriented, and segregated during the following mitosis, as long as Dis2 is present. However, if kinetochores are detached from a spindle pole early in anaphase B, then these sister chromatids become missegregated. These data suggest Nsk1 ensures accurate chromosome segregation by promoting the tethering of kinetochores to spindle poles during anaphase B.
INTRODUCTION
Proper segregation of sister chromatids to opposite poles of the cell during mitosis is essential for cell proliferation. Defects in chromosome segregation are implicated in human diseases such as cancers and congenital disorders, which are characterized by chromosome instability and aneuploidy (Holland and Cleveland, 2009). Segregation of sister chromatids depends on forces generated by the microtubules that attach to kinetochores. For accurate chromosome segregation, kinetochores must be captured by spindle microtubules and properly aligned on the spindle before anaphase onset (Tanaka, 2010). As cells enter mitosis, one of the two kinetochores of a sister-chromatid pair is initially captured by the lateral surface of a microtubule nucleated from one of the two spindle poles. Chromosomes are primarily retrieved to the spindle pole along the lateral surface of the microtubule by interaction of the kinetochore with kinesin motors. In yeast, chromosomes can be retrieved not only on the lateral surface of the microtubule by kinesin motors, but also by end-on attachment to the depolymerizing microtubule end. The latter process requires coupling of the kinetochore to the microtubule end by the DASH complex (Rieder and Alexander, 1990; Tanaka et al., 2005, 2007; Franco et al., 2007).
Following transport to the spindle pole, sister kinetochores are captured by microtubules from either the opposite or the same spindle pole. In the latter case, kinetochore–microtubule attachment must be reset until proper biorientation is established. Kinetochore–microtubule interactions are governed by dynamic changes in phosphorylation of key proteins at the kinetochore–microtubule interface. The chromosomal passenger complex (CPC) component Aurora B kinase promotes chromosome biorientation by phosphorylating kinetochore components to disestablish incorrect microtubule attachments (Cheeseman et al., 2002; Tanaka et al., 2002; Yang et al., 2008; Liu et al., 2010). Turnover of the kinetochore–microtubule attachment stops once tension is generated across sister kinetochores upon the establishment of correct biorientation (Zhai et al., 1995; Mallavarapu et al., 1999). Inter-kinetochore tension separates outer kinetochore Aurora B targets from the centromere-associated CPC, promoting kinetochore association of protein phosphatase 1 (PP1). This is thought to reverse Aurora B–dependent phosphorylation of the outer kinetochore, thus stabilizing tension-bearing kinetochore–microtubule attachments and silencing the spindle checkpoint (Liu et al., 2010; Meadows et al., 2011; Rosenberg et al., 2011). Lowered Aurora B kinase activity at the outer kinetochore also promotes the recruitment of a complex composed of astrin, SKAP/kinastrin, and dynein light chain (Tctex-1/LC8), which further stabilizes correct kinetochore–microtubule interactions in mammalian cells (Thein et al., 2007; Manning et al., 2010; Schmidt et al., 2010).
Once all sister kinetochores are correctly bioriented on the spindle, cohesion between sister chromatids is removed, allowing sister chromatids to segregate to opposite spindle poles during anaphase. Importantly, end-on, kinetochore–microtubule attachments must be maintained both during anaphase A, as sister kinetochores are pulled toward the opposite poles by depolymerization of kinetochore microtubules, and anaphase B, as the distance between poles increases, driven by microtubule polymerization and sliding at the spindle midzone (Nabeshima et al., 1998; Khodjakov et al., 2004; Civelekoglu-Scholey and Scholey, 2010). Altered kinetochore and microtubule behavior in anaphase requires reversal of early mitotic phosphorylation events. Dephosphorylation of cyclin B/Cdk1 substrates is catalyzed by both Cdc14-like and type 2A (PP2A) phosphatases (Minshull et al., 1996; Visintin et al., 1998). Budding yeast Cdc14 promotes chromosome segregation by stabilizing microtubule dynamics at anaphase onset, largely through influencing the behavior of interpolar microtubules (Higuchi and Uhlmann, 2005). The Cdc14-like phosphatase, Clp1, is also required for accurate chromosome segregation in fission yeast. This was initially thought to be due to altered localization of Aurora B, but has since been disputed (Trautmann et al., 2004; Bohnert et al., 2009; J. C. Meadows, unpublished data). As such, the mechanisms by which Clp1 promotes accurate chromosome segregation in fission yeast are largely unknown.
Yeast kinetochores normally cluster in a region of the nuclear envelope underlying the spindle pole body (SPB) during interphase in a Rabl-like formation (Funabiki et al., 1993; Jin et al., 1998, 2000). It is unclear whether Rabl formation aids the efficiency of kinetochore capture or enforces a geometric arrangement of kinetochores that promotes chromosome biorientation. Our finding that the fission yeast DASH complex is required for the retrieval of kinetochores that have become unclustered suggests that the pathways required to establish chromosome biorientation are influenced by intranuclear kinetochore position (Franco et al., 2007). Although kinetochore clustering is established at the end of anaphase A, the mechanisms by which it is maintained during interphase differ between yeasts. Budding yeast kinetochores remain attached to the old SPB throughout the cell cycle via short microtubules. After S phase, both sister kinetochores initially bind to microtubules from the same (old) SPB and therefore need to be reoriented during mitosis to establish biorientation (Biggins et al., 1999; Kitamura et al., 2007). By contrast, in fission yeast, the SPB defenestrates from the nuclear envelope at the end of anaphase B, yet kinetochores remain clustered near the SPB via a nonmicrotubule-based linkage (Ding et al., 1997). The molecular nature of this linkage is unclear but is thought to involve the transmembrane protein Ima1, which binds indirectly to the heterochromatic region of centromeres, and other, as-yet-unknown proteins that link the outer kinetochore to the inner face of the SPB (King et al., 2008). In this paper, we reveal that association of kinetochores to SPBs during anaphase B is necessary to maintain accurate chromosome segregation, and we identify a novel substrate of the Clp1 phosphatase, Nsk1, that binds to the kinetochore–SPB junction during anaphase B to facilitate this process.
RESULTS
Nsk1 is required for accurate chromosome segregation and timely anaphase onset in fission yeast
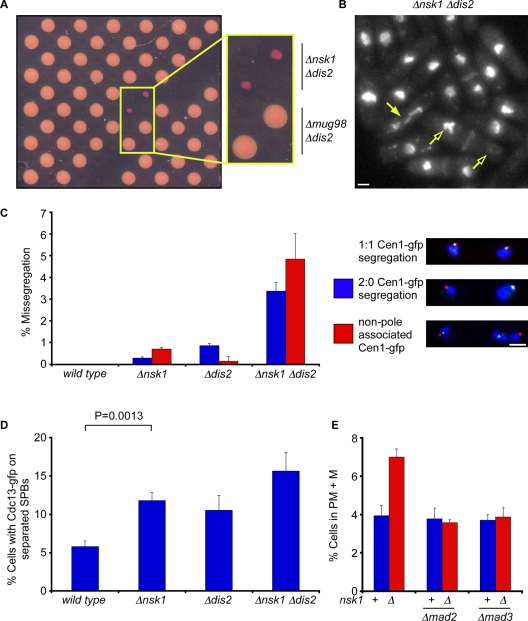
Two PP1 catalytic subunits exist in fission yeast, namely Dis2 and Sds21. Dis2, but not Sds21, binds the kinetochore and is required for correct chromosome segregation and spindle checkpoint silencing (Ohkura et al., 1989; Vanoosthuyse and Hardwick, 2009). To identify additional factors required for accurate chromosome segregation, we screened a genome-wide deletion library for alleles that perturbed the growth of Δdis2 mutants (Figure 1A). Thirty-eight strong hits were identified (Supplemental Table SI). Since Δnsk1 Δdis2 double mutants displayed mitotic abnormalities, we chose Nsk1 for further analysis (Figure 1B). Nsk1 does not contain any domains of known function, but sequence analyses revealed Nsk1 homologues in the closely related fission yeast, Schizosaccharomyces cryophilus and Schizosaccharomyces octosporus, but not in Schizosaccharomyces japonicus (Supplemental Figure S1). Nevertheless, we found that both Δnsk1 and Δdis2 cells missegregate chromosome 1, and that this is exacerbated in Δnsk1 Δdis2 double mutants (Figure 1C). Similar results were observed when the accuracy of chromosome segregation was monitored using a minichromosome loss assay (Table 1).
FIGURE 1:
Nsk1 is required for accurate chromosome segregation and timely anaphase onset. (A) Plate containing selective antibiotics and 1 mg/ml phloxine B from a genome-wide screen for mutants that are slow growing in the absence of Dis2. Phloxin B stains dead cells. (B) Log-phase Δnsk1 Δdis2 cells were fixed and stained with DAPI and calcofluor to stain chromatin and septa, respectively. Closed arrow indicates a cell undergoing abnormal chromosome segregation in mitosis. Open arrows indicate a cell in which all chromosomes have been segregated to one of the two daughter cells. Scale bars: 2 μm. (C) G2-synchronized wild-type, Δnsk1, Δdis2, and Δnsk1 Δdis2 cells expressing lys1:lacO gfp-NLS-lacI cdc11-cfp were isolated from a 10–40% lactose gradient and segregation of chromosome 1 was monitored in the following anaphase. The percentage of missegregation was calculated as the total number of cells displaying either nondisjunction (blue bar) or non-SPB–associated sister chromatids (red bar). Error bars are the SD from the mean of three independent experiments. (D) Log-phase wild-type, Δnsk1, Δdis2, and Δnsk1 Δdis2 cells expressing Cdc13-gfp were fixed, and the percentage of cells with spindle- and SPB-associated Cdc13 assessed. Error bars are the SD from the mean of three independent experiments. (E) Log-phase cultures of wild-type, Δmad2, Δmad3, Δnsk1, Δnsk1 Δmad2, and Δnsk1 Δmad3 cells expressing dad1-gfp and sid4-tdtomato were fixed, and the percentage of prometaphase and metaphase (PM + M) cells were assessed. Error bars are the SD from the mean of three independent experiments.
TABLE 1:
Fidelity of chromosome segregation by minichromosome loss assay
| Minichromosome loss per division (%) | |
|---|---|
| wild type | 0.01 ± 0.03 |
| Δnsk1 | 1.06 ± 0.59 |
| Δdis2 | 1.37 ± 0.15 |
| Δnsk1 Δdis2 | 27.00 ± 5.83 |
| clp1(C286S) | 1.12 ± 0.34 |
| Δnsk1 clp1(C286S) | 1.59 ± 0.52 |
| Δdis2 clp1(C286S) | 24.48 ± 3.66 |
| Δmad2 | 0.15 ± 0.07 |
| Δnsk1 Δmad2 | 2.46 ± 0.32 |
| Δdlc1 | 0.04 ± 0.02 |
| Δnsk1 Δdlc1 | 0.85 ± 0.07 |
| nsk1-gfp | 0.05 ± 0.01 |
| nsk1-gfp Δdlc1 | 0.13 ± 0.01 |
Defects in kinetochore–microtubule attachment are recognized by the spindle assembly checkpoint (SAC), and this delays the onset of anaphase. To assess whether loss of nsk1 delays anaphase, we monitored both Cdc13 (cyclin B) localization to SPBs and the percentage of cells in prometaphase and metaphase. We found that both Δnsk1 and Δdis2 cells are delayed in the timing of anaphase onset and that this effect is exacerbated in Δnsk1 Δdis2 double mutants (Figure 1D). The delay in anaphase onset observed in Δnsk1 cells is abolished by deletion of either mad2 or mad3, indicating that this is due either to activation of, or failure to silence, the SAC (Figure 1E). To distinguish between these possibilities, we monitored Cdc13 (Cyclin B) destruction in single cells following chemical inactivation of Ark1 (Aurora B) kinase in mitotically arrested nda3-KM311 cells. The nda3-KM311 allele encodes a cold-sensitive β-tubulin protein that causes cells to arrest in mitosis at 18°C with no microtubules (Hiraoka et al., 1984). We found that loss of nsk1 had no effect in this assay, indicating that Nsk1 is not required to silence the SAC (Figure S2A). Indeed, we found that Δdis2, but not Δnsk1, mutants suppress the proliferation defect of a temperature-sensitive ark1-T7 mutant, indicating that Nsk1 does not counteract Ark1 function (Figure S2B). Nevertheless, the chromosome loss rate was considerably worse in Δnsk1 Δmad2 cells than in Δnsk1 single mutant (Table 1). These results establish Nsk1 as a novel factor required for accurate chromosome segregation and timely anaphase onset in fission yeast.
Nsk1 binds the kinetochore–SPB junction during anaphase B
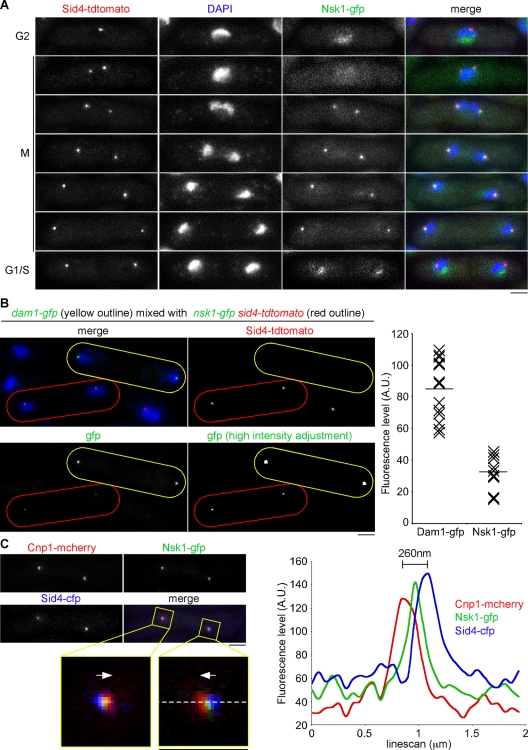
To gain further insight into the function of Nsk1, we monitored the localization of Nsk1 in both fixed and live cells. Nsk1 was tagged at its C-terminus with green fluorescent protein (GFP) in sid4-tdtomato cells. Sid4 is a constitutively bound component of the SPB outer plaque (Krapp et al., 2001). The nsk1-gfp allele does not display any growth defect in the absence of dis2, nor any delay in anaphase onset or elevated rate of minichromosome loss, indicating that tagging the C-terminus of Nsk1 with GFP does not noticeably alter Nsk1 function (unpublished data). We found that Nsk1 localizes predominantly in the nucleolus during interphase, as judged by colocalization with fission yeast fibrillarin, Fib1 (Beauregard et al., 2009; Figures 2A and S3A). During prometaphase and metaphase, Nsk1 localizes broadly throughout the nucleoplasm, suggesting it is released from the nucleolus at the G2/M transition (Figure 2A). During anaphase B, Nsk1 appears as two prominent dots that seem to colocalize with SPBs, before it returns to the nucleolus at the end of anaphase B, indicating that Nsk1 undergoes cell cycle–dependent changes in its localization (Figure 2A). An identical localization pattern was observed in nmt1:gfp-nsk1 cells under repressive conditions, although Nsk1 decorates the whole anaphase B spindle when overexpressed (Figure S3B).
FIGURE 2:
Nsk1 localizes at the kinetochore–SPB interface in a cell cycle–dependent manner. (A) Log-phase cultures of nsk1-gfp sid4-tdtomato cells were fixed and analyzed by fluorescence microscopy. A series of images of cells at different stages of the cell cycle are depicted. Scale bars: 2 μm. (B) Equal volumes of log-phase dam1-gfp (yellow outline) and nsk1-gfp sid4-tdtomato (red outline) cells were mixed and fixed. Left panels show a representative image containing both cell types. Bottom, right image shows the GFP channel with a high-intensity adjustment. Far right panel shows the fluorescence intensities of Dam1-gfp and Nsk1-gfp signals from these mixed-population experiments. Horizontal bars indicate the mean fluorescence levels. (C) Fluorescence levels over a 2-μm distance were calculated for the three channels in a nsk1-gfp cnp1-mcherry sid4-cfp cell. Top, left panel shows the cell under examination with the analyzed regions boxed (yellow). Enlargements are shown below. Arrows indicate the direction of the cell middle. The pixel line (white, dashed) used to measure fluorescence levels is shown. Right panel is a graph generated by this line scan.
We noted that Nsk1-gfp signals are very weak. To quantify this, we compared anaphase Nsk1-gfp fluorescence with anaphase Dam1-gfp fluorescence in a mixed population of cells. Dam1 is a component of the DASH complex that is estimated to be present at 6–7 copies per kinetochore in fission yeast (Joglekar et al., 2008). We found that the Dam1-gfp signal is ∼2.5 times stronger than the Nsk1-gfp signal, implying that ∼7–8 copies of Nsk1 are present at each SPB-associated dot during anaphase B (Figure 2B).
To examine the localization of Nsk1 during anaphase B more carefully, we analyzed the relative positions of Nsk1; Sid4; and the fission yeast CENP-A homologue, Cnp1, in fixed nsk1-gfp sid4-cfp cnp1-mcherry cells. In all analyzed cells (n > 20), we found the peak of Nsk1-gfp fluorescence occurs between the Cnp1-mcherry and Sid4-cfp peaks, suggesting that Nsk1 binds the junction between kinetochores and the inner face of the SPB (Figure 2C).
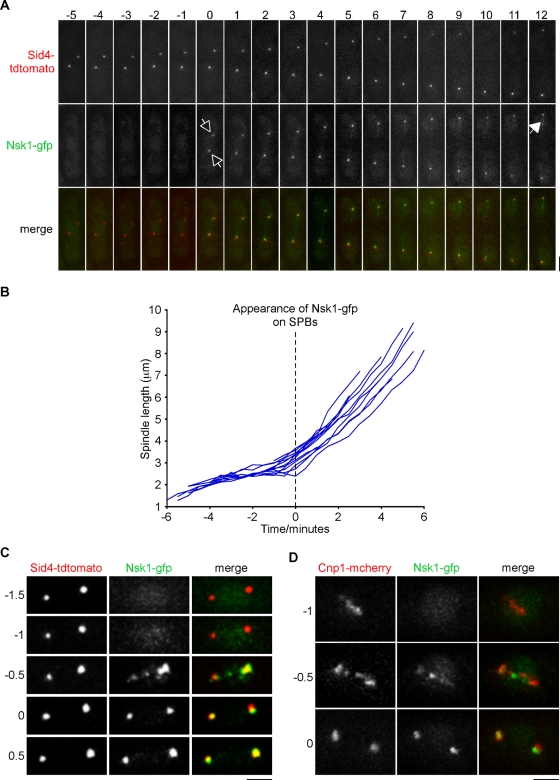
As seen by live-cell imaging, Nsk1 appears at SPBs coincident with the onset of spindle elongation (Figure 3, A and B). Some Nsk1 binds weakly to the elongating anaphase B spindle, but it does not accumulate at the spindle midzone (Figure 3A). To more precisely determine the timing of Nsk1 localization during mitosis, we filmed nsk1-gfp sid4-tdtomato and nsk1-gfp cnp1-mcherry cells at a higher temporal resolution. We found that Nsk1 localizes in puncta along the axis of the spindle during anaphase A and then associates to the kinetochore–SPB interface from the end of anaphase A and during anaphase B (Figure 3, C and D, and Supplemental Movie S1).
FIGURE 3:
Nsk1 loading occurs at anaphase onset. (A) Log-phase cultures of nsk1-gfp sid4-tdtomato cells were grown on agar pads at 30°C and analyzed by fluorescence microscopy. A series of images of a mitotic cell at 1-min intervals is depicted. The time at which Nsk1 appears at the kinetochore–SPB junction (open arrows) and late anaphase B spindle (closed arrow) is highlighted. Scale bars: 2 μm. (B) Graph of spindle length against time for multiple nsk1-gfp sid4-tdtomato cells. Spindle length is the distance between two Sid4-tdtomato dots. Time zero corresponds to the appearance of Nsk1 at the pole. (C & D) Log-phase cultures of nsk1-gfp sid4-tdtomato (C) or nsk1-gfp cnp1-mcherry (D) cells were grown on agar pads at 30°C and analyzed by fluorescence microscopy. A series of images from two representative cells at 30 s intervals are depicted. Note that in each case Nsk1 appears before the end of anaphase A. Time 0 corresponds to the appearance of Nsk1 at the poles.
Nsk1 does not bind to lagging sister kinetochores during anaphase B
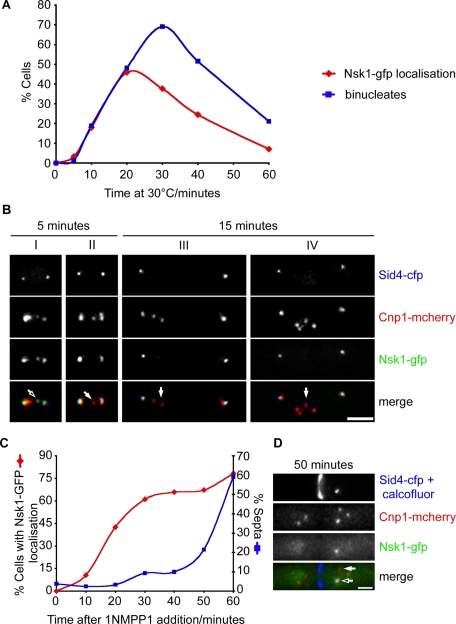
We were unable to determine whether Nsk1 binds kinetochores or the spindle during anaphase A by live-cell analysis, since anaphase A only lasts 40–45 s at this temperature (Meadows and Millar, 2008). Imaging at even higher temporal resolution was not possible, due to the very low abundance of Nsk1. To circumvent this problem, we arrested nda3-KM311 nsk1-gfp sid4-cfp cnp1-mcherry cells in mitosis and examined the localization of Nsk1 in fixed-cell populations following release into anaphase. Importantly, lagging sister chromatids are frequently observed on release of nda3-KM311 cells to the permissive temperature (Trautmann et al., 2004). At the arrest point, we did not observe discrete dots of Nsk1, indicating that Nsk1 does not localize to kinetochores or SPBs before anaphase onset (Figure 4A). This is not due to the absence of microtubules, as similar results were observed when cells were arrested in mitosis by overexpression of Mph1 (MPS1) kinase, which ectopically activates the spindle checkpoint (K. M. May and K. G. Hardwick, unpublished data). However, following release of nda3-KM311 nsk1-gfp sid4-cfp cnp1-mcherry cells to the permissive temperature, Nsk1 appeared at the kinetochore–SPB junction at the end of anaphase A (Figure 4, A and B). At early time points, we found that Nsk1 occasionally colocalizes with a single lagging Cnp1 dot on the anaphase A spindle (Figure 4B, cell I). In other cells with lagging kinetochores in anaphase A (∼50%), no Nsk1 localization could be detected (Figure 4B, cell II). At later time points, we found that Nsk1 always binds to the kinetochore–SPB junction, so long as at least one kinetochore is associated with the SPB. However, we never observed association of Nsk1 to the kinetochores of lagging sister chromatids (Figure 4B, cells III and IV).
FIGURE 4:
Nsk1 associates to the kinetochore–spindle junction during anaphase B in the absence of microtubules. (A) Log-phase cultures of nda3-KM311 nsk1-gfp cnp1-mcherry sid4-cfp cells were arrested in mitosis by incubation at 18°C for 6 h. Following release at 30°C, cells were fixed and the percentage of cells with Nsk1 at SPBs (red diamonds) or two separated nuclei (blue squares) was analyzed by fluorescence microscopy. (B) Images of cells from the experiment in (A) either 5 min (cells I and II) or 15 min (cells III and IV) following release to the permissive temperature. Localization of SPBs (blue), kinetochores (red), and Nsk1 (green) are shown both separately and merged. Arrows highlight representative kinetochores with (open arrow) and without (closed arrows) Nsk1. Scale bars: 2 μm. (C) Log-phase cultures of nda3-KM311 ark1-as3 nsk1-gfp cnp1-mcherry sid4-cfp were arrested in mitosis by incubation at 18°C for 6 h. Following addition of 1NMPP1 and continued incubation at 18°C, cells were fixed at intervals and the percentage with SPB-associated Nsk1 (red diamonds) or septa (blue squares) was determined by fluorescence microscopy. (D) Images of a cell from the experiment in (C) 50 min after addition of 1NMPP1. Localization of SPBs (blue), kinetochores (red), and Nsk1 (green) are shown separately and merged. Arrows highlight representative kinetochores with (open arrow) and without (closed arrow) Nsk1.
Recruitment of Nsk1 to the kinetochore–SPB junction does not require microtubules
To determine whether localization of Nsk1 to the kinetochore–SPB junction requires spindle microtubules, we took advantage of the observation that inactivation of Ark1 (Aurora B) kinase in arrested nda3-KM311 ark1-as3 cells by addition of a cell-permeable ATP analogue (1NMPP1) silences the SAC. This leads to cyclin B (Cdc13) destruction and the onset of cytokinesis, even in the absence of microtubules (Vanoosthuyse and Hardwick, 2009). We found that, following addition of 1NMPP1 to arrested nda3-KM311 ark1-as3 nsk1-gfp sid4-cfp cnp1-mcherry cells, Nsk1 appeared at the kinetochore–SPB junction in the absence of microtubules (Figure 4C). Notably, in arrested nda3-KM311 cells, many but not all kinetochores became detached from the SPB. We found that Nsk1 appears strongly at kinetochores that remain bound to SPBs, but not kinetochores that are not associated to SPBs (Figure 4D). We never observed Nsk1 at SPBs that are not associated with kinetochores. These results indicate that Nsk1 binds to the kinetochore–SPB junction after anaphase onset but, paradoxically, loss of Nsk1 influences the timing of anaphase onset. This paradox is resolved later in the paper in Figures 7–9 and the associated text.
Dephosphorylation of Nsk1 by Clp1 controls its relocalization at anaphase onset
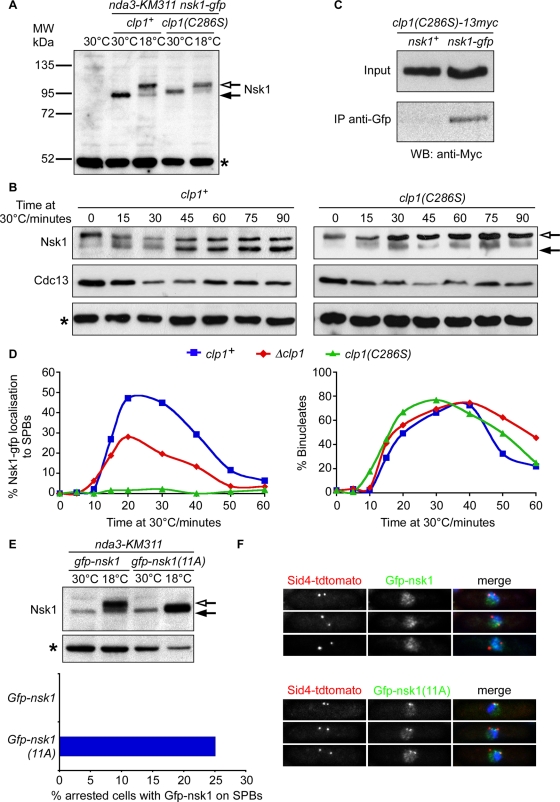
To determine how the localization of Nsk1 changes at anaphase onset, we first examined whether Nsk1 is differentially modified during mitosis. Cell lysates were prepared from either log-phase (30°C) or mitotically arrested nda3-KM311 nsk1-gfp cells (18°C) and analyzed by Western blotting. In arrested cells, additional species of Nsk1 that migrated with a slower electrophoretic mobility were observed by SDS–PAGE, consistent with Nsk1 being phosphorylated (Figure 5A). These slower-migrating species become scarcer, coincidental with reduction in Cdc13 levels, as mitotically arrested nda3-KM311 nsk1-gfp cells enter anaphase following release to the permissive temperature (Figure 5B). This indicates that Nsk1 may be dephosphorylated during anaphase. To test whether Nsk1 is dephosphorylated by the proline-directed Cdc14-like phosphatase Clp1, cell cycle–dependent modification of Nsk1 was examined in nda3-KM311 nsk1-gfp clp1(C286S) cells, which express a substrate-trapping allele of Clp1 that binds, but does not release, its phosphosubstrates (Jia et al., 1995; Chen et al., 2008). In these cells, the proportion of faster-migrating Nsk1 increases only very slowly after release into anaphase, suggesting that Clp1 dephosphorylates Nsk1 (Figure 5, A and B). Consistent with this, we found that Clp1(C286S) coimmunoprecipitates with Nsk1 from cell lysates (Figure 5C).
FIGURE 5:
Dephosphorylation of Nsk1 by Clp1 promotes its association to the kinetochore–SPB junction. (A) Total-cell extracts were prepared from log-phase or mitotically arrested cultures of wild-type, nda3-KM311 nsk1-gfp clp1+, or nda3-KM311 nsk1-gfp clp1(C286S) cells. Proteins were separated by SDS–PAGE and Western blotting and probed with anti-GFP antibodies. Closed arrow indicates major Nsk1-gfp species observed in log-phase extracts. Open arrow indicates slower-migrating species observed in mitotically arrested extracts. Asterisk indicates a 50-kDa nonspecific band also found in the wild-type strain and used as a loading control throughout. (B) Log-phase cultures of nda3-KM311 nsk1-gfp clp1+ or nda3-KM311 nsk1-gfp clp1(C286S) cells were arrested in mitosis by incubation at 18°C for 6 h and then released to the permissive temperature. At the times indicated, total-cell extracts were prepared and probed by Western blotting using anti-GFP antibodies to detect Nsk1-gfp and anti-Cdc13 antibodies to monitor mitotic exit. Open arrow indicates slower-migrating Nsk1-gfp. Closed arrow indicates more rapidly migrating species. Some slower-migrating Nsk1-gfp remains after 90 min at permissive temperature, but note that ∼20% of cells do not enter anaphase under this regime. (C) Extracts were prepared from log-phase cultures of clp1(C286S)-13myc or nsk1-gfp clp1(C286S)-13myc cells and incubated in the presence of anti-GFP antibodies. Extracts and immunoprecipitates were probed by Western blotting using anti-Myc antibodies. (D) Log-phase cultures of nda3-KM311 nsk1-gfp sid4-tdtomato, nda3-KM311 nsk1-gfp sid4-tdtomato Δclp1, and nda3-KM311 nsk1-gfp sid4-tdtomato clp1(C286S) cells were arrested in mitosis by incubation at 18°C for 6 h and then released to the permissive temperature. At each time point, cells were fixed with formaldehyde and the percentage of cells displaying SPB-associated Nsk1 (left panel) or two separated nuclei (right panel) was measured. (E) Log-phase cultures of nmt1:nsk1-gfp Δnsk1 sid4-tdtomato nda3-KM311 or nmt1:nsk1-11A-gfp Δnsk1 sid4-tdtomato nda3-KM311 cells were grown in rich medium (containing thiamine) either at 30°C or arrested in mitosis by incubation at 18°C for 6 h. Total-cell extracts were prepared, and proteins were separated by SDS–PAGE and Western blotting and probed with anti-GFP antibodies as in (A). Alternatively, mitotically arrested cells were fixed, and the percentage of cells with Nsk1 associated to spindle poles was assessed by fluorescence microscopy. (F) Log-phase cultures of nmt1:nsk1-gfp Δnsk1 sid4-tdtomato or nmt1:nsk1-11A-gfp Δnsk1 sid4-tdtomato cells grown in rich medium (containing thiamine) at 30°C were fixed and analyzed by fluorescence microscopy.
To examine whether Clp1 influences the localization of Nsk1, we monitored Nsk1 localization following the release of mitotically arrested nda3-KM311 nsk1-gfp clp1+, nda3-KM311 nsk1-gfp Δclp1, and nda3-KM311 nsk1-gfp clp1(C286S) cells to the permissive temperature. Whereas Nsk1 appeared as two dots coincident with the onset of anaphase in wild-type cells, fewer Nsk1 dots were detected in cells lacking Clp1, and no signal whatsoever was detected in the clp1(C286S) mutant, even though all three strains exited mitosis with approximately the same kinetics (Figure 5D). This suggests that dephosphorylation of Nsk1 by Clp1 is necessary for its relocalization at anaphase onset. However, we noted that Nsk1 migrated at an intermediate mobility in log-phase clp1(C286S) cells between that observed in log-phase, wild-type cells and cells arrested in mitosis, suggesting that another phosphatase may inefficiently dephosphorylate Nsk1 when Clp1 is absent or inactive (Figure 5A).
Cells lacking Clp1 display defects in chromosome biorientation (Trautmann et al., 2004). To assess whether this is due to a failure to trigger kinetochore localization of Nsk1, we monitored the fidelity of chromosome segregation in Δnsk1, Δdis2, and clp1(C286S) single mutants and Δnsk1 clp1(C286S), Δnsk1 Δdis2, and Δdis2 clp1(C286S) double mutants, using a minichromosome loss assay. We observed minichromosome loss in all single mutants and striking synergistic defects in Δnsk1 Δdis2 and Δdis2 clp1(C286S) cells, but not in Δnsk1 clp1(C286S) cells (Table 1). This strongly suggests that the Dis2 and Clp1 phosphatases act in parallel to promote accurate chromosome segregation in fission yeast and that regulated association of Nsk1 to the kinetochore–SPB junction partly explains the role of Clp1 in this process.
Nsk1 contains 11 consensus phosphorylation sites ([ST]PX[RK]) for Cdk1/Cdc2 kinase, several of which are phosphorylated in vivo (Koch et al., 2011; Figure S4). To examine the role of Cdk1/Cdc2 phosphorylation on Nsk1 localization, these phosphorylation sites were mutated to nonphosphorylatable alanine residues, and this nsk1-11A mutant was expressed in cells lacking endogenous nsk1. We found that additional, slower-migrating species of Nsk1 were not observed when nda3-KM311 nsk1-11A cells were blocked in mitosis (Figure 5E). Importantly, unlike wild-type Nsk1, the Nsk1-11A protein appeared at the kinetochore–SPB junction in 25.1% of mitotically arrested nda3-KM311 cells (Figure 5E). In log-phase cultures, Nsk1-11A appeared as punctate dots along the spindle in 41.8% of preanaphase mitotic cells (Figure 5F). These data indicate that Nsk1 is phosphorylated by Cdk1/Cdc2 kinase on multiple sites in early mitosis, and this prevents Nsk1 localization to the spindle and kinetochore–SPB interface until it is dephosphorylated by Clp1 at anaphase onset.
Nsk1 stability is regulated by dynein light chain
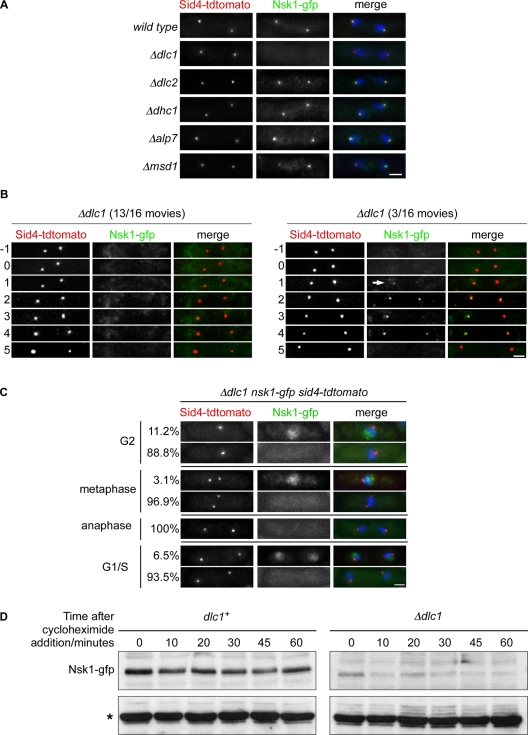
We next examined whether accumulation of Nsk1 at the kinetochore–SPB interface is dependent on known SPB-binding proteins, including Msd1, Alp7, and dynein light chain (Dlc1; Miki et al., 2002; Oliferenko and Balasubramanian, 2002; Sato et al., 2004; Toya et al., 2007). To our surprise, although loss of Msd1 or Alp7 had little or no effect on Nsk1 localization, we were unable to observe Nsk1 at the spindle pole in the absence of Dlc1 in fixed-cell preparations (Figure 6A). This effect was specific to Dlc1, since absence of Dlc2 (another dynein light chain protein) or Dhc1 (dynein heavy chain) had no effect on Nsk1 localization (Figure 6A). To examine this more closely, we filmed Δdlc1 nsk1-gfp sid4-tdtomato cells during anaphase B. Although we were unable to detect Nsk1 at the kinetochore–SPB junction in most movies (13/16), we detected weak Nsk1 signal in other movies (3/16) (Figure 6B and Movie S2). Furthermore, in fixed cells, we also observed substantially reduced localization of Nsk1-gfp to both the nucleolus during interphase and to the nucleus during prometaphase and metaphase, indicating that loss of Dlc1 effects Nsk1 localization throughout the cell cycle (Figure 6C). Importantly, we found that the steady-state level of Nsk1 was lower in Δdlc1 cells than in wild-type cells (Figure S5A). On addition of cycloheximide, Nsk1-gfp levels remained relatively constant over 1 h in the presence of Dlc1 but were further reduced in its absence (Figure 6D). This suggests that Dlc1 is primarily required for the stability of the Nsk1 protein, rather than for its subcellular localization. Consistent with this, overexpressing Nsk1 restores nuclear and spindle localization in the absence of Dlc1 (Figure S5B). Notably, we found an artificial minichromosome is missegregated in 0.04% of Δdlc1 cells, compared with 1.06% of Δnsk1 cells, but only 0.01% of wild-type cells, suggesting that there is sufficient Nsk1 protein in Δdlc1 cells to promote accurate chromosome segregation (Table 1). By contrast, an artificial minichromosome is missegregated in 0.05 ± 0.01% of nsk1-gfp and 0.13 ± 0.02% of nsk1-gfp Δdlc1 cells (Table 1), indicating that although the Nsk1-gfp protein is functional, its stability is more sensitive to loss of Dlc1 than the wild-type Nsk1 protein.
FIGURE 6:
Nsk1 stability requires Dlc1. (A) Log-phase nsk1-gfp sid4-tdtomato cells that were either wild-type or deleted for Dlc1 (Δdlc1), Dlc2 (Δdlc2), Dhc1 (Δdhc1), Alp7/Mia1 (Δalp7), or Msd1 (Δmsd1) were fixed, and Nsk1 localization was examined in anaphase B cells. Scale bars: 2μm. (B) Images from two representative movies of Δdlc1 nsk1-gfp sid4-tdtomato cells during anaphase B taken at 1-min intervals. Arrow indicates weak Nsk1-gfp signal in Δdlc1 cells. Time zero is the end of anaphase A. (C) Log-phase Δdlc1 nsk1-gfp sid4-tdtomato cells were fixed, and Nsk1-gfp localization was assessed at different cell cycle stages. (D) Log-phase nsk1-gfp and Δdlc1 nsk1-gfp cells were treated with 100 μg/ml cycloheximide. Samples were taken at the times indicated and analyzed by Western blotting. Asterisk indicates nonspecific 50-kDa band used as a loading control.
Nsk1 promotes the association of kinetochores to SPBs during anaphase B
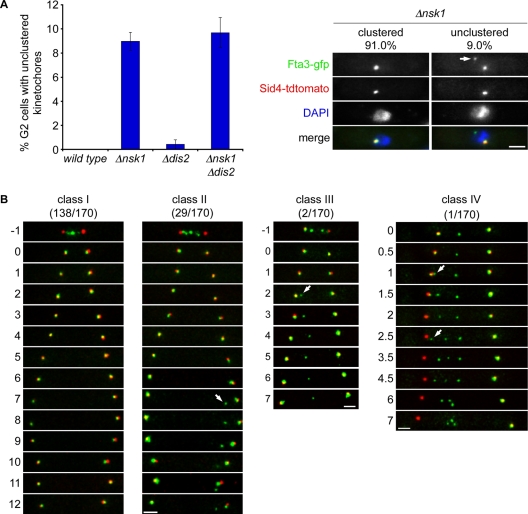
To understand how Nsk1 influences chromosome segregation, we next examined kinetochore (Fta3) and SPB (Sid4) position in fixed-cell populations. In 9.0% of interphase Δnsk1 cells, one pair of sister kinetochores failed to cluster near the SPB during interphase (Figure 7A). By contrast loss of Dis2 had only a marginal effect on kinetochore clustering in the presence or absence of Nsk1 (Figure 7A). Similar results were obtained when kinetochore position was assessed with other GFP-tagged kinetochore proteins and in clp1(C286S) mutant, in which relocalization of Nsk1 to the kinetochore–SPB junction is impaired (Figure S6A). To investigate how unclustered kinetochores are generated in the absence of Nsk1, we filmed Δnsk1 fta3-gfp sid4-tdtomato cells during anaphase B, since this is when Nsk1 can be observed at the kinetochore–SPB junction. In 138/170 (81.2%) movies no obvious defect in kinetochore behavior could be detected, compared with wild-type cells (Figure 7B, class I, and Movie S3). However, in 29/170 (17.1%) movies, sister-chromatid separation and early anaphase B were normal, but one kinetochore became detached from one of the two SPBs late in anaphase B and remained detached during the following interphase (Figures 7B, class II, and S6B and Movie S4). This phenotype, which has not been observed in more than 200 movies of wild-type cells, accounts for the percentage of unclustered kinetochores observed in log-phase populations of Δnsk1 cells (Figure 7A). More strikingly, in two movies (1.2%), sister-chromatid separation occurred normally, but one kinetochore became detached from the SPB in early anaphase B and was not segregated to either pole (Figure 7B, class III, and Movie S5). In one extreme example, we filmed a cell in which all three kinetochores became sequentially detached from one of the two SPBs (Figure 7B, class IV, and Movie S6). This strongly suggests that Nsk1 is required for accurate chromosome segregation, promoting the tethering of kinetochores to SPBs during anaphase B (see Discussion).
FIGURE 7:
Nsk1 promotes association of kinetochores to SPBs during anaphase B. (A) Log-phase cultures of wild-type, Δnsk1, Δdis2, and Δnsk1 Δdis2 cells expressing fta3-gfp, and sid4-tdtomato cells were fixed, and the percentage of interphase cells with kinetochores that were not clustered near the SPB was determined by fluorescence microscopy (left panel). Images of Δnsk1 fta3-gfp sid4-tdtomato cells with either clustered (left column) or unclustered (right column) kinetochores. Arrow indicates an unclustered kinetochore. Scale bars: 2 μm. (B) Images from four representative movies of individual Δnsk1 fta3-gfp sid4-tdtomato cells during anaphase B. In the first three movies, time zero is the end of anaphase A. In the fourth movie, imaging started partway through anaphase B. Arrows indicate the time of initial detachment of kinetochores from SPBs. Time is shown in minutes.
Unclustered kinetochores are retrieved in the absence of Nsk1, but this process delays the onset of anaphase
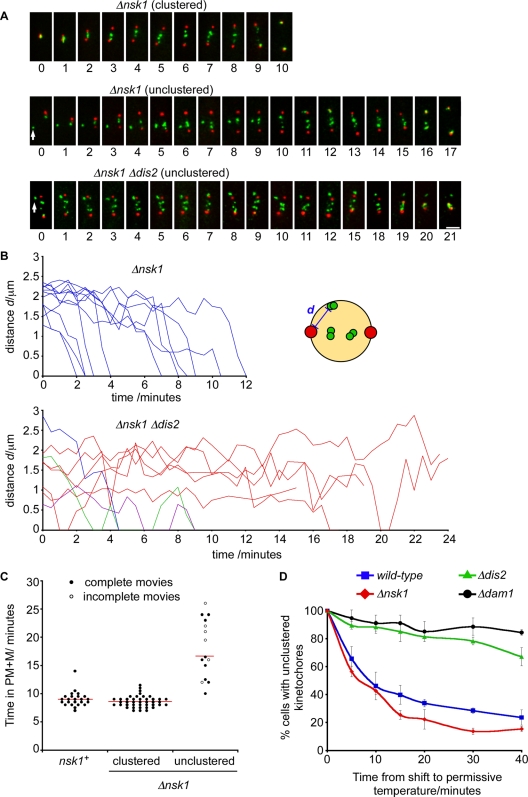
To examine the fate of unclustered kinetochores, we filmed fta3-gfp sid4-tdtomato cells and Δnsk1 fta3-gfp sid4-tdtomato cells that had both clustered and unclustered kinetochores (Figure 8A and Movies S7 and S8, respectively). Without exception, we found that, in the absence of Nsk1, unclustered kinetochores were retrieved to one or the other spindle pole with kinetics comparable to those reported in other studies (0.6–2.0 μm/min; Figure 8B; Grishchuk and McIntosh, 2006; Franco et al., 2007). However, we noted that Δnsk1 cells that had unclustered kinetochores during interphase spent on average more time in prometaphase and metaphase than Δnsk1 cells with clustered kinetochores (Figure 8, A and C). Conversely, Δnsk1 cells with clustered kinetochores spent on average the same time in prometaphase and metaphase as wild-type cells (Figure 8C). This strongly suggests that the delay in anaphase onset observed in log-phase Δnsk1 cultures (Figure 1, D and E) is due to the extra time required to capture, retrieve, and biorient unclustered kinetochores during the subsequent mitosis. These data also demonstrate that Nsk1 is not required for the initial capture, retrieval, or biorientation of kinetochores during prometaphase or metaphase, consistent with the observation that Nsk1 only binds the kinetochore–SPB junction after anaphase onset.
FIGURE 8:
Retrieval of unclustered kinetochores delays the onset of anaphase and requires PP1Dis2. (A) Images from two representative movies of mitotic Δnsk1 fta3-gfp sid4-tdtomato cells that entered mitosis with either clustered kinetochores (top panels) or an unclustered kinetochore (middle panels). Time zero is the point of pole separation. Images from a representative movie of a mitotic Δnsk1 Δdis2 fta3-gfp sid4-tdtomato cell that entered mitosis with an unclustered kinetochore pair (bottom panels). This cell was already in mitosis when imaging began. Images were taken at 1-min intervals. Arrows indicate unclustered kinetochores. (B) Movies of Δnsk1 fta3-gfp sid4-tdtomato and Δdis2 Δnsk1 fta3-gfp sid4-tdtomato cells with unclustered kinetochores during mitosis were analyzed, and the distance (“d” in the schematic) from kinetochore (green) to SPB (red) was measured in each frame. Blue lines represent retrieval events in cells that subsequently go through anaphase. Red lines represent the paths of kinetochores in cells where spindles subsequently collapse (see Figure 8A and Movie S5). Green and purple lines represent kinetochores that are retrieved but subsequently lost and retrieved again before proceeding into anaphase. Note: we cannot confirm whether the same kinetochores are lost in each case. (C) The length of time individual fta3-gfp sid4-tdtomato and Δnsk1 fta3-gfp sid4-tdtomato cells spent in prometaphase and metaphase (PM + M) was determined by live-cell imaging. Cells lacking Nsk1 were divided into those that entered mitosis with at least one unclustered kinetochore (unclustered) and those that did not (clustered). Closed circles represent movies where mitotic entry and exit can be assessed. Open circles represent movies where either entry or exit from mitosis are absent. The mean duration of completed mitoses from each class is represented by a horizontal line. (D) Log-phase cultures of wild-type, Δnsk1, Δdis2, and Δdam1 cells expressing nda3-KM311 dad1-gfp sid4-tdtomato were arrested in mitosis, as before. Kinetochore recapture was assayed as described in Materials and Methods. Error bars are the SD from the mean of three independent experiments.
Dis2 is required for the retrieval of unclustered kinetochores
We initially identified Nsk1 based on the additive growth defect of Δnsk1 Δdis2 cells (Figure 1A and Table 1). Although both Δnsk1 and Δdis2 cells individually display defects in chromosome segregation, we wished to explain this synthetic phenotype. To do this, we filmed Δnsk1 Δdis2 cells during mitosis. We found that unclustered kinetochores are seldom retrieved to the SPB in mitotic Δnsk1 Δdis2 cells. In cells that fail to retrieve their kinetochores, anaphase does not take place and spindles eventually collapse (Figure 8, A and B, and Movie S9). Interestingly, in some Δnsk1 Δdis2 cells, unclustered kinetochores appear to be retrieved, lost, and retrieved again (Figure 8B). These data suggest that Dis2, like the DASH complex, is required both for kinetochore retrieval and for the establishment of end-on attachments at the spindle pole (Franco et al., 2007; Gachet et al., 2008; Vanoosthuyse et al., 2009). Consistent with this, Δnsk1 Δdam1 cells display comparable growth defects to Δnsk1 Δdis2 (unpublished data). To further clarify this, we monitored kinetochore and SPB position following release of mitotically arrested nda3-KM311 cells to the permissive temperature. At the arrest point, kinetochores were displaced from SPBs and needed to be recaptured and retrieved to the spindle pole before they were bioriented on the mitotic spindle. Whereas cells lacking Nsk1 displayed no defect in kinetochore retrieval, cells lacking either Dis2 or Dam1 were substantially impaired (Figure 8D). Thus the failure to recapture unclustered kinetochores in Δnsk1 Δdis2 cells is likely to be a major contributory factor to the slow-growth phenotype of this double mutant.
DISCUSSION
PP1 antagonizes Aurora B at the kinetochore to stabilize tension-bearing kinetochore–microtubule attachments and silence the SAC (Pinsky et al., 2006, 2009; Vanoosthuyse and Hardwick, 2009; Liu et al., 2010). We found that fission yeast Δdis2 cells, which lack one of two type 1 phosphatases, suppress the temperature sensitivity of ark1-T7 mutants, are defective in kinetochore retrieval, missegregate chromosomes at high frequency, and are delayed in the timing of anaphase onset. We screened for new factors that exacerbate these effects and identified Nsk1, a novel protein that promotes kinetochore–SPB association in anaphase. Nsk1 localizes to the nucleolus during interphase and redistributes throughout the nucleoplasm from mitotic entry until anaphase onset. Nsk1 appears on the axis of the anaphase A spindle and then at the inner face of SPBs during anaphase B. Importantly, we did not see Nsk1 on kinetochores of lagging sister chromatids, indicating Nsk1 is not a kinetochore protein sensu stricto. Likewise, Nsk1 never colocalizes with SPBs during anaphase B, unless they are associated with at least one kinetochore, indicating Nsk1 is not simply an SPB-associated protein. Rather, we found Nsk1 accumulates at the kinetochore–SPB junction during anaphase B.
Our data demonstrate that Nsk1 is phosphorylated in mitosis and dephosphorylated during anaphase, coincident with its relocalization to the kinetochore–SPB junction. Both dephosphorylation of Nsk1 and its relocalization are disrupted in clp1(C286S) cells, suggesting that dephosphorylation of Nsk1 on proline-directed sites controls its relocalization. Consistent with this, recent phosphoproteome analysis has shown that Nsk1 is highly phosphorylated during mitosis and, although only 38% of the Nsk1 sequence is covered in this study, many of the reported Nsk1 phosphorylation sites correspond to Cdk1/Cdc2 consensus sites (Koch et al., 2011; Figure S4). We demonstrate that mutation of these sites to nonphosphorylatable alanine residues largely abolishes the mobility shift of Nsk1 on SDS–PAGE and causes premature localization of Nsk1 to the preanaphase mitotic spindle and SPB. These data strongly suggest that phosphorylation of Nsk1 on consensus Cdk1/Cdc2 sites prevents its localization during prometaphase and metaphase and dephosphorylation of Nsk1 on these sites, at least in part by Clp1, promotes its association to the spindle and kinetochore–SPB junction during anaphase B. At present, it is unclear why Nsk1 is sequestered in the nucleolus during interphase or what triggers its export to the nucleoplasm at the G2/M transition or its return to the nucleolus at the end of anaphase B.
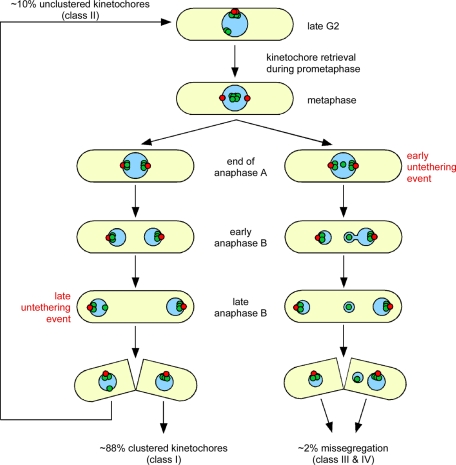
Δnsk1 cells display an increased rate of chromosome missegregation. However, unlike Dis2, Nsk1 is not required for the capture, retrieval, or biorientation of sister chromatids and does not oppose Ark1 activity (Figure S7). So why is the SAC activated in some Δnsk1 cells? In wild-type cells, kinetochores cluster in an area underlying the SPB during interphase (Funabiki et al., 1993). This clustering is disrupted in around 9% of Δnsk1 cells. We found that only Δnsk1 cells that enter mitosis with unclustered kinetochores are delayed in the timing of anaphase, compared with wild-type cells. This suggests unclustered kinetochores activate the SAC in early mitosis until they are captured, retrieved, and bioriented by spindle microtubules. We conclude that the delay in anaphase onset observed in some Δnsk1 cells results from a failure to properly cluster kinetochores during the previous mitosis (see model in Figure 9).
FIGURE 9:
Model to explain the different fates of clustered and unclustered kinetochores (green) relative to the position of spindle poles (red) in the absence of Nsk1. Please refer to the text for an explanation.
In most Δnsk1 cells, chromosomes segregate normally to spindle poles during anaphase B. However, in ∼17% of cells, one or more kinetochores become detached from one of the two SPBs. If detachment occurs late in anaphase B, the chromosome remains in the correct nucleus, but has to be captured and retrieved during the following prometaphase (Figure 9). Infrequently, kinetochores become detached from SPBs in early anaphase B, causing chromosome missegregation (Figure 9). These data suggest the primary role of Nsk1 in accurate chromosome segregation is in promoting association of kinetochores with SPBs during anaphase B. Other factors, such as Ima1, must be necessary for maintenance of kinetochore clustering during interphase, since spindles disassemble, SPBs defenestrate from the nuclear envelope, and Nsk1 returns to the nucleolus at the end of anaphase B (Hagan and Hyams, 1988; Ding et al., 1997; King et al., 2008). We found that retrieval of unclustered kinetochores requires Dis2. This is likely to be a major contributory factor to the slow-growth phenotype of Δdis2 Δnsk1 double mutants. The frequency of detachment of kinetochores from SPBs during early anaphase B may also be increased in Δnsk1 Δdis2 mutants; however, the percentage of dead cells in log-phase cultures makes this difficult to assess by live-cell imaging. Regardless, these data suggest that kinetochore clustering aids chromosome biorientation in yeast.
So what is the molecular function of Nsk1? One possibility is that Nsk1 physically tethers kinetochores to SPBs. This seems unlikely, as Nsk1 is a low-abundance protein. Indeed, the stability of Nsk1 requires Dlc1, and Nsk1-gfp protein is barely detectable by fluorescence microscopy in the absence of Dlc1. Nevertheless, Δdlc1 cells display relatively mild chromosome segregation and kinetochore-unclustering defects, suggesting even a small amount of Nsk1 at the kinetochore–SPB junction is sufficient for its function. This is incongruous with the idea of Nsk1 being a physical tether. An alternative possibility is that Nsk1 acts to regulate a tether. Nsk1 shares some phenotypic similarities with S. cerevisiae protein Fin1, which is negatively regulated by Cdk1-dependent phosphorylation (Woodbury and Morgan, 2007). Fin1 targets PP1 to kinetochores and ectopically silences the SAC when overexpressed (Akiyoshi et al., 2009). Interestingly, Nsk1 and its S. cryophilus and S. octosporus homologues contain consensus PP1-binding motifs ([K/R]X(0–1)VX[F/W]) in their C-termini (Rhind et al., 2011; Figure S1). However, we could not detect interaction between Nsk1 and Dis2 by coimmunoprecipitation or yeast two-hybrid screening. Moreover, mutation of this site does not influence Nsk1 function in vivo (Figure S7). The mechanism of Nsk1 action therefore remains unclear. Identifying additional Nsk1-interacting partners will be key for determining how Nsk1 links kinetochores to SPBs during anaphase B.
Although no obvious structural Nsk1 homologues exist outside fission yeasts, it is interesting to speculate whether Nsk1 function is conserved. In mammalian cells, a complex containing astrin, SKAP (also known as kinastrin), and dynein light chain (Tctex-1/LC8) is recruited to the kinetochore upon the establishment of correct biorientation to selectively stabilize microtubule attachments at these kinetochores (Thein et al., 2007; Manning et al., 2010; Schmidt et al., 2010). Separation of segregated chromosomes is largely dependent on microtubule-mediated anaphase A movement in mammalian cells. In yeast, elongation of the anaphase B spindle, which increases from 2 to 14 μm in fission yeast, has a more major role, and this is thought to aid the resolution of merotelic kinetochore–microtubule attachments (Courtheoux et al., 2009). We suggest that the kinetochores of segregated sister chromatids in yeast need to be tightly tethered, not only to microtubules, but also to SPBs, to prevent chromosome missegregation during early anaphase B. Although Nsk1 is not a direct astrin or SKAP homologue, we suggest regulated recruitment of Nsk1 to the kinetochore may serve an analogous role. It is intriguing in this respect that fission yeast Dlc1 is the direct homologue of Tctex-1/LC8, a component of the astrin/SKAP complex (Miki et al., 2002; Schmidt et al., 2010). Regardless, our finding that association of Nsk1 to the kinetochore is dependent on Clp1 provides at least a partial explanation why this conserved phosphatase is required for accurate chromosome segregation in fission yeast.
MATERIALS AND METHODS
Genome-wide screen for synthetic sick interactions with Δdis2 mutants
The PEM-2 (Pombe Epistasis Mapper-2) method (Roguev et al., 2007) was applied with some minor modifications (see Strain Construction) to carry out genetic crosses between a strain lacking PP1Dis2 (dis2::hygR) and the collection of all deletions of nonessential genes in S. pombe (Bioneer, version 1). A Singer RoToR HAD station (kindly made available by the Tyers Lab, Wellcome Trust Center for Cell Biology, Edinburgh) was used to array the cells so that each cross was duplicated (768 independent spots per plate). After mating on SPAS for 4 d at 30°C, cells were transferred onto YES (yeast extract with supplements) medium containing 100 μg/ml G418, 20 μg/ml hygromycin B, and 100 μg/ml cycloheximide. After 2–3 d at 30°C, cells were transferred onto YES medium containing 100 μg/ml G418, 100 μg/ml hygromycin B, 100 μg/ml cycloheximide, and 1 mg/ml phloxin B and grown for another 2 d at 30°C. We found that addition of phloxin B helped with the identification of synthetic sick interactions, in which the double mutant is viable but grows poorly. The screen was repeated three times with Δdis2 as bait and a list of the 38 genes whose deletion gave a strong synthetic sick phenotype is shown in Table SI.
Cell culture
Media, growth, and maintenance of strains were as previously described (Moreno et al., 1991). Strains used in this study are listed in Table SII. All experiments were performed at 30°C, unless otherwise stated.
Strain construction
Deletion of nsk1 and carboxy-terminal tagging of Nsk1 with GFP were performed by two-step PCR-based gene targeting, as previously described (Bahler et al., 1998; Krawchuk and Wahls, 1999). Amino-terminal tagging of Nsk1 was achieved using Gateway Technology (Invitrogen, Carlsbad, CA). pDONR was used as the entry vector and pLYS1U-HFG1c was used as the destination vector. Correct clones were verified by sequencing. Integration and verification were as previously described (Matsuyama et al., 2008). nsk1-11A was synthesized by GeneArt (Invitrogen), introducing alanine residues at the following sites: S48, S71, S103, S113, S140, S171, S195, S236, S304, T381, and T437 and the open reading frame (ORF) cloned into pLYS1U-HFG1c using Gateway Technology. The nsk1-V390A, W392A mutation was made using the Phusion site-directed mutagenesis kit (Thermo Scientific, Lafayette, CO) according to the manufacturer's instructions. In all gene deletions (Δ), the entire ORF was removed. The genotypes of the strains used in this study are detailed in Table SII. A list of oligonucleotides used is provided in Table SIII.
Fluorescence microscopy
Analysis of live cells was performed in an imaging chamber (CoverWell PCI-2.5; Grace Bio-Labs, Bend, OR) filled with 1 ml of 1% agarose in minimal medium and sealed with a 22-mm × 22-mm glass coverslip. Fluorescence imaging was performed on a Nikon TE-2000 inverted microscope with a 100×, 1.49 numerical aperture objective lens equipped with a Photometrics Coolsnap-HQ2 liquid-cooled CCD camera (Photometrics, Tucson, AZ). Images were collected and analyzed using MetaMorph (version 7.5.2.0; Molecular Devices, Sunnyvale, CA). Stacks of six z-sections (0.6 μm apart) were taken at each time point, and exposure times of 1 s for GFP, tdTomato, or mCherry and 0.25 s for DAPI were used. Maximum-intensity projections were made for each time point, which was followed by intensity adjustments and conversion to 24-bit TIFF images.
Fluorescence levels for high-resolution localization were measured in MetaMorph using the linescan option set at one pixel width for a 2-μm length. Chromatic aberrations were minimized by using a single dichroic filter for all channels and by observing spindle poles and kinetochores in various orientations to correct for any pixel shift. The pattern of localization was maintained in all imaged cells. Fluorescence levels for relative intensities were measured using Metamorph by setting an active region circle with a diameter of five pixels (90 nm2). Average intensities within this region were calculated, and the average cytoplasmic fluorescence background was subtracted.
Measurement of mitotic index
Mid-log cdc13-gfp, fta4-gfp sid4-tdtomato, or dad1-gfp sid4-tdtomato cells were fixed in 3.7% formaldehyde for 10 min and mounted in medium containing 4′,6-diamidino-2-phenylindole (DAPI; to label DNA). Stacks of 16 z-sections (0.2 μm apart) were taken and projected images were made for each time point, which was followed by intensity adjustments. The percentage of cells with Cdc13-gfp on separated SPBs or the percentage of cells with kinetochores in between separated SPBs was determined.
Minichromosome loss assay
Minichromosome loss in cells bearing the Ch16 (ade6-M216) minichromosome and ade6-M210 allele was assayed as previously described (Niwa et al., 1989). Cells were grown to mid-log phase in Edinburgh minimal medium (EMM) lacking adenine and then plated onto yeast extract agar containing no additional adenine for 3 d at 30°C. Half-sectored, or greater than half-sectored, pink colonies were scored.
Chromosome retrieval assay
The retrieval of kinetochores following release of mitotically arrested nda3-KM311 cells to the permissive temperature was measured exactly as previously described (Vanoosthuyse et al., 2009).
Microtubule-independent localization assay
Mid-log-phase ark1-as3 nda3-KM311 nsk1-gfp cnp1-mcherry sid4-cfp cells were arrested in early mitosis in liquid cultures by shifting to 18°C for 6 h followed by addition of 5 μM 1NMPP1 (1-naphthylmethyl-4-amino-1-tert-butyl-3-(p-methylphenyl)pyrazolo[3,4-d]pyrimidine). Cells were fixed in 3.7% formaldehyde mounted in medium containing DAPI (to label DNA) and calcofluor (to label cell walls and septa). For each treatment condition and for each time point, 200–300 cells were analyzed.
Spindle checkpoint silencing assay
Spindle checkpoint silencing assay was done exactly as described (Meadows et al., 2011).
Immunoprecipitation and Western blotting
Proteins were extracted in HB buffer (25 mM MOPS, pH 7.2, 150 mM KCl, 15 mM MgCl2, 15 mM ethylene glycol tetraacetic acid, 1 mM dithiothreitol, 0.1% Triton, 0.1 mM sodium vanadate, 60 mM β-glycerophosphate, complete protease inhibitor cocktail [EDTA free; Roche, Indianapolis, IN], 1 mM phenylmethylsulfonyl fluoride) from ∼2 × 108 cells. Cleared extracts were then incubated for 3 h at 4°C with sheep anti-GFP antibody prebound to protein G. The beads were then washed three times with HB buffer and once with phosphate-buffered saline containing 0.02% Tween. The immunoprecipitated complexes were then analyzed by immunoblot using anti-Myc antibody (9E10; Cambridge Bioscience, Cambridge, UK). For Western blotting of total-cell extracts, cells were lysed in 20% trichloroacetic acid and precipitated proteins were solubilized in SDS sample buffer. Mitotic exit was assayed using purified anti-Cdc13 antibodies (a gift from Paul Nurse).
Supplementary Material
Acknowledgments
We thank Hille Tekotte and Mike Tyers for assistance and use of the Singer RoToR HAD station. We thank Mohan Balasubramanian, Iain Hagan, Silke Hauf, Xiangwei He, Yasushi Hiraoka, Jean-Paul Javerzat, Dan McCollum, Paul Nurse, Luis Rockeach, Paul Russell, Takashi Toda, and Mitsuhiro Yanagida for strains and reagents, and Jacqueline Hayles for deriving the haploid deletion collection. We thank Silke Hauf and Kathy Gould for sharing data prior to publication. This work was supported by a program grant from the Medical Research Council to J.B.A.M. and a Wellcome Trust program grant to K.G.H.
Abbreviations used:
- Clp1
Cdc14-like phosphatase
- CPC
chromosomal passenger complex
- DAPI
4′,6-diamidino-2-phenylindole
- Dlc1
dynein light chain
- GFP
green fluorescent protein
- ORF
open reading frame
- PP1
type 1 phosphatase
- PP2A
type 2A phosphatases
- SAC
spindle assembly checkpoint
- SPB
spindle pole body
- YES
yeast extract with supplements
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E11-07-0608) on September 30, 2011.
REFERENCES
- Akiyoshi B, Nelson CR, Ranish JA, Biggins S. Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 2009;23:2887–2899. doi: 10.1101/gad.1865909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu JQ, Longtine MS, Shah NG, McKenzie A III, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Beauregard PB, Guerin R, Turcotte C, Lindquist S, Rokeach LA. A nucleolar protein allows viability in the absence of the essential ER-residing molecular chaperone calnexin. J Cell Sci. 2009;122:1342–1351. doi: 10.1242/jcs.040949. [DOI] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohnert KA, Chen JS, Clifford DM, Vander Kooi CW, Gould KL. A link between aurora kinase and Clp1/Cdc14 regulation uncovered by the identification of a fission yeast borealin-like protein. Mol Biol Cell. 2009;20:3646–3659. doi: 10.1091/mbc.E09-04-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR III, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Chen CT, Feoktistova A, Chen JS, Shim YS, Clifford DM, Gould KL, McCollum D. The SIN kinase Sid2 regulates cytoplasmic retention of the S. pombe Cdc14-like phosphatase Clp1. Curr Biol. 2008;18:1594–1599. doi: 10.1016/j.cub.2008.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G, Scholey JM. Mitotic force generators and chromosome segregation. Cell Mol Life Sci. 2010;67:2231–2250. doi: 10.1007/s00018-010-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courtheoux T, Gay G, Gachet Y, Tournier S. Ase1/Prc1-dependent spindle elongation corrects merotely during anaphase in fission yeast. J Cell Biol. 2009;187:399–412. doi: 10.1083/jcb.200902093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R, West RR, Morphew DM, Oakley BR, McIntosh JR. The spindle pole body of Schizosaccharomyces pombe enters and leaves the nuclear envelope as the cell cycle proceeds. Mol Biol Cell. 1997;8:1461–1479. doi: 10.1091/mbc.8.8.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco A, Meadows JC, Millar JB. The Dam1/DASH complex is required for the retrieval of unclustered kinetochores in fission yeast. J Cell Sci. 2007;120:3345–3351. doi: 10.1242/jcs.013698. [DOI] [PubMed] [Google Scholar]
- Funabiki H, Hagan I, Uzawa S, Yanagida M. Cell cycle-dependent specific positioning and clustering of centromeres and telomeres in fission yeast. J Cell Biol. 1993;121:961–976. doi: 10.1083/jcb.121.5.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet Y, Reyes C, Courtheoux T, Goldstone S, Gay G, Serrurier C, Tournier S. Sister kinetochore recapture in fission yeast occurs by two distinct mechanisms, both requiring Dam1 and Klp2. Mol Biol Cell. 2008;19:1646–1662. doi: 10.1091/mbc.E07-09-0910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grishchuk EL, McIntosh JR. Microtubule depolymerization can drive poleward chromosome motion in fission yeast. EMBO J. 2006;25:4888–4896. doi: 10.1038/sj.emboj.7601353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan IM, Hyams JS. The use of cell division cycle mutants to investigate the control of microtubule distribution in the fission yeast Schizosaccharomyces pombe. J Cell Sci. 1988;89:343–357. doi: 10.1242/jcs.89.3.343. [DOI] [PubMed] [Google Scholar]
- Higuchi T, Uhlmann F. Stabilization of microtubule dynamics at anaphase onset promotes chromosome segregation. Nature. 2005;433:171–176. doi: 10.1038/nature03240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiraoka Y, Toda T, Yanagida M. The NDA3 gene of fission yeast encodes β-tubulin: a cold-sensitive nda3 mutation reversibly blocks spindle formation and chromosome movement in mitosis. Cell. 1984;39:349–358. doi: 10.1016/0092-8674(84)90013-8. [DOI] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Barford D, Flint AJ, Tonks NK. Structural basis for phosphotyrosine peptide recognition by protein tyrosine phosphatase 1B. Science. 1995;268:1754–1758. doi: 10.1126/science.7540771. [DOI] [PubMed] [Google Scholar]
- Jin Q, Trelles-Sticken E, Scherthan H, Loidl J. Yeast nuclei display prominent centromere clustering that is reduced in nondividing cells and in meiotic prophase. J Cell Biol. 1998;141:21–29. doi: 10.1083/jcb.141.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin QW, Fuchs J, Loidl J. Centromere clustering is a major determinant of yeast interphase nuclear organization. J Cell Sci. 2000;113:1903–1912. doi: 10.1242/jcs.113.11.1903. [DOI] [PubMed] [Google Scholar]
- Joglekar AP, Bouck D, Finley K, Liu X, Wan Y, Berman J, He X, Salmon ED, Bloom KS. Molecular architecture of the kinetochore-microtubule attachment site is conserved between point and regional centromeres. J Cell Biol. 2008;181:587–594. doi: 10.1083/jcb.200803027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, La Terra S, Chang F. Laser microsurgery in fission yeast; role of the mitotic spindle midzone in anaphase B. Curr Biol. 2004;14:1330–1340. doi: 10.1016/j.cub.2004.07.028. [DOI] [PubMed] [Google Scholar]
- King MC, Drivas TG, Blobel G. A network of nuclear envelope membrane proteins linking centromeres to microtubules. Cell. 2008;134:427–438. doi: 10.1016/j.cell.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura E, Tanaka K, Kitamura Y, Tanaka TU. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007;21:3319–3330. doi: 10.1101/gad.449407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch A, Krug K, Pengelley S, Macek B, Hauf S. Mulitple substrates of the mitotic kinase Aurora with roles in chromatin regulation identified through quantitative phosphoproteomics of fission yeast. Sci Signal. 2011;4:rs6. doi: 10.1126/scisignal.2001588. [DOI] [PubMed] [Google Scholar]
- Krapp A, Schmidt S, Cano E, Simanis V. S. pombe cdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr Biol. 2001;11:1559–1568. doi: 10.1016/s0960-9822(01)00478-x. [DOI] [PubMed] [Google Scholar]
- Krawchuk MD, Wahls WP. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast. 1999;15:1419–1427. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1419::AID-YEA466>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Vleugel M, Backer CB, Hori T, Fukagawa T, Cheeseman IM, Lampson MA. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J Cell Biol. 2010;188:809–820. doi: 10.1083/jcb.201001006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallavarapu A, Sawin K, Mitchison T. A switch in microtubule dynamics at the onset of anaphase B in the mitotic spindle of Schizosaccharomyces pombe. Curr Biol. 1999;9:1423–1426. doi: 10.1016/s0960-9822(00)80090-1. [DOI] [PubMed] [Google Scholar]
- Manning AL, Bakhoum SF, Maffini S, Correia-Melo C, Maiato H, Compton DA. CLASP1, astrin and Kif2b form a molecular switch that regulates kinetochore-microtubule dynamics to promote mitotic progression and fidelity. EMBO J. 2010;29:3531–3543. doi: 10.1038/emboj.2010.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama A, Shirai A, Yoshida M. A novel series of vectors for chromosomal integration in fission yeast. Biochem Biophys Res Commun. 2008;374:315–319. doi: 10.1016/j.bbrc.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Meadows JC, Millar J. Latrunculin A delays anaphase onset in fission yeast by disrupting an Ase1-independent pathway controlling mitotic spindle stability. Mol Biol Cell. 2008;19:3713–3723. doi: 10.1091/mbc.E08-02-0164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows JC, Shepperd LA, Vanoosthuyse V, Lancaster TC, Sochaj AM, Buttrick GJ, Hardwick KG, Millar JBA. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin 8 motors. Dev Cell. 2011;20:739–750. doi: 10.1016/j.devcel.2011.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki F, Okazaki K, Shimanuki M, Yamamoto A, Hiraoka Y, Niwa O. The 14-kDa dynein light chain-family protein Dlc1 is required for regular oscillatory nuclear movement and efficient recombination during meiotic prophase in fission yeast. Mol Biol Cell. 2002;13:930–946. doi: 10.1091/mbc.01-11-0543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshull J, Straight A, Rudner AD, Dernburg AF, Belmont A, Murray AW. Protein phosphatase 2A regulates MPF activity and sister chromatid cohesion in budding yeast. Curr Biol. 1996;6:1609–1620. doi: 10.1016/s0960-9822(02)70784-7. [DOI] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nabeshima K, Nakagawa T, Straight AF, Murray A, Chikashige Y, Yamashita YM, Hiraoka Y, Yanagida M. Dynamics of centromeres during metaphase-anaphase transition in fission yeast: Dis1 is implicated in force balance in metaphase bipolar spindle. Mol Biol Cell. 1998;9:3211–3225. doi: 10.1091/mbc.9.11.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa O, Matsumoto T, Chikashige Y, Yanagida M. Characterization of Schizosaccharomyces pombe minichromosome deletion derivatives and a functional allocation of their centromere. EMBO J. 1989;8:3045–3052. doi: 10.1002/j.1460-2075.1989.tb08455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H, Kinoshita N, Miyatani S, Toda T, Yanagida M. The fission yeast dis2+ gene required for chromosome disjoining encodes one of two putative type 1 protein phosphatases. Cell. 1989;57:997–1007. doi: 10.1016/0092-8674(89)90338-3. [DOI] [PubMed] [Google Scholar]
- Oliferenko S, Balasubramanian MK. Astral microtubules monitor metaphase spindle alignment in fission yeast. Nat Cell Biol. 2002;4:816–820. doi: 10.1038/ncb861. [DOI] [PubMed] [Google Scholar]
- Pinsky BA, Kotwaliwale CV, Tatsutani SY, Breed CA, Biggins S. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol Cell Biol. 2006;26:2648–2660. doi: 10.1128/MCB.26.7.2648-2660.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky BA, Nelson CR, Biggins S. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr Biol. 2009;19:1182–1187. doi: 10.1016/j.cub.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhind N, et al. Comparative functional genomics of the fission yeasts. Science. 2011;332:930–936. doi: 10.1126/science.1203357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder CL, Alexander SP. Kinetochores are transported poleward along a single astral microtubule during chromosome attachment to the spindle in newt lung cells. J Cell Biol. 1990;110:81–95. doi: 10.1083/jcb.110.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Wiren M, Weissman JS, Krogan NJ. High-throughput genetic interaction mapping in the fission yeast Schizosaccharomyces pombe. Nat Methods. 2007;4:861–866. doi: 10.1038/nmeth1098. [DOI] [PubMed] [Google Scholar]
- Rosenberg JS, Cross FR, Funabiki H. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr Biol. 2011;21:942–947. doi: 10.1016/j.cub.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M, Vardy L, Angel Garcia M, Koonrugsa N, Toda T. Interdependency of fission yeast Alp14/TOG and coiled coil protein Alp7 in microtubule localization and bipolar spindle formation. Mol Biol Cell. 2004;15:1609–1622. doi: 10.1091/mbc.E03-11-0837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt JC, Kiyomitsu T, Hori T, Backer CB, Fukagawa T, Cheeseman IM. Aurora B kinase controls the targeting of the Astrin-SKAP complex to bioriented kinetochores. J Cell Biol. 2010;191:269–280. doi: 10.1083/jcb.201006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Kitamura E, Kitamura Y, Tanaka TU. Molecular mechanisms of microtubule-dependent kinetochore transport toward spindle poles. J Cell Biol. 2007;178:269–281. doi: 10.1083/jcb.200702141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Mukae N, Dewar H, van Breugel M, James EK, Prescott AR, Antony C, Tanaka TU. Molecular mechanisms of kinetochore capture by spindle microtubules. Nature. 2005;434:987–994. doi: 10.1038/nature03483. [DOI] [PubMed] [Google Scholar]
- Tanaka TU. Kinetochore-microtubule interactions: steps towards bi-orientation. EMBO J. 2010;29:4070–4082. doi: 10.1038/emboj.2010.294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka TU, Rachidi N, Janke C, Pereira G, Galova M, Schiebel E, Stark MJ, Nasmyth K. Evidence that the Ipl1-Sli15 (Aurora kinase-INCENP) complex promotes chromosome bi-orientation by altering kinetochore-spindle pole connections. Cell. 2002;108:317–329. doi: 10.1016/s0092-8674(02)00633-5. [DOI] [PubMed] [Google Scholar]
- Thein KH, Kleylein-Sohn J, Nigg EA, Gruneberg U. Astrin is required for the maintenance of sister chromatid cohesion and centrosome integrity. J Cell Biol. 2007;178:345–354. doi: 10.1083/jcb.200701163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya M, Sato M, Haselmann U, Asakawa K, Brunner D, Antony C, Toda T. γ-tubulin complex-mediated anchoring of spindle microtubules to spindle-pole bodies requires Msd1 in fission yeast. Nat Cell Biol. 2007;9:646–653. doi: 10.1038/ncb1593. [DOI] [PubMed] [Google Scholar]
- Trautmann S, Rajagopalan S, McCollum D. The S. pombe Cdc14-like phosphatase Clp1p regulates chromosome biorientation and interacts with Aurora kinase. Dev Cell. 2004;7:755–762. doi: 10.1016/j.devcel.2004.10.006. [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V, Hardwick KG. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr Biol. 2009;19:1176–1181. doi: 10.1016/j.cub.2009.05.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Meadows JC, van der Sar SJ, Millar JB, Hardwick KG. Bub3p facilitates spindle checkpoint silencing in fission yeast. Mol Biol Cell. 2009;20:5096–5105. doi: 10.1091/mbc.E09-09-0762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visintin R, Craig K, Hwang ES, Prinz S, Tyers M, Amon A. The phosphatase Cdc14 triggers mitotic exit by reversal of Cdk-dependent phosphorylation. Mol Cell. 1998;2:709–718. doi: 10.1016/s1097-2765(00)80286-5. [DOI] [PubMed] [Google Scholar]
- Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- Yang Y, et al. Phosphorylation of HsMis13 by Aurora B kinase is essential for assembly of functional kinetochore. J Biol Chem. 2008;283:26726–26736. doi: 10.1074/jbc.M804207200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Y, Kronebusch PJ, Borisy GG. Kinetochore microtubule dynamics and the metaphase-anaphase transition. J Cell Biol. 1995;131:721–734. doi: 10.1083/jcb.131.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.