Abstract
The tumor microenvironment modifies the malignancy of tumors. In solid tumors, this environment is populated by many macrophages that, in genetic studies that depleted these cells from mouse models of breast cancer, were shown to promote tumor progression to malignancy and increase metastatic potential. Mechanistic studies showed that these tumor-promoting effects of macrophages are through the stimulation of tumor cell migration, invasion, intravasation, and enhancement of angiogenesis. Using an in vivo invasion assay, it was demonstrated that invasive carcinoma cells are a unique subpopulation of tumor cells whose invasion and chemotaxis is dependent on the comigration of tumor-associated macrophages (TAMs) with obligate reciprocal signaling through an epidermal growth factor–CSF-1 paracrine loop. In this study, these invasion-promoting macrophages were isolated and subjected to analysis of their transcriptome in comparison with TAMs isolated indiscriminately to function using established macrophage markers. Unsupervised analysis of transcript patterns showed that the invasion-associated TAMs represent a unique subpopulation of TAMs that, by gene ontology criteria, have gene expression patterns related to tissue and organ development. Gene set enrichment analysis showed that these macrophages are also specifically enriched for molecules involved in Wnt-signaling. Previously, it was shown that macrophage-derived Wnt molecules promote vascular remodeling and that tumor cells are highly motile and intravasate around perivascular TAM clusters. Taken together, we conjecture that invasive TAMs link angiogenesis and tumor invasion and that Wnt-signaling plays a role in mediating their activity.
Tumor-associated macrophages (TAM) have been shown to perform a number of different roles in the tumor microenvironment to facilitate tumor progression (1–4). For example, in human breast cancer an increased density of TAMs has correlated with poor prognosis including increased lymph node involvement and decreased survival (5–7). In previous work in the polyoma middle T (PyMT) mouse model of human breast cancer (8) genetic depletion of the key macrophage growth factor, CSF-1 using the mice homozygous for the Csf1op null allele resulted in significantly reduced TAM density. In these animals, a significant delay in tumor progression to metastatic disease was observed, indicating that TAMs are required for efficient tumor metastasis (9).
Within the primary tumor microenvironment, at least two mechanisms are proposed by which TAMs facilitate tumor metastasis. The first relates to the demonstrated ability of TAMs to secrete proteases within the tumor microenvironment, such as urokinase plasminogen activator (10) (uPA), cathepsins B and D (11, 12), and matrix metallopeptidases 2 and 9 (13). It is believed that these TAM-derived enzymes digest the tumor basement membrane, facilitating tumor cell escape. Indeed, there are increased TAM numbers overlying sites of basement membrane breakdown in PyMT tumors (8). Moreover, there is enrichment of numerous other extracellular proteases in the tumor stroma of PyMT animals (14), coinciding with the localization of most TAMs in the tumor microenvironment (5, 15).
A second mechanism by which TAMs facilitate tumor metastasis is through directly enhancing an early stage of the metastatic cascade (16): carcinoma cell invasion. In vitro evidence demonstrated that macrophage production and release of the Wnt-ligand Wnt5a can stimulate the noncanonical planar cell polarity Wnt signaling pathway in carcinoma cells to promote invasion (17). In addition, an in vivo invasion assay (15, 18) similarly showed that TAMs promote carcinoma cell motility and invasion, but through a different mechanism that involves a paracrine signaling loop between tumor cells and TAMs.
In the in vivo invasion assay, a growth factor or chemokine was held within a 33-gauge microneedle containing Matrigel (BD Biosciences, San Jose, CA) and then positioned and stabilized within the primary tumor to collect responsive cells (18). It was noted that whether the tumor cell chemokine epidermal growth factor (EGF) or the macrophage chemokine CSF-1 was placed into the needles, both tumor cells and macrophages migrated in response (18). Moreover, pharmacologic or Ab functional inhibition of EGF/EGFR or CSF-1/CSF-1R signaling halted the movement of both cell types (15, 19, 20). Further experiments confirming the results of the in vivo invasion assay demonstrated that significantly fewer cells were collected in Csf1op/op animals (21), but cell numbers increased when CSF-1 was transgenically restored to the mammary fat pad. Furthermore, preinjection of wild type tumors with CSF-1 significantly increases collected cell numbers (19). These results are in agreement with human breast cancer studies in which increased levels of circulating CSF-1 (22) and/or increased tumor CSF-1R staining (23) correlate with poor prognosis and the observed expression of EGF by TAMs in breast cancer (24).
Isolation and expression profiling of invasive carcinoma cells demonstrated that these are a unique subpopulation of tumor cells (25, 26) that are highly motile, but less proliferative and apoptotic compared with the general carcinoma cell population (26, 27). Overall, these invasive cells have a survival advantage and increased resistance to chemotherapeutic agents (28). Although these studies were originally performed in tumor xenografts, the results are conserved in the transgenic PyMT animals (29).
We hypothesized that, similar to the invasive carcinoma cells, invasive TAMs are a unique subpopulation of TAMs found at sites of tumor invasion. We therefore aimed to collect and compare these cells to general TAMs (GTs) collected indiscriminately to specific function, using cell markers and flow cytometry. The studies herein identify a gene expression signature for invasive TAMs and provide the first example of an isolated subpopulation of TAMs. These invasive cells are compelling therapeutic targets because they represent a population of cells caught at an early stage of metastasis. Considering the invasive carcinoma cell requirement of comigrating TAMs, targeting these nongenomically transformed— and therefore less therapy resistant—host cells could provide a novel opportunity for therapeutic benefits.
Materials and Methods
Mice
All procedures involving mice were conducted in accordance with National Institutes of Health regulations concerning the use and care of experimental animals. The study of mice was approved by the Albert Einstein College of Medicine animal use committee. The FVB/N-Tg(MMTV-PyVmT)Mul (PyMT) transgenic mice were provided by Dr. W.J. Muller (McGill University, Montreal, Quebec, Canada) and have been described previously (8, 30). Tg(Csf1r-Gfp)Hume (MacGreen) mice have also been described previously (31). Male PyMT mice on an FVB background were bred to homozygous MacGreen female mice on a mixed background to generate PyMT mice that produce tumors with GFP-labeled macrophages. All genotyping was done by PCR. Tumors were allowed to grow until 14–16 wk to ensure late-stage carcinomas for TAM isolation by flow cytometry or in vivo invasion assay.
Collection of general TAMs (flow cytometry)
PyMT female mice with late-stage tumors of ~2 cm diameter were injected in the lateral tail-vein with 200 µl 10 mg/ml 70 kDa dextran conjugated to the Texas Red fluorphore (Invitrogen, Eugene, Oregon) resuspended in PBS. Two hours postinjection, animals were anesthetized with isofluorane and perfused intracardiacally with ice cold PBS. Following sacrifice, all subsequent steps were performed at 4°C. Tumors were minced and filtered four times through graded nylon filters, centrifuged at 1200 RPM for 5 min, and resuspended in erythrocyte lysis buffer (Beckman-Coulter, Marseille, France). Cells were washed three times in nuclease-free PBS containing 2% BSA and incubated with the macrophage-specific Alexa-fluor 488 conjugated F4/80 monoclonal Ab (Invitrogen, Carlsbad, CA). Dextran/F4/80 double-positive cells were sorted on a DakoCytomation MoFlo High-Speed Cell Sorter (DakoCytomation, Fort Collins, Colorado) at 23 pounds per square inch. into PBS + 2% BSA. Dextran/Csf1r-enhanced GFP (EGFP) double positive cells were isolated similarly as previously described (4).
Collection of invasive TAMs (in vivo invasion assay)
The collection of invasive cells via the in vivo invasion assay was originally designed, validated, and described by Wyckoff et al. (18) and Wang et al. (25). A solution of 25 nM purified EGF and 0.01 mM EDTA (pH 7.4) in Leibowitz’s L-15 media and 10% BSA was freshly made the day of collection. Matrigel was added to 10% total volume (needle mixture). This mixture was then filled into six 33-gauge microneedles and kept at 4°C until the animal was prepared.
A mouse with two solid tumors of ~2 cm diameter was anesthetized using isoflurane, and a small flap of skin was removed to expose the tumor. A micromanipulator connected to a needle holder (consisting of three 25-gauge needles) was used to stably guide the insertion of three guide wires ~2–5 mm into the surface of the tumors. Two micromanipulators were used per animal in separate tumors. Gently, the guide wires were removed and replaced with the filled collection needles one by one. The animal was kept under monitored anesthesia for 4 h and sacrificed by cervical dislocation at termination of collection.
Prior to sacrifice, the six collection needles were removed and the contents extruded using ~100 µl per needle of Leibowitz’s L-15 media and 10% BSA into an RNase-free 1.5 ml Eppendorf tube. Five percent total volume was stained by DAPI for cell count. Ten microliters CD11b (Mac-1) magnetic beads (Miltenyi-Biotec, Gladbach, Germany) were added to the remaining volume and incubated at 4°C with gentle agitation for 30 min. The sample was next placed onto a magnetic block for 30 min. and the remaining invasive carcinoma cells in the supernatant were removed. Two hundred microliters guanidine thiocyanate containing Buffer RLT (Qiagen, Valencia, CA) was added to remaining beads and then incubated over ice for 10 m. Beads were centrifuged and the supernatant containing RNA was transferred into an RNAse-free 1.5 ml Eppendorf tube. RNA isolation was performed on a cohort of four animals, resulting in RNA from ~6000 invasive TAMs per final array sample.
RNA extraction, amplification, and cDNA preparation
Total RNA was extracted from flow-sorted GTs and in vivo invasion assay isolated invasive TAMs using RNeasy Micro Kits (Qiagen) according to the manufacturer’s instruction. Amplification-grade DNase 1 treatment was performed on the RNA elution column to remove potential genomic DNA contamination. Approximate yields were 150 ng. Quality was determined using a nanobiosizing assay (Agilent Bioanalyzer; Agilent Technologies, Palo Alto, CA).
Approximately 100 ng RNA from samples was resuspended into 11 µl RNase/DNase-free water, and a single round of linear amplification was performed by the in vitro transcription T7 promoter method as outlined by the manufacturer’s protocol (Ambion’s Message Amp T7 Kit; Ambion, Austin, TX). For microarray samples, a second round of linear amplification was performed with 200 ng first round amplified material. At all steps, yield and quality were established using spectrophotometry and an Agilent Bioanalyzer.
For samples to be used for microarray hybridization, Superscript III (Invitrogen) reverse transcriptase was used to prepare 5 µg cDNA from amplified RNA. Random primers (Invitrogen) were used to prime the reaction. Second-strand cDNA synthesis was performed using Escherichia coli DNA Ligase (Invitrogen), E. coli DNA Polymerase 1 (Invitrogen) and T4 DNA Polymerase (Invitrogen). RNase H (Invitrogen) treatment was additionally performed. The reaction was stopped with 0.5 M EDTA and purified using a PCR purification kit (Qiagen) following the manufacturer’s protocol. Samples were resuspended to ~200 ng/µl.
Gene expression arrays
Five micrograms of double-stranded cDNA from each invasive TAM and GT sample was used for gene expression array processing. The expression array chip used contained 385,000 60-mer probes representing 42,586 genes (average nine probes per target; NimbleGen, Reykjavik, Iceland). A total of five independent samples for each macrophage population were prepared. At NimbleGen, quality and yield were verified before DNA end-labeling, hybridization, and scanning. Raw data files for each sample were normalized, background-corrected, and saved to logarithmic scale using a robust multi-array analysis (32) as implemented by NimbleScan software, version 2.2.33 (Nimblegen, Reykjavik, Iceland). Raw and normalized data available at the National Center for Biotechnology Information Gene Expression Omnibus (www.ncbi.nlm.nih.gov/geo/), series record GSE18295. Normalized data were analyzed and presented using R project (33, 34). Several packages developed for R through Bioconductor (35)—an open-source, open-development software project—were used for array analysis. These packages include limma (36), marray, siggenes and RColorBrewer. Significance analysis of microarray (SAM) (37) was implemented for stringent gene selection criteria using the siggenes package. A δ value of 3.859 called 1457 significantly regulated transcripts with a false discovery rate (FDR) of 1% when comparing invasive TAM to GT.
Five biologic repeats were prepared for gene expression arrays when comparing F4/80+/dextran+ FVB background TAMs to Csf1r-EGFP+/dextran+ mixed background TAMs as described above. By SAM, a δ value of 1.4 called 41 significantly regulated transcripts with a 10% FDR.
All transcripts assayed by the microarrays were subjected to hierarchical clustering, and a sample dendrogram was produced using the R package pvclust (38). The required R commands are included in the text file “TAMpvclust.r” (http://tinyurl.com/yg3l8gr) and the input data are the file “TAM.txt” (http://tinyurl.com/yg5j2qn).
Bioinformatics
The Ingenuity Pathways Knowledge Base (IPA) version 6.3 (www.ingenuity.com/products/pathways_analysis.html) was used to identify enriched cellular and molecular functions and canonical pathways among differentially regulated transcripts. p Values were calculated through IPA 6.3 using a right-tailed Fisher exact test and related to the likelihood of enrichment of specific function within a queried gene-set. Oncomine (www.oncomine.org) was used to mine human breast cancer microarray data as previously described (39, 40).
Gene set enrichment analysis (GSEA) (www.broad.mit.edu/gsea/) was used to identify KEGG pathways upregulated in invasive TAMs. KEGG gene sets containing between 15 and 500 genes were considered, and statistical significance was assessed using 5000 gene set permutations. Additional details about the GSEA run can be found here: http://tinyurl.com/yh2rzed; the files needed to recreate the analysis are here: http://tinyurl.com/ybe4dc5.
Quantitative real time PCR
Four subsequent biologic repeats of each GT and invasive TAM sample that had been single-round amplified were used for quantitative real-time polymerase chain reaction (QRT-PCR). Needle control bone marrow macrophage experiments (described below) did not require amplification. Superscript III (Invitrogen) reverse transcriptase was used to prepare 200 ng cDNA from RNA or amplified RNA. Random nonamers (a gift from Dr. Sumanta Goswami, Yeshiva University, New York, NY) were used to prime the reaction. Relative transcript abundance was detected by SybrGreen (Applied Biosystems, Foster City, CA) on the ABI 7900HT thermal cycler using gene-specific primers. Gene expression was normalized to the housekeeping gene, cyclophilin A (Ppia), and expressed values in invasive TAMs relative to GTs were determined using the ΔΔCT method (41). In needle control experiments, gene expression was reported as normalized to the housekeeping gene.
Cultured flow cytometry–sorted macrophages
TAMs from PyMT females and splenic macrophages from non–tumor-bearing littermate controls were isolated as previously described (4). Resulting cells were seeded onto bacterial plastic plates and allowed to adhere for 12 h. Cells were washed and the RNA was extracted using the RNeasy Micro kit (Qiagen) per the manufacturer’s protocol. RNA was quantitated before semiquantitative PCR for Wnt7b expression and then normalized to the housekeeping gene GAPDH.
Results
Invasive TAMs collected via the in vivo invasion assay are a unique subpopulation of TAM
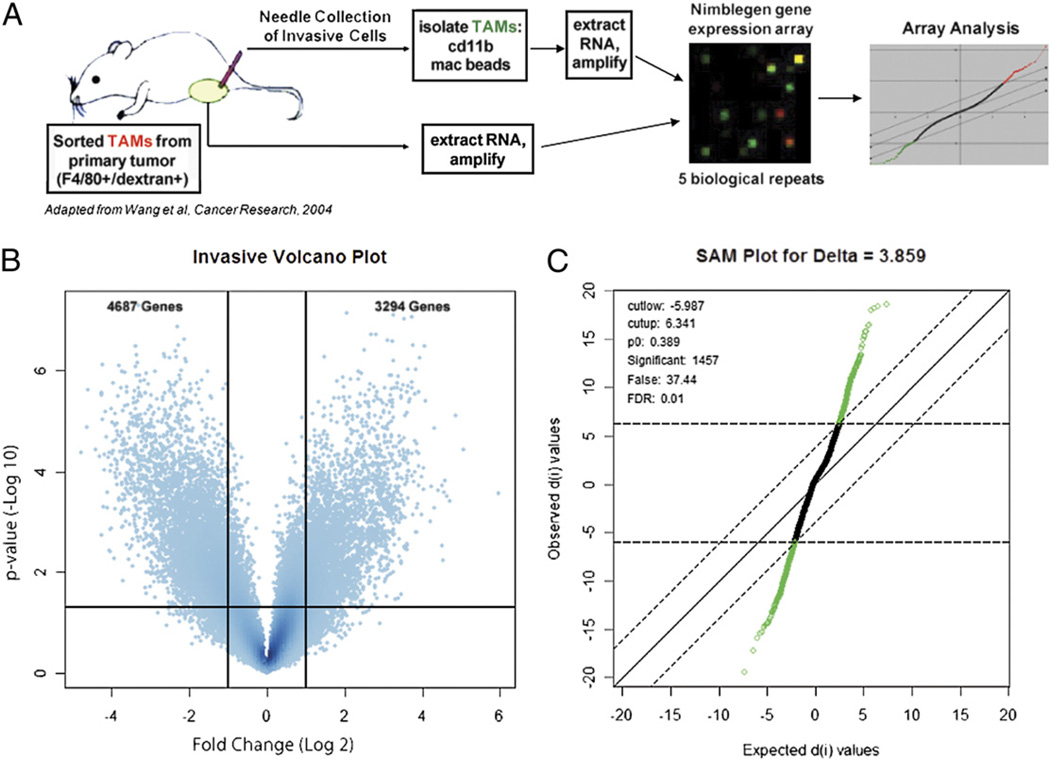
Invasive TAMs were isolated from PyMT animals on the FVB background with late stage tumors as previously described (18, 25, 26). In brief, 33-gauge microneedles containing a mixture including 25 nM EGF and the matrix, Matrigel, were placed into the primary tumors of anesthetized animals. Over a 4-h collection ~1000 carcinoma cells and macrophages migrate into a single needle in an ~3:1 ratio (26). Six needles were used per animal, yielding ~1500 invasive TAMs. Positive selection for invasive TAMs was performed using CD11b (Mac-1:integrin αM subunit) magnetic beads to separate TAMs from the invasive carcinoma cells as previously described (25). To obtain sufficient material for efficient RNA isolation and amplification for gene expression array, invasive TAMs from four separate animals were pooled for each sample. Five of these biologic replicates were obtained and compared by microarray with five biologic replicates of GTs, which were isolated by flow cytometry based on their expression of the macrophage-specific marker F4/80 and their ability to macropinocytose fluorochrome conjugated 70 kDa dextran (Fig. 1A). These sorted cells are uniformly CD11b positive and Gr1 negative, defining them further as macrophages (4, 42).
FIGURE 1.
Isolation of a functionally defined subpopulation of invasive TAMs for gene expression analysis. A, Schematic of array experimental protocol comparing gene expression profile of invasive TAMs to GTs (26). B, Volcano plot illustrating fold change (log base 2) compared with p value (−log base 10) between invasive TAMs and GTs. Horizontal bar at y ≈ 1.303 represents a significance level of p = 0.05 significance level. C, SAM for δ 3.859, FDR 1%.
Previously, it has been shown that TAMs are not compromised in their ability to secrete EGF or migrate into microneedles following ingestion of 70 kDa dextran (15), and that invasive TAMs uniformly express F4/80 (19). However, in the absence of transgenic labeling, F4/80 and dextran phagocytosis are minimally needed to isolate a pure population of TAMs from the heterogeneous tumor microenvironment (4). Therefore, although commercially produced and validated CD11b magnetic beads were sufficient for separating the two invasive cell populations that migrate into the needle, more stringent criteria were required for flow cytometrically obtaining the GT population. Of note, by defining markers for macrophages, these two populations are identical; therefore, microarrays were used to determine variance in gene expression between all TAMs and the functionally defined invasive subpopulation.
A volcano plot illustrates the large number of transcript abundance differences between the invasive TAM and GT populations. 4687 genes were identified as downregulated (Log2Ratio < 1; p < 0.05), and 3294 genes were upregulated (Log2Ratio > 1; p < 0.05; Fig. 1B). To obtain a more stringently selected population of differentially regulated transcripts, SAM (37) was used with a δ value of 3.859, corresponding to a 1% FDR. This criterion resulted in 1457 genes called as significantly differentially regulated (Fig. 1C).
An early concern in the analysis of the invasive TAM microarrays had been the large number of transcripts called as differentially regulated. Previous work comparing the gene expression profiles of macrophages from two separate tissues (tumor and spleen) yielded 460 differentially regulated genes by SAM using a standard FDR of 10% (4). In this study, 1457 transcripts were called with an FDR of 1% when comparing macrophages isolated from the same tissue. Therefore, to proceed with the analysis it was first necessary to explore the arrays to validate their utility. Results summarized in Supplemental Fig. 1 indicate that all samples were successfully labeled, hybridized, scanned, and globally reproducible. Excess differences between GTs and invasive TAMs could also be due to the increased irreducible contamination of carcinoma cells present in the invasive TAM sample, which has been demonstrated previously to be ~10% (25), and was unchanged in our hands (data not shown). To assess whether tumor cell contamination was significant in the data set, the previously published invasive carcinoma cell signature from PyMT tumors (29) was compared with that for the invasive TAM. Of the 901 differentially regulated transcripts (absolute fold change >2) in invasive carcinoma cells, 55 were also represented in the 1457 differentially regulated transcripts of the invasive TAMs. Only 17 transcripts were differentially regulated in the same direction (i.e., up in both samples or down in both samples). Of the 901 genes from the invasive carcinoma cell study, 188 were not able to be annotated because of expired annotations, and therefore may have been missed. However, overall we found no evidence of significant contamination of carcinoma cell transcripts in our samples.
Although no specific confounder was identified that caused the differences between general and invasive TAMs, in our data analysis we used unsupervised analytical techniques for broad generalizations of invasive TAM physiological functions in combination with carefully supervised analyses to establish that sufficient precedent exists to ascribe specific results to invasive TAM activity.
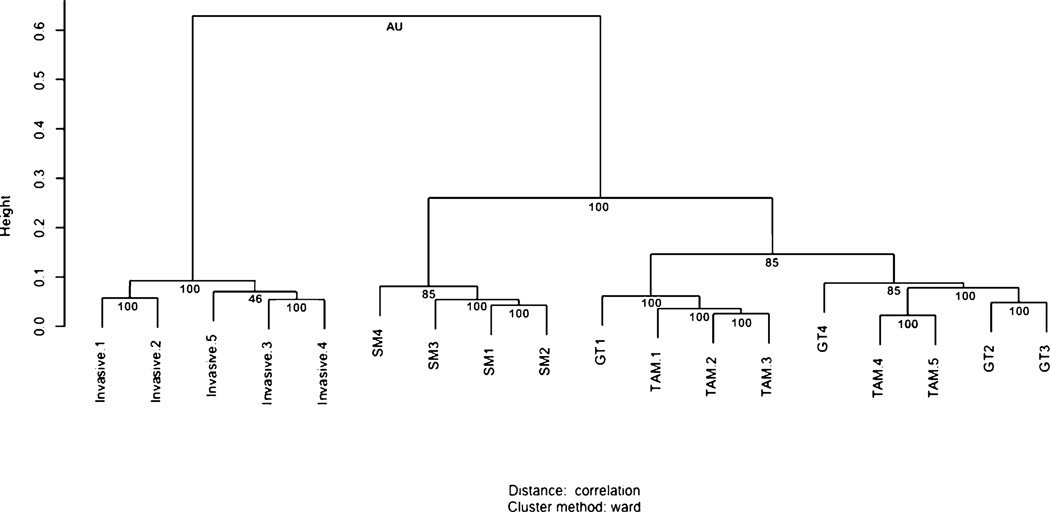
Consistent with the volcano plot and SAM analyses, hierarchical clustering (43) of all transcripts assayed on the gene expression chip demonstrates clear separation between the invasive TAM and GT samples (Fig. 2). Hierarchical clustering and a sample dendrogram was produced using the R package pvclust (38). The approximately unbiased scores give an indication of clade reliability. Clades supported by scores near 100 can be considered reliable.
FIGURE 2.
Invasive TAMs are a unique subpopulation of TAMs Unsupervised clustering of data from several macrophage populations shows that invasive TAMs group together in a clade with strong statistical support. A total of 18 macrophage samples were used including the five invasive TAM samples (Invasive) and five GT samples from the FVB mouse strain used in this study, and the four TAM and 4 splenic macrophage (SM) samples from mice on a mixed background, as previously published (4). Approximately unbiased (AU) scores are listed below individual pairs. Values close to 100 indicate a reliable clade. Larger heights on the vertical axis are indicative of a greater distance between clades.
Interestingly, the GT samples from the pure FVB mice used in this study did not separate from the TAM samples sorted by flow cytometry from MacGreen mice, a mixed strain. The MacGreen animals express EGFP via the colony stimulating factor 1 receptor (Csf1r) promoter, in granulocytes, monocytes and macrophages (4, 31, 44). Previously we demonstrated that a pure population of TAMs can be isolated based on EGFP expression (myeloid lineage) and Texas Red dextran uptake (macropinocytic ability) from these tumor-bearing animals (4). This is consistent with the DNA microarray comparison of both these samples that demonstrated only a small number of differences between the samples, despite strain differences and methods of isolation. These hierarchical clustering data are consistent with microarray comparison of these samples, demonstrating minimal changes despite differences in strain and markers for isolation. It further confirms that GTs are distinct compared with splenic macrophages. It is now also shown that the invasive TAM subpopulation is unique to each.
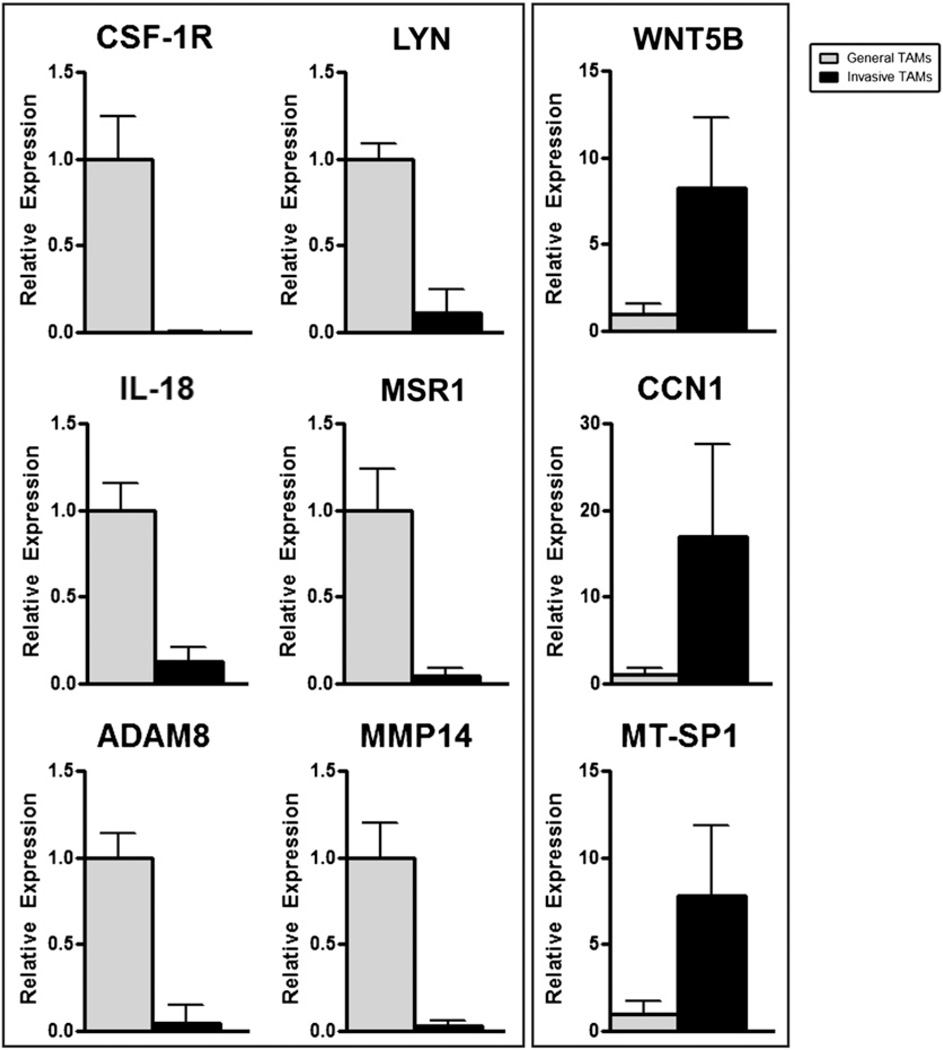
The final list of 1457 identified differentially regulated transcripts is listed in Supplemental Table I. QRT-PCR was used to confirm the results from the gene expression arrays, and all transcripts tested were validated in their relative expression pattern by this assay (Fig. 3). mRNA transcripts of decreased abundance in the invasive TAMs include the Csf1r, the src-family kinase Lyn, the proinflammatory cytokine IL-18, a macrophage scavenger receptor (Msr1), and two metalloproteases (Adam8 and Mmp14). Transcripts more abundantly expressed in invasive TAMs include a serine protease (Mtsp1), a proangiogenic molecule (Ccn1/Cyr61) and a member of the Wnt-family of ligands, Wnt5b. Notably, a second Wnt-family ligand, Wnt7b, was also identified as significantly increased in invasive TAMs; however, the baseline transcript abundance in GTs was not sufficient for detection by QRT-PCR and could not be tested for comparison. However, semiquantitative RT-PCR was used to validate increased Wnt7b expression (Fig. 4E).
FIGURE 3.
QRT-PCR validates gene expression array data for downregulated (A) and upregulated (B) transcripts. Invasive TAMs (black bars) and GTs (gray bars) normalized to expression of housekeeping gene cyclophilin A (Ppia).
FIGURE 4.
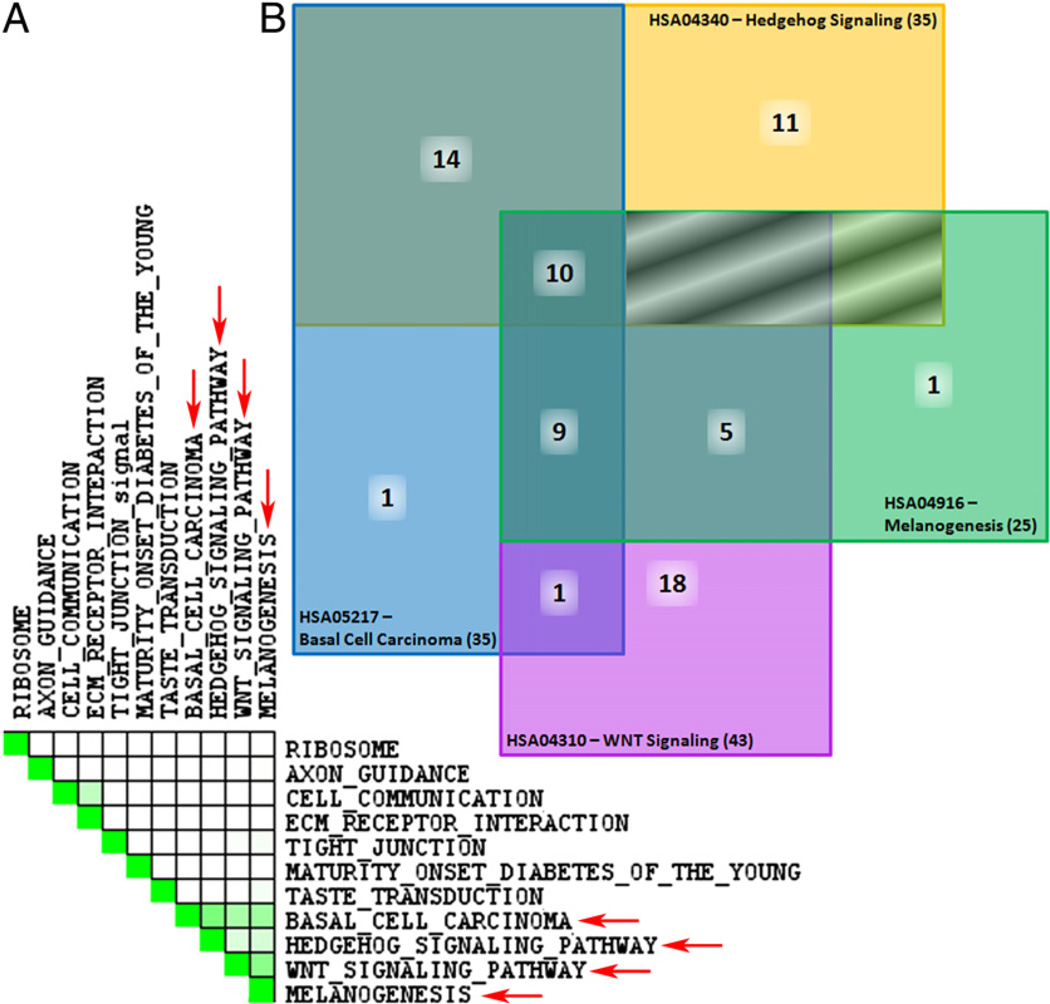
KEGG pathways enriched in invasive TAMs are related through Wnts. A, Set-to-set diagram, created through performing a leading edge analysis using GSEA, indicates the overlap between significantly enriched KEGG pathways listed in Table II. Dark green cells indicate that the gene sets have leading edge genes in common, whereas a white cell indicates no leading edge gene overlap. Four sets demonstrate overlap (red arrows). B, Venn diagram of overlapping molecules between enriched KEGG pathways. Notably, 10 molecules are common to all four pathways including: Wnts 1, 3, 4, 5B, 6, 7B, 9A, 9B, 10A and 16. Boxes shaded using diagonal lines have no overlap between pathways. Official human KEGG pathway signifiers are listed.
Gene expression pattern of invasive TAMs
Using the Ingenuity Pathway Analysis system (IPA 6.3), differentially regulated transcripts were analyzed for physiological functions and corresponding significance. To gain an appreciation for tissue trophic functions of this TAM population, Table I ranks the enriched functional categories relating to physiologic system and development when comparing those regulated transcripts that are upregulated or downregulated. Embryonic development and tissue development were the two functions determined to be most enriched in invasive TAMs. Previously, TAMs generically have been shown to have an expression signature similar to that of macrophages in developing embryos (4, 45). These results suggest that invasive TAMs are a TAM subpopulation that is expressly engaged in tissue trophic functions most similarly to embryonic phagocytes.
Table I.
Categories relating to physiologic system development and function enriched in invasive TAMs
| Rank | Category | p Value |
|---|---|---|
| Upregulated | ||
| 1 | Embryonic development | 2.87 × 10−4 – 4.14 × 10−2 |
| 2 | Tissue development | 2.87 × 10−4 – 4.14 × 10−2 |
| 3 | Tissue morphology | 2.87 × 10−4 – 4.14 × 10−2 |
| 4 | Nervous system development and function | 1.21 × 10−3 – 4.14 × 10−2 |
| 5 | Immune response | 1.29 × 10−3 – 4.14 × 10−2 |
| Downregulated | ||
| 1 | Immune response | 2.18 × 10−10 – 2.04 × 10−3 |
| 2 | Immune and lymphatic system development and function | 5.08 × 10−9 – 2.19 × 10−3 |
| 3 | Hematologic system development and function | 8.02 × 10−9 – 2.19 × 10−3 |
| 4 | Tissue development | 2.36 × 10−7 – 2.00 × 10−3 |
| 5 | Reproductive system development and function | 7.38 × 10−7 – 7.38 × 10−7 |
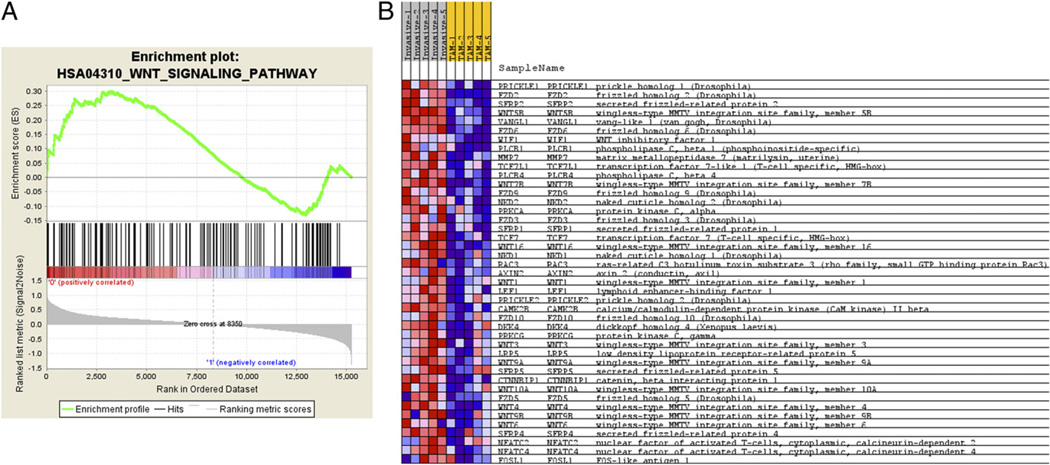
To further explore the dataset, GSEA (46) was used to identify groups of functionally related genes with expression patterns that correlate with the invasive TAM phenotype. The dataset was collapsed to a single expression value per HUGO gene symbol, so that the probe with the highest average expression value across all the samples was selected as the representative for that gene in the dataset. Genes were ranked based on differential expression between invasive TAMs and GTs with genes high in invasive TAMs at the top of the list and those high in GTs at the bottom. The GSEA method was then used to examine the distribution of the 148 functionally related human KEGG pathway gene sets (47) within the ranked gene list to gain an appreciation for functional motifs mediating invasive TAM activity. Eleven gene sets were enriched in the invasive TAM phenotype using a 25% false discovery rate cut-off (Fig. 4A, Table II). GSEA leading edge analysis was then performed to compare the sets of genes responsible for the enrichment observations. Two clusters of relationship were observed (Fig. 4A). The first contains the cell communication gene set (HSA01430) and the ECM receptor interaction gene set (HSA04512) and is driven by a group of laminin, collagen, and thrombospondin genes (http://tinyurl.com/yg3dhrf). The second set consists of four gene sets: basal cell carcinoma (HSA05217), Hedgehog signaling pathway (HAS04340), Wnt signaling pathway (HSA04310), and melanogenesis (HAS04916; Fig. 4B). This relationship is driven by a group 10 Wnt family members that are present in all four of these gene sets (WNT 1, 4, 5, 5B, 6, 7B, 9A, 9B, 10 and 16; http://tinyurl.com/yg3dhrf). This Wnt-containing set is of particular interest because of the recognized role of the Wnt-signaling pathway in development and its identified role in macrophage function (17, 48, 49). Besides the Wnt-ligands, several frizzled receptors and molecules that modulate Wnt-signaling were also present in these enriched gene sets, pointing toward a larger role for macrophage-mediated Wnt activity in the tumor microenvironment. Using GSEA, the genes associated with a role in the Wnt pathway were graphed (Fig. 5A) (46). The enrichment score of the Wnt-signaling pathway is 0.299 and the nominal p value is 0.021 (Table II). Of the 140 pathway-affiliated genes, there are groups of genes positively (red) and negatively (blue) correlated with invasive TAM expression. Those positively correlated genes that most-contributed to Wnt pathway enrichment are depicted with the associated expression normograms to demonstrate the consistency between samples (Fig. 5B).
Table II.
KEGG gene sets enriched in invasive TAMs and associated statistics
| Gene Set | Gene Set Size |
Enrichment Score |
Normalized Enrichment Score |
Nominal p Value |
FDR q Value |
Rank at Max |
|---|---|---|---|---|---|---|
| HSA01430_CELL_COMMUNICATION | 121 | 0.59563637 | 2.6974535 | 0 | 0 | 3511 |
| HSA05217_BASAL_CELL_CARCINOMA | 55 | 0.5553169 | 2.1564224 | 0 | 4.06 × 10−4 | 4580 |
| HSA04512_ECM_RECEPTOR_INTERACTION | 83 | 0.45781794 | 1.9435273 | 0 | 0.00513476 | 2211 |
| HSA04530_TIGHT_JUNCTION | 124 | 0.39952913 | 1.808554 | 0 | 0.0177126 | 3194 |
| HSA03010_RIBOSOME | 61 | 0.44803682 | 1.7929574 | 8.62 × 10−4 | 0.01682182 | 6356 |
| HSA04340_HEDGEHOG_SIGNALING_PATHWAY | 54 | 0.44944367 | 1.7346396 | 8.31 × 10−4 | 0.02487251 | 5313 |
| HSA04950_MATURITY_ONSET_DIABETES_OF_THE_YOUNG | 25 | 0.4811138 | 1.5646982 | 0.0249273 | 0.08812157 | 4153 |
| HSA04742_TASTE_TRANSDUCTION | 39 | 0.4193784 | 1.5260606 | 0.02017655 | 0.10253292 | 3603 |
| HSA04360_AXON_GUIDANCE | 124 | 0.31950954 | 1.4521579 | 0.0087222 | 0.15447515 | 2581 |
| HSA04310_WNT_SIGNALING_PATHWAY | 140 | 0.29928747 | 1.3827081 | 0.02131439 | 0.22107984 | 3271 |
| HSA04916_MELANOGENESIS | 96 | 0.31248757 | 1.3567276 | 0.03821103 | 0.23827241 | 2987 |
FIGURE 5.
GSEA analysis for Wnt-signaling pathway and positively enriched associated molecules. A, Distribution of the 142 Wnt-signaling pathway molecules amid the total ranked list of all transcripts analyzed by GSEA (46). Enrichment score of the pathway of 0.299 is identified by determining the peak of positively correlated transcripts. B, Normogram for the core enriched Wnt-signaling pathway related molecules in the array dataset. Increased expression (red), decreased expression (blue).
The Wnt-signaling pathway has a role in TAM activity
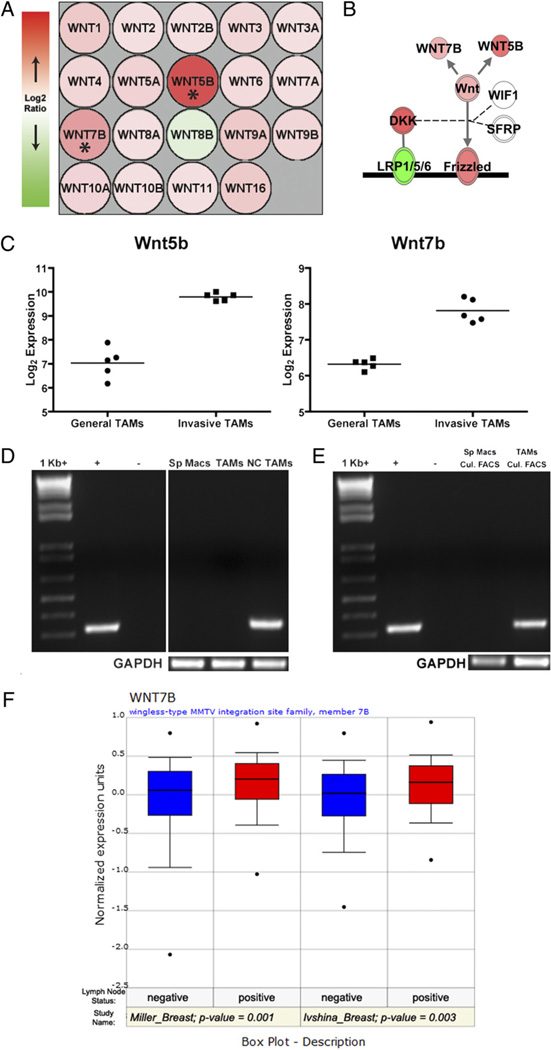
Returning to the IPA, graphing the log2ratio for every Wnt ligand demonstrates that all but Wnt8b were increased in invasive TAMs (Fig. 6A). However, as previously mentioned, only Wnt5b and Wnt7b were identified by stringent criteria (SAM) as independently statistically significant. Those significant molecules called by SAM were superimposed on the Wnt pathway, and similar to the GSEA results, other molecules related to Wnt activity were represented in this gene set (Fig. 6B). Wnt5b and Wnt7b were reproducible between gene expression chips (Fig. 6C). Wnt5b expression was validated by QRT-PCR in Fig. 3. Wnt7b is of very low transcript abundance and therefore nested semiquantitative RT-PCR, a two-step PCR protocol, is required for detection. Fig. 6D illustrates that Wnt7b is not produced in splenic macrophages (Sp Macs), but is slightly increased in GTs and highly enriched in invasive TAMs (NC TAMs).
FIGURE 6.
Wnt7b in TAMs and human breast cancer. A, Expression pattern for all Wnt ligands queried by Nimblegen gene expression arrays. Most Wnt ligands were found to be increased (red) in invasive TAMs compared with general TAMs, except for Wnt8b which was decreased (green). Asterisks indicate Wnt5b and Wnt7b that were called as significantly differentially regulated. B, Generalized representation of relative abundance of molecules driving Wnt-signaling in invasive TAMs. Double outlined molecules (Frizzled, LRP1/5/6, DKK, and WNT) contain multiple members that are not individually depicted. Shading represents the trend for the entire group (e.g., note expression of individual Wnt ligands in A versus total WNT expression). Diagram generated using IPA 6.3. C, Normalized array expression intensities (log base 2) for Wnt5b and Wnt7b for each biologic replicate depict reproducibility between samples. The line indicates average expression for each set of experiments.D, Semiquantitative PCR for Wnt7b expression in splenic macrophages (Sp Macs), GTs (TAMs) and invasive, needle-collected TAMs (NC TAMs). E, Representative semiquantitative PCR for Wnt7b in splenic macrophages (Sp Macs) and TAMs following flow cytometric sorting and 12-h culture to ensure pure macrophage populations. F, Higher expression of Wnt7b in human breast cancer correlates with advanced disease. Two separate studies were mined using Oncomine (39) and indicate that breast cancer metastatis to lymph nodes has significantly higher WNT7B expression.
Because Wnt7b is a confirmed macrophage gene product (48, 49), it was selected for more stringent confirmation of expression in TAMs, which has not previously been demonstrated. Considering there is a known contamination of invasive carcinoma cells in the invasive TAM preparations (25), it was desired to attribute the expression of Wnt7b to the TAMs directly. Wnt7b was indicated to be enriched in GTs, and although GTs show decreased expression compared with invasive TAMs, we aimed to determine whether Wnt7b expression could be detected in a pure population of GTs. It has been previously established that using a protocol exploiting the adhesive property of macrophages, a pure population of TAMs can be isolated (50). Dextran+ Csf1r-EGFP+ splenic macrophages and TAMs were sorted by flow cytometry and plated onto bacterial plastic. After 12 h incubation, macrophages were washed and RNA was extracted. Following semiquantitative RT-PCR, Wnt7b expression was present only in the TAM population (Fig. 6E). It is noted that this method is not specific for invasive TAMs whose paucity precludes this type of analysis; however, by establishing that Wnt7b was present in a pure population of TAMs, the microarray data already presented suggest that this expression would be enriched in invasive TAMs.
Lastly, to ascertain the significance of Wnt7b expression in human breast cancer, Oncomine (www.oncomine.org) (39) was used to analyze previously published human breast cancer microarray studies for its expression (Fig. 6F). Oncomine compiles and organizes human tumor microarray studies, allowing the data to be analyzed by independent researchers. In these prior studies, tumor samples are generally from whole-tissue biopsies; therefore, a heterogeneous assortment of cell types from the tumor microenvironment can be carefully assessed as previously demonstrated in follicular lymphoma (51) and human breast cancer (52). In our screening of the database, increased expression of WNT7B was found to be significantly correlated with positive lymph node metastasis (53, 54). Although this analysis interrogates the expression of every cell present in the tumor biopsy and not TAMS alone, when analyzed in conjunction with the other data, it provides support that this increased WNT7B expression in advanced disease is due to the increased expression in present TAMs.
Discussion
As an early step in tumor metastasis, targeting carcinoma cell motility and invasion within the primary tumor microenvironment presents a unique opportunity to impede tumor progression. Previous clinical data have indicated that high density of TAMs within a tumor microenvironment correlates with poor prognosis (55). Furthermore, recent experimental data has provided compelling evidence indicating TAMs in the direct facilitation of tumor invasion (15, 19, 20). The studies herein describe the in vivo functional isolation of a subpopulation of TAMs caught in the act of promoting carcinoma cell motility; their gene expression profile, compared with a GT population, isolated indiscriminately to function by flow cytometry using the macrophage known macropinocytic capacity and cell-surface expression of F4/80 as markers. It has previously been shown that phagocytosis of dextran does not compromise a macrophage’s motility or its ability to secrete EGF in response to CSF-1 (15). Furthermore, it has previously been shown that invasive TAMs are a uniform population that express F4/80 and are macropinocytic (15, 19). Therefore, these experiments are specific for comparing the gene expression of those TAMs actively promoting tumor cell invasion versus the vast majority of TAMs that are not.
Previously, gene expression studies of invasive tumor cells collected through the in vivo invasion assay have yielded mechanistic insight into the cell biology–namely motility (26, 27, 29, 56) and viability (28)–of this subpopulation of tumor cells. Considering an inherent requirement for TAMs, we hypothesized that gene expression studies of invasive TAMs would also provide useful insight into the physiology of TAM promotion of tumor progression. The invasive TAM arrays indicated a considerable number of transcripts as differentially regulated, despite use of highly stringent gene selection criteria. Therefore, we interrogated possible artifactual explanations that could account for these differences. In-depth examination of the array data failed to identify consistent biases or deviations from the norm; therefore, technical artifacts could not account for why so many genes were differentially regulated (Supplemental Fig. 1). Moreover, QRT-PCR on subsequent biologic repeats confirmed all tested results from the gene expression arrays (Fig. 3). We were also concerned that the large number of differences would be due to an irreducible contamination of carcinoma cells, but significant overlap in expression data was not found when the results were compared with invasive carcinoma cell expression data. Taken together, these results indicate that this invasive macrophage population has a unique transcriptome significantly different from the GT population. Useful data can be extracted from the microarray data; however, given that some of these differences may be caused by the experimental perturbation required to collect these cells, caution must be exercised and mechanistic experiments will be needed to confirm that the results are physiologically relevant.
IPA 6.3 was used to embark on a bioinformatic analysis of the enriched functional categories within the stringently identified 1457 transcripts called as differentially regulated by SAM. Physiologic system development and function categories were ranked by significance of enrichment for clues as to how this invasive TAM population was affecting the tumor microenvironment (Table I). It was anticipated that, similar to invasive carcinoma cells (25), invasive TAMs would distinctly demonstrate enrichment in molecules involved in the paracrine loop required for tumor cell migration and invasion–EGF and CSF-1R in macrophages (19)– and those mediating macrophage motility to be specifically enriched in invasive TAMs. In fact, the opposite was found wherein Csf1r transcript expression, whose biochemical function is critical to the paracrine loop expression, was diminished, as were molecules critical to normal macrophage migration (Fig. 3). Unlike tumor cells, macrophages are nongenomically transformed immune cells in which exquisite transcriptional, translational, and posttranslational control is maintained to prevent a shift in balance of immune activity to an overly exaggerated inflammatory response. Therefore, although the results were not anticipated, we do not suggest that they conflict with the established protein requirement for these molecules in the paracrine loop and macrophage motility. The CSF-1R, while not only being on the cell surface, is maintained in a large cryptic intracellular pool from where it can be rapidly mobilized to the cell surface (57). Furthermore, it has recently been shown that Csf1r expression is decreased in macrophages following stimulation by lipopolysaccharide (58, 59), and it is feasible that other molecules communicating between TAMs and tumor cells during their active comigration may produce a similar effect.
Conversely, embryonic and tissue development were enriched ontologic designations of genes overexpressed in invasive TAMs. This finding was interesting, considering that macrophages are well known to have essential tissue trophic functions during development (1, 60), and especially during mammary gland development (21, 61, 62). TAMs may be recapitulating a number of these tissue trophic functions executed during development to promote tumor progression (63, 64). The annotation of transcriptional profiles to function was further explored using GSEA (46), an analytical method that examines the distribution of functionally related genes in an ordered gene expression dataset without the selection of upregulated or downregulated genes. This method was used to examine the KEGG pathways that are present in humans and enriched in the invasive TAM dataset. Notably, the Wnt-signaling pathway, a pathway critical to normal tissue development was identified as highly enriched (Fig. 4, Table II). Although a KEGG pathway does not exist specifically for breast cancer, it was noted that the invasive TAM gene set was enriched for molecules frequently overexpressed in human basal cell carcinoma (Fig. 4B). Except for Wnt-signaling being a common denominator, the significance of this association is unclear. However, through population studies it is known that the diagnosis of numerous skin cancers, including basal cell carcinoma, confers an increased relative risk of the development of breast cancer (65, 66). Moreover, in human basal cell carcinoma, increased TAM density correlates with increased depth of invasion and microvessel density (67). This finding suggests that perhaps the KEGG gene set attributed to basal cell carcinoma derives in part from the TAMs present within the tumor micronenvironment.
We were compelled to further study the significant over-expression of Wnt7b in TAMs because of its known production by macrophages (48, 49) and the known role of Wnt signaling in development (68, 69) and cancer (70). The Wnt7b ligand is a notoriously low-abundant transcript, and it was necessary to use nested primers for semiquantitative PCR to detect message (48). Genetic tools to transgenically label cells producing Wnt7b, described in studies of macrophage contribution to vascular remodeling of hyaloid vessels in the developing mouse eye (48), will be useful in future tumor studies although conclusive studies may be hampered when transitioning these models from the relatively hypocellular developing mouse eye to the hypercellular tumor microenvironment. Therefore, although our gene expression studies originally identified Wnt7b as being increased in invasive TAMs compared with GTs, we aimed to provide evidence that in general, TAMs express Wnt7b.
Semiquantitative PCR confirmed that Wnt7b is minimally enriched in TAMs and further enriched in invasive TAMs (Fig. 6D). We considered that if carcinoma cells produced the majority of Wnt7b, a profile similar to that seen in Fig. 6D would be produced, because flow cytometric sorting generally isolates a 97–99% pure population on postsort, whereas cell separation following the in vivo invasion assay yields a slightly higher contamination (25). Therefore, a second validated (50) macrophage isolation step following flow cytometry was used to more purely study the Wnt7b expression in adherent TAMs. After isolation of TAMs and splenic macrophages by flow cytometry, they were allowed to adhere to petri dishes. RNA was subsequently extracted from both pure macrophage populations and Wnt7b expression was only detected in TAMs.
As mentioned above, in the developing mouse eye, macrophages direct vascular remodeling of hyaloid vessels by producing Wnt7b (48, 49). Interestingly, recent work (49) has proposed a mechanism in this model whereby macrophages produce Wnt7b that stimulates the canonical Wnt signaling pathway in vascular endothelial cells (VECs), which express Wnt coreceptors Lrp5 and Frizzled, in a paracrine fashion. This signaling cascade eventually results in the stabilization of β-catenin and VEC entry into cell cycle. Furthermore, in the developing mouse eye, angiopoietin-2 production by vessel-related pericytes provides an apoptotic signal to VECs, leading to their death and overall vascular remodeling. This would suggest that in the absence of an apoptotic signal to VECs, this model could provide a mechanism for macrophage stimulation of VEC proliferation, such as in tumor angiogenesis.
Perivascular TAM clusters are indicated to most efficiently promote carcinoma cell motility in an EGF/CSF-1 paracrine loop dependent manner in PyMT mammary tumors. Consequently, these perivascular TAM clusters have been proposed to be an invasive hotspot (15). In human breast cancer tissue sections, triple staining for juxtaposed clusters of macrophages, vascular endothelial cells, and a specific protein marker found in invasive carcinoma cells indicated that more of these clusters correlated with human metastatic disease (71). We propose that these macrophages located along the tumor vasculature and associated with invasive tumor cells are the invasive TAM subpopulation and that these TAMs function, in part, through modulating Wnt-signaling in the local microenvironment. It would not be insignificant that TAM-derived Wnt7b may also have a role in vasculogenesis, as seen in the developing eye (48, 49) because there is considerable evidence of extensive angiogenesis at the sites of leukocytic invasion in human breast tumors (6). Furthermore, it is believed that these angiogenic beds are also the sites of tumor cell invasion (3, 63). Our results linking Wnt7b and tumor cell invasion are consistent with this hypothesis. Not insignificantly, human breast tumors that have higher expression levels of WNT7B are associated with advanced disease as defined by tumor-positive lymph nodes (Fig. 6F).
This article describes the gene expression signature of a unique invasive TAM subpopulation that putatively has roles in tumor progression through promotion of tumor metastasis and angiogenesis, with the role of Wnt signaling as a common denominator via TAM-derived Wnt7b. Moreover, we submit that with careful mining, the results from the gene expression arrays described in this study will provide a useful tool for further insight into mechanisms of TAM promotion of tumor progression.
Supplementary Material
Acknowledgments
This research was supported by National Institutes of Health Grants CA131270 (J.W.P), CA100324 (J.S.C. and J.W.P), and CA126511 (J.S.C.); the Einstein Cancer Center Grant P30 133330 (J.W.P., J.S.C.); and the Massachusetts Institute of Technology Cancer Center Grant P30-CA14051 (C.A.W.). L.S.O. was supported by the Medical Sciences Training Grant T32 GM 07288. J.W.P. is the Louis Goldstein Swan Chair in Women’s Cancer Research.
Abbreviations used in this paper
- AU
approximately unbiased
- EGF
epidermal growth factor
- EGFP
enhanced GFP
- FDR
false discovery rate
- GSEA
gene set enrichment analysis
- GT
general tumor-associated macrophage
- IPA
Ingenuity Pathways Knowledge Base
- PyMT
polyoma middle T
- QRT-PCR
quantitative real-time polymerase chain reaction
- SAM
significance analysis of microarray
- SM
splenic macrophage
- TAM
tumor-associated macrophage
- VEC
vascular endothelial cell
Footnotes
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Pollard JW. Trophic macrophages in development and disease. Nat. Rev. Immunol. 2009;9:259–270. doi: 10.1038/nri2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nat. Rev. Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat. Rev. Cancer. 2004;4:71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 4.Ojalvo LS, King W, Cox D, Pollard JW. High-density gene expression analysis of tumor-associated macrophages from mouse mammary tumors. Am. J. Pathol. 2009;174:1048–1064. doi: 10.2353/ajpath.2009.080676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lewis CE, Pollard JW. Distinct role of macrophages in different tumor microenvironments. Cancer Res. 2006;66:605–612. doi: 10.1158/0008-5472.CAN-05-4005. [DOI] [PubMed] [Google Scholar]
- 6.Leek RD, Landers RJ, Harris AL, Lewis CE. Necrosis correlates with high vascular density and focal macrophage infiltration in invasive carcinoma of the breast. Br. J. Cancer. 1999;79:991–995. doi: 10.1038/sj.bjc.6690158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsutsui S, Yasuda K, Suzuki K, Tahara K, Higashi H, Era S. Macrophage infiltration and its prognostic implications in breast cancer: the relationship with VEGF expression and microvessel density. Oncol. Rep. 2005;14:425–431. [PubMed] [Google Scholar]
- 8.Lin EY, Jones JG, Li P, Zhu L, Whitney KD, Muller WJ, Pollard JW. Progression to malignancy in the polyoma middle T oncoprotein mouse breast cancer model provides a reliable model for human diseases. Am. J. Pathol. 2003;163:2113–2126. doi: 10.1016/S0002-9440(10)63568-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin EY, Nguyen AV, Russell RG, Pollard JW. Colony-stimulating factor 1 promotes progression of mammary tumors to malignancy. J. Exp. Med. 2001;193:727–740. doi: 10.1084/jem.193.6.727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hildenbrand R, Wolf G, Böhme B, Bleyl U, Steinborn A. Urokinase plasminogen activator receptor (CD87) expression of tumor-associated macrophages in ductal carcinoma in situ, breast cancer, and resident macrophages of normal breast tissue. J. Leukoc. Biol. 1999;66:40–49. doi: 10.1002/jlb.66.1.40. [DOI] [PubMed] [Google Scholar]
- 11.Vasiljeva O, Papazoglou A, Krüger A, Brodoefel H, Korovin M, Deussing J, Augustin N, Nielsen BS, Almholt K, Bogyo M, et al. Tumor cell-derived and macrophage-derived cathepsin B promotes progression and lung metastasis of mammary cancer. Cancer Res. 2006;66:5242–5250. doi: 10.1158/0008-5472.CAN-05-4463. [DOI] [PubMed] [Google Scholar]
- 12.Wolf M, Clark-Lewis I, Buri C, Langen H, Lis M, Mazzucchelli L. Cathepsin D specifically cleaves the chemokines macrophage inflammatory protein-1 α, macrophage inflammatory protein-1 β, and SLC that are expressed in human breast cancer. Am. J. Pathol. 2003;162:1183–1190. doi: 10.1016/s0002-9440(10)63914-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hagemann T, Robinson SC, Schulz M, Trümper L, Balkwill FR, Binder C. Enhanced invasiveness of breast cancer cell lines upon co-cultivation with macrophages is due to TNF-α dependent up-regulation of matrix metalloproteases. Carcinogenesis. 2004;25:1543–1549. doi: 10.1093/carcin/bgh146. [DOI] [PubMed] [Google Scholar]
- 14.Pedersen TX, Pennington CJ, Almholt K, Christensen IJ, Nielsen BS, Edwards DR, Rømer J, Danø K, Johnsen M. Extracellular protease mRNAs are predominantly expressed in the stromal areas of microdissected mouse breast carcinomas. Carcinogenesis. 2005;26:1233–1240. doi: 10.1093/carcin/bgi065. [DOI] [PubMed] [Google Scholar]
- 15.Wyckoff JB, Wang Y, Lin EY, Li JF, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67:2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 16.Fidler IJ. The pathogenesis of cancer metastasis: the ‘seed and soil’ hypothesis revisited. Nat. Rev. Cancer. 2003;3:453–458. doi: 10.1038/nrc1098. [DOI] [PubMed] [Google Scholar]
- 17.Pukrop T, Klemm F, Hagemann T, Gradl D, Schulz M, Siemes S, Trümper L, Binder C. Wnt 5a signaling is critical for macrophage-induced invasion of breast cancer cell lines. Proc. Natl. Acad. Sci. USA. 2006;103:5454–5459. doi: 10.1073/pnas.0509703103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyckoff JB, Segall JE, Condeelis JS. The collection of the motile population of cells from a living tumor. Cancer Res. 2000;60:5401–5404. [PubMed] [Google Scholar]
- 19.Wyckoff J, Wang W, Lin EY, Wang Y, Pixley F, Stanley ER, Graf T, Pollard JW, Segall J, Condeelis J. A paracrine loop between tumor cells and macrophages is required for tumor cell migration in mammary tumors. Cancer Res. 2004;64:7022–7029. doi: 10.1158/0008-5472.CAN-04-1449. [DOI] [PubMed] [Google Scholar]
- 20.Goswami S, Sahai E, Wyckoff JB, Cammer M, Cox D, Pixley FJ, Stanley ER, Segall JE, Condeelis JS. Macrophages promote the invasion of breast carcinoma cells via a colony-stimulating factor-1/epidermal growth factor paracrine loop. Cancer Res. 2005;65:5278–5283. doi: 10.1158/0008-5472.CAN-04-1853. [DOI] [PubMed] [Google Scholar]
- 21.Lin EY, Gouon-Evans V, Nguyen AV, Pollard JW. The macrophage growth factor CSF-1 in mammary gland development and tumor progression. J. Mammary Gland Biol. Neoplasia. 2002;7:147–162. doi: 10.1023/a:1020399802795. [DOI] [PubMed] [Google Scholar]
- 22.Kacinski BM. CSF-1 and its receptor in ovarian, endometrial and breast cancer. Ann. Med. 1995;27:79–85. doi: 10.3109/07853899509031941. [DOI] [PubMed] [Google Scholar]
- 23.Scholl SM, Pallud C, Beuvon F, Hacene K, Stanley ER, Rohrschneider L, Tang R, Pouillart P, Lidereau R. Anti-colony-stimulating factor-1 antibody staining in primary breast adenocarcinomas correlates with marked inflammatory cell infiltrates and prognosis. J. Natl. Cancer Inst. 1994;86:120–126. doi: 10.1093/jnci/86.2.120. [DOI] [PubMed] [Google Scholar]
- 24.O’Sullivan C, Lewis CE, Harris AL, McGee JO. Secretion of epidermal growth factor by macrophages associated with breast carcinoma. Lancet. 1993;342:148–149. doi: 10.1016/0140-6736(93)91348-p. [DOI] [PubMed] [Google Scholar]
- 25.Wang W, Wyckoff JB, Wang Y, Bottinger EP, Segall JE, Condeelis JS. Gene expression analysis on small numbers of invasive cells collected by chemotaxis from primary mammary tumors of the mouse. BMC Biotechnol. 2003;3:13. doi: 10.1186/1472-6750-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W, Goswami S, Lapidus K, Wells AL, Wyckoff JB, Sahai E, Singer RH, Segall JE, Condeelis JS. Identification and testing of a gene expression signature of invasive carcinoma cells within primary mammary tumors. Cancer Res. 2004;64:8585–8594. doi: 10.1158/0008-5472.CAN-04-1136. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Mouneimne G, Sidani M, Wyckoff J, Chen X, Makris A, Goswami S, Bresnick AR, Condeelis JS. The activity status of cofilin is directly related to invasion, intravasation, and metastasis of mammary tumors. J. Cell Biol. 2006;173:395–404. doi: 10.1083/jcb.200510115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goswami S, Wang W, Wyckoff JB, Condeelis JS. Breast cancer cells isolated by chemotaxis from primary tumors show increased survival and resistance to chemotherapy. Cancer Res. 2004;64:7664–7667. doi: 10.1158/0008-5472.CAN-04-2027. [DOI] [PubMed] [Google Scholar]
- 29.Wang W, Wyckoff JB, Goswami S, Wang Y, Sidani M, Segall JE, Condeelis JS. Coordinated regulation of pathways for enhanced cell motility and chemotaxis is conserved in rat and mouse mammary tumors. Cancer Res. 2007;67:3505–3511. doi: 10.1158/0008-5472.CAN-06-3714. [DOI] [PubMed] [Google Scholar]
- 30.Guy CT, Cardiff RD, Muller WJ. Induction of mammary tumors by expression of polyomavirus middle T oncogene: a transgenic mouse model for metastatic disease. Mol. Cell. Biol. 1992;12:954–961. doi: 10.1128/mcb.12.3.954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasmono RT, Oceandy D, Pollard JW, Tong W, Pavli P, Wainwright BJ, Ostrowski MC, Himes SR, Hume DA. A macrophage colony-stimulating factor receptor-green fluorescent protein transgene is expressed throughout the mononuclear phagocyte system of the mouse. Blood. 2003;101:1155–1163. doi: 10.1182/blood-2002-02-0569. [DOI] [PubMed] [Google Scholar]
- 32.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 33.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 34.Ihaka R, Gentleman R. R: A Language for Data Analysis and Graphics. J. Comput. Graph. Statist. 1996;5:299–314. [Google Scholar]
- 35.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smyth GK. Limma: linear models for microarray data. In: Gentleman VCR, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 37.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci. U.S.A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 39.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Δ Δ C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 42.Qian B, Deng Y, Im JH, Muschel RJ, Zou Y, Li J, Lang RA, Pollard JW. A distinct macrophage population mediates metastatic breast cancer cell extravasation, establishment and growth. PLoS One. 2009;4:e6562. doi: 10.1371/journal.pone.0006562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sasmono RT, Ehrnsperger A, Cronau SL, Ravasi T, Kandane R, Hickey MJ, Cook AD, Himes SR, Hamilton JA, Hume DA. Mouse neutrophilic granulocytes express mRNA encoding the macrophage colony-stimulating factor receptor (CSF-1R) as well as many other macrophage-specific transcripts and can transdifferentiate into macrophages in vitro in response to CSF-1. J. Leukoc. Biol. 2007;82:111–123. doi: 10.1189/jlb.1206713. [DOI] [PubMed] [Google Scholar]
- 45.Pucci F, Venneri MA, Biziato D, Nonis A, Moi D, Sica A, Di Serio C, Naldini L, De Palma M. A distinguishing gene signature shared by tumor-infiltrating Tie2-expressing monocytes (TEMs), blood “resident” monocytes and embryonic macrophages suggests common functions and developmental relationships. Blood. 2009;114:901–914. doi: 10.1182/blood-2009-01-200931. [DOI] [PubMed] [Google Scholar]
- 46.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kanehisa M, Goto S, Hattori M, Aoki-Kinoshita KF, Itoh M, Kawashima S, Katayama T, Araki M, Hirakawa M. From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 2006;34(Database issue):D354–D357. doi: 10.1093/nar/gkj102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lobov IB, Rao S, Carroll TJ, Vallance JE, Ito M, Ondr JK, Kurup S, Glass DA, Patel MS, Shu W, et al. WNT7b mediates macrophage-induced programmed cell death in patterning of the vasculature. Nature. 2005;437:417–421. doi: 10.1038/nature03928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rao S, Lobov IB, Vallance JE, Tsujikawa K, Shiojima I, Akunuru S, Walsh K, Benjamin LE, Lang RA. Obligatory participation of macrophages in an angiopoietin 2-mediated cell death switch. Development. 2007;134:4449–4458. doi: 10.1242/dev.012187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Biswas SK, Gangi L, Paul S, Schioppa T, Saccani A, Sironi M, Bottazzi B, Doni A, Vincenzo B, Pasqualini F, et al. A distinct and unique transcriptional program expressed by tumor-associated macrophages (defective NF-kappaB and enhanced IRF-3/STAT1 activation) Blood. 2006;107:2112–2122. doi: 10.1182/blood-2005-01-0428. [DOI] [PubMed] [Google Scholar]
- 51.Dave SS, Wright G, Tan B, Rosenwald A, Gascoyne RD, Chan WC, Fisher RI, Braziel RM, Rimsza LM, Grogan TM, et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N. Engl. J. Med. 2004;351:2159–2169. doi: 10.1056/NEJMoa041869. [DOI] [PubMed] [Google Scholar]
- 52.Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 53.Miller LD, Smeds J, George J, Vega VB, Vergara L, Ploner A, Pawitan Y, Hall P, Klaar S, Liu ET, Bergh J. An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. U.S.A. 2005;102:13550–13555. doi: 10.1073/pnas.0506230102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ivshina AV, George J, Senko O, Mow B, Putti TC, Smeds J, Lindahl T, Pawitan Y, Hall P, Nordgren H, et al. Genetic reclassification of histologic grade delineates new clinical subtypes of breast cancer. Cancer Res. 2006;66:10292–10301. doi: 10.1158/0008-5472.CAN-05-4414. [DOI] [PubMed] [Google Scholar]
- 55.Bingle L, Brown NJ, Lewis CE. The role of tumour-associated macrophages in tumour progression: implications for new anticancer therapies. J. Pathol. 2002;196:254–265. doi: 10.1002/path.1027. [DOI] [PubMed] [Google Scholar]
- 56.Lapidus K, Wyckoff J, Mouneimne G, Lorenz M, Soon L, Condeelis JS, Singer RH. ZBP1 enhances cell polarity and reduces chemotaxis. J. Cell Sci. 2007;120:3173–3178. doi: 10.1242/jcs.000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guilbert LJ, Stanley ER. The interaction of 125I-colony-stimulating factor-1 with bone marrow-derived macrophages. J. Biol. Chem. 1986;261:4024–4032. [PubMed] [Google Scholar]
- 58.Bonifer C, Hume DA. The transcriptional regulation of the Colony-Stimulating Factor 1 Receptor (csf1r) gene during hematopoiesis. Front. Biosci. 2008;13:549–560. doi: 10.2741/2700. [DOI] [PubMed] [Google Scholar]
- 59.Sester DP, Trieu A, Brion K, Schroder K, Ravasi T, Robinson JA, McDonald RC, Ripoll V, Wells CA, Suzuki H, et al. LPS regulates a set of genes in primary murine macrophages by antagonising CSF-1 action. Immunobiology. 2005;210:97–107. doi: 10.1016/j.imbio.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 60.Rae F, Woods K, Sasmono T, Campanale N, Taylor D, Ovchinnikov DA, Grimmond SM, Hume DA, Ricardo SD, Little MH. Characterisation and trophic functions of murine embryonic macrophages based upon the use of a Csf1r-EGFP transgene reporter. Dev. Biol. 2007;308:232–246. doi: 10.1016/j.ydbio.2007.05.027. [DOI] [PubMed] [Google Scholar]
- 61.Gouon-Evans V, Lin EY, Pollard JW. Requirement of macrophages and eosinophils and their cytokines/chemokines for mammary gland development. Breast Cancer Res. 2002;4:155–164. doi: 10.1186/bcr441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gouon-Evans V, Rothenberg ME, Pollard JW. Postnatal mammary gland development requires macrophages and eosinophils. Development. 2000;127:2269–2282. doi: 10.1242/dev.127.11.2269. [DOI] [PubMed] [Google Scholar]
- 63.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124:263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 64.Ingman WV, Wyckoff J, Gouon-Evans V, Condeelis J, Pollard JW. Macrophages promote collagen fibrillogenesis around terminal end buds of the developing mammary gland. Dev. Dyn. 2006;235:3222–3229. doi: 10.1002/dvdy.20972. [DOI] [PubMed] [Google Scholar]
- 65.Frisch M, Hjalgrim H, Olsen JH, Melbye M. Risk for subsequent cancer after diagnosis of basal-cell carcinoma. A population-based, epidemiologic study. Ann. Intern. Med. 1996;125:815–821. doi: 10.7326/0003-4819-125-10-199611150-00005. [DOI] [PubMed] [Google Scholar]
- 66.Soerjomataram I, Louwman WJ, Lemmens VE, Coebergh JW, de Vries E. Are patients with skin cancer at lower risk of developing colorectal or breast cancer? Am. J. Epidemiol. 2008;167:1421–1429. doi: 10.1093/aje/kwn077. [DOI] [PubMed] [Google Scholar]
- 67.Tjiu JW, Chen JS, Shun CT, Lin SJ, Liao YH, Chu CY, Tsai TF, Chiu HC, Dai YS, Inoue H, et al. Tumor-associated macrophage-induced invasion and angiogenesis of human basal cell carcinoma cells by cyclooxygenase-2 induction. J. Invest. Dermatol. 2009;129:1016–1025. doi: 10.1038/jid.2008.310. [DOI] [PubMed] [Google Scholar]
- 68.Moon RT, Brown JD, Torres M. WNTs modulate cell fate and behavior during vertebrate development. Trends Genet. 1997;13:157–162. doi: 10.1016/s0168-9525(97)01093-7. [DOI] [PubMed] [Google Scholar]
- 69.Schwab KR, Patterson LT, Hartman HA, Song N, Lang RA, Lin X, Potter SS. Pygo1 and Pygo2 roles in Wnt signaling in mammalian kidney development. BMC Biol. 2007;5:15. doi: 10.1186/1741-7007-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Logan CY, Nusse R. The Wnt signaling pathway in development and disease. Annu. Rev. Cell Dev. Biol. 2004;20:781–810. doi: 10.1146/annurev.cellbio.20.010403.113126. [DOI] [PubMed] [Google Scholar]
- 71.Robinson BD, Sica GL, Liu YF, Rohan TE, Gertler FB, Condeelis JS, Jones JG. Tumor microenvironment of metastasis in human breast carcinoma: a potential prognostic marker linked to hematogenous dissemination. Clin. Cancer Res. 2009;15:2433–2441. doi: 10.1158/1078-0432.CCR-08-2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.