Abstract
Charles Darwin has proposed the theory that evolution of live organisms is based on random variation and natural selection. Jacques Monod in his classic book Chance and Necessity, published 40 years ago, presented his thesis “that the biosphere does not contain a predictable class of objects or events, but constitutes a particular occurrence, compatible indeed with the first principles, but not deducible from those principals and therefore, essentially unpredictable.” Recent discoveries in eye evolution are in agreement with both of these theses. They confirm Darwin's assumption of a simple eye prototype and lend strong support for the notion of a monophyletic origin of the various eye types. Considering the complexity of the underlying gene regulatory networks the unpredictability is obvious. The evolution of the Hox gene cluster and the specification of the body plan starting from an evolutionary prototype segment is discussed. In the course of evolution, a series of similar prototypic segments gradually undergoes cephalization anteriorly and caudalization posteriorly through diversification of the Hox genes.
Keywords: eye evolution, Jacques Monod, chance, necessity
Introduction
In his classic book Chance and Necessity, Jacques Monod (1970) puts forward the thesis “that the biosphere does not contain a predictable class of objects or events, but constitutes a particular occurrence, compatible indeed with the first principles, but not deducible from those principles and therefore essentially unpredictable.” Darwin’s theory of evolution is based on these same principles—random variation and natural selection. One of the greatest difficulties with his theory was to explain the origins of organs of extreme perfection, like the eyes of eagles, which seem to be designed for a given purpose. Darwin (1872) found a way out of this difficulty by assuming a very simple prototypic eye consisting of just two cells, a photoreceptor cell (“nerve”) and a pigment cell, shielding the light from one side, thus allowing the animal to detect the direction of the incoming light which confers a great selective advantage. As selection can only become effective when an organ is functional, at least to a small extent, he considered the assembly of the prototypic eye as a merely stochastic event due to random variation. The pioneering work of Gregor Mendel showed that random variations were caused by mutations and that the predictions for the outcome of genetic crosses could be no more than statistical. Neo-Darwinists introduced genetics into evolutionary biology, but eye evolution remained enigmatic because it was assumed that the evolution of the various eye types occurred 40–60 times independently in the different animal phyla. This was essentially incompatible with Darwin’s ideas, which assumed a rare stochastic event to give rise to prototypic eye. Recent molecular genetic work strongly supports the notion that the various eye types arose monophyletically. Pax6 was identified as a master control gene for eye morphogenesis and found to be involved in eye development of all bilaterian animals (Halder et al. 1995). This suggests that Pax6 was selected as a master control gene in the last common ancestor of all bilateria and maintained in all bilaterian phyla to control eye development. Because there are no functional constraints on Pax6 to control eye morphogenesis, the reason for Pax6 being involved in the genetic control of eye morphogenesis in all bilaterian phyla is a reflection of its evolutionary history and due to common descent.
Darwin’s Concept of Eye Evolution
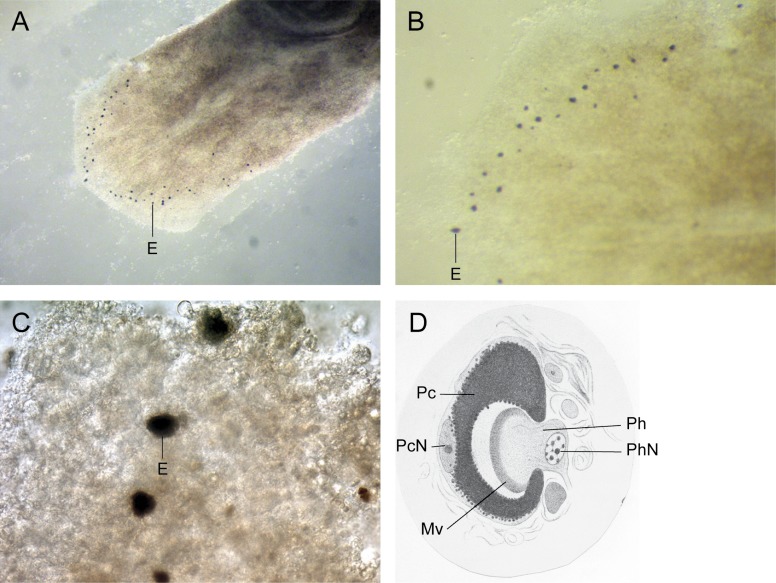
For Charles Darwin, eye evolution was a difficult problem to explain in terms of his evolutionary theory, and he devoted an entire chapter of “On the Origin of Species” to the discussion of the difficulties of his theory in which organs of extreme perfection like the eye figure prominently (Darwin 1872). He found a way out by proposing the existence of a very simple prototypic eye consisting of two cells only: a photoreceptor cell, which he called a nerve, and a pigment cell shielding the photoreceptor cell from one side, which allows for directional vision and confers a considerable selective advantage. Such a prototypic eye was found later in planarians (fig. 1) and in trochophora larvae of many annelids and mollusks. On the basis of structural and functional differences among the various eye types, neo-Darwinists assumed that the eye evolved 40–60 times independently in the various taxa (Salvini-Plawen and Mayr 1977). Because Darwin admitted that selection could only set in once the prototype had evolved, it has to be assumed that the origin of the prototype must have been a rare stochastic event. This is incompatible with its independent occurrence 40–60 times. The finding of Pax6 as a universal master control gene for eye development has resolved this paradox.
FIG. 1.—
The planarian eye. (A) Polycelis auricularia with prototypic eyes; (B and C), P. auricularia eyes at higher magnification; (D) histological section across the eye of Planaria torva (from Hesse 1897): E, eye; Pc, pigment cell; PcN, pigment cell nucleus; Mv, microvilli; Ph, photoreceptor cell; PhN, photoreceptor cell nucleus.
Pax6 as a Universal Master Control Gene for Eye Morphogenesis and Eye Evolution in Bilateria
The serendipitous cloning of the Drosophila homolog of the mouse Pax6 gene (Quiring et al. 1994) indicated that in both mammals and insects the gene essential for eye development and that it is expressed from early stages of eye development at least to the end of morphogenesis. Loss-of-function mutants have an eyeless phenotype. These findings led me to the idea that—contrary to the neo-Darwinists' dogma—Pax6 might be a universal master control gene for eye development. To test this idea, we constructed gain-of-function mutants in Drosophila which allow the ectopic expression of both the Drosophila Pax6 (eyeless) gene and the mouse Pax6 gene using the yeast gal 4 system (Halder et al. 1995) and genomic enhancers identified by enhancer trapping (O'Kane and Gehring 1987).
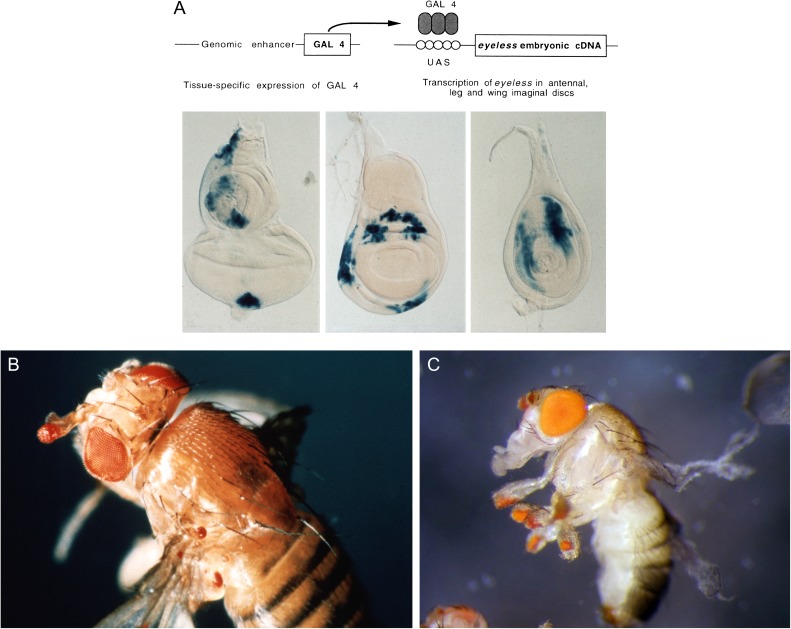
As shown in figure 2, ectopic eyes can be induced on the antennae, wings, and legs of the fly. However, the timing of Pax6 expression is crucial. The enhancer has to function before the larvae have reached the early third instar when cell fate determination occurs. After that time point, the discs are difficult to reprogram. Interestingly, the mouse Pax6 gene can substitute for the Drosophila gene indicating a strong evolutionary conservation of the gene. Subsequent studies have shown that in Drosophila, but not in mouse or human, a gene duplication has occurred and, besides eyeless (ey), a second gene twin of eyeless (toy) was found (Czerny et al. 1999), which in loss-of-function mutants leads to a headless pupal lethal phenotype. Pax6 mice are eyeless, noseless, and lack a large part of the brain and are, therefore, lethal (Hill et al. 1991; Walther and Gruss 1991). Similarly, toy flies lack the entire head capsule including eyes, antennae (nose), and maxillary palps. It is generally assumed that a master control gene for eye development should be expressed specifically in the eyes, but Pax6 is expressed region specifically in the head region, including the brain, into which visual axons project. The eye is not formed in isolation and inserted into the head, but it has to be localized properly and integrated into the body plan. This is accomplished by a region-specific complex pattern of gene expression.
FIG. 2.—
Targeted expression of eyeless (ey) and induction of ectopic eyes in Drosophila (after Halder et al. 1995). (A) Targeted expression of ey cDNA using a genomic enhancer to induce the yeast gal4 transcription factor driving ey cDNA in various imaginal discs. Gal4 binds to its upstream activating sequences (UASs) and drives the expression of ey into the respective areas of the eye-antennal, wing, and leg discs (blue). (B) Ectopic induction of eyes on the antenna and mesothorax. (C) Ectopic induction of eyes on all six legs by using the decapentaplegic enhancer.
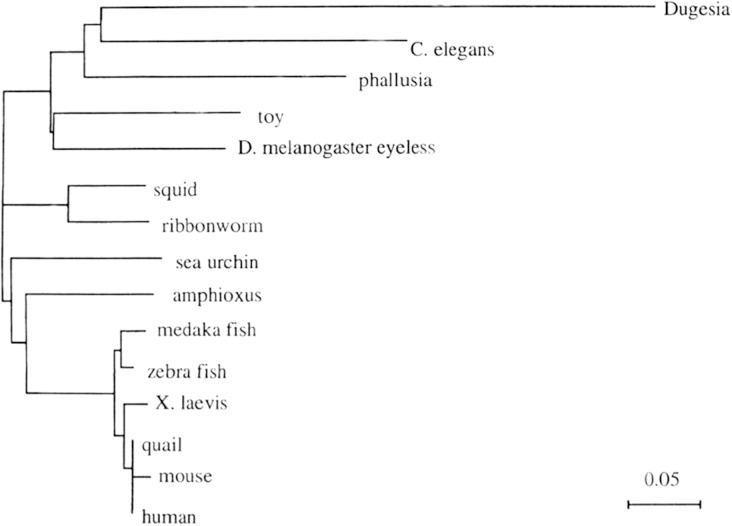
Subsequently, we isolated a Pax6 homolog from squid (Tomarev et al. 1997), which for decades was considered to be a classic case of convergent eye evolution. However, the finding of Pax6 in all bilaterian phyla argues strongly for a monophyletic origin of the various eye types and subsequent divergent, parallel, or convergent evolution (see below). The gene structure of Pax6 indicates that it encodes a transcription factor—a gene regulatory protein—containing two types of DNA-binding domains, a paired domain (PD), and a homeodomain (HD). Using the DNA sequences of the PDs known at the time, we calculated a phylogenetic tree using the neighbor-joining method (fig. 3) (from Gehring and Ikeo 1999). This pedigree reflects the known phylogenetic tree rather well, with the exception of fast evolving species that branch off too early. The nematode Caenorhabditis elegans, whose genome has been sequenced entirely, requires some explanation. More primitive marine nematodes have evolved eyes, whereas C. elegans lives inside rotting fruit and has lost its eyes, like many other animals living in the soil or in caves. The reason is the lack of selection pressure. However, C. elegans retains its Pax6 gene, because Pax6 is pleiotropic and not only required for eye development but also for the formation of the nose and the brain, which C. elegans retains. However, the worm has lost its rhodopsin genes, which have only one function, light perception. In this case, the lack of selective pressure leads to the loss of the respective genes.
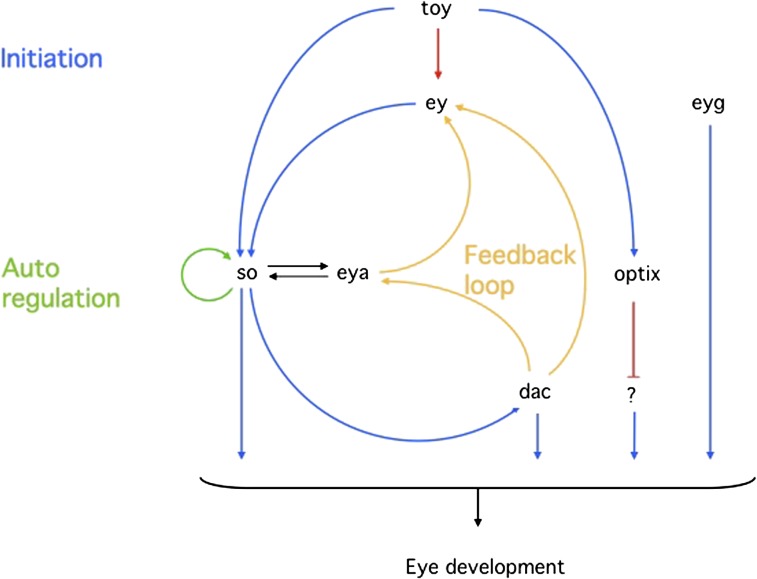
FIG. 3.—
Regulatory scheme on the top of the eye developmental pathway. Twin of eyeless (toy), eyeless (ey), and possibly eyegone (eyg), three Pax genes, are master control genes on the top of the hierarchy. Sine oculis (so), eyes absent (eya), dachshund (dac), and optix are second-order transcription factors regulated by the master control genes. Note that the pathway is not linear but rather a network interconnected by feedback loops.
We have tried to decipher the eye developmental genetic program in Drosophila. Pax6, alias ey and toy, initiates the eye development program by activating a set of subordinate transcription factors designated as the Retinal Determination Gene Network (RDGN), illustrated in figure 4. The most important member of these subordinate transcription factors is sine oculis (so), a member of the six gene family (fig. 5). Loss-of-function mutants of so lack both compound eyes and ocelli.
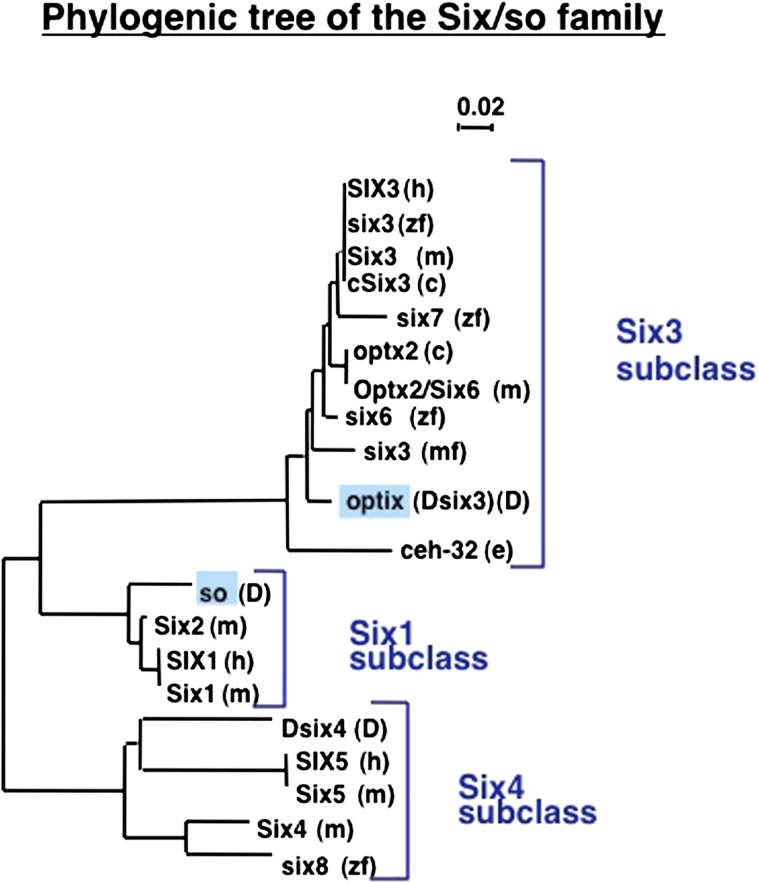
FIG. 4.—
Phylogenetic tree of the Six/so family. The phylogenetic analysis shows that Drosophila sine oculis (so) belongs to the six 1 subclass, and optix (opt) resides in the six 3 subclass, both of which are also expressed in eye development of vertebrates. In contrast, Dsix4 belongs to the six 4 subclass which is expressed during myogenesis.
FIG. 5.—
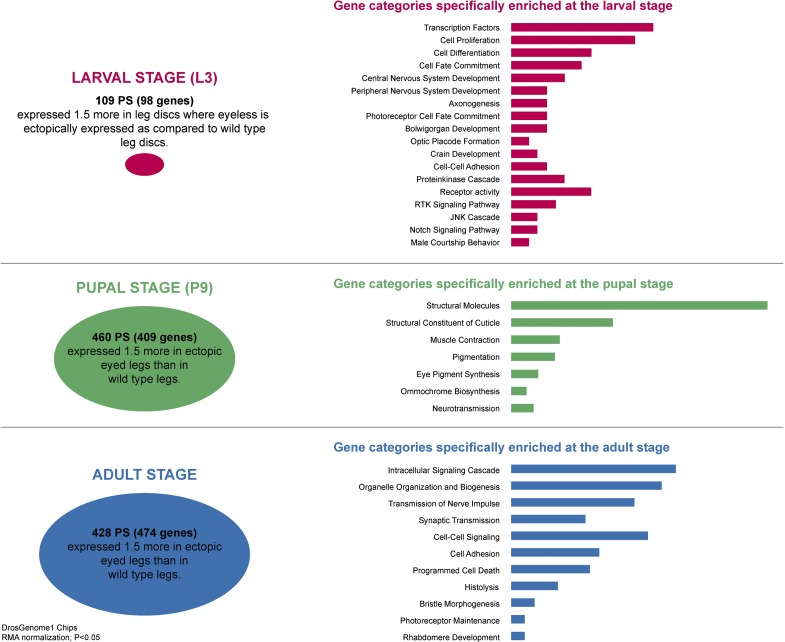
Expression of eye-specific genes at the larval, pupal, and adult stages of Drosophila (courtesy of Lydia Michaut). The transcriptomes of leg imaginal discs, pupal, and adult legs were compared with those in which an eye field was induced by ectopic ey expression. The respective gene categories and number of genes are indicated.
Both ey and toy activate so, the enhancer of which contains five Pax6 binding sites (Niimi et al. 1999). Two are specific for toy and three are “recognized” by ey and toy. This is typical for cis-regulatory elements of transcription factors, they contain multiple binding sites for multiple transcription factors constituting a regulatory network. The six gene family can be subdivided into three subclasses (fig. 5). The six1 subclass includes so, optix (Dsix3) belongs to the six3 subclass and also belongs to the RDGN (see fig. 4), whereas Drosophila six4 (Dsix4) belongs to subclass 4 and is involved in myogenesis. Also the mammalian members of subclasses 1 and 3 are involved in eye morphogenesis, whereas the members of subclass 4 participate in myogenesis. This illustrates that a given transcription can control any set of target genes as long as they contain the appropriate cis-regulatory elements, and there are no functional constraints on genes like Pax6 and so to control eye development. Therefore, the use by organisms as distantly related as mouse and Drosophila of Pax6 to control eye development reflects their evolutionary history; Pax6 was used to initiate the eye developmental pathway in their last common ancestor and has been maintained ever since. This must have been true already in the Precambrium, before chordates and ecdysiozoa separated. In general, important innovations occur rarely during evolution.
Besides Pax6, the genes of the RDGN are also highly conserved in evolution and found in all Bilateria, and more recently they were found to be present in Cnidaria as well. Highly developed eyes are found sporadically in some medusae of Hydrozoa and Scyphozoa which have only about four Pax genes rather than nine found in higher metazoa. They do not possess a bona fide Pax6 gene, but two other Pax genes have been identified which are expressed in the eyes and capable of inducing ectopic eyes in Drosophila, PaxB in the Scyphozoan Tripedalia (Kozmic et al. 2003) and PaxA in the Hydrozoan Cladonema (Suga et al. 2010). PaxB may be considered as a precursor of Pax6 and Pax2, but PaxA may indicate that Pax genes are flexibly deployed in eye development in Cnidarians. However, recent studies have identified all the members of RDGN in cnidarians: sine oculis (six), dachshund (dac), and eyes absent (eya) (Graziussi DF, Suga H, Schmid V, Gehring WJ, unpublished data) supporting the notion of a monophyletic origin of the eyes.
We continued to decipher the eye developmental program by using microarrays and gene chips (Michaut et al. 2003 and Michaut L, unpublished data) to identify the eye-specific transcripts at larval, pupal, and adult stages. In order to have a suitable reference, we used developing eyes which were ectopically induced in leg imaginal discs and subtracted the RNAs expressed in normal leg discs, which served as a control. As shown in figure 6, about 100 genes are differentially expressed in the eye field induced in leg imaginal discs of third instar larvae that were not induced in control leg discs. This number depends on where you set the threshold. We have used a factor of 1.5 by which the transcripts have to be overexpressed relative to the control discs. Ey mainly induces genes acting early in retinal differentiation in particularly transcription factors involved in cell fate determination. At the pupal stage, approximately 400 genes were identified mostly encoding structural proteins and proteins involved in the synthesis of eye pigments. In the adult eyes, around 500 genes were found to be actively involved mainly in the visual process and the functioning of the eye. On the basis of these findings, we assume at least a thousand genes to be involved in the development of compound eyes with Pax6 initiating network.
FIG. 6.—
Phylogenetic tree of the Pax6 genes. The neighbor-joining method was used to generate a phylogenetic tree of the Pax6 genes of various metazoa based on the sequences of the respective paired boxes. Note that eyeless and twin of eyeless of Drosophila are closely related. The scale shows the number of amino acid substitutions per site. The monophyly of the eyeless/Pax6 group of genes is strongly supported by the phylogenetic analysis of Jacobs et al. (1998).
Conservation of Target Sequences
If Pax6 is so highly conserved in evolution, its target sequences must also be conserved. We have tested this prediction in various ways, and one of the more convincing experiments was carried out on the chicken δ crystalline gene, encoding a lens-specific protein that is found in birds and reptiles only, due to their evolutionary relatedness. Experiments in Hsiato Kondo’s laboratory had previously shown that this gene is regulated by Pax6 and Sox2, another transcription factor. The minimal enhancer consists of only about 25 bp and includes a Pax6 binding site adjacent to a Sox2 binding site. Upon fusion of multiple enhancers to green fluorescent protein (GFP) and transfer into transgenic flies, GFP was indeed expressed into the eyes of the transgenic flies, whereas mutations in either the Pax6 or the Sox2 binding site abolished this capacity (Blanco et al. 2005). Drosophila has two kinds of lenses, a cuticular lens and a liquid lens underneath, which is secreted by a set of cone cells. Because GFP is not a secreted protein, we had to attach a signal peptide to GFP in order to find out whether this reporter protein is secreted into the liquid lens. Indeed the liquid lenses of these transgenic flies show green fluorescence, whereas the controls are negative. This indicates that the cis-regulatory target sites from the chicken can be correctly interpreted by Drosophila despite of the fact that these two species have been separated by more than 540 My.
The Origin of the Darwinian Prototypic Eye from Single Cells
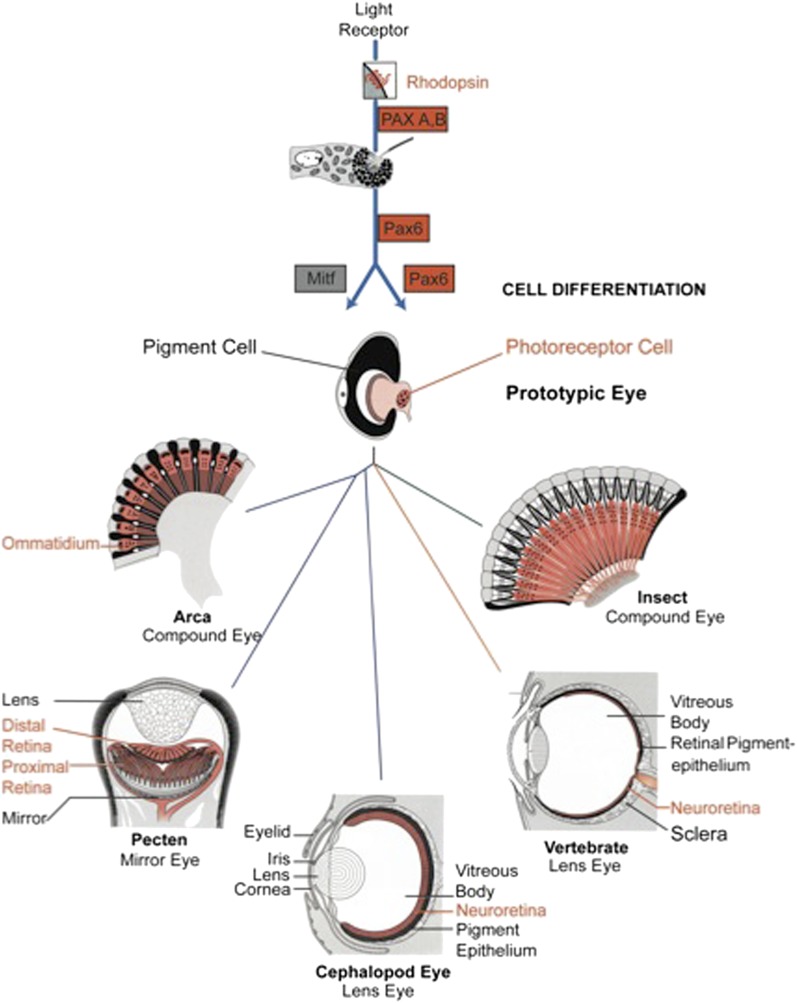
At the transition of unicellular to multicellular organisms, a major new mechanism for generating increasing complexity evolved: cell differentiation, the formation of different cell types originating from the zygote. Ultimately, the eye originated from a single cell. In principle, the Darwinian prototype consisting of a photoreceptor and pigment cell could be formed by assembly of these two cell types into a primitive eye. Alternatively, the two cell types could have arisen by cell differentiation from a common precursor cell. The second hypothesis is strongly supported by developmental studies in planarians (Takeda et al. 2009; Watanabe K, unpublished data) indicating that the two-celled prototypic eyes of Polycelis auricularia (fig. 1) arise by differential cell division from a common precursor cell. The gene that serves as a pigment cell determinant was first identified in mice and designated as microphthalmia transcription factor (Mitf) (Hertwig 1942). Mutations in this gene affect mainly eye size (microphthalmy) and pigment cells: The melanocytes of the skin, which are derived from the neural crest, and the retinal pigment epithelial cells of the eye, which are derived from the brain of the embryo. Mitf mutant mice are white and have reduced red eyes, because they lack both types pigment cells. The Mitf gene encodes a helix-loop-helix zipper transcription factor (Tachibana et al. 1992; Hodgkinson et al. 1993; Hughes et al. 1993). Several lines of evidence indicate that Mitf is a pigment cell determinant. Tachibana et al. (1992) were able to convert fibroblasts into melanocytes by stably transfecting NIH3T3 cells with human Mitf cDNA. Similarly, chicken neuroretinal cells were converted into melanocytes by transfecting with mouse Mitf cDNA (Planque et al. 1999). On the basis of several lines of evidence, Arnheiter (1998) postulated a common evolutionary origin of pigment cells and photoreceptor cells. We conclude from these considerations that the Darwinian eye prototype arose from a single cell by cellular differentiation, Pax6 determining the photoreceptor cell and Mitf the pigment cell, as outlined in figure 7.
FIG. 7.—
General scheme of eye evolution. The first step in eye evolution is the evolution of a light receptor molecule which in all metazoa is rhodopsin. In the most ancestral metazoa, the sponges, a single Pax gene, but no opsin gene has been found. In the cubozoan jellyfish Tripedalia, a unicellular photoreceptor has been described in the larva. The adult jellyfish has complex lens eyes which form under the control of PaxB, whereas the eyes of a hydrozoan jellyfish (Cladonema) are controlled by PaxA. We propose that from the unicellular photoreceptor cell the prototypic eye postulated by Darwin originated by a first step of cellular differentiation into photoreceptor cell and pigment cell controlled by Pax6 and Mitf, respectively. From this prototype, all the more complex eye types arose monophyletically. As a mechanism, we propose intercalary evolution of progressively more genes such as lens genes into the eye developmental pathway.
Single celled photoreceptors have indeed been described in the planula larva of the Scyphozoan Tripedalia which forms several types of lens eyes and slit eyes later in development of the medusa (Nordström et al. 2003; Parkefelt et al. 2005; Skogh et al. 2006). The larva possesses unicellular photoreceptors containing pigment granules, microvilli, which presumably contain rhodopsin, and a flagellum that allows it to be phototactic. It will be interesting to find out whether the microvilli indeed contain rhodopsin, which would indicate that these are rhabdomeric (i.e., microvillar) photoreceptors, whereas the photoreceptors of the adult jellyfish are of the ciliary type. The unicellular cellular photoreceptors of metazoa might eventually be traced back all the way to cyanobacteria (see Gehring 2005).
Positioning of the Eye on the Body Plan
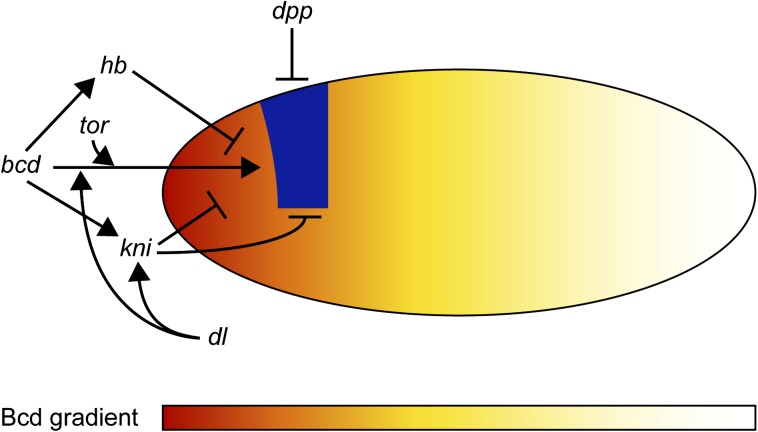
The eye developmental program has to be initiated by Pax6 at a specific position in the body plan. We have studied the positioning by analyzing the regulation of the twin of eyeless (toy) gene which is the first gene to be expressed zygotically in the embryonic primordia of the eye. As shown in figure 8, the initial expression domain of toy at the cellular blastoderm stage is cooperatively regulated by the three maternal patterning systems (the anterior, terminal, and dorsoventral systems) cooperatively with the zygotically active gap genes. Bicoid, Dorsal and Torso act synergistically as activators of toy, whereas Hunchback, Knirps, and Decapentaplegic function as repressors. The repressor acting from the posterior remains to be identified. Again, the positioning of the eye primordia is not deducible from the known function of these genes and therefore, unpredictable in the sense of Jacques Monod.
FIG. 8.—
Positioning of the eye in the head region in Drosophila. Proposed model for the onset of toy expression at cellular blastoderm. Arrows represent activating activities and bars repressing activities of the indicated genes. For details, see text (after Blanco and Gehring 2008).
Evolution of the Hox Gene Cluster and Specification of the Body Plan
Ordered Hox gene clusters are found in all the major superphyla, ecdysozoa, lophotrochozoa, and chordates (Gehring et al. 2009), which implies that they arose prior to the Cambrian “explosion” when all these taxa were already present. The Hox genes are arranged in the same order along the chromosome as they are expressed along the anteroposterior body axis as first found by E. B. Lewis (1978). This is a unique universal principle underlying animal development and it was designated as the “colinearity rule.” The genes specifying the dorsoventral axis are also conserved, but to a much smaller extent, taking an inversion of the dorsoventral body plan in chordates into account. Based on his studies of bithorax complex (BX-C) in Drosophila which includes the posterior half of the Hox gene complex only, Lewis considered the mesothoracic segment (T2) to be the ground state, because the deletion of all the genes of the BX-C leads to a transformation of all segments from T3 to A9 (the last abdominal segment) into T2 segments. T. C. Kaufman (Kaufman et al. 1980) has extended the idea of a homeotic gene cluster to the Antennapedia complex (ANTC) specifying the anterior thoracic and the posterior head segments. This raised the question of whether the ground state was the anterior most head segment or in T2. We define the developmental ground state genetically, by assuming that loss-of-function mutants lead to transformations toward the ground state, whereas gain-of-function mutants lead to homeotic transformations in the opposite direction away from the ground state. By this definition, T2 also represents the developmental ground state, if one includes the anterior genes from the ANTC.
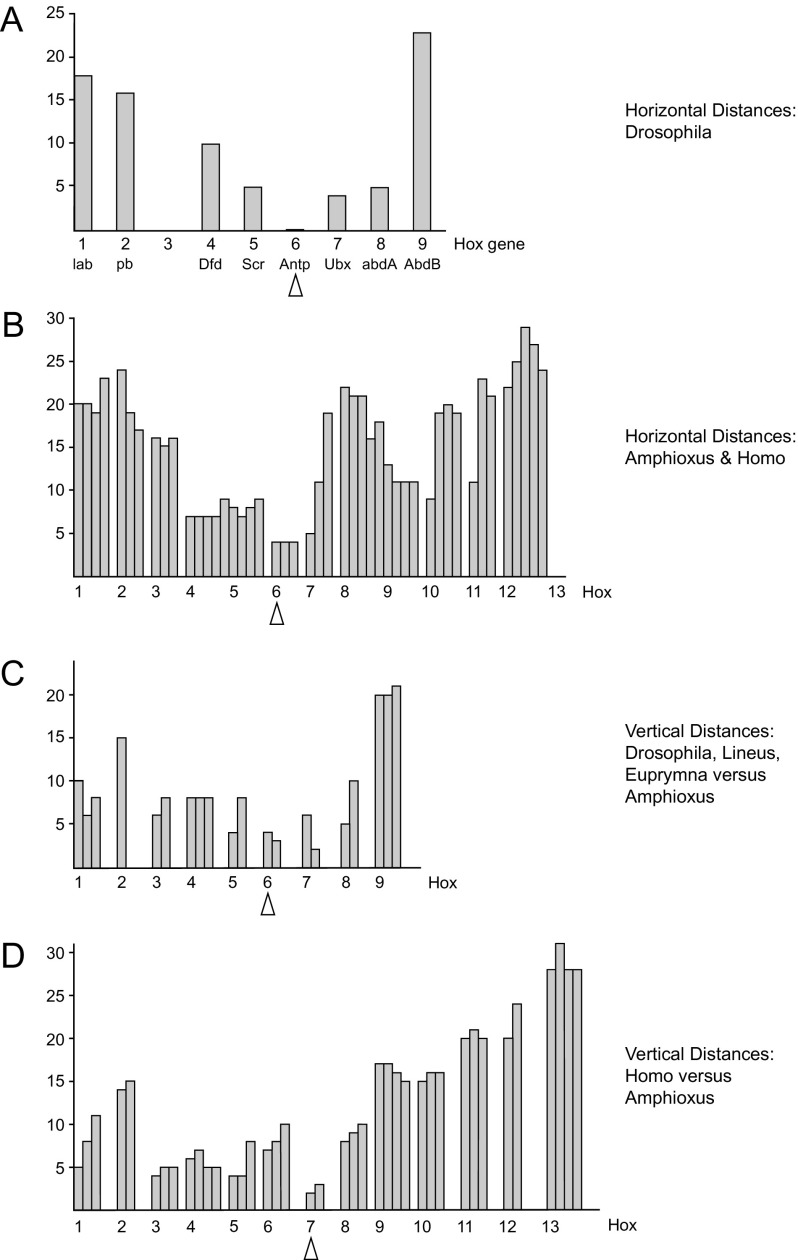
We have attempted to reconstruct the evolution of the Hox cluster on the basis of known genetic mechanisms. Starting from an Urhox gene (fig. 9A), a first unequal crossing-over presumably mediated by pairing of two copies of a transposon, flanking the Urhox gene on either side and in the same orientation leads to a gene duplication (and the respective deletion) (fig. 9B). This mechanism for generating gene duplications has been demonstrated experimentally for the white gene (Goldberg et al. 1983). The two genes are assumed to diverge and specify anterior versus posterior body parts (fig. 9C). The second crossover can occur upon displaced pairing between the anterior gene on one parental chromosome and the posterior gene on the other, yielding an anterior/posterior fusion gene, specifying the middle of the animal and resembling the Urhox gene more closely (fig. 9D). During the following rounds of unequal crossover, the outermost genes are not affected by crossover and therefore, they have the longest time to diverge; the new genes are added in the interior of the cluster leading to the ordered arrangement of the genes. This hypothesis can be tested by calculating the horizontal evolutionary distances (in terms of amino acid substitutions) between the homeoboxes (fig. 10A and B). The horizontal distances increase progressively from the center of the cluster, that is, Antennapedia (Hox6) in Drosophila and Hox6 in Amphioxus and the four human Hox clusters toward the anterior (Hox1) and the posterior end (Hox9 and Hox13, respectively). This is in line with the notion that Antp specifies the ground state. Therefore, we can assume that the developmental ground state also represents the evolutionary ground state.
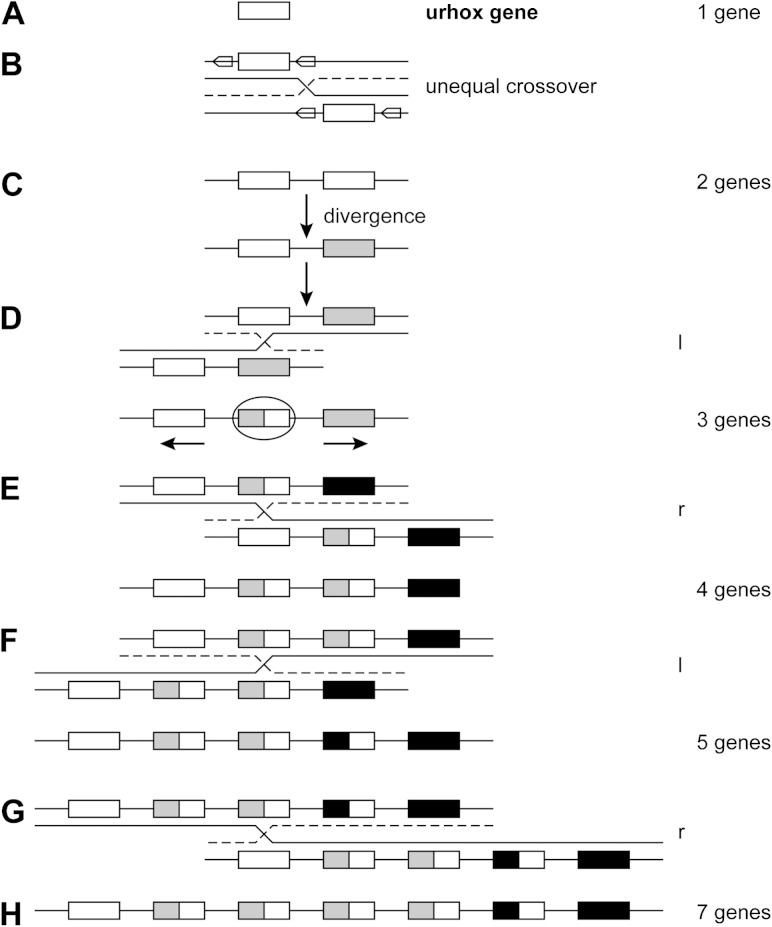
FIG. 9.—
Generation of the Hox gene cluster by unequal crossover (from Gehring et al. 2009). Generation of the Hox cluster by unequal crossover. (A) Urhox gene. (B) A transposon (arrow) flanking the Urhox gene on either side and in the same orientation allows for displaced chromosome pairing and unequal crossover generating a first gene duplication. (C) The two first Hox genes diverge into an anterior (white) and a posterior (gray) gene. (D) Displaced pairing between the duplicated genes generates a third gene which is a hybrid between the anterior and posterior genes (encircled) which resembles the original Urhox gene most closely. The outer genes are not affected by the unequal crossover and continue to diverge in the anterior and posterior direction (arrows), respectively, leading to an anterior (white) gene, an intermediate hybrid gene, and a posterior (black) gene. (E) The next unequal crossover leads to four genes. The pairing can be displaced either to the left (l) as in (D) or to the right as in (E). The probability for displacement to the left and to the right is the same. (F) The new genes are always added in the middle of the cluster, and the flanking anterior (white) and posterior (black) genes, which arose first during evolution, are not affected. They have the longest time to diverge. The intermediate genes in the middle of the cluster are homogenized by unequal crossover. Therefore, their sequences most closely resemble those of the Urhox gene, even though they arose later in evolution. (G) The chromosome pairing can also be displaced by two genes leading from five genes to seven genes in (H). The clusters of protostomes generally contain nine genes, whereas the chordates have 13 genes per cluster or 14 in the case of Amphioxus.
FIG. 10.—
Horizontal and vertical evolutionary distances between the Hox gene complexes of Drosophila, Lineus, Euprymna, Amphioxus, and the four human complexes. (A) Cumulative horizontal distances between the amino acid sequences of the eight homeodomains of Drosophila relative to Antennapedia: lab, Labial (Hox1); pb, Proboscipedia (Hox2); Dfd, Deformed (Hox4); Scr, Sex combs reduced (Hox5); Antp, Antennapedia (Hox6); Ubx, Ultrabithorax (Hox7); abdA, Abdominal-A (Hox8); and Abd-B, Abdominal-B (Hox9). There is no Hox3 homolog with a homeotic gene function in Drosophila, because zerknullt (zen and zen2) and bicoid (bcd) have diverged and serve a different function. The horizontal distances increase progressively from Antp to both the anterior and the posterior end, that is, Hox1 and Hox9, respectively. (B) Horizontal distances between the homeodomains of neighboring Hox genes of Amphioxus and the four human Hox complexes. The human clusters have undergone some gene losses, so that there are between three and five values per Hox gene. The low point is at Hox6 and the distances increase progressively toward both ends of Hox complex. (C) Vertical distances between the orthologous homeodomains of Drosophila, Lineus, and Euprymna relative to the Amphioxus sequences. The low point is at Hox6 (Antp) and the number of amino acid substitutions increases toward both ends of the Hox complex. (D) Vertical distances between the homeodomains of the chordate Hox genes relative to those of Amphioxus. The low point is located at Hox7 and the number of amino acid substitutions increases toward both ends Hox1/2 and Hox13. Abscissa: Hox gene number. Ordinate: number of amino acid substitutions.
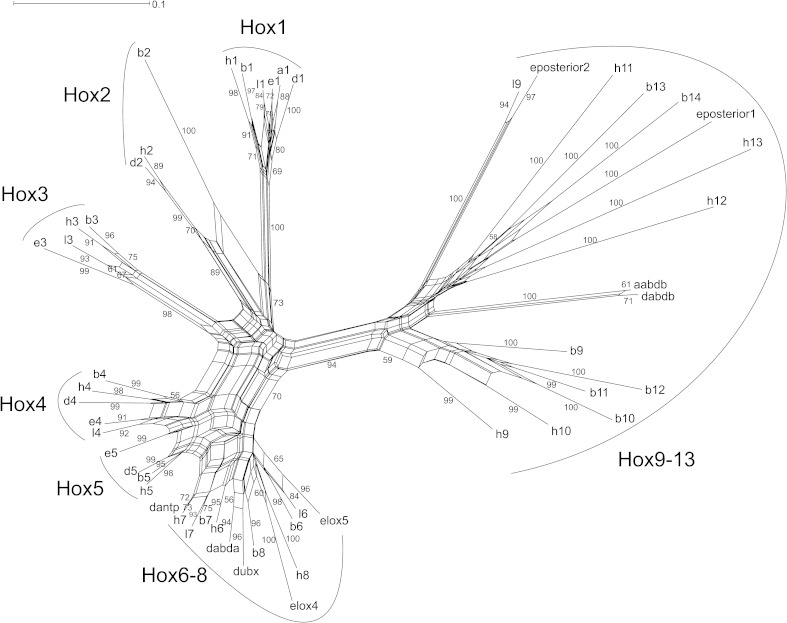
By using the split-tree program (Huson and Bryant 2006), we have reconstructed the phylogenetic network of the Hox genes from Drosophila (ecdysozoa), from Lineus and Euprymna (Lophotrochozoa), and from Amphioxus and Homo (chordata) (fig. 11). The anterior Hox genes (Hox1 to Hox5) clearly cluster together according to their position in the cluster. However, the intermediate genes (Hox6–8) are not resolved. The posterior genes (Hox9–13) have diverged to the largest extent, indicating that the first split has occurred between the anterior and the posterior genes as predicted by our model (fig. 9).
FIG. 11.—
Phylogenetic network of the Hox cluster genes using the split-tree program (Huson and Bryant 2006). The anterior (Hox1–8) and posterior Hox genes (Hox9–13) are clearly separated. Whereas the Hox1–5 genes are clearly defined according to their position in the cluster, the intermediate genes Hox6–8 are not resolved. This is in line with the assumption that intermediate Hox genes have arisen more recently in evolution and, therefore, have diverged the least.
Evolution of the Segmental Body Plan in Arthropods
In tracing back the evolution of the body plan of insects back to their putative ancestors, we have to assume that these were homonomously segmented arthropods, presumably crustaceans with a uniform series of segments with a pair of legs on each segment. In apterygot insects, the rudiments of these legs are still visible. In insects, the legs were removed from the abdomen. Deletion of the posteriormost Hox gene Abdominal B (AbdB) in genital disc cells leads to homeotic transformation of the genital disc structures into legs, that is, reversion to the prototypic T2 segment. Similarly, proboscipedia loss-of-function mutants lead to formation of legs on the proboscis. This indicates that the prototypic thoracic segment with a pair of legs was modified in the anterior direction to form the mouthparts and posteriorly to remove the legs from the abdomen and to form the genitalia. In addition, new structures evolved on the thorax, the wings. In fossil Paleodictyoptera, there are three pairs of wings, the prothoracic winglets being smaller, and full-sized wings on the meso- and metathorax. The latter were retained in most insects, with the exception of diptera which have reduced the metathoric pair of wings converting them into halteres through the action of the Ultrabithorax (Ubx) gene (=Hox7). The anterior and posterior most segments are specified by homeobox-containing genes located outside of the hox cluster; orthodenticle (otd) and empty spiracles (ems) in the anterior head segments and caudal (cad) in the tail. These genes and their spatial expression patterns are also conserved in mammals (Reichert and Simeone 1999).
Onychophora are among the closest ancestors of the arthropods. They are also segmented with one pair of legs per segment, but the process of cephalization has not proceeded as far as in insects in which the brain consists of three neuromeres, the proto-, deuto-, and tritocerebrum. In Onychophora, only two neuromeres are formed (Mayer et al. 2010). Today, Onychophora are represented only by a small number of species, but among the earliest fossils, there are some interesting records (see below). It is worth noting that in Amphioxus, a primitive chordate, cephalization has hardly begun and head and brain evolution starts only in vertebrates.
The Prototypic Body Segment and Eye Evolution in Early Cambrian Fossils
We are used to finding the eyes on the head of an animal, but there are notable exceptions to this rule. For example, in scallops and ark shells, the eyes are located at the edge of the mantle. In annelids, the localization of the eyes varies greatly; in the tubeworm Branchiomma, one eye is located on each of the tentacle gills. Amphiglena mediterranea has three eyes on either side of the brain and others at the end of the tail (Plate 1924).
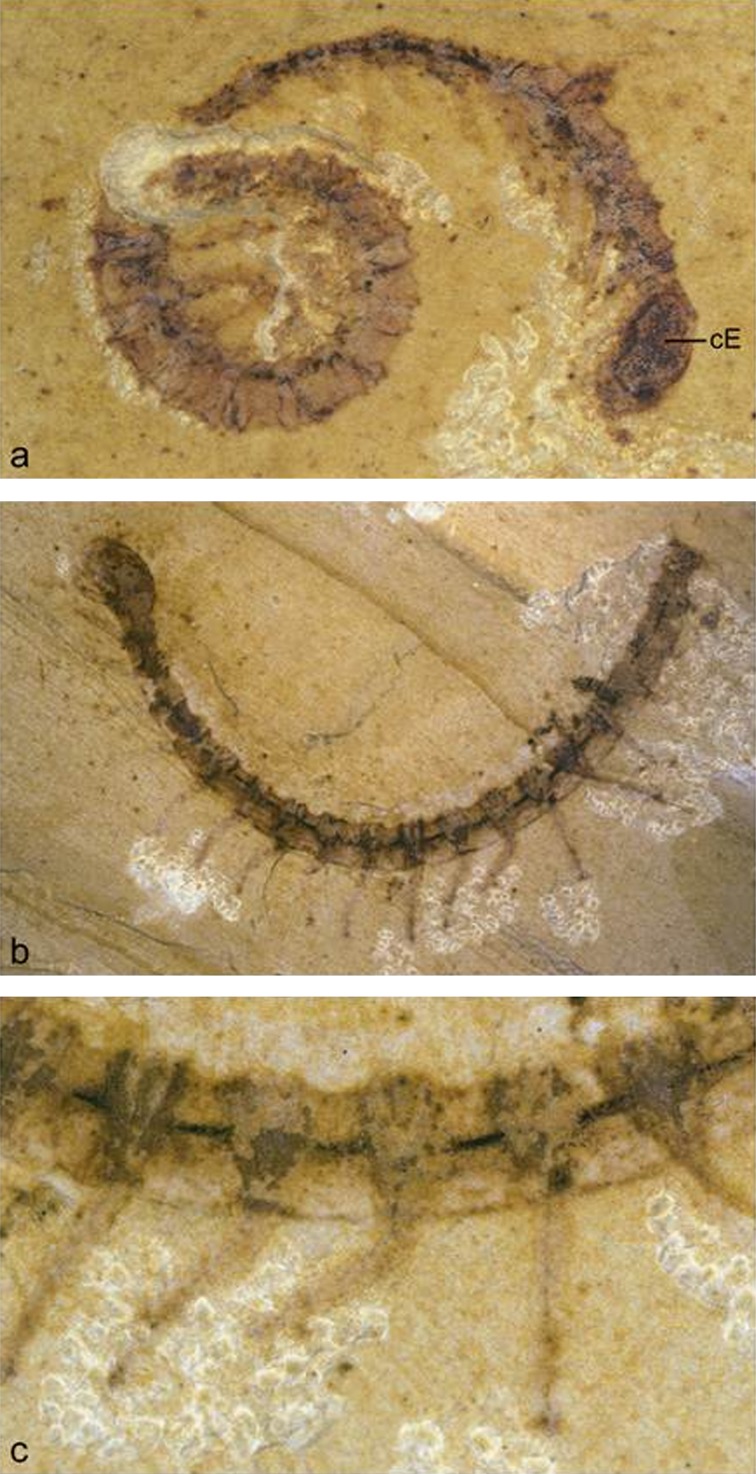
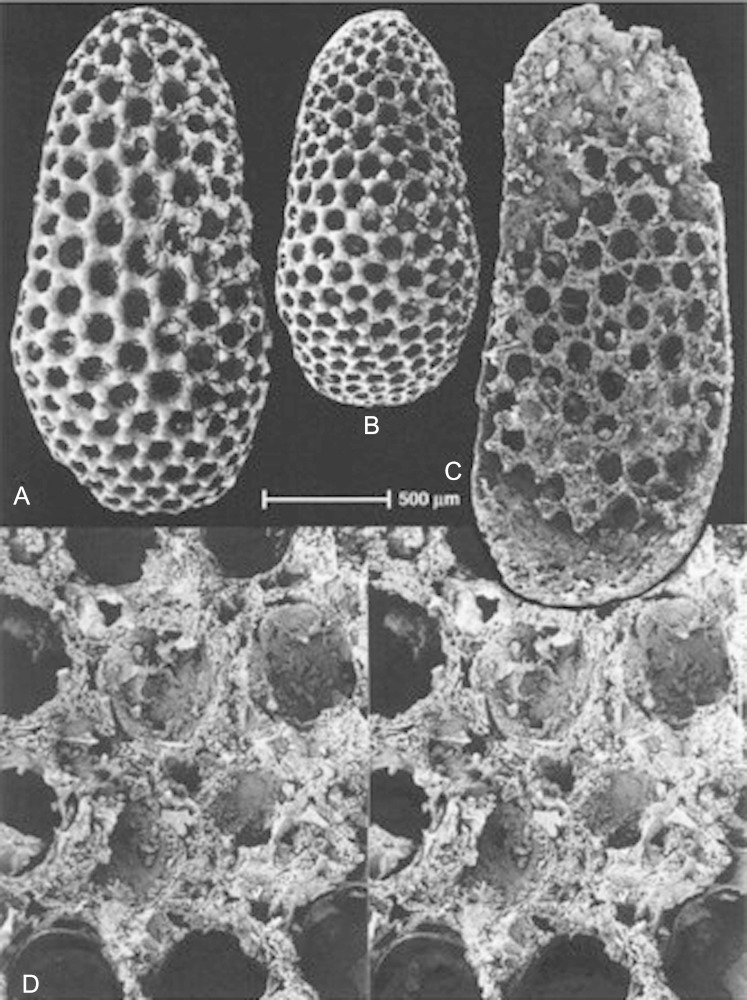
The fossil record shows that the eye evolved already during the precambrium and cambrian trilobites possess highly structured compound eyes with calcite lenses. Most recently, exceptionally well-preserved fossil eyes have been reported from the early Cambrium (ca. 515 Ma) from Australia, indicating that some of the earliest arthropods possessed highly advanced compound eyes, with over 3,000 ommatidial lenses. These were presumably cuticular (nonbiomineralized) lenses (Lee et al. 2011). Another site which has been a gold mine for palaeontologists is Chengjiang, China. Its fossil fauna have been described in Hou et al. (2007), giving a survey of the Cambrian fossils found so far on this site. Of particular interest, with respect to arthropod and eye evolution, are the Lobopodia. These fossil animals are related to the recent Onychophora, of which Peripatus is the best-known representative. Peripatus is an annulated, caterpillar-like terrestrial animal with 14–43 pairs of legs, depending on the species, with paired segmental secretory organs, a pair of antennae, an annelid-like eye behind each antenna, and a tracheal system for respiration, linking them to the arthropods (Clarkson 1998). Lobopodia were small marine animals with a worm-like body consisting of soft tissue, a lightly sclerotized cuticle, and uniramous leg-like appendages. Some fossil forms have isolated, partially mineralized plates in the trunk region. They are known almost exclusively from Cambrian rocks, the only others being from Carboniferous and Tertiary deposits (Hou et al. 2007). In going through the Chengjiang collection represented in Hou et al. (2007), I was struck by the fact that Cardiodictyon catenulum appears to have a compound eye with many ommatidia on the head (fig. 12), whereas Microdictyon sinicum seems to have a pair of these compound eyes on every annulus (segment) above each of the nine pairs of legs (fig. 13). There has been some debate about the nature of these “net-like sclerites” and whether they are indeed compound eyes. Dzik (2003), on the basis of the “sclerites” he found in Kazachstan, considers them likely to be compound eyes and possibly homologous to arthropod eyes. After microscopic inspection of these fossils shown in figure 14, I convinced myself that these “sclerites” are in fact compound eyes with hexagonally structured ommatidia that strikingly resemble those of modern insects.
FIG. 12.—
Cardiodictyon catenulum, a Cambrian Lobopodian fossil from Chengjiang (China) with a compound eye on the head (reproduced with permission from Hou et al. 2007). (a) Lateral view cE, compound eye (5.3×), (b) lateral view, and (c) detail of trunk region.
FIG. 13.—

Microdictyon sinicum, a Cambrian Lobopodian fossil from Chengjiang (China) with compound eye on every annulus (segment) above every leg. Top: reconstruction; bottom, lateral view (3.4×) (reproduced with permission from Hou et al. 2007).
FIG. 14.—
Sclerites from Microdictyon effusum Bengtson et al. in Missarzhevsky and Mambetov (1981) from Malyi Karatan, Kazakhstan, early Cambrian. (A–C) Sclerites showing a hexagonal structure like that of ommatidia in compound eyes of arthropods. (D) An exuviated sclerite showing remnants of a few lenticular bodies (from Dzik 2003).
Because in Microdictyon a pair of compound eyes is found in every segment, the prototype segment may have been endowed with a pair of walking legs and a pair of visual sense organs. As mentioned above, we are used to finding the eyes on the head, but it should be remembered that spiders, for example, have a pair of ears associated with each pair of legs. Microdictyon sinicum may represent an early stage of evolution, when all of the segments formed a pair of legs and a pair of eyes. Subsequently, cephalization took place, and the eyes were retained in the head segment only, as in C. catenulum. The study of eye evolution keeps raising the most interesting perspectives.
Conclusions
The relatively simple prototypic eye postulated by Darwin has been found in flat worms and in many trochophora larvae. The discovery of Pax6 as a master control gene for eye development strongly supports the idea that the various eye types originated monophyletically from such a prototype. What is still surprising is the rapidity of eye evolution, because compound eyes with over 3,000 ommatidia were discovered in the early Cambrium, some 515 Ma, in early arthropods. The analysis of the complex gene regulatory networks specifying eye development supports the unpredictability thesis of Monod.
The phylogenetic analysis of the Hox gene cluster and the colinearity rule led to the concept of an evolutionary and developmental ground state which specifies a prototypic body segment. In the course of evolution, a series of similar prototypic body segments is converted in a stepwise manner into anterior head and posterior abdominal segments. This cephalization and caudalization result from the progressive divergence of the Hox genes and is found not only in arthropod evolution but also in the evolution of chordates, in which the most primitive amphioxus shows very little cephalization and caudalization, as compared with vertebrates, in which cephalization plays a particularly important role. In insects, the anterior walking legs were converted into antennae and mouthparts, whereas the posterior walking legs are either removed from the abdomen or converted into genitalia. Upon deletion of the anterior or posterior Hox genes, the respective segments form legs again, turning the wheel of evolution backwards.
Acknowledgments
This work was supported by a grant from the Swiss National Science Foundation and the Kantons of Basel-Stadt and Basel-Landschaft, for which we are grateful.
References
- Arnheiter H. Evolutionary biology. Eyes viewed from the skin. Nature. 1998;391:632–633. doi: 10.1038/35487. [DOI] [PubMed] [Google Scholar]
- Blanco J, Gehring WJ. Analysis of twin of eyeless during early embryogenesis in Drosophila melanogaster. Gene Expr Patterns. 2008;8:523–527. doi: 10.1016/j.gep.2008.06.002. [DOI] [PubMed] [Google Scholar]
- Blanco J, Girard F, Kamachi Y, Kondoh H, Gehring WJ. Functional analysis of the chicken δ1-crystallin enhancer activity in Drosophila reveals remarkable evolutionary conservation between chicken and fly. Development. 2005;132:1895–1905. doi: 10.1242/dev.01738. [DOI] [PubMed] [Google Scholar]
- Clarkson ENK. Invertebrate palaeontology and evolution. 4th ed. Oxford: Blackwell; 1998. [Google Scholar]
- Czerny T, et al. Twin of eyeless, a second Pax-6 gene of Drosophila, acts upstream of eyeless in the control of eye development. Mol Cell. 1999;3:297–307. doi: 10.1016/s1097-2765(00)80457-8. [DOI] [PubMed] [Google Scholar]
- Darwin CR. The origin of species by means of natural selection, or the preservation of favoured races in the struggle for life. 6th ed. London: John Murray; 1872. [PMC free article] [PubMed] [Google Scholar]
- Dzik J. Early Cambrian lobopodian sclerites and associated fossils from Kazakhstan. Palaeontology. 2003;46:93–112. [Google Scholar]
- Gehring WJ. New perspectives on eye development and the evolution of eyes and photoreceptors. J Hered. 2005;96:171–184. doi: 10.1093/jhered/esi027. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Ikeo K. Pax6: Mastering eye morphogenesis and eye evolution. Trends Genet. 1999;15:371–377. doi: 10.1016/s0168-9525(99)01776-x. [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Kloter U, Suga H. Evolution of the Hox gene complex from an evolutionary ground state. Curr Topics Develop Biol. 2009;88:35–61. doi: 10.1016/S0070-2153(09)88002-2. [DOI] [PubMed] [Google Scholar]
- Goldberg ML, Sheen JY, Gehring WJ, Green MM. Unequal crossing-over associated with asymmetrical synapsis between nomadic elements in the Drosophila melanogaster genome. Proc Natl Acad Sci U S A. 1983;80:5017–5021. doi: 10.1073/pnas.80.16.5017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziussi DF, Suga H, Schmid V, Gehring WJ. Eyes absent in the eye bearing hydrozoan jelly fish Cladonema radiatum: conservation of the retinal determination network (submitted) 2011 doi: 10.1002/jez.b.22442. [DOI] [PubMed] [Google Scholar]
- Halder G, Callaerts P, Gehring WJ. Induction of ectopic eyes by targeted expression of the eyeless gene in Drosophila. Science. 1995;267:1788–1792. doi: 10.1126/science.7892602. [DOI] [PubMed] [Google Scholar]
- Hertwig P. Neue Mutationen und Kopplungsgruppen bei der Hausmaus. Z Indukt Abstammungs Vererbungslehre. 1942;80:220–246. [Google Scholar]
- Hesse R. Untersuchungen über die Organe der Lichtempfindung bei niederen Tieren. II. Die Augen der Plathelminthen. Z Wiss Zool. 1897;62:527–582. [Google Scholar]
- Hill RE, et al. Mouse small eye result from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Hodgkinson CA, et al. Mutations at the mouse microphthalmia locus are associated with defects in a gene encoding a novel basic helix-loop-helix zipper protein. Cell. 1993;74:395–404. doi: 10.1016/0092-8674(93)90429-t. [DOI] [PubMed] [Google Scholar]
- Hou XG, et al. The Cambrian fossils of Chengjiang China, the flowering of early animal life. Oxford: Blackwell; 2007. [Google Scholar]
- Hughes MJ, Lingrel JB, Krakowsky JM, Anderson KP. A helix-loop-helix transcription factor-like gene is located at the mi locus. J Biol Chem. 1993;268:20687–20690. [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jacobs DK, et al. The history of development through the evolution of molecules: gene trees, hearts, eyes and dorsoventral inversion. In: De Salle R, Schierwater B, editors. Molecular approaches to ecology and evolution. Basel (Switzerland): Birkhäuser; 1998. pp. 323–357. [Google Scholar]
- Kaufman TC, Lewis R, Wakimoto B. Cytogenetic analysis of chromosome 3 in Drosophila melanogaster: the homoeotic gene complex in polytene chromosome interval 84A-B. Genetics. 1980;94:115–133. doi: 10.1093/genetics/94.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozmik Z, et al. Role of Pax genes in eye evolution: A cnidarian Pax B gene uniting Pax2 and Pax6 functions. Developmental Cell. 2003;5:773–785. doi: 10.1016/s1534-5807(03)00325-3. [DOI] [PubMed] [Google Scholar]
- Lee MS, et al. Modern optics in exceptionally preserved eyes of early Cambrian arthropods from Australia. Nature. 2011;474:631–634. doi: 10.1038/nature10097. [DOI] [PubMed] [Google Scholar]
- Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276:565–570. doi: 10.1038/276565a0. [DOI] [PubMed] [Google Scholar]
- Mayer G, Whitington PM, Sunnucks P, Pflüger HJ. A revision of brain composition in Onychophora (velvet worms) suggests that the tritocerebrum evolved in arthropods. BMC Evol Biol. 2010;10:255–264. doi: 10.1186/1471-2148-10-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaut L, et al. Analysis of the eye developmental pathway in Drosophila using DNA microarrays. Proc Natl Acad Sci U S A. 2003;100:4024–4029. doi: 10.1073/pnas.0630561100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missarzhevsky VV, Mabetow AM. Stratigrafia i fauna pogranichnych sloiev kembria i dokembria Malogo Karatau. Trudy Geologicheskogo Instituta AN SSSR. 1981;326:1–91. [Google Scholar]
- Monod J. Le hazard et la nécessité. Paris: Edition du Seuil; 1970. [Google Scholar]
- Niimi T, Seimiya M, Kloter U, Flister S, Gehring WJ. Direct regulatory interaction of the eyeless protein with an eye-specific enhancer in the sine oculis gene during eye induction in Drosophila. Development. 1999;126:2253–2260. doi: 10.1242/dev.126.10.2253. [DOI] [PubMed] [Google Scholar]
- Nordström K, Wallen R, Seymour J, Nilsson D. A simple visual system without neurons in jellyfish larvae. Proc R Soc Lond B Biol Sci. 2003;270:2349–2354. doi: 10.1098/rspb.2003.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Kane CJ, Gehring WJ. Detection in situ of genomic regulatory elements in Drosophila. Proc Natl Acad Sci U S A. 1987;84:9123–9127. doi: 10.1073/pnas.84.24.9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkefelt L, Skogh C, Nilsson DE, Ekström P. Bilateral symmetric organization of neural elements in the visual system of a coelenterate, Tripedalia cystophora (Cubozoa) J Comp Neurol. 2005;492:251–262. doi: 10.1002/cne.20658. [DOI] [PubMed] [Google Scholar]
- Planque N, et al. Expression of the microphthalmia-associated basic helix-loop-helix leucine zipper transcription factor Mi in avian neuroretina cells induces a pigmented phenotype. Cell Growth Differ. 1999;10:525–536. [PubMed] [Google Scholar]
- Plate L. Allgemeine Zoologie und Abstammungslehre. Zweiter Teil: Die Sinnesorgane der Tiere. Jena (Germany): Gustav Fischer Verlag; 1924. p. 444. [Google Scholar]
- Quiring R, Walldorf U, Kloter U, Gehring WJ. Homology of the eyeless gene of Drosophila to the small eye gene in mice and Aniridia in humans. Science. 1994;265:785–789. doi: 10.1126/science.7914031. [DOI] [PubMed] [Google Scholar]
- Reichert H, Simeone A. Conserved usage of gap and homeotic genes in patterning the CNS. Curr Opin Neurobiol. 1999;9:589–595. doi: 10.1016/S0959-4388(99)00002-1. [DOI] [PubMed] [Google Scholar]
- Salvini-Plawen L, Mayr E. On the evolution of photoreceptors and eyes. New York: Plenum; 1977. [Google Scholar]
- Skogh C, Garm A, Nilsson DE, Ekstörm P. Bilaterally symmetrical rhopalial nervous system of the box jellyfish Tripedalia cystophora. J Morphol. 2006;267:1391–1405. doi: 10.1002/jmor.10472. [DOI] [PubMed] [Google Scholar]
- Suga H, et al. Flexibly deployed Pax genes in eye development at the early evolution of animals demonstrated by studies on a hydrozoan jelly fish. Proc Natl Acad Sci U S A. 2010;107:14263–14268. doi: 10.1073/pnas.1008389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachibana M, et al. Cochlear disorder associated with melanocyte anomaly in mice with a transgenic insertional mutation. Mol Cell Neurosci. 1992;3:433–445. doi: 10.1016/1044-7431(92)90055-7. [DOI] [PubMed] [Google Scholar]
- Takeda H, Nishimura K, Agata K. Planarians change their body size by maintaining a constant ratio of different cell types using stem cell system. Zool Sci. 2009;26:805–813. doi: 10.2108/zsj.26.805. [DOI] [PubMed] [Google Scholar]
- Tomarev SI, et al. Squid Pax-6 and eye development. Proc Natl Acad Sci U S A. 1997;94:2421–2426. doi: 10.1073/pnas.94.6.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther C, Gruss P. Pax6, a murine paired box gene, is expressed in the developing CNS. Development. 1991;113:1435–1449. doi: 10.1242/dev.113.4.1435. [DOI] [PubMed] [Google Scholar]