Abstract
The complex [Re(CO)3(dppz)(py′-OR)]+ (dppz = dipyrido[3,2-a:2′,3′-c]phenazine; py′-OR = 4-functionalized pyridine) offers IR sensitivity and can oxidize DNA directly from the excited state, making it a promising probe for the study of DNA-mediated charge transport (CT). The behavior of several covalent and noncovalent Re-DNA constructs was monitored by time-resolved IR (TRIR) and UV/visible spectroscopies, as well as biochemical methods, confirming the long-range oxidation of DNA by the excited complex. Optical excitation of the complex leads to population of MLCT and at least two distinct intraligand states. Experimental observations that are consistent with charge injection from these excited states include similarity between long-time TRIR spectra and the reduced state spectrum observed by spectroelectrochemistry, the appearance of a guanine radical signal in TRIR spectra, and the eventual formation of permanent guanine oxidation products. The majority of reactivity occurs on the ultrafast timescale, although processes dependent on slower conformational motions of DNA, such as the accumulation of oxidative damage at guanine, are also observed. The ability to measure events on such disparate timescales, its superior selectivity in comparison to other spectroscopic techniques, and the ability to simultaneously monitor carbonyl ligand and DNA IR absorption bands makes TRIR a valuable tool for the study of CT in DNA.
INTRODUCTION
The ability of DNA to mediate charge transport (CT) has been established using a variety of redox-active probes and in a great diversity of experimental systems.1–3 The efficiency of DNA-mediated CT is affected by several factors, including the extent of electronic coupling between the probe and the DNA base stack, coupling within the base stack itself, the driving force of the CT reaction, and the base sequence. DNA CT has been observed over long molecular distances with little attenuation,4–6 suggesting its utility in molecular-scale devices7–9 and in biological systems.2,10–13 Many of the properties of DNA CT have been elucidated in experiments involving the slow accumulation of oxidative damage at low potential guanine sites. While such methods remain useful in the investigation of DNA CT, a general probe for direct, time-resolved monitoring of these processes remains elusive.
Time-resolved infrared (TRIR) spectroscopy offers several advantages over other time-resolved methods for the study of CT events.14 With the proper choice of IR-active probe and solvent medium, changes in the absorption pattern of well-resolved, transient IR bands provide kinetic information on specific photophysical, chemical, and biochemical processes, together with structural characterization of the excited states and reaction intermediates involved. One common family of probes are coordination complexes of the type [Re(CO)3(N,N)(L)]n, where N,N stands for an α-diimine ligand such as 2,2′-bipyridine (bpy), phenanthroline (phen), or dppz (dipyrido[3,2-a:2′,3′-c]phenazine) and L represents an axial ligand, often Cl (n = 0) or functionalized pyridine (n = 1+).15–23 Photophysical or photochemical reactions involving these Re complexes are manifested in TRIR spectra as changes in the intensities and positions (energies) of absorption bands due to CO stretching vibrations of the Re(CO)3 group, ν(C≡O). Variation of the N,N and L ligands affords fine control over the excited-state characters and energetics.16,18–20,22–27 These complexes have also proven useful as biochemical probes for fluorescence imaging,28 for monitoring the dynamics of structural fluctuations,29,30 and especially, for triggering photoinduced electron transfer (ET).31 Information on ET kinetics and intermediates provided by TRIR is more direct than that obtained using UV/visible time-resolved spectroscopic methods due to their low specificity. Recently, the presence of tryptophan along the ET pathway in Re(CO)3(4,7-dimethyl-1,10-phenanthroline)-modified azurin was shown to increase the rate of ET.32–34 Although other coordination complexes, such as dicarbonyl Ru species, W(CO)5(4-cyanopyridine), and [Ru(bpy)(CN)4]2− have been employed as TRIR probes, tricarbonyl Re complexes have been studied much more extensively.14,16,35 Most studies have focused on using TRIR to monitor protein ET. TRIR can also be used to monitor changes in the vibrational frequencies and IR band intensities of organic functionalities in ET assemblies.36 Of particular interest, TRIR spectra were recorded following the 267 nm excitation of the four canonical nucleotides and of poly(dG-dC)·poly(dG-dC) and poly(dA-dT)·poly(dA-dT).37 In that work, the lifetimes of the transient states of the free nucleotides ranged from 2.2 to 4.7 ps, while those of the polymers were an order of magnitude longer. Upon 200 nm photoionization of 5′-dGMP and poly(dG-dC)·poly(dG-dC), evidence for the formation of the guanine radical was observed by TRIR as the growth of a transient band at 1702 cm−1.38 In other experiments, TRIR was used to observe the triplet state of thymine and of 2′-dT,39 as well as to unravel the pH-dependent photophysics of 5′-G, 5′-GMP, and poly(G).40 Importantly, these studies indicate that TRIR can be used to monitor photoinduced changes of DNA and of [Re(CO)3(N,N)(L)]n simultaneously, making it possible to investigate both the donor and the acceptor sites of Re-DNA CT assemblies. Although interactions between Re complexes and DNA have been studied by UV/visible spectroscopy,41,42 these interactions had not been investigated by vibrational methods until very recently.43,44
Here, TRIR spectroscopy is used in conjunction with other methods to observe the DNA-mediated oxidation of guanine in DNA by photoexcited [Re(CO)3(dppz)(py′-OR)]+, where py′-OR represents pyridine functionalized at the 4 position (Figure 1). The influence of guanine on the photochemical behavior of the Re complex bound to DNA is investigated by comparing results obtained in four different DNA contexts, including two in which the complex is covalently tethered to specific locations on the duplex. The data presented show that the photoexcited Re complex can oxidize guanine at a distance of several bases away by DNA-mediated CT and that this process can be monitored on the ps to μs timescale by TRIR. The results of this study, in which TRIR is used for the first time to observe DNA-mediated CT between photooxidants and guanine in well-defined covalent constructs, shows that the DNA sequence surrounding the metal complex binding site has a large influence on the photophysics and photochemistry of the system.
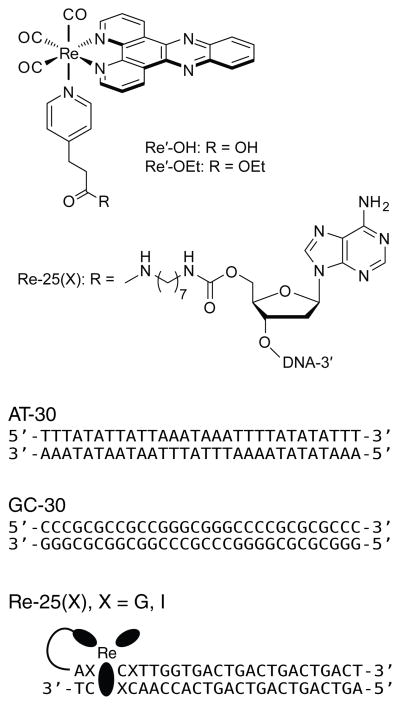
Figure 1.
Schematic illustration of [Re(CO)3(dppz)(py′-OR)]+, the covalent linker, and the DNA sequences used for studies of guanine oxidation. Experiments involving AT-30 and GC-30 were conducted in the presence of the free complex Re′-OH. In the covalent assemblies Re-25(G) and Re-25(I), the Re photooxidant is tethered to the 5′ end of one strand via a peptide linkage.
EXPERIMENTAL SECTION
Materials
Most reagents for metal complex synthesis and coupling were purchased from Sigma-Aldrich unless otherwise indicated. 3-(pyridin-4-yl)propanoic acid (py′-OH) was purchased from Chess GmbH (Mannheim, Germany). Reagents for DNA synthesis were purchased from Glen Research (Sterling, VA). All reagents were used as received.
Complex and Conjugate Synthesis
Preparation of [Re(CO)3(dppz)(py′-OH)](PF6) was adapted from previously described methods.41 Following the synthesis, the PF6− counter anion was exchanged (QAE Sephadex A-25 resin, GE Healthcare) for chloride ion in order to increase the solubility of the complex in aqueous media. Because facile proton loss from the carboxylic acid-modified pyridine ligand results in an overall neutral zwitterionic species, altering the extent of electrostatic repulsion between complex molecules and of electrostatic attraction to DNA, the protected ethyl ester version of the complex, [Re(CO)3(dppz)(py′-OEt)]+ (py′-OEt = ethyl 3-(pyridin-4-yl)propanoate), was used for some experiments.
Oligonucleotide Synthesis and Modification
Oligonucleotides were synthesized using standard solid-phase phosphoramidite chemistry on an Applied Biosystems 3400 DNA synthesizer. Covalent tethers were appended to the 5′-OH termini of resin-bound oligonucleotides as described by Holmlin.45 The alkyl tether was added to the DNA strand by successive treatment with carbonyldiimidazole and diaminononane. Agitation of the resin-bound, amine modified DNA strands in the presence of excess (5 mg) [Re(CO)3(dppz)(py′-OH)]Cl, O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU), 1-hydroxybenzotriazole hydrate (HOBT), and diisopropylethylamine (DIEA) in anhydrous DMF for 24 hrs resulted in covalent attachment of the metal complex to the DNA. Cleavage from the resin was effected by incubation in NH4OH at 60 °C for 6 h. Oligonucleotides were purified by reversed-phase HPLC and characterized by MALDI-TOF mass spectrometry (see Supporting Information). Oligonucleotide concentrations were determined by UV/visible spectrophotometry (Beckman DU 7400). Annealing was accomplished by incubating solutions containing equimolar amounts of complementary strands in buffer (10 mM NaPi, 50 mM NaCl buffer; pH 7.0) at 90 °C for 5 min followed by slow cooling over 90 min to ambient temperature.
Assay for Oxidative DNA Damage
Oxidative DNA cleavage experiments were performed using a protocol adapted from Zeglis46 with the following adjustments. Oligonucleotides were labeled at the 3′-end by incubating a mixture of 2 μL single-stranded DNA (4 μM), 5 μL [α-32P]-dTTP (Perkin Elmer), 2 μL terminal transferase (TdT; New England Biolabs), 5 μL CoCl2 solution (included with TdT), and 5 μL terminal transferase reaction buffer (included with TdT) for 2 h at 37 °C. Before gel purification, strands were incubated at 90 °C for 20 min in 100 μL 10% aqueous piperidine to induce cleavage of damaged strands. Following purification and annealing, samples (10 μL, 2 μM) were irradiated in parallel for 2 hrs using a solar simulator (Oriel Instruments) fitted with a 340 nm internal long pass filter. Samples were then treated with 0.2 units calf thymus DNA and 10% piperidine (v/v), heated for 30 min at 90 °C, and dried in vacuo. After gel electrophoresis, oxidative damage was quantified by phosphorimagery (ImageQuant). Sample counts are reported as % of total counts per lane and were corrected by subtracting the dark control.
Spectroelectrochemistry
UV/visible spectroelectrochemistry was carried out using a custom-built, optically transparent, thin-layer electrode (OTTLE) cell (path length = 0.1 mm) consisting of vapor-deposited platinum working and pseudoreference electrodes and a Pt-wire auxiliary electrode.47 The potential of the cell was controlled by a potentiostat (CH Instruments Model 650A electrochemical workstation). Samples consisted of saturated solutions of metal complexes in dry acetonitrile with 0.1 M Bu4NPF6 electrolyte. Samples were degassed by bubbling argon and introduced into the optical cell using a gas-tight syringe prior to measurement. The cell was held at a reducing potential, and spectra were acquired on a Hewlett Packard 8452A diode array spectrophotometer every 4 s until the sample was fully reduced.
UV/Visible Emission and Transient Absorption Spectroscopy
Steady-state emission spectra were recorded on a Fluorolog-3 spectrofluorometer (Jobin Yvon) using 2 mm slits. Scattered excitation light was rejected from the detector by appropriate filters. Reported spectra are averages of at least five consecutive measurements.
All time-resolved UV/visible spectroscopic measurements were carried out at the Beckman Institute Laser Resource Center. Nanosecond luminescence decay measurements and transient absorption (TA) measurements were performed using the third harmonic (355 nm) of a 10 Hz, Q-switched Nd:YAG laser (Spectra-Physics Quanta-Ray PRO-Series) as the excitation source (8 ns pulse width, 5 mJ/pulse). Probe light was provided by a synchronized, pulsed 75 W Hg-Xe arc lamp (PTI model A 1010), and detection was accomplished using a photomultiplier tube (Hamamatsu R928) following wavelength selection by a double monochromator (Instruments SA DH-10). Scattered light was rejected using suitable filters. The samples were held in 1 cm path length quartz cuvettes (Starna) equipped with stir bars. TA measurements were made with and without excitation, and were corrected for background light, scattering, and fluorescence.
Picosecond emission decay measurements48–51 were performed using the third harmonic of a regeneratively amplified mode-locked Nd:YAG laser (355 nm, 1 ps pulse width after amplification) as the excitation source and a picosecond streak camera (Hamamatsu C5680, photon-counting mode) as the detector. Emission was observed under magic angle conditions using a 550 nm long-pass cutoff filter.
TRIR Spectroscopy
The ULTRA instrument at the STFC Rutherford Appleton Laboratory has been used. It is described in detail elsewhere.52 In brief, a titanium sapphire laser-based regenerative amplifier (Thales) produces 800 nm, ~50 fs pulses at a 10 kHz repetition rate. The laser output is split in two parts, one of which is either frequency doubled or used to drive an OPA (Light Conversion, TOPAS) equipped with SHG and SFG units to produce a pump beam at 400 or 355 nm, respectively. The second pumps a TOPAS OPA, yielding signal and idler beams that are difference frequency mixed to generate ~400 cm−1 broad mid IR probe pulses. An optical delay line is used to introduce a delay between the pump and probe beams, and the mid IR probe spectrum is recorded at a given time delay using two 128 element HgCdTe detectors (Infrared Associates). For ns-μs measurements, the sample was pumped with 355 nm, 0.7 ns FWHM pulses (AOT, AOT-YVO-20QSP/MOPO), and probed with electronically synchronized 50 fs IR pulses.53 The sample solutions were placed in a round dip ca. 0.75 mm deep, drilled into a CaF2 plate and tightly covered with a polished CaF2 window. The cell was scanned-rastered across the area of the dip in two dimensions to prevent laser heating and decomposition of the sample. FTIR spectra measured before and after the experiment demonstrated sample stability.
Fitting Methods
TRIR data were simulated at each time delay as a series of Gaussian terms in order to extract kinetic data from overlapping transient bands. The area of each Gaussian was calculated, and kinetic decays were constructed as the change in area with delay time. Nanosecond time-resolved emission, TRIR, and TA data were fit by nonlinear least-squares analysis using IGOR Pro software (Wavemetrics). Model functions consisted of a linear series of exponential terms of the form y(t) = Σ ai exp[−t/τi], where ai and τi are the pre-exponential factor and lifetime, respectively, of the ith term. Up to three exponential terms were included until reasonable fits were obtained. For time-resolved emission data, the percent relative contribution reported in Table 1 represents the number of photons emitted at the probe wavelength by each emissive population, and is calculated as anτn/Σaiτi (the area under the decay for the nth exponential term normalized to the total area under the decay curve). For TRIR and TA data, the percent relative contribution represents the change in absorbance of species n extrapolated to time t = 0, and is calculated as an/Σai.
Table 1.
Decay lifetimes from least-squares fits (λex = 355 nm).
| Technique | Detail | Sample | Lifetimes, seconds (% Relative Contributiona)
|
|||||
|---|---|---|---|---|---|---|---|---|
| 10−9 | 10−8 | 10−7 | 10−6 | 10−5 | >10−5b | |||
| TRIRc | Re(CO)3 Bleach Recovery | AT-30 + Re′-OH | 1.4 (28) | 3.0 (31) | 2.5 (41) | |||
| GC-30 + Re′-OH | 6.3 (43) | 5.5 (49) | 1.1 (8) | |||||
| Re-25(I) | 2.6 (16) | 4.8 (30) | 2.8 (54) | |||||
| Re-25(G) | 4.8d | 2.9 (65) | 9.1 (35) | |||||
|
| ||||||||
| MLCT (2071 cm−1) | AT-30 + Re′-OH | 8.8 (38) | 5.6 (38) | Long (23) | ||||
| Re-25(I) | 3.2 (36) | 8.9 (44) | Long (20) | |||||
|
| ||||||||
| IL (2030 cm−1) | AT-30 + Re′-OH | 1.5 (31) | 2.4 (69) | |||||
| GC-30 + Re′-OH | 1.8 (51) | 1.0 (42) | 2.2 (7) | |||||
| Re-25(I) | 3.0 (34) | Long (66) | ||||||
| Re-25(G) | 4.0d | 3.0 (58) | 9.2 (42) | |||||
|
| ||||||||
| G•+/G• (1702 cm−1) | GC-30 + Re′-OH | 2.1d | 5.8 | |||||
|
| ||||||||
| DNA Bleach Recovery | AT-30 + Re′-OH | 1.4 (48) | 5.0 (18) | Long (35) | ||||
| GC-30 + Re′-OH | 5.9 (31) | 9.6 (69) | ||||||
| Re-25(I) | 3.1 (88) | Long (12) | ||||||
| Re-25(G) | 2.9d | 8.4 (53) | 1.0 (47) | |||||
|
| ||||||||
| NS Visible TAe | λprobe = 475 nm | AT-30 + Re′-OH | 4.9 (17) | 2.7 (83) | ||||
| GC-30 + Re′-OH | 2.7 (42) | 2.0 (58) | ||||||
| Re-25(I) | 9.6 (42) | 2.0 (58) | ||||||
| Re-25(G) | 4.4 (37) | 1.4 (63) | ||||||
|
| ||||||||
| NS Emissione,f | λprobe = 570 nm | AT-30 + Re′-OH | 2.9 (34) | 2.4 (23) | 5.7 (43) | |||
| GC-30 + Re′-OH | 3.2 (39) | 2.7 (35) | 2.4 (26) | |||||
| Re-25(I) | 5.3 (17) | 2.6 (21) | 5.4 (62) | |||||
| Re-25(G) | 3.9 (42) | 2.1 (30) | 4.5 (28) | |||||
Determined by different methods for absorption and emission; see Experimental Section.
“Long” indicates incomplete decay.
Uncertainty estimated at 20%.
These values reflect an increase in intensity.
Uncertainty estimated at 10%.
Processes faster than 8 ns are convoluted with instrument response.
Picosecond emission data were collected at 1 ns, 5 ns, and 50 ns time ranges and spliced together before fitting. Data were compressed logarithmically in time prior to fitting in order to decrease the bias of long time data on the fit. These data could not be fit well to a series of exponential terms and were instead analyzed by the maximum entropy method using a MATLAB (MathWorks) routine written at Caltech.48–51
RESULTS
Research Strategy and Design of Re-DNA CT Assemblies
With the aim to establish DNA oxidation by electronically excited rhenium tricarbonyl-diimine complexes, we have employed a newly developed Re sensitizer, [Re(CO)3(dppz)(py′-OR)]+ (R = H, Re′-OH; or R = CH2CH3, Re′-OEt), which can be covalently linked to DNA (Figure 1). Three design elements make this a promising probe for the study of DNA-mediated CT. The first is the incorporation of TRIR-active carbonyl ligands. Re carbonyl-diimine complexes are useful probes in TRIR spectroscopic experiments due to the intense and well-resolved bands corresponding to carbonyl stretching modes. These modes are extremely sensitive to changes in electron density distribution, molecular structure and environment.17,24,30,32,54,55 The second design element is the inclusion of the planar dppz ligand. By incorporating dppz on the metal center, we ensure effective electronic coupling with the DNA base stack. Indeed, the binding constants for intercalating dppz complexes such as [Ru(bpy)2(dppz)]2+ and [Ru(phen)2(dppz)]2+ are greater than 106 M−1.56 While the binding of complexes like [Re(CO)3(dppz)(py′-OR)]+ is weaker (105 M−1)41,42,57 due to its lower electrostatic charge, the decrease of the molar absorptivity of its near-UV absorption band (i.e., hypochromicity) upon incubation with DNA, as well as an increase in the melting temperature of the bound DNA duplex by approximately 5 °C (depending on the sequence), indicate that this Re complex indeed binds by intercalation. The third design element is the ability to covalently attach the complex to DNA via carboxyalkyl-modified pyridine incorporated at the axial coordination site. The covalent link between the complex and the DNA strand, while flexible, restricts diffusion of the unbound complex, ensuring a higher percentage bound than if the complex were allowed to diffuse freely. In addition, the covalent link enables us to define the DNA sequence at the binding region, eliminating sequence effects as a variable. Physical models suggest that in the equilibrium geometry (disregarding frayed end effects) tethering restricts binding to the region within three base pairs from the end of the duplex.
The DNA duplexes used were designed to test for the effect of the DNA sequence on the efficiency of DNA oxidation. For systems in which guanine, an effective hole trap, is placed near the expected binding site of the Re complex, charge injection may be followed by facile back electron transfer (BET). Such nonproductive reactions are competitive with permanent charge trapping at guanine sites.58–60 The frequency of nonproductive events can be reduced by replacing guanine at the Re binding site with inosine (I), a base analog that has a higher oxidation potential than guanine [E° (I•+/I) ≈1.5 V vs. NHE; E° (G•+/G) = 1.29 V vs. NHE].60–64 With these considerations in mind, four DNA sequences were designed (Figure 1). Two of them contain only adenine and thymine (AT-30) or guanine and cytosine (GC-30) and are expected to reveal the effect of the absence or presence, respectively, of strong guanine thermodynamic hole traps on DNA oxidation by noncovalently bound Re′-OH. Two DNA sequences were also designed to test for the effect of neighboring guanine on the efficiency of long range DNA oxidation by covalently-bound Re. These are Re-25(G), which contains guanine next to the Re binding site, and Re-25(I), in which guanine is replaced by inosine.
Sensitizer Characterization
The photophysics of [Re(CO)3(dppz)(py′-OH)]+ and [Re(CO)3(dppz)(py′-OEt)]+ are very similar, suggesting that modification at the py′ carbonyl has little effect on the energetics of the complex. For example, each complex exhibits absorption maxima at 364 and 382 nm (ε ≈ 11,000 M−1 cm−1),41,65 with a tail that extends into the visible region.66 The emission spectra of both complexes show maxima at 554 and 595 nm. At 570 nm, Re′-OH and Re′-OEt each show a biexponential emission decay in acetonitrile, with lifetimes on the order of 200 ns (~10%) and 10 μs (~90%), tentatively attributed to emission from different 3IL states.66 Tethering the Re species to DNA, therefore, is expected to have negligible influence on the energetics of the complex.
The reduction potential of the emissive 3IL state(s), E° (*Re+/Re0), of the Re label can be estimated as the sum of the ground state reduction potential, E° (Re+/Re0), and the zero-zero excited-state energy, (E0,0).67 The exact value of E0,0 is unknown, but it is estimated to lie between the energy at which the excitation and emission spectra coincide (480 nm, 2.58 eV) and the energy of the emission maximum in aqueous solution (570 nm, 2.18 eV). For Re′-OEt in acetonitrile, E° (Re+/Re0) was reported as −850 mV vs. NHE,66 predicting the excited-state reduction potential to lie between 1.33 and 1.73 eV. As an oxidant, electronically excited Re′-OEt is clearly strong enough to oxidize guanine, and it may be strong enough to oxidize adenine [E° (A•+/A) = 1.42 V vs. NHE].68 The latter reaction, however, is expected to be slower due to the lower driving force.
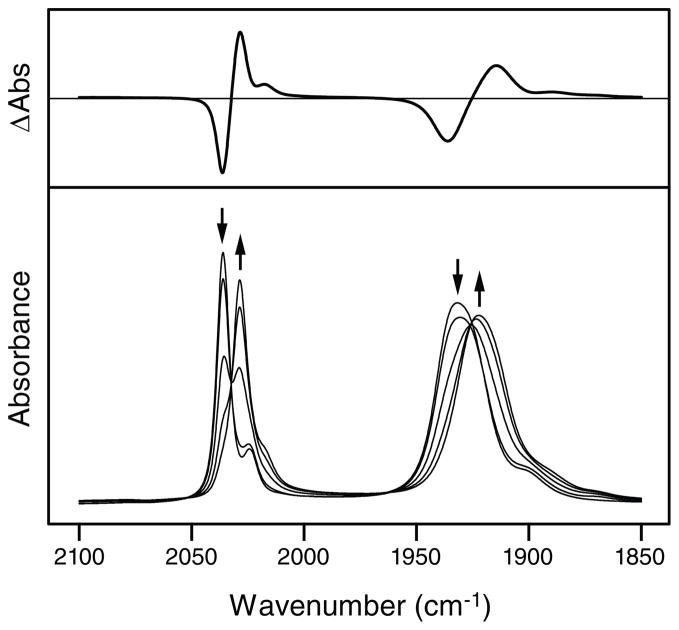
Hole injection into the DNA base stack must coincide with reduction of the metal complex. In order to characterize this reduced state independently, IR spectroelectrochemical reduction of saturated Re′-OEt in acetonitrile was carried out (Figure 2). Before reduction, the spectrum exhibits a band at 2036 cm−1 assigned to the totally symmetric in-phase ν(C≡O) vibration A′(1), and a band at 1932 cm−1 due to quasidegenerate totally symmetric out-of-phase A′(2) and equatorial CO antisymmetric A″ ν(C≡O) vibrations.17,54,69 Reduction results in a bathochromic shift of these bands to 2029 cm−1 and 1922 cm−1, respectively. This shift is similar to that observed previously23 upon reduction of [ReCl(CO)3(dppz)] and its small magnitude is consistent with occupation of the phenazine π* orbital of the dppz ligand in the [ReI(CO)3(dppz•−)(py′-OEt)] reduction product.24 Subsequent regeneration of the initial species via reoxidation was 95% complete, suggesting partial irreversible decomposition of the electrogenerated product; however, these decomposition products are not expected to interfere in time-resolved spectroscopic experiments employing fast photocycles. An attempt was made to duplicate the experiment in D2O buffer (10 mM NaPi, 50 mM NaCl, pD 7.0) in order to generate spectra that would be more directly comparable to TRIR measurements conducted in D2O buffer. Although the low solubility of the complex and the strong background absorbance of the solvent in this energy region prevented precise analysis, band positions, widths, and relative intensities were similar to those observed in acetonitrile solutions.
Figure 2.
Steady-state FTIR spectra (bottom) of saturated Re′-OEt in acetonitrile recorded during bulk reduction using an OTTLE cell. Arrows indicate spectral changes that occur upon reduction. The difference in absorbance between the fully reduced species and the initial species is also shown (top).
Oxidative Damage Pattern of Re-25(G) and Re-25(I) Observed by PAGE
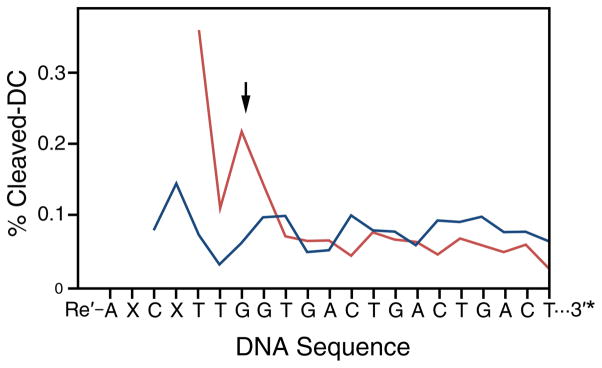
Figure 3 shows DNA-mediated oxidative damage in 2 μM solutions of Re-25(G) and Re-25(I) observed after 2 hours of broadband (λ > 340 nm) irradiation and 20% PAGE analysis. Damage occurs as base radicals, formed following hole injection by the excited Re complex, react with solution species such as H2O or O2 to form irreversible products.70,71 Subsequent treatment of the 3′-[32P]-labeled DNA with piperidine induces cleavage at damage sites. For both Re-25(G) and Re-25(I), damage is observed primarily at the 5′-G site of the 5′-GG-3′ doublet, several bases distant from the Re complex binding site predicted from physical models. Importantly, the low concentrations used in these experiments preclude interstrand damage (i.e., it is unlikely that the Re moiety of one construct will intercalate into the base stack of another). The observation of damage at the 5′-GG-3′ site indicates that long-range photoinduced hole injection from the Re label to DNA indeed occurs, consistent with results obtained for a similar Re-DNA conjugate.66 However, the extent of damage is consistently greater in the case of Re-25(I) than Re-25(G).
Figure 3.
Quantification of oxidative damage observed for Re-25(I) (X = I; red) or Re-25(G) (X = G; blue) by PAGE analysis. Aqueous samples containing 3′-[α-32P]-radiolabelled (indicated by *) Re-DNA constructs (2 μM) were irradiated for 2 hrs and treated with piperidine to induce cleavage at damaged bases. Cleavage products were separated by 20% PAGE and imaged by phosphorimagery. Quantitation was accomplished by normalizing counts at each site to total counts per lane. Traces were corrected for false positives by subtracting the dark control (DC). The arrow indicates the 5′-guanine of a 5′-GG-3′ doublet. Re is expected to bind 2–3 bases in from the 5′-end of the duplex.
Emission Measurements
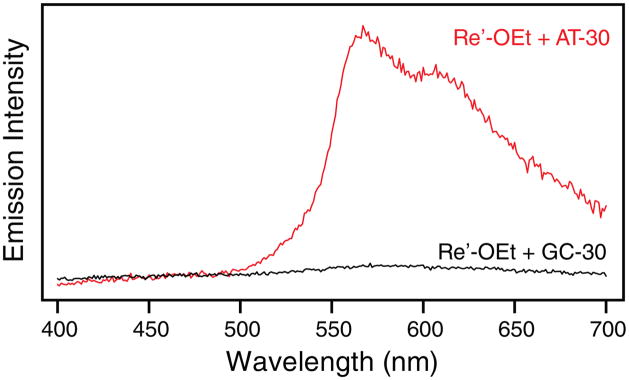
Many Re tricarbonyl complexes of dppz behave as DNA light switches,18–23,41,42 much like their Ru counterparts,72 and the complexes studied here are no exception. In the absence of DNA, negligible emission is observed from an aqueous solution of Re′-OH or Re′-OEt; however, in the presence of AT-30 and in the Re-25(I) sample, a prominent emission band is observed that exhibits a maximum at 570 nm and a shoulder near 600 nm, resembling the emission spectrum seen for similar Re complexes in organic solvents.19,21,22,41,42,57,65 By comparison with the emission of [Ru(bpy)3]2+ in deaerated acetonitrile, the emission quantum yield of Re′-OEt in the presence of AT-30 in buffer is 0.008 (compared to 0.062 for [Ru(bpy)3]2+).73 In the presence of GC-30 and in the Re-25(G) sample, the emission is much less intense, the maxima are shifted to 585 nm, and no shoulder is observed. Steady-state emission spectra of AT-30 alone and in the presence of Re′-OEt are shown in Figure S1. Interestingly, the DNA oligomers used in this study are themselves emissive under 355 nm excitation, giving rise to a broad band near 450 nm that tails into the visible region. All efforts were made to ensure that this is not an effect of the instrument, solvent, scattering, or impurities. Such emission, ascribed to excitons or charge transfer excited states, has previously been observed in DNA oligomers but not in calf thymus DNA.74–76 The Re-loaded AT-30 sample shows overlapping DNA and Re′-OEt emission. By scaling and subtracting the emission band due to DNA alone, it is possible to isolate emission from only the intercalated complexes. Significantly, emission from Re′-OEt becomes strongly quenched on going from AT-30 to GC-30 (Figure 4). A similar decrease is observed for Re-25(G) compared to Re-25(I). The concentrations of DNA and of the Re complex are the same in all of the samples, but the intensity of emission decreases as AT-30 ≈ Re-25(I) > Re-25(G) ≈ GC-30.
Figure 4.
Steady-state emission spectra of 20 μM Re′-OEt and 0.5 mM DNA (base pairs) in D2O buffer (10 mM NaPi, 50 mM NaCl; pD 7.0) solution following excitation at 355 nm. Emission spectra of Re′-OEt with AT-30 (red) or GC-30 (black) have been corrected for emission from DNA alone.
Differences in emission intensity are also observed in time-resolved measurements carried out on the nanosecond timescale with a PMT detector (response time 8 ns) and on the picosecond timescale using a streak camera (response time 55 ps). On the nanosecond timescale, the time-integrated emission intensity of Re-25(G) at 570 nm is 14% that of Re-25(I), and the intensity of GC-30 is 12% that of AT-30, following the trend observed in stationary spectra. Even on the picosecond timescale, the instantaneous emission intensity extrapolated to t = 0 is lower in the GC-30 and Re-25(G) samples than in the AT-30 and Re-25(I) samples, respectively. In addition, on this timescale the time-integrated emission intensity of Re-25(G) is 79% that of Re-25(I), and the intensity of GC-30 is 69% that of AT-30. These observations clearly indicate reaction(s) between electronically excited Re complex and DNA occurring on the picosecond to nanosecond timescale. Based on results of the PAGE experiment, hole transfer from *Re to G is most likely a prominent contributing reaction pathway.
The emission decay of the four DNA samples is highly multiexponential, with lifetimes varying over four orders of magnitude, from ~100 ps to ~500 ns. The present data do not allow us to attribute individual emission decay components to particular species present in the solution, although steady-state results suggest that DNA excimer emission contributes significantly (~20 %) to the total decay. After accounting for DNA excimer emission, which decays with a lifetime of only a few ns,76 about half of the Re emission decays within 50 ns, and the remainder persists for hundreds of ns. Maximum entropy fitting of the emission decays yields several distributions of rate constants (Figure S2),. The lifetime distributions vary only slightly between samples, and in every sample, the majority component has a lifetime of less than 1 ns. Notably, while most of the lifetimes are shortened slightly on going from AT-30 to GC-30 and from Re-25(I) to Re-25(G), no decay component is observed that corresponds to quenching of the excited Re sensitizer by guanine. Considering the significant quenching in steady-state measurements of the GC-30 and Re-25(G) samples, it seems that quenching at the reactive binding site(s) is ultrafast, probably tens of picoseconds or faster, but involves only a fraction of the excited population.
Time-Resolved Infrared (TRIR) Spectra
Whereas emission spectra provide evidence for ultrafast hole injection from electronically excited Re into the GC-30 and Re-25(G) samples, TRIR has the potential to characterize the reacting state(s) of the Re complex and to detect products and intermediates. To this effect, TRIR spectra were investigated in the picosecond (1–100 ps) and nanosecond-to-microsecond time domains in the regions of the Re(CO)3 ν(C≡O) and DNA organic carbonyl vibrations.
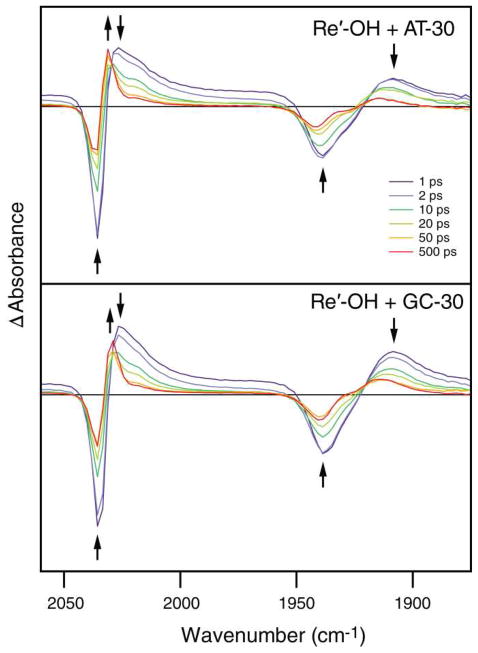
Typical picosecond TRIR spectra obtained in the ν(C≡O) region after 355 nm excitation are shown in Figure 5 for AT-30 and GC-30. The spectra measured 1 ps after excitation show negative bands due to bleaching of the ground state absorption (2036 and 1939 cm−1) and broad transient bands at 2026 and 1908 cm−1. Over the course of time, both features decay in intensity while a sharp band grows in at 2031 cm−1 (overlapping with the 2036 cm−1 bleach) together with a broad band between 1915 and 1935 cm−1. These new transients partially overlap with the parent bleaches at 2036 and 1939 cm−1; hence, the growth of the transients is accompanied by a decrease in the intensities of both bleaches and a distortion of the band shape of the 1939 cm−1 bleach. The down-shift in the energies of the transient bands from the ground-state positions is typical of ππ* 3IL(dppz) excited states.19–23,26,27,54 Tentatively, we attribute the initially formed 2026 and 1908 cm−1 transient bands to a hot 3IL state localized at the phen part of the dppz ligand, 3IL(phen). Subsequent electron density reorganization and cooling produce another 3IL state localized predominantly at the phenazine part, 3IL(phz), manifested as the sharp 2036 cm−1 band. The 3IL(phz) IR spectrum is more similar to that of the ground states than the 3IL(phen) spectrum since the electronic changes in 3IL(phz) occur further away from the Re center. The excited-state conversion is largely completed in the first 100 ps. The spectra measured at 100 and 500 ps also show a shoulder at ~2020 cm−1 that probably corresponds to a residual population of the IL(phen) state. The ps spectra do not show any bands attributable to Re→dppz MLCT states, which are expected to occur at higher energies.
Figure 5.
Picosecond-timescale TRIR difference spectra of 4.8 mM (base pairs) AT-30 (top) or GC-30 (bottom) with 0.5 mM Re′-OH measured at specified time delays after 355 nm, 50 fs excitation. Each probe data point is separated by ca. 2.1 cm−1. Arrows indicate changes in the spectra with time. Delay times displayed are a subset of the data collected. (Similar TRIR spectra of Re-25(I) and Re-25(G) are shown in Figure S3.)
The GC-30 sample shows very similar behavior (Figure 5, bottom); however, there is one important difference: the ~2031 cm−1 3IL(phz) feature at longer time delays (>50 ps) is much weaker relative to the initially formed transient than in the case of AT-30. In accordance with the ultrafast GC-30 emission intensity quenching, we attribute this deficiency to a partial picosecond quenching of the 3IL state(s) by CT with guanine to produce [ReI(CO)3(dppz•−)(py′-OH)] and G•+. The lack of IR features in the TRIR spectra due to the reduced Re complex is likely caused by two factors. The first is very close similarity with the spectrum of the 3IL(phz) state (compare with Figure 2); the second is very fast BET that regenerates the ground state and keeps the concentration of the reduced state low. The persistence of the 2031 cm−1 band of GC-30 into the nanosecond-to-microsecond domain (see below) demonstrates that the relaxed 3IL(phz) state of Re′-OH shows little reactivity, if any. This spectral feature could also correspond to a population of Re complexes that are protected from solvent quenching by DNA binding but are not well coupled to the base stack.
The picosecond TRIR spectrum of AT-30 in the DNA region is very similar to that measured in the nanosecond time domain (Figure 6). The spectra show instantaneous formation of bleach bands at ca. 1618 (weak), 1635, 1660, and 1690 (weak) cm−1 that are not accompanied by the formation of transients. These bleaches originate from a decrease in the intensity of the nucleobase carbonyl IR bands upon excitation, rather than band shifts, and they compare well with bleaches observed upon direct 267 nm photoexcitation of nucleic acid polymers.37 The GC-30 sample shows strong bleaches at about 1577, 1619 (weak), 1648 and 1679 cm−1, again without the formation of transients. Notably, on the picosecond timescale there is no evidence of a transient due to oxidized G•+ or G•, which would be expected at ~1700 cm−1.38,43,44 The absence of such a transient is consistent with the ultrafast BET proposed above.
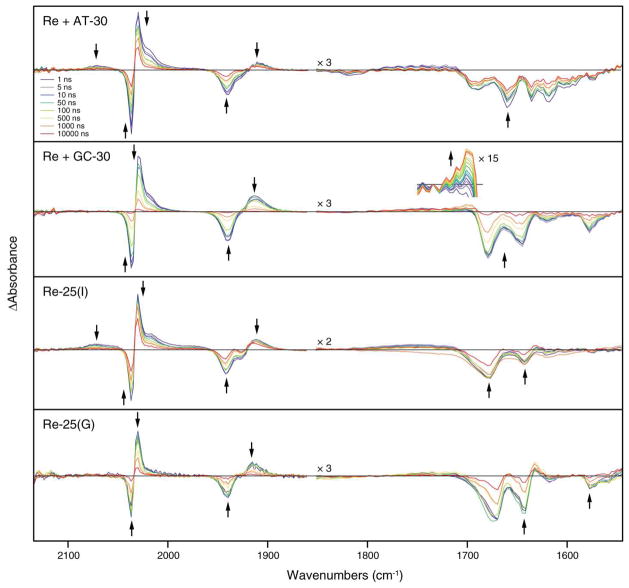
Figure 6.
Nanosecond-timescale TRIR difference spectra showing changes in the IR absorbance of systems containing 0.5 mM [Re(CO)3(dppz)(py′-OR)]+ and 4.8 mM DNA (base pairs) following 355 nm excitation. Both the Re(CO)3 ν(C≡O) (1860–2150 cm−1) and the DNA C=O stretching (1550–1850 cm−1) regions are shown. Arrows indicate changes in the spectra with time. Delay times displayed are a subset of the data collected. The growth of the signal at ~1700 cm−1 in the GC-30 sample is shown in the inset.
Picosecond TRIR spectra (Figure S3) of the Re-25(I) and Re-25(G) samples in both the Re(CO)3 ν(C≡O) and the DNA carbonyl regions closely resemble those of the AT-30 and GC-30 samples, respectively. Importantly, the 3IL(phz) band intensity at 100–500 ps is much lower for Re-25(G) than Re-25(I) relative to the initial transient, again indicating ultrafast *Re→G CT. Absence of any [ReI(CO)3(dppz•−)(py′-OH)] or G•+/G• IR features suggests ultrafast BET, as in the case of GC-30.
TRIR spectra recorded between 1 ns and 10 μs after photoexcitation are shown in Figure 6. The spectral patterns are very similar to those obtained in picosecond experiments at 100 ps and longer: The 3IL(phz) bands, as well as the bleaches in the DNA region, appear prominently in all four samples. Despite these similarities, closer examination reveals several important spectral differences. The AT-30 and Re-25(I) samples both show a weak isolated positive band at 2070 cm−1 and a broad, positive absorbance near 1980 cm−1. The 2070 cm−1 band can be assigned definitively to the MLCT excited state based on analyses of related complexes.19–23,25–27,54,69,77 This assignment predicts two additional low-intensity absorption bands near 2015 cm−1 and 1960 cm−1 due to hypsochromic shift of the A′(2) and A″ modes upon excitation of the complex into the MLCT state. These features are probably encompassed by the broad unresolved absorption between 1960 cm−1 and 1990 cm−1 and eclipsed by the much stronger absorption of IL states at higher energies. The MLCT features are absent in the GC-30 and Re-25(G) spectra. The AT-30 and Re-25(I) samples also exhibit a pronounced shoulder near 2020 cm−1 that is weaker for GC-30 and nearly absent in the Re-25(G) sample. This shoulder grows in intensity with increasing sample irradiation during the experiment, so it is in part related to transient absorption of a side photoproduct. However, its greater intensity in the AT-30 and Re-25(I) samples may be due to the presence of an underlying MLCT band or residual population of the 3IL(phen) state, as observed in the picosecond experiments (see above). Importantly, on the nanosecond timescale, TRIR spectra of GC-30 in the DNA region show a growing band at ~1700 cm−1 attributable to the oxidized guanine radical, G•+ or G•. This transient is very similar to that observed at 1702 cm−1 in both 5′-dGMP and poly(dG-dC)·poly(dG-dC) upon 200 nm photoionization, which was assigned to oxidized guanine (although the particular ionic state of this radical was not determined).38,43,44
The nanosecond kinetic behavior of the four samples differs substantially in several ways (Table 1). (i) The bleach recoveries and 3IL(phz) decays of the AT-30 and Re-25(I) are largely composed of long-lived components (≥ 20 μs) with smaller contributions on the timescale of tens to hundreds of nanoseconds. The occurrence of such slow microsecond processes, which have no counterparts in emission decays, indicates the presence of long-lived, non-emissive 3IL excited states or transient species. (ii) The AT-30 and Re-25(I) MLCT band at ~2070 cm−1 is fully formed in the 1 ns spectra and decays monotonically over time with lifetimes of 9 and 32 ns, respectively. In general, the lifetimes of the MLCT bands are significantly different than those of the IL bands, showing that the 3IL and 3MLCT states are not equilibrated. Importantly, the 2070 cm−1 MLCT band is completely absent in the spectra of the GC-30 and Re-25(G) samples, probably due to very fast quenching of the 3MLCT excited state by guanine. (iii) Compared with AT-30 and Re-25(I), both GC-30 and Re-25(G) show faster 3IL decay and bleach recovery (iv) Direct IR evidence for G•+/G• formation was obtained for the GC-30 sample, where a band appears with a lifetime of 210 ns in the DNA spectral region at ~1700 cm−1 and then decays over ~20 μs with a lifetime estimated roughly as 6 μs (Figure S4).
As in the picosecond TRIR spectra, we do not see any distinct signals attributable to the reduced Re sensitizer [ReI(CO)3(dppz•−)(py′-OH)] in any of the samples. This again is because its IR spectrum is nearly identical with that of the 3IL(phz) state; moreover, the yield of reduced Re species is low due to efficient BET.
Visible TA
Transient absorption decay in the visible spectral range at 475 nm was investigated in order to compare the TRIR kinetics specific to the Re(CO)3 moiety with those of the dppz part of the chromophore. A single exponential term was a poor model for the transient decay, indicating that more than one transient species exists during the course of the measurement. Biexponential fit parameters for the transient decays are shown in Table 1. It should be noted that the TA experiments were performed with a time resolution of about 10 ns, so they only provide information on the slower kinetics and longer-lived intermediates. Still, the TA decay lifetimes for each sample are comparable to the decay of the TRIR band near 2030 cm−1, including the lifetime shortening upon guanine incorporation near the Re binding site. It follows that the same states and processes are monitored by both methods. In a similar system, the TA spectrum of the reduced state following 355 nm excitation of a Re-DNA conjugate could not be distinguished from the spectrum of the excited state, presumably due the greater concentration of the excited state and the strong similarity between the two spectra.66 However, a change in the lifetime of the transient upon DNA binding suggested that DNA-mediated quenching by guanine was taking place. A similar effect is expected for the conjugates studied here.
DISCUSSION
Interactions Between [Re(CO)3(dppz)(py′-OR)]+ and DNA
Strong interactions between intercalating metal complexes and DNA are well known. As observed with several other dppz-bearing cationic metal complexes, incubation with DNA results in hypochromicity of the electronic spectrum and increased luminescence of the complex.78–80 Certainly, the light switch effect is a strong indicator of intercalative binding. Biexponential emission decays observed for other light switch complexes bound to DNA, such as dppz complexes of Ru, have been attributed to the existence of two different intercalative binding modes: a perpendicular mode, in which the metal-phenazine axis of the dppz ligand lies along the DNA dyad axis, and a side-on mode, in which the metal-phenazine axis lies along the long axis of the base pairs.81 In a similar way, the multiexponential emission decays observed for the Re complexes are probably due in part to the existence of several binding modes. Emission decay lifetimes of intercalated complexes are also affected by the DNA sequence to which they are bound.82–84 Although the range of DNA binding sites in the tethered complexes is limited, the tether is flexible enough to allow for binding at any of several locations, each of which may have a different effect on the luminescence lifetime. Similarly, for non-tethered samples, slight variations in the sequence at the binding site may contribute differently to the overall decay. DNA sequence effects, therefore, also contribute to the multiexponential emission decay kinetics of bound complexes.
The bleaches observed in the organic carbonyl stretching region (1600 cm−1 to 1700 cm−1) of the TRIR spectra could be another indication of the strong interaction between the complexes and DNA. It is possible that such bleach signals arise from direct photoexcitation of DNA, but excited states thus generated are expected to persist for only a few nanoseconds.76 On the contrary, the μs DNA bleach recovery lifetimes, commensurate with the Re excited-state lifetimes observed herein, indicate that the bleached signals originate from perturbation of the bases upon photoexcitation of the electronically coupled Re chromophore. A similar effect was observed previously upon 400 nm photoexcitation of [Ru(dppz)(tap)2]2+ intercalated nonspecifically into poly(dG-dC)·poly(dG-dC).85 In that work, a series of overlapping bleach and transient signals in the organic carbonyl stretching region at short times (2 ps to 2 ns) was attributed to guanine oxidation by excited Ru via a proton-coupled electron transfer (PCET) mechanism. Such a mechanism seems unlikely in our system because of the absence of TRIR transients that could be assigned to changes in cytosine carbonyl stretching frequency.
Guanine Oxidation by *[Re(CO)3(dppz)(py′-OR)]+
Previous work has shown that extended irradiation of mixtures of [Re(CO)3(dppz)(py)]+ and supercoiled plasmid DNA at λ > 350 nm results in nicks in the DNA backbone.42 In that work, the yield of cleavage did not depend on the concentration of singlet oxygen, suggesting that cleavage is the result of direct oxidation of guanine by the excited complex. The experimental results described here provide further evidence for the oxidation of guanine in DNA duplexes by photoexcited [Re(CO)3(dppz)(py′-OR)]+. In PAGE experiments, oxidation was observed preferentially at the the 5′-guanine of the 5′-GG-3′ doublet. Importantly, the observation of oxidation at this site, at least three base pairs removed from the Re binding site, indicates that long-range DNA-mediated CT has occurred. The preferential oxidation of the 5′-guanine of the doublet is typical for long-range DNA-mediated CT processes.86,87 This pattern is due to localization of the injected hole at guanine, the site of lowest oxidation potential.68 Once localized on guanine, proton transfer with base-paired cytosine results in the formation of the neutral guanine radical (k ≈ 107 s−1).38 In this state, the radical is quite stable, and can persist for > 1 ms.88 In the present study, a greater yield of guanine damage was observed by PAGE at the guanine doublet in Re-25(I) than in Re-25(G). This result can be attributed to the effect of the flanking guanines in Re-25(G). For each photon absorbed, CT may occur to any low potential guanine site that is well-coupled to the probe. Statistically, transfer to and trapping at the guanine doublet is more probable in Re-25(I) than in Re-25(G) since CT to inosine is expected to be thermodynamically less favorable. The long-range DNA-mediated oxidation of guanine observed in the gel experiment is not surprising, given the favorable driving force and strong electronic coupling between the complex and DNA.
The spectroscopic data are also consistent with guanine oxidation. By both steady-state and time-resolved emission, the luminescence intensity of each AT-30 and Re-25(I) is greater than that for GC-30 and Re-25(G), respectively. In early work, a similar disparity in the emission intensity of [Re(CO)3(dppz)(py)]+, a known DNA light-switch complex, bound to poly(dA)·poly(dT) versus poly(dG)·poly(dC) was ascribed to steric inhibition of binding to the latter duplex.42 Such an interpretation falls short on several accounts. First, it cannot explain the difference in emission intensity observed between Re-25(I) and Re-25(G); exchanging guanine for inosine at the Re binding site is expected to present a negligible change in steric interactions between the complex and the duplex. Second, it is not consistent with the equal degree of hypochromicity observed in the electronic spectrum of a similar Re complex when bound to either poly(dG-dC)·poly(dG-dC) or poly(dA-dT)·poly(dA-dT).57 Finally, it contradicts the strong luminescence observed from the bulkier light switch complex [Ru(bpy)2(dppz)]2+ bound to poly(dG-dC)·poly(dG-dC).72,89 A more consistent explanation put forth involves facile quenching of the Re excited state by guanine.43,44,57 CT from excited Re to guanine accounts well for our observation that the Re-25(G) and GC-30 samples, in which guanine neighbors the intercalation site, show less emission than the Re-25(I) and AT-30 samples, in which direct interaction between the complex and guanine is prevented.
TRIR spectra reported above provide further information on the rate and mechanism of guanine oxidation in GC-30 and Re-25(G). The reduced yield of the IL(phz) state relative to AT-30 and Re-25(I) suggests that *Re→G CT involves the IL(phen) state and occurs on a comparable timescale as the IL(phen)→IL(phz) conversion, namely a few tens of picoseconds. In addition, the absence of MLCT features in spectra observed on the nanosecond timescale shows that parallel CT involving the MLCT state occurs with a sub-nanosecond lifetime. Under some circumstances, IL(phz) could be reactive as well, but we have no direct evidence for a process involving this state. The rate of *Re→G CT cannot be determined exactly by TRIR because the spectral patterns of the IL-excited and reduced states cannot be distinguished. Nevertheless, the picosecond-timescale CT rates are further corroborated by comparison of the instantaneous (t = 0) emission intensity between samples. On the nanosecond timescale, the four samples give similar emission decay rates, although the integrated emission intensity is much less for Re-25(G) and GC-30 than for Re-25(I) and AT-30.
The reason for the absence of a guanine oxidation signal in TRIR spectra of Re-25(G) and Re-25(I) is unclear, but it may be an effect of the mixed base sequence used in these constructs. In previous studies of guanine oxidation by [RuIII(phen)2(dppz)]3+, a strong transient was observed in the visible region that was attributed to the neutral guanine radical when the complex was intercalated in poly(dG)·poly(dC) or poly(dG-dA)·poly(dC-dT), but no signal was seen when the complex was intercalated in poly(dG-dT)·poly(dC-dA).88 This difference was attributed to sequence dependent variations in the redox potential of guanine or structural variations, which would alter the coupling in the system.
Long-Lived Transient States
In addition to the reactive Re states, TRIR and TA measurements indicate that one or more non-emissive transient states persists long after the emissive species has been depleted. We have established that the long-lived transients are composed primarily of mixtures of Re in the 3IL(phz) excited state and in the reduced state, [Re(CO)3(dppz•−)(py′-OR)]. The long-lifetime decay processes observed by these absorption methods therefore contain contributions from the decay of these two states. From the 3IL(phz) state, the decay is likely due to internal conversion to the ground state. From the reduced state, the decay is caused by charge recombination, i.e., BET. The observation of long-lived transients in the AT-30 sample and the possibility for the oxidation of adenine by excited Re indicate that some amount of charge injection may occur in the absence of guanine. However, the lack of evidence for the formation of A•+ and the relatively strong emission observed in the AT-30 system suggest that if CT with adenine occurs, it is slow, minimally competitive with emission, and followed by fast BET.
Suggested Mechanism of DNA-Mediated Guanine Oxidation
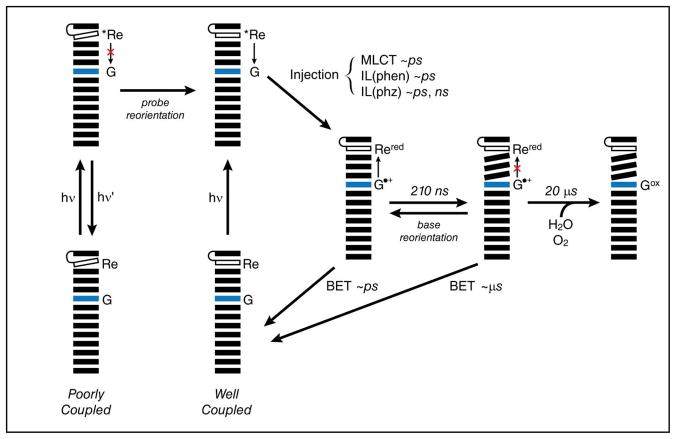
Based on spectroscopic evidence, a model can be generated for the oxidation of guanine by excited [Re(CO)3(dppz)(py′-OR)]+ (Figure 7). Photoexcitation of the complex populates a mixture of close lying IL(phen), IL(phz) and MLCT excited states, presumably spin-triplets, that are clearly observed by TRIR. (Such a mixture of states has been observed experimentally in several rhenium tricarbonyl complexes and has been verified in computational models.16,17,19,21–23,26,27,41,90) Based on our TRIR results, it appears that different excited states are more or less likely to participate in DNA-mediated CT. The MLCT state in particular, which is not observed in samples where the excited complex is in direct contact with guanine, seems to be more easily quenched than the IL states. The CT reactivity appears to decrease in the order MLCT > IL(phen) > IL(phz). It is also possible that conversion between excited states affects the apparent rates and yields observed for charge injection or emission. The reaction pathways from the excited state are also governed by the extent of electronic coupling in the system, which itself is determined by the dynamics of the probe and of the bases themselves.66 At the instant of excitation, two major populations exist. The first involves complexes which are poorly bound or which are bound to DNA in orientations that are not conducive to electron transfer. In this population, the mechanism of relaxation involves either quenching by water, as is observed for dppz complexes of Ru in polar, protic solvents,56,91 or emission. Emission is expected to occur primarily from the 3IL state, as reported for [Re(CO)3(dppz)(py)]+ in acetonitrile.19 In the second population, the excited complex is well coupled to the DNA. Here, excited state quenching via positive charge (i.e., hole) injection into the DNA duplex is the preferred reaction pathway. Indeed, primarily coherent CT at a distance of ten base pairs was observed in systems utilizing 2-aminopurine as a hole donor.60 Such processes are rapid. In systems involving DNA-bound ethidium, DNA-mediated CT over distances of several bases was observed to occur in 5 ps.92 Further, emission quenching is not limited to the population that exists in a CT-active configuration at the moment of excitation; reorientation of the bound oxidant to generate such a configuration may occur within the lifetime of the excited state. The rate of reorientation for DNA-bound ethidium is 75 ps,92 although for a larger molecule such as [Re(CO)3(dppz)(py′-OR)]+, this rate may be slower. Following charge separation, charge recombination (BET) may occur. After all, the ground state oxidation of [ReI(CO)3(dppz•−)(py′-OH)]0 [E° (Re+/Re0) = −0.85 V vs. NHE] by G•+ [E° (G•+/G) = 1.29 V vs. NHE]68 is thermodynamically favorable, and immediately after charge separation, the system exists in a CT-active state. Back reaction along this pathway is consistent with the absence of a guanine signal at short times in TRIR experiments. While this non-productive reaction pathway can be invoked to explain some of the experimental observations, additional pathways must be operative; quantitative deactivation of the charge separated state via short-range BET would prevent the eventual formation of permanent oxidative damage. A third population, then, involves molecules that are well coupled during charge injection, but that lose coupling before BET can take place due to reorientational motion of either the probe or the bases. The holes thus isolated within the base stack are quite stable and can migrate away from the site of injection, further reducing the probability for BET to occur and increasing the yield of permanent oxidative damage.60 Charge migration is limited in rate by stacking and destacking motions of the duplex, which form transient delocalized electronic domains.93,94 The 210 ns rate of formation for the guanine radical signal observed at ~1700 cm−1 by TRIR in the GC-30 sample may therefore reflect the rate of this conformational gating.
Figure 7.
The proposed model for the oxidation of guanine by photoexcited [Re(CO)3(dppz)(py′-OR)]+. Photoexcitation in the poorly coupled system results in emission (hν′) or non-radiative decay to the ground state. Photoexcitation in the well-coupled system results in charge injection over an arbitrary distance to form reduced [Re(CO)3(dppz)(py′-OR)]0 (Rered) and the guanine radical cation (G•+). During the excited state lifetime of the complex, the poorly coupled system may undergo reorientation, allowing charge injection. From the charge-separated state, facile back electron transfer (BET) competes with charge migration and trapping, resulting either in no reaction or the formation of permanent oxidation products. Base motions may result in isolation of the injected charge, favoring the trapping pathway.
CONCLUDING REMARKS
Complexes that contain IR-active moieties show promise as probes for the study of DNA CT. In this work, we have used PAGE and time-resolved spectroscopy to observe the oxidation of guanine in DNA by photoexcited [Re(CO)3(dppz)(py′-OR)]+. Although no direct evidence for this reaction is afforded by UV/visible methods, fast excited-state quenching by guanine provides indirect evidence that oxidation is taking place. Direct evidence for the formation of guanine oxidation products is observed biochemically by PAGE analysis and spectroscopically by TRIR following photoexcitation of Re′-OH in the presence of GC-30. Similarities between the spectral features and kinetics of this system with those of other DNA sequences containing guanine allow us to conclude that the photochemical processes observed in the GC-30 sample are general. In these systems, the rate of guanine oxidation (herein 210 ns) is dictated largely by motions of the bases, which allow for long-range charge separation and prevent BET, rather than by the intrinsic photophysics of the photosensitizer complex. In this respect, the role of Re′-OH is similar to that of other photooxidants that have been used in DNA CT studies.60,93,94
Unlike the well-known [Ru(phen)2(dppz)]2+ DNA “light-switch,” rhenium(I) tricarbonyl-dppz complexes are strong enough photooxidants to inject positive charge into DNA directly from their electronically excited state(s), i.e., without the use of an external quencher and involvement of diffusion-controlled steps. This allows for ultrafast charge injection, with possible applications in mechanistic studies of DNA-mediated CT and in development of DNA-based photonic devices. However, the present study indicates that charge injection by [Re(CO)3(dppz)(py′-OR)]+ preferentially involves the initially populated IL(phen) and the minor MLCT states, with the long-lived 3IL(phz) state showing little reactivity, if any. This, together with fast BET, limits the reaction yield. From the experimental point of view, Re tricarbonyl-diimines have the advantage of being both ET phototriggers and probes by virtue of their sensitive IR spectral responses to changes in the electron density distribution.32,34 However, in the particular case of dppz complexes, the TRIR spectral analysis is complicated by the close resemblance of 3IL(phz) and reduced-state spectral patterns that renders the two species indistinguishable. It is suggested that optimization of the Re-photooxidant structure will improve both the charge injection efficiency and the IR spectral response.
A complete picture of DNA CT requires the observation of processes on very different timescales. At the instant of photoexcitation, the extent of coupling between the probe and the base stack, and between the bases themselves, defines two populations of DNA: one that is CT-active and one that is CT-inactive. The outcomes of fast processes, such as fluorescence and charge injection, are determined based on the relative sizes of these populations. At longer times, base motions change the energetic landscape, offering alternative reaction pathways, such as charge migration and trapping, that were not available immediately after excitation. TRIR allows for the observation of processes at all of these timescales, making it a valuable addition to the methods employed for the study of DNA-mediated CT.
Supplementary Material
Acknowledgments
We are grateful to the NIH (GM49216) for financial support. E.D.O. also thanks G.E. Keller, N.D. Ford, and J.R. Winkler for assistance with ps emission measurements and M.G. Hill for assistance with spectroelectrochemical measurements.
Footnotes
Supporting Information. Strand sequences and MALDI data; Steady-state emission spectra of AT-30 and AT-30 with Re′-OEt before correction for emission from DNA; histograms from maximum entropy analysis of ps time-resolved emission data; ps TRIR difference spectra of Re-25(G) and Re-25(I) samples; detailed view of ns TRIR difference spectra in the DNA spectral region for the GC-30 + Re′-OH sample. This material is available free of charge via the internet at http://pubs.acs.org.
References
- 1.Genereux JC, Barton JK. Chem Rev. 2010;110:1642–1662. doi: 10.1021/cr900228f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barton JK, Olmon ED, Sontz PA. Coord Chem Rev. 2010;255:619–634. doi: 10.1016/j.ccr.2010.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schuster GB. Topics in Current Chemistry: Long-Range Charge Transfer in DNA I. Springer-Verlag; New York: 2004. [Google Scholar]
- 4.Núñez ME, Hall DB, Barton JK. Chem Biol. 1999;6:85–97. doi: 10.1016/S1074-5521(99)80005-2. [DOI] [PubMed] [Google Scholar]
- 5.Slinker JD, Muren NB, Renfrew SE, Barton JK. Nature Chem. 2011;3:228–233. doi: 10.1038/nchem.982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Augustyn KE, Genereux JC, Barton JK. Angew Chem Int Ed. 2007;46:5731–5733. doi: 10.1002/anie.200701522. [DOI] [PubMed] [Google Scholar]
- 7.Gorodetsky AA, Buzzeo MC, Barton JK. Bioconjugate Chem. 2008;19:2285–2296. doi: 10.1021/bc8003149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guo X, Gorodetsky AA, Hone J, Barton JK, Nuckolls C. Nature Nanotech. 2008;3:163–167. doi: 10.1038/nnano.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slinker JD, Muren NB, Gorodetsky AA, Barton JK. J Am Chem Soc. 2010;132:2769–2774. doi: 10.1021/ja909915m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genereux JC, Boal AK, Barton JK. J Am Chem Soc. 2010;132:891–905. doi: 10.1021/ja907669c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boal AK, Genereux JC, Sontz PA, Gralnick JA, Newman DK, Barton JK. Proc Natl Acad Sci USA. 2009;106:15237–15242. doi: 10.1073/pnas.0908059106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee PE, Demple B, Barton JK. Proc Natl Acad Sci USA. 2009;106:13164–13168. doi: 10.1073/pnas.0906429106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Augustyn KE, Merino EJ, Barton JK. Proc Natl Acad Sci USA. 2007;104:18907–18912. doi: 10.1073/pnas.0709326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Turner JJ, George MW, Johnson FPA, Westwell JR. Coord Chem Rev. 1993;125:101–114. [Google Scholar]
- 15.Kumar A, Sun S-S, Lees AJ. Top Organomet Chem. 2010;29:1–35. [Google Scholar]
- 16.Stufkens DJ, Vlček A. Coord Chem Rev. 1998;177:127–179. [Google Scholar]
- 17.Vlček A. Top Organomet Chem. 2010;29:73–114. [Google Scholar]
- 18.Dyer J, Grills DC, Matousek P, Parker AW, Towrie M, Weinstein JA, George MW. Photochem Photobiol Sci. 2002:872–873. doi: 10.1039/b200586g. [DOI] [PubMed] [Google Scholar]
- 19.Dyer J, Blau WJ, Coates CG, Creely CM, Gavey JD, George MW, Grills DC, Hudson S, Kelly JM, Matousek P, McGarvey JJ, McMaster J, Parker AW, Towrie M, Weinstein JA. Photochem Photobiol Sci. 2003;2:542–554. doi: 10.1039/b212628a. [DOI] [PubMed] [Google Scholar]
- 20.Kuimova MK, Alsindi WZ, Dyer J, Grills DC, Jina OS, Matousek P, Parker AW, Porius P, Sun XZ, Towrie M, Wilson C, Yang J, George MW. Dalton Trans. 2003:3996–4006. [Google Scholar]
- 21.Kuimova MK, Sun Z, Matousek P, Grills DC, Parker AW, Towrie M, George MW. Photochem Photobiol Sci. 2007;6:1158–1163. doi: 10.1039/b705002j. [DOI] [PubMed] [Google Scholar]
- 22.Dyer J, Creely CM, Penedo JC, Grills DC, Hudson S, Matousek P, Parker AW, Towrie M, Kelly JM, George MW. Photochem Photobiol Sci. 2007;6:741–748. doi: 10.1039/b618651c. [DOI] [PubMed] [Google Scholar]
- 23.Kuimova MK, Alsindi WZ, Blake AJ, Davies ES, Lampus DJ, Matousek P, McMaster J, Parker AW, Towrie M, Sun X-Z, Wilson C, George MW. Inorg Chem. 2008;47:9857–9869. doi: 10.1021/ic800753f. [DOI] [PubMed] [Google Scholar]
- 24.Vlček A, Záliš S. Coord Chem Rev. 2007;251:258–287. [Google Scholar]
- 25.George MW, Turner JJ. Coord Chem Rev. 1998;177:201–217. [Google Scholar]
- 26.Vlček A, Busby M. Coord Chem Rev. 2006;250:1755–1762. [Google Scholar]
- 27.Busby M, Matousek P, Towrie M, Vlček A. Inorg Chim Acta. 2007;360:885–896. [Google Scholar]
- 28.Lo KK-W, Louie M-W, Zhang KY. Coord Chem Rev. 2010;254:2603–2622. [Google Scholar]
- 29.Blanco-Rodríguez AM, Busby M, Grădinaru C, Crane BR, Di Bilio AJ, Matousek P, Towrie M, Leigh BS, Richards JH, Vlček A, Gray HB. J Am Chem Soc. 2006;3:3552–3557. doi: 10.1021/ja057451+. [DOI] [PubMed] [Google Scholar]
- 30.Blanco-Rodríguez AM, Busby M, Ronayne K, Towrie M, Grădinaru C, Sudhamsu J, Sýkora J, Hof M, Záliš S, Di Bilio AJ, Crane BR, Gray HB, Vlček A. J Am Chem Soc. 2009;3:11788–11800. doi: 10.1021/ja902744s. [DOI] [PubMed] [Google Scholar]
- 31.Gray HB, Winkler JR. Chem Phys Lett. 2009;483:1–9. doi: 10.1016/j.cplett.2009.10.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Blanco-Rodríguez AM, Di Bilio AJ, Shih C, Museth AK, Clark IP, Towrie M, Cannizzo A, Sudhamsu J, Crane BR, Sýkora J, Winkler JR, Gray HB, Záliš S, Vlček A. Chem Eur J. 2011;17:5350–5361. doi: 10.1002/chem.201002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Connick WB, Di Bilio AJ, Hill MG, Winkler JR, Gray HB. Inorg Chim Acta. 1995;240:169–173. [Google Scholar]
- 34.Shih C, Museth AK, Abrahamsson M, Blanco-Rodríguez AM, Di Bilio AJ, Sudhamsu J, Crane BR, Ronayne KL, Towrie M, Vlček A, Richards JH, Winkler JR, Gray HB. Science. 2008;320:1760–1762. doi: 10.1126/science.1158241. [DOI] [PubMed] [Google Scholar]
- 35.Encinas S, Morales AF, Barigelletti F, Barthram AM, White CM, Couchman SM, Jeffery JC, Ward MD, Grills DC, George MW. J Chem Soc, Dalton Trans. 2001:3312–3319. [Google Scholar]
- 36.Towrie M, Doorley GW, George MW, Parker AW, Quinn SJ, Kelly JM. Analyst. 2009;134:1265–1273. doi: 10.1039/b902108f. [DOI] [PubMed] [Google Scholar]
- 37.Kuimova MK, Dyer J, George MW, Grills DC, Kelly JM, Matousek P, Parker AW, Sun XZ, Towrie M, Whelan AM. Chem Commun. 2005:1182–1184. doi: 10.1039/b414450c. [DOI] [PubMed] [Google Scholar]
- 38.Kuimova MK, Cowan AJ, Matousek P, Parker AW, Sun XZ, Towrie M, George MW. Proc Natl Acad Sci USA. 2006;103:2150–2153. doi: 10.1073/pnas.0506860103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hare PM, Middleton CT, Mertel KI, Herbert JM, Kohler B. Chem Phys. 2008;347:383–392. doi: 10.1016/j.chemphys.2007.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McGovern DA, Doorley GW, Whelan AM, Parker AW, Towrie M, Kelly JM, Quinn SJ. Photochem Photobiol Sci. 2009;8:542–548. doi: 10.1039/b817756b. [DOI] [PubMed] [Google Scholar]
- 41.Stoeffler HD, Thornton NB, Temkin SL, Schanze KS. J Am Chem Soc. 1995;117:7119–7128. [Google Scholar]
- 42.Yam VW-W, Lo KK-W, Cheung K-K, Kong RY-C. J Chem Soc, Dalton Trans. 1997;14:2067–2072. [Google Scholar]
- 43.Smith JA, George MW, Kelly JM. Coord Chem Rev. 2011 doi: 10.1016/j.ccr.2011.04.007. [DOI] [Google Scholar]
- 44.Cao Q, Creeley CM, Dyer J, Easun T, Grills DC, Hudson S, McGovern DA, McMaster J, Pitchford J, Smith JA, Sun X-Z, Kelly JM, George MW. Photochem Photobiol Sci. 2011 doi: 10.1039/C1PP05050H. [DOI] [PubMed] [Google Scholar]
- 45.Holmlin RE, Dandliker PJ, Barton JK. Bioconjugate Chem. 1999;10:1122–1130. doi: 10.1021/bc9900791. [DOI] [PubMed] [Google Scholar]
- 46.Zeglis BM, Barton JK. Nat Protoc. 2007;2:357–371. doi: 10.1038/nprot.2007.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Boyle PD, Boyd DC, Mueting AM, Pignolet LH. Inorg Chem. 1988;27:4424–4429. [Google Scholar]
- 48.Pletneva EV, Gray HB, Winkler JR. J Molec Biol. 2005;345:855–867. doi: 10.1016/j.jmb.2004.10.085. [DOI] [PubMed] [Google Scholar]
- 49.Pletneva EV, Gray HB, Winkler JR. Proc Natl Acad Sci USA. 2005;102:18397–18402. doi: 10.1073/pnas.0509076102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kimura T, Lee JC, Gray HB, Winkler JR. Proc Natl Acad Sci USA. 2007;104:117–122. doi: 10.1073/pnas.0609413103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kimura T, Lee JC, Gray HB, Winkler JR. Proc Natl Acad Sci USA. 2009;106:7834–7839. doi: 10.1073/pnas.0902562106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Greetham GM, Burgos P, Cao Q, Clark IP, Codd PS, Farrow RC, George MW, Kogimtzis M, Matousek P, Parker AW, Pollard MR, Robinson DA, Xin Z-J, Towrie M. Appl Spectrosc. 2010;64:1311–1319. doi: 10.1366/000370210793561673. [DOI] [PubMed] [Google Scholar]
- 53.Towrie M, Gabrielsson A, Matousek P, Parker AW, Blanco-Rodríguez AM, Vlček A. Appl Spectrosc. 2005;59:467–473. doi: 10.1366/0003702053641397. [DOI] [PubMed] [Google Scholar]
- 54.Dattelbaum DM, Omberg KM, Hay PJ, Gebhart NL, Martin RL, Schoonover JR, Meyer TJ. J Phys Chem A. 2004;108:3527–3536. [Google Scholar]
- 55.Gamelin DR, George MW, Glyn P, Grevels F-W, Johnson FPA, Klotzbücher W, Morrison SL, Russell G, Schaffner K, Turner JJ. Inorg Chem. 1994;33:3246–3250. [Google Scholar]
- 56.Turro C, Bossmann SH, Jenkins Y, Barton JK, Turro NJ. J Am Chem Soc. 1995;117:9026–9032. [Google Scholar]
- 57.Ruiz GT, Juliarena MP, Lezna RO, Wolcan E, Feliz MR, Ferraudi G. Dalton Trans. 2007;3:2020–2029. doi: 10.1039/b614970g. [DOI] [PubMed] [Google Scholar]
- 58.Dohno C, Stemp EDA, Barton JK. J Am Chem Soc. 2003;125:9586–9587. doi: 10.1021/ja036397z. [DOI] [PubMed] [Google Scholar]
- 59.Williams TT, Dohno C, Stemp EDA, Barton JK. J Am Chem Soc. 2004;126:8148–8158. doi: 10.1021/ja049869y. [DOI] [PubMed] [Google Scholar]
- 60.Genereux JC, Wuerth SM, Barton JK. J Am Chem Soc. 2011;133:3863–3868. doi: 10.1021/ja107033v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoo J, Delaney S, Stemp EDA, Barton JK. J Am Chem Soc. 2003;125:6640–6641. doi: 10.1021/ja034326u. [DOI] [PubMed] [Google Scholar]
- 62.O’Neill MA, Dohno C, Barton JK. J Am Chem Soc. 2004;126:1316–1317. doi: 10.1021/ja037802p. [DOI] [PubMed] [Google Scholar]
- 63.Genereux JC, Augustyn KE, Davis ML, Shao F, Barton JK. J Am Chem Soc. 2008;130:15150–15156. doi: 10.1021/ja8052738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.The redox potentials of the canonical bases described in this work were determined by pulse radiolysis of the free nucleosides and are therefore estimates of the potentials of the bases in the DNA polymer environment. For a summary of experimentally-determined guanine redox potentials in different contexts, see Ref. 1.
- 65.Yam VW-W, Lo KK-W, Cheung K-K, Kong RY-C. J Chem Soc, Chem Commun. 1995;3:1191–1193. [Google Scholar]
- 66.Olmon ED, Hill MG, Barton JK. 2011 submitted. [Google Scholar]
- 67.Juris A, Balzani V, Barigelletti F, Campagna S, Belser P, Von Zelewsky A. Coord Chem Rev. 1988;84:85–277. [Google Scholar]
- 68.Steenken S, Jovanovic SV. J Am Chem Soc. 1997;119:617–618. [Google Scholar]
- 69.Dattelbaum DM, Omberg KM, Schoonover JR, Martin RL, Meyer TJ. Inorg Chem. 2002;41:6071–6079. doi: 10.1021/ic020400i. [DOI] [PubMed] [Google Scholar]
- 70.Burrows CJ, Muller JG. Chem Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 71.Williams TT, Odom DT, Barton JK. J Am Chem Soc. 2000;122:9048–9049. [Google Scholar]
- 72.Friedman AE, Chambron J-C, Sauvage J-P, Turro NJ, Barton JK. J Am Chem Soc. 1990;112:4960–4962. [Google Scholar]
- 73.Caspar JV, Meyer TJ. J Am Chem Soc. 1983;105:5583–5590. [Google Scholar]
- 74.Kwok W-M, Ma C, Phillips DL. J Phys Chem B. 2009;113:11527–11534. doi: 10.1021/jp906265c. [DOI] [PubMed] [Google Scholar]
- 75.Markovitsi D, Gustavsson T, Vayá I. J Phys Chem Lett. 2010;1:3271–3276. [Google Scholar]
- 76.Banyasz A, Vayá I, Changenet-Barret P, Gustavsson T, Douki T, Markovitsi D. J Am Chem Soc. 2011;133:5163–5165. doi: 10.1021/ja110879m. [DOI] [PubMed] [Google Scholar]
- 77.Kuimova MK, Gordon KC, Howell SL, Matousek P, Parker AW, Sun X-Z, Towrie M, George MW. Photochem Photobiol Sci. 2006;5:82–87. doi: 10.1039/b508335d. [DOI] [PubMed] [Google Scholar]
- 78.Hiort C, Lincoln P, Nordén B. J Am Chem Soc. 1993;115:3448–3454. [Google Scholar]
- 79.Holmlin RE, Barton JK. Inorg Chem. 1995;34:7–8. doi: 10.1021/ic970869r. [DOI] [PubMed] [Google Scholar]
- 80.Shao F, Elias B, Lu W, Barton JK. Inorg Chem. 2007;46:10187–10199. doi: 10.1021/ic7014012. [DOI] [PubMed] [Google Scholar]
- 81.Hartshorn RM, Barton JK. J Am Chem Soc. 1992;114:5919–5925. [Google Scholar]
- 82.Jenkins Y, Friedman AE, Turro NJ, Barton JK. Biochemistry. 1992;31:10809–10816. doi: 10.1021/bi00159a023. [DOI] [PubMed] [Google Scholar]
- 83.Holmlin RE, Stemp EDA, Barton JK. Inorg Chem. 1998;37:29–34. doi: 10.1021/ic970869r. [DOI] [PubMed] [Google Scholar]
- 84.Stemp EDA, Holmlin RE, Barton JK. Inorganica Chimica Acta. 2000;297:88–97. [Google Scholar]
- 85.Elias B, Creely C, Doorley GW, Feeney MM, Moucheron C, Kirsch-DeMesmaeker A, Dyer J, Grills DC, George MW, Matousek P, Parker AW, Towrie M, Kelly JM. Chem Eur J. 2008;14:369–375. doi: 10.1002/chem.200700564. [DOI] [PubMed] [Google Scholar]
- 86.Saito I, Takayama M, Sugiyama H, Nakatani K, Tsuchida A, Yamamoto M. J Am Chem Soc. 1995;117:6406–6407. [Google Scholar]
- 87.Hall DB, Holmlin RE, Barton JK. Nature. 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 88.Stemp EDA, Arkin MR, Barton JK. J Am Chem Soc. 1997;119:2921–2925. [Google Scholar]
- 89.Murphy CJ, Arkin MR, Ghatlia ND, Bossmann S, Turro NJ, Barton JK. Proc Natl Acad Sci USA. 1994;91:5315–5319. doi: 10.1073/pnas.91.12.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.MacQueen DB, Schanze KS. J Am Chem Soc. 1991;113:7470–7479. [Google Scholar]
- 91.Olson EJC, Hu D, Hörmann A, Jonkman AM, Arkin MR, Stemp EDA, Barton JK, Barbara PF. J Am Chem Soc. 1997;7863:11458–11467. [Google Scholar]
- 92.Wan C, Fiebig T, Kelley SO, Treadway CR, Barton JK, Zewail AH. Proc Natl Acad Sci USA. 1999;96:6014–6019. doi: 10.1073/pnas.96.11.6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.O’Neill MA, Becker H-C, Wan C, Barton JK, Zewail AH. Angew Chem Int Ed. 2003;42:5896–5900. doi: 10.1002/anie.200352831. [DOI] [PubMed] [Google Scholar]
- 94.Shao F, O’Neill MA, Barton JK. Proc Natl Acad Sci USA. 2004;101:17914–17919. doi: 10.1073/pnas.0408128101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.