Abstract
One of the most important problems in evolutionary biology is to understand how new species are generated in nature. In the past, it was difficult to study this problem because our lifetime is too short to observe the entire process of speciation. In recent years, however, molecular and genomic techniques have been developed for identifying and studying the genes involved in speciation. Using these techniques, many investigators have already obtained new findings. At present, however, the results obtained are complex and quite confusing. We have therefore attempted to understand these findings coherently with a historical perspective and clarify the roles of mutation and natural selection in speciation. We have first indicated that the root of the currently burgeoning field of plant genomics goes back to Hugo de Vries, who proposed the mutation theory of evolution more than a century ago and that he unknowingly found the importance of polyploidy and chromosomal rearrangements in plant speciation. We have then shown that the currently popular Dobzhansky–Muller model of evolution of reproductive isolation is only one of many possible mechanisms. Some of them are Oka’s model of duplicate gene mutations, multiallelic speciation, mutation-rescue model, segregation-distorter gene model, heterochromatin-associated speciation, single-locus model, etc. The occurrence of speciation also depends on the reproductive system, population size, bottleneck effects, and environmental factors, such as temperature and day length. Some authors emphasized the importance of natural selection to speed up speciation, but mutation is crucial in speciation because reproductive barriers cannot be generated without mutations.
Keywords: chromosomal mutation, Dobzhansky–Muller model, hybrid sterility, hybrid inviability, Oka model, polyploidy
Introduction
In the history of evolutionary biology, Hugo de Vries is known as a proponent of the mutation theory of evolution, in which new species are believed to arise by single mutational events (de Vries 1901–1903, 1909, 1910). This theory is based on the breeding experiment he conducted for 13 years with the evening primrose Oenothera lamarckiana and its mutant descendants. In this experiment, he discovered a number of phenotypic variants, which bred true or segregated variant types in addition to the parental type. Because some of these variants were so different from the original O. lamarckiana, he called them elementary species (meaning incipient species) and assigned new species names. In addition, he observed many minor variants, which may be called individual variations or varieties. Because his work was the first experimental study of evolution in a large scale, de Vries’ mutation theory was widely accepted when it was proposed (Allen 1969).
However, his theory was later questioned because O. lamarckiana was apparently a permanent heterozygote for chromosomal complexes and most of de Vries’ mutants were chromosomal rearrangements derived from this unusual genetic form (Davis 1912; Renner 1917; Cleland 1923). The fact that a number of Oenothera species contained these chromosomal complexes was a new discovery in genetics at that time, and therefore, much attention was given to this discovery rather than to de Vries’ mutation theory. For this reason, de Vries’ work is now often regarded as a failed attempt to modify Darwin’s theory of origin of species (Mayr 1980). This view was partially due to the fact that Thomas Morgan and his colleagues found a large number of genic mutations in the 1910’s and 1920’s, and many geneticists used the word mutation to indicate only the genetic changes of single genes. However, at the time of de Vries, the genetic cause of mutations was not known, and he regarded any heritable changes of phenotypic characters as mutations. Later studies showed that at least one of his elementary species was a tetraploid (see below) and it established itself as a new species in self-fertilizing evening primrose. Therefore, he was right in his proposal of mutation theory. In fact, recent genomic data abundantly support his theory of origin of species by chromosomal changes.
In general, however, the formation of new species by chromosomal mutations appears to be rare, and most speciation events are regarded to be due to the establishment of genic sterility or inviability of hybrids between different species. The evolutionary mechanism of genic speciation is complicated, and there are many different ways. In this area too, genomic data are playing important roles in clarifying the mechanisms of speciation. In the case of genic speciation, however, many investigators have emphasized the importance of natural selection rather than mutation (e.g., Presgraves et al. 2003; Coyne and Orr 2004; Maheshwari et al. 2008). Some authors implied that adaptive evolution of incompatibility genes is important in speeding up speciation. In our view, the crucial event of speciation is the development of reproductive barriers between species, and this is accomplished by mutation.
In this review, we first discuss the roles of chromosomal variation in speciation in the light of recent genomic data and then discuss various mechanisms of speciation by means of genic mutation and selection. We will consider both theory and experimental data that support or do not support a particular speciation model. In this article, we will not consider geographical and ecological factors because of space limitation. Our primary purpose is to clarify the roles of mutation and selection in the evolution of reproductive isolation and show that the molecular basis of speciation is more complicated than generally thought at present.
Speciation by Chromosomal Mutations
Formation of New Species by Polyploidization
Soon after de Vries reported various mutants derived from O. lamarckiana, a number of investigators studied their chromosomal numbers and chromosomal segregation at meiosis (Cleland 1972). They found many aneuploids and trisomics, but there was one elementary species (O. gigas), which was bigger and more vigorous than O. lamarckiana. This was later shown to be a tetraploid (Lutz 1907; Gates 1908; Davis 1943). Furthermore, cytogenetic studies of flowering plants (angiosperms) in the mid 20th century showed that 20–40% of the species had experienced polyploidization in their origin (Stebbins 1950; Grant 1981). At this stage, it was clear that the chromosomal mutation called polyploidization is an important mechanism of creating new species in angiosperms. As is well known, polyploid plants establish a sterility barrier from their parental species immediately after their occurrence because the hybrids between them have an abnormal segregation of chromosomes at meiosis and, consequently, they are sterile. Yet, many plant geneticists did not realize that polyploidy was important in plant evolution. For example, Stebbins (1966, p. 129) stated “the large amount of gene duplication dilutes the effects of mutations and gene combinations to such an extent that polyploids have great difficulty evolving truly new adaptive gene complexes.”
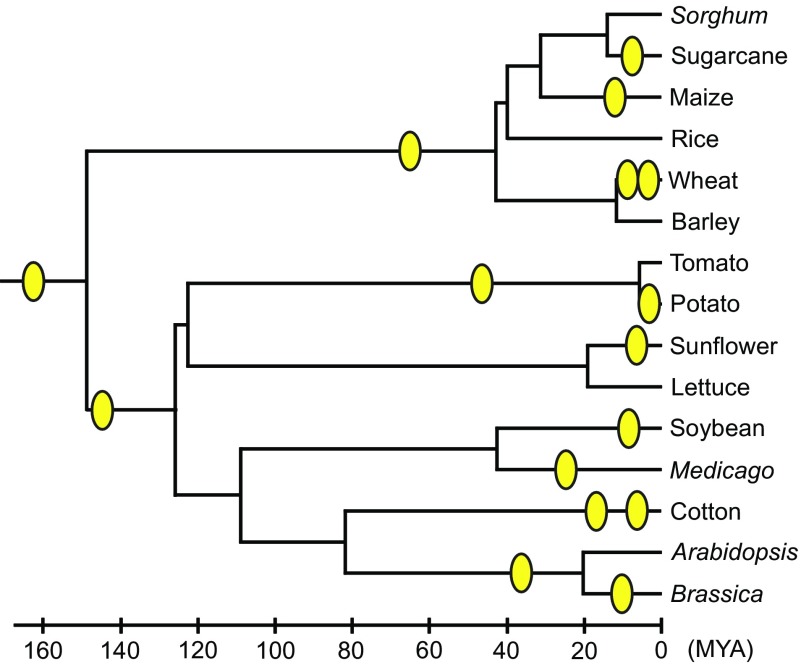
In recent years, our knowledge of polyploid evolution expanded enormously because of the availability of genomic sequences of many different organisms. Statistical analyses of these sequences have shown that polyploidization or genome duplication has occurred quite often particularly in flowering plants. Doyle et al. (2008) state that the genomes of flowering plants are fundamentally polyploid and most species in plants have experienced polyploidization far more frequently than previously suspected. Adams and Wendel (2005) and De Bodt et al. (2005) believe that angiosperms underwent two genome duplication events in the early stage of evolution (fig. 1). This indicates that de Vries’ view of species formation by single mutational events is valid, though the extent of chromosomal variation is not necessarily as high as in Oenothera species. Polyploid species are also abundant in ferns (Grant 1981; Wood et al. 2009). They are also known to exist in yeasts (Wolfe and Shields 1997; Kellis et al. 2004) and some insect species (Otto and Whitton 2000).
FIG. 1.—
Inferred polyploidization events during the evolution of angiosperms. Circles indicate suspected genome duplication events. Approximate time scale is shown below the tree. Modified from Adams and Wendel (2005).
In animals, genome duplication occurs much less frequently than in plants, apparently because sex is often determined by the XY or the ZW chromosomal system in animals and polyploidization would disturb this sex determination (Muller 1925). However, comparison of genome sizes of different groups of animals suggests that polyploidization has occurred quite frequently before the sex determination evolved (Nei 1969). In fact, Ohno (1970, 1998) proposed that two rounds of genome duplication occurred in the early stage of vertebrate evolution. Genome duplications have also been reported in Xenopus (Hirsch et al. 2002) and teleost fish (Jaillon et al. 2004). Therefore, polyploidization may have been an important mechanism of speciation in early stages of animal evolution.
Changes of Genomic Structures and Speciation
We have seen that genome duplication is an important mechanism of speciation. Genome duplication occurs when autotetraploids are formed by duplication of the genome of an organism or when allotetraploids are formed by duplication of the genome of a hybrid between two different species. In either case, the new polyploid species exhibits a sterility barrier from the parental species. Therefore, polyploidization establishes a new species as proposed by de Vries (1901–1903).
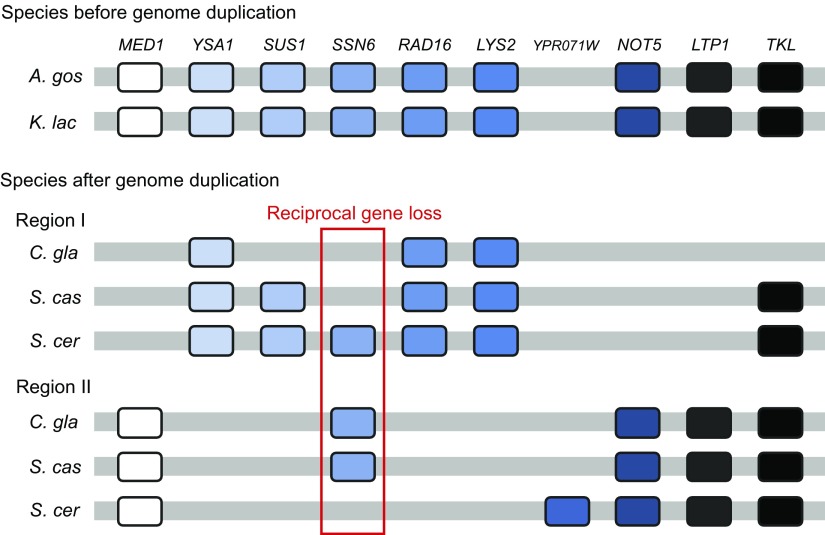
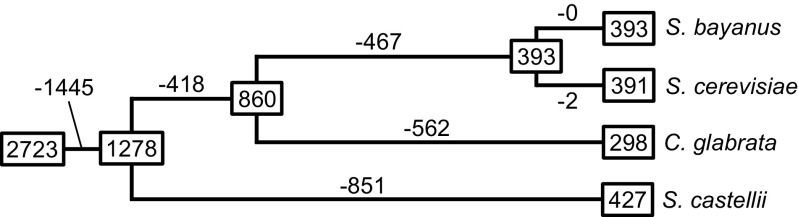
However, it was recently discovered that the number of genes in polyploid species does not necessarily increase in proportion to the number of genome duplications (Wendel 2000; Adams and Wendel 2005; Doyle et al. 2008). Some chromosomes or genes are often lost after polyploidization, and therefore, the new species established may not have twice the number of genes of the parental species (fig. 2). The loss of genes is usually species specific or gene family specific (Rensing et al. 2008; Flagel and Wendel 2009). At the same time, the number of gene copies in some gene family may increase. If this type of increase and decrease of gene number occurs, this process may generate a situation which may not be easily distinguishable from the case of evolution by segmental genomic duplication and deletion. If this is the case, de Vries’ mutation theory, which encompasses any type of hereditary mutations including chromosomal rearrangements, would not be as unrealistic as generally thought. In other words, tetraploids, aneuploids, or trisomics, which de Vries identified as varieties or elemental species, may become new species. In fact, Scannell et al. (2006) showed that an ancestral species of the yeast Saccharomyces cerevisiae apparently experienced polyploidization and then generated at least four well-established species with a reduced number of genes (fig. 3).
FIG. 2.—
Gene order relationships in the region around Saccharomyces cerevisiae SSN6 and its homologs in the species before and after genome duplication. Each ortholog is shown by a different color. Gene names are given at the top in italics. Reciprocal gene loss shown by a red box supports the Oka model of speciation. Abbreviations are as follows: A. gos, Ashbya gossypii; K. lac, Kluyveromyces lactis; C. gla, Candida glabrata; S. cas, S. castellii; and S. cer, S. cerevisiae. Modified from Scannell et al. (2006).
FIG. 3.—
Loss of duplicate genes after genome duplication in four species of yeasts. The numbers in squares represent the numbers of loci, which were derived by genome duplication in the ancestral species and have been retained in the genome. Numbers (–) on branches indicate the numbers of loci, in which one of the duplicate genes was lost. In total, 2,723 duplicate loci were analyzed. Modified from Scannell et al. (2006).
Chromosomal Rearrangements and Speciation
As mentioned above, de Vries did not know the chromosomal structure of O. lamarckiana and simply compiled various forms of morphological mutations. However, it is interesting to note that O. lamarckiana apparently had several sets of reciprocal translocations of chromosomes (Cleland 1923). It was soon realized that this type of plants generates gametes with balanced and unbalanced sets of chromosomes and only those with balanced sets are fertile. It was also noted that individuals with different sets of balanced chromosomes will be separated by reproductive barriers because the hybrids between them will be partially or completely sterile. Similar situations are known to occur when telomeric inversions or other chromosomal rearrangements are generated and recombination occurs (White 1969; Brown and O'Neill 2010).
However, population geneticists such as Wright (1941) showed that the probability of fixation of these chromosomal rearrangements is so low that they would not be easily established in the population unless population size is very small (say less than 10). For this reason, the idea that new species are formed by chromosomal rearrangements was almost abandoned. In selfing plants like O. lamarckiana, however, the chance of fixation of new chromosomal rearrangements would not be very small because the effective population size can be very small. This suggests that some of the elementary species de Vries discovered in his experimental farm might have been reproductively isolated from others by this mechanism even if they were not tetraploids. It should also be noted that chromosomal rearrangements can be fixed even in a randomly mating population if it goes through bottlenecks multiple times.
In fact, recent studies of speciation suggest that this form of speciation is quite common in plants (Rieseberg 2001; Badaeva et al. 2007; Rieseberg and Willis 2007). Plant populations are usually sedentary and often reproduce asexually or by selfing. These reproductive systems enhance the chance of fixation of chromosomal rearrangements, and therefore, the speciation by this process should be reconsidered. This type of speciation by chromosomal rearrangements is also known to occur in yeasts and mammals (Delneri et al. 2003; Brown and O'Neill 2010) (table 1). Of course, de Vries (1901–1903) did not have any idea about chromosomal variation, but his study of morphological mutations stimulated other workers to study the chromosomal mutations and their importance in speciation. Unfortunately, this type of speciation is still underappreciated in the current literature.
Table 1.
Molecular Studies of Speciation Genes and a Few Related Examples
| Gene | Gene Function | Outcome | Species | Referencee |
| Duplicate gene mutations (Oka model) | ||||
| DPL1/DPL2 | Pollen germination | F1 pollen sterility | Rice | Mizuta et al. (2010) |
| RPL27/RPL27 | Mitochondrial ribosomal protein | F1 pollen sterility | Rice | Yamagata et al. (2010) |
| HPA1/HPA2 | Histidinol-phosphate amino-transferase | F1 inviability | Arabidopsis | Bikard et al. (2009) |
| Multiple genesa | Gene losses | New species | Yeast | Scannell et al. (2006) |
| Incompatibility genes (DM model) | ||||
| SaF/SaM | E3 ligase/F-box protein | F1 pollen sterility | Rice | Long et al. (2008) |
| MRS1/COX1 (mt gene) | RNA binding/cytochrome oxidase | F2 sterility | Yeast | Chou et al. (2010) |
| AEP2/OLI1 (mt gene) | RNA binding/ATP synthase | F2 sterility | Yeast | Lee et al. (2008) |
| Incompatibility genes (DM model): possible | ||||
| Nup96/genes on Xc | Nucleoporin/unknown | F1 male inviability | Drosophila | Presgraves et al. (2003) |
| Nup160/genes on Xc | Nucleoporin/unknown | F1 male inviability | Drosophila | Tang and Presgraves (2009) |
| Hmr/Lhrb | DNA binding/protein–protein binding | F1 male inviability | Drosophila | Brideau et al. (2006) |
| Ovd/unknownc | DNA-binding/unknown | F1 male sterility | Drosophila | Phadnis and Orr (2009) |
| tmyc/broadiec | Unknown/unknown | F1 male sterility | Drosophila | Tao et al. (2001) |
| zeel-1/peel-1c | Ubiquitin ligase/unknown | F2 inviability | Nematode | Seidel et al. (2008) |
| CKI1/NBS-LRR(s) | Casein kinase/pathogen detection | F2 sterility | Rice | Yamamoto et al. (2010) |
| DM1 (NBS-LRR)/DM2(NBS-LRRs?)c | Toll interleukin receptor (TIR)/TIR | F1 necrosis | Arabidopsis | Bomblies et al. (2007) |
| AIM22/mt gene(s)c | Lipoate-protein ligase/unknown | F2 sterility | Yeast | Chou et al. (2010) |
| Multiallelic complementary genes model: suggestive | ||||
| Lysin/VERL | Envelope dissolution protein/envelope receptor | No fertilization | Abalone | Lyon and Vacquier (1999) |
| Bindin/EBR1 | Envelope dissolution protein/envelope receptor | No fertilization | Sea urchin | Kamei and Glabe (2003) |
| ADAM2/ITGA9-ITGB7 (α9β7) | Metalloprotease/transmembrane protein | No fertilization | Mammal | Desiderio et al. (2010) |
| ZP3/unknownc | Egg glycoprotein/sperm ZP3-receptor | No fertilization | Mammal | Evans and Florman (2002) |
| Izumo/CD9 | Immunoglobulin/membrane protein | No fertilization | Mouse | Inoue et al. (2005) |
| Mutation-rescue model: possible | ||||
| Xmrk/Rc | Melanoma receptor tyrosine kinase/unknown | F2 inviability | Platyfish | Schartl (2008) |
| Chimeric mt genec/PPR(s)c | Transcript modification/NADH dehydrogenase | F2 anther sterility | Monkeyflower | Barr and Fishman (2010) |
| Chimeric mtgenec/PPR | Unknown/RNA binding | F2 pollen sterility | Petunia | Bentolila et al. (2002) |
| Atp6 (mt gene)/PPRb | ATP synthase/RNA binding? | F2 pollen sterility | Rice | Kazama and Toriyama (2003) |
| Mt gene(s)c/RMS | Unknown/ACPS-like protein | F2 pollen sterility | Rice | Fujii and Toriyama (2009) |
| Segregation distortion model | ||||
| Ovd/unknownc | DNA binding/unknown | Sex ratio distortion (SRD) | Drosophila | Phadnis and Orr (2009) |
| Dox/Nmy | Unknown/noncoding siRNA (?) | SRD/suppression of SRD | Drosophila | Tao et al. (2007) |
| Unknownc/Tmyc | Unknown/unknown | SRD/suppression of SRD | Drosophila | Tao et al. (2001) |
| Heterochromatin-associated speciation: suggestive | ||||
| Zhr (359bp repeats on X)/unknownc | Satellite DNA/maternal cytoplasm | F1 female inviability | Drosophila | Ferree and Barbash (2009) |
| OdsH/heterochromatin of Y | DNA binding/unknown | F1 male sterility | Drosophila | Bayes and Malik (2009) |
| Lhr/HP1 | Protein–protein binding/heterochromatin protein | F1 male inviability | Drosophila | Brideau et al. (2006) |
| Prdm9/satellite DNA? | Histone H3 methyltransferase/unknown | F1 male sterility | Mouse | Oliver et al. (2009) |
| Single-locus mutations | ||||
| S5 | Aspartic protease | F1 embryo-sac sterility | Rice | Chen et al. (2008) |
| FLC1 | MADS-box transcription factor | Flowering time change | Cabbage | Yuan et al. (2009) |
| AN2 | Transcription factor | Pollinator change | Petunia | Hoballah et al. (2007) |
| F3’h | Flavonoid hydroxylase | Pollinator change | Morning glory | Des Marais and Rausher (2010) |
| Style2.1 | Transcription factor | Allogamy to autogamy | Tomato | Chen et al. (2007) |
| Gene and chromosomal translocation | ||||
| JYAlpha | Adenosine triphosphatase | F2 sterility | Drosophila | Masly et al. (2006) |
| Chromosomal translocation | — | F1 sterility | Yeast | Delneri et al. (2003) |
| Chromosomal translocationd | — | F1 sterility or inviability | Plant | Rieseberg (2001) |
This study identified hundreds of reciprocal duplicate gene losses in yeasts.
Both genes have been identified, but molecular interaction remains unclear.
The responsible genes has not really been identified.
This study reviewed the models of chromosomal rearrangements and showed several examples.
Only one paper which seems to be most relevant is listed due to space limitation. See also the references therein.
Evolution of Reproductive Isolation by Genic Mutation
According to the biological species concept (Dobzhansky 1937; Mayr 1963), a group of individuals is called a species when they are isolated from other groups of individuals by premating or postmating isolation mechanisms. It is therefore important to know how the reproductive barrier is generated at the genetic level. In the case of polyploidization, the reproductive barrier is instantly generated in self-fertilizing organisms because the hybrid of a new polyploid and its parental species is generally sterile as mentioned above. However, how does the reproductive barrier arise in the absence of chromosomal rearrangements? There are various genetic models that can explain the evolution of reproductive isolation. Here, we would like to discuss only the genetic models that have been studied empirically at the molecular level. In practice, hybrid sterility or inviability is a complex character and is controlled by a large number of genes (Coyne 1992), and it is difficult to study the effect of all these genes simultaneously. Therefore, most experimentalists extract a small number of major genes and then study the mechanism of reproductive isolation at the molecular level. This approach is certainly important, but we should not forget that it may lead to biased conclusions. Note also that the biological species concept is not always applicable to plants or fungi because these organisms often reproduce by selfing or asexual reproduction and populations are not well definable (Rieseberg and Willis 2007). Initial reproductive isolation is also generally achieved by prezygotic isolation rather than postzygotic isolation. For this reason, speciation occurs more easily in plants and fungi than in animals.
During the last dozen years, many investigators have used the so-called Dobzhansky–Muller (DM) model (see below) as a guideline for conducting experimental studies and interpreting their results. In practice, however, this is only one of the many possible models for the evolution of reproductive isolation as will be mentioned below.
Oka Model of Speciation by Duplicate Gene Mutations
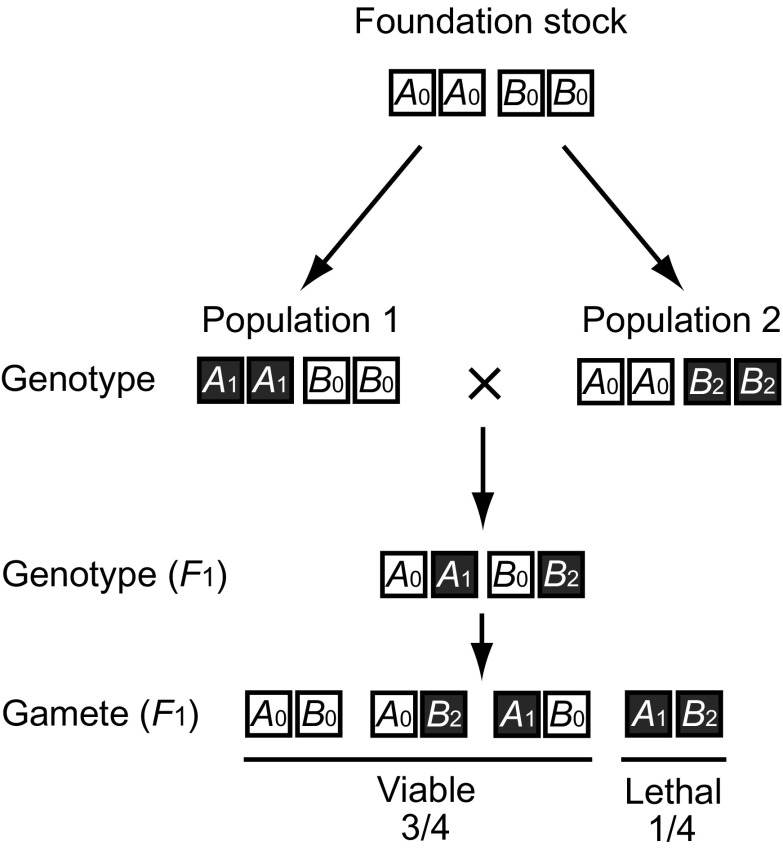
One of the simplest models is Oka’s (1953, 1957, 1974) speciation model by lethal mutations occurring in duplicate genes. Being apparently unaware of Oka’s papers, Werth and Windham (1991) and Lynch and Force (2000) proposed essentially the same model, which is better known in the United States. In this model, the foundation stock is assumed to diverge into two geographically isolated populations (populations 1 and 2) and these populations evolve independently (fig. 4). It is also assumed that the original foundation stock contains two duplicate genes (alleles), A0 and B0, which have redundant functions and that in population 1 allele A0 mutates to a lethal allele, A1, and in population 2 allele B0 mutates to another lethal allele, B2 (see fig. 4). If these evolutionary events occur and populations 1 and 2 are crossed, the hybrid genotype will be A0A1B0B2. This genotype will produce gamete A0B0, A1B0, A0B2, and A1B2 each with a probability of 1/4 if the two loci are unlinked. Therefore, one quarter (A1B2) of them will be sterile.
FIG. 4.—
Oka model of speciation by duplicate gene mutations. A and B are duplicate genes. A0 and B0 are the original normal alleles, and A1 and B2 are lethal mutations.
Drosophila experiments have shown that the rate of lethal mutations per locus is about 10−5 per generation. Therefore, the probability of occurrence of hybrid sterility would not be very small. Note that the rate of fixation of a recessive lethal mutation in one of the two duplicate loci is nearly equal to the mutation rate when the effective sizes of local populations are relatively small (Nei and Roychoudhury 1973). These considerations make it likely for a reproductive barrier to develop in this way.
The extent of gamete sterility obviously increases when there are many such sets of duplicate loci. In fact, when there are n independent sets of duplicate loci that control the formation of sperm or eggs, the expected proportion of sterile gametes will be 1 – (3/4)n, which becomes 0.9 for n = 8 and 0.99 for n = 16. Therefore, this type of gamete sterility is likely to occur in the progeny of a newly generated polyploid, where there are a large number of duplicate genes. In recent years, however, it has been found that even nonpolyploid organisms contain a large number of small-scale duplicate genes (copy number variation) in their genomes (e.g., Redon et al. 2006). Therefore, the Oka model is likely to apply to virtually all species. In practice, the relationship between the number of lethal genes and the extent of male sterility would not be as simple as mentioned above. Lynch and Force (2000) suggested that the functional divergence of duplicate genes may enhance the probability of occurrence of hybrid sterility.
In rice, Oryza sativa, there are two subspecies called Japonica and Indica, which diverged about 400,000 years ago. These subspecies have two duplicate genes DPL1 and DPL2, which encode highly conserved plant-specific small proteins and are highly expressed in mature anther. Mizuta et al. (2010) showed that Japonica carries a functional (DPL1+) and nonfunctional (DPL2−) alleles at the DPL1 and DPL2 loci, respectively. By contrast, Indica has a nonfunctional (DPL1−) and functional (DPL2+) alleles at the two loci. The inactivation of allele DPL1− is caused by a transposon insertion in one of the exons of the gene, whereas the nonfunctionality of DPL2− is due to the A → G mutation at an intron splicing site. Alleles DPL1+, DPL1−, DPL2+, and DPL2− correspond to alleles A0, A1, B0, and B2 in figure 4, respectively, and therefore, the partial sterility of the hybrid between Japonica and Indica can be explained by the Oka model. A similar reproductive isolation caused by duplicate gene mutations has been observed between O. sativa and its related species O. glumaepatula (Yamagata et al. 2010). In this case, the genes involved are the duplicate gene copies S27 and S28 encoding mitochondrial ribosomal protein L27. It was shown that the S27 gene is absent in O. glumaepatula and the S28 gene from O. sativa contains nonfunctional mutations. Therefore, a quarter of hybrid pollen do not have any functional gene, and therefore it causes pollen sterility. Another example of this type of reproductive isolation has been reported in Arabidopsis (Bikard et al. 2009, see table 1).
Actually, using classical genetic techniques, Oka (1953, 1974) had identified a number of hybrid sterility genes, which apparently occurred by duplicate gene mutations. In his time, however, no molecular techniques were available to study the evolutionary changes of genes, and therefore his conclusions have remained as conjectures. In this sense, recent molecular studies have provided solid empirical evidence for his theory. Actually, Oka (1974) was aware of the possibility of ancient polyploidization of rice based on the cytogenetic study by Sakai (1935) and Nandi (1936).
At this point, it should be noted that A1 and B2 in figure 4 represented lethal mutations but they may also represent the loss of the duplicate genes A0 and B0, respectively, because they have the same effect as that of lethal mutations in generating reproductive isolation. In fact, the formation of new species in yeasts after the genome duplication in their ancestral species (figs. 2 and 3) can be explained by the Oka model (Scannell et al. 2006). It should also be noted that most authors who studied the duplicate gene mutation hypothesis mistakenly called it the DM model instead of the Oka model (e.g., Werth and Windham 1991; Lynch and Force 2000; Mizuta et al. 2010). In the Oka model, lethal mutations or gene losses are the causal factors, and there is no need of interaction between A1 and B2. In the DM model, however, A1 and B2 are functional genes and a special form of gene interaction between alleles A1 and B2 is assumed to exist, as will be discussed below. In the DM model, the fixation of alleles A1 and B2 by positive selection is also often assumed.
Some authors are not enthusiastic about the importance of the Oka model of speciation. Coyne and Orr (2004) stated that polyploidization does not occur so often in animal species and this minimizes the importance of this model. As mentioned above, however, recent genomic studies indicate that small-scale gene duplications are abundant, and there is no reason to believe that the Oka model is less important in animals than in plants. Coyne and Orr also stated that the ultimate fate of duplicate genes is to acquire new gene functions rather than nonfunctionality. Actually, this statement is incorrect. Duplicate genes become pseudogenes much more frequently than gain new functions (Lynch and Force 2000; Nei and Rooney 2005). For these reasons, the Oka model may play an important role in speciation in both plants and animals.
DM Model of Evolution of Reproductive Isolation
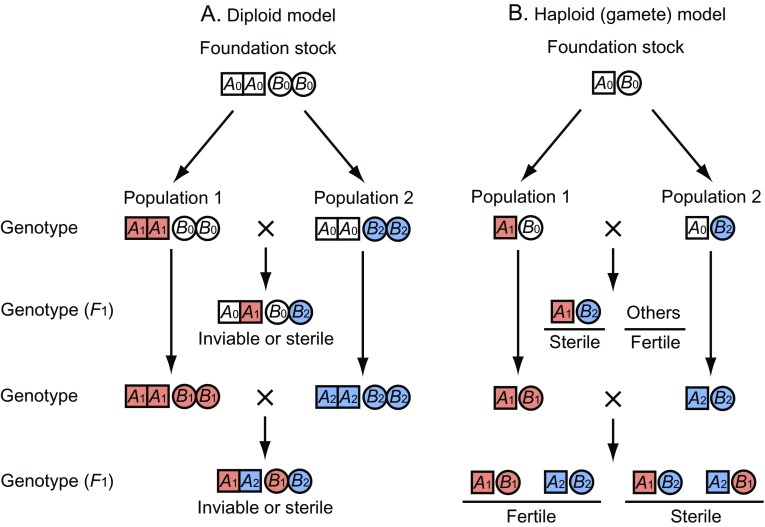
In the Oka model of speciation, it is necessary to have duplicate genes. However, reproductive isolation may be developed without duplicate genes if there are two or more genes that interact with each other negatively when they are brought together in hybrids. One of such models is the so-called DM model (Dobzhansky 1937; Muller 1940, 1942). The essence of this model is presented in figure 5A. In this figure, two loci, A and B, are considered, and A0A0B0B0 represents the genotype for these loci in the foundation stock from which populations 1 and 2 were derived. If these two populations are geographically or ecologically isolated, it is possible that A0 mutates to A1 in population 1 and this mutant allele is fixed in the population by natural selection or genetic drift. Genotype A0A0B0B0 may then be replaced by A1A1B0B0 without loss of viability and fertility (fig. 5A). Similarly, B0 may mutate to B2 in population 2 and the mutant allele may be fixed. However, if there is gene interaction such that any combination of mutant genes A1 and B2 in an individual results in inviability or sterility, the hybrids (A0A1B0B2) between the two populations will be inviable or sterile. In figure 5A, we assumed that the foundation stock had genotype A0A0B0B0. Theoretically, however, it is possible to assume that the ancestral genotype is A1A1B1B1 and that this genotype remained unchanged in population 1 but it changed to A2A2B2B2 in population 2.
FIG. 5.—
DM model of evolution of reproductive isolation. A0, A1, and A2 represent alleles at the A locus, whereas B0, B1, and B2 represent alleles at the B locus. (A) Diploid model. (B) Haploid (gamete) model.
Orr (1996) argued that the first person who proposed the DM model is neither Dobzhansky nor Muller but Bateson (1909) and Bateson’s model was identical with that of Dobzhansky and Muller. In our view, this argument is disputable. It is certainly true that Bateson considered a two-locus model of complementary genes to explain hybrid sterility, but he never considered how such a system can evolve. By contrast, Dobzhansky and Muller spelled out the evolutionary process of hybrid sterility genes, albeit very crudely. In evolutionary biology, it is important to understand the process of evolution. For this reason, we will refer to the model as the DM model in this paper. However, Dobzhansky and Muller presented only a verbal argument and never explained why only A1 is fixed in population 1 and B2 is fixed in population 2. Theoretically, the B0 → B1 mutation may also happen in population 1 and the A0 → A2 mutation may occur in population 2 (fig. 5A). How is then only A1 fixed in population 1 and only B2 fixed in population 2? Both Dobzhansky and Muller argued that allele A1 may affect a secondary character through the pleiotropic effect and this effect may confer a selective advantage for A1 over A0 in population 1. Similarly, B2 may have a selective advantage over B0 in population 2 because of the pleiotropic effect.
The first mathematical study of this problem was conducted by Nei (1976). Here, let us present a summary of his results. For simplicity, we consider the haploid model given in figure 5B instead of the diploid model because essentially the same result is obtained by both models. Note also that the haploid model directly applies to sperm or egg fertility. In the haploid model, four possible genotypes may be generated for the two alleles at each of loci A and B, and we assign the fitnesses for the four genotypes as given in table 2. Here, x and y represent the frequencies of alleles A1 and B2, respectively, whereas sA and sB are selective advantages conferred by pleiotropy for alleles A1 and B2, respectively, and t is the selective disadvantage of genotype A1B2, which becomes 1 when the interpopulational hybrids are completely sterile. Note that alleles A0, A1, B0, and B2 are all vitally important in this model. Here, we have assumed no linkage disequilibrium for simplicity.
Table 2.
Fitnesses and Frequencies of the Four Genotypes for the Two Incompatibility Loci in the Haploid Model
| Alleles | A0 | A1 | |
| B0 | Fitness | 1 | 1 + sA |
| Frequency | (1 – x)(1 – y) | x (1 – y) | |
| B2 | Fitness | 1 + sB | 1 – t |
| Frequency | (1 – x)y | xy |
If we use this model, the amounts of changes (Δx and Δy) of allele frequencies x and y per generation are given by
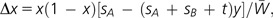 |
(1) |
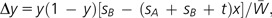 |
(2) |
where 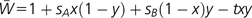 (Nei 1976). Therefore, x increases if y is smaller than
(Nei 1976). Therefore, x increases if y is smaller than 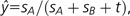 , whereas it decreases if y is greater than
, whereas it decreases if y is greater than 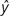 . Similarly, y increases if x is smaller than
. Similarly, y increases if x is smaller than 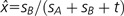 but decreases if x is greater than
but decreases if x is greater than 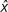 . This means that if mutant allele A1 occurs before B2 and starts to increase in frequency allele A1 tends to be fixed in the population, whereas mutant allele B2 would be fixed if it occurs first and starts to increase before the occurrence of A1. Therefore, selection is exclusive, and in any population, either A1 or B2 may be fixed depending on the allele that starts to increase in frequency earlier than the other. Because the fixation of A1 or B2 occurs at random, the probability that two populations show hybrid sterility or inviability is 1/2. However, if there are many loci controlling reproductive isolations, any pair of populations would eventually develop reproductive isolation.
. This means that if mutant allele A1 occurs before B2 and starts to increase in frequency allele A1 tends to be fixed in the population, whereas mutant allele B2 would be fixed if it occurs first and starts to increase before the occurrence of A1. Therefore, selection is exclusive, and in any population, either A1 or B2 may be fixed depending on the allele that starts to increase in frequency earlier than the other. Because the fixation of A1 or B2 occurs at random, the probability that two populations show hybrid sterility or inviability is 1/2. However, if there are many loci controlling reproductive isolations, any pair of populations would eventually develop reproductive isolation.
One problem here is whether alleles A1 and B2 have selective advantage (sA > 0 and sB > 0) conferred by pleiotropic effects or not. Generally speaking, it is very difficult to identify any character affected by pleiotropic effects of speciation genes A1 and B2, and even if a character is identified, the selection coefficient sA and sB are unlikely to remain constant for the entire process of fixation of alleles A1 and B2. However, even if sA and sB are 0, alleles A0 and B0 may be replaced by A1 and B2 in populations 1 and 2, respectively, by the effect of genetic drift. In this case too, only A1 or B2 must be fixed in a population, and the average replacement time will be 1/v + 2N generations, where v and N are the mutation rate and the effective population size (Nei 1976). Therefore, it will take a long time for alleles A0 and B0 to be replaced by A1 and B2, respectively. Even if A1 and B2 are selected with positive values of sA and sB, the replacement time will not be much shorter because it primarily depends on the mutation rate (Li and Nei 1977).
It should also be noted that the mutation rate v refers only to those mutations that enjoy selective advantage because of the pleiotropic effect within populations but generate strong deleterious effects when they are brought together in hybrid individuals. No one has measured the mutation rate for this type of mutations, but the rate must be very low because only special mutations would be able to produce such dual gene effects. We know that the DM model is currently very popular (e.g., Coyne and Orr 2004), but there are only a small number of experimental data that support the model in the strict sense (table 1). Orr’s (1995) paper is often cited as the theoretical justification of the model. In reality, he assumed the validity of the model from the beginning and simply studied the possibility of continuous accumulation of incompatibility genes. He conceived that reproductive isolation is developed by positive Darwinian selection caused by their pleiotropic effects. This is in contrast to the Oka model, where reproductive isolation is assumed to occur due to deleterious mutations in duplicate genes.
Let us now examine some recent experimental data that have been regarded to support the DM model. The first data set we consider is that of Presgraves et al.’s (2003) paper, in which the evolutionary change of a nuclear pore protein (nucleoporin), Nup96, has been studied in D. melanogaster and D. simulans. Nuclear pores are large protein complexes that cross the nuclear envelope and allow the transport of water-soluble molecules such as RNAs, DNA polymerases, and carbohydrates between the nucleus and the cytoplasm. This nuclear pore is composed of a large molecular structure called the nuclear pore complex, which contains about 30 different protein components, each with multiple copies (Presgraves and Stephan 2007). One of the proteins is the nucleoporin Nup96, and Presgraves et al. (2003) showed that this protein is involved in causing hybrid male inviability between the two Drosophila species. This hybrid inviability occurred only when the D. simulans Nup96 gene is associated with the D. melanogaster X chromosome. They therefore assumed that the hybrid inviability occurs when the D. simulans Nup96 gene negatively interacts with one or more genes of the D. melanogaster X chromosome. Furthermore, McDonald and Kreitman (MK)’s (1991) test of neutrality suggested that the Nup96 gene evolved by positive selection after divergence of the two species. They then concluded that their observations support the DM model of speciation, and the hybrid inviability is a consequence of adaptive evolution at the Nup96 locus. A similar study was conducted by Tang and Presgraves (2009), who identified another nucleoporin gene, Nup160, involved in the hybrid male inviability between the two Drosophila species. This gene in D. simulans was inferred to interact negatively with the D. melanogaster X chromosome genes as well as with the D. simulans Nup96 gene.
However, there are a few problems with their conclusions. First, they have not really identified the D. melanogaster X chromosome genes that are supposed to interact with the D. simulans Nup96 or Nup160. This identification is critical because otherwise we do not know how the interaction between the two genes leads to hybrid male inviability. Theoretically, the X chromosome genes may not be protein-coding genes but the heterochromatin that is often involved in hybrid inviability (e.g., Ferree and Barbash 2009, see below). Second, Presgraves and his colleagues obtained a signature of positive selection for the increase in frequency of Nup96 and Nup160 by using the MK test. However, the MK test depends on a number of simplifying assumptions, and it may give erroneous conclusions when these assumptions are not satisfied (Nei et al. 2010).
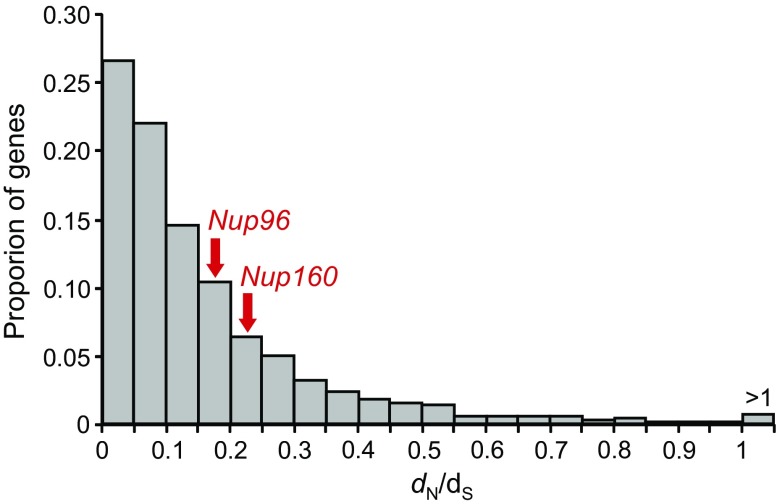
We therefore examined the extent of positive selection by using the modified Nei-Gojobori method (Zhang et al. 1998) of DNA sequence comparison. In this method, the ratio of the number of nonsynonymous nucleotide substitutions per site (dN) to that of synonymous nucleotide substitutions (dS) is computed, and the extent of positive selection is measured by dN /dS. If dN /dS is greater than 1, positive selection is suggested, whereas dN /dS < 1 indicates negative or purifying selection. In addition, this analysis tells us whether the nucleoporin genes evolve more rapidly than other genes as often claimed by Presgraves and others. When we computed this ratio for Nup96 and Nup160 using the D. melanogaster and D. simulans sequences, we obtained only 0.19 and 0.25, respectively. These values indicate that the two genes have not evolved particularly fast among the 5,314 genes examined (0.16 on average) and are likely under purifying selection (fig. 6).
FIG. 6.—
Distribution of dN/dS ratio between the Drosophila melanogaster and D. simulans genes. A total of 5,314 protein-coding genes having one-to-one orthologs among 12 Drosophila species were used. The dN and dS values were computed by the modified Nei-Gojobori method (Zhang et al. 1998) with a transition/transversion ratio of 2.
However, what is important here is not to know whether positive selection has occurred for the new alleles but to understand how these genes generate hybrid inviability. Some may argue that positive selection is important because it would speed up the speciation process. In reality, there is no need for any organism to have fast speciation. Reproductive isolation occurs merely as a consequence of a more general evolutionary change of morphological or physiological characters, and therefore, it must be a passive process, as was emphasized by Darwin (1859, p. 245).
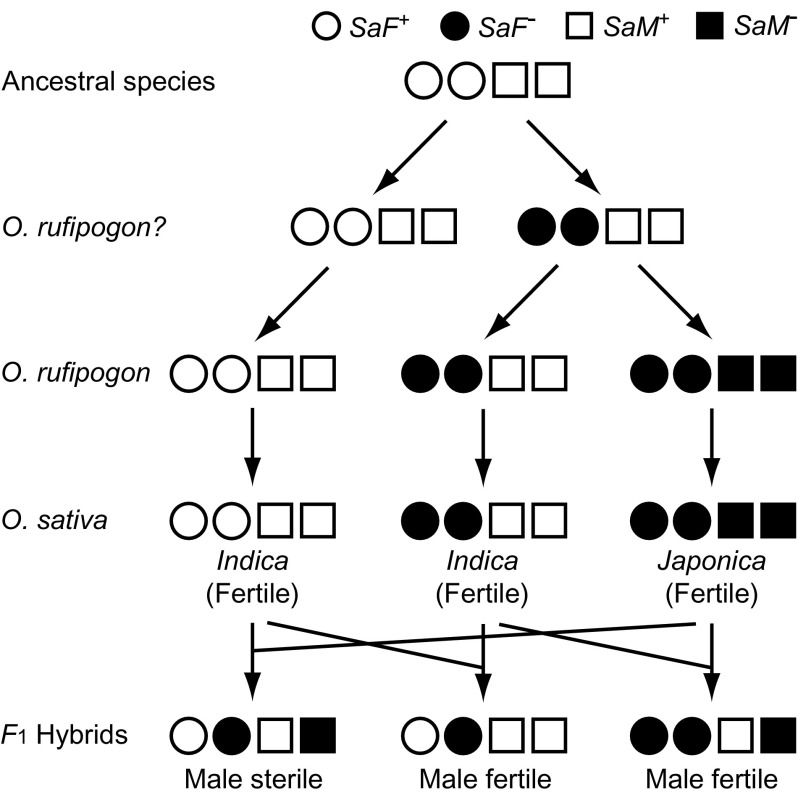
However, there are a few data sets that apparently support the DM model. Long et al. (2008) discovered that a pair of closely linked loci SaF and SaM in rice contain different alleles in subspecies Japonica (SaF− and SaM−) and Indica (SaF+ and SaM+), and the males of their hybrids are sterile. Gene SaF encodes an F-box protein involved in protein degradation, whereas SaM produces a small ubiquitin-like modifier E3 ligase-like protein. The protein encoded by SaF is 476 amino acid long, and there is only one amino acid difference between alleles SaF+ and SaF−. By contrast, SaM+ and SaM− encode proteins with 257 amino acids and 217 amino acids, respectively, the latter being a truncated protein. Alleles SaF+ and SaM+ in Indica are considered as the ancestral genes, and SaF− and SaM− are regarded as mutants generated in the process of evolution of Japonica (fig. 7). The haplotype SaF−;SaM+ that is found in Indica could be the ancestor of the haplotype SaF−;SaM− in Japonica. It hybridizes both with Indica and Japonica without any problem (fig. 7). If this is the case, this haplotype represents an intermediate stage in the process of evolution of SaF−;SaM− in Japonica. These evolutionary changes of SaF+;Sam+ to SaF−;SaM− are consistent with the DM model. However, the molecular basis of the gene interaction to generate the hybrid sterility is still unknown.
FIG. 7.—
Male sterility caused by different combinations of alleles at the SaF and SaM loci in rice. Modified from Long et al. (2008).
Another data set that supports the DM model is that of Chou et al. (2010), who studied a pair of genes causing the F2 sterility between the yeasts S. cerevisiae and S. paradoxus. The genes studied are the nuclear-encoded mitochondrial RNA splicing gene (MRS1) and the mitochondria-encoded cytochrome oxidase 1 gene (COX1). In S. paradoxus, the introns of COX1 are properly spliced out by its own MRS1. In S. cerevisiae, however, one (M1) of the introns has been lost from COX1. This mutation is most likely to have been neutral because the loss of intron did not affect the gene or protein function of COX1. Subsequently, the MRS1 lost its splicing function, which also seems to have been neutral. However, the hybrids between the two species show sterility because the MRS1 protein in S. cerevisiae cannot splice the M1 intron of COX1 from S. paradoxus. This scheme of evolution of reproductive isolation is consistent with the DM model, and in this case, it is likely that the evolutionary changes of the genes have occurred primarily by mutation and genetic drift. A similar evolutionary change has been reported to explain the hybrid sterility caused by the AEP2 and OLI1 genes between S. cerevisiae and S. bayanus (Lee et al. 2008, see table 1).
There are many other papers that have claimed to support the DM model (Coyne and Orr 2004; Wu and Ting 2004, see table 1). However, close examination of the papers indicates that the authors often misunderstood the concept of the model or the demonstration is incomplete. Therefore, more careful studies are necessary about the genes reported in these papers (table 1). Note that even when some genes completely follow the DM model, they may have nothing to do with speciation but they just became incompatible simply as a by-product of species divergence.
Multiallelic Complementary Genes Model
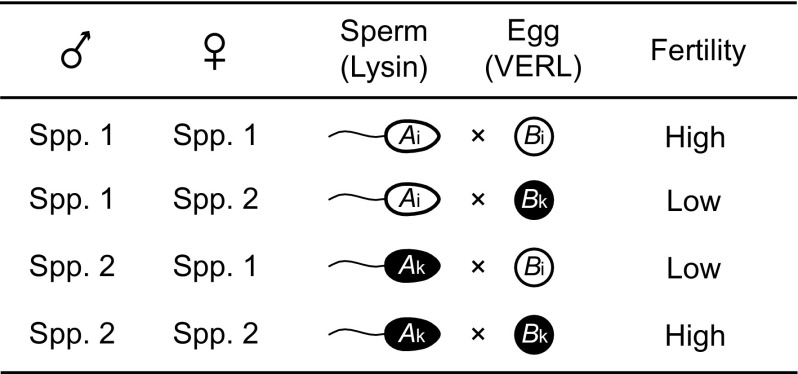
Nei et al. (1983) proposed an extended version of the DM model to explain the species-specific gene compatibility and other reproductive isolation. As a concrete example, let us consider the evolutionary changes of sperm protein lysin and its egg receptor VERL in abalone species. In abalone, the eggs are enclosed by a vitelline envelope, and sperm must penetrate this envelope to fertilize the egg (Shaw et al. 1995). The receptor VERL for lysin is a long acidic glycoprotein composed of 22 tandem repeats of 153 amino acids and about 40 molecules of lysin bind to one molecule of VERL (Galindo et al. 2003). The interaction between lysin and VERL is species-specific, and therefore, this pair of proteins apparently controls species-specific mating. Figure 8 shows a genetic model explaining the species specificity between the lysin and VERL genes. Within a species (species 1 or 2), the lysin and VERL genes are compatible, so that mating occurs freely. However, if species 1 and 2 are hybridized, lysin and VERL are incompatible, and therefore, the fertilization is blocked. This guarantees the species-specific mating when the two species are mixed.
FIG. 8.—
A model of species specificity of gamete recognition between lysin and VERL in abalone. Modified from Nei and Zhang (1998).
However, it is not very simple to produce the gene for species 2 from that for species 1 or those for species 1 and 2 from their common ancestral genes by a single mutation at the lysin and VERL loci because a mutation (Ai → Ak) at the lysin locus makes the lysin gene incompatible with the wild-type allele (Bi) at the VERL locus. A mutation (Bi → Bk) at the VERL locus also results in the incompatibility with the wild-type allele (Ai) at the lysin locus. Therefore, these mutations would not increase in frequency in the population. Of course, if mutations Ai → Ak and Bi → Bk occur simultaneously, lysin Ak and VERL Bk may become compatible. However, the chance that these mutants meet with each other in a large population would be very small.
For this reason, Nei et al. (1983) and Nei and Zhang (1998) proposed that the evolutionary change of allele Ai (or Bi) to Ak (or Bk) occurs through intermediate alleles and that closely related alleles have similar functions and therefore compatible. For example, Ai may mutate first to Aj and then to Ak, whereas Bi may mutate to Bj and then to Bk. Suppose Ai is compatible with Bi and Bj but not with Bk and Bi is compatible with Ai and Aj but not with Ak. If Aj is compatible with Bi, Bj, and Bk and if Bj is compatible with Ai, Aj, and Ak, then it is possible to generate the species-specific combination of alleles at the lysin and VERL loci in each species (i and k, respectively) by means of mutation and genetic drift without positive selection that is often assumed by the DM model.
There are several other examples of ligand and receptor gene incompatibilities involved in fertilization or reproduction. For example, sea urchin protein bindin mediates the fertilization of a sperm to an egg. The receptor of bindin is called EBR1, and its interaction with bindin is species specific (Kamei and Glabe 2003). In mammals, a protein called ADAM2 (or fertilin β) plays a role of sperm ligand for the egg plasma membrane receptors, integrins (Evans and Florman 2002; Desiderio et al. 2010), and the interactions between these proteins seem to be species specific. The protein–protein interaction in various biochemical processes required for development and physiology is also often complementary. Similarly, the control of expression of protein-coding genes by cis-regulatory elements is complementary by nature.
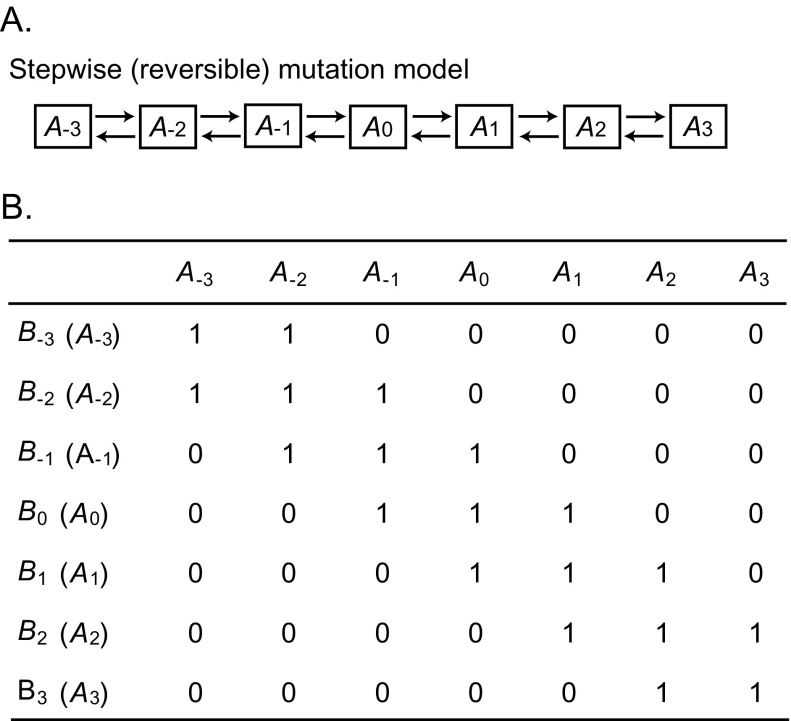
Nei et al. (1983) developed several models of evolution of reproductive isolation by means of multiallelic complementarity genes. They considered both one-locus and two-locus models. Mutation was assumed to occur following either the stepwise or the infinite-allele model (Kimura 1983), and the fitness of a genotype was assumed to be either 1 or 0 depending on the mutation model and the genotype generated (fig. 9). Premating and postmating isolations were also considered. Their conclusions are summarized in the following way. 1) The single-locus model generates speciation more quickly than the two-locus model. 2) The infinite-allele model generates speciation more quickly than the step-wise mutation model. 3) With the epistatic gene model used (fig. 9), the evolution of reproductive isolation occurs more rapidly in small populations than in large populations. 4) Generally speaking, the time to the occurrence of speciation is very long and is roughly proportional to the inverse of mutation rate.
FIG. 9.—
Stepwise mutation model for hybrid sterility (or inviability) genes. (A) In the stepwise mutation model, the forward and backward mutation may occur. (B) The fertilities for various haplotypes for loci A and B (two-locus model) and genotypes for locus A (one-locus model) are given by 0 (infertile) or 1 (fertile). Distantly related haplotypes or genotypes are infertile.
However, these results are model dependent, and we cannot apply the results to natural populations without qualifications. For example, the single-locus model, which will be considered below, may be applicable only to certain characters such as the flowering time in plants and developmental time in animals. At present, we also do not know which of the stepwise and infinite-allele models is more realistic than the other, though we believe that the latter model is more realistic because reproductive isolation is controlled by a large number of genes controlling different phenotypic characters. Our results imply that speciation occur more rapidly through bottlenecks. This conclusion is in agreement with Mayr’s (1963) theory of founder principle, which has been criticized by many authors (e.g., Coyne and Orr 2004). However, Nei et al.’s (1983) study was done by using specific mathematical models of epistatic gene interaction, unlike Mayr’s verbal argument without any genetic model. This would also means that self-fertilizing organisms may undergo more rapid evolution than random mating populations.
In general, however, speciation occurs very slowly, and it takes millions to tens of millions of years for well-established species to be developed (Coyne and Orr 2004, pp. 419–421). This suggests that Nei et al.’s conclusion about speciation time may not be so unrealistic.
Mutation-Rescue Model of Speciation
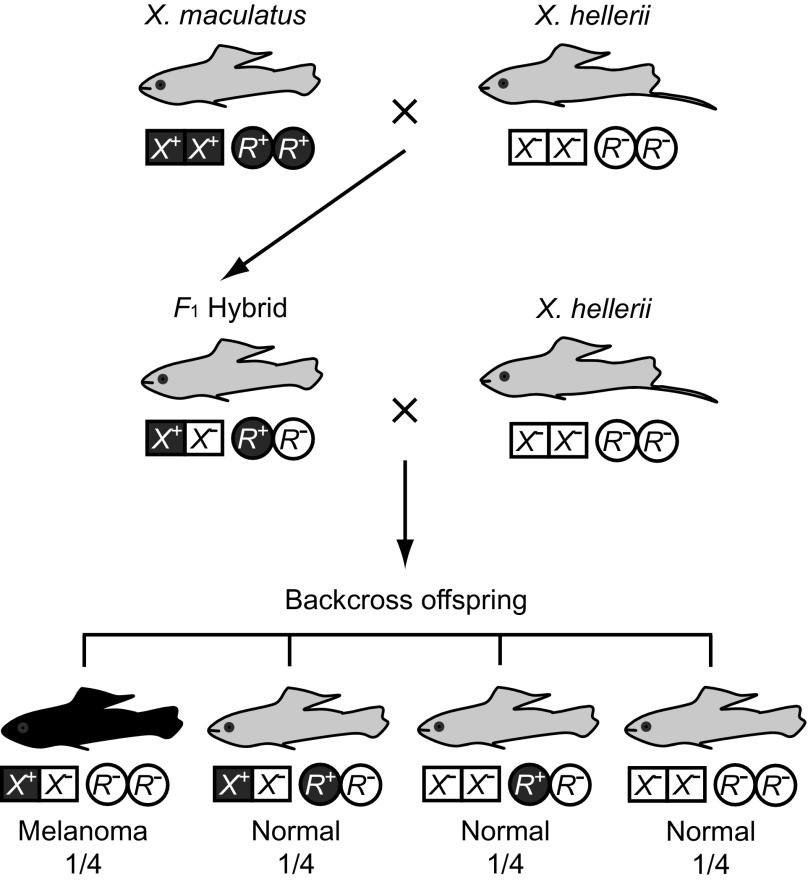
One example of hybrid inviability genes that is often cited as support of the DM model is the melanoma formation genes in the fish species belonging to Xiphophorus (Orr and Presgraves 2000; Orr et al. 2004; Wu and Ting 2004). Hybrids between the species of this genus often produce malignant melanoma. This tumor is developed when a certain type of crossing experiments is conducted, and one of them is presented in figure 10. In this figure, X+ and X− represent the presence and absence of the tumorigenic gene Xmrk, respectively, whereas R+ and R− denote the presence and absence of the regulatory gene (R), respectively.
FIG. 10.—
Typical crossing experiments in Xiphophorus species. X+ and X− represent the presence and absence of the Xmrk gene, respectively. R+ and R− represent the presence and absence of the R gene. F1 hybrids (X+X−, R+R−) express benign melanoma, but they are shown with that of the normal type in this figure. One quarter of backcross offspring will develop melanoma. Modified from Schartl (2008).
Gene Xmrk is a duplicate copy of an epidermal growth factor receptor gene and has acquired the ability of generating melanoma. However, this gene is expressed only when the suppressor gene R is absent in the genome. Therefore, the species with genes Xmrk and R (X. maculatus in fig. 10) does not show melanoma. Similarly, the species with no Xmrk and R genes (X. helleri) is free of melanoma. However, the genotype (X+X−; R−R−) with one Xmrk gene but no R gene in the backcross offspring develops the melanoma cancer (Schartl 2008). If this genotype shows a low fitness, reproductive isolation may be developed.
However, the genetic basis of this hybrid inviability is very different from that of the DM model. Note that genotypes (X+X−; R+R−) and (X−X−; R+R−) are normal unlike the case of the DM model. In the present case, the mutant gene Xmrk is inherently deleterious (fig. 10), but within each species, its deleterious effect is diminished by the R gene. This means that Xmrk (X+) is a deleterious mutation and R+ rescues the deleterious effect of the mutation and therefore the individuals with genes X+ and X+ become normal. We therefore call this hybrid incapacity system the mutation-rescue model. Noting that the deleterious effect of gene X+ on fitness is not serious in nature, Schartl (2008) questions the relevance of this system to speciation. However, we would like to use this system as an example of the mutation-rescue model with the anticipation that similar examples will probably be found in the future.
At present, there are several data sets that can be explained by the mutation-rescue model, but they are not as straightforward as in the above example. In plants, there are many examples of cytoplasmic male sterility (CMS) (e.g., Zeh JA and Zeh DW 2005; Chase 2007 for reviews), and the hybrid sterility caused by CMS is believed to occur when some specific cytotypes, probably specific mitochondrial genes, interact with nuclear genes (cytonuclear gene interaction). Interestingly, some mutant alleles of the nuclear genes can rescue the male fertility, although the molecular basis of this rescue is not well understood. For example, in the cross of two monkeyflower species, Mimulus guttatus and M. nasutus, Case and Willis (2008) showed that the gene involved in CMS is one of the open reading frames which are cotranscribed with the mitochondrial NADH dehydrogenase gene (NAD6), whereas Barr and Fishman (2010) showed that the nuclear rescue element is the genes belonging to a very large gene family called the pentatricopeptide repeat (PPR) family. The gene copy number of this family has expanded enormously in plants, and Arabidopsis has ∼450 PPR genes scattered all over the genome (Lurin et al. 2004). The function of this gene family is not well understood, but it has been predicted that about a half of the PPR proteins are targeted to mitochondria (Chase 2007), and several authors have reported the PPR genes as rescue genes (table 1). However, how this large gene family interacts with CMS elements to reduce deleterious effects of mitochondrial mutations is a mystery. Furthermore, the evolutionary process of the mitochondrial and nuclear genes has not been examined in these studies, so that it is unclear whether this system really fits the mutation-rescue model. Obviously, more detailed study is necessary. The molecular study of this system seems to be even behind the study of the segregation distorter gene model, which will be discussed below.
Segregation Distorters and Speciation
In diploid organisms, a male heterozygote (Aa) for a locus produces two different types of sperm (A and a) with an equal frequency. However, there are genes that distort the Mendelian segregation ratio in their favor so that their frequency in sperm is much higher than 50% (sometimes nearly 100%). These genes are called segregation distorters (D−). The segregation distortion occurs because the distorter gene destroys a high proportion of chromosomes carrying the opposite allele in the process of spermatogenesis (Hartl 1969; Wu and Hammer 1991; Kusano et al. 2003). The distorter gene is often located on the X chromosome, and therefore, the sex ratio in the offspring is distorted (Presgraves 2008). Because these males produce more sperm with the X chromosome than sperm with the Y, there will be more female offspring than male offspring, and this distorted sex ratio is disadvantageous for the species. Furthermore, distorter genes themselves are often deleterious but their frequencies may increase drastically by segregation distortion.
Interestingly, the expression of D− genes is often suppressed by suppressor genes (S−). Therefore, if a new distorter mutation (D−) occurs in a population, its frequency would initially increase rapidly because of segregation distortion despite its deleterious effects. However, this increase in frequency may be stopped if a new suppressor mutation (S−) arises and suppress the deleterious effect of the D− gene. The D− and S− genes may then be fixed in a species simultaneously. After the fixation of these mutations, there will be no segregation distortion and no deleterious effects of the D− gene (Wu et al. 1988; Frank 1991; Lyttle 1991; Tao et al. 2001; Phadnis and Orr 2009).
However, if this new species with mutant genes D− and S− is crossed with its sibling species with wild-type alleles D+ and S+, the effect of D− may reappear in the F1 hybrids if S− is not dominant over S+ or in the F2 hybrids if D− is recombined with S+ and genotypes D−D−S+S+ or D−D+S+S+ are produced (Frank 1991; Hurst and Pomiankowski 1991; Tao et al. 2001). If these events reduce the fitness of hybrid individuals, it will constitute a new way of generating reproductive barriers between the two species. The genetic nature of suppressor genes is not well known. However, in the case of the Segregation Distorter (D−) haplotype first reported by Sandler et al. (1959) in D. melanogaster, the S locus (Responder) consists of a large number of about 120-bp repeats, and it was shown that the segregation distortion becomes stronger as the number of repeats increases and that when the suppressor locus contains a small number of repeats no segregation distortion occurs (Wu et al. 1988).
D− genes are present on autosomal chromosomes as well as on the Y, but they seem to be less frequent than those of the X chromosome, as discussed by Frank (1991) and Jaenike (2001). This observation provides an explanation for Haldane’s (1922) rule, which states that when two species or subspecies are intercrossed the heterogametic sex with XY or ZW chromosomes are sterile or inviable more often than the homogametic sex with XX or ZZ chromosomes (Frank 1991; Hurst and Pomiankowski 1991).
Dozens of distorter genes have been reported in insects, mammals, and plants, though the molecular basis of the distortion is not well understood (Jaenike 2001). In D. simulans, at least three D− loci have been identified and each S− locus corresponds to each D− locus (Presgraves 2008). In these cases, however, the molecular mechanism of the function of S− loci is unclear.
Heterochromatin-Associated Hybrid Incapacity
A number of investigators (e.g., Henikoff and Malik 2002; Brideau et al. 2006; Bayes and Malik 2009) have reported that repeat DNA elements in the heterochromatin regions of genomes are often associated with hybrid sterility or inviability. One of the interesting observations is the hybrid sterility caused by the zygote hybrid rescue (Zhr) locus in Drosophila. When D. simulans females are crossed with D. melanogaster males, hybrid females die in early embryogenesis. However, a mutant allele (Zhr1) of Zhr is known to rescue the female viability (Sawamura et al. 1993). Ferree and Barbash (2009) showed that the wild-type allele Zhr represents a region of 359 nucleotide repeats in the D. melanogaster X chromosome that interact with some cytoplasmic factors of D. simulans. The number of 359 nucleotide repeats is small in D. simulans, so that the fertility within this species is high. In D. melanogaster, the number of DNA repeats is high, but this species also shows a high fertility apparently because cytoplasmic factors are different from those of D. simulans. At present, however, the cytoplasmic elements have not been identified, and therefore, the molecular basis of the interaction between the DNA and the cytoplasmic factors remains unclear. Yet, it appears that the number of DNA repeats is species-specific and can change relatively rapidly due to concerted or birth-and-death evolution (Henikoff and Malik 2002). Although little is known about the evolutionary mechanism of cytoplasmic elements, it is possible that both DNA repeat elements and cytoplasmic factors coevolve as in the case of two-locus multiallelic complementary genes model discussed earlier.
Another example is the Odysseus homeobox (OdsH) gene, which causes hybrid male sterility when D. mauritiana females are crossed with D. simulans males (Ting et al. 1998; Sun et al. 2004). In this case, the receptor for this transcription factor gene is not well defined. Recently, however, Bayes and Malik (2009) discovered that the OdsH protein produced from D. mauritiana localizes to the heterochromatic Y chromosome from D. simulans but the OdsH protein from D. simulans does not. They then proposed that the overexpression of the OdsH protein in the Y heterochromatin is responsible for the male sterility. Again, however, the molecular mechanism of the interaction remains unclear. A similar mechanism appears to be operating with the Prdm9 gene in mice (Oliver et al. 2009, see table 1).
Single-Locus Speciation
It is often stated that reproductive isolation cannot be achieved by single-locus mutations because a deleterious mutation occurring at a locus cannot be fixed in any population. However, if multiallelic mutations occur at a locus and they are compatible with one another when they are closely related but mutant alleles become incompatible when they are distantly related, hybrid sterility or inviability may be generated at a single locus. Figure 9 shows one such example, where allele A0 is compatible with allele A−1, A0, and A1 but not with other alleles (ignore the alleles at locus B). Thus, genotype A2A2 is not compatible with genotype A0A0 in either mating ability or zygotic viability or sterility. The two populations composed of A0A0 and A2A2 individuals will then be reproductively isolated.
In rice, there is one example that supports this model at the molecular level. The reproductive barrier between Indica and Japonica is controlled by many genes. One of them is the S5 gene that encodes an aspartic protease determining embryonic sac fertility. The protein encoded by this gene in Indica (S5i) and Japonica (S5j) are different at two amino acid sites (Chen et al. 2008). One of these differences (F273L) in Japonica seems to be responsible for the sterility of the hybrid between Indica and Japonica. Interestingly, however, there are strains that produce fertile hybrids both with Indica and Japonica. The S5 gene (S5n) in these strains encodes a protein with a deletion of a fragment of 115 amino acids, and this might have been an intermediate allele between S5i and S5j. This type of single-locus speciation is plausible particularly in self-fertilizing organisms like rice.
Reproductive isolation by single-loci may also occur by mutations controlling flowering time in plants. This would occur easily in self-fertilizing plants. At the molecular level, flowering time is controlled by a large number of loci, many of which are duplicate genes. Environmental factors such as photoperiodicity and vernalization also affect flowering time. In recent years, many genes involved in the regulation of flowering time have been identified in Arabidopsis (Simpson et al. 1999; Boss et al. 2004; Pouteau et al. 2008). Although flowering time is controlled by many genes, a single mutation may change flowering time drastically and may produce reproductive isolation. One of the interesting cases is a mutation that occurred at the flowering locus (FLC), a repressor of flowering involved in the vernalization pathway. The genomes of Brassica species contain several FLC paralogous genes. Yuan et al. (2009) discovered that one of the FLC genes, FLC1 in Brassica rapa, is polymorphic with respect to flowering time in nature, and this polymorphism is caused by the mutation of splicing site (G → A) in intron 6. This mutation was then shown to change flowering time. Reproductive isolation due to heterogeneity in developmental time also occurs in insects (Tauber CA and Tauber MJ 1977). However, the molecular basis of evolution of this type of reproductive isolation has not been investigated.
Other Mechanisms of Evolution of Reproductive Isolation
There are many other speciation models that are based on relatively small number of observations or of which the theoretical basis is unclear. For example, gene transposition may also cause hybrid incompatibility. Masly et al. (2006) showed that the JYAlpha gene encoding the alpha subunit of Na+ and K+ adenosine triphosphatase was transposed from chromosome 4 to chromosome 3R in D. simulans after the divergence from D. melanogaster. Consequently, when these two species are crossed, some of the F2 individuals have no copy of JYAlpha in the genome and become sterile. This is a special case of classical reciprocal translocation of chromosomes discussed earlier. However, gene transposition or translocation may occur more frequently than chromosomal translocation, simply because there are more genes than chromosomes in the genome and transposons may mediate gene transfer. Several authors (Henikoff and Malik 2002; Brown and O'Neill 2010) have suggested that rapid concerted evolution of DNA repeat elements at the centromeric chromatin may generate speciation by distorting chromosomal segregation. The logic behind this argument is not very clear, but it is interesting to note that repeat elements are often involved in hybrid sterility and inviability. It should also be noted that epigenetic factors controlling photoperiodicity and vernalization in plants are apparently involved in speciation, though molecular study of these problems are still in its infancy.
Another speciation model proposed is speciation by antirecombination. In yeasts, the recombination machinery checks DNA sequence identity between homologous chromosomes. If the identity is low, the machinery suppresses recombination. Consequently, a hybrid between two yeast species would have a reduced number of recombination events, which causes chromosome nondisjunction in meiosis and aneuploidy, which in turn results in a reduced fertility of the hybrid (Hunter et al. 1996; Greig et al. 2003; Liti et al. 2006). Note that in this mechanism mutation apparently plays an important role because this mechanism exclusively relies on genomic sequence divergence between two populations, where no natural selection is necessary.
Discussion
We have seen that polyploidization and chromosomal rearrangements play important roles for generating new species and even segmental gene duplication may lead to the formation of new species. We have also seen that O. gigas discovered by de Vries was actually a polyploid. Therefore, de Vries’ assertion that new species may arise by mutational events has been vindicated. However, there are many different ways of evolution of reproductive isolation when genic mutations are considered.
It is well known that Charles Darwin had a difficulty to explain hybrid sterility or inviability by natural selection. Some authors had suggested that hybrid sterility or inviability might be enhanced by natural selection because the mixing of two incipient species by hybridization is disadvantageous in the formation of new species. However, Darwin rejected this idea after examination of various cases of species hybridization. He then concluded that “hybrid sterility is not a specially acquired or endowed quality but is incidental on other acquired characters” (Darwin 1859, p. 245).
Yet, many investigators have tried to understand speciation by means of natural selection. Natural selection may occur when the genes involved in hybrid sterility undergo evolutionary changes in different allopatric populations. In recent years, many authors have argued that this type of natural selection has speeded up the development of hybrid sterility or inviability (see Coyne and Orr 2004). However, this type of selection has nothing to do with the development of sterility because hybrid sterility is caused by mutations that have no deleterious effects within species but have deleterious effects in interspecific hybrids. Furthermore, the idea of accelerated evolution of reproductive isolation is teleological because there is no need for any organism to speed up reproductive isolation. Natural populations evolve without purpose so that they will manifest whatever happened in their genomic structure.
As we have seen, there are various kinds of hybrid sterility genes, and they are always accumulating in the genome of any species without being noticed until hybridization occurs artificially or naturally. In other words, hybrid sterility or inviability need not be the results of natural selection. It is quite likely to be a mere consequence of the evolutionary change of interactive genetic systems within species, as indicated by Darwin.
Some authors have suggested that hybrid sterility genes that are expressed in early stages of speciation would be more important in speeding up speciation than those that are expressed in later stages. This view is not acceptable because we know that any hybrid sterility genes are mere consequence of mutations that disturb the gene interaction systems in interspecific hybrids. If a pair of species are kept isolated for a long evolutionary time, both early or late stage expression genes should have developed sterility barriers.
We would also expect that hybrid sterility mutations would increase with evolutionary time and in the long run any pair of species would not be able to mate and produce offspring. For example, macaques and mice would never be able to mate and produce any offspring because they have accumulated so many mutations affecting important gene interaction systems. By contrast, some species of mice may be able to produce offspring because the extent of disturbance of gene interaction systems would not be so large.
The above consideration suggests that the identification of hybrid sterility genes for any pair of species is complicated. If we study distantly related species, there may be a large number of sterility genes involved, but their detection may be difficult because many sets of gene interaction would be compounded. By contrast, if we study closely related species, there may not be many hybrid sterility genes, but once they are identified, it would be easy to study the nature of the hybrid sterility genes.
As was mentioned in the beginning, the purpose of this review is to understand the roles of mutation and selection in speciation. We believe that we have shown that mutation is essential for the evolution of reproductive isolation though selection, particularly deleterious epistatic selection, is necessary.
Acknowledgments
We thank Sayaka Miura for her help on our data analysis. We also thank Dan Hartl and Chung-I Wu for their comments on earlier versions of the manuscript. This work was supported by National Institute of Health grant GM020293 to M. Nei and Japan Society for the Promotion of Science to M. Nozawa.
References
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Curr Opin Plant Biol. 2005;8:135–141. doi: 10.1016/j.pbi.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Allen GE. Hugo de Vries and the reception of the “mutation theory”. J Hist Biol. 1969;2:55–87. [Google Scholar]
- Badaeva ED, et al. Chromosomal rearrangements in wheat: their types and distribution. Genome. 2007;50:907–926. doi: 10.1139/g07-072. [DOI] [PubMed] [Google Scholar]
- Barr CM, Fishman L. The nuclear component of a cytonuclear hybrid incompatibility in Mimulus maps to a cluster of pentatricopeptide repeat genes. Genetics. 2010;184:455–465. doi: 10.1534/genetics.109.108175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateson W. Heredity and variation in modern lights. In: Seward AC, editor. Darwin and modern science. Cambridge: Cambridge University Press; 1909. pp. 85–101. [Google Scholar]
- Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;326:1538–1541. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc Natl Acad Sci U S A. 2002;99:10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D, et al. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science. 2009;323:623–626. doi: 10.1126/science.1165917. [DOI] [PubMed] [Google Scholar]
- Bomblies K, et al. Autoimmune response as a mechanism for a Dobzhansky-Muller-type incompatibility syndrome in plants. PLoS Biol. 2007;5:e236. doi: 10.1371/journal.pbio.0050236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. Plant Cell. 2004;16:S18–S31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, et al. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- Brown JD, O'Neill RJ. Chromosomes, conflict, and epigenetics: chromosomal speciation revisited. Annu Rev Genomics Hum Genet. 2010;11:291–316. doi: 10.1146/annurev-genom-082509-141554. [DOI] [PubMed] [Google Scholar]
- Case AL, Willis JH. Hybrid male sterility in Mimulus (Phrymaceae) is associated with a geographically restricted mitochondrial rearrangement. Evolution. 2008;62:1026–1039. doi: 10.1111/j.1558-5646.2008.00360.x. [DOI] [PubMed] [Google Scholar]
- Chase CD. Cytoplasmic male sterility: a window to the world of plant mitochondrial–nuclear interactions. Trends Genet. 2007;23:81–90. doi: 10.1016/j.tig.2006.12.004. [DOI] [PubMed] [Google Scholar]
- Chen J, et al. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc Natl Acad Sci U S A. 2008;105:11436–11441. doi: 10.1073/pnas.0804761105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen KY, Cong B, Wing R, Vrebalov J, Tanksley SD. Changes in regulation of a transcription factor lead to autogamy in cultivated tomatoes. Science. 2007;318:643–645. doi: 10.1126/science.1148428. [DOI] [PubMed] [Google Scholar]
- Chou JY, Hung YS, Lin KH, Lee HY, Leu JY. Multiple molecular mechanisms cause reproductive isolation between three yeast species. PLoS Biol. 2010;8:e1000432. doi: 10.1371/journal.pbio.1000432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland RE. Chromosome arrangements during meiosis in certain Oenotherae. Am Nat. 1923;57:562–566. [Google Scholar]
- Cleland RE. Oenothera: cytogenetics and evolution. New York: Academic Press; 1972. [Google Scholar]
- Coyne JA. Genetics and speciation. Nature. 1992;355:511–515. doi: 10.1038/355511a0. [DOI] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. Speciation. Sunderland (MA): Sinauer Associates; 2004. [Google Scholar]
- Darwin C. On the origin of species. London: Murray; 1859. [Google Scholar]
- Davis BM. Genetical studies on Oenothera III. Further hybrids of Oenothera biennis and O. grandiflora that resemble O. lamarckiana. Am Nat. 1912;46:377–427. [Google Scholar]
- Davis BM. An amphidiploid in the F1 generation from the cross Oenothera Franciscana × Oenothera Biennis, and its progeny. Genetics. 1943;28:275–285. doi: 10.1093/genetics/28.4.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Bodt S, Maere S, Van de Peer Y. Genome duplication and the origin of angiosperms. Trends Ecol Evol. 2005;20:591–597. doi: 10.1016/j.tree.2005.07.008. [DOI] [PubMed] [Google Scholar]
- de Vries H. Die mutationstheorie. Vol. I and II. Leipzig (Germany): Verlag von Veit and Company; 1901–1903. [Google Scholar]
- de Vries H. The mutation theory: experiments and observations on the origin of species in the vegetable kingdom. Vol. I. The origin of species by mutation. English translation by J. B. Farmer and A. D. Darbishire. Chicago (IL): Open Court Publishing Company; 1909. [Google Scholar]
- de Vries H. The mutation theory: experiments and observations on the origin of species in the vegtable kingdom. Vol. II. The origin of varieties by mutation. English translation by J. B. Farmer and A. D. Darbishire. Chicago (IL): Open Court Publishing Company; 1910. [Google Scholar]
- Delneri D, et al. Engineering evolution to study speciation in yeasts. Nature. 2003;422:68–72. doi: 10.1038/nature01418. [DOI] [PubMed] [Google Scholar]
- Des Marais MD, Rausher DL. Parallel evolution at multiple levels in the origin of hummingbird pollinated flowers in Ipomoea. Evolution. 2010;64:2044–2054. doi: 10.1111/j.1558-5646.2010.00972.x. [DOI] [PubMed] [Google Scholar]
- Desiderio UV, Zhu X, Evans JP. ADAM2 interactions with mouse eggs and cell lines expressing alpha4/alpha9 (ITGA4/ITGA9) integrins: implications for integrin-based adhesion and fertilization. PLoS One. 2010;5:e13744. doi: 10.1371/journal.pone.0013744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobzhansky T. Genetics and the origin of species. New York: Columbia University Press; 1937. [Google Scholar]
- Doyle JJ, et al. Evolutionary genetics of genome merger and doubling in plants. Annu Rev Genet. 2008;42:443–461. doi: 10.1146/annurev.genet.42.110807.091524. [DOI] [PubMed] [Google Scholar]
- Evans JP, Florman HM. The state of the union: the cell biology of fertilization. Nat Cell Biol. 2002;4(Suppl):s57–s63. doi: 10.1038/ncb-nm-fertilityS57. [DOI] [PubMed] [Google Scholar]
- Ferree PM, Barbash DA. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 2009;7:e1000234. doi: 10.1371/journal.pbio.1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- Frank SA. Divergence of meiotic drive-suppression systems as an explanation for sex-biased hybrid sterility and inviability. Evolution. 1991;45:262–267. doi: 10.1111/j.1558-5646.1991.tb04401.x. [DOI] [PubMed] [Google Scholar]
- Fujii S, Toriyama K. Suppressed expression of retrograde-regulated male sterility restores pollen fertility in cytoplasmic male sterile rice plants. Proc Natl Acad Sci U S A. 2009;106:9513–9518. doi: 10.1073/pnas.0901860106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo BE, Vacquier VD, Swanson WJ. Positive selection in the egg receptor for abalone sperm lysin. Proc Natl Acad Sci U S A. 2003;100:4639–4643. doi: 10.1073/pnas.0830022100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates RR. The chromosomes of Oenothera. Science. 1908;27:193–195. doi: 10.1126/science.27.683.193-a. [DOI] [PubMed] [Google Scholar]
- Grant V. Plant speciation. New York: Columbia University Press; 1981. [Google Scholar]
- Greig D, Travisano M, Louis EJ, Borts RH. A role for the mismatch repair system during incipient speciation in Saccharomyces. J Evol Biol. 2003;16:429–437. doi: 10.1046/j.1420-9101.2003.00546.x. [DOI] [PubMed] [Google Scholar]
- Haldane JB. Sex-ratio and unidirectional sterility in hybrid animals. J Genet. 1922;12:101–109. [Google Scholar]
- Hartl DL. Dysfunctional sperm production in Drosophila melanogaster males homozygous for the segregation distorter elements. Proc Natl Acad Sci U S A. 1969;63:782–789. doi: 10.1073/pnas.63.3.782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Malik HS. Centromeres: selfish drivers. Nature. 2002;417:227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]
- Hirsch N, Zimmerman LB, Grainger RM. Xenopus, the next generation: X. tropicalis genetics and genomics. Dev Dyn. 2002;225:422–433. doi: 10.1002/dvdy.10178. [DOI] [PubMed] [Google Scholar]
- Hoballah ME, et al. Single gene-mediated shift in pollinator attraction in Petunia. Plant Cell. 2007;19:779–790. doi: 10.1105/tpc.106.048694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N, Chambers SR, Louis EJ, Borts RH. The mismatch repair system contributes to meiotic sterility in an interspecific yeast hybrid. Embo J. 1996;15:1726–1733. [PMC free article] [PubMed] [Google Scholar]
- Hurst LD, Pomiankowski A. Causes of sex ratio bias may account for unisexual sterility in hybrids: a new explanation of Haldane’s rule and related phenomena. Genetics. 1991;128:841–858. doi: 10.1093/genetics/128.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue N, Ikawa M, Isotani A, Okabe M. The immunoglobulin superfamily protein Izumo is required for sperm to fuse with eggs. Nature. 2005;434:234–238. doi: 10.1038/nature03362. [DOI] [PubMed] [Google Scholar]
- Jaenike J. Sex chromosome meiotic drive. Annu Rev Ecol Syst. 2001;32:25–49. [Google Scholar]
- Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Kamei N, Glabe CG. The species-specific egg receptor for sea urchin sperm adhesion is EBR1, a novel ADAMTS protein. Genes Dev. 2003;17:2502–2507. doi: 10.1101/gad.1133003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazama T, Toriyama K. A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of cytoplasmic male-sterile rice. FEBS Lett. 2003;544:99–102. doi: 10.1016/s0014-5793(03)00480-0. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kusano A, Staber C, Chan HY, Ganetzky B. Closing the (Ran) GAP on segregation distortion in Drosophila. Bioessays. 2003;25:108–115. doi: 10.1002/bies.10222. [DOI] [PubMed] [Google Scholar]
- Lee HY, et al. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell. 2008;135:1065–1073. doi: 10.1016/j.cell.2008.10.047. [DOI] [PubMed] [Google Scholar]
- Li WH, Nei M. Persistence of common alleles in two related populations or species. Genetics. 1977;86:901–914. doi: 10.1093/genetics/86.4.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G, Barton DB, Louis EJ. Sequence diversity, reproductive isolation and species concepts in Saccharomyces. Genetics. 2006;174:839–850. doi: 10.1534/genetics.106.062166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Y, et al. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc Natl Acad Sci U S A. 2008;105:18871–18876. doi: 10.1073/pnas.0810108105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lurin C, et al. Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell. 2004;16:2089–2103. doi: 10.1105/tpc.104.022236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz AM. A preliminary note on the chromosomes of Oenothera Lamarckiana and one of its mutants, O. gigas. Science. 1907;26:151–152. doi: 10.1126/science.26.657.151. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. Gene duplication and the origin of interspecific genomic incompatibility. Am Nat. 2000;156:590–605. doi: 10.1086/316992. [DOI] [PubMed] [Google Scholar]
- Lyon JD, Vacquier VD. Interspecies chimeric sperm lysins identify regions mediating species-specific recognition of the abalone egg vitelline envelope. Dev Biol. 1999;214:151–159. doi: 10.1006/dbio.1999.9411. [DOI] [PubMed] [Google Scholar]
- Lyttle TW. Segregation distorters. Annu Rev Genet. 1991;25:511–557. doi: 10.1146/annurev.ge.25.120191.002455. [DOI] [PubMed] [Google Scholar]
- Maheshwari S, Wang J, Barbash DA. Recurrent positive selection of the Drosophila hybrid incompatibility gene Hmr. Mol Biol Evol. 2008;25:2421–2430. doi: 10.1093/molbev/msn190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masly JP, Jones CD, Noor MA, Locke J, Orr HA. Gene transposition as a cause of hybrid sterility in Drosophila. Science. 2006;313:1448–1450. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- Mayr E. Animal species and evolution. Cambridge: Harvard University Press; 1963. [Google Scholar]
- Mayr E. Prologue: some thoughts on the history of the evolutionary synthesis. In: Mayr E, Provine WB, editors. The evolutionary synthesis. Cambridge (MA): Harvard University Press; 1980. pp. 1–48. [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Mizuta Y, Harushima Y, Kurata N. From the Cover: rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc Natl Acad Sci U S A. 2010;107:20417–20422. doi: 10.1073/pnas.1003124107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller HJ. Why polyploid is rarer in animals than in plants. Am Nat. 1925;59:346–353. [Google Scholar]
- Muller HJ. Bearing of the Drosophila work on systematics. In: Huxley J, editor. The new systematics. Oxford: Clarendon Press; 1940. [Google Scholar]
- Muller HJ. Isolating mechanisms, evolution and temperature. Biol Symp. 1942;6:71–125. [Google Scholar]
- Nandi HK. The chromosome morphology, secondary association and origin of cultivated rice. J Genet. 1936;33:316–336. [Google Scholar]
- Nei M. Gene duplication and nucleotide substitution in evolution. Nature. 1969;221:40–42. doi: 10.1038/221040a0. [DOI] [PubMed] [Google Scholar]
- Nei M. Mathematical models of speciation and genetic distance. In: Karlin S, Nevo E, editors. Population genetics and ecology. New York: Academic Press; 1976. pp. 723–765. [Google Scholar]
- Nei M, Maruyama T, Wu CI. Models of evolution of reproductive isolation. Genetics. 1983;103:557–579. doi: 10.1093/genetics/103.3.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Roychoudhury AK. Probability of fixation of nonfunctional genes at duplicate loci. Am Nat. 1973;107:362–372. [Google Scholar]
- Nei M, Suzuki Y, Nozawa M. The neutral theory of molecular evolution in the genomic era. Annu Rev Genomics Hum Genet. 2010;11:265–289. doi: 10.1146/annurev-genom-082908-150129. [DOI] [PubMed] [Google Scholar]
- Nei M, Zhang J. Molecular origin of species. Science. 1998;282:1428–1429. doi: 10.1126/science.282.5393.1428. [DOI] [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. Berlin (Germany): Springer; 1970. [Google Scholar]
- Ohno S. The notion of the Cambrian pananimalia genome and a genomic difference that separated vertebrates from invertebrates. Prog Mol Subcell Biol. 1998;21:97–117. doi: 10.1007/978-3-642-72236-3_5. [DOI] [PubMed] [Google Scholar]
- Oka HI. The mechanisms of sterility in the intervarietal hybrids. Phylogenetic differentiation of cultivated rice. VI. (In Japanese with English summary) Japan J Breed. 1953;2:217–224. [Google Scholar]
- Oka HI. Genic analysis for the sterility of hybrids between distantly related varieties of cultivated rice. J Genet. 1957;55:397–409. [Google Scholar]
- Oka HI. Analysis of genes controlling F1 sterility in rice by the use of isogenic lines. Genetics. 1974;77:521–534. doi: 10.1093/genetics/77.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, et al. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet. 2009;5:e1000753. doi: 10.1371/journal.pgen.1000753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. The population genetics of speciation: the evolution of hybrid incompatibilities. Genetics. 1995;139:1805–1813. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA. Dobzhansky, Bateson, and the genetics of speciation. Genetics. 1996;144:1331–1335. doi: 10.1093/genetics/144.4.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr HA, Masly JP, Presgraves DC. Speciation genes. Curr Opin Genet Dev. 2004;14:675–679. doi: 10.1016/j.gde.2004.08.009. [DOI] [PubMed] [Google Scholar]
- Orr HA, Presgraves DC. Speciation by postzygotic isolation: forces, genes and molecules. Bioessays. 2000;22:1085–1094. doi: 10.1002/1521-1878(200012)22:12<1085::AID-BIES6>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Otto SP, Whitton J. Polyploid incidence and evolution. Annu Rev Genet. 2000;34:401–437. doi: 10.1146/annurev.genet.34.1.401. [DOI] [PubMed] [Google Scholar]
- Phadnis N, Orr HA. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science. 2009;323:376–379. doi: 10.1126/science.1163934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouteau S, et al. Diversification of photoperiodic response patterns in a collection of early-flowering mutants of Arabidopsis. Plant Physiol. 2008;148:1465–1473. doi: 10.1104/pp.108.127639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. Sex chromosomes and speciation in Drosophila. Trends Genet. 2008;24:336–343. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC, Balagopalan L, Abmayr SM, Orr HA. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature. 2003;423:715–719. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- Presgraves DC, Stephan W. Pervasive adaptive evolution among interactors of the Drosophila hybrid inviability gene, Nup96. Mol Biol Evol. 2007;24:306–314. doi: 10.1093/molbev/msl157. [DOI] [PubMed] [Google Scholar]
- Redon R, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renner O. Versuche uber die gametische Konstitution der Oenotheren. Z Ind Abst Vererb. 1917;18:121–294. [Google Scholar]
- Rensing SA, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Rieseberg LH, Willis JH. Plant speciation. Science. 2007;317:910–914. doi: 10.1126/science.1137729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai K. Chromosome study of Oryza sativa L. I. The secondary association of hte meiotic chromosomes. Japan J Genet. 1935;44:149–156. [Google Scholar]
- Sandler L, Hiraizumi Y, Sandler I. Meiotic drive in natural populations of Drosophila melanogaster. I. The cytogenetic basis of segregation-distortion. Genetics. 1959;44:233–250. doi: 10.1093/genetics/44.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawamura K, Yamamoto MT, Watanabe TK. Hybrid lethal systems in the Drosophila melanogaster species complex. II. The zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics. 1993;133:307–313. doi: 10.1093/genetics/133.2.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell DR, Byrne KP, Gordon JL, Wong S, Wolfe KH. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature. 2006;440:341–345. doi: 10.1038/nature04562. [DOI] [PubMed] [Google Scholar]
- Schartl M. Evolution of Xmrk: an oncogene, but also a speciation gene? Bioessays. 2008;30:822–832. doi: 10.1002/bies.20807. [DOI] [PubMed] [Google Scholar]
- Seidel HS, Rockman MV, Kruglyak L. Widespread genetic incompatibility in C. elegans maintained by balancing selection. Science. 2008;319:589–594. doi: 10.1126/science.1151107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw A, Fortes PA, Stout CD, Vacquier VD. Crystal structure and subunit dynamics of the abalone sperm lysin dimmer: egg envelopes dissociate dimmers, the monomer is the active species. J Cell Biol. 1995;130:1117–1125. doi: 10.1083/jcb.130.5.1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson GG, Gendall AR, Dean C. When to switch to flowering. Annu Rev Cell Dev Biol. 1999;15:519–550. doi: 10.1146/annurev.cellbio.15.1.519. [DOI] [PubMed] [Google Scholar]
- Stebbins GL. Variation and evolution in plants. New York: Columbia University Press; 1950. [Google Scholar]
- Stebbins GL. Processes of organic evolution. Upper Saddle River (NJ): Prentice-Hall; 1966. [Google Scholar]
- Sun S, Ting CT, Wu CI. The normal function of a speciation gene, Odysseus, and its hybrid sterility effect. Science. 2004;305:81–83. doi: 10.1126/science.1093904. [DOI] [PubMed] [Google Scholar]
- Tang S, Presgraves DC. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science. 2009;323:779–782. doi: 10.1126/science.1169123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, et al. A sex-ratio meiotic drive system in Drosophila simulans. II: An X-linked distorter. PLoS Biol. 2007;5:e293. doi: 10.1371/journal.pbio.0050293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Hartl DL, Laurie CC. Sex-ratio segregation distortion associated with reproductive isolation in Drosophila. Proc Natl Acad Sci U S A. 2001;98:13183–13188. doi: 10.1073/pnas.231478798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tauber CA, Tauber MJ. Sympatric speciation based on allelic changes at three loci: evidence from natural populations in two habitats. Science. 1977;197:1298–1299. doi: 10.1126/science.197.4310.1298. [DOI] [PubMed] [Google Scholar]
- Ting CT, Tsaur SC, Wu ML, Wu CI. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science. 1998;282:1501–1504. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- Wendel JF. Genome evolution in polyploids. Plant Mol Biol. 2000;42:225–249. [PubMed] [Google Scholar]
- Werth CR, Windham MD. A model for divergent, allopatric speciation of polyploid pteridophytes resulting from silencing of duplicate-gene expression. Am Nat. 1991;137:515–526. [Google Scholar]
- White MJD. Chromosomal rearrangements and speciation in animals. Annu Rev Genet. 1969;3:75–98. [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Wood TE, et al. The frequency of polyploid speciation in vascular plants. Proc Natl Acad Sci U S A. 2009;106:13875–13879. doi: 10.1073/pnas.0811575106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. On the probability of fixation of reciprocal translocations. Am Nat. 1941;75:513–522. [Google Scholar]
- Wu CI, Hammer MF. Molecular evolution of ultraselfish genes of meiotic drive systems. In: Selander RK, Whittam TS, Clark AG, editors. Evolution at the molecular level. Sunderland (MA): Sinaur Press; 1991. pp. 177–203. [Google Scholar]
- Wu CI, Lyttle TW, Wu ML, Lin GF. Association between a satellite DNA sequence and the responder of segregation distorter in D. melanogaster. Cell. 1988;54:179–189. doi: 10.1016/0092-8674(88)90550-8. [DOI] [PubMed] [Google Scholar]
- Wu CI, Ting CT. Genes and speciation. Nat Rev Genet. 2004;5:114–122. doi: 10.1038/nrg1269. [DOI] [PubMed] [Google Scholar]
- Yamagata Y, et al. Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc Natl Acad Sci U S A. 2010;107:1494–1499. doi: 10.1073/pnas.0908283107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto E, et al. Gain of deleterious function causes an autoimmune response and Bateson-Dobzhansky-Muller incompatibility in rice. Mol Genet Genomics. 2010;283:305–315. doi: 10.1007/s00438-010-0514-y. [DOI] [PubMed] [Google Scholar]
- Yuan YX, et al. A naturally occurring splicing site mutation in the Brassica rapa FLC1 gene is associated with variation in flowering time. J Exp Bot. 2009;60:1299–1308. doi: 10.1093/jxb/erp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeh JA, Zeh DW. Maternal inheritance, sexual conflict and the maladapted male. Trends Genet. 2005;21:281–286. doi: 10.1016/j.tig.2005.03.006. [DOI] [PubMed] [Google Scholar]
- Zhang J, Rosenberg HF, Nei M. Positive Darwinian selection after gene duplication in primate ribonuclease genes. Proc Natl Acad Sci U S A. 1998;95:3708–3713. doi: 10.1073/pnas.95.7.3708. [DOI] [PMC free article] [PubMed] [Google Scholar]