Abstract
Bacteria withstand starvation during long-term stationary phase through the acquisition of mutations that increase bacterial fitness. The evolution of the Growth Advantage in Stationary Phase (GASP) phenotype results in the ability of bacteria from an aged culture to outcompete bacteria from a younger culture when the two are mixed together. The GASP phenotype was first described for Escherichia coli but has not been examined for an environmental bacterial pathogen which must balance long-term survival strategies that promote fitness in the outside environment with those that promote fitness within the host. Listeria monocytogenes is an environmental bacterium that lives as a saprophyte in soil but is capable of replicating within the cytosol of mammalian cells. Here we demonstrate the ability of L. monocytogenes to express GASP via the acquisition of mutations during long-term stationary growth. L. monocytogenes GASP occurred through mechanisms that were both dependent and independent of the stress responsive alternative sigma factor SigB. Constitutive activation of the central virulence transcriptional regulator PrfA interfered with the development of GASP, however L. monocytogenes GASP cultures retained full virulence in mice. These results indicate that L. monocytogenes can accrue mutations that optimize fitness during long-term stationary growth without negatively impacting virulence.
Keywords: PrfA, bacterial virulence, stationary phase survival, SigB, GASP
Introduction
Bacteria exhibit a remarkable ability to adapt to disparate conditions that would otherwise limit growth. A simple yet compelling example of bacterial adaptation can be observed during the distinct phases of growth in liquid culture. The lag, logarithmic, and stationary phases of bacterial growth have been well described (Perry & Staley, 1997), however the phases of growth following stationary phase have only recently been investigated in detail. Following entry into stationary phase, a death phase occurs during which a greater than 90% loss of bacterial viability is observed (Perry & Staley, 1997). The amount of viable bacteria then levels off and remains relatively constant. This second stable stationary phase is known as the long-term stationary phase (Finkel et al., 2000; Steinhaus & Birkeland, 1939). The timing of bacterial growth phases varies depending on the growth medium and on the bacterial species being studied. For Escherichia coli in LB, the death phase starts 2–3 days after the initial inoculation and lasts for about a day while the long-term stationary phase of growth lasts for several weeks or longer (Finkel, 2006). Studies of long term stationary phase growth and survival of E. coli led to the discovery of the growth advantage in stationary phase or GASP phenotype, which reflects the ability of bacteria from an aged culture to outcompete the same strain of bacteria from a younger culture when the two are grown together (Zambrano et al., 1993). For E. coli grown in LB, the aged culture must be at least 8-days old and in the long-term stationary phase of growth to effectively outcompete a younger 1-day old culture (Finkel, 2006; Zambrano et al., 1993; Zambrano & Kolter, 1993). The GASP phenotype of E. coli results from a dynamic and continuous acquisition of mutations that increase bacterial fitness during periods of long-term stationary growth (Farrell & Finkel, 2003; Zambrano et al., 1993; Zambrano & Kolter, 1993; Zinser & Kolter, 1999; Zinser & Kolter, 2000; Zinser et al., 2003; Zinser & Kolter, 2004).
Listeria monocytogenes is a Gram-positive environmental bacterial pathogen that has evolved to survive in disparate environments both inside and outside of mammalian hosts (Czuprynski, 2005; Gray et al., 2006; Vazquez-Boland et al., 2001). As an intracellular pathogen, the bacterium invades mammalian cells, escapes from host cell phagosomes, replicates within the cytosol, and spreads into neighboring cells (Freitag et al., 2009; Hamon et al., 2006). A number of bacterial factors are required for L. monocytogenes intracellular replication and cell-to-cell spread (Goebel et al., 2000; Vazquez-Boland et al., 2001), and the expression of a majority of these gene products is regulated by the transcriptional regulator known as PrfA (Kreft & Vazquez-Boland, 2001; Scortti et al., 2007). The fitness of L. monocytogenes inside of the host is severely compromised in the absence of PrfA (Freitag, 2006).
Outside of mammalian hosts, L. monocytogenes is widely distributed and is believed to live as a saprophyte off of decaying plant material (Czuprynski, 2005; Freitag et al., 2009; Gray & Killinger, 1966; Vazquez-Boland et al., 2001). L. monocytogenes has been isolated from soil, silage, ground water, sewage, and vegetation (Thevenot et al., 2006) and, although it does not form spores, the bacterium can become firmly established in food processing environments and persist for long periods of time, even years (Orsi et al., 2011; Lunden et al., 2002). Based upon an anticipated requirement for L. monocytogenes to be able to balance survival under nutrient poor conditions in the outside environment with life within the infected host, we assessed the bacterium for its ability to adapt to periods of long-term stationary phase growth through the development of GASP. Our results indicate that L. monocytogenes is capable of stably adapting itself for long-term survival without compromising its ability to cause disease.
Materials and Methods
Bacterial strains
The bacterial strains and plasmids are listed in Table 1. Antibiotics were used as follows: erythromycin (1 μg/ml), chloramphenicol (10 μg/ml), and streptomycin (200 μg/ml).
Table 1.
Bacterial strains and plasmid used in this study.
| Strain | Description/Genotype | Designation | Reference |
|---|---|---|---|
| SM10 | E. coli strain for harboring plasmids | ||
| NF-L100 | 10403S | (Bishop & Hinrichs, 1987) | |
| NF-L476 | NF-L100 actA-gus-plcB | (Shetron-Rama et al., 2002) | |
| NF-L1124 | NF-L100 actA-gus-neo-plcB | WT | (Miner et al., 2008) |
| NF-L1177 | NF-L1124 prfAG145S | prfA G145S | (Miner et al., 2008) |
| NF-L1006 | NF-L476 tRNAArg::pPL2 | WT camR | |
| NF-E1000 | SM10 with pPL2 | ||
| FSL A1-254 | 10403S ΔsigB | ΔsigB | (Wiedmann et al., 1998) |
| NF-L1823 | FSL A1-254 tRNAArg::pPL2 | ΔsigB camR | This study |
| NF-L1824 | NF-L1177 tRNAArg::pPL2 | prfA G145S camR | This study |
| DP-L3903 | 10403S with Tn917 insertion | WT ermR | (Auerbuch et al., 2001) |
|
| |||
| Plasmid | Description/Genotype | Designation | Reference |
|
| |||
| pPL2 | Site-specific phage integration vector | (Lauer et al., 2002) | |
Monoculture growth experiments
Overnight cultures grown in BHI were added at a 1:250 (vol:vol) dilution to fresh BHI and incubated at 37°C with aeration. CFU/mL were determined by plating dilutions of culture aliquots on BHI agar.
Stationary phase mixing experiments
Competitive indices of mixed bacterial cultures during stationary phase were performed as previously described (Bruno & Freitag, 2010; Finkel et al., 2000; Zambrano et al., 1993) (Fig 1A). Aliquots of 12-day old cultures were stored at −80°C. For each experiment, an aliquot of frozen cells was thawed and 50 μL was added to 12.5 mL of BHI and grown overnight at 37°C. 125 μL of the overnight 12-day old culture was added to 12.5 mL of a 1-day old culture at a ratio of 1:100 and incubated at 37°C for 10 days. 12-day old and 1-day old were distinguished based on chloramphenicol resistance of the 1-day old cultures containing the site-specific integration vector pPL2 which conferred chloramphenicol resistance without influencing bacterial growth (Lauer et al., 2002). Every 24 hours, an aliquot of the mixed culture was removed, diluted, and plated onto BHI agar to enumerate bacterial CFUs. 150 of the resulting colonies were then patched onto BHI agar containing chloramphenicol, selecting for the original 1-day old chloramphenicol resistant bacteria; this was found to be the most reliable method for clearly distinguishing drug resistant colonies. The competitive index (CI) value was determined as follows: CI = (test strain CFU)/(reference strain CFU).
Figure 1. Overview of stationary phase culture mixing experiments and of repeated cycles of culture dilution and outgrowth.
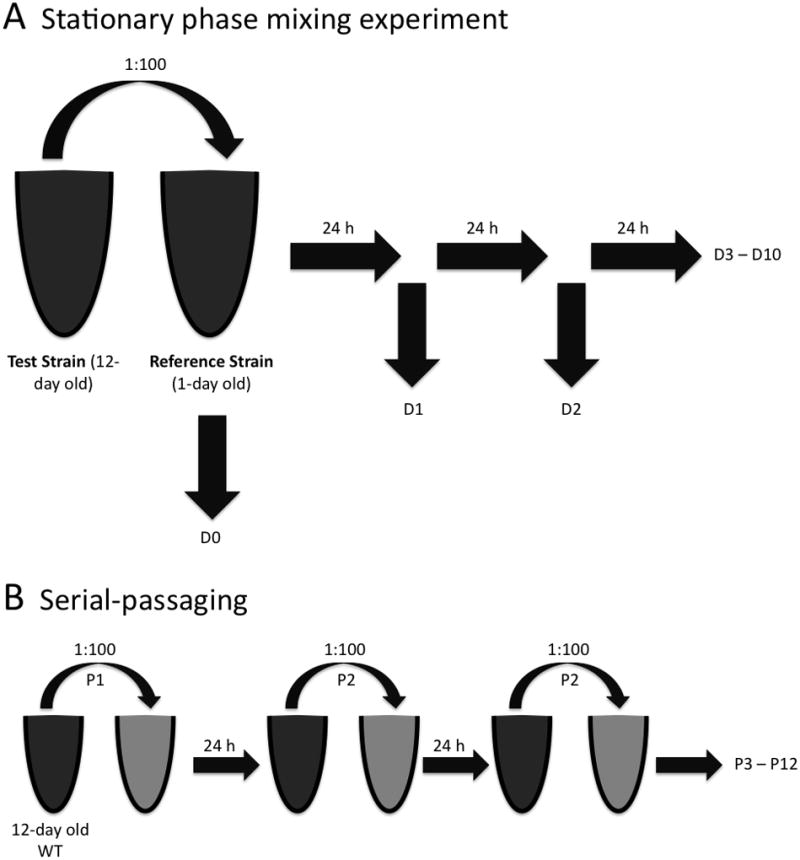
(A) Stationary phase mixing experiments. 125 μL of a test strain culture was added to a 12.5 mL of a reference strain culture. Every 24 hours, the CI value of the mixed culture was determined as described in Materials and Methods, with Day 0 (D0) representing the initial mixture of the two cultures. For experiments that assessed the capacity of a strain to express the GASP phenotype, the test strain culture was an aged (12-day old) culture and the reference strain culture was a younger (1-day old) culture of the same bacterial strain. (B) Culture dilution and outgrowth. 125 μL of a culture was added to 12.5 mL of fresh media and grown for 24 hours. This process constituted one cycle of growth (or passage). Then, 125 μL of the culture was added to another 12.5 mL of fresh media and grown for 24 hours. This repeated cycle of inoculating fresh media with a fraction of a culture and propagating the fraction is referred to as ‘serial-passaging’. Bacteria from a 12-day old L. monocytogenes culture that was serially-passaged for a total of 12 passages are referred to as passaged 12-day old L. monocytogenes (12d-P) in subsequent figures.
Intravenous mouse infections
Mid-log L. monocytogenes were washed and diluted in PBS to a final concentration of 1×105 CFU/ml. 7–8 week old ND4 Swiss Webster mice (Harlan Laboratories, Inc., Madison, WI) were infected via tail vein with 2×104 CFU. Forty-eight hours post infection homogenized tissue dilutions were plated on BHI agar to determine CFU/organ.
For competitive index experiments, mice were infected via tail vein with a 1:1 mixture of a reference and test strains. The reference strain was DP-L390, a wild type strain with a Tn917-LTV3 insertion that confers erythromycin resistance and has been confirmed to have no affect L. monocytogenes virulence [(Auerbuch et al., 2001) and Fig 5B]. Strains were grown to mid-log phase and mixed together in PBS. 200 μL of 2×104 CFU mixed bacterial suspension was used for infection. After 48 hours, livers and spleens were harvested and homogenized. The CI value for each organ was determined as previously described (Auerbuch et al., 2001).
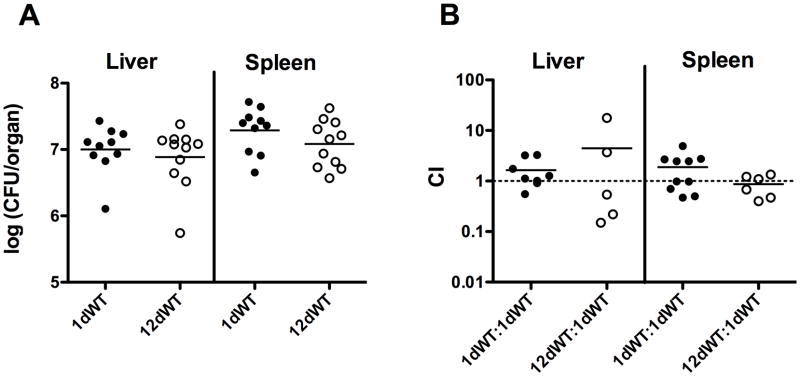
Figure 5. The expression of GASP does not significantly impact L. monocytogenes virulence.
(A) Growth of bacteria from 12-day old L. monocytogenes cultures in the livers and spleens of intravenously infected mice. 7–8 week old ND4 Swiss Webster mice were infected with 2×104 CFU of bacteria from either 1-day old or 12-day old L. monocytogenes cultures via tail vein injections. 48 hours post infection, the bacterial loads of the livers and spleens were determined. A Student’s T-test was performed to compare the bacterial loads of mice infected with 12-day old bacteria to those of mice infected with 1-day old bacteria. Two-tailed p-values were greater than 0.05 for both the liver and spleen, indicating an absence of statistically significant difference. (B) 12-day old L. monocytogenes GASP cultures exhibit no significant competitive defect in comparison to bacteria derived from 1-day old cultures. Erythromycin-sensitive bacteria from either 1-day old or 12-day old L. monocytogenes cultures and erythromycin-resistant bacteria from 1-day old L. monocytogenes cultures were mixed 1:1 for a total bacterial suspension of 2×104 CFU and then intravenously injected into mice. 48 hours post infection, the mice were sacrificed and the competitive index (CI) values of the livers and spleens were determined as described in Materials and Methods. Horizontal lines represent the mean CI values. A one-sided Student’s T-test was performed to determine if any statistically significant difference was present between a mean CI value and 1. All two-tailed p-values were greater than 0.05, indicating an absence of statistically significant differences.
Statistics
Statistical analysis was performed using Prism Software (GraphPad v.2.0). Where appropriate, a Student’s T-test was used to identify statistically significant differences. In all cases, a p-value <0.05 was considered significant.
Results and Discussion
L. monocytogenes expresses GASP
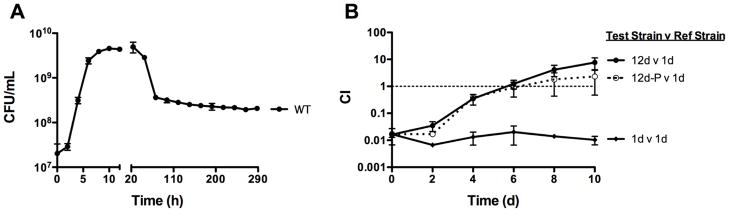
The GASP phenotype reflects bacterial adaptation to long term nutrient starvation (Finkel, 2006) and, as an environmental organism, L. monocytogenes would be anticipated to encounter periods of sustained nutrient deprivation. The development of the GASP phenotype is marked by the ability of bacteria from an aged culture to outcompete bacteria from a younger culture during long-term stationary phase growth (Finkel, 2006). GASP thus requires that a bacterial strain be capable of surviving for an extended period of time following its inoculation into growth medium. To measure the survival of L. monocytogenes during nutrient starvation, bacteria grown in nutrient-rich broth (BHI) were assessed for viability following incubation for 12 days at 37°C. Cultures exhibited a characteristic lag, logarithmic, and stationary growth phase during the first 24 hours of growth (Fig 2A). After remaining in stationary phase for 1–2 days, L. monocytogenes entered a death phase during which an approximate 90% loss of cell viability was observed over 24 hours. The subsequent bacterial population then maintained a stable cell density representative of a long-term stationary growth phase that persisted for the remaining days (Fig 2A).
Figure 2. Long-term growth and the expression of the GASP phenotype by L. monocytogenes.
(A) Long term culture growth curve of L. monocytogenes in BHI at 37°C. Bacterial growth and survival was determined by measuring CFU/mL at the indicated time points. Data shown represents the mean ± the standard error of three independent experiments. (B) L. monocytogenes expression of GASP. Bacteria from a 12-day old (12d), a passaged 12-day old (12d-P), or a 1-day old (1d) L. monocytogenes culture (test strains) were added to a 1-day old L. monocytogenes culture (reference strain). Mixed bacterial cultures were incubated in BHI at 37°C. Passaged 12-day old bacteria are bacteria from a culture that was aged 12 days and then diluted 1:100 in fresh BHI every 24 hours for a total of 12 passages (Fig 1B). The reference strains contained the stable integrated plasmid vector pPL2, which conferred chloramphenicol resistance. The test strains were chloramphenicol sensitive. The competitive index (CI) values were determined at the indicated time points as described in Materials and Methods, with Day 0 being the time point immediately after bacteria from a test strain culture were added to the reference strain culture. The data represent the means ± standard errors of three independent experiments.
The ability of L. monocytogenes to express the GASP phenotype was next assessed. As E. coli cultures need to be at least 8 days old (when cultured in LB under aerobic conditions) to express the GASP phenotype (Finkel, 2006; Zambrano et al., 1993), we aged L. monocytogenes cultures for 12 days prior to the assessment for GASP as an arbitrary staring point. Bacteria from a L. monocytogenes 12-day old culture were added to a 1-day old culture at a ratio of 1:100 (Fig 1). Bacteria from the 12-day old culture outcompeted bacteria of the 1-day old culture over 10 days, such that the ratio at day 10 was 10:1 of 12-day old cells to 1-day old cells (Fig 2B). In contrast, when bacteria from a 1-day old culture of L. monocytogenes were added to another 1-day old culture at a ratio of 1:100, no change in this ratio was observed over 10 days (Fig 2B). The competitive advantage exhibited by the bacteria from a 12-day old culture was thus reflective of culture age, and indicated that L. monocytogenes is capable of expressing GASP.
To determine if the L. monocytogenes GASP phenotype was the result of a stable genetic change, bacteria from a 12-day old culture were grown in BHI to a high cell density, diluted 1:100 into fresh media, and once again grown to high cell density. This process was repeated every 24 hours for a total of 12 cycles of dilution and outgrowth or passages (Fig 1B). Bacteria from the passaged 12-day old culture were then added to a 1-day old culture of wild type L. monocytogenes at a ratio of 1:100. Just as with bacteria from a non-passaged 12-day old culture, bacteria from the passaged 12-day old culture outcompeted bacteria of the 1-day old culture over 10 days (Fig 2B), thus indicating that L. monocytogenes GASP resulted from a stable genetic change.
Constitutive activation of PrfA interferes with GASP expression
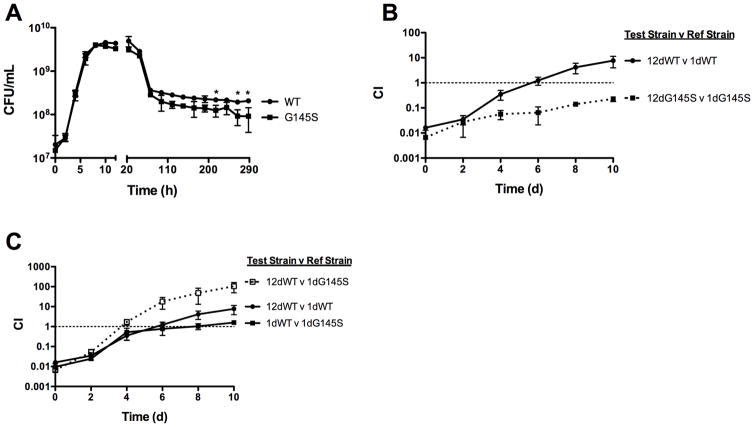
The transcriptional activator PrfA regulates the expression of most of the gene products that have been associated with L. monocytogenes pathogenesis (Scortti et al., 2007). PrfA exists in both low activity and high activity forms, and constitutive activation of PrfA via prfA* mutations enhances L. monocytogenes virulence while compromising the fitness of bacteria in broth culture (Bruno & Freitag, 2010). To evaluate the impact of PrfA activation on L. monocytogenes long-term survival, the mutationally activated prfA* G145S mutant was grown for 12 days in BHI at 37°C. Cultures of the prfA G145S mutant exhibited death and long-term stationary growth phases (Fig 3A), indicating that the L. monocytogenes prfA* mutant was capable of long-term survival. However, cultures of the prfA G145S mutant exhibited final bacterial cell densities that were two to three-fold lower than those of wild type cultures in the same growth phase (Fig 3A). The constitutive activation of PrfA thus reduced the overall numbers of L. monocytogenes that were capable of surviving long-term in exhausted media.
Figure 3. Constitutive PrfA activation reduces bacterial fitness during long-term stationary growth and impairs the expression of the GASP phenotype.
(A) Long term stationary phase growth of wild type and prfA* L. monocytogenes. Bacterial growth in BHI at 37°C was measured by enumerating CFU/mL at the indicated time points. Each growth curve represents the mean ± standard error of three independent experiments with the degree of statistical significance indicated for the last four time points (* = p < 0.05). (B) Mutationally activated prfA G145S mutants express a reduced GASP phenotype. Bacteria from 12-day old chloramphenicol sensitive cultures (test strains) were added to a 1-day old chloramphenicol resistant cultures (reference strains). Mixed cultures were maintained in BHI at 37°C and the competitive index (CI) values were determined at the indicated time points. The data represent the means ± standard errors of three independent experiments. (C) Mutational activation of prfA interferes with the development of GASP. Bacteria from 12-day old wild type chloramphenicol sensitive cultures (test strains) were added to a 1-day old wild type or prfA G145S chloramphenicol resistant cultures (reference strains). Mixed cultures were maintained in BHI at 37°C and the competitive index (CI) values were determined at the indicated time points. The data represent the means ± standard errors of three independent experiments.
To determine if constitutive activation of PrfA affected the development of GASP, prfA G145S mutant bacteria from a 12-day old culture were added to a 1-day old culture of prfA G145S at a ratio of 1:100 (Fig 3B). Over the course of 10 days, bacteria from the prfA* 12-day old culture outcompeted the prfA* 1-day old culture such that the ratio at day 10 was a little less than 1:10 (Fig 3B). While the competitive advantage of the aged culture indicates that the L. monocytogenes prfA* mutant was indeed capable of exhibiting a GASP phenotype, the phenotype was weaker than that exhibited by wild type bacteria (Fig 3B).
The failure of the prfA G145S mutant to express a robust GASP phenotype could reflect an impaired ability of bacteria to develop GASP, or may indicate that PrfA activation contributed to the development of a partial GASP phenotype in the 1-day old cultures. To help distinguish whether the presence of the prfA* mutation impaired or enhanced the expression of GASP, the competitive index between wild type 12-day old cultures and 1-day old wild type or prfA G145S cultures was assessed. Because prfA* mutants exhibit a competitive defect with wild type strains during short periods of growth in BHI [(Bruno & Freitag, 2010) and Fig. 3C], this fitness defect would be anticipated to contribute to the magnitude of any GASP-related fitness effect observed between 12-day old wild type and 1-day old prfA* cultures. If the prfA G145S mutant expresses a partial GASP phenotype as the result of PrfA activation, then the competitive advantage of a wild type 12-day old culture should be less in comparison to 1-day prfA* than in comparison to 1-day old wild type. Alternatively, if prfA* interferes with GASP, the overall defect observed between wild type 12-day old cultures and 1-day old prfA* mutants should reflect both the prfA*-associated fitness defect in BHI broth culture as well as an impaired GASP phenotype. When bacteria from a 12-day old wild type culture were added to a 1-day old culture of the prfA G145S mutant at a ratio of 1:100, after 10 days the ratio of wild type to mutant shifted from 1:100 to 100:1, representing a 10,000-fold increase in 12-day aged wild type bacteria (Fig. 3C). This shift was larger than the 1,000-fold increase in 12-day aged bacteria observed when bacteria from a 12-day old wild type culture were added to a 1-day wild type old culture (Fig. 3C). This enhanced fitness advantage was nearly equal to the sum of the fitness advantage observed for wild type versus prfA* strains for one day old cultures (Fig. 3C, 1dWT v 1dG145S) plus the magnitude of wild type GASP expression (Fig. 3C, 1dWT v 12dWT), suggesting that PrfA activation impedes the development of GASP.
Activation of PrfA via a prfA* mutation has been shown to influence the metabolic capacity of L. monocytogenes, enhancing bacterial growth in the presence of some carbon sources while decreasing growth in the presence of others (Bruno & Freitag, 2010; Chico-Calero et al., 2002; Deutscher et al., 2005; Deutscher et al., 2006; Goetz et al., 2001; Joseph et al., 2006; Joseph & Goebel, 2007; Joseph et al., 2008). It is possible that the metabolic shift that occurs in L. monocytogenes as a result of PrfA activation interferes with efficient nutrient acquisition during the conditions of long-term stationary phase. However, activation of PrfA has also been shown to increase the sensitivity of L. monocytogenes to osmotic and acid stresses (Bruno & Freitag, 2010), thus there may be multiple mechanisms functioning simultaneously to reduce bacterial fitness during long term stationary phase. Finally, as the prfA* strains exhibited a 2 to 3-fold lower cell density at stationary phase, it is possible that the reduced GASP phenotype reflects a reduction in overall cell numbers available for the accumulation of potential GASP mutations.
The stress related alternative sigma factor SigB contributes to long-term stationary phase survival and influences the development of GASP
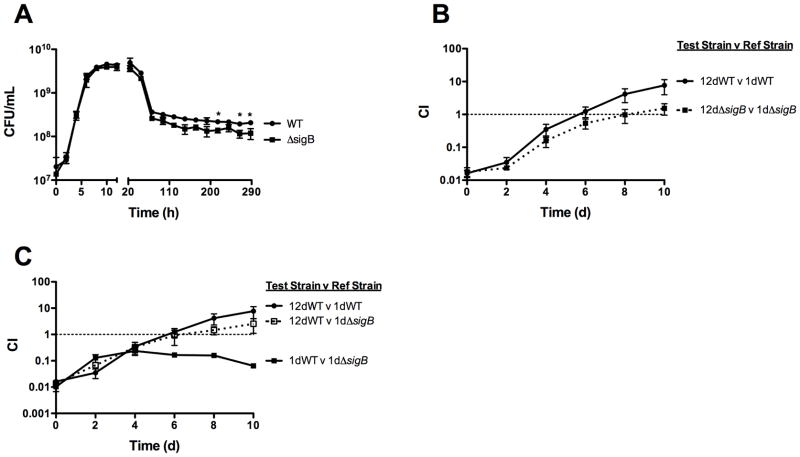
The most common mutations resulting in the E. coli GASP phenotype are mutations within rpoS (Farrell & Finkel, 2003; Finkel & Kolter, 1999; Hengge-Aronis, 2000; Zinser & Kolter, 2004), which encodes a member of the σ70 family of sigma factors that contribute to bacterial stress responses in E. coli and other bacteria (Hengge-Aronis, 2000; Loewen et al., 1998; Zinser & Kolter, 2004). rpoS is not essential for the expression of the E. coli GASP phenotype, as aged ΔrpoS mutants out-compete younger ΔrpoS mutants (Finkel, 2006) and mutations associated with GASP have been mapped to other genes unrelated to rpoS (Zinser & Kolter, 1999; Zinser & Kolter, 2000; Zinser et al., 2003). However, the most common mutations associated with E. coli GASP are mutations within rpoS that result in the attenuation of RpoS activity; these mutations are sufficient to confer the GASP phenotype (Farrell & Finkel, 2003; Finkel & Kolter, 1999; Hengge-Aronis, 2000; Zinser & Kolter, 2004). L. monocytogenes harbors a stress-responsive σ70 sigma factor, known as SigB (Abram et al., 2008; Chaturongakul & Boor, 2006; Garner et al., 2006; Kazmierczak et al., 2003; O’Byrne & Karatzas, 2008), thus it seemed logical to assess if SigB function contributed to the development of L. monocytogenes GASP. When examined for long-term survival in culture, a ΔsigB mutant exhibited the expected death and long-term stationary growth phases during the course of a 12-day incubation in BHI at 37°C (Fig 4A). Similar to the prfA* mutant, ΔsigB long-term stationary phase cultures exhibited final stable bacterial CFU numbers that were approximately two-fold lower than those maintained by wild type L. monocytogenes (Fig 4A). SigB is thus required for the optimal fitness of L. monocytogenes during the long-term stationary growth phase.
Figure 4. SigB contributes to bacterial fitness during long-term stationary growth but negatively impacts the development of GASP.
(A) Comparison of long term stationary phase growth and survival of wild type and ΔsigB cultures. Bacterial growth in BHI at 37°C was measured by enumerating CFU/mL at the indicated time points. Each growth curve represents the mean ± standard error of three independent experiments with the degree of statistical significance indicated for the last four time points (* = p < 0.05). (B) ΔsigB mutants express a reduced GASP phenotype. Bacteria from 12-day old chloramphenicol sensitive cultures (test strains) were added to a 1-day old chloramphenicol resistant cultures (reference strains). Mixed cultures were maintained in BHI at 37°C and the competitive index (CI) values were determined at the indicated time points. The data represent the means ± standard errors of three independent experiments. (C) Loss of SigB function contributes to GASP. Bacteria from 12-day old wild type chloramphenicol sensitive cultures (test strains) were added to a 1-day old wild type or ΔsigB chloramphenicol resistant cultures (reference strains). Mixed cultures were maintained in BHI at 37°C and the competitive index (CI) values were determined at the indicated time points. The data represent the means ± standard errors of three independent experiments.
ΔsigB mutant bacteria from 12-day old cultures were added to 1-day old mutant cultures at a final ratio of 1:100. Over 10 days, bacteria from the 12-day old culture outcompeted bacteria of the 1-day old culture such that the ratio at day 10 was 1:1 (Fig 4B), indicating that the ΔsigB mutant retained its ability to express the GASP phenotype. However, similar to the phenotype expressed by the prfA* mutant, the GASP phenotype exhibited by the ΔsigB strain was not as robust as that exhibited by wild type L. monocytogenes (Fig. 4B). Whereas bacteria derived from 12-day old wild type cultures increased 1,000-fold in comparison to 1-day old wild type bacteria (Fig. 4B), the bacterial numbers of a 12-day old ΔsigB culture increased approximately 100-fold in comparison to those of the 1-day old ΔsigB culture (Fig 4B).
Similar to the situation described above for prfA* strains, the failure of the ΔsigB mutant to express a robust GASP phenotype could reflect an impaired ability to develop GASP, or may indicate that the loss of SigB contributed to a partial GASP phenotype for 1-day old cultures. To distinguish between these two possibilities, the competitive index between wild type 12-day old cultures and 1-day old wild type or ΔsigB cultures was assessed. If the ΔsigB mutant expresses a partial GASP phenotype as the result of the loss of SigB, then the competitive advantage of a wild type 12-day old culture should be less in comparison to 1-day old ΔsigB than in comparison to 1-day old wild type. Interestingly, the difference in the competitive advantage of wild type 12-day old cultures observed versus 1-day old wild type or 1-day old ΔsigB was minimal (Fig. 4C). SigB contributes to L. monocytogenes fitness in broth culture, based on the competitive advantage of 1-day old wild type strains versus 1-day old ΔsigB mutants (Fig. 4A). Thus, in spite of ΔsigB mutants exhibiting a broth culture fitness defect, the overall magnitude of the competitive defect observed between 12-day old wild type L. monocytogenes and 1-day old wild type strains and ΔsigB mutants was similar rather than exacerbated for ΔsigB, suggesting that the loss of SigB may indeed contribute to the development of the GASP phenotype. Taken together, these data indicate a role for SigB in L. monocytogenes long-term stationary phase survival and in the expression of GASP.
GASP cultures of L. monocytogenes remain fully virulent
The GASP mutation(s) that enable L. monocytogenes to adapt to long-term stationary growth and to nutrient starvation could potentially impact other aspects of L. monocytogenes physiology, including those relating to bacterial virulence. As an environmental pathogen, L. monocytogenes would presumably encounter conditions in which long-term stationary growth survival would be required prior to human or animal infection. To determine if adaptation to nutrient starvation affected the virulence of L. monocytogenes, bacteria from 12-day old cultures were used to intravenously infect mice. At forty-eight hours post infection, the bacterial loads of the livers and spleens from mice infected with bacteria from 12-day old wild type L. monocytogenes cultures were not statistically different from those of mice infected with bacteria from 1-day old L. monocytogenes cultures (Fig 5A). To further examine the age-adapted bacteria for subtle fitness defects in vivo that might be detectable in comparison to 1-day old bacterial cells, competition experiments were performed (Fig. 5B). Mice were intravenously infected with a 1:1 mixed bacterial suspension of bacteria from 12-day old and 1-day old cultures, and 48 hours post infection the competitive index (CI) values for bacteria isolated from the murine livers and spleens were determined. CI values remained very close to 1 (Fig 5B), indicating that genetic alterations that promote L. monocytogenes long-term stationary phase survival under nutrient limited conditions do not appear to impact bacterial virulence in systemic models of animal infection.
Based on observations made with E. coli (Finkel & Kolter, 1999; Finkel, 2006), the bacteria from 12-day old L. monocytogenes cultures likely reflect dynamic and evolving populations of cells. If a GASP mutation within a sub-population of cells attenuates bacterial virulence, the presence of the other bacteria with different mutational adaptations could potentially mask sub-population defects. It has recently been reported that the phenomena of GASP is complex, with mutant and wild type strains cooperating within the population to maximize bacterial fitness (Keymer et al., 2008). Cooperation between GASP mutant and wild type bacteria may thus ensure that L. monocytogenes effectively adapts for long-term stationary phase survival while maintaining bacterial virulence under nutrient poor conditions.
Supplementary Material
Acknowledgments
We thank Dr. Kathryn Boor for providing the ΔsigB deletion mutant in 10403S (FSL A1-254) and members of the Freitag lab for helpful discussions. We thank the reviewers of this manuscript for helpful comments and suggestions. This work was supported by Public health service grant AI41816 (N.E.F) from NIAID. The contents of the article are solely the responsibility of the authors and do not necessarily represent the official views of the funding sources.
References
- Abram F, Starr E, Karatzas KA, Matlawska-Wasowska K, Boyd A, Wiedmann M, Boor KJ, Connally D, O’Byrne CP. Identification of components of the sigma B regulon in Listeria monocytogenes that contribute to acid and salt tolerance. Appl Environ Microbiol. 2008;74:6848–6858. doi: 10.1128/AEM.00442-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auerbuch V, Lenz LL, Portnoy DA. Development of a competitive index assay to evaluate the virulence of Listeria monocytogenes actA mutants during primary and secondary infection of mice. Infect Immun. 2001;69:5953–5957. doi: 10.1128/IAI.69.9.5953-5957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DK, Hinrichs DJ. Adoptive transfer of immunity to Listeria monocytogenes: The influence of in vitro stimulation on lymphocyte subset requirements. J Immunol. 1987;139:2005–2009. [PubMed] [Google Scholar]
- Bruno JC, Jr, Freitag NE. Constitutive activation of PrfA tilts the balance of Listeria monocytogenes fitness towards life within the host versus environmental survival. PLoS One. 2010;5:e15138. doi: 10.1371/journal.pone.0015138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturongakul S, Boor KJ. SigmaB activation under environmental and energy stress conditions in Listeria monocytogenes. Appl Environ Microbiol. 2006;72:5197–5203. doi: 10.1128/AEM.03058-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico-Calero I, Suarez M, Gonzalez-Zorn B, Scortti M, Slaghuis J, Goebel W, Vazquez-Boland JA European Listeria Genome Consortium. Hpt, a bacterial homolog of the microsomal glucose-6-phosphate translocase, mediates rapid intracellular proliferation in Listeria. Proc Natl Acad Sci U S A. 2002;99:431–436. doi: 10.1073/pnas.012363899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czuprynski CJ. Listeria monocytogenes: silage, sandwiches and science. Anim Health Res Rev. 2005;6:211–217. doi: 10.1079/ahr2005111. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Herro R, Bourand A, Mijakovic I, Poncet S. P-Ser-HPr--a link between carbon metabolism and the virulence of some pathogenic bacteria. Biochim Biophys Acta. 2005;1754:118–125. doi: 10.1016/j.bbapap.2005.07.029. [DOI] [PubMed] [Google Scholar]
- Deutscher J, Francke C, Postma PW. How phosphotransferase system-related protein phosphorylation regulates carbohydrate metabolism in bacteria. Microbiol Mol Biol Rev. 2006;70:939–1031. doi: 10.1128/MMBR.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell MJ, Finkel SE. The growth advantage in stationary-phase phenotype conferred by rpoS mutations is dependent on the pH and nutrient environment. J Bacteriol. 2003;185:7044–7052. doi: 10.1128/JB.185.24.7044-7052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel SE, Kolter R. Evolution of microbial diversity during prolonged starvation. Proc Natl Acad Sci U S A. 1999;96:4023–4027. doi: 10.1073/pnas.96.7.4023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel SE, Zinser E, Kolter R. Long-term survival and evolution in stationary phase. In: Hengge-Aronis GSaR., editor. Bacterial Stress Responses. Washington, DC: ASM Press; 2000. pp. 231–238. [Google Scholar]
- Finkel SE. Long-term survival during stationary phase: evolution and the GASP phenotype. Nat Rev Microbiol. 2006;4:113–120. doi: 10.1038/nrmicro1340. [DOI] [PubMed] [Google Scholar]
- Freitag NE. From hot dogs to host cells: how the bacterial pathogen Listeria monocytogenes regulates virulence gene expression. Future Microbiol. 2006;1:89–101. doi: 10.2217/17460913.1.1.89. [DOI] [PubMed] [Google Scholar]
- Garner MR, Njaa BL, Wiedmann M, Boor KJ. Sigma B contributes to Listeria monocytogenes gastrointestinal infection but not to systemic spread in the guinea pig infection model. Infect Immun. 2006;74:876–886. doi: 10.1128/IAI.74.2.876-886.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel W, Kreft J, Bockmann R. Regulation of virulence genes in pathogenic Listeria spp. In: Fischetti VA, Novick RP, Ferretti JJ, Portnoy DA, Rood JI, editors. Gram-Positive pathogens. Washington D. C: ASM Press; 2000. pp. 499–506. [Google Scholar]
- Goetz M, Bubert A, Wang G, Chico-Calero I, Vazquez-Boland JA, Beck M, Slaghuis J, Szalay AA, Goebel W. Microinjection and growth of bacteria in the cytosol of mammalian host cells. Proc Natl Acad Sci U S A. 2001;98:12221–12226. doi: 10.1073/pnas.211106398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray MJ, Freitag NE, Boor KJ. How the bacterial pathogen Listeria monocytogenes mediates the switch from environmental Dr. Jekyll to pathogenic Mr. Hyde. Infect Immun. 2006;74:2505–2512. doi: 10.1128/IAI.74.5.2505-2512.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray ML, Killinger AH. Listeria monocytogenes and listeric infections. Bacteriol Rev. 1966;30:309–382. doi: 10.1128/br.30.2.309-382.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamon M, Bierne H, Cossart P. Listeria monocytogenes: a multifaceted model. Nat Rev Microbiol. 2006;4:423–434. doi: 10.1038/nrmicro1413. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. The general stress response in Escherichia coli. In: Hengge-Aronis GSaR., editor. Bacterial Stress Responses. Washington, DC: ASM Press; 2000. pp. 161–178. [Google Scholar]
- Joseph B, Przybilla K, Stuhler C, Schauer K, Slaghuis J, Fuchs TM, Goebel W. Identification of Listeria monocytogenes genes contributing to intracellular replication by expression profiling and mutant screening. J Bacteriol. 2006;188:556–568. doi: 10.1128/JB.188.2.556-568.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Goebel W. Life of Listeria monocytogenes in the host cells’ cytosol. Microbes Infect. 2007;9:1188–1195. doi: 10.1016/j.micinf.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Joseph B, Mertins S, Stoll R, Schar J, Umesha KR, Luo Q, Muller-Altrock S, Goebel W. Glycerol metabolism and PrfA activity in Listeria monocytogenes. J Bacteriol. 2008;190:5412–5430. doi: 10.1128/JB.00259-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmierczak MJ, Mithoe SC, Boor KJ, Wiedmann M. Listeria monocytogenes sigma B regulates stress response and virulence functions. J Bacteriol. 2003;185:5722–5734. doi: 10.1128/JB.185.19.5722-5734.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keymer JE, Galajda P, Lambert G, Liao D, Austin RH. Computation of mutual fitness by competing bacteria. Proc Natl Acad Sci USA. 2008;105:20269–20273. doi: 10.1073/pnas.0810792105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreft J, Vazquez-Boland JA. Regulation of virulence genes in Listeria. Int J Med Microbiol. 2001;291:145–157. doi: 10.1078/1438-4221-00111. [DOI] [PubMed] [Google Scholar]
- Lauer P, Chow MY, Loessner MJ, Portnoy DA, Calendar R. Construction, characterization, and use of two Listeria monocytogenes site-specific phage integration vectors. J Bacteriol. 2002;184:4177–4186. doi: 10.1128/JB.184.15.4177-4186.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loewen PC, Hu B, Strutinsky J, Sparling R. Regulation in the rpoS regulon of Escherichia coli. Can J Microbiol. 1998;44:707–717. doi: 10.1139/cjm-44-8-707. [DOI] [PubMed] [Google Scholar]
- Lunden JM, Autio TJ, Korkeala HJ. Transfer of persistent Listeria monocytogenes contamination between food-processing plants associated with a dicing machine. J Food Prot. 2002;65:1129–1133. doi: 10.4315/0362-028x-65.7.1129. [DOI] [PubMed] [Google Scholar]
- Miner MD, Port GC, Freitag NE. Functional impact of mutational activation on the Listeria monocytogenes central virulence regulator PrfA. Microbiology. 2008;154:3579–3589. doi: 10.1099/mic.0.2008/021063-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Byrne CP, Karatzas KA. The role of sigma B (σB) in the stress adaptations of Listeria monocytogenes: overlaps between stress adaptation and virulence. Adv Appl Microbiol. 2008;65:115–140. doi: 10.1016/S0065-2164(08)00605-9. [DOI] [PubMed] [Google Scholar]
- Orsi RH, den Bakker HC, Wiedmann M. Listeira monocytogenes lineages: genomics, evolution, ecology, and phenotypic characterizatics. International J Med Microbiol. 2011;301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Perry JJ, Staley JT. Microbiology: dynamics and diversity. Orlando, FL: Saunders College Publishing; 1997. [Google Scholar]
- Scortti M, Monzo HJ, Lacharme-Lora L, Lewis DA, Vazquez-Boland JA. The PrfA virulence regulon. Microbes Infect. 2007;9:1196–1207. doi: 10.1016/j.micinf.2007.05.007. [DOI] [PubMed] [Google Scholar]
- Shetron-Rama LM, Marquis H, Bouwer HGA, Freitag NE. Intracellular induction of Listeria monocytogenes actA expression. Infect Immun. 2002;70:1087–1096. doi: 10.1128/IAI.70.3.1087-1096.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinhaus EA, Birkeland JM. Studies on the life and death of bacteria: I. The senescent phase in aging cultures and the probable mechanisms involved. J Bacteriol. 1939;38:249–261. doi: 10.1128/jb.38.3.249-261.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thevenot D, Dernburg A, Vernozy-Rozand C. An updated review of Listeria monocytogenes in the pork meat industry and its products. J Appl Microbiol. 2006;101:7–17. doi: 10.1111/j.1365-2672.2006.02962.x. [DOI] [PubMed] [Google Scholar]
- Vazquez-Boland JA, Kuhn M, Berche P, Chakraborty T, Dominguez-Bernal G, Goebel W, Gonzalez-Zorn B, Wehland J, Kreft J. Listeria pathogenesis and molecular virulence determinants. Clin Microbiol Rev. 2001;14:584–640. doi: 10.1128/CMR.14.3.584-640.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeiser B, Pepper ED, Goodman MF, Finkel SE. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc Natl Acad Sci U S A. 2002;99:8737–8741. doi: 10.1073/pnas.092269199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano MM, Kolter R. Escherichia coli mutants lacking NADH dehydrogenase I have a competitive disadvantage in stationary phase. J Bacteriol. 1993;175:5642–5647. doi: 10.1128/jb.175.17.5642-5647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zambrano MM, Siegele DA, Almiron M, Tormo A, Kolter R. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science. 1993;259:1757–1760. doi: 10.1126/science.7681219. [DOI] [PubMed] [Google Scholar]
- Zinser ER, Kolter R. Mutations enhancing amino acid catabolism confer a growth advantage in stationary phase. J Bacteriol. 1999;181:5800–5807. doi: 10.1128/jb.181.18.5800-5807.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Kolter R. Prolonged stationary-phase incubation selects for lrp mutations in Escherichia coli K-12. J Bacteriol. 2000;182:4361–4365. doi: 10.1128/jb.182.15.4361-4365.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Schneider D, Blot M, Kolter R. Bacterial evolution through the selective loss of beneficial genes. Trade-offs in expression involving two loci. Genetics. 2003;164:1271–1277. doi: 10.1093/genetics/164.4.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinser ER, Kolter R. Escherichia coli evolution during stationary phase. Res Microbiol. 2004;155:328–336. doi: 10.1016/j.resmic.2004.01.014. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.