Abstract
The discovery that several inherited human diseases are caused by mtDNA depletion has led to an increased interest in the replication and maintenance of mtDNA. We have isolated a new mutant in the lopo (low power) gene from Drosophila melanogaster affecting the mitochondrial single-stranded DNA-binding protein (mtSSB), which is one of the key components in mtDNA replication and maintenance. lopo1 mutants die late in the third instar before completion of metamorphosis because of a failure in cell proliferation. Molecular, histochemical, and physiological experiments show a drastic decrease in mtDNA content that is coupled with the loss of respiration in these mutants. However, the number and morphology of mitochondria are not greatly affected. Immunocytochemical analysis shows that mtSSB is expressed in all tissues but is highly enriched in proliferating tissues and in the developing oocyte. lopo1 is the first mtSSB mutant in higher eukaryotes, and its analysis demonstrates the essential function of this gene in development, providing an excellent model to study mitochondrial biogenesis in animals.
INTRODUCTION
Mitochondria are critical for their central role in energy metabolism, but they also play important roles in aging, apoptosis, and a variety of genetically inherited human diseases (Larsson and Clayton, 1995; Grossman and Shoubridge, 1996; Zeviani et al., 1997). Whereas there is a wealth of information on the structure and function of mitochondria, very little is known about the regulation of their biogenesis. Eukaryotic cells contain not only highly variable numbers of mitochondria per cell but mitochondria also differ substantially in their mtDNA copy number, where the number of mtDNA genomes varies depending on the energy requirements of a particular cell (Shadel and Clayton, 1997). A complex process of interactions between proteins encoded in the nuclear and mtDNA genomes regulates mitochondrial biogenesis. mtDNA generally encodes 22 tRNAs, 2 rRNAs, and 13 polypeptides that are all components of the respiratory chain enzyme complexes. All of the remaining mitochondrial proteins are encoded by nuclear genes and are targeted specifically to mitochondria (Schatz, 1996). In particular, enzymes required for mtDNA replication and transcription are all encoded by the nuclear genome.
The complexity of interactions between the nucleus and mitochondria is also apparent in the variety of human diseases resulting from defects in mtDNA: a large number of mutations (either point mutations or mtDNA rearrangements) have been identified as the cause of severe human diseases. There are, however, autosomal inherited genetic defects that lead to mtDNA depletion (Larsson and Clayton, 1995; Zeviani et al., 1997). Obvious candidate genes for the latter are nuclear genes involved in mtDNA replication or maintenance, and among them are those encoding the mtDNA polymerase, pol γ, which consists of two subunits and is the key enzyme in mtDNA replication (Wernette and Kaguni, 1986; Shadel and Clayton, 1997). At least four other proteins that are required for proper replication and/or maintenance of mtDNA have been identified, including two DNA helicases (Lahaye et al., 1993; Sedman et al., 2000), the mitochondrial transcription factor A (Larsson et al. 1998), and mitochondrial single-stranded DNA-binding protein (mtSSB).
mtSSB is a conserved molecule of 13–18 kDa identified in a variety of eukaryotic organisms ranging from yeast to humans (Pavco and Van Tuyle, 1985, Hoke et al., 1990; van Dyck et al., 1992; Tiranti et al. 1993, Stroumbakis et al., 1994; Thömmes et al., 1995; Li and Williams, 1997). mtSSB binds to single-stranded DNA (ssDNA), coating the displaced ssDNA, which is the template for lagging DNA strand synthesis in mtDNA replication, and preventing it from renaturation (Van Tuyle and Pavco, 1985). mtSSB also stimulates greatly the activity of pol γ from various species (Mignotte et al., 1988; Hoke et al. 1990; Williams and Kaguni, 1994; Thömmes et al., 1995). Although the mtSSB gene was shown to be essential in yeast (van Dyck et al. 1992), its role in mitochondrial biogenesis in animals has not been explored, despite the fact that it represents a candidate gene for inherited human mtDNA depletion.
In this report, we present the molecular and phenotypic analysis of a new mutation in the Drosophila mtSSB gene, which we have called lopo (low power). We show in this first mtSSB mutant in animals that the absence of mtSSB results in the loss of mtDNA and hence a loss of respiration in mitochondria. Furthermore, lopo is absolutely required for proper development because flies die prematurely during the larval and pupal stages. The Drosophila mtSSB mutant might therefore serve as an excellent model system to study mtDNA maintenance and replication in higher eukaryotes.
MATERIALS AND METHODS
Fly Stocks, P-Element Excision, and Transgenic Rescue Analysis
The line P[mini-w+;lacW]0046/13 was originally isolated as a third chromosomal lethal mutation (Deák et al. 1997). Remobilization was achieved using the Δ2–3 Sb chromosome as a transposase source. Excision events were collected as white-eyed flies. From 109 established lines, 99 lines were still lethal, whereas 10 lines showed a reversion of the lethal phenotype. Rescue of lopo1 lethality was evaluated at 25°C in crosses of lopo1 heterozygotes carrying independent insertions of a wild-type mtSSB transgene in the second chromosome and under the control of the hsp70 promoter (lines B and D). Whereas ∼25% of the progeny of a control cross of lopo1 heterozygotes lacking a wild-type mtSSB transgene die in the late third larval instar, 14.9 and 9.8% of the anticipated 25% moribund larvae are recovered as adults in crosses of lopo1 heterozygous lines B and D.
Cloning and Molecular Analysis of lopo
DNA flanking the P-element insertion was isolated using the plasmid-rescue method (Wilson et al., 1989) and sequenced with an Alf-sequencing apparatus (Pharmacia, Piscataway, NJ). A 3.2-kb genomic XhoI fragment from the P1 phage DS07224(Hartl et al., 1994) was identified as the site of the lopo gene, subcloned in pKS and further characterized using standard molecular methods (Sambrook et al., 1989). Several different methods of DNA extraction were tested for mtDNA analysis. Best results were obtained by the protocol described by Pflugfelder et al. (1990).
Western Blots
Embryos of wild-type and heterozygous lopo flies were collected for 0–16 h on yeasted grape agar plates, washed with 0.01% Triton X-100 and 0.7% NaCl, and allowed to develop on grape agar in Petri dishes for 5–6 d. The heterozygous (tubby) larvae were separated from the homozygous (thin) lopo larvae at third instar and, in parallel with the wild-type larvae, were washed five times with 0.01% Triton X-100 and 0.7% NaCl and then frozen in liquid nitrogen and stored at −80°C. To prepare larval extracts, 300 larvae (∼0.5 g) were thawed on ice for 30 min and then suspended in 1.4 ml of extraction buffer (50 mM Tris-HCl, pH 7.5, 10% glycerol, 8 mM EDTA, 0.1% Triton X-100, 0.5 M NaCl) at 3°C and homogenized by 10 strokes in a Dounce homogenizer. The homogenate was filtered through a 75-mm Nitex screen; the retentate was rehomogenized in the same buffer (0.5 ml) and filtered. The combined homogenates were centrifuged at 17, 500 × g for 30 min at 3°C, and the resulting supernatant fraction was used as the larval extract. Larval extracts were fractionated on Blue Sepharose (Pharmacia) essentially as described by Farr et al. (1999), except that batch loading and elution were used. The larval extracts derived from 300 larvae were mixed with a 50% slurry of Blue Sepharose (140 ml) in buffer containing 30 mM Tris-HCl, pH 7.5/10% glycerol/100 mM NaCl/2 mM EDTA/2 mM dithiothreitol/1 mM phenylmethylsulfonyl fluoride/10 mM sodium metabisulfite/2 mg/ml leupeptin. The beads were incubated overnight at 3°C with gentle agitation and then collected by centrifugation at 3000 × g for 30 s. The beads were then twice washed for 10 min with 2 volumes (140 ml) of the same buffer containing 800 mM NaCl and recentrifuged. The bound protein was eluted twice with 2 volumes each of buffer lacking NaCl and containing 0.5 and 1.0 M sodium thiocyanate. Finally, mtSSB was eluted twice for 2 h with 2 volumes of buffer lacking NaCl and containing 1.5 M sodium thiocyanate.
Oxygen Measurements
Five third instar larvae were immobilized in liquid nitrogen, homogenized in 100 μl of buffer (0.9% NaCl, 20 mM Tris-HCl, pH 8.0, 20 mM glucose), and stored on ice. Oxygen consumption was determined by measuring oxygen concentration (in percentage saturation) with optoelectronic sensors kindly provided by PreSens Precision Sensing (Regensburg, Germany). These electrodes measure oxygen due to the dynamic quenching of luminescence that depends on oxygen concentration (Preininger et al. 1994). Measurements can be done in volumes as small as 10 μl and were preferred over oxygen electrodes which require several milliliters of volume with large oxygen buffer capacity. Samples were adjusted to 20°C, 10 μl of 1 M NADH was added, and oxygen consumption was measured for at least 5 min or until oxygen concentration was below 10%. The dependence of oxygen consumption on mitochondrial activity was tested by the ability to inhibit the reaction with 1 μl of 1 mM KCN. Wild-type and mutant homogenates were measured alternatively to ensure that samples were unaffected by storage (up to 30 min on ice). Protein concentrations were determined by the Bradford (1976) method. Oxygen concentrations are given in nanomoles, assuming that 100% saturation at 20°C corresponds to 0.276 μmol O2/ml.
Histological Analysis
Third instar larval sections (10 μm thick) were used for mitochondrial enzyme histochemistry. For succinate dehydrogenase (SDH) activity, sections were stained unfixed according to the method of Lojda et al. (1979). Nitroblue tetrazolium was used as the color reagent, and the staining specificity was evaluated by omission of the enzyme substrate (succinate). For cytochrome c oxidase (COX) histochemistry, sections were fixed for 15 min in 1% glutaraldehyde in phosphate-buffered saline (PBS) before staining. Staining specificity was tested by staining in the presence of 0.0065% KCN. 5-Bromodeoxyuridine (BUdR) staining was performed according to the method of Truman and Bate (1988), except for a 15 min 2% paraformaldehyde fixation instead of Carnoy.
Immunocytochemistry
Embryonic, larval or adult tissue was frozen in OCT (Tissue-tek, O.C.T. compound, Sakura Finetek Europe, Netherlands) and cut at 10 μm in a cryostat. Tissue was fixed for 10 min in 4% paraformaldehyde in PBS, washed with PBS, and incubated with the primary antiserum (anti-mtSSB; diluted 1:100; Farr et al. 1999) overnight at 4°C. The secondary antibody was a goat anti-rabbit immunoglobulin G-CY3 (Jackson Laboratories, West Grove, PA) used at a 1:1000 dilution for 2 h. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI).
Electron Microscopy
Midgut tissue from third instar larvae was dissected in ice-cold fixative (4% glutaraldehyde in 0.1 M phosphate buffer, pH 7.4) and fixed overnight at 4°C in the same fixative. Tissue samples were washed in phosphate buffer, postfixed in ice-cold 1% Os O4 in phosphate buffer for 1 h, and washed again in phosphate buffer. The specimens were dehydrated in an alcohol series and embedded in Durcupan (Fluka Chemicals, Deisenhofen, Germany). Gray sections were cut on an Ultracut E (Reichert-Jung, Heidelberg, Germany), stained with uranyl acetate (2% in 50% alcohol, 15 min) and lead citrate (2 min; Reynolds, 1963), and examined with an EM10 electron microscope (Zeiss, Thornwood, NY).
RESULTS
Isolation and Molecular Characterization of lopo (Low Power)
lopo1 was isolated in a screen for lethal mutants with defects in the developing visual system. It originates from a collection of P-element–induced lethal mutations on the third chromosome (Deák et al., 1997). Molecular analysis of this line led to the identification of a single P-element inserted in the chromosomal region 89B12. Flanking genomic DNA was isolated by plasmid rescue and was subsequently sequenced, which placed the P-element within the third intron of the gene encoding the mtSSB (Figure 1; Ruiz de Mena et al., 2000). Drosophila mtSSB is a small polypeptide of ∼14 kDa that forms a homotetramer (Thömmes et al., 1995) and is characterized by a mitochondrial presequence and a DNA-binding domain that is conserved among mtSSBs from yeast (van Dyck et al., 1992) to humans (Tiranti et al., 1993).
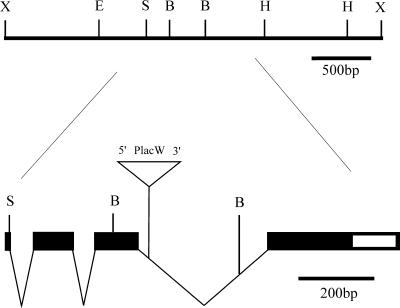
Figure 1.
Molecular map of lopo. The gene was mapped within a 3.2-kb genomic XhoI fragment isolated from the P1-phage DS07224 (Hartl et al., 1994). The open reading frame is indicated by the filled box. It starts at the SphI site and is disrupted by three introns. The P-element insertion in lopo1 is located within the third intron. B, BamHI; E, EcoRI; H, HindIII; S, SphI; X, XhoI.
To prove that the lopo1 mutation is indeed induced by the P-element insertion in the mtSSB gene, the P-element was remobilized. In several independent lines, a precise excision was detected by genomic Southern blots and polymerase chain reaction, which led to a complete reversion of the lethality and all observed phenotypic effects of lopo1 (Maier, Poeck, Vogel, Fischer, and Schneuwly, unpublished results). Moreover, we were able to rescue the lethal phenotype at 25°C, using transformed lines carrying wild-type mtSSB transgenes under the control of the hsp70 promoter (see MATERIALS AND METHODS). We therefore conclude that lopo1 is indeed a lethal P-element–induced mutation in the mtSSB gene.
Lopo Is an Essential Gene Required for Cell Proliferation in Development
To determine the major cause for the lethality in lopo1 mutants, we pursued a phenotypic analysis to determine the lethal phase in this line. As shown in Figure 2, most flies enter the first larval instar, but only 30% are able to start pupariation and eventually die prematurely. The late lethality is perhaps surprising for an essential gene in mitochondrial function (see later) but could be explained by a strong maternal contribution. To test this hypothesis, we performed immunocytochemical analysis with rabbit antiserum against mtSSB. As expected, mtSSB is present in all tissues of adult flies (male and female) and shows a typical mitochondrial spot-like cellular distribution (Figure 3). However, it is not expressed uniformly in all tissues. One of the tissues with a high concentration of mtSSB is the CNS, and in particular the neuropil region where axons are located, whereas only limited expression is seen in neuronal cell bodies (Figure 3A). Other tissues with high mtSSB levels are the digestive tract and ovaries: mtSSB is synthesized highly in nurse cells (Figure 3B) and is transported into the oocyte (Figure 3C). This is consistent with the fact that most mtSSB cDNAs have been isolated from ovarian cDNA libraries (Stroumbakis et al., 1994) and confirms the hypothesis of a strong maternal contribution.
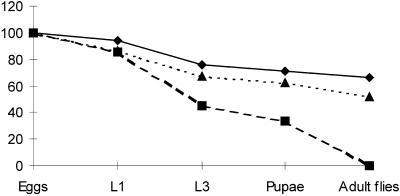
Figure 2.
Effective lethal phase of lopo1. Eggs of wild-type Canton S (bold line), lopo1 (hatched line), and a lopo1 revertant lopo1R47 (dotted line) were collected on apple juice plates and staged accordingly. The graph shows the percentage of surviving individuals at various stages of development (L1, first larval instar; L3, third larval instar). The effective lethal phase is during late third instar and early pupation. Thirty percent of lopo1 larvae enter pupation but never reach adulthood. The revertant lopo1R47 shows a wild-type developmental profile.
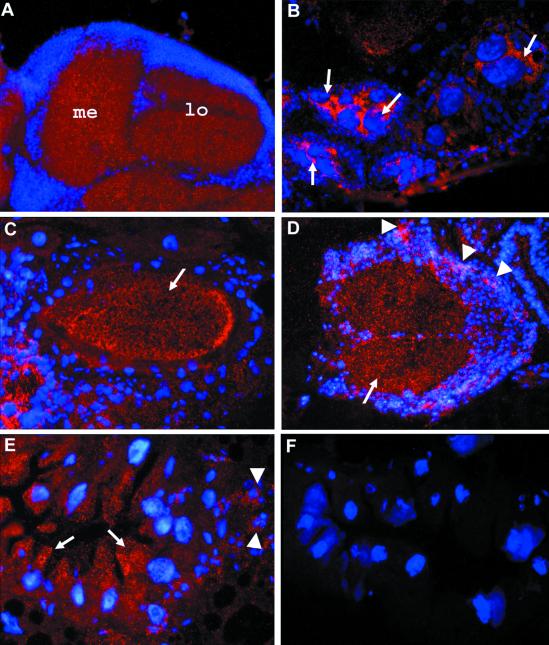
Figure 3.
Immunohistochemical localization of mtSSB. Cryosections of various tissues and stages (10 μm thick) were subjected to immunofluorescence analysis using a rabbit mtSSB antiserum. The tissue was counterstained with DAPI to visualize nuclei. (A) Horizontal section through the visual system of an adult head. Strong labeling is seen in the neuropil region. (B and C) Horizontal sections through the abdomen of an adult female. In B, a high level of mtSSB is seen in nurse cells (arrows). (C) The protein is transported into the oocyte (arrow). (D) A cross-section through a third instar larval ventral ganglion shows expression of mtSSB in the neuropil region (arrow) and elevated expression in proliferating neuroblasts (arrowheads). (E) Cross-section through the midgut of a third instar larvae. A high concentration of mtSSB is found in the apical region of epithelial cells facing the gut lumen (arrows) and in islets of small imaginal cells, giving rise to the adult midgut tissue (arrowheads). (F) Comparable section as in E from a homozygous lopo1 larva. No specific staining is seen.
During early embryogenesis, the protein is evenly distributed, and in late third instar larvae, a high concentration of mtSSB is detected in proliferating tissues such as neuroblasts of the ventral ganglion of the CNS (Figure 3D), in islets of small imaginal cells, which give rise to the adult midgut epithelium (Figure 3E), and in imaginal discs (Maier and Schneuwly, unpublished results), documenting the higher demand for mtSSB in proliferating tissues. Subcellular localization of mtSSB shows its association with mtDNA, as indicated by double staining with DAPI (Maier and Schneuwly, unpublished results). Staining specificity was verified by examination of lopo1 mutant tissue from third instar larvae, which shows no mitochondrial staining (Figure 3F).
These results together with the late onset of lethality suggest that defects in cell proliferation during metamorphosis are likely the major cause for lethality. Cell proliferation can be visualized by BUdR labeling in developing larvae as shown in Figure 4. Indeed, aberrant and drastically reduced proliferation are detected in lopo1. These data also explain the disturbance in visual system development, which was the primary screen used in the identification of this mutant (Maier, Poeck, and Schneuwly, unpublished results).
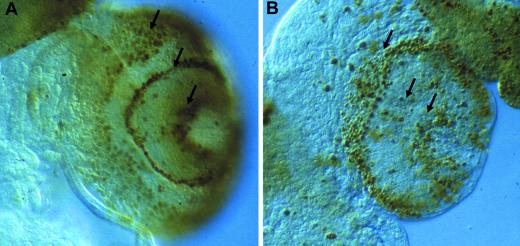
Figure 4.
Proliferation defects in lopo1. BUdR labeling is shown in the third instar larval CNS of wild-type (A) and lopo1 (B). Proliferating cells are organized in proliferation centers (arrows). In lopo1, the number of labeled cells is reduced drastically.
Lopo1 Is Defective in mtDNA Replication and Maintenance Resulting in Loss of Mitochondrial Respiration
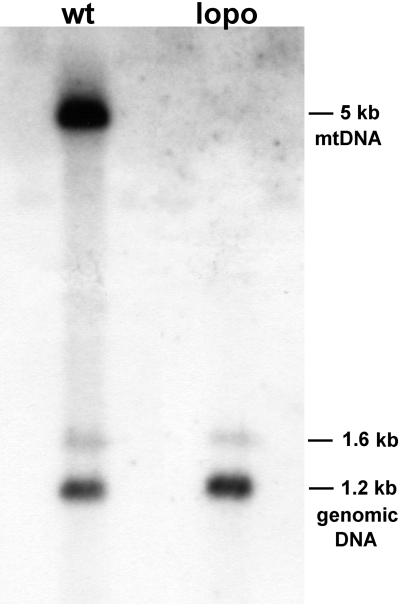
What are the molecular causes for premature lethality and aberrant cell proliferation in lopo1? Lopo encodes Drosophila mtSSB, which binds ssDNA (Stroumbakis et al., 1994; Thömmes et al., 1995) and stimulates dramatically the activity of mtDNA polymerase (Thömmes et al., 1995; Farr et al., 1999). It was therefore important to evaluate mtDNA content and integrity in lopo1. Figure 5 shows a quantitative Southern blot of wild-type and lopo1 DNA hybridized with an mtDNA probe and with a single-copy genomic probe as a control. These experiments reproducibly fail to detect any mtDNA and thus demonstrate severe mtDNA depletion in lopo1. Similar results were obtained by performing quantitative polymerase chain reaction (Maier, Vogel, and Schneuwly, unpublished results).
Figure 5.
mtDNA depletion in lopo1. A quantitative Southern blot was generated containing equal amounts of total DNA from wild-type Canton S (wt) and lopo1 (lopo) digested with EcoRI (equivalent of 100 mg of third instar larvae). mtDNA was visualized using a 5-kb fragment of mtDNA (Garesse, 1988). No mtDNA is detected in lopo1. Efficiency of loading and blotting was verified by hybridizing the blot with a single-copy nuclear gene (aos cDNA; Kretzschmar et al., 1992). The corresponding 1.2-kb fragment (and a weaker 1.6-kb fragment) show equal intensities in both lanes.
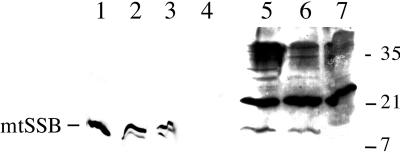
To determine the level of mtSSB protein in larvae, extracts were examined by immunoblot analysis. Protein extracts were prepared from wild-type, heterozygous, and homozygous lopo1 larvae at the third instar stage and subjected to SDS-PAGE followed by immunoblot analysis with rabbit antiserum against native mtSSB from Drosophila embryos (Figure 6). The immunoblot analysis shows the presence of mtSSB migrating as a polypeptide of ∼12 kDa in the wild-type and heterozygote extracts, whereas mtSSB is entirely absent in the corresponding homozygote extract (Figure 6, lane 7). Quantitation of three independent experiments indicated that the level of mtSSB protein is ∼0.5 μg/g of larvae in the wild-type and heterozygote extracts, a value comparable to that determined previously for mtSSB from wild-type embryos (Thömmes et al., 1995). We then purified the mtSSB from the soluble cell extracts by a chromatographic step in which the protein is bound to Blue Sepharose under stringent conditions (in the presence of 0.8 M NaCl) and eluted with increasing concentrations of sodium thiocyanate (Farr et al., 1999). The 1.5 M sodium thiocyanate eluate demonstrates clearly the enrichment of mtSSB over immunoreactive contaminants in both the wild-type and heterozygote extracts (Figure 6, lanes 2 and 3, respectively). Nonetheless, a polypeptide corresponding to mtSSB was still absent in the homozygote lopo1 eluate (Figure 6, lane 4). The absence of mtDNA in the lopo1 mutant and the complete absence of mtSSB support further the conclusion that lopo encodes a gene affecting the replication and maintenance of mtDNA.
Figure 6.
Larval expression and fractionation of Drosophila mtSSB protein. Protein fractions as described in MATERIALS AND METHODS were denatured and electrophoresed in a 17% SDS-polyacrylamide gel, transferred to nitrocellulose, and detected by immunoblotting with rabbit antiserum against D. melanogaster mtSSB at a 1:1000 dilution using the ECL method (Amersham Life Sciences, Arlington Heights, IL). Lane 1, recombinant Drosophila mtSSB (10 ng); lanes 2–4, Blue Sepharose eluates (60 μl) from wild-type, heterozygous, and homozygous lopo1 larvae, respectively; lanes 5–7, larval extracts (∼200 μg of total protein) from wild-type, heterozygous, and homozygous lopo larvae, respectively.
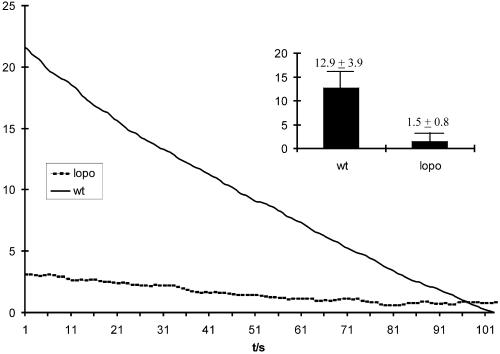
To analyze the effect of mtDNA depletion on mitochondrial physiology, we measured the O2 consumption of mitochondria from third instar larval tissue. Figure 7 shows a typical curve for wild-type and lopo1 mitochondria, documenting that respiration is reduced by 90% in the latter. Thus, whereas lopo1 flies are able to survive embryogenesis most likely through a maternal contribution of mtSSB, they exhibit a nearly complete loss of oxygen consumption in late larvae. This is consistent with the developmental profile, in which lopo1 larvae require almost twice as long to reach pupation, which can apparently occur under nearly anaerobic conditions. However, with the requirement for a high level of cell proliferation during metamorphosis, they eventually die.
Figure 7.
Oxygen consumption in larval tissues. The graph shows a typical curve of oxygen consumption in wild type (wt; straight line) and lopo1 (dotted line). The oxygen consumption is given in nanomoles per microgram of protein; the x axis defines time in seconds (t/s). Typically the oxygen consumption in lopo1 is reduced to ∼10% of the wild-type level. The inset shows a comparison of the mean relative oxygen consumption per min (three measurements each).
Functionally Impaired lopo1 Mitochondria Are Largely Normal in Number and Structure
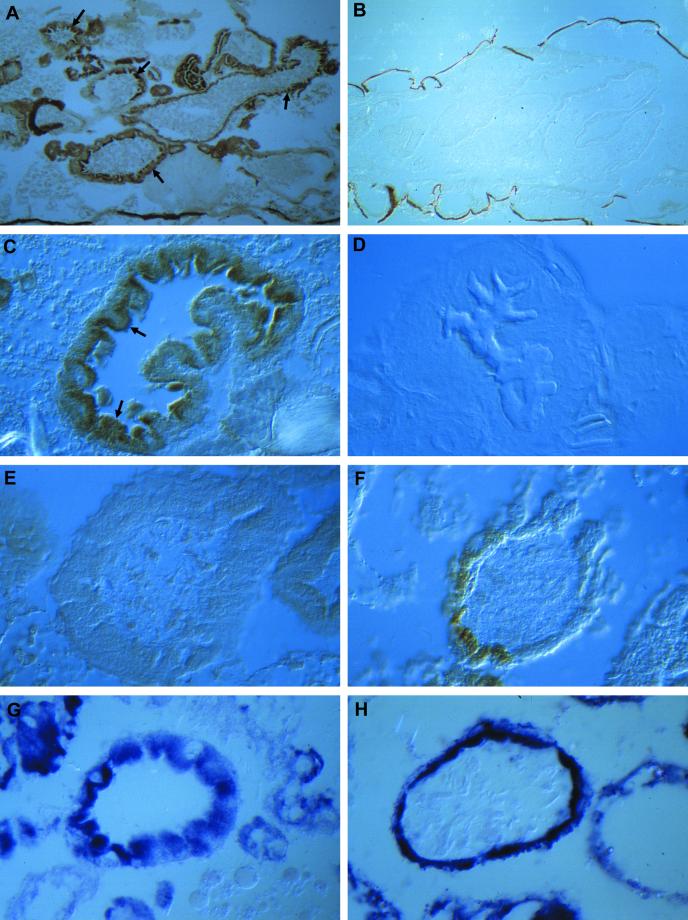
The observed mtDNA depletion and the reduced physiological function of mitochondria in lopo1 raise several questions about the status of mitochondria in this mutant. Is the number of mitochondria in lopo1 reduced, which would indicate that there is feedback control of mitochondrial biogenesis via mtDNA replication? Alternatively, are there mitochondria present without mtDNA and hence not able to perform respiration? To answer these questions, we performed a histological analysis in lopo1. One mechanism to discriminate mitochondria containing mtDNA from those that do not is differential histochemical staining for the enzymes COX and SDH. Because three subunits of COX are encoded by the mitochondrial genome, functional COX staining is observed only in mitochondria containing mtDNA, whereas, because SDH is encoded exclusively by nuclear genes, it should be functional. Figure 8 shows the digestive tract of third instar larvae, where high numbers of mitochondria are found, especially in the midgut (Figure 8, A and C). As expected, COX staining is absent in lopo1, indicating either the absence of mtDNA or the absence of mitochondria per se (Figure 8, B and E). In contrast, SDH staining in larval gut tissue (Figure 8, G and H) shows the presence of SDH enzyme activity in wild type and lopo1. This indicates that there are indeed mitochondria in lopo1 mutants, but they are not able to perform respiration because of the absence of mtDNA. Only a very limited number of cells (Figure 8F) still show fully functional mitochondria, probably because of the mtDNA remaining from the maternal contribution. Finally, in an ultrastructural analysis we found that, for the most part, mitochondria in lopo1 exhibit normal morphology (Figure 9) and number, although some variation might occur among different tissues. No changes at all are seen in the CNS, whereas an increase in mitochondria can be seen in the midgut (Figure 9), where we observed some cells in which a large number of aberrant mitochondria appeared to be in various stages of disintegration or engulfment by lysosomes. These cells are most likely apoptotic.
Figure 8.
Mitochondrial histochemistry on third instar larvae. (A–F) Histochemical staining for COX. (A) Wild-type larvae: note the strong staining in all parts of the digestive tract (arrows). (B) Similar plane of section from lopo1. No staining is detected. (C) Cross-section through the midgut. COX staining is enriched highly in endodermal cells facing the gut lumen (arrows).( D) Control of staining specificity. No COX staining is visible in the presence of KCN. (E and F) Cross-sections through the midgut of lopo1. No COX staining is visible in E but partial staining can be detected in some cells of the midgut (F). (G and H) SDH staining on cross-sections through the midgut of wild-type (G) and lopo1 (H) larvae. Both strains show equally strong staining.
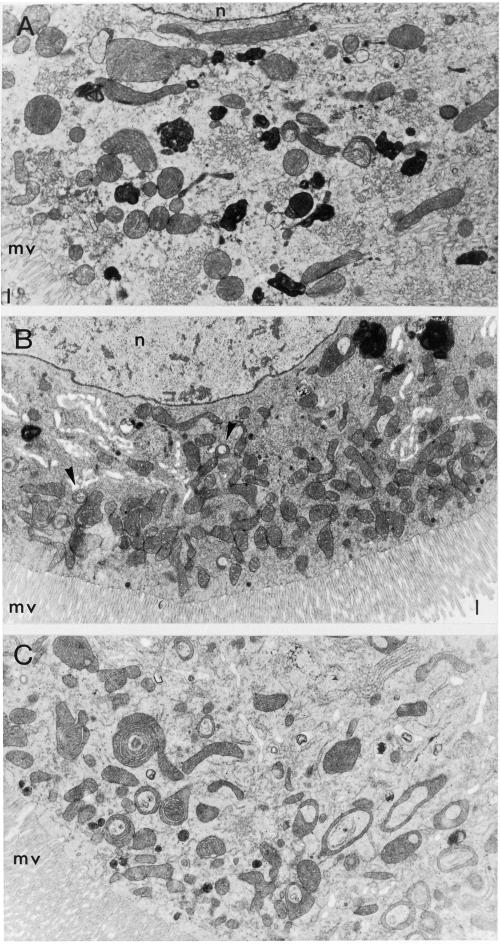
Figure 9.
Ultrastructure of mitochondria in lopo1. Third instar larval midgut tissue from wild-type (A) and lopo1 (B and C) is shown. lopo1 mutants show at least equal numbers of mitochondria as compared with wild type. The morphology of mutant mitochondria is normal (B) except for a few degenerating mitochondria (arrows). However, some cells (C) show an extremely high number of degenerating mitochondria. L, gut lumen; N, nucleus; mv, microvilli.
DISCUSSION
The isolation of a new mutant in the mtSSB gene allowed us to study the function of mtSSB in the context of the complex process of mitochondrial biogenesis in higher eukaryotes. Rather surprisingly, lopo1 is a late third instar larval-lethal mutation, despite the fact that mtSSB is thought to be an essential mitochondrial protein. The late larval lethality can be explained by a strong maternal contribution, which was identified through an immunocytochemical analysis of adult ovaries. This maternal source of mtSSB appears to be sufficient to allow development through embryogenesis, where mtDNA replication begins 10–12 h after egg laying (Rubenstein et al., 1977). However, the maternal source of mtSSB is insufficient to allow the larvae to develop through metamorphosis, where extensive cell proliferation is required and, indeed as shown by BUdR labeling, cell proliferation is disturbed drastically and appears to be the major cause of lethality. This is consistent with the immunocytochemical experiments in wild-type larvae, where we showed that, in addition to a constitutive level of mtSSB protein in all tissues, there is a high level of expression of mtSSB in proliferating cells, especially in imaginal discs, in the CNS and in the gut. The proliferation defect also explains the aberrant development of the visual system in lopo1 (Maier, Poeck, and Schneuwly, unpublished results), a phenotype that is very similar to that of the recently isolated mutant tamas, which encodes the catalytic subunit of mtDNA polymerase (Iyengar et al., 1999). This highlights the functional interactions of mtSSB and pol γ that have been demonstrated in biochemical studies (Farr et al., 1999). Biochemical analysis of Drosophila mtSSB and the analysis of a yeast mtSSB mutant (van Dyck et al., 1992) document the important role of mtSSB in mtDNA replication and maintenance. Our study of lopo1 demonstrates that, in the absence of mtSSB, severe mtDNA depletion occurs and, as a consequence, the respiration of mitochondria as measured by oxygen consumption is reduced drastically to <10%. Thus, the third instar larvae appear to survive nearly anaerobically during the last days of this developmental phase.
One of the fundamental problems in mitochondrial biogenesis is the regulation of the number of mitochondria per cell and the mtDNA copy number per mitochondrion. To examine this in lopo1, a detailed histochemical and electron microscopic analysis was performed. Our results show that the number and the ultrastructural morphology of mitochondria are not altered in lopo1 mutants, although we found that the majority of mitochondria have lost respiratory function due to the absence of mtDNA and hence functional COX. However, there are tissue-specific differences; at least in the midgut of third instar larvae, we can see alterations both in morphology and in number, whereas no changes can be detected in the CNS. However, one of the major causes of altered mitochondrial morphology might be the status of these cells, which undergo cell death. These results would exclude mtSSB as a major determinant in controlling mitochondrial biogenesis and indicate no link between mtDNA synthesis or copy number and mitochondrial biogenesis. In contrast, we have shown recently that mtSSB gene expression provides a link between nuclear and mtDNA replication in Drosophila: the mtSSB gene is regulated coordinately with a number of nuclear genes encoding key proteins in nuclear DNA replication (Ruiz de Mena et al., 2000).
Interestingly, whereas no phenotypic or molecular defects were observed in lopo1 heterozygotes, mitochondrial transcription factor A mutants show a strong dose dependence, which leads to reduced numbers of functional mitochondria in heterozygotes (Larsson et al., 1998). This difference is potentially important, and both mutants may now be used as model systems to study mtDNA replication and maintenance. Furthermore, the availability of lopo1, together with the potential for a combined approach of molecular, biochemical, genetic, and developmental analyses in Drosophila, should allow a thorough evaluation of mtSSB function in vivo.
ACKNOWLEDGMENTS
We thank P. Deak (University of Cambridge, Cambridge, UK) for the fly stock 46-13, P. Tolias (International Center for Public Health, Newark, NJ) for the lopo cDNA, M. Mlodzik (EMBL, Heidelberg, Germany) for the P1-phage, J. Stangelmayer (University of Regensburg, Regensburg, Germany) for providing the oxygen electrodes and advice regarding measurements, Ingolf Koch for the in situ localization of the P-element insertion on chromosomes, and R. Garesse (Consejo Superior de Investigaciones Científicas-Universidad Autónoma, Madrid, Spain) for the mtDNA probe. Also many thanks to U. Roth and G. Thiess for excellent technical assistance. This work was supported by funding from the DFG (to S.S.) and the National Institutes of Health (grants GM45295 and HL59656; to L.S.K).
REFERENCES
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;7:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Deák P, Omar MM, Saunders RDC, Pál M, Komonyi O, Szidonya J, Maróy P, Zhang Y, Ashburner M, Benos P, Savakis Ch, Siden-Kiamos I, Louis Ch, Bolshakov VN, Kafatos FC, Madueno E, Modolell J, Glover DM. P-element insertion alleles of essential genes on the third chromosome of Drosophila melanogaster: correlation of physical and cytogenetic maps in chromosomal region 86E–87F. Genetics. 1997;147:1697–1722. doi: 10.1093/genetics/147.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farr CL, Wang Y, Kaguni LS. Functional interactions of mitochondrial DNA polymerase and single-stranded DNA-binding protein: template-primer DNA binding and initiation and elongation of DNA strand synthesis. J Biol Chem. 1999;274:14779–14785. doi: 10.1074/jbc.274.21.14779. [DOI] [PubMed] [Google Scholar]
- Garesse R. Drosophila melanogaster mitochondrial DNA: gene organization and evolutionary considerations. Genetics. 1988;118:649–663. doi: 10.1093/genetics/118.4.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman LI, Shoubridge EA. Mitochondrial genetics and human disease. Bioessays. 1996;18:983–991. doi: 10.1002/bies.950181208. [DOI] [PubMed] [Google Scholar]
- Hartl DL, Nurminsky DI, Jones RW, Lozovskaya ER. Genome structure and evolution in Drosophila: applications of the framework P1 map. Proc Natl Acad SciUSA. 1994;91:6824–6829. doi: 10.1073/pnas.91.15.6824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoke GD, Pavco PA, Ledwith BJ, Van Tuyle GC. Structural and functional studies of the rat mitochondrial single strand DNA binding protein P16. Arch Biochem Biophys. 1990;282:116–124. doi: 10.1016/0003-9861(90)90094-f. [DOI] [PubMed] [Google Scholar]
- Iyengar B, Roote J, Campos AR. The tamas gene, identified as a mutation that disrupts larval behavior in Drosophila melanogaster, codes for the mitochondrial DNA polymerase catalytic subunit (DNApol-γ125) Genetics. 1999;153:1809–1824. doi: 10.1093/genetics/153.4.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar D, Brunner A, Wiersdorff V, Pflugfelder GO, Heisenberg M, Schneuwly S. Giant lens, a gene involved in cell determination and axon guidance in the visual system of Drosophila melanogaster. EMBO J. 1992;11:2531–2539. doi: 10.1002/j.1460-2075.1992.tb05318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahaye A, Leterme S, Foury F. PIF1 DNA helicase from Saccharomyces cerevisiae: biochemical characterization of the enzyme. J Biol Chem. 1993;268:155–161. [PubMed] [Google Scholar]
- Larsson NG, Clayton DA. Molecular genetic aspects of human mitochondrial disorders. Annu Rev Genet. 1995;29:151–178. doi: 10.1146/annurev.ge.29.120195.001055. [DOI] [PubMed] [Google Scholar]
- Larsson N-G, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nat Genet. 1998;18:231–236. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Li K, Williams RS. Tetramerization and single-stranded DNA binding properties of native and mutated forms of murine mitochondrial single-stranded DNA-binding proteins. J Biol Chem. 1997;272:8686–8694. doi: 10.1074/jbc.272.13.8686. [DOI] [PubMed] [Google Scholar]
- Lojda Z, Gossrau R, Schiebler TA. Enzyme histochemistry. A laboratory manual. Berlin, Heidelberg, New York: Springer-Verlag; 1979. [Google Scholar]
- Mignotte B, Marsault J, Barat-Gueride M. Effects of the Xenopus laevis mitochondrial single-stranded DNA-binding protein on the activity of DNA polymerase gamma. Eur J Biochem. 1988;174:479–484. doi: 10.1111/j.1432-1033.1988.tb14123.x. [DOI] [PubMed] [Google Scholar]
- Pavco PA, Van Tuyle GC. Purification and general properties of the DNA-binding protein (P16) from rat liver mitochondria. J Cell Biol. 1985;100:258–264. doi: 10.1083/jcb.100.1.258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugfelder GO, Schwarz H, Roth H, Poeck B, Sigl A, Kerscher S, Jonschker B, Pak WL, Heisenberg M. Genetic and molecular characterization of the optomotor-blind gene locus in Drosophila melanogaster. Genetics. 1990;126:91–104. doi: 10.1093/genetics/126.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preininger C, Klimant I, Wolfbeis OS. Optical fiber sensor for biological oxygen demand. Anal Chem. 1994;88:1841–1846. [Google Scholar]
- Reynolds ES. The use of lead citrate at high pH as an electron opaque stain in electron microscopy. J Cell Biol. 1963;17:202–212. doi: 10.1083/jcb.17.1.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL, Brutlag D, Clayton DA. The mitochondrial DNA of Drosophila melanogaster exists in two distinct and stable superhelical forms. Cell. 1977;12:471–482. doi: 10.1016/0092-8674(77)90123-4. [DOI] [PubMed] [Google Scholar]
- Ruiz de Mena I, Lefai E, Garesse R, Kaguni LS. Regulation of mitochondrial single-stranded DNA-binding protein gene expression links nuclear and mitochondrial DNA replication in Drosophila. J Biol Chem. 2000;275:13628–13636. doi: 10.1074/jbc.275.18.13628. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- Sedman T, Kuusk S, Kivi S, Sedman J. A DNA. helicase required for maintenance of the functional mitochondrial genome in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:1816–1824. doi: 10.1128/mcb.20.5.1816-1824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shadel GS, Clayton DA. Mitochondrial DNA maintenance in vertebrates. Annu Rev Biochem. 1997;66:409–435. doi: 10.1146/annurev.biochem.66.1.409. [DOI] [PubMed] [Google Scholar]
- Stroumbakis ND, Li Z, Tolias PP. RNA- and single-stranded DNA-binding (SSB) proteins expressed during Drosophila melanogaster oogenesis: a homolog of bacterial and eukaryotic mitochondrial SSBs. Gene. 1994;143:171–177. doi: 10.1016/0378-1119(94)90093-0. [DOI] [PubMed] [Google Scholar]
- Thömmes P, Farr CL, Marton RF, Kaguni LS, Cotterill S. Mitochondrial single-stranded DNA-binding protein from Drosophila embryos: physical and biochemical characterization. J Biol Chem. 1995;270:21137–21143. doi: 10.1074/jbc.270.36.21137. [DOI] [PubMed] [Google Scholar]
- Tiranti V, Rocchi M, DiDonato S, Zeviani M. Cloning of human and rat cDNAs encoding the mitochondrial single-stranded DNA-binding protein (SSB) Gene. 1993;126:219–225. doi: 10.1016/0378-1119(93)90370-i. [DOI] [PubMed] [Google Scholar]
- Truman JW, Bate M. Spatial and temporal patterns of neurogenesis in the central nervous system of Drosophila melanogaster. Dev Biol. 1988;125:145–157. doi: 10.1016/0012-1606(88)90067-x. [DOI] [PubMed] [Google Scholar]
- van Dyck E, Foury F, Stillman B, Brill SJ. A single-stranded DNA binding protein required for mitochondrial DNA replication in S. cerevisiae is homologous to E. coli SSB. EMBO J. 1992;11:3421–3430. doi: 10.1002/j.1460-2075.1992.tb05421.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Tuyle GC, Pavco PA. The rat liver mitochondrial DNA-protein complex: displaced single strands of replicative intermediates are protein coated. J Cell Biol. 1985;100:251–257. doi: 10.1083/jcb.100.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernette CM, Kaguni LS. A mitochondrial DNA polymerase from embryos of Drosophila melanogaster: purification, subunit structure, and partial characterization. J Biol Chem. 1986;261:14764–14770. [PubMed] [Google Scholar]
- Williams AJ, Kaguni LS. Stimulation of Drosophila mitochondrial DNA polymerase by single-stranded DNA-binding protein. J Biol Chem. 1994;270:860–865. doi: 10.1074/jbc.270.2.860. [DOI] [PubMed] [Google Scholar]
- Wilson C, Kurth-Pearson R, Bellen HJ, O'Kane CJ, Grossniklaus U, Gehring WJ. P-element–mediated enhancer detection: an efficient method for isolating and characterizing developmentally regulated genes in Drosophila. Genes Dev. 1989;3:1301–1313. doi: 10.1101/gad.3.9.1301. [DOI] [PubMed] [Google Scholar]
- Zeviani M, Petruzzella V, Carrozzo R. Disorders of nuclear-mitochondrial intergenomic signaling. J Bioenerg Biomembr. 1997;29:121–130. doi: 10.1023/a:1022633912917. [DOI] [PubMed] [Google Scholar]