Abstract
DNA ‘barcoding’ relies on a short fragment of mitochondrial DNA to infer identification of specimens. The method depends on genetic diversity being markedly lower within than between species. Closely related species are most likely to share genetic variation in communities where speciation rates are rapid and effective population sizes are large, such that coalescence times are long. We assessed the applicability of DNA barcoding (here the 5′ half of the cytochrome c oxidase I) to a diverse community of butterflies from the upper Amazon, using a group with a well-established morphological taxonomy to serve as a reference. Only 77% of species could be accurately identified using the barcode data, a figure that dropped to 68% in species represented in the analyses by more than one geographical race and at least one congener. The use of additional mitochondrial sequence data hardly improved species identification, while a fragment of a nuclear gene resolved issues in some of the problematic species. We acknowledge the utility of barcodes when morphological characters are ambiguous or unknown, but we also recommend the addition of nuclear sequence data, and caution that species-level identification rates might be lower in the most diverse habitats of our planet.
Keywords: DNA barcoding, Amazon, biodiversity, Lepidoptera, mimicry
1. Introduction
There has been considerable recent interest in the use of short sequence tags known as ‘barcodes’ for the documentation and identification of a species (Blaxter 2003; Hebert et al. 2003a,b; Hebert & Gregory 2005; Savolainen et al. 2005; Dasmahapatra & Mallet 2006; Hajibabaei et al. 2007. In arthropods and vertebrates the DNA sequence used as a barcode is the 5′ half of the mitochondrial gene cytochrome c oxidase I (CoI). The utility of barcodes to accurately pigeonhole species has been successfully demonstrated in a number of studies (e.g. birds, Hebert et al. 2004b; Kerr et al. 2007), and has been offered as a tool for the discovery of cryptic butterfly and dipteran species (Hebert et al. 2004a; Smith et al. 2006, 2007; van Velzen et al. 2007). However, barcoding has been criticized from both theoretical and practical perspectives (Will & Rubinoff 2004; DeSalle et al. 2005; Ebach & Holdrege 2005; Meyer & Paulay 2005; Will et al. 2005; Brower 2006; Meier et al. 2006). Notably, successful barcode identification depends upon genetic diversity being markedly lower within than between species (Hebert et al. 2004b). This assumption is likely to be broken sometimes, especially when rates of speciation are greater than coalescence times of the gene in question (Pamilo & Nei 1988; Brower et al. 1996; Monaghan et al. 2006; Nielsen & Matz 2006; Wiemers & Fiedler 2007). While the success rate of barcoding undoubtedly varies among groups, some taxa and ecosystems are particularly likely to be subject to difficulties—in particular, groups in which recent speciation rates are high and effective population sizes large and reasonably stable, as is probable in many tropical insects. Here we carry out one of the first community-level barcoding studies in the most diverse terrestrial ecosystem in the world—the upper Amazon basin.
Our study group are the Ithomiinae, an entirely Neotropical subfamily of butterflies containing approximately 360 species confined to moist forest habitats. Adults are distasteful to predators and have warning wing colour patterns, and virtually all species are involved in Müllerian mimicry (Müller 1879; Brown 1984). Mimicry is likely to cause speciation (Jiggins et al. 2001, 2006) and thus to be involved in the diversification of ithomiine genera. The alpha taxonomy of ithomiines has a relatively long history of study and seems to be fairly well resolved, being largely based on abundant morphological characters including wing pattern, venation, androconia and genitalic structures (Lamas 2004; Willmott & Freitas 2006). The Ithomiinae are thus a good test case to assess the effectiveness of barcoding.
Our primary aim is to assess the utility of DNA barcoding in the identification of specimens from an exceptionally diverse ithomiine community in eastern lowland Ecuador. Previous studies based on a larger fragment of mitochondrial DNA (mtDNA) revealed that some ithomiine species in Amazonian Peru were not monophyletic (Whinnett et al. 2005), thus questioning the utility of barcoding in this group. Here we measure the frequency of misidentification at the species and genus levels, based on CoI barcode sequences only. To take into account geographical diversity, we include also conspecifics from more distant sampling sites, and congeners and other ithomiine genera not represented in the community. We also investigate whether the accuracy of identification is improved by increasing the length of mtDNA sequence and by including a nuclear gene.
2. Material and methods
(a) Study sites and sampling
Our main sampling effort is focused on two study sites located in eastern Ecuador, across the approximately 1 km wide Río Napo—Garza Cocha/La Selva Jungle Lodge near the Quechua communities of Pilche and Sani Isla on the north bank (0° 29.87′ S, 76° 22.45′ W, 6 km2 surveyed) and around the Añangu Quechua community/Napo Wildlife Center on the south bank (0° 31.41′ S, 76° 23.73′ W, 15 km2 surveyed). Ithomiine populations were studied between 2000 and 2007 at Garza Cocha and in 2005 and 2007 at Añangu. We collected all the 58 species present locally (table 1, electronic supplementary material 1). Specimens were identified by the authors using morphology of wing pattern, venation and genitalia, and were collected during each field season for genetic analyses. Bodies or legs were preserved in salt -saturated DMSO (20% DMSO, 0.25 M EDTA, saturated with NaCl), and wings were kept in envelopes. Rarely, individuals were dried as complete specimens and a single leg was used for genetic analysis. Two to nine local individuals per species were used in the analyses (except for the rare species Brevioleria seba and Dircenna dero, table 1).
Table 1.
Checklist and number of local and geographically distant conspecifics, support values (bootstrap or Bayesian posterior probabilities, in percentage) of the three clustering methods of the barcode sequences for genera and species represented by more than one species or individual.
| genus (ML, Bayesian and NJ support values) | species | local subspecies | no. local individuals | no. distant conspecifics | support values | ||
|---|---|---|---|---|---|---|---|
| ML | Bayes | NJ | |||||
| Aeria (only one species) | eurimedia | negricola | 6 | 3 | 100 | 100 | 100 |
| Brevioleria (no, no, no) | arzalia | ssp. | 5 | 2 | no | 78 | 98 |
| seba | oculata | 1 | no | ||||
| Callithomia (100, 100, 100) | alexirrhoe | butes | 2 | 2 | no | no | no |
| lenea | zelie | 5 | 3 | no | no | no | |
| Ceratinia (no, no, no) | tutia | poecila | 6 | 4 | 99 | 100 | 99 |
| Dircenna (only one species) | dero | 1 | 1 | 100 | 100 | 100 | |
| Episcada (no, no, 54) | sulphurea | ssp. 1 | 4 | 1 | 99 | 99 | 98 |
| Forbestra (98, 100, 84) | equicola | equicoloides | 3 | 1 | 97 | 100 | 100 |
| olivencia | juntana | 6 | 2 | no | no | no | |
| proceris | 2 | 1 | no | no | no | ||
| Godyris (95, 100, 99) | zavaleta | matronalis | 5 | 3 | 95 | 97 | 96 |
| Heterosais (only one species) | nephele | nephele | 5 | 1 | 95 | 100 | 100 |
| Hyalyris (no, no, no) | sp. | 4 | no | 90 | 100 | 100 | |
| Hypoleria (only one species) | lavinia | chrysodonia | 5 | 1 | 100 | 100 | 100 |
| Hyposcada (no, no, 21) | anchiala | ssp. | 5 | 2 | 84 | 100 | 98 |
| illinissa | ida | 5 | 5 | 99 | 100 | 99 | |
| kena | kena | 2 | 1 | 100 | 100 | 100 | |
| Hypothyris (no, no, no) | anastasia | honesta | 5 | 2 | 100 | 100 | 100 |
| euclea | intermedia | 5 | 1 | 100 | 100 | 99 | |
| fluonia | berna | 5 | 2 | 100 | 100 | 100 | |
| mamercus | mamercus | 7 | 2 | 100 | 100 | 100 | |
| moebiusi | moebiusi | 5 | no | 100 | 100 | 100 | |
| semifulva | satura | 5 | 1 | 95 | 100 | 100 | |
| Ithomia (27, 50, 29) | agnosia | agnosia | 6 | 1 | 100 | 100 | 100 |
| amarilla | 6 | no | 100 | 100 | 100 | ||
| salapia | salapia | 5 | 3 | 100 | 100 | 100 | |
| Mechanitis (100, 100, 99) | lysimnia | roqueensis | 6 | 1 | 80 | 84 | 96 |
| mazaeus | mazaeusa | 5 | 2 | 100 | 100 | 100 | |
| ‘mazaeus’ | deceptusa | 5 | 2 | no | no | no | |
| polymnia | ssp. | 7 | 4 | no | no | no | |
| Melinaea (100, 100, 100) | marsaeus | macaria/mothone | 9 | 3 | no | no | no |
| menophilus | menophilus | 5 | 3 | no | no | no | |
| satevis | maeonis | 6 | 3 | no | no | no | |
| Methona (100, 100, 98) | confusa | confusa | 3 | 5 | 98 | 100 | 100 |
| curvifascia | 6 | 1 | 100 | 100 | 100 | ||
| grandior | ssp. | 5 | no | 100 | 100 | 100 | |
| Napeogenes (66, 100, 72) | achaea | achaea | 3 | no | 100 | 100 | 100 |
| duessa | ssp. | 6 | 1 | 95 | 100 | 92 | |
| inachia | pozziana | 5 | 4 | 69 | 69 | 99 | |
| ‘larina’ | aethrab | 5 | no | 100 | 100 | 100 | |
| pharo | pharo | 5 | 1 | 91 | 100 | 100 | |
| quadrilis | 4 | no | 100 | 100 | 100 | ||
| sylphis | corena | 5 | 2 | 94 | no | 99 | |
| Oleria (18, 100, no) | agarista | agarista | 2 | 1 | no | no | no |
| assimilis | assimilis | 7 | no | 100 | 100 | 100 | |
| gunilla | lota | 5 | 1 | 100 | 100 | 100 | |
| ilerdina | lerida | 4 | no | 100 | 100 | 100 | |
| onega | ssp. | 5 | 2 | 90 | 100 | 96 | |
| sexmaculata | sexmaculata | 3 | no | 100 | 100 | 100 | |
| Pseudoscada (no, no, no) | florula | aureola | 4 | 1 | 100 | 100 | 100 |
| timna | utilla | 5 | 4 | no | 59 | 27 | |
| Pteronymia (65, 100, 56) | primula | primula | 5 | 1 | no | no | no |
| sao | ssp. | 4 | 1 | 100 | 100 | 100 | |
| vestilla | sparsa | 5 | 1 | no | no | no | |
| Scada (100, 100, 99) | zibia | batesi | 5 | 1 | 100 | 100 | 100 |
| Thyridia (only one species) | psidii | ino | 2 | 2 | 100 | 100 | 100 |
| Tithorea (only one species) | harmonia | hermias | 6 | 2 | 100 | 100 | 100 |
| species that form clusters (all species)a | 43 | 44 | 44 | ||||
| species that form clusters (species with foreign conspecifics and congeners)a,b | 27 | 28 | 28 | ||||
Mechanitis mazaeus is here regarded as two species, represented by M. mazaeus mazaeus and M. mazaeus deceptus.
Napeogenes larina aethra is here considered a species different from N. larina otaxes.
(b) DNA extraction and sequencing, and GenBank sequences
DNA was extracted using the QIAGEN DNeasy Kit, according to the manufacturer's protocol. Two hundred and seventy three local specimens and 80 non-local specimens belonging to the same species or genera (electronic supplementary material 1) were sequenced at the arthropod ‘barcode’ region, namely the 5′ half of the mitochondrial gene CoI (653 bp fragment). Primers, PCR and sequencing reaction conditions are given in the electronic supplementary material 2. In addition, 70 sequences of conspecifics, congeners or other ithomiine genera were downloaded from GenBank and aligned with our sequences (electronic supplementary material 1). Tellervo zoilus (Tellervinae) was used as an out-group in the analyses detailed below.
To test whether increasing information improves the accuracy of identification, a larger mitochondrial fragment (the entire CoI, 1464 bp, tRNA-Leucine, 62 bp and CoII, 716 bp) and a fragment of a nuclear gene, Elongation factor 1α (Ef1α, 1215 bp), were sequenced or downloaded from GenBank for a subset of the previous specimens (electronic supplementary material 1). Sequences were aligned using CodonCode Aligner v. 1.6.3 (CodonCode Corporation) and checked for reading frame errors using MacClade v. 4.07 (Maddison & Maddison 1997). All sequences have been deposited in GenBank under accession numbers EU068763–EU069266.
(c) Data analyses
Relationships among barcode sequences were inferred with three clustering methods. A neighbour-joining (NJ) tree based on Kimura 2-parameter (K2P) distances was computed using the software Mega v. 3.1 (Kumar et al. 2004). Branch support was assessed with 1000 bootstrap replicates. A maximum likelihood (ML) tree was generated with the program Phyml (Guindon & Gascuel 2003) using a GTR+Γ substitution model. Branch support was assessed with 100 bootstrap replicates. We also performed a Bayesian analysis using the program MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001). We performed two runs of four simultaneous Markov chains each for 1 000 000 generations starting from random initial trees and under a GTR+I+Γ model, and sampled a tree every 100 generations. Data from the first 900 000 generations (9000 trees) were discarded, after confirming that likelihood values had stabilized well before that. The consensus tree and posterior probability of nodes were calculated from the remaining 1000 trees.
Accuracy of identification was also tested by using the BLAST algorithm (Altschul et al. 1997) as follows. A FASTA file of the barcode data was first formatted as a BLAST database and a local BLASTn search (v. 2.2.15) was carried out on the entire dataset against this database.
Specimens that did not cluster with conspecifics or congeners in our barcode analysis were re-examined carefully to confirm identification (real specimen or photo) and were sometimes sequenced again to confirm initial results.
Two phylogenetic trees based on CoI–CoII and EF1α were obtained with MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001) as described previously. Mitochondrial sequence data were divided into eight partitions: CoI codon positions 1, 2 and 3, CoII codon positions 1, 2 and 3, tRNA-leucine, and non-coding DNA upstream of CoI; EF1α sequence data were partitioned by codon position. The best model of substitution for each region was selected using MrModelTest v. 2.2 (Nylander 2004; electronic supplementary material 3). ML and parsimony analyses were also performed, and they showed identical clustering patterns at the genus and species levels (results not shown).
3. Results
(a) Analyses of barcode sequences
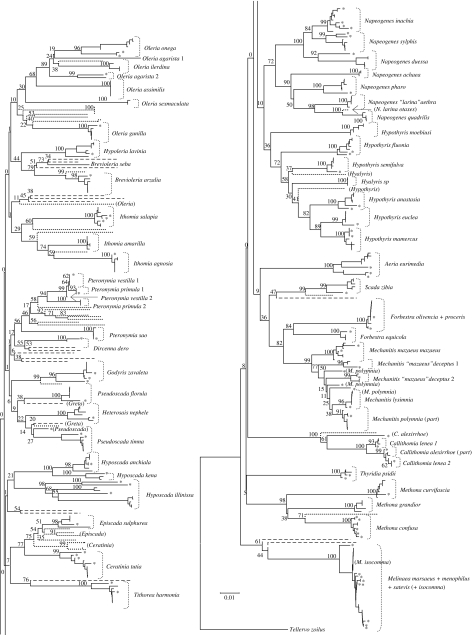
The three clustering methods (NJ, ML and Bayesian) gave comparable results for all but three species: Brevioleria arzalia, Napeogenes sylphis and Pseudoscada timna (table 1). Up to 42 out of 56 species present locally and represented by more than one individual (75%) formed well-supported monophyletic groups (bootstrap or Bayesian probability greater than 50%, table 1, figure 1, electronic supplementary material 4a-c). Species monophyly increased to 77% (44 out of 57) when Napeogenes larina aethra and Mechanitis mazaeus deceptus were considered species separate from Napeogenes larina otaxes and Mechanitis mazaeus mazaeus, respectively, as these and other data suggest (Elias et al. 2007; R. I. Hill et al. 2006–2007, unpublished data). Consequently, Napeogenes ‘larina’ aethra and M. ‘mazaeus’ deceptus will be considered ‘good’ species in the analyses below. Considering only those species from the community dataset represented by more than one congener and including remote conspecifics in the analysis (i.e. situations where barcoding would be most useful), only 28 out of 41 species (68%) were monophyletic. Similar results were achieved using the BLAST algorithm. In 13 of the species (23% of all the species, 32% of the species with congeners and remote conspecifics), at least one non-conspecific individual showed a higher BLAST bit score as compared with other conspecifics in the dataset.
Figure 1.
Neighbour-joining clustering of barcode sequences, with bootstrap values shown on the nodes. Geographically distant conspecifics are represented by an asterisk. Dotted and dashed lines represent congeners and genera that are not represented in the Río Napo community, respectively. Names of non-local specimens are indicated in brackets in case of ambiguity or particular interest. Complete trees obtained with the three clustering methods are available in the electronic supplementary material 4a–c.
In the genus Melinaea, the three local species were entirely undifferentiated, confirming previous observations from Peru (Whinnett et al. 2005). Similarly, sister species pairs such as Forbestra olivencia and Forbestra proceris, Pteronymia vestilla and Pteronymia primula, Callithomia lenea and Callithomia alexirrhoe were partly or completely undifferentiated with respect to one another. In contrast, M. ‘mazaeus’ deceptus and Oleria agarista were split into several lineages, thus becoming para- or polyphyletic with respect to closely related taxa. Failure to form monophyletic groups occurred between local conspecifics in 10 species, while only divergence between geographical populations was involved in the remaining three cases (figure 1). There was no differentiation between the two banks of the Río Napo, the only exception being O. agarista (electronic supplementary material 4a-c).
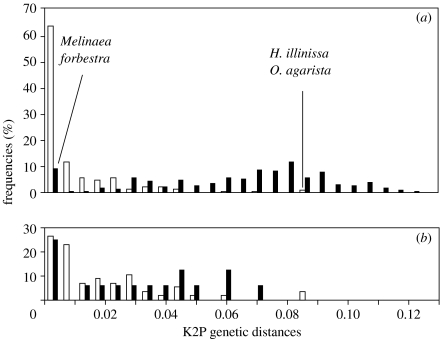
Genetic distances within species present locally (0.0085±0.0137) were on average much lower than distances between congeneric species (0.0602±0.0292, figure 2a). However, the distributions overlapped, with intraspecific distances over 0.08 in Oleria and Hyposcada contrasting with no interspecific divergence in Melinaea and Forbestra (figure 2a,b).
Figure 2.
Distribution of within-species (white) and between congeneric species (black) K2P distances of barcode sequences. Only species and genera represented locally are considered. (a) All pairwise distances and (b) only maximum intraspecific and minimum interspecific distances.
At the generic level, 9 (ML) to 11 (Bayesian analysis) of the 18 genera present locally and represented by more than one species (50–61%) were monophyletic (figure 1, table 1, electronic supplementary material 4a-c). Paraphyletic genera were Brevioleria, Ceratinia, Episcada, Hyalyris, Hyposcada, Hypothyris, Ithomia, Oleria and Pseudoscada.
(b) Improved accuracy by using more genes
Inclusion of the entire CoI–CoII region resulted in the same relationships within species as with the barcode alone (electronic supplementary material 5a). The only exception to this was O. agarista, which was monophyletic in the full mtDNA dataset. The nuclear gene EF1α did contribute more to identification accuracy (electronic supplementary material 5b). Pteronymia primula, F. proceris, F. olivencia and O. agarista were monophyletic, sequences of the last species being virtually identical. All M. mazaeus clustered together, but there was no distinction between M. mazaeus mazaeus and M. ‘mazaeus’ deceptus.
At the generic level, Ceratinia remained paraphyletic with respect to Episcada with both gene regions. Species from the genera Greta and Pseudoscada were intermixed in the mtDNA analysis, but with EF1α Pseudoscada species formed a monophyletic group, close to a paraphyletic Greta. Oleria became monophyletic with the longer mitochondrial region but not with EF1α. Hyalyris, Hyposcada and Ithomia formed monophyletic groups using each of the additional regions.
4. Discussion
Barcoding has triggered a passionate debate between proponents who aim to ‘tag’ the diversity of life (Hebert et al. 2003a,b; Hebert & Gregory 2005; Savolainen et al. 2005; Hajibabaei et al. 2007), and detractors who highlight the pitfalls of the single-gene approach and are concerned about competition for funds with traditional taxonomy (Will & Rubinoff 2004; Ebach & Holdrege 2005; Wheeler 2005; Will et al. 2005; Brower 2006; Cameron et al. 2006; Rubinoff 2006). It has been claimed that mtDNA barcodes achieve an accuracy close to 100% in delimiting species in some groups (Hebert et al. 2004b; Hajibabaei et al. 2006a; Clare et al. 2007; Ekrem et al. 2007; Kerr et al. 2007), although in some cases it has proved less successful (Meyer & Paulay 2005; Meier et al. 2006; Wiemers & Fiedler 2007; Whitworth et al. 2007). Our own study shows a rather poor success of DNA barcoding, with only 77% of species unambiguously identified.
Previous barcoding work in the tropics has mainly focused on the Guanacaste National Park in Costa Rica (Hebert et al. 2004a; Janzen et al. 2005; Hajibabaei et al. 2006a; Smith et al. 2006, 2007), a mosaic of mainly dry forest and cloud forest habitats which is less diverse than the habitat studied here. For example, in the whole of Costa Rica (51 100 km2) there are 61 species of ithomiine butterflies (DeVries 1997), similar to the diversity found in the 21 km2 sampled in the Napo community. The relatively aseasonal and continuous habitat of the upper Amazon probably maintains large and stable population sizes of these butterflies, with correspondingly longer coalescence times when compared with more seasonal habitats such as dry forest, or temperate regions where populations have been reduced to small refugia during glacial periods. Moreover, butterfly genera of the upper Amazon tend to contain more species (Lamas 2004). Both factors point to a greater challenge for gene-based identification in the Amazon. Not surprisingly, the six species without congeners in our analysis were accurately diagnosed. However, adding congeners and geographical populations decreased the success of barcoding: 32% of such species could not be diagnosed. In other words, the barcode method of identification becomes significantly less reliable when groups of closely related (congeneric) species are examined, or geographical populations of the same species are included, exactly the circumstances where barcoding would be most useful as a tool. Thus, biogeographic and evolutionary history also plays a role in the success of the identification of specimens using barcoding.
Hebert et al. (2004b) suggested the use of a threshold in sequence divergence in the discovery of new species, claiming that barcoding could be an outstanding tool for this purpose (Gomez et al. 2007). Yet the existence of the so-called ‘barcode gap’ has been challenged both on theoretical (Hickerson et al. 2006) and empirical grounds (Cognato 2006; Wiemers & Fiedler 2007). The pattern of barcode evolution in ithomiine butterflies, with highly variable levels of divergence within different species complexes and genera (Whinnett et al. 2005; figure 2), adds fuel to these criticisms. We agree that barcoding should not be relied on as the sole basis for species discovery, except as a preliminary effort in the case where ecological, morphological or additional genetic data are absent.
(a) Generic level identification
At best 61% of currently recognized genera represented by more than one species formed stable clusters. Some generic circumscriptions are likely to be revised in the near future on the basis of ongoing morphological and molecular work (Brower et al. 2006; Willmott & Freitas 2006; K. R. Willmott 2001–2007, unpublished data), and some taxa could be moved from one genus to another. This might affect the pairs of genera Pseudoscada and Greta, Hypothyris and Hyalyris, and Ceratinia and Episcada. In the most optimistic hypothesis where these pairs of genera in our study are pooled, only 11–13 of the rearranged genera represented locally (69–81%) are monophyletic. Thus, even at the generic level, the barcode performs rather poorly in assigning names to specimens.
(b) Limitations of existing databases
DNA barcodes cannot be a useful identification tool without a comprehensive and reliable reference database (Meyer & Paulay 2005; Brower 2006; Hajibabaei et al. 2006b; Scheffer et al. 2006). To investigate the extent to which existing databases can be used to identify specimens in our study, we tested the Barcode of Life Data Systems identification engine (BoLD-ID) in August 2007 (Ratnasingham & Hebert 2007; http://www.barcodinglife.com/views/idrequest.php), with mixed results. We used a subset of 61 sequences of local specimens representing all species and major lineages within species. While 74% of the specimens were correctly identified at the genus level using the full database (but only 41% using the reference database), a number of specimens (18 and 44%, using the full and reference databases, respectively) were assigned to an incorrect (and unrelated) genus with a probability of placement of 1. The remaining specimens could not be assigned to any genus. In the latter cases, the paucity of ithomiine sequences in the database (15 and 9 ithomiine genera found in the full and reference databases, respectively) explains the failure to hit the correct genera. Moreover, the decision system that generates the probabilities of placement tends to overestimate these values for taxa poorly represented in the database. Thus the BoLD-ID engine is currently not suitable for a reliable identification of ithomiines (and that of other relatively poorly represented groups), although the identification accuracy should improve when the sequences of this study are included in the BoLD database. As a comparison when BLASTing the same sequences against the GenBank nucleotide database, where all the tested genera are represented, the correct genus was identified in 88% of the cases.
(c) Analytical methods for barcode data
As no consensus has been reached yet concerning the analysis of barcode data, there is a growing literature testing and describing analytical methods (e.g. Little & Stevenson 2007). A number of authors have proposed or tested novel methods based on diagnostic molecular characters relying either on the presence/absence of short fragments of sequence (DasGupta et al. 2005; Little & Stevenson 2007) or on parsimony informative sites or combinations of sites referred to as ‘characteristic attributes’ (Sarkar et al. 2002; Kelly et al. 2007). We have not used any of these methods as we believe that the use of diagnostic molecular characters to identify species is likely to result in lower accuracy due to incomplete conspecific reference material (see previous paragraph), especially from across the geographical ranges of the species (Little & Stevenson 2007). To establish a sequence database for the Ithomiinae over the entire range of the subfamily would require collecting and sequencing more than 1600 morphologically divergent subspecific taxa, which is a daunting task.
Here, we compared three alignment-based clustering methods and an alignment-free similarity method, which gave similar results. Since the NJ clustering and the similarity method (BLAST) perform considerably faster than the others, we believe they are a good choice for barcode analyses (Little & Stevenson 2007). NJ clustering has indeed been used in the great majority of published barcoding studies. However, the most promising area for future research is methods that take into account population size and therefore coalescence times in the estimation of barcoding probabilities (Nielsen & Matz 2006).
(d) The value of nuclear data
Despite extensive use of the mitochondrial gene CoI in vertebrates, arthropods and, more recently, fungi (Seifert et al. 2007), there are potential drawbacks to using the mitochondrial genome (Rubinoff et al. 2006). Notably, maternal inheritance, the absence of recombination or the spread of cytoplasmically inherited symbionts can lead to misleading identification using the mtDNA barcode (Hurst & Jiggins 2005; Rubinoff et al. 2006; Whitworth et al. 2007).
Our data have demonstrated the value of adding nuclear sequence data to the mtDNA barcode. Despite the relatively slow rate of evolution at EF1α, it has nonetheless resolved several cases where the mtDNA barcode failed. Notably, species with very divergent mtDNA sequences between geographical populations showed nearly identical EF1α sequences. As already suggested (Dasmahapatra & Mallet 2006; Smith et al. 2006, 2007; Gomez et al. 2007) we would therefore recommend that the mtDNA barcode be supplemented where possible by one or more nuclear genes. EF1α has been used here as it is widely studied in insect phylogenetics and easily amplified across many insect groups (Wahlberg et al. 2005; Danforth et al. 2006), although its slow rate of evolution means that other genes, such as rapidly evolving introns (Beltrán et al. 2002), might be more informative for closely related species.
5. Conclusion
Our work on ithomiine butterflies has demonstrated that the barcode method of identification, as currently applied, may be significantly less successful in certain circumstances than proponents have suggested. In particular, diverse tropical invertebrate faunas with multiple congeneric species and geographical races, in other words the bulk of biodiversity, pose a particular challenge. Despite the limitations of barcoding for species identification and discovery, the use of mitochondrial sequence data is clearly a valuable tool for taxonomy. Insect molecular systematics employing mtDNA has enjoyed great success for two decades (DeSalle et al. 1987; Brower 1994; Caterino et al. 2000). Unexpected mtDNA CoI patterns have led to the discovery of cryptic species (Brower 1996; Hebert et al. 2004a; Mallarino et al. 2005; Smith et al. 2006, 2007). Similarly, barcode patterns have encouraged us to search for covarying morphological, genetic and ecological characters in the genus Mechanitis (R. I. Hill et al. 2006–2007, unpublished data) and in the species Napeogenes larina (Elias et al. 2007), where divergent lineages probably represent unrecognized cryptic species. We are also using barcode sequences to recover the identity of immature stages and thus host plant records, where larvae were collected in the field but died before reaching the adult stage (K. R. Willmott et al. 2005–2007, unpublished data), and to identify the sources of blood meals from engorged mosquitoes (Townzen et al. 2005, unpublished data). Finally, mitochondrial barcode sequences are sometimes the best sequence data that can be obtained from old, degraded museum specimens. The growing public databases containing a homologous sequence region from a diversity of taxa will undoubtedly continue to be a valuable tool for the study of biodiversity.
Acknowledgments
We thank Raul Aldaz, Julia Robinson Willmott, Aniko Zölei, Gabor Papp, Alexander Toporov and Carlos Sanchez for their help in the field. We thank Gerardo Lamas, Mathieu Joron, Mark Blaxter and two anonymous referees for their useful comments and taxonomic advice and Fraser Simpson and Lisa Leadbeater for providing specimens. We thank the Ministerio del Ambiente and the Museo Ecuatoriano de Ciencias Naturales in Ecuador, and the Instituto Nacional de Recursos Naturales (INRENA), the Museo de Historia Natural and the Universidad Nacional Mayor de San Marcos in Peru for collecting permits and support. The Napo Wildlife Centre and La Selva Jungle Lodge provided logistic support for our fieldwork. This research was funded by the Leverhulme trust and the Royal Society (M.E., K.R.W. and C.D.J.), by the Margaret C. Walker fund for teaching and research in systematic entomology (R.I.H.), NERC (K.K.D. and J.M.), and by the US National Science Foundation grants DEB 0089886 and DEB 0640301 (A.V.Z.B.).
Supplementary Material
Complete list of samples used with locality data
Primers, PCR and sequencing reaction conditions
Models of substitution selected for each partition (Bayesian analyses)
Barcode trees obtained from (a) maximum likelihood, (b) Bayesian analyses and (c) neighbour-joining
Phylogenetic trees obtained with (a) CoI–CoII sequences and (b) EF1α sequences
References
- Altschul S.F, Madden T.L, Schaffer A.A, Zhang J.H, Zhang Z, Miller W, Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. doi:10.1093/nar/25.17.3389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltrán M.S, Jiggins C.D, Bull V, Linares M, Mallet J, McMillan W.O, Bermingham E. Phylogenetic discordance at the species boundary: comparative gene genealogies among rapidly radiating Heliconius butterflies. Mol. Biol. Evol. 2002;19:2176–2190. doi: 10.1093/oxfordjournals.molbev.a004042. [DOI] [PubMed] [Google Scholar]
- Blaxter M. Molecular systematics—counting angels with DNA. Nature. 2003;421:122–124. doi: 10.1038/421122a. doi:10.1038/421122a [DOI] [PubMed] [Google Scholar]
- Brower A.V.Z. Phylogeny of Heliconius butterflies inferred from mitochondrial DNA sequences (Lepidoptera: Nymphalidae) Mol. Phylogenet. Evol. 1994;3:159–174. doi: 10.1006/mpev.1994.1018. doi:10.1006/mpev.1994.1018 [DOI] [PubMed] [Google Scholar]
- Brower A.V.Z. A new mimetic species of Heliconius (Lepidoptera: Nymphalidae), from southeastern Colombia, as revealed by cladistic analysis of mitochondrial DNA sequences. Zool. J. Linn. Soc. 1996;116:317–332. doi:10.1006/zjls.1996.0022 [Google Scholar]
- Brower A.V.Z. Problems with DNA barcodes for species delimitation: ‘ten species’ of Astraptes fulgerator reassessed (Lepidoptera: Hesperiidae) Syst. Biodiv. 2006;4:127–132. doi:10.1017/S147720000500191X [Google Scholar]
- Brower A.V.Z, DeSalle R, Vogler A.P. Gene trees, species trees and systematics: a cladistic perspective. Annu. Rev. Ecol. Syst. 1996;27:423–450. doi:10.1146/annurev.ecolsys.27.1.423 [Google Scholar]
- Brower A.V.Z, Freitas A.V.L, Lee M.M, Silva-Brandao K.L, Whinnett A, Willmott K.R. Phylogenetic relationships among the Ithomiini (Lepidoptera: Nymphalidae) inferred from one mitochondrial and two nuclear gene regions. Syst. Entomol. 2006;31:288–301. doi:10.1111/j.1365-3113.2006.00321.x [Google Scholar]
- Brown K.S., Jr Adult-obtained pyrrolizidine alkaloids defend ithomiine butterflies against a spider predator. Nature. 1984;309:707–709. doi:10.1038/309707a0 [Google Scholar]
- Cameron S, Rubinoff D, Will K. Who will actually use DNA barcoding and what will it cost? Syst. Biol. 2006;55:844–847. doi: 10.1080/10635150600960079. doi:10.1080/10635150600960079 [DOI] [PubMed] [Google Scholar]
- Caterino M.S, Cho S, Sperling F.A.H. The current state of insect molecular systematics: a thriving tower of Babel. Annu. Rev. Entomol. 2000;45:1–54. doi: 10.1146/annurev.ento.45.1.1. doi:10.1146/annurev.ento.45.1.1 [DOI] [PubMed] [Google Scholar]
- Clare E.L, Lim B.K, Engstrom M.D, Eger J.L, Hebert P.D.N. DNA barcoding of Neotropical bats: species identification and discovery within Guyana. Mol. Ecol. Notes. 2007;7:184–190. doi:10.1111/j.1471-8286.2006.01657.x [Google Scholar]
- Cognato A.I. Standard percent DNA sequence difference for insects does not predict species boundaries. J. Econ. Entomol. 2006;99:1037–1045. doi: 10.1603/0022-0493-99.4.1037. [DOI] [PubMed] [Google Scholar]
- Danforth B.N, Sipes S, Fang J, Brady S.G. The history of early bee diversification based on five genes plus morphology. Proc. Natl Acad. Sci. USA. 2006;103:15 118–15 123. doi: 10.1073/pnas.0604033103. doi:10.1073/pnas.0604033103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DasGupta B, Konwar K.M, Măndoiu I.I, Shvartsman A.A. DNA-BAR: distinguisher selection for DNA barcoding. Bioinformatics. 2005;21:3424–3426. doi: 10.1093/bioinformatics/bti547. doi:10.1093/bioinformatics/bti547 [DOI] [PubMed] [Google Scholar]
- Dasmahapatra K.K, Mallet J. DNA barcodes: recent successes and future prospects. Heredity. 2006;97:254–255. doi: 10.1038/sj.hdy.6800858. doi:10.1038/sj.hdy.6800858 [DOI] [PubMed] [Google Scholar]
- DeSalle R, Freedman T, Prager E.M, Wilson A.C. Tempo and mode of sequence evolution in mitochondrial DNA of Hawaiian Drosophila. J. Mol. Evol. 1987;26:157–164. doi: 10.1007/BF02111289. doi:10.1007/BF02111289 [DOI] [PubMed] [Google Scholar]
- DeSalle R, Egan M.G, Siddall M. The unholy trinity: taxonomy, species delimitation and DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1905–1916. doi: 10.1098/rstb.2005.1722. doi:10.1098/rstb.2005.1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeVries P.J. Papilionidae, Pieridae, Nymphalidae. vol. I. Princeton University Press; Princeton, NJ: 1997. The butterflies of Costa Rica and their natural history. [Google Scholar]
- Ebach M.C, Holdrege C. DNA barcoding is no substitute for taxonomy. Nature. 2005;434:697–697. doi: 10.1038/434697b. doi:10.1038/434697b [DOI] [PubMed] [Google Scholar]
- Ekrem T, Willassen E, Stur E. A comprehensive DNA sequence library is essential for identification with DNA barcodes. Mol. Phylogenet. Evol. 2007;43:530–542. doi: 10.1016/j.ympev.2006.11.021. doi:10.1016/j.ympev.2006.11.021 [DOI] [PubMed] [Google Scholar]
- Elias, M., et al 2007 Phylogenetic hypothesis, pattern of speciation and evolution of wing pattern in neotropical Napeogenes butterflies (Lepidoptera: Nymphalidae). In Seventh Int. Workshop on the Molecular Biology and Genetics of the Lepidoptera 20–26 August 2006, Orthodox Academy of Crete, Kolympari, Crete, Greece, (eds K. Iatrou & P. Couble), pp. 13–14. J. Insect Sci.,7, 29. [DOI] [PMC free article] [PubMed]
- Gomez A, Wright P.J, Lunt D.H, Cancino J.M, Carvalho G.R, Hughes R.N. Mating trials validate the use of DNA barcoding to reveal cryptic speciation of a marine bryozoan taxon. Proc. R. Soc. B. 2007;274:199–207. doi: 10.1098/rspb.2006.3718. doi:10.1098/rspb.2006.3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. doi:10.1080/10635150390235520 [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Janzen D.H, Burns J.M, Hallwachs W, Hebert P.D.N. DNA barcodes distinguish species of tropical Lepidoptera. Proc. Natl Acad. Sci. USA. 2006a;103:968–971. doi: 10.1073/pnas.0510466103. doi:10.1073/pnas.0510466103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajibabaei M, Singer G.A.C, Hickey D.A. Benchmarking DNA barcodes: an assessment using available primate sequences. Genome. 2006b;49:851–854. doi: 10.1139/g06-025. doi:10.1139/G06-025 [DOI] [PubMed] [Google Scholar]
- Hajibabaei M, Singer G.A.C, Hebert P.D.N, Hickey D.A. DNA barcoding: how it complements taxonomy, molecular phylogenetics and population genetics. Trends Genet. 2007;23:167–172. doi: 10.1016/j.tig.2007.02.001. doi:10.1016/j.tig.2007.02.001 [DOI] [PubMed] [Google Scholar]
- Hebert P.D.N, Gregory T.R. The promise of DNA barcoding for taxonomy. Syst. Biol. 2005;54:852–859. doi: 10.1080/10635150500354886. doi:10.1080/10635150500354886 [DOI] [PubMed] [Google Scholar]
- Hebert P.D.N, Cywinska A, Ball S.L, DeWaard J.R. Biological identifications through DNA barcodes. Proc. R. Soc. B. 2003a;270:313–321. doi: 10.1098/rspb.2002.2218. doi:10.1098/rspb.2002.2218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Ratnasingham S, deWaard J.R. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. Proc. R. Soc. B. 2003b;270:S96–S99. doi: 10.1098/rsbl.2003.0025. doi:10.1098/rsbl.2003.0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Penton E.H, Burns J.M, Janzen D.H, Hallwachs W. Ten species in one: DNA barcoding reveals cryptic species in the neotropical skipper butterfly Astraptes fulgerator. Proc. Natl Acad. Sci. USA. 2004a;101:14 812–14 817. doi: 10.1073/pnas.0406166101. doi:10.1073/pnas.0406166101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert P.D.N, Stoeckle M.Y, Zemlak T.S, Francis C.M. Identification of birds through DNA barcodes. PLoS Biol. 2004b;2:1657–1663. doi: 10.1371/journal.pbio.0020312. doi:10.1371/journal.pbio.0020312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickerson M, Meyer C, Moritz C. DNA barcoding will often fail to discover new animal species over broad parameter space. Syst. Biol. 2006;55:729–739. doi: 10.1080/10635150600969898. doi:10.1080/10635150600969898 [DOI] [PubMed] [Google Scholar]
- Huelsenbeck J.P, Ronquist F. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. doi:10.1093/bioinformatics/17.8.754 [DOI] [PubMed] [Google Scholar]
- Hurst G.D.D, Jiggins F.M. Problems with mitochondrial DNA as a marker in population, phylogeographic and phylogenetic studies: the effects of inherited symbionts. Proc. R. Soc. B. 2005;272:1525–1534. doi: 10.1098/rspb.2005.3056. doi:10.1098/rspb.2005.3056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janzen D.H, Hajibabaei M, Burns J.M, Hallwachs W, Remigio E, Hebert P.D.N. Wedding biodiversity inventory of a large and complex Lepidoptera fauna with DNA barcoding. Phil. Trans. R. Soc. B. 2005;1462:1835–1846. doi: 10.1098/rstb.2005.1715. doi:10.1098/rstb.2005.1715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiggins C.D, Naisbit R.E, Coe R.L, Mallet J. Reproductive isolation caused by colour pattern mimicry. Nature. 2001;411:302–305. doi: 10.1038/35077075. doi:10.1038/35077075 [DOI] [PubMed] [Google Scholar]
- Jiggins C.D, Mallarino R, Willmott K.W, Bermingham E. The phylogenetic pattern of speciation and wing pattern change in neotropical Ithomia butterflies (Lepidoptera; Nymphalidae) Evolution. 2006;60:1454–1466. doi: 10.1554/05-483.1. [DOI] [PubMed] [Google Scholar]
- Kelly R.P, Sarkar I.N, Eernisse D.J, Desalle R. DNA barcoding using chitons (genus Mopalia) Mol. Ecol. Notes. 2007;7:177–183. doi:10.1111/j.1471-8286.2006.01641.x [Google Scholar]
- Kerr K.C.R, Stoeckle M.Y, Dove C.J, Weigt L.A, Francis C.M, Hebert P.D.N. Comprehensive DNA barcode coverage of North American birds. Mol. Ecol. Notes. 2007;7:535–543. doi: 10.1111/j.1471-8286.2007.01670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. Mega3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. doi:10.1093/bib/5.2.150 [DOI] [PubMed] [Google Scholar]
- Lamas, G. 2004 Ithomiinae. In Atlas of neotropical Lepidoptera. Checklist: Part 4A. Hesperioidea–Papilionoidea (ed. J. B. Heppner), pp. 172–191. Gainesville, FL: Association for Tropical Lepidoptera/Scientific Publishers.
- Little D.P, Stevenson D.W. A comparison of algorithms for the identification of specimens using DNA barcodes: examples from gymnosperms. Cladistics. 2007;23:1–21. doi: 10.1111/j.1096-0031.2006.00126.x. doi:10.1111/j.1096-0031.2006.00126.x [DOI] [PubMed] [Google Scholar]
- Maddison W.P, Maddison D.R. Sinauer Associates; Sunderland, MA: 1997. MacClade: analysis of phylogeny and character evolution. [DOI] [PubMed] [Google Scholar]
- Mallarino R, Bermingham E, Willmott K.R, Whinnett A, Jiggins C.D. Molecular systematics of the butterfly genus Ithomia (Lepidoptera: Ithomiinae): a composite phylogenetic hypothesis based on seven genes. Mol. Phylogenet. Evol. 2005;34:625–644. doi: 10.1016/j.ympev.2004.10.021. doi:10.1016/j.ympev.2004.10.021 [DOI] [PubMed] [Google Scholar]
- Meier R, Shiyang K, Vaidya G, Ng P.K.L. DNA barcoding and taxonomy in Diptera: a tale of high intraspecific variability and low identification success. Syst. Biol. 2006;55:715–728. doi: 10.1080/10635150600969864. doi:10.1080/10635150600969864 [DOI] [PubMed] [Google Scholar]
- Meyer C.P, Paulay G. DNA barcoding: error rates based on comprehensive sampling. PLoS Biol. 2005;3:2229–2238. doi: 10.1371/journal.pbio.0030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan M.T, Balke M, Pons J, Vogler A.P. Beyond barcodes: complex DNA taxonomy of a south pacific island radiation. Proc. R. Soc. B. 2006;273:887–893. doi: 10.1098/rspb.2005.3391. doi:10.1098/rspb.2005.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller F. Ituna and Thyridia: a remarkable case of mimicry in butterflies. Trans. Entomol. Soc. London. 1879;1879:xx–xxix. [Google Scholar]
- Nielsen R, Matz M. Statistical approaches for DNA barcoding. Syst. Biol. 2006;55:162–169. doi: 10.1080/10635150500431239. doi:10.1080/10635150500431239 [DOI] [PubMed] [Google Scholar]
- Nylander J.A.A. Evolutionary Biology Center; Uppsala University, Sweden: 2004. MrModeltest v2. Program distributed by the author. [Google Scholar]
- Pamilo P, Nei M. Relationships between gene trees and species trees. Mol. Biol. Evol. 1988;5:568–583. doi: 10.1093/oxfordjournals.molbev.a040517. [DOI] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert P.D.N. BOLD: the Barcode of Life Data System. Mol. Ecol. Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. http://www.barcodinglife.org [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinoff D. Utility of mitochondrial DNA barcodes in species conservation. Conserv. Biol. 2006;20:1026–1033. doi: 10.1111/j.1523-1739.2006.00372.x. doi:10.1111/j.1523-1739.2006.00542.x [DOI] [PubMed] [Google Scholar]
- Rubinoff D, Cameron S, Will K. A genomic perspective on the shortcomings of mitochondrial DNA for “barcoding” identification. J. Hered. 2006;97:581–594. doi: 10.1093/jhered/esl036. doi:10.1093/jhered/esl036 [DOI] [PubMed] [Google Scholar]
- Sarkar I.N, Joseph W.T, Paul J.P, David H.F, Bernd S, Rob D. An automated phylogenetic key for classifying homeoboxes. Mol. Phylogenet. Evol. 2002;24:388–399. doi: 10.1016/s1055-7903(02)00259-2. doi:10.1016/S1055-7903(02)00259-2 [DOI] [PubMed] [Google Scholar]
- Savolainen V, Cowan R.S, Vogler A.P, Roderick G.K, Lane R. Towards writing the encyclopaedia of life: an introduction to DNA barcoding. Phil. Trans. R. Soc. B. 2005;360:1805–1811. doi: 10.1098/rstb.2005.1730. doi:10.1098/rstb.2005.1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffer S.J, Lewis M.L, Joshi R.C. DNA barcoding applied to invasive leafminers (Diptera: Agromyzidae) in The Philippines. Ann. Entomol. Soc. Am. 2006;99:204–210. doi:10.1603/0013-8746(2006)099[0204:DBATIL]2.0.CO;2 [Google Scholar]
- Seifert K.A, Samson R.A, Dewaard J.R, Houbraken J, Levesque C.A, Moncalvo J.M, Louis-Seize G, Hebert P.D.N. Prospects for fungus identification using C01 DNA barcodes, with Penicillium as a test case. Proc. Natl Acad. Sci. USA. 2007;104:3901–3906. doi: 10.1073/pnas.0611691104. doi:10.1073/pnas.0611691104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.A, Woodley N.E, Janzen D.H, Hallwachs W, Hebert P.D.N. DNA barcodes reveal cryptic host-specificity within the presumed polyphagous members of a genus of parasitoid flies (Diptera: Tachinidae) Proc. Natl Acad. Sci. USA. 2006;103:3657–3662. doi: 10.1073/pnas.0511318103. doi:10.1073/pnas.0511318103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M.A, Wood D.M, Janzen D.H, Hallwachs W, Hebert P.D.N. DNA barcodes affirm that 16 species of apparently generalist tropical parasitoid flies (Diptera, Tachinidae) are not all generalists. Proc. Natl Acad. Sci. USA. 2007;104:4967–4972. doi: 10.1073/pnas.0700050104. doi:10.1073/pnas.0700050104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velzen R, Bakker F.T, van Loon J.J.A. DNA barcoding reveals hidden species diversity in Cymothoe (Nymphalidae) Proc. Neth. Entomol. Soc. Meet. 2007;18:95–103. [Google Scholar]
- Wahlberg N, Brower A.V.Z, Nylin S. Phylogenetic relationships and historical biogeography of tribes and genera in the subfamily Nymphalinae (Lepidoptera: Nymphalidae) Biol. J. Linn. Soc. 2005;86:227–251. doi:10.1111/j.1095-8312.2005.00531.x [Google Scholar]
- Wheeler Q.D. Losing the plot: DNA “barcodes” and taxonomy. Cladistics. 2005;21:405–407. doi: 10.1111/j.1096-0031.2005.00075.x. doi:10.1111/j.1096-0031.2005.00075.x [DOI] [PubMed] [Google Scholar]
- Whinnett A, Zimmermann M, Willmott K.R, Herrera N, Mallarino R, Simpson F, Joron M, Lamas G, Mallet J. Strikingly variable divergence times inferred across an Amazonian butterfly ‘suture zone’. Proc. R. Soc. B. 2005;272:2525–2533. doi: 10.1098/rspb.2005.3247. doi:10.1098/rspb.2005.3247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitworth T.L, Dawson R.D, Magalon H, Baudry E. DNA barcoding cannot reliably identify species of the blowfly genus Protocalliphora (Diptera: Calliphoridae) Proc. R. Soc. B. 2007;274:1731–1739. doi: 10.1098/rspb.2007.0062. doi:10.1098/rspb.2007.0062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiemers M, Fiedler K. Does the DNA barcoding gap exist?—a case study in blue butterflies (Lepidoptera: Lycaenidae) Front. Zool. 2007;4:8. doi: 10.1186/1742-9994-4-8. doi:10.1186/1742-9994-4-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will K.W, Rubinoff D. Myth of the molecule: DNA barcodes for species cannot replace morphology for identification and classification. Cladistics. 2004;20:47–55. doi: 10.1111/j.1096-0031.2003.00008.x. doi:10.1111/j.1096-0031.2003.00008.x [DOI] [PubMed] [Google Scholar]
- Will K.W, Mishler B.D, Wheeler Q.D. The perils of DNA barcoding and the need for integrative taxonomy. Syst. Biol. 2005;54:844–851. doi: 10.1080/10635150500354878. doi:10.1080/10635150500354878 [DOI] [PubMed] [Google Scholar]
- Willmott K.R, Freitas A.V.L. Higher-level phylogeny of the Ithomiinae (Lepidoptera: Nymphalidae): classification, patterns of larval hostplant colonization and diversification. Cladistics. 2006;22:297–368. doi: 10.1111/j.1096-0031.2006.00108.x. doi:10.1111/j.1096-0031.2006.00108.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Complete list of samples used with locality data
Primers, PCR and sequencing reaction conditions
Models of substitution selected for each partition (Bayesian analyses)
Barcode trees obtained from (a) maximum likelihood, (b) Bayesian analyses and (c) neighbour-joining
Phylogenetic trees obtained with (a) CoI–CoII sequences and (b) EF1α sequences