Abstract
Background
Myotonic dystrophy type 1 (DM1) is the most common muscle dystrophy in adults. The disease is caused by a triplet expansion in the 3'end of the myotonic dystrophy protein kinase (DMPK) gene. In order to develop a human cell model for investigation of possible effects of antisense and RNAi effector molecules we have used lentiviral mediated myoD-forced myogenesis of DM1 patient fibroblasts.
Findings
Transduced fibroblasts show a multinuclear phenotype and express the differentiation marker myogenin. Furthermore, fluorescence in situ hybridization (FISH) analysis revealed a statistical significant increase in the amount of nuclear foci in DM1 patient fibroblasts after myogenesis. Finally, no nuclear foci were found after treatment with oligonucleotides targeting the repeat expansions.
Conclusions
The abundance of nuclear foci in DM1 patient fibroblasts increase following myogenesis, as visualized by FISH analysis. Foci were eradicated after treatment with antisense oligonucleotides. Thus, we propose that the current cell model is suitable for testing of novel treatment modalities.
Background
Myotonic dystrophy 1 (DM1) is a multisystemic dominant disease and it is the most common muscular dystrophy in adults [1]. The symptoms include muscle wasting (muscular dystrophy), cataract, heart conduction defects, insulin resistance, and myotonia. The current treatment is insufficient, ranging from muscle exercise to breathing assistance. The genetic cause of DM1 is a (CTG)n repeat in the 3'-untranslated region of the dystrophia myotonica protein kinase gene, DMPK [2] . Current evidence supports an RNA-gain-of-function pathogenesis [1]. Indeed, mutant DMPK mRNA localizes to distinct foci in the nucleus and sequesters multiple proteins, among these the alternative splicing regulator muscleblind-like protein 1 (MBNL1). This results in a depletion of MBNL1 in the nucleus, leading to multiple events of aberrant splicing. Other factors affected by the accumulation of foci include CUG-binding protein 1 (CUG-BP1) which is another alternative splicing regulator. Both MBNL1 and CUG-BP1 were recently shown to regulate the alternative splicing of numerous genes [3-5]. The importance of the nuclear foci has been underlined by the discovery that reduction of the number of foci is associated with normalized splice patterns in DM1 cells [6,7]. Foci abundancy and brightness has been reported to increase during myogenesis, but statistical analysis of the number of foci per cell was not performed [8].
To study DM1 pathogenesis in vitro, human DM1 myocytes can be used as model system. However, DM1 patient muscle cells are a scarce resource for research as a muscle biopsy is required to collect each sample. Transfer of the myoD gene has previously been shown to convert fibroblasts into myoblasts [9]. The myoD protein activates several transcription factors including myogenin. Inducible overexpression of myoD combined with a chloride channel luciferase minigene reporter system has been described recently for drug screening in a cell line [10]. Moreover, immortalized skin fibroblasts from a Duchenne Muscular Dystrophy patient have been used in a cell model, where cells were transduced with an inducible myoD-construct [11]. Here, we have characterized the reprogramming of DM1 patient fibroblasts to muscle cells by demonstrating a muscular phenotype and a statistical significant increase in the number of RNA foci per cell. Furthermore, we have used the cells for evaluating the treatment effect of previously described antisense oligonucleotides [7,12].
Methods
Cells and media
Normal human dermal fibroblasts (NHDF) were obtained from ATCC, USA. DM1 fibroblasts (GM03132) were from Coriell Institute, USA. Southern blotting analysis showed that the expanded allele contained approx. 2250 CTG repeats in the DMPK gene. Standard medium: DMEM from Invitrogen™ with 10% fetal calf serum (Sigma-Aldrich), glutamine, streptomycin and penicillin. Low serum medium (HS): F12 medium from Invitrogen™ containing 3% horse serum, glutamine, streptomycin and penicillin.
Lentiviral production and transduction
The lentiviral vector encoding myoD was generated by replacing the puro gene in pCCL-WPS-PGK-puro-WHV [13] with myoD cDNA. For lentiviral production, 293T cells were seeded at 3 × 106 cells/p10 dish in standard medium, which was refreshed one hour prior to transfection. Cells were transfected by a CaPO4 co-precipitation method with 3.75 μg pMD.2G, 3 μg pRSV-Rev, 13 μg pMDGP-Lg/RRE and 13 μg of transfer vector (either pCCL-WPS-PGK-MyoD-WHV or pCCL-WPS-PGK-GFP-WHV). The medium was refreshed 24 hours post-transfection. One day later, supernatant containing the viral vector was filtered through a 0.45 μm pore filter, and polybrene added to a final concentration of 8 μg/ml. The medium was diluted 1:3 with standard medium and transferred to NHDF and DM1 fibroblasts. Medium was refreshed 24 hours after transduction.
Quantitative RT-PCR
RNA was isolated and cDNA was synthesized according to manufacturer's protocol (Sigma® and BioRad®, respectively). Before use, the cDNA was thawed and diluted appropriately; in this range of experiments a dilution of 1:4 was used. Furthermore, dilutions were made to set up a standard curve for the reactions. 2 μl of cDNA 1:4 dilution of each sample was added to 23 μl solution consisting of 12.5 μl TaqMan® universal PCR mastermix, 1.25 μl TaqMan mRNA specific primer set and 9.25 μl H2O. Reaction plates were analyzed by an ABI Prism® 7000 sequence detection system (Applied Biosystems). All samples were analyzed as triple determinations. The following TaqMan® assays were used: myogenin (MYOG, Hs01072232_m1, Applied Biosystems) and myoD (MYOD1, Hs00159528_m1, Applied Biosystems).
Immunofluorescence (IF)
Human primary fibroblasts were grown in either standard medium or F12 with 3% horse serum (HS medium) at 37°C in 5% CO2. The cells were grown to appropriate confluence and fixed in 4% formaldehyde (Lilly's solution) for 10 minutes. After fixation, the cells were washed 3 times in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM NaH2PO4 (pH 7.2)). To lower the non-specific binding, the slide was incubated in 100 μl 1% BSA in PBS (blocking buffer) prior to antibody addition. Slides were washed in PBS and incubated with 100 μl primary antibody diluted (1:100) in blocking buffer (BSA-PBS) for 2 hours. 3× wash in PBS followed before 100 μl secondary antibody (1:400) diluted in blocking buffer, was added. The slide was incubated with secondary antibody for 1 hour before it was washed and 100 μl antifade, with or without DAPI (4',6-diamidino-2-phenylindole) was added. Primary antibodies were mouse monoclonal anti-myoD and anti-myogenin (both Santa Cruz Biotechnology). The secondary antibodies were Alexa Flour® 488 conjugated goat anti mouse (Invitrogen). All secondary antibodies were diluted 1:400 in 1% BSA PBS solution.
Fluorescence in situ hybridization (FISH)
FISH was performed according to protocol from SingerLab Online [14].
Probes were RP-HPLC purified and labeled with Cy3 (red) purchased from DNA Technology A/S, Denmark. The Cy3-labeled probe consisted of a (CAG)10-sequence with a fluorophore at the 5'-end [15]. All reagents used in the above protocol were nuclease free and dissolved/diluted in nuclease free/DEPC-treated water or PBS unless otherwise stated. The numbers of RNA foci were counted microscopically and compared using Student's t-test.
Quantification of foci
The numbers of foci per cell were quantified by two different methods giving essentially similar results. The first method was based on direct counting, using a fluorescence microscope. Microscope fields were chosen randomly, using the DAPI filter, and dots were counted in all cells where the complete cell was present in the field. In another method images were acquired. This was done (in the Cy3 channel), using multidimensional acquisition, and images were stacked into a 2d-picture. The counting was performed by allowing the software to count the number of foci based on an arbitrary threshold value for minimum light intensity. Software used in the image acquisition was MetaMorph, and in the image processing, including counting, ImageJ was used.
Combined IF and FISH
Cells were rehydrated and permeabilized according to the FISH procedure described above. Next, cells were blocked in 1%BSA solution in a humidified hybridization chamber for 1 hour at room temperature. Afterwards, cells were transferred to a new hybridization chamber and incubated with primary antibodies for 1.5 hours at 37°C. Following this, cells were washed three times 5 minutes in PBS containing 5 mM MgCl2 at room temperature and thereafter incubated in secondary antibodies for 45 minutes at 37°C in a humidified hybridization chamber. Subsequently, cells were washed five times 5 minutes in PBS containing 5 mM MgCl2 at room temperature. Cells were fixed in 4% paraformaldehyde for 20 min at room temperature and the FISH procedure was performed as described above.
Transfection with RNA oligonucleotides
Oligonucleotides used in the present study comprised 21 nucleotides. They were chemically modified, as previously described [7] with a phosphorothioate backbone (PT) and methylated at 2'O (2'OMe). The antisense oligonucleotide had the sequence (CAG)7, targeting the pathogenic repeat. The control oligonucleotide had the following sequence: GUAGCGACUAAACACAUCAAG.
Cells were transferred to glass cover slips in a 12-well plate with a confluency of 1.5 × 105 cells/well. The following day, cells were transfected with the oligonucleotide, using polyethyleneimine (PEI) aided by hyaluronic acid (HA) as transfection reagent (Sigma-Aldrich). Transfection was performed as previously described [16].
Oligonucleotides (600 μg/ml), hyaluronic acid (HA) (Sigma-Aldrich; 3.5 mg/ml) and polyethyleneimine (PEI) (Sigma-Aldrich; 937.5 μg/ml) were mixed in a 1:2:1 ratio (by volume). This suspension was diluted in an equal volume of doubly concentrated PBS. The DNA/HA/PEI/PBS solution was incubated for 30 minutes at room temperature. Immediately before adding the complex to the cells, the medium was refreshed (ordinary DMEM) and the complex was added to the cells. Cells were incubated for 4 hours at 37°C with the transfection complex and the medium was subsequently replaced with fresh DMEM. Cells were fixed for FISH analysis 24 hours post-transfection.
Results and discussion
Analysis of myoD-transduced primary human fibroblasts
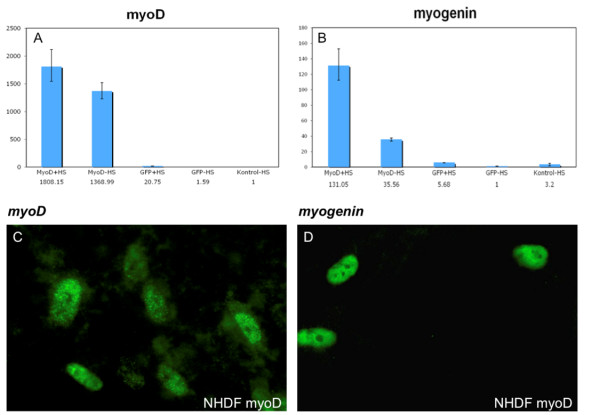
Lentiviral mediated gene transfer was used for expression of myoD in normal human dermal fibroblasts (NHDF). Quantitative PCR (qPCR) analysis was used to determine the expression of myoD and myogenin in transduced NHDFs. Cell differentiation was enhanced using low serum medium (HS) containing 3% horse serum (see materials and methods) [17]. We observed that myoD was expressed in fibroblasts 6 days after lentiviral transduction (Figure 1A). Cells transduced with GFP and non-transduced cells expressed negligible quantities of myoD relative to cells transduced with myoD. The levels of myoD RNA were further increased in the myoD-transduced cells cultured in HS medium. Analysis by qPCR likewise showed that the differentiation marker myogenin is expressed in cells transduced with myoD, and further increased by cultivation in HS medium (Figure 1B). Immunofluorescence revealed that both myoD and myogenin could be detected in myoD-transduced fibroblasts (Figure 1C, D). As expected, both proteins were located in the nuclei. Neither myoD nor myogenin were observed in non-transduced or GFP-transduced cells (data not shown).
Figure 1.
Expression of myoD and myogenin in normal human dermal fibroblasts (NHDF). (A and B) The cells were cultured for 6 days before harvest for quantitative RT-PCR or (C and D) IF: myoD and myogenin expression relative to beta-actin. +HS indicate cells grown in low serum medium; -HS designates cells grown in standard DMEM medium. Control designates no transduction. (C and D) Immunofluorescence of myoD and myogenin proteins, respectively. Original magnification × 630.
myoD-transduction of DM1 fibroblasts leads to a muscle-like phenotype and increased numbers of nuclear RNA foci
Having shown myogenesis of normal cells, myoD-transduced DM1 fibroblasts were cultivated in low-serum medium (HS) for 2-3 weeks. Cellular differentiation was mediated by lentiviral delivery of the transcription factor myoD. As seen in Figure 2, myoD-transduced cells display multinuclear tubular morphology and express myogenin (Figure 2B, D), respectively, compared to non-transduced cells (Figure 2A, B). Fluorescence in situ hybridization (FISH) using a Cy3-labeled (CAG)10 oligonucleotide probe revealed distinct nuclear foci in DM1 patient fibroblasts (Figure 2G and 2H), whereas no foci were found in NHDF (Figure 2E, F). Statistical analysis of the number of foci in myoD-transduced DM1 cells revealed that transduction with myoD lead to increased numbers of Cy3-foci per nucleus (Figure 3). The increased foci abundancy was only seen in myoD-transduced cells with a myogenic phenotype (p < 0.01; Student's t-test) (Figure 3F).
Figure 2.
Characterization of DM1 patient fibroblasts (PT) transduced with myoD. (A and B) Light microscopy of PT cells cultured in low serum medium for 21 days post-transduction. (A) Non-transduced (NT) PT cells display a fibroblast phenotype, (B) PT cells transduced with myoD display elongated tubular morphology. (D) IF of myogenin in DM1 patient fibroblasts (PT), 12-days post-transduction PT cells express myogenin and exhibit multinuclear morphology, (C) while non-transduced PT cells do not. (E-H) myoD induced myogenesis increase the number of nuclear foci in DM1 patient fibroblasts. (G-H) DM1 patient fibroblasts and (E-F) normal fibroblasts were cultured 10 days in low serum medium following transduction with (E and G) GFP or (F and H) myoD. Cells were analyzed by FISH with a Cy3-labeled (CAG)10 probe (red) targeting the pathogenic CUG repeat expansion of DMPK mRNA. (E-F) Normal fibroblasts (NHDF) displayed no nuclear foci when transduced with GFP or myoD. (G-H) DM1 patient fibroblasts exhibited an increased number of nuclear foci in fibroblasts transduced with myoD relative to GFP transduced fibroblasts. (H) The high number of foci was only observed in myoD transduced fibroblasts with myogenic morphology. (A-B) Original magnification × 400 and (C-H) × 630.
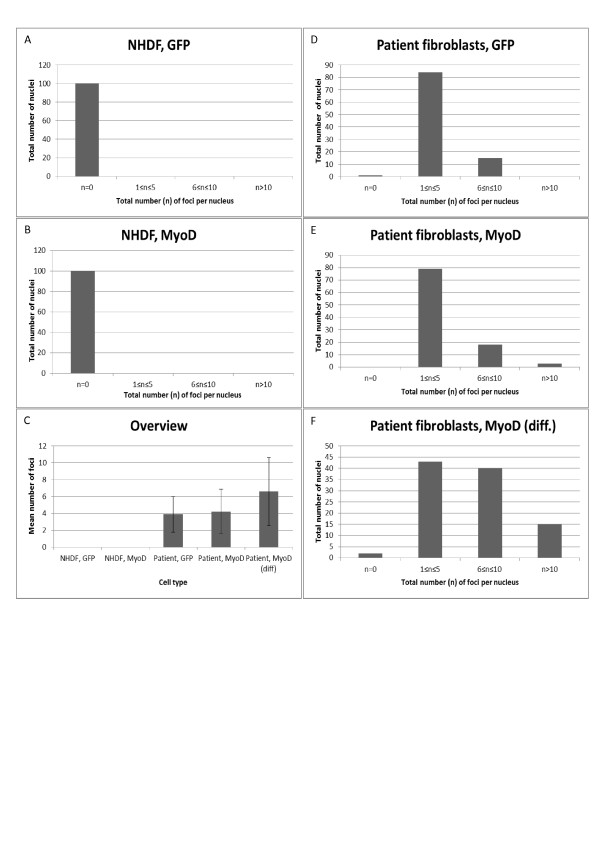
Figure 3.
Quantification of foci in DM1 patient cells and NHDF. (A and D) Fibroblasts were transduced with GFP or (B, E-F) myoD. Nuclear foci in 100 cell nuclei were counted for each sample. (A and B) NHDF cell nuclei contained no foci following tranduction with GFP or myoD. (D and E) PT cells transduced with GFP or myoD showed foci in cell nuclei. (F) Differentiated (myoD) patient fibroblasts displayed markedly changed morphology and exhibited an increase in the average as well as the maximum number of nuclear foci per cell.
Combined immunostaining and fluorescence in situ hybridization (IF/FISH) revealed that only cells transduced with myoD had multiple nuclei, tubular morphology and an increased number of nuclear foci (Figure 4E-H). Furthermore, myogenin was expressed in all nuclei of cells with multiple nuclei and tubular morphology (Figure 4F, H). Cell fusion was only found between cells in which all nuclei expressed myogenin (Figure 4F, H). The morphology of myoD-transduced normal fibroblasts was similar to that observed in myoD-transduced DM1 patient fibroblasts. Thus, combined immunostaining and fluorescence in situ hybridization confirmed that myogenesis leads to significantly increased numbers of nuclear RNA foci per cell.
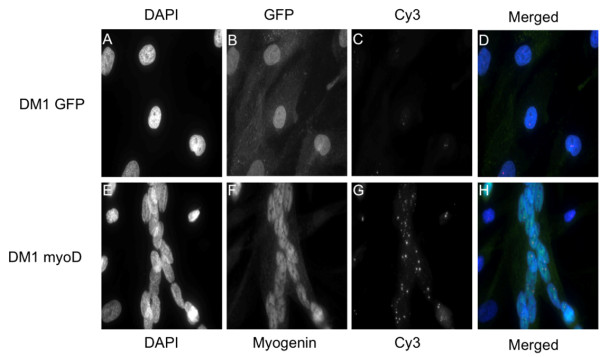
Figure 4.
IF/FISH targeting myogenin (IF) and the (CAG)-repeat (FISH) of transduced DM1 fibroblasts. (A and E) Panels display DAPI counterstain, (B and F) FITC/GFP channel, (C and G) Cy3 channel, (D and H) merge of DAPI/GFP/Cy3. (A-D) Cells transduced with GFP display no change in morphology or in the number of Cy3-foci (red) top panel. (E-H) Cells transduced with myoD display a multinuclear morphology, signs of cell fusion (syncytia), and an increased number of Cy3-foci per nucleus. Primary antibodies target myogenin, secondary antibodies are Alexa Fluor 488-labeled. The probe is a Cy3-labeled (CAG)10 DNA oligonucleotide. Original magnification × 630.
myoD-transduced DM1 patient fibroblasts can be used for analysis of treatment effects of antisense oligonucleotides
DM1 patient cells were transfected with antisense oligonucleotides (AONs) targeting the CUG repeat in dmpk mRNA. Already one day after treatment, FISH revealed elimination of the nuclear foci, both in non-transduced and myoD-transduced cells transfected with the AONs (Figure 5B, D). Transfection with control oligonucleotides with the same length and chemical modifications did not affect the number of foci (data not shown). It has previously been shown that the present AONs reduce the levels of mutant DMPK RNA [7]. However, although evaluation of treatment effects 2 days after AON transfection gave similar results (data not shown), it cannot be completely ruled out that the observed eradication of foci could be a consequence of competition between the therapeutic AONs and the labeled FISH probe.
Figure 5.

Treatment of DM1 cells with antisense oligonucleotides. Cells were transfected with 1 μg of oligonucleotides (AON) targeting the CUG triplet repeat, and RNA foci visualized using FISH with a Cy3-labeled CAG probe (red dots). (C and D) Differentiated DM1 cells cultivated in HS medium, 2 weeks post-myoD-transduction. (A and B) DM1 fibroblasts. (B and D) Cells transfected with AONs. (A and C) Untransfected. Original magnification × 630.
Conclusions
In DM1 muscle cells the transcription factor myoD is downregulated, and poor muscle differentiation has been described [18,19]. However, normal myogenin expression has been shown in developing myoblasts isolated from DM1 patient biopsies [20]. We here show that lentiviral mediated myoD-transduction leads to myoD and myogenin expression in both normal and DM1 fibroblasts, indicating that the impaired differentiation can be overcome at least to some extent. Myogenin regulates myotube formation and accordingly we detected myogenin in all nuclei of cells with a myotubular phenotype (Figure 4H).
Foci of nuclear mutant DMPK is a characteristic manifestation of the DM1 disease phenotype. These foci were visualized by FISH analysis, using a Cy3-labeled CAG-probe [15,20-25]. The appearance of the foci differed considerably, both in shape, intensity and area (size). As previously described, the vast majority were localized to the nucleus and the intranuclear position was seemingly random [15]. We observed an increase in the number of foci in differentiated cells, displaying a mature muscle phenotype (Figure 3). This is in agreement with earlier reports using retrovirally transduced DM1 fibroblasts [8]. Both NHDF and DM1 patient cells were found to be responsive to myoD-forced myogenesis (Figure 1 and 2) as has been reported previously [8-10,25]. However, the myogenic capability and increase in foci abundancy is slightly controversial, since earlier reports indicate decreased differentiation in DM1 cells [18,19]. In contrast, a more recent study reported normal myogenesis, but increased apoptosis in DM1 cells [20]. Indeed, we observed normal myogenesis in the DM1 cells, and no obvious signs of increased cell death were seen in the differentiated DM1 fibroblasts compared to the differentiated NHDF fibroblasts. Transfection with antisense oligonucleotides (AONs) can lead to reversal of RNA toxicity in a DM1 cell model from transgenic mice through elimination of RNA foci [7]. To ascertain that differentiated DM1 cells are applicable as a human cell model, we reproduced foci reduction in differentiated human DM1 cells (Figure 5), by transfecting with antisense oligonucleotides previously shown to be capable of rescuing the phenotype [12]. Thus, myoD-transduced DM1 cells are capable of myogenic differentiation and can be used as a cell model for in vitro experiments studying the cellular pathophysiology and possible effects of therapeutic compounds on the DM1 phenotype.
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
JL and OJP contributed equally to this work, carried out the molecular genetic studies and drafted the manuscript. MJ constructed the lentiviral vector. RT consulted on the immunoassays and in situ hybridizations. CBP and NG consulted on the RTQ-PCR assay. JMH and TJC participated in study design and helped to draft the manuscript. TGJ conceived of the study, and participated in its design and coordination and helped to draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jan Larsen, Email: janvlarsen@gmail.com.
Olof J Pettersson, Email: olofjp@gmail.com.
Maria Jakobsen, Email: maria@humgen.au.dk.
Rune Thomsen, Email: rune@humgen.au.dk.
Christina B Pedersen, Email: cbak@ki.au.dk.
Jens M Hertz, Email: jens.michael.hertz@ouh.regionsyddanmark.dk.
Niels Gregersen, Email: nig@ki.au.dk.
Thomas J Corydon, Email: tjc@humgen.au.dk.
Thomas G Jensen, Email: thomas@humgen.au.dk.
Acknowledgements
This work was supported by the Danish Research Council (FSS) and the Karen Elise Jensen Foundation.
References
- Lee JE, Cooper TA. Pathogenic mechanisms of myotonic dystrophy. Biochem Soc Trans. 2009;37:1281–1286. doi: 10.1042/BST0371281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook JD, McCurrach ME, Harley HG, Buckler AJ, Church D, Aburatani H, Hunter K, Stanton VP, Thirion JP, Hudson T, Sohn R, Zemelman B, Snell RG, Rundle SA, Crow S, Davies J, Shelbourne P, Buxton J, Jones C, Juvonen V, Johnson K, Harper PS, Shaw DJ, Housman DE. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3' end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. [DOI] [PubMed] [Google Scholar]
- Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, Ares M Jr. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- Ho TH, Bundman D, Armstrong DL, Cooper TA. Transgenic mice expressing CUG-BP1 reproduce splicing mis-regulation observed in myotonic dystrophy. Hum Mol Genet. 2005;14:1539–1547. doi: 10.1093/hmg/ddi162. [DOI] [PubMed] [Google Scholar]
- Warf MB, Nakamori M, Matthys CM, Thornton CA, Berglund JA. Pentamidine reverses the splicing defects associated with myotonic dystrophy. Proc Natl Acad Sci USA. 2009;106:18551–18556. doi: 10.1073/pnas.0903234106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulders SA, van den Broek WJ, Wheeler TM, Croes HJ, van Kuik-Romeijn P, de Kimpe SJ, Furling D, Platenburg GJ, Gourdon G, Thornton CA, Wieringa B, Wansink DG. Triplet-repeat oligonucleotide-mediated reversal of RNA toxicity in myotonic dystrophy. Proc Natl Acad Sci USA. 2009;106:13915–13920. doi: 10.1073/pnas.0905780106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis BM, McCurrach ME, Taneja KL, Singer RH, Housman DE. Expansion of a CUG trinucleotide repeat in the 3' untranslated region of myotonic dystrophy protein kinase transcripts results in nuclear retention of transcripts. Proc Natl Acad Sci USA. 1997;94:7388–7393. doi: 10.1073/pnas.94.14.7388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RL, Weintraub H, Lassar AB. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell. 1987;51:987–1000. doi: 10.1016/0092-8674(87)90585-x. [DOI] [PubMed] [Google Scholar]
- O'Leary DA, Vargas L, Sharif O, Garcia ME, Sigal YJ, Chow SK, Schmedt C, Caldwell JS, Brinker A, Engels IH. HTS-Compatible Patient-Derived Cell-Based Assay to Identify Small Molecule Modulators of Aberrant Splicing in Myotonic Dystrophy Type 1. Curr Chem Genomics. 2010;4:9–18. doi: 10.2174/1875397301004010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaouch S, Mouly V, Goyenvalle A, Vulin A, Mamchaoui K, Negroni E, Di Santo J, Butler-Browne G, Torrente Y, Garcia L, Furling D. Immortalized skin fibroblasts expressing conditional MyoD as a renewable and reliable source of converted human muscle cells to assess therapeutic strategies for muscular dystrophies: validation of an exon-skipping approach to restore dystrophin in Duchenne muscular dystrophy cells. Hum Gene Ther. 2009;20:784–790. doi: 10.1089/hum.2008.163. [DOI] [PubMed] [Google Scholar]
- Wheeler TM, Sobczak K, Lueck JD, Osborne RJ, Lin X, Dirksen RT, Thornton CA. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen M, Stenderup K, Rosada C, Moldt B, Kamp S, Dam TN, Jensen TG, Mikkelsen JG. Amelioration of psoriasis by anti-TNF-alpha RNAi in the xenograft transplantation model. Mol Ther. 2009;17:1743–1753. doi: 10.1038/mt.2009.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SingerLab Protocols. http://www.singerlab.org/protocols/insitu_mammalian.htm
- Taneja KL. Localization of trinucleotide repeat sequences in myotonic dystrophy cells using a single fluorochrome-labeled PNA probe. Biotechniques. 1998;24:472–476. doi: 10.2144/98243rr02. [DOI] [PubMed] [Google Scholar]
- Ito T, Yoshihara C, Hamada K, Koyama Y. DNA/polyethyleneimine/hyaluronic acid small complex particles and tumor suppression in mice. Biomaterials. 2010;31:2912–2918. doi: 10.1016/j.biomaterials.2009.12.032. [DOI] [PubMed] [Google Scholar]
- Bergstrom DA, Penn BH, Strand A, Perry RL, Rudnicki MA, Tapscott SJ. Promoter-specific regulation of MyoD binding and signal transduction cooperate to pattern gene expression. Mol Cell. 2002;9:587–600. doi: 10.1016/s1097-2765(02)00481-1. [DOI] [PubMed] [Google Scholar]
- Lee KS, Smith K, Amieux PS, Wang EH. MBNL3/CHCR prevents myogenic differentiation by inhibiting MyoD-dependent gene transcription. Differentiation. 2008;76:299–309. doi: 10.1111/j.1432-0436.2007.00209.x. [DOI] [PubMed] [Google Scholar]
- Amack JD, Reagan SR, Mahadevan MS. Mutant DMPK 3'-UTR transcripts disrupt C2C12 myogenic differentiation by compromising MyoD. J Cell Biol. 2002;159:419–429. doi: 10.1083/jcb.200206020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loro E, Rinaldi F, Malena A, Masiero E, Novelli G, Angelini C, Romeo V, Sandri M, Botta A, Vergani L. Normal myogenesis and increased apoptosis in myotonic dystrophy type-1 muscle cells. Cell Death Differ. 2010;17:1315–1324. doi: 10.1038/cdd.2010.33. [DOI] [PubMed] [Google Scholar]
- Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum Mol Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–3088. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]