Abstract
Tight junctions are intercellular junctions localized at the most apical end of the lateral plasma membrane. They consist of four kinds of transmembrane proteins (occludin, claudins, junctional adhesion molecules, and tricellulin) and huge numbers of scaffolding proteins and contribute to the paracellular barrier and fence function. The mutation and deletion of these proteins impair the functions of tight junctions and cause various human diseases. In this paper, we provide an overview of recent studies on transmembrane proteins of tight junctions and highlight the functional significance of tight junctions, extracellular matrix, and nuclear receptors in epithelial differentiation.
1. Introduction
The epithelial tissue in various organs (e.g., lungs, intestines, and skin) is composed of a seat of epithelial cells that separate the biological compartments in the body with different internal environments. The intercellular adhesion complex between epithelial cells consists of tight junctions, adherens junctions, and desmosomes and is fundamental to the construction of the epithelial cell seat and the maintenance of cellular polarity [1, 2].
Within the intercellular adhesion complex, tight junctions possess two distinct functions in the epithelium tissue (Figure 1(a)). They function as a barrier controlling molecular penetration of ions, solutes, water, and cells through intercellular space and act as a fence dividing apical and basolateral domains to compartmentalize the plasma membrane [3]. These characteristics of tight junctions allow epithelium to prevent pathogens and foreign substances from invading and to facilitate directional exchange of materials.
Figure 1.
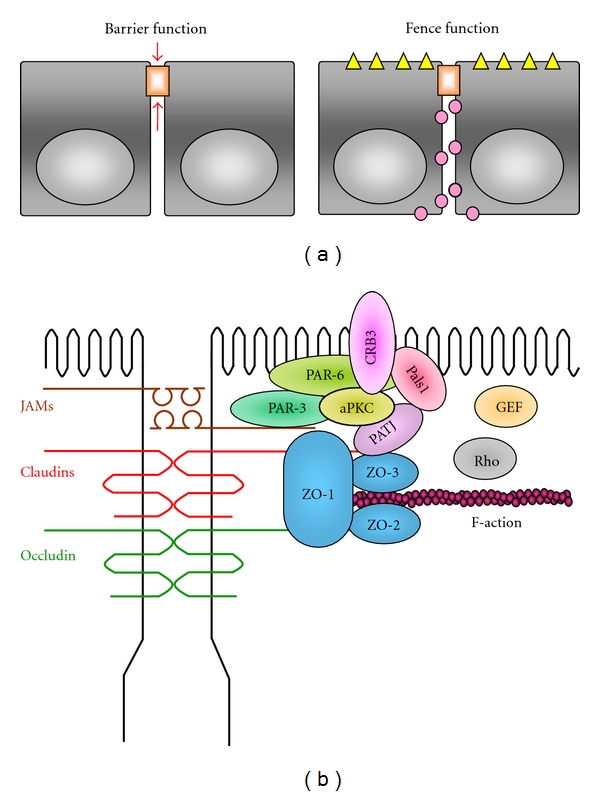
Functions of tight junctions and molecular components. (a) Functions of tight junctions. Barrier function: tight junctions limit the penetration of intercellular material and impose selective permeability. Fence function: tight junctions restrict the diffusion of proteins and lipids in a cell membrane. (b) Schematic drawing of bicellular junction proteins. Transmembrane and scaffold proteins of tight junctions and polarity proteins are presented. These drawings are modified from a previously published review [4].
Tight junctions comprised 4 kinds of transmembrane proteins: occludin, claudins, junctional adhesion molecules (JAMs), and tricellulin as well as numerous cytosolic proteins (Figure 1(b)). The cytosolic proteins are roughly divided into two groups depending on the presence or absence of PDZ (PSD-95, Dlg, and ZO-1) domains: the PDZ proteins (ZO-1, -2, -3, Par-3, -6, and membrane-associated guanylase kinase protein [MAGI]-1, -2, -3, etc.) and the non-PDZ proteins (cingulin, heteromeric G proteins, atypical PKC [aPKC], rab-3b, -13, PTEN, etc.) [5, 6]. Thus, a growing body of studies has clarified the molecular components of tight junctions, but it is still obscure how they accumulate and form tight junctions.
The intercellular adhesion complex and cell polarity should be established during differentiation of stem cells into epithelial cells. In this paper, we focus on transmembrane proteins of tight junctions and highlight the participation of tight junctions, extracellular matrix, and nuclear receptors in epithelial differentiation.
2. Transmembrane Proteins of Tight Junctions and Their Involvement in Epithelial Differentiation
2.1. Occludin
Occludin is a tetraspan membrane protein with two extracellular loops (EC1 and EC2), a short intracellular turn, and N- and C-terminal cytoplasmic domains [7]. Among these domains, the long C-terminal domain is phosphorylated at serine, threonine, and tyrosine residues by various protein kinases including src family kinase and CK2 [8, 9]. The C-terminal region of occludin also directly binds to ZO-1 [10], and the phosphorylation of some tyrosine residues prevents both the interaction with ZO-1 and the assembly at tight junctions [11]. In addition, the phosphorylation of some threonine and serine residues enhances occludin trafficking to tight junctions and the barrier function [9]. Although there are four splicing variants in occludin [12], the biological significance of each variant in tight junctions is unclear.
Overexpression of full-length and mutated occludin gene in Madin-Darby canine kidney cells or Xenopus cells [13] suggests that occludin contributes to the barrier function of tight junctions. By contrast, occludin-deficient embryonic stem cells differentiate into polarized epithelial cells with well-developed tight junctions [14]. Moreover, occludin-null mice are born with normal structure and barrier function of tight junctions in the intestinal epithelial tissue [15], but they exhibit various phenotypes, such as growth retardation, hyperplasia of gastric mucosa, absence of cytoplasmic granules in striated duct cells of the salivary gland, thinning of the compact bone, brain calcification, and testicular atrophy. These phenotypes imply that occludin may be involved in neither epithelial differentiation nor the barrier function but in other unknown roles.
Occludin interacts with a variety of cellular signaling molecules and may contribute to the signal transduction. For example, occludin binds to transforming growth factor [TGF]-β type I receptor and regulates TGF-β-dependent disassembly of tight junctions during epithelial-to-mesenchymal transition [16]. It also associates with nonreceptor tyrosine kinase c-Yes, aPKC, PI-3 kinase, and protein phosphatases 2A [6, 17]. Furthermore, occludin regulates the organization of actin and the directional migration in epithelial cells [18]. In addition, it is reported that occludin is concerned with apoptosis via activation of the small GTPase RhoA, mitogen-activated protein kinase (MAPK), and Akt signaling pathways [19, 20].
2.2. Claudins
The claudin family consists of 24 distinct members in human and mice with three others recently identified [4, 21, 22]. They are 18- to 27-kDa tetraspan proteins with N- and C-terminal cytoplasmic domains and two extracellular loop domains, and are expressed in tissue- and cell type-specific manners [3, 23]. In addition, isoforms of some claudins (e.g., claudin-10 and -18) are generated by alternative splicing and exhibit different expression patterns and functions [24, 25].
Claudin genes have few introns, and several pairs of them are located in close proximity in human and mouse genome. For example, in humans, Claudin-3 and Claudi-4 are located on chromosome 7, Claudin-6 and Claudin-9 on chromosome 16, Claudin-8 and Claudin-17 on chromosome 21, and Claudin-22 and Claudin-24 on chromosome 4 [26]. Gene duplication is assumed in these claudins, and the coordinated expression is reported at least for Claudin-3 and Claudin-4 genes [27].
Claudins are indeed the backbone of tight junctions, since they can reconstitute tight-junction strands even in fibroblasts [28]. All claudins except for claudin-12 possess a PDZ-binding motif and are capable of direct interaction with the proteins containing PDZ domain such as ZO-1, -2, -3, multi-PDZ domain protein (MUPP-1), and PALS-1 associated TJ protein via the cytoplasmic tail (PATJ) [29–32]. Hence, claudins are linked to the actin cytoskeleton through binding to the scaffolding proteins (ZO-1 and ZO-2), stabilizing tight junctions [33].
It should be noted that the posttranslational modification of claudins influences their localization and function. First, the phosphorylation of claudins is associated with tight-junctions assembly and paracellular permeability. For example, phosphorylation of claudin-3, -5, and -16 by protein kinase A facilitates tight-junctions assembly and functionality [34, 35]. Phosphorylation of claudin-1 and/or claudin-4 by MAPK and aPKC is also reported [36]. Second, palmitoylation of claudins at the dicysteine motif, which is conserved through the claudin family, is thought to increase claudin accumulation in tight junctions and enable their translocation to detergent-resistant plasma membranes (lipid rafts) [37]. Third, O-glycosylation in some claudins, claudin-1, -3, and -4 by O-linked N-acetylglucosamine transferase is predicted, but the function of this modification is unknown [38].
Physiological roles of claudins have been clarified from studies of transgenic and knockout mice or human diseases [39]. Mutations in claudin-1 gene cause neonatal ichthyosis and sclerosing cholangitis in humans, and claudin-1-deficient mice result in neonatal death due to the disturbance of skin permeability [40, 41]. Claudin-2 knockout mice show no obvious phenotypic changes (e.g., of growth and behavior) [42]. The deletion of claudin-2 gene causes the decrease in Na+ paracellular permeability [43]. Claudin-7-null mice die within 12 days of birth owing to severe salt wasting, chronic dehydration, and growth retardation, which are caused by the breakdown of NaCl homeostasis [44]. Claudin-11 (also known as oligodendrocyte-specific protein) KO mice exhibit slowed CNS nerve conduction, markedly hindlimb weakness, and sterility in males due to the absence of tight-junction strands in CNS myelin and between Sertoli cells [45]. Mutations of claudin-14 gene lead to autosomal recessive deafness in mice and humans, suggesting that claudin-14 is associated with the cation-restrictive barrier in outer hair cells of the cochlea in the ear [46, 47]. Claudin-15-deficient mice show uneventful birth and growth but manifest an enlarged upper small intestine due to increased proliferation of normal cryptic cells [48]. Moreover, in the adult small intestine of Claudin-15-deficient mice, luminal Na+ and K+ homeostasis is disturbed, and glucose absorption is significantly decreased [43]. Various mutations in claudin-16 gene are seen in patients of familial hypomagnesemia with hypercalciuria and nephrocalcinosis, an autosomal recessive disease with severe Mg2+ and Ca2+ wasting [49]. In fact, claudin-16 is expressed in epithelial cells of the thick ascending loop of Henle [50]. Taken together with the finding that mutations in claudin-19 gene also show similar phenotypes with abnormal Mg2+ reabsorption [51], it appears that claudin-16 and -19 modulate paracellular absorption of Mg2+ and Ca2+ in the kidney.
2.3. Junctional Adhesion Molecules (JAMs)
JAMs belong to the immunoglobulin (Ig) superfamily and are N-glycosylated transmembrane proteins of tight junctions [5, 52]. They consist of two extracellular Ig-like domains, a single transmembrane region, and a C-terminal cytoplasmic domain that ends in a canonical PDZ domain-binding site. JAMs are classified into two subgroups by their sequence and structure. JAM-A (also referred as JAM/JAM-1/106 antigen/F11R), JAM-B (also known as VE-JAM/mJAM-3/hJAM-2), and JAM-C (also known as mJAM-2/hJAM-3) have a class II PDZ-binding domain at C-terminal ends, which directly interact with proteins containing PDZ domains: ZO-1 [53], AF-6/afadin [54], MUPP1 [30], and PAR-3 [55]. On the other hand, coxsackie and adenovirus receptor (CAR), endothelial cell-selective adhesion molecule (ESAM), and JAM4 contain a class I PDZ domain-binding motif at their C-terminal ends, associate with Ligand-of-Numb protein X1 and MAGI-1 [56–59]. In addition, it has been reported that JAM-A, JAM-B, and JAM-C interact with integrins αLβ2 (LFA-1), α4β1 (VAL-4), and αMβ2 (Mac-1), respectively, via the extracellular domains [60, 61].
JAM-A contributes to the formation of tight junctions and cell polarity through homophilic binding and interaction with PAR-3/aPKC/PAR-6 complex in epithelial cells [62, 63]. JAM-B and JAM-C also associate with PAR-3 via PDZ-binding domain [52]. JAM-B and JAM-C are expressed in Sertoli cells and spermatids, and JAM-C is essential for the polarization of round spermatids [64]. By contrast, ESAM is observed in endothelial cells and platelets, and the deletion of ESAM gene increases VEGF-induced permeability in mice endothelial cells [65–67]. Interestingly, JAM-C is expressed in undifferentiated embryonic stem cells more abundantly than in differentiated cells [68]. However, JAM-C mutant mice do not show developmental defects. Further studies are required to elucidate the role of JAMs in epithelialization.
2.4. Tricellulin
Tricellulin, a tetraspan protein that concentrates at tricellular contacts of epithelial cells, was identified by using gene chip analysis to compare parental epithelial cells and cells undergoing EMT [69]. It is phosphorylated by CK1, and its expression is repressed by SNAI1, a zinc-finger type transcription factor that plays an important role in EMT [70]. In addition, tricellulin concentrates at tricellular tight junctions in cochlear vestibular epithelial cells and the recessive mutations of tricellulin gene cause nonsyndromic deafness [71]. Moreover, tricellulin is related to the epithelial barrier and organization of both bicellular and tricellular tight junctions. Interestingly, tricellulin is incorporated into claudin-based tight junctions independently of ZO-1 binding and is translocated from bicellular to tricellular tight junctions in the presence of occludin [72].
3. Possible Involvement of Claudins in Epithelial Differentiation
During embryonic development in mice, claudin-6 is first detected in the epithelial zone of embryonic bodies at least in part via the bone morphogenic protein-signaling pathway [73], and afterwards observed in several types of epithelial tissues such as the epiblast and hypoblast (to E7.5), the definitive endoderm (to E8.5), the entire gut, optic vesicles, and a small region of the forebrain (at E9.5) [74]. Although these results imply that claudin-6 is involved in epithelialization, the examined claudin-6-null mice show no abnormality [74]. Other claudins might compensate for the function of claudin-6.
The epithelial-mesenchymal transition (EMT) is a process during which epithelial cells convert to mesenchymal cell morphology. This occurs in normal developmental processes including mesoderm and neural-crest formation and in the invasive process in tumors of epithelial origin. EMT has two steps: loss of intercellular adhesion (adherens and tight junctions) and acquiring cell motility [75]. Snail directly binds to the promoter of E-cadherin, claudin-3, claudin-4, and claudin-7 and represses the expression of those genes [76], thereby inducing EMT. Therefore, not only E-cadherin, but also claudins are possibly related to EMT.
In the development of gut tube, endodermal cells differentiate into gut epithelial cells and form a lumen as the cells polarize. Claudin-15 associates with single lumen formation and forms ion-permeable pores from the study of knockdown of it using morpholino in zebrafish [77]. These findings also suggest that claudins directly relate to epithelial differentiation.
4. Involvement of Nuclear Receptors in Epithelial Differentiation
Retinoids have numerous biological effects on vertebrate development, differentiation, proliferation, and homeostasis through two types of nuclear receptors, retinoic acid receptors (RARs) and retinoid X receptors (RXRs) [78, 79]. Using a mouse F9 stem-cell line, we previously showed that various RXR/RAR heterodimers exerted both specific and redundant functions in endodermal (epithelium-like cells) differentiation [80]. In addition, we found that the RXRα/RARγ pair mediated the induced expression of tight-junctions molecules (occludin, claudin-6, and claudin-7) and the establishment of both polarized epithelial morphology and functional tight junctions (Table 1) [81]. It is also reported that retinoid receptors and the kinase IKK1 cooperatively regulate claudin-23 expression in keratinocytes and participate in the formation of the epidermal barrier (Table 1) [82].
Table 1.
Nuclear receptors induce the expression of claudins.
| Nuclear receptors | Induced claudin expression | Cells | References |
|---|---|---|---|
| Retinoic acid receptor | Cldn1, Cldn4 | Human nasal epithelium | [83] |
| Cldn3 | Human urothelium | [84] | |
| Cldn6, Cldn7 | Mouse F9 stem cell | [81] | |
| Cldn23 | Mouse epidermis | [82] | |
| Hepatocyte nuclear factor 4α (HNF4α) | Cldn6, Cldn7 | Mouse F9 stem cell | [85, 86] |
| Cldn1 | Mouse hepatocyte | [87] | |
| Androgen receptor | Cldn3 | Mouse Seltoli cell | [88] |
| Progesterone receptor | Cldn3, Cldn4 | Mouse amniotic epithelium | [89] |
| Corticoid receptor | Cldn3, Cldn4 | Gill epithelium | [90] |
| Vitamin D receptor | Cldn2, Cldn12 | Mouse intestinal Caco-2 cell | [91] |
Hepatocyte nuclear factor 4α (HNF4α), another member of the nuclear receptor superfamily, transcriptionally controls the expression of a large number of target genes involved in nutrient and drug metabolism, hematopoiesis, and blood coagulation [92–94]. It is initially detected in primitive endoderm and subsequently in visceral endoderm (VE) during early mouse development [95]. In adults, HNF4α is expressed in a variety of epithelial cells of the liver, kidney, intestine, pancreas, and stomach. HNF4α also participates in the differentiation of VE cells and hepatocytes [96, 97]. Interestingly, we demonstrated that HNF4α triggered the expression of tight-junction molecules (occludin, claudin-6, and claudin-7) and translocated tight junction proteins (ZO-1, -2, -3, JAM-B, and JAM-C) and cell polarity proteins (PAR-3, PAR6, aPKC, CRB3, Pals1, and PATJ) to tight junctions as well as the formation of functional tight-junctions and epithelial polarity [85, 86] (Figure 2 and Table 1). HNF4α also induced the expression of ezrin/radixin/moesin- (ERM-) binding phosphoprotein 50 and phosphorylation and apical concentration of ERM proteins [98] (Figure 2). Thus, HNF4α initiates the formation of tight-junctions and microvillus and induces differentiation in epithelial cells.
Figure 2.
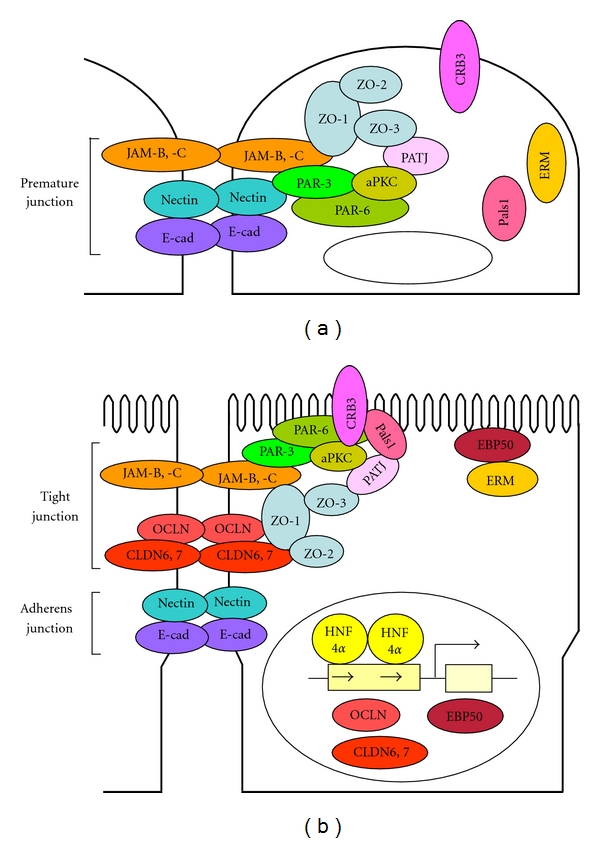
Models of HNF4α-triggered formation of tight junctions and microvilli in F9 cells. (a) In an undifferentiated state, several junctional proteins are accumulated to premature junctions. (b) HNF4α provokes the formation of junctional complexes and microvilli via induction of expression of occludin [OCLN], claudin-6 [CLDN6], claudin-7 [CLDN7], and ezrin/radixin/moesin-binding phosphoprotein 50 [EBP50].
Androgen and estrogen receptors induce the differentiation of prostate and mammary gland epithelium, respectively [99]. Several other nuclear receptors, including those of progesterone, corticoid, vitamin D, and PPARgamma, induce the expression of claudins in various cells (Table 1) [83, 84, 89–91]. Taken collectively, it is strongly suggested that nuclear receptors induce the expression of tight junctions and cell-polarity proteins and provoke epithelial differentiation with the formation of cell junctions and cell polarity.
5. Involvement of Extracellular Matrix in Epithelial Differentiation
Extracellular matrix (ECM, also known as basement membrane) is composed of various proteins, such as collagen, laminin, fibronectin, heparan sulfate proteoglycan, and nidogen, and these proteins are expressed in an organ- and development-dependent manner [100, 101]. In epithelial tissue, ECM functions to support epithelial cells and to stimulate their proliferation. ECM also seems to be essential for the differentiation of epithelial cells [102]. For instance, laminin and collagen type IV promote the differentiation of intestinal epithelial cells [103], and ECM functions to increase the ability of proliferation and differentiation of human renal cells and to maintain the differentiated epithelial cells for a long term [104].
Concerning the relationship between ECM and tight junctions, ECM proteins, especially fibronectin, increase the expression of claudin-18 and occludin along the plasma membrane in alveolar epithelial cells, and enhance the barrier function [105]. In turn, claudins recruit certain types of matrix metalloproteinases (MMPs, which serve as proteinases for ECM) onto cell surfaces and enhance the activation of MMP [106]. Thus, ECM and tight junctions influence the expression and function of each other's components in ways that suggest exciting avenues for further research.
6. Conclusion
Our knowledge of the molecular nature of tight junctions is still expanding. For instance, MarvelD3 has similar structure to occludin and tricellulin and is able to partially compensate for the deletion of those genes [107]. Bves/Pop1a, which belongs to the Popeye family Popdc, modulates RhoA and ZONAB/DbpA, a y-box transcription factor, and associates with tight-junctions proteins via ZO-1 in epithelial cells [4, 108–110]. In addition, a novel RhoGEF (p114RhoGEF) is identified to be involved in maturation of tight junctions via restricted activation of RhoA [111]. Thus, a variety of extracellular and intracellular proteins appear to participate in the formation and function of tight junctions in epithelial cells.
However, there remain several open questions in tight junctions. (1) Where are tight-junctions proteins assembled (e.g., cytoplasmic versus junctional assembly)? (2) How are they recruited into the junctions (e.g., do cargo proteins exit)? (3) What accounts for the selectivity of heteromeric or homomeric claudin-claudin interactions? (4) What is the functional relevance of posttranslational modification of tight-junctions proteins? In addition, it should also be determined how tight-junctions proteins regulate epithelial differentiation.
Acknowledgments
The authors are grateful to Dr. Kenneth Nollet in Fukushima Medical University School of Medicine for his critical reading of the paper. Part of this work was supported by Grants-in-Aid from Ministry of Education, Culture, Sports, Science, and Technology of Japan and the Naito Foundation and Grant-in-Aid for Young Scientists (B) in Japan.
References
- 1.Chen X, Gumbiner BM. Crosstalk between different adhesion molecules. Current Opinion in Cell Biology. 2006;18(5):572–578. doi: 10.1016/j.ceb.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 2.Matter K, Balda MS. Epithelial tight junctions, gene expression and nucleo-junctional interplay. Journal of Cell Science. 2007;120(9):1505–1511. doi: 10.1242/jcs.005975. [DOI] [PubMed] [Google Scholar]
- 3.Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nature Reviews Molecular Cell Biology. 2001;2(4):285–293. doi: 10.1038/35067088. [DOI] [PubMed] [Google Scholar]
- 4.Mineta K, Yamamoto Y, Yamazaki Y, et al. Predicted expansion of the claudin multigene family. FEBS Letters. 2011;585(4):606–612. doi: 10.1016/j.febslet.2011.01.028. [DOI] [PubMed] [Google Scholar]
- 5.Chiba H, Osanai M, Murata M, Kojima T, Sawada N. Transmembrane proteins of tight junctions. Biochimica et Biophysica Acta. 2008;1778(3):588–600. doi: 10.1016/j.bbamem.2007.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Matter K, Aijaz S, Tsapara A, Balda MS. Mammalian tight junctions in the regulation of epithelial differentiation and proliferation. Current Opinion in Cell Biology. 2005;17(5):453–458. doi: 10.1016/j.ceb.2005.08.003. [DOI] [PubMed] [Google Scholar]
- 7.Furuse M, Hirase T, Itoh M, et al. Occludin: a novel integral membrane protein localizing at tight junctions. Journal of Cell Biology. 1993;123(6):1777–1788. doi: 10.1083/jcb.123.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kale G, Naren AP, Sheth P, Rao RK. Tyrosine phosphorylation of occludin attenuates its interactions with ZO-1, ZO-2, and ZO-3. Biochemical and Biophysical Research Communications. 2003;302(2):324–329. doi: 10.1016/s0006-291x(03)00167-0. [DOI] [PubMed] [Google Scholar]
- 9.Raleigh DR, Boe DM, Yu D, et al. Occludin S408 phosphorylation regulates tight junction protein interactions and barrier function. Journal of Cell Biology. 2011;193(3):565–582. doi: 10.1083/jcb.201010065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuse M, Itoh M, Hirase T, et al. Direct association of occludin with ZO-1 and its possible involvement in the localization of occludin at tight junctions. Journal of Cell Biology. 1994;127(6 I):1617–1626. doi: 10.1083/jcb.127.6.1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elias BC, Suzuki T, Seth A, et al. Phosphorylation of Tyr-398 and Tyr-402 in occludin prevents Its interaction with ZO-1 and destabilizes Its assembly at the tight junctions. Journal of Biological Chemistry. 2009;284(3):1559–1569. doi: 10.1074/jbc.M804783200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mankertz J, Stefan Waller J, Hillenbrand B, et al. Gene expression of the tight junction protein occludin includes differential splicing and alternative promoter usage. Biochemical and Biophysical Research Communications. 2002;298(5):657–666. doi: 10.1016/s0006-291x(02)02487-7. [DOI] [PubMed] [Google Scholar]
- 13.Muresan Z, Paul DL, Goodenough DA. Occludin 1B, a variant of the tight junction protein occludin. Molecular Biology of the Cell. 2000;11(2):627–634. doi: 10.1091/mbc.11.2.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saitou M, Fujimoto K, Doi Y, et al. Occludin-deficient embryonic stem cells can differentiate into polarized epithelial cells bearing tight junctions. Journal of Cell Biology. 1998;141(2):397–408. doi: 10.1083/jcb.141.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitou M, Furuse M, Sasaki H, et al. Complex phenotype of mice lacking occludin, a component of tight junction strands. Molecular Biology of the Cell. 2000;11(12):4131–4142. doi: 10.1091/mbc.11.12.4131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barrios-Rodiles M, Brown KR, Ozdamar B, et al. High-throughput mapping of a dynamic signaling network in mammalian cells. Science. 2005;307(5715):1621–1625. doi: 10.1126/science.1105776. [DOI] [PubMed] [Google Scholar]
- 17.Jain S, et al. PKCzeta phosphorylates occludin and promotes assembly of epithelial tight junctions. Biochemical Journal. 2011;437(2):289–299. doi: 10.1042/BJ20110587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Du D, Xu F, Yu L, et al. The tight junction protein, occludin, regulates the directional migration of epithelial cells. Developmental Cell. 2010;18(1):52–63. doi: 10.1016/j.devcel.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Yu ASL, McCarthy KM, Francis SA, et al. Knockdown of occludin expression leads to diverse phenotypic alterations in epithelial cells. American Journal of Physiology. 2005;288(6):C1231–C1241. doi: 10.1152/ajpcell.00581.2004. [DOI] [PubMed] [Google Scholar]
- 20.Murata M, Kojima T, Yamamoto T, et al. Down-regulation of survival signaling through MAPK and Akt in occludin-deficient mouse hepatocytes in vitro. Experimental Cell Research. 2005;310(1):140–151. doi: 10.1016/j.yexcr.2005.07.017. [DOI] [PubMed] [Google Scholar]
- 21.Turksen K, Troy TC. Barriers built on claudins. Journal of Cell Science. 2004;117(12):2435–2447. doi: 10.1242/jcs.01235. [DOI] [PubMed] [Google Scholar]
- 22.Furuse M, Tsukita S. Claudins in occluding junctions of humans and flies. Trends in Cell Biology. 2006;16(4):181–188. doi: 10.1016/j.tcb.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 23.Furuse M, Fujita K, Hiiragi T, Fujimoto K, Tsukita S. Claudin-1 and -2: novel integral membrane proteins localizing at tight junctions with no sequence similarity to occludin. Journal of Cell Biology. 1998;141(7):1539–1550. doi: 10.1083/jcb.141.7.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.van Itallie CM, Rogan S, Yu A, Vidal LS, Holmes J, Anderson JM. Two splice variants of claudin-10 in the kidney create paracellular pores with different ion selectivities. American Journal of Physiology. 2006;291(6):F1288–F1299. doi: 10.1152/ajprenal.00138.2006. [DOI] [PubMed] [Google Scholar]
- 25.Niimi T, Nagashima K, Ward JM, et al. Claudin-18, a novel downstream target gene for the T/EBP/NKX2.1 homeodomain transcription factor, encodes lung- and stomach-specific isoforms through alternative splicing. Molecular and Cellular Biology. 2001;21(21):7380–7390. doi: 10.1128/MCB.21.21.7380-7390.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lal-Nag M, Morin PJ. The claudins. Genome biology. 2009;10(8):p. 235. doi: 10.1186/gb-2009-10-8-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hewitt KJ, Agarwal R, Morin PJ. The claudin gene family: expression in normal and neoplastic tissues. BMC Cancer. 2006;6, article 186 doi: 10.1186/1471-2407-6-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furuse M, Sasaki H, Fujimoto K, Tsukita S. A single gene product, claudin-1 or -2, reconstitutes tight junction strands and recruits occludin in fibroblasts. Journal of Cell Biology. 1998;143(2):391–401. doi: 10.1083/jcb.143.2.391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itoh M, Furuse M, Morita K, Kubota K, Saitou M, Tsukita S. Direct binding of three tight junction-associated MAGUKs, ZO-1, ZO-2, and ZO-3, with the COOH termini of claudins. Journal of Cell Biology. 1999;147(6):1351–1363. doi: 10.1083/jcb.147.6.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamazaki Y, Itoh M, Sasaki H, Furuse M, Tsukita S. Multi-PDZ domain protein 1 (MUPP1) is concentrated at tight junctions through its possible interaction with claudin-1 and junctional adhesion molecule. Journal of Biological Chemistry. 2002;277(1):455–461. doi: 10.1074/jbc.M109005200. [DOI] [PubMed] [Google Scholar]
- 31.Roh MH, Liu CJ, Laurinec S, Margolis B. The carboxyl terminus of zona occludens-3 binds and recruits a mammalian homologue of discs lost to tight junctions. Journal of Biological Chemistry. 2002;277(30):27501–27509. doi: 10.1074/jbc.M201177200. [DOI] [PubMed] [Google Scholar]
- 32.van Itallie CM, Anderson JM. Claudins and epithelial paracellular transport. Annual Review of Physiology. 2006;68:403–429. doi: 10.1146/annurev.physiol.68.040104.131404. [DOI] [PubMed] [Google Scholar]
- 33.Umeda K, Ikenouchi J, Katahira-Tayama S, et al. ZO-1 and ZO-2 independently determine where claudins are polymerized in tight-junction strand formation. Cell. 2006;126(4):741–754. doi: 10.1016/j.cell.2006.06.043. [DOI] [PubMed] [Google Scholar]
- 34.D'Souza T, Agarwal R, Morin PJ. Phosphorylation of Claudin-3 at threonine 192 by cAMP-dependent protein kinase regulates tight junction barrier function in ovarian cancer cells. Journal of Biological Chemistry. 2005;280(28):26233–26240. doi: 10.1074/jbc.M502003200. [DOI] [PubMed] [Google Scholar]
- 35.Ikari A, Matsumoto S, Harada H, et al. Phosphorylation of paracellin-1 at Ser217 by protein kinase A is essential for localization in tight junctions. Journal of Cell Science. 2006;119(9):1781–1789. doi: 10.1242/jcs.02901. [DOI] [PubMed] [Google Scholar]
- 36.Fujibe M, Chiba H, Kojima T, et al. Thr203 of claudin-1, a putative phosphorylation site for MAP kinase, is required to promote the barrier function of tight junctions. Experimental Cell Research. 2004;295(1):36–47. doi: 10.1016/j.yexcr.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 37.Nusrat A, Parkos CA, Verkade P, et al. Tight junctions are membrane microdomains. Journal of Cell Science. 2000;113(10):1771–1781. doi: 10.1242/jcs.113.10.1771. [DOI] [PubMed] [Google Scholar]
- 38.Butt AM, et al. Role of post translational modifications and novel crosstalk between phosphorylation and O-beta-GlcNAc modifications in human claudin-1, -3 and -4. doi: 10.1007/s11033-011-0870-7. Molecular Biology Reports. In press. [DOI] [PubMed] [Google Scholar]
- 39.Findley MK, Koval M. Regulation and roles for claudin-family tight junction proteins. IUBMB Life. 2009;61(4):431–437. doi: 10.1002/iub.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hadj-Rabia S, Baala L, Vabres P, et al. Claudin-1 gene mutations in neonatal sclerosing cholangitis associated with ichthyosis: a tight junction disease. Gastroenterology. 2004;127(5):1386–1390. doi: 10.1053/j.gastro.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 41.Furuse M, Hata M, Furuse K, et al. Claudin-based tight junctions are crucial for the mammalian epidermal barrier: a lesson from claudin-1-deficient mice. Journal of Cell Biology. 2002;156(6):1099–1111. doi: 10.1083/jcb.200110122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muto S, Hata M, Taniguchi J, et al. Claudin-2-deficient mice are defective in the leaky and cation-selective paracellular permeability properties of renal proximal tubules. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(17):8011–8016. doi: 10.1073/pnas.0912901107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura A, Hayashi H, Imasato M, et al. Loss of claudin-15, but not claudin-2, causes Na+ deficiency and glucose malabsorption in mouse small intestine. Gastroenterology. 2011;140(3):913–923. doi: 10.1053/j.gastro.2010.08.006. [DOI] [PubMed] [Google Scholar]
- 44.Tatum R, Zhang Y, Salleng K, et al. Renal salt wasting and chronic dehydration in claudin-7-deficient mice. American Journal of Physiology: Renal Physiology. 2010;298(1):F24–F34. doi: 10.1152/ajprenal.00450.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gow A, Southwood CM, Li JS, et al. CNS Myelin and sertoli cell tight junction strands are absent in OSP/claudin-11 null mice. Cell. 1999;99(6):649–659. doi: 10.1016/s0092-8674(00)81553-6. [DOI] [PubMed] [Google Scholar]
- 46.Wattenhofer M, Reymond A, Falciola V, et al. Different mechanisms preclude mutant CLDN14 proteins from forming tight junctions in vitro. Human Mutation. 2005;25(6):543–549. doi: 10.1002/humu.20172. [DOI] [PubMed] [Google Scholar]
- 47.Ben-Yosef T, Belyantseva IA, Saunders TL, et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Human Molecular Genetics. 2003;12(16):2049–2061. doi: 10.1093/hmg/ddg210. [DOI] [PubMed] [Google Scholar]
- 48.Tamura A, Kitano Y, Hata M, et al. Megaintestine in claudin-15-deficient mice. Gastroenterology. 2008;134(2):523–534. doi: 10.1053/j.gastro.2007.11.040. [DOI] [PubMed] [Google Scholar]
- 49.Weber S, Schneider L, Peters M, et al. Novel paracellin-1 mutations in 25 families with familial hypomagnesemia with hypercalciuria and nephrocalcinosis. Journal of the American Society of Nephrology. 2001;12(9):1872–1881. doi: 10.1681/ASN.V1291872. [DOI] [PubMed] [Google Scholar]
- 50.Simon DB, Lu Y, Choate KA, et al. Paracellin-1, a renal tight junction protein required for paracellular Mg2+ resorption. Science. 1999;285(5424):103–106. doi: 10.1126/science.285.5424.103. [DOI] [PubMed] [Google Scholar]
- 51.Konrad M, Schaller A, Seelow D, et al. Mutations in the tight-junction gene claudin 19 (CLDN19) are associated with renal magnesium wasting, renal failure, and severe ocular involvement. American Journal of Human Genetics. 2006;79(5):949–957. doi: 10.1086/508617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ebnet K, Suzuki A, Ohno S, Vestweber D. Junctional adhesion molecules (JAMs): more molecules with dual functions? Journal of Cell Science. 2004;117(1):19–29. doi: 10.1242/jcs.00930. [DOI] [PubMed] [Google Scholar]
- 53.Ebnet K, Suzuki A, Horikoshi Y, et al. The cell polarity protein ASIP/PAR-3 directly associates with junctional adhesion molecule (JAM) EMBO Journal. 2001;20(14):3738–3748. doi: 10.1093/emboj/20.14.3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebnet K, Schulz CU, Meyer Zu Brickwedde MK, Pendl GG, Vestweber D. Junctional adhesion molecule interacts with the PDZ domain-containing proteins AF-6 and ZO-1. Journal of Biological Chemistry. 2000;275(36):27979–27988. doi: 10.1074/jbc.M002363200. [DOI] [PubMed] [Google Scholar]
- 55.Itoh M, Sasaki H, Furuse M, Ozaki H, Kita T, Tsukita S. Junctional adhesion molecule (JAM) binds to PAR-3: a possible mechanism for the recruitment of PAR-3 to tight junctions. Journal of Cell Biology. 2001;154(3):491–497. doi: 10.1083/jcb.200103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sollerbrant K, Raschperger E, Mirza M, et al. The Coxsackievirus and adenovirus receptor (CAR) forms a complex with the PDZ domain-containing protein Ligand-of-Numb protein-X (LNX) Journal of Biological Chemistry. 2003;278(9):7439–7444. doi: 10.1074/jbc.M205927200. [DOI] [PubMed] [Google Scholar]
- 57.Kansaku A, Hirabayashi S, Mori H, et al. Ligand-of-numb protein X is an endocytic scaffold for junctional adhesion molecule 4. Oncogene. 2006;25(37):5071–5084. doi: 10.1038/sj.onc.1209468. [DOI] [PubMed] [Google Scholar]
- 58.Hirabayashi S, Tajima M, Yao I, Nishimura W, Mori H, Hata Y. JAM4, a junctional cell adhesion molecule interacting with a tight junction protein, MAGI-1. Molecular and Cellular Biology. 2003;23(12):4267–4282. doi: 10.1128/MCB.23.12.4267-4282.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wegmann F, Ebnet K, Du Pasquier L, Vestweber D, Butz S. Endothelial adhesion molecule ESAM binds directly to the multidomain adaptor MAGI-1 and recruits it to cell contacts. Experimental Cell Research. 2004;300(1):121–133. doi: 10.1016/j.yexcr.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 60.Bazzoni G. The JAM family of junctional adhesion molecules. Current Opinion in Cell Biology. 2003;15(5):525–530. doi: 10.1016/s0955-0674(03)00104-2. [DOI] [PubMed] [Google Scholar]
- 61.Weber C, Fraemohs L, Dejana E. The role of junctional adhesion molecules in vascular inflammation. Nature Reviews Immunology. 2007;7(6):467–477. doi: 10.1038/nri2096. [DOI] [PubMed] [Google Scholar]
- 62.Suzuki A, Ishiyama C, Hashiba K, Shimizu M, Ebnet K, Ohno S. aPKC kinase activity is required for the asymmetric differentiation of the premature junctional complex during epithelial cell polarization. Journal of Cell Science. 2002;115(18):3565–3573. doi: 10.1242/jcs.00032. [DOI] [PubMed] [Google Scholar]
- 63.Yamanaka T, Horikoshi Y, Suzuki A, et al. PAR-6 regulates aPKC activity in a novel way and mediates cell-cell contact-induces formation of the epithelial junctional complex. Genes to Cells. 2001;6(8):721–731. doi: 10.1046/j.1365-2443.2001.00453.x. [DOI] [PubMed] [Google Scholar]
- 64.Gliki G, Ebnet K, Aurrand-Lions M, Imhof BA, Adams RH. Spermatid differentiation requires the assembly of a cell polarity complex downstream of junctional adhesion molecule-C. Nature. 2004;431(7006):320–324. doi: 10.1038/nature02877. [DOI] [PubMed] [Google Scholar]
- 65.Hirata K, Ishida T, Penta K, et al. Cloning of an immunoglobulin family adhesion molecule selectively expressed by endothelial cells. Journal of Biological Chemistry. 2001;276(19):16223–16231. doi: 10.1074/jbc.M100630200. [DOI] [PubMed] [Google Scholar]
- 66.Nasdala I, Wolburg-Buchholz K, Wolburg H, et al. A transmembrane tight junction protein selectively expressed on endothelial cells and platelets. Journal of Biological Chemistry. 2002;277(18):16294–16303. doi: 10.1074/jbc.M111999200. [DOI] [PubMed] [Google Scholar]
- 67.Wegmann F, Petri B, Khandoga AG, et al. ESAM supports neutrophil extravasation, activation of Rho, and VEGF-induced vascular permeability. Journal of Experimental Medicine. 2006;203(7):1671–1677. doi: 10.1084/jem.20060565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sakaguchi T, Nishimoto M, Miyagi S, et al. Putative "stemness" gene Jam-B is not required for maintenance of stem cell state in embryonic, neural, or hematopoietic stem cells. Molecular and Cellular Biology. 2006;26(17):6557–6570. doi: 10.1128/MCB.00729-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ikenouchi J, Furuse M, Furuse K, Sasaki H, Tsukita S, Tsukita S. Tricellulin constitutes a novel barrier at tricellular contacts of epithelial cells. Journal of Cell Biology. 2005;171(6):939–945. doi: 10.1083/jcb.200510043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dörfel MJ, Westphal JK, Huber O. Differential phosphorylation of occludin and tricellulin by CK2 and CK1. Annals of the New York Academy of Sciences. 2009;1165:69–73. doi: 10.1111/j.1749-6632.2009.04043.x. [DOI] [PubMed] [Google Scholar]
- 71.Riazuddin S, Ahmed ZM, Fanning AS, et al. Tricellulin is a tight-junction protein necessary for hearing. American Journal of Human Genetics. 2006;79(6):1040–1051. doi: 10.1086/510022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ikenouchi J, Sasaki H, Tsukita S, Furuse M, Tsukita S. Loss of occludin affects tricellular localization of tricellulin. Molecular Biology of the Cell. 2008;19(11):4687–4693. doi: 10.1091/mbc.E08-05-0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turksen K, Troy TC. Claudin-6: a novel tight junction molecule is developmentally regulated in mouse embryonic epithelium. Developmental Dynamics. 2001;222(2):292–300. doi: 10.1002/dvdy.1174. [DOI] [PubMed] [Google Scholar]
- 74.Anderson WJ, Zhou Q, Alcalde V, et al. Genetic targeting of the endoderm with Claudin-6CreER. Developmental Dynamics. 2008;237(2):504–512. doi: 10.1002/dvdy.21437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Guarino M. Epithelial-mesenchymal transition and tumour invasion. International Journal of Biochemistry and Cell Biology. 2007;39(12):2153–2160. doi: 10.1016/j.biocel.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 76.Ikenouchi J, Matsuda M, Furuse M, Tsukita S. Regulation of tight junctions during the epithelium-mesenchyme transition: direct repression of the gene expression of claudins/occludin by Snail. Journal of Cell Science. 2003;116(10):1959–1967. doi: 10.1242/jcs.00389. [DOI] [PubMed] [Google Scholar]
- 77.Bagnat M, Cheung ID, Mostov KE, Stainier DYR. Genetic control of single lumen formation in the zebrafish gut. Nature Cell Biology. 2007;9(8):954–960. doi: 10.1038/ncb1621. [DOI] [PubMed] [Google Scholar]
- 78.de Luca LM. Retinoids and their receptors in differentiation, embryogenesis, and neoplasia. FASEB Journal. 1991;5(14):2924–2933. [PubMed] [Google Scholar]
- 79.Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB Journal. 1996;10(9):940–954. [PubMed] [Google Scholar]
- 80.Kraft AS, Anderson WB. Characterization of cytosolic calcium-activated phospholipid-dependent protein kinase activity in embryonal carcinoma cells. Effect of retinoic acid-induced differentiation of F9 cells to parietal endoderm. Journal of Biological Chemistry. 1983;258(15):9178–9183. [PubMed] [Google Scholar]
- 81.Kubota H, Chiba H, Takakuwa Y, et al. Retinoid X receptor α and retinoic acid receptor γ mediate expression of genes encoding tight-junction proteins and barrier function in F9 cells during visceral endodermal differentiation. Experimental Cell Research. 2001;263(1):163–172. doi: 10.1006/excr.2000.5113. [DOI] [PubMed] [Google Scholar]
- 82.Gareus R, Huth M, Breiden B, et al. Normal epidermal differentiation but impaired skin-barrier formation upon keratinocyte-restricted IKK1 ablation. Nature Cell Biology. 2007;9(4):461–469. doi: 10.1038/ncb1560. [DOI] [PubMed] [Google Scholar]
- 83.Ogasawara N, Kojima T, Go M, et al. PPARγ agonists upregulate the barrier function of tight junctions via a PKC pathway in human nasal epithelial cells. Pharmacological Research. 2010;61(6):489–498. doi: 10.1016/j.phrs.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 84.Varley CL, Southgate J. Effects of PPAR agonists on proliferation and differentiation in human urothelium. Experimental and Toxicologic Pathology. 2008;60(6):435–441. doi: 10.1016/j.etp.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 85.Chiba H, Gotoh T, Kojima T, et al. Hepatocyte nuclear factor (HNF)-4α triggers formation of functional tight junctions and establishment of polarized epithelial morphology in F9 embryonal carcinoma cells. Experimental Cell Research. 2003;286(2):288–297. doi: 10.1016/s0014-4827(03)00116-2. [DOI] [PubMed] [Google Scholar]
- 86.Satohisa S, Chiba H, Osanai M, et al. Behavior of tight-junction, adherens-junction and cell polarity proteins during HNF-4α-induced epithelial polarization. Experimental Cell Research. 2005;310(1):66–78. doi: 10.1016/j.yexcr.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 87.Battle MA, Konopka G, Parviz F, et al. Hapatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(22):8419–8424. doi: 10.1073/pnas.0600246103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Meng J, Holdcraft RW, Shima JE, Griswold MD, Braun RE. Androgens regulate the permeability of the blood-testis barrier. Proceedings of the National Academy of Sciences of the United States of America. 2005;102(46):16696–16700. doi: 10.1073/pnas.0506084102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kobayashi K, Miwa H, Yasui M. Progesterone maintains amniotic tight junctions during midpregnancy in mice. Molecular and Cellular Endocrinology. 2011;337(1-2):36–42. doi: 10.1016/j.mce.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 90.Kelly SP, Chasiotis H. Glucocorticoid and mineralocorticoid receptors regulate paracellular permeability in a primary cultured gill epithelium. Journal of Experimental Biology. 2011;214(14):2308–2318. doi: 10.1242/jeb.055962. [DOI] [PubMed] [Google Scholar]
- 91.Fujita H, Sugimoto K, Inatomi S, et al. Tight junction proteins claudin-2 and -12 are critical for vitamin D-dependent Ca2+ absorption between enterocytes. Molecular Biology of the Cell. 2008;19(5):1912–1921. doi: 10.1091/mbc.E07-09-0973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sladek FM, Zhong W, Lai E, Darnell JE. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes and Development. 1990;4(12 B):2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 93.Hayhurst GP, Lee YH, Lambert G, Ward JM, Gonzalez FJ. Hepatocyte nuclear factor 4α (Nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Molecular and Cellular Biology. 2001;21(4):1393–1403. doi: 10.1128/MCB.21.4.1393-1403.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tirona RG, Lee W, Leake BF, et al. The orphan nuclear receptor HNF4α determines PXR- and CAR-mediated xenobiotic induction of CYP3A4. Nature Medicine. 2003;9(2):220–224. doi: 10.1038/nm815. [DOI] [PubMed] [Google Scholar]
- 95.Duncan SA, Manova K, Chen WS, et al. Expression of transcription factor HNF-4 in the extraembryonic endoderm, gut, and nephrogenic tissue of the developing mouse embryo: HNF-4 is a marker for primary endoderm in the implanting blastocyst. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(16):7598–7602. doi: 10.1073/pnas.91.16.7598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chen WS, Manova K, Weinstein DC, et al. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and impaired gastrulation of mouse embryos. Genes and Development. 1994;8(20):2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- 97.Li J, Ning G, Duncan SA. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genes and Development. 2000;14(4):464–474. [PMC free article] [PubMed] [Google Scholar]
- 98.Chiba H, Sakai N, Murata M, et al. The nuclear receptor hepatocyte nuclear factor 4α acts as a morphogen to induce the formation of microvilli. Journal of Cell Biology. 2006;175(6):971–980. doi: 10.1083/jcb.200608012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cunha GR, Cooke PS, Kurita T. Role of stromal-epithelial interactions in hormonal responses. Archives of Histology and Cytology. 2004;67(5):417–434. doi: 10.1679/aohc.67.417. [DOI] [PubMed] [Google Scholar]
- 100.Kruegel J, Miosge N. Basement membrane components are key players in specialized extracellular matrices. Cellular and Molecular Life Sciences. 2010;67(17):2879–2895. doi: 10.1007/s00018-010-0367-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yurchenco PD, Patton BL. Developmental and pathogenic mechanisms of basement membrane assembly. Current Pharmaceutical Design. 2009;15(12):1277–1294. doi: 10.2174/138161209787846766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kleinman HK, Graf J, Iwamoto Y, et al. Role of basement membranes in cell differentiation. Annals of the New York Academy of Sciences. 1987;513:134–145. doi: 10.1111/j.1749-6632.1987.tb25004.x. [DOI] [PubMed] [Google Scholar]
- 103.Hahn U, Stallmach A, Hahn EG, Riecken EO. Basement membrane components are potent promoters of rat intestinal epithelial cell differentiation in vitro. Gastroenterology. 1990;98(2):322–335. doi: 10.1016/0016-5085(90)90821-h. [DOI] [PubMed] [Google Scholar]
- 104.Zhang H, Tasnim F, Ying JY, Zink D. The impact of extracellular matrix coatings on the performance of human renal cells applied in bioartificial kidneys. Biomaterials. 2009;30(15):2899–2911. doi: 10.1016/j.biomaterials.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 105.Koval M, Ward C, Findley MK, Roser-Page S, Helms MN, Roman J. Extracellular matrix influences alveolar epithelial claudin expression and barrier function. American Journal of Respiratory Cell and Molecular Biology. 2010;42(2):172–180. doi: 10.1165/rcmb.2008-0270OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Agarwal R, D'Souza T, Morin PJ. Claudin-3 and claudin-4 expression in ovarian epithelial cells enhances invasion and is associated with increased matrix metalloproteinase-2 activity. Cancer Research. 2005;65(16):7378–7385. doi: 10.1158/0008-5472.CAN-05-1036. [DOI] [PubMed] [Google Scholar]
- 107.Raleigh DR, Marchiando AM, Zhang Y, et al. Tight junction-associated MARVEL proteins marvelD3, tricellulin, and occludin have distinct but overlapping functions. Molecular Biology of the Cell. 2010;21(7):1200–1213. doi: 10.1091/mbc.E09-08-0734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Osler ME, Chang MS, Bader DM. Bves modulates epithelial integrity through an interaction at the tight junction. Journal of Cell Science. 2005;118(20):4667–4678. doi: 10.1242/jcs.02588. [DOI] [PubMed] [Google Scholar]
- 109.Osler ME, Smith TK, Bader DM. Bves, a member of the Popeye domain-containing gene family. Developmental Dynamics. 2006;235(3):586–593. doi: 10.1002/dvdy.20688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Russ PK, Pino CJ, Williams CS, Bader DM, Haselton FR, Chang MS. Bves modulates tight junction associated signaling. PLoS ONE. 2011;6(1, article e14563) doi: 10.1371/journal.pone.0014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Terry SJ, Zihni C, Elbediwy A, et al. Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nature Cell Biology. 2011;13(2):159–166. doi: 10.1038/ncb2156. [DOI] [PMC free article] [PubMed] [Google Scholar]


