Abstract
The complex changes in the life cycle of Clostridium acetobutylicum, a promising biofuel producer, are not well understood. During exponential growth, sugars are fermented to acetate and butyrate, and in the transition phase, the metabolism switches to the production of the solvents acetone and butanol accompanied by the initiation of endospore formation. Using phosphate-limited chemostat cultures at pH 5.7, C. acetobutylicum was kept at a steady state of acidogenic metabolism, whereas at pH 4.5, the cells showed stable solvent production without sporulation. Novel proteome reference maps of cytosolic proteins from both acidogenesis and solventogenesis with a high degree of reproducibility were generated. Yielding a 21% coverage, 15 protein spots were specifically assigned to the acidogenic phase, and 29 protein spots exhibited a significantly higher abundance in the solventogenic phase. Besides well-known metabolic proteins, unexpected proteins were also identified. Among these, the two proteins CAP0036 and CAP0037 of unknown function were found as major striking indicator proteins in acidogenic cells. Proteome data were confirmed by genome-wide DNA microarray analyses of the identical cultures. Thus, a first systematic study of acidogenic and solventogenic chemostat cultures is presented, and similarities as well as differences to previous studies of batch cultures are discussed.
Electronic supplementary material
The online version of this article (doi:10.1007/s00253-010-2741-x) contains supplementary material, which is available to authorized users.
Keywords: Phosphate-limited chemostat, Acidogenesis, Solventogenesis, Proteome reference maps, Transcriptome, CAP0037 and CAP0036
Introduction
The obligate anaerobe Clostridium acetobutylicum is well known for its acetone–butanol (AB) fermentation (Dürre 2008; Jones 2001; Jones and Woods 1986; Lee et al. 2008). Transition from exponential to stationary growth typically comprises a metabolic change from the formation of acetate and butyrate towards the formation of acetone and butanol as the dominant fermentation products. Due to its past importance for the industrial production of the bulk chemicals acetone and butanol (Jones 2001; Jones and Woods 1986) and its potential for future solvent production, e.g., especially with respect to n-butanol as promising bio-based liquid fuel with several advantages over ethanol (Dürre 2007; Ni and Sun 2009), much research has focused on the regulation of the metabolic “shift”. Since the genome sequence of C. acetobutylicum was published (Nölling et al. 2001), transcriptomic and proteomic studies were included (Alsaker and Papoutsakis 2005; Jones et al. 2008; Sullivan and Bennett 2006; Tomas et al. 2003). Nevertheless, the regulation of solvent formation remains largely unknown yet. Although much valuable information was provided, all these studies generally exhibited poor reproducibility as a major drawback because all experimental data resulted from simple batch fermentations. As a prerequisite for further comparative analyses, defined growth conditions without constantly changing environmental conditions are required. Continuous cultivation constitutes a preferred fermentation method to accomplish standardized conditions with a maximum degree of reproducibility because cells are kept in a steady state with constant endogenous as well as exogenous parameters, such as growth rate, substrate, and product concentrations.
Almost 30 years ago, chemostat cultivation of C. acetobutylicum was performed to analyze various parameters affecting butanol production. Using a synthetic medium and phosphate limitation, C. acetobutylicum could be kept in a steady state of acidogenesis when the pH was >5 and in a steady state of solventogenesis at pH 4.3 for at least several months (Bahl et al. 1982a, b). Although this process was not considered to be competitive with industrial butanol production in the batch or fed-batch mode (Jones et al. 1982), chemostat cultures are valuable scientific applications in analyzing specific parameters with limited or ideally without any perturbations.
Systematic “omic” analyses of chemostat cells are still at their beginning. The aim of this study was to comprehensively compare C. acetobutylicum acidogenic and solventogenic steady-state cells to provide new insights into the metabolic changes. Stable growth rates and constant exogenous parameters during the chemostat fermentation process enabled homogeneity of bacterial cells, and the pH as single parameter was changed to switch from acidogenesis to solventogenesis. Thus, a “master” fermentation was established and used for detailed analyses of the proteome and transcriptome of the identical culture of steady-state cells growing at pH 5.7 and pH 4.5, respectively.
Material and methods
Organism and growth conditions
C. acetobutylicum ATCC824 (COSMIC strain) was grown under anaerobic conditions at 37°C according to a standard operation procedure developed for the COSMIC consortium (http://www.sysmo.net). Precultures starting from spore stocks were prepared as previously described (Fischer et al. 2006). The phosphate-limited chemostat experiments (n = 2) with 0.5 mM KH2PO4 and 4% (wt/vol) glucose in the supplying medium (Fiedler et al. 2008) were performed in a BiostatB 1.5-l fermenter system (BBI, Melsungen, Germany) at 37°C. The dilution rate (respective generation time) was D = 0.075 h−1. After inoculation, cell growth caused a natural decrease of the pH. At pH 5.7, pH control was turned on and pH was kept constant by automatic addition of 2 M KOH until stable acidogenic steady state was reached and first sampling occurred. Then the pH control was temporarily turned off to induce solvent formation and was started again at pH 4.5, keeping the cells in stable solventogenic steady state when the second sampling took place (for further details, refer to “Results” section).
OD, phosphate concentration, and fermentation products
The measurement of the optical density at 600 nm (OD600), phosphate concentration, and the analysis of the fermentation products were accomplished as described elsewhere (Fischer et al. 2006).
Isolation of total RNA, microarray experiments, and Northern blot analysis
The isolation of total RNA and DNase treatment was performed as previously described (Fischer et al. 2006). The integrity of the RNA samples was verified by gel electrophoresis. RNA concentrations were determined by their specific absorptions at A260 nm. DNA-free RNA aliquots were transcribed into cDNA and cy3- or cy5-labeled, respectively. Microarray experiments using a DNA chip containing 3,840 spotted oligonucleotides of protein coding genes of C. acetobutylicum were performed as published recently (Hillmann et al. 2009). Microarray experiments were performed twice (n = 2), and duplicate arrays were hybridized with reverse-labeled samples (dye-flip: pH 5.7-Cy3/pH 4.5-Cy5 and pH 5.7-Cy5/pH 4.5-Cy3). Full lists of transcribed genes, functional information, and regulation are available in supplementary data SD5 and SD6. Microarray design and data are also available from the ArrayExpress database under accession no. E-MEXP-2503 (http://www.ebi.ac.uk/microarray-as/aer/entry).
Probes for the Northern hybridization were PCR-generated using standard protocols with Pwo polymerase (Peqlab, Biotechnology, Erlangen, Germany) and chromosomal DNA of C. acetobutylicum as template. Amplification of gene specific probes (cap0037 542 bp, cap0036 881 bp) based on the primers Cap0037_fw (5′-GAACAAGAGATGCTGTATTTTC-3′) and Cap0037_rev (5′-ACTTCTTGAGATAACTCATATTTC-3′) and Cap0036_fw (5′-TGTTTTATCATAGAAAGCCTTTTG-3′) and Cap0036_rev (5′-GAAAATACAGCATCTCTTGTTC-3′). The 16S rRNA probe (1,510 bp) amplified with the forward primer (Eub1, 5′-GAGTTTGATCCTGGCTCAG-3′) and the reverse primer (Eub2, 5′-AGAAAGGAGGTGATCCAGCC-3′) served as an internal control to ensure integrity and amount equity of the total RNA used. Labeling with digoxigenin (DIG) and Northern blot hybridization at 42°C (overnight) were performed according to the directions of the manufacturer (DIG DNA labeling and detection kit; Roche Diagnostic, Mannheim, Germany). Detection of chemiluminescence was performed as described previously (Fischer et al. 2006).
Two-dimensional gel electrophoresis
The soluble intracellular protein fractions of C. acetobutylicum cells were illustrated by two-dimensional polyacrylamide gel electrophoresis as described in detail by Schwarz et al. (2007). Proteins were visualized by colloidal Coomassie staining, and gels were documented by scanning (UMAX 2100, Biostep GmbH, Jahnsdorf, Germany). Gel images were transferred to Delta 2D software (version 3.5) for detailed analysis. All detected protein spots were identified by MALDI-TOF MS/MS.
MALDI-TOF MS/MS
Mass spectrometry was performed in a Proteomics Analyzer 4800 (Applied Biosystems, Foster City, USA) as published by Voigt et al. (2006), except that the signal to noise ratio for the TOF-TOF measurements was raised to 10. Proteins were identified using the Mascot search engine with a C. acetobutylicum databank (Matrix Science, London, UK; Voigt et al. 2006).
Results
Master fermentation
Cells of chemostat cultures are strongly recommended for “omic” analyses in contrast to batch cultures because exponentially growing cells can be kept at specifically defined physiological conditions without any transient changes in the environment. Thus, chemostat cultivation optimally allows analyses of the effects of single external parameter changes. In this study, acidogenic or solventogenic metabolism of C. acetobutylicum chemostat cells was directed exclusively by the external pH values of pH 5.7 or pH 4.5, respectively. Under both steady-state conditions, only vegetative cells and no other differentiated cell forms like clostridial stages, prespores containing cells, or mature spores were detected.
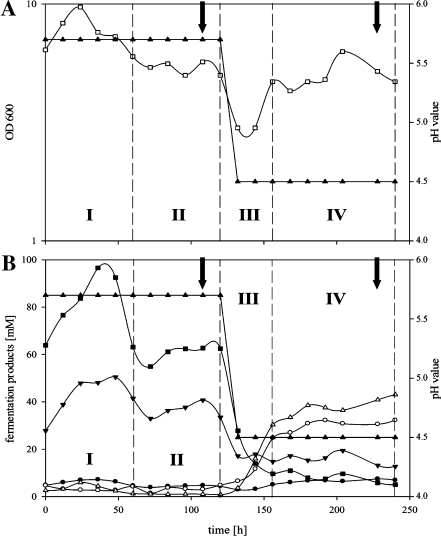
A typical continuous chemostat culture referred to as “master” fermentation is described in Fig. 1. The time course of a master fermentation was subdivided into four parts (I–IV). Phase I reflects the time period after starting the medium pump, the transition of batch growth to continuous pH-controlled growth at pH 5.7. Typically, acidogenic cells of phase I exhibited highest cell densities of up to OD 10 and consequently the highest butyrate and acetate concentrations measured throughout the master fermentation. After approximately 3 days (t = 65–80 h), stable cell densities (OD 5 ± 0.5) and fermentation products (acetate ~35 mM, butyrate ~60 mM) indicated the beginning of phase II, balanced steady-state growth at pH 5.7. To ensure the steady state, sampling took place at least 24 h later (usually 1 to 2 days later, respective 4 to 5 days (t = 96–120 h) after starting continuous growth). Thereafter, the pH control was turned off, initializing phase III. Promptly, the external pH decreased. This exclusive external signal induced reorganization of the metabolism of C. acetobutylicum from acidogenesis to solventogenesis, and at pH 4.5 (~15–20 h later), the pH control was turned on again. Lowering of the pH was typically accompanied by a transient drop of the cell densities documented by minimal OD values of about 3. Steady-state growth at pH 4.5 was usually reached within 48 h after the drop of the pH, the beginning of phase IV. At least 3 days later (around 10 days or 240 h after starting the continuous cultivation), the second sets of samples were drawn for the analyses of solventogenic cells.
Fig. 1.
“Master” fermentation of C. acetobutylicum. Monitored 240 h after starting the medium supply at t0. a shows pH (filled upright triangles) and optical density (empty squares); b illustrates the pH (filled upright triangles) and the fermentation products butyrate (filled squares), acetate (filled inverse triangles), butanol (empty upright triangles), acetone (empty circles), and ethanol (filled circles). Roman numbers highlight the four different phases, I: starting of continuous culture; II: establishing of steady-state growth at pH 5.7; III: switch of pH from 5.7 to 4.5; IV: establishing of steady-state growth at pH 4.5 of the fermentation. Arrows indicated sampling points for proteome and transcriptome analyses at the end of phase II and phase IV
Proteome analysis
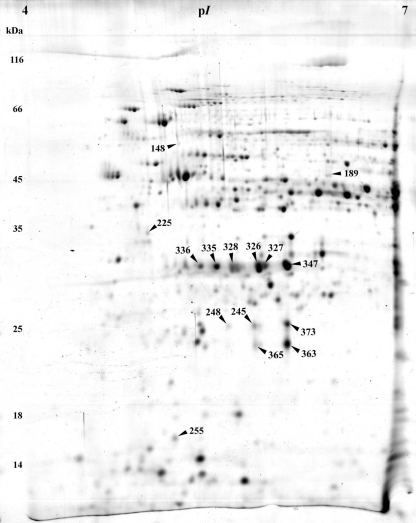
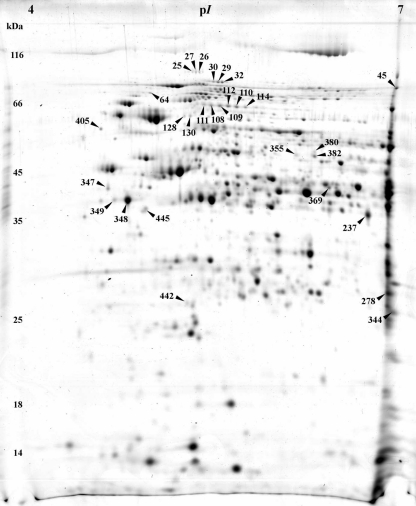
Representative 2-D gels of acidogenic steady-state and solventogenic steady-state cells growing at pH 5.7 or pH 4.5, respectively, are shown in Figs. 2 and 3. The complete novel proteome reference maps are presented in supplementary data file SD1 (Figs. S1 and S2) in combination with files SD2 and SD3. The cytoplasmic protein extracts revealed a coverage of more than 21% of proteins with calculated isoelectric points between pH 4 and pH 7 and molecular weights between 10 and 200 kDa (Hiller et al. 2006). Protein extracts of acidogenic cells yielded 357 spots with 178 identified different proteins whereas in solventogenic cells 205 different proteins were represented in 415 spots.
Fig. 2.
Representative colloidal Coomassie-stained 2-D gel of cytosolic proteins from cells grown at pH 5.7. The numbers highlight protein spots with higher abundance in acidogenic cells; respective proteins are listed in Table 1
Fig. 3.
Representative colloidal Coomassie-stained 2-D gel of cytosolic proteins from cells grown at pH 4.5. The numbers highlight protein spots with higher abundance in solventogenic cells; respective proteins are listed in Table 2
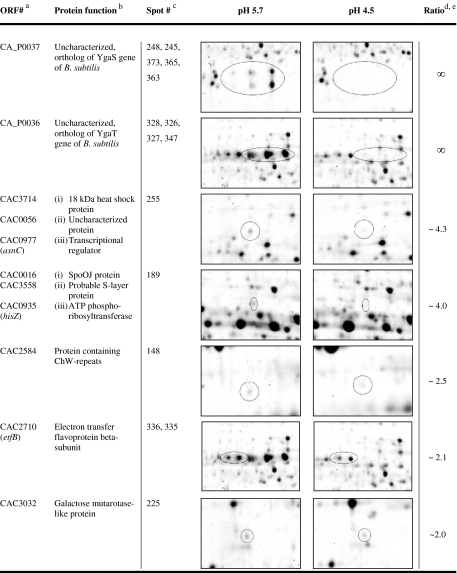
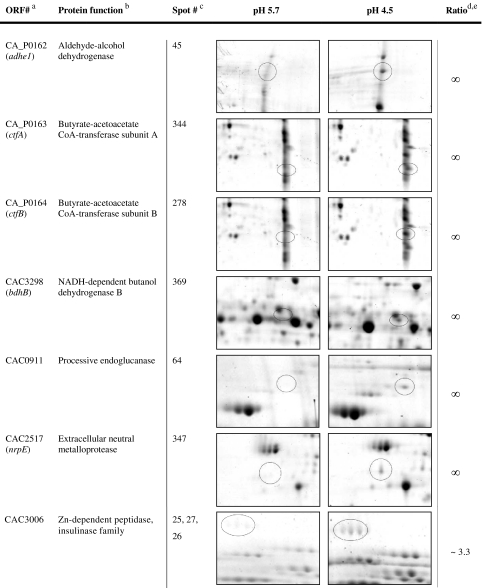
For the elucidation of characteristic proteins of cells grown at pH 5.7 and at pH 4.5, respectively, the data were screened for proteins with enhanced appearance (supplementary data files SD1 with Figs. S1 and S2, SD2, and SD3). Significance was defined as an at least twofold increase in spot intensity in comparison to the spots in the gels of the compared pH. Accordingly, 2-D gels of acidogenic cells at pH 5.7 revealed 15 enhanced spots as highlighted in Fig. 2 and further described in Table 1.
Table 1.
Proteins with increased (≥2.0) abundance in the acidogenesis at pH 5.7
aProteins of spots with more than one polypeptide are listed in their identification order
bNames based on Nölling et al. (2001)
cCorresponding spot numbers in Fig. 2
dIf more than one protein spot was detected, the average of all protein spots is shown (SD 4)
eThe sign for nonterminating (∞) indicates that the protein was not detectable at pH 4.5
Most noticeable were the proteins CAP0037 and CAP0036. Their genes are located on the megaplasmid pSOL1 (Cornillot et al. 1997), and both were annotated as uncharacterized gene products with unknown functions (Nölling et al. 2001). Both proteins were present in multiple spots, up to five in the case of CAP0037 and at least up to four in the case of CAP0036 (Fig. 2, Table 1). None of these spots were detectable in 2-D gels of solventogenic cells grown at pH 4.5. Their horizontal spot distributions indicated three different isoelectric points for each protein due to different protein charges. Additionally, CAP0037 revealed two different apparent molecular masses of 22 and 25 kDa, indicating posttranslational modifications, such as phosphorylation, acetylation, glycosylation, or methylation, which have been reported to occur in C. acetobutylicum (Balodimos et al. 1990; Lyristis et al. 2000; Sullivan and Bennett 2006) and other prokaryotic organisms (Rosen and Ron 2002; Rosen et al. 2004).
Another interesting spot with increased abundance (~2.1-fold) at pH 5.7 revealed electron transfer flavoprotein beta-subunit (EtfB, CAC2710). The etfB gene is part of the etfA–etfB–bcd–crt operon (CAC2709–CAC2712) whose mRNA transcript was also significantly elevated at pH 5.7 (~3.0–3.5-fold, Table 3 and supplementary data file SD1, Table S2). EtfA, EtfB, and Bcd showed multiple spots in the 2-D gels (supplementary data file SD1, Table S1), but only EtfB revealed a significantly higher abundance.
Table 3.
Genes with a significant induction at the transcript level at pH 5.7
| ORF# | Gene | Protein functiona | 1. array | 2. array | 3. array | 4. array | Average fold reg. | SD | COGb |
|---|---|---|---|---|---|---|---|---|---|
| CAC0029 | Distantly related to cell wall-associated hydrolase, similar to yycO B. subtilis | 2.1 | 1.5 | 7.5 | 3.2 | 3.6 | 2.7 | S | |
| CAC0149 | Hypothetical protein | 5.9 | 5.2 | 14.5 | 10.1 | 8.9 | 4.3 | – | |
| CAC0164 | ABC transporter, ATP binding protein | 2.4 | 2.0 | 9.4 | 2.9 | 4.2 | 3.5 | V | |
| CAC0231 | Transcriptional regulator of sugar metabolism | 3.5 | 2.7 | 12.1 | 3.7 | 5.5 | 4.4 | K,G | |
| CAC0232 | fruB | 1-phosphofructokinase | 2.2 | 1.9 | 11.8 | 2.9 | 4.7 | 4.8 | G |
| CAC0233 | PTS system, IIA component | 2.5 | 2.0 | 18.2 | 2.3 | 6.2 | 8.0 | G,T | |
| CAC0234 | PTS system, fructoso-specific IIBC component | 2.2 | 2.1 | 8.1 | 3.0 | 3.9 | 2.9 | G | |
| CAC0360 | Transcriptional regulator | 3.1 | 2.6 | 6.5 | 5.6 | 4.5 | 1.9 | K | |
| CAC0407 | PP2C phosphatase family | 2.7 | 2.1 | 4.6 | 3.5 | 3.2 | 1.1 | T | |
| CAC0409 | Hypothetical protein | 2.1 | 1.9 | 5.7 | 3.2 | 3.2 | 1.8 | – | |
| CAC0410 | Hypothetical protein | 2.0 | 1.8 | 5.6 | 2.6 | 3.0 | 1.8 | S | |
| CAC0411 | Hypothetical protein | 2.8 | 2.3 | 7.3 | 5.3 | 4.4 | 2.3 | S | |
| CAC0412 | TPR-repeat-containing protein | 2.0 | 1.7 | 5.5 | 3.9 | 3.3 | 1.8 | R | |
| CAC0427 | Glycerol-3-phosphate ABC transporter, permease component | 3.1 | 3.1 | 5.7 | 2.9 | 3.7 | 1.3 | G | |
| CAC0428 | Sugar permease | 4.2 | 2.7 | 11.2 | 3.5 | 5.4 | 3.9 | G | |
| CAC0429 | Glycerol-3-phosphate ABC transporter, periplasmic component | 4.6 | 3.3 | 9.2 | 5.7 | 5.7 | 2.5 | G | |
| CAC0430 | Glycerophosphoryl diester phosphodiesterase | 2.2 | 3.5 | 7.1 | 6.8 | 4.9 | 2.4 | C | |
| CAC0742 | Phosphatase domain-containing protein | 2.2 | 1.8 | 6.3 | 3.1 | 3.3 | 2.0 | I | |
| CAC0946 | ComE-like protein | 2.8 | 2.4 | 4.6 | 3.9 | 3.4 | 1.0 | R | |
| CAC1081 | Hypothetical protein | 2.1 | 2.1 | 13.0 | 5.6 | 5.7 | 5.1 | S | |
| CAC1230 | Hypothetical protein | 2.6 | 2.6 | 6.9 | 2.3 | 3.6 | 2.2 | – | |
| CAC1231 | Predicted dehydrogenase | 2.0 | 1.7 | 7.1 | 3.8 | 3.6 | 2.5 | R | |
| CAC1547 | trxA | Thioredoxin | 4.1 | 3.4 | 8.3 | 6.8 | 5.6 | 2.3 | O |
| CAC1548 | trxB | Thioredoxin reductase | 3.3 | 3.6 | 6.9 | 5.2 | 4.8 | 1.7 | O |
| CAC1549 | bsaA | Glutathione peroxidase | 2.8 | 2.7 | 8.3 | 3.2 | 4.2 | 2.7 | O |
| CAC1583 | Predicted P-loop ATPase | 2.4 | 2.2 | 3.5 | 5.3 | 3.3 | 1.4 | E | |
| CAC1702 | Hypothetical protein | 3.2 | 3.0 | 4.9 | 5.0 | 4.0 | 1.1 | – | |
| CAC1703 | Methyl-accepting chemotaxis protein (fragment) | 2.4 | 2.5 | 5.2 | 4.4 | 3.6 | 1.4 | N,T | |
| CAC1704 | Hypothetical protein | 2.8 | 2.8 | 4.9 | 5.0 | 3.9 | 1.2 | – | |
| CAC2252 | Alpha-glucosidase | 1.9 | 2.3 | 7.1 | 5.2 | 4.1 | 2.5 | G | |
| CAC2342 | Predicted membrane protein | 4.1 | 4.3 | 2.1 | 3.4 | 3.5 | 1.0 | R | |
| CAC2365 | sspA | Small acid-soluble spore protein | 13.1 | 13.9 | 6.1 | – | 11.0 | 4.3 | – |
| CAC2438 | Predicted phosphatase | 5.3 | 4.7 | 3.4 | 4.8 | 4.5 | 0.8 | – | |
| CAC2601 | S-adenosylmethionine decarboxylase | 1.9 | 2.1 | 5.2 | 3.2 | 3.1 | 1.5 | E | |
| CAC2702 | Possible signal transduction protein | 4.0 | 3.7 | 3.6 | 5.0 | 4.1 | 0.6 | T | |
| CAC2709 | etfA | Electron transfer flavoprotein alpha-subunit | 2.3 | 2.0 | 5.4 | 2.4 | 3.0 | 1.6 | C |
| CAC2711 | bcd | Butyryl-CoA dehydrogenase | 2.0 | 1.8 | 6.1 | 2.4 | 3.1 | 2.0 | I |
| CAC2712 | crt | Enoyl-CoA hydratase | 2.1 | 1.8 | 7.2 | 2.9 | 3.5 | 2.5 | I |
| CAC2810 | Glucoamylase family protein | 2.7 | 2.2 | 10.1 | 4.9 | 5.0 | 3.6 | G | |
| CAC2873 | thlA | Acetyl-CoA acetyltransferase | 2.3 | 1.7 | 8.1 | 3.5 | 3.9 | 2.9 | I |
| CAC2938 | Hypothetical protein | 2.4 | 1.9 | 6.5 | 5.2 | 4.0 | 2.2 | R | |
| CAC3075 | buk | Butyrate kinase | 4.0 | 2.8 | 8.1 | 5.7 | 5.1 | 2.3 | C |
| CAC3076 | ptb | Phosphate butyryltransferase | 3.4 | 2.5 | 9.5 | 5.4 | 5.2 | 3.1 | C |
| CAC3236 | Transcriptional regulator | 2.7 | 2.1 | 13.2 | 8.5 | 6.6 | 5.3 | T,Q | |
| CAC3237 | msmX | Sugar ABC transporter, ATP binding protein | 2.5 | 2.2 | 10.9 | 8.1 | 5.9 | 4.3 | G |
| CAC3379 | Hypothetical protein | 3.6 | 3.5 | 5.6 | 6.3 | 4.7 | 1.4 | S | |
| CA_P0036 | Uncharacterized, ortholog of YgaT gene of B. subtilis | 90.4 | 19.0 | 270.0 | 200.0 | 144.9 | 111.8 | M | |
| CA_P0037 | Uncharacterized, ortholog of YgaS gene of B. subtilis | 114.9 | 54.1 | 451.9 | 250.0 | 217.7 | 176.3 | – | |
| CA_P0038 | Uncharacterized conserved protein, YCII family | 4.2 | 3.4 | 3.2 | 2.5 | 3.3 | 0.7 | S | |
| CA_P0072 | Hypothetical protein | 2.0 | 2.1 | 6.0 | 2.0 | 3.0 | 2.0 | – | |
| CA_P0073 | ABC ATPase containing transporter | 2.7 | 2.4 | 7.6 | 2.1 | 3.7 | 2.6 | V | |
| CA_P0074 | Hypothetical protein | 2.9 | 2.2 | 8.3 | 2.0 | 3.9 | 3.0 | – | |
| CA_P0149 | Xre family DNA-binding domain and TRP-repeat-containing protein | 2.6 | 2.6 | 4.2 | 3.2 | 3.1 | 0.8 | K |
Two spots with ~4.3-fold and ~4.0-fold increased abundances at pH 5.7 contained mixtures of three different polypeptides each. The first spot comprised three proteins with similar molecular masses (~17.5–18 kDa) and isoelectrical points (pI ~ 4.95–5.27): Hsp18, an 18-kDa heat shock protein (CAC3714), CAC0056, a putative uncharacterized protein, and a transcriptional regulator of the Lrp family (AsnC, CAC0977). Thereof, only cac0056 showed a higher transcript level (~1.5-fold) under these conditions (SD5). The second inhomogeneous spot contained a protein identified as related to the HTH domain of SpoOJ/ParA/ParB/RepB family, involved in chromosome partitioning (CAC0016), a putative S-layer protein (CAC3558), and an ATP phosphoribosyltransferase regulatory subunit (HisZ, CAC0935). The proteins of the latter spot exhibit similar molecular masses (~46–48 kDa) but considerable different isoelectrical points of 5.78 (CAC0016), 6.32 (CAC0935), and 8.26 (CAC3558). Since the pI of CAC3558 was out of the pH range of 4–7 of the used strip, its occurrence was not expected and remains unexplainable.
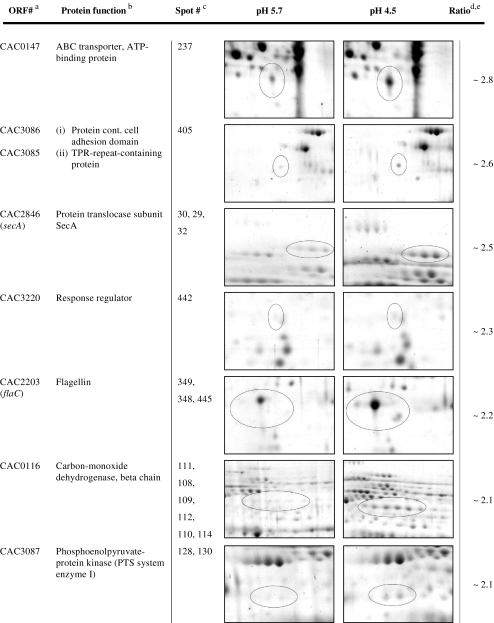
On the other hand, extracts of solventogenic cells grown at pH 4.5 revealed a total of 29 protein spots to be significantly increased (which are highlighted in Fig. 3 and further information is given in Table 2). Not surprisingly, the gene products of the sol operon (CAP0162–CAP0164), aldehyde alcohol dehydrogenase 1 (AdhE1), and the two subunits of the acetoacetyl coenzyme A:acetate/butyrate:coenzyme A transferase (“CoA-transferase,” CtfA, CtfB) were not detectable at pH 5.7, representing the acidogenic growth. This finding was not unexpected since the respective enzymes are involved in butanol formation, a metabolic pathway which is completely “turned off” (uninduced or repressed) in acidogenic cells. In addition, our array data (see transcriptomic section below) indicated highly elevated transcript levels at pH 4.5 of ~125-fold of the corresponding genes (Table 4 and supplementary data file SD1, Table S2). The protein spots were found in a typical “protein lane” at the right border of the 2-D gels caused by pI values of above 7.0 (AdhE1, 8.44; CtfA, 8.99; CtfB, 7.79; supplementary data file SD3). Thus, during separation in the first dimension, proteins accumulated at the right edge of the strip because it covered a pI range between 4 and 7 only. AdhE1 was detected additionally in numerous smaller “smearing” spots between pI 4.5 and pI 7.0 (SD1, Fig. S2; spot numbers 33, 34, 38–44, 46–47, 50–53 in combination with SD3).
Table 2.
Proteins with increased (≥2.0) abundance in the solventogenesis at pH 4.5
Table 4.
Genes with a significant induction at the transcript level in the solventogenesis
| ORF# | Gene | Protein functiona | 1. array | 2. array | 3. array | 4. array | Average fold reg. | SD | COGb |
|---|---|---|---|---|---|---|---|---|---|
| CAC0186 | Xre family DNA-binding domain and TPR-repeat-containing protein | 0.19 | 0.15 | 0.24 | 0.19 | 0.19 | 0.04 | – | |
| CAC0254 | nifHD | Nitrogen regulatory protein PII (nitrogen fixation nifHD) | 0.21 | 0.17 | 0.53 | 0.43 | 0.33 | 0.17 | E |
| CAC0255 | nifHD | Nitrogen regulatory protein PII (nitrogen fixation nifHD) | 0.22 | 0.20 | 0.41 | 0.46 | 0.32 | 0.13 | E |
| CAC0256 | nifD | Nitrogenase molybdenum–iron protein, alpha chain (nitrogenase component I) | 0.21 | 0.16 | 0.42 | 0.47 | 0.32 | 0.15 | C |
| CAC0392 | Peptidoglycan-binding domain | 0.30 | 0.28 | 0.25 | 0.36 | 0.30 | 0.04 | M | |
| CAC0538 | ChW repeat-containing mannanase ManB | 0.35 | 0.34 | 0.24 | 0.22 | 0.29 | 0.07 | G | |
| CAC0561 | Cellulase CelE-like protein | 0.04 | 0.05 | 0.11 | 0.07 | 0.07 | 0.03 | – | |
| CAC0574 | Pectate lyase H (FS) | 0.08 | 0.08 | 0.19 | 0.14 | 0.12 | 0.05 | – | |
| CAC0717 | Predicted membrane protein | 0.10 | 0.12 | 0.15 | 0.10 | 0.12 | 0.02 | – | |
| CAC0746 | Secreted protease metal-dependent protease | 0.14 | 0.15 | 0.27 | 0.25 | 0.20 | 0.07 | S | |
| CAC0826 | Endoglucanase family 5 | 0.16 | 0.16 | 0.29 | 0.19 | 0.20 | 0.06 | G | |
| CAC0910 | Cellulosomal scaffolding protein | 0.007 | 0.007 | 0.012 | 0.007 | 0.008 | 0.003 | G | |
| CAC0911 | Processive endoglucanase | 0.007 | 0.007 | 0.012 | 0.010 | 0.009 | 0.002 | – | |
| CAC0912 | Nonprocessive endoglucanase | 0.007 | 0.006 | 0.013 | 0.007 | 0.008 | 0.003 | G | |
| CAC0913 | Nonprocessive endoglucanase | 0.010 | 0.008 | 0.012 | 0.008 | 0.009 | 0.002 | – | |
| CAC0914 | Cellulosome integrating cohesin-containing protein, secreted | 0.009 | 0.008 | 0.009 | – | 0.008 | – | – | |
| CAC0915 | Endoglucanase A | 0.015 | 0.016 | 0.010 | 0.008 | 0.012 | 0.004 | – | |
| CAC0918 | Nonprocessive endoglucanase | 0.009 | 0.007 | 0.008 | 0.006 | 0.008 | 0.001 | – | |
| CAC0919 | Sialidase | 0.009 | 0.013 | 0.013 | 0.013 | 0.012 | 0.002 | – | |
| CAC1214 | Xre family DNA-binding domain and TPR-repeat-containing protein | 0.30 | 0.25 | 0.38 | 0.36 | 0.32 | 0.06 | – | |
| CAC1314 | Hypothetical protein | 0.03 | 0.03 | 0.04 | 0.04 | 0.03 | 0.002 | – | |
| CAC1315 | Peptidoglycan-binding domain-containing protein | 0.03 | 0.03 | 0.04 | 0.03 | 0.03 | 0.004 | M | |
| CAC1322 | glpA | Glycerol-3-phosphate dehydrogenase | 0.06 | 0.09 | 0.15 | 0.08 | 0.09 | 0.04 | R |
| CAC1323 | NAD(FAD)-dependent dehydrogenase | 0.13 | 0.16 | 0.29 | 0.19 | 0.19 | 0.07 | O | |
| CAC1324 | Hypothetical protein | 0.13 | 0.15 | 0.14 | 0.19 | 0.15 | 0.03 | S | |
| CAC1554 | Heavy metal-binding domain-containing protein | 0.32 | 0.34 | 0.30 | 0.24 | 0.30 | 0.05 | S | |
| CAC1611 | Cation efflux pump (multidrug resistance protein) | 0.34 | 0.33 | 0.27 | 0.26 | 0.30 | 0.04 | V | |
| CAC1673 | gltA | Large subunit of NADH-dependent glutamate synthase | 0.22 | 0.26 | 0.36 | 0.43 | 0.32 | 0.09 | E |
| CAC1968 | Pectate lyase related enzyme | 0.12 | 0.11 | 0.14 | 0.20 | 0.14 | 0.04 | – | |
| CAC1977 | Predicted membrane protein | 0.16 | 0.19 | 0.18 | 0.17 | 0.02 | – | ||
| CAC1980 | Predicted ATPase involved in pili biogenesis | 0.25 | 0.26 | 0.28 | 0.28 | 0.27 | 0.01 | U | |
| CAC2052 | DNA-dependent RNA polymerase sigma subunit | 0.24 | 0.19 | 0.47 | – | 0.30 | 0.15 | K | |
| CAC2053 | Hypothetical protein | 0.27 | 0.19 | 0.43 | 0.26 | 0.29 | 0.10 | – | |
| CAC2293 | Hypothetical protein | 0.11 | 0.19 | 0.21 | 0.28 | 0.20 | 0.07 | – | |
| CAC2392 | ABC transporter, ATPase component | 0.18 | 0.19 | 0.34 | 0.22 | 0.23 | 0.07 | V | |
| CAC2393 | ABC transporter, ATPase component | 0.21 | 0.18 | 0.40 | 0.18 | 0.24 | 0.10 | V | |
| CAC2396 | Xylanase/chitin deacetylase | 0.25 | 0.29 | 0.29 | 0.21 | 0.26 | 0.04 | G | |
| CAC2404 | Glycosyltransferase | 0.21 | 0.17 | 0.20 | 0.13 | 0.18 | 0.03 | R | |
| CAC2405 | Glycosyltransferase | 0.18 | 0.14 | 0.28 | 0.14 | 0.19 | 0.07 | M | |
| CAC2406 | O-antigen transporter permease | 0.12 | 0.16 | 0.20 | 0.15 | 0.16 | 0.03 | R | |
| CAC2407 | CheY-like domain-containing protein | 0.18 | 0.15 | 0.20 | 0.14 | 0.17 | 0.03 | T | |
| CAC2408 | Glycosyltransferase | 0.14 | 0.12 | 0.24 | 0.18 | 0.17 | 0.05 | M | |
| CAC2450 | Desulfoferrodoxin | 0.28 | 0.35 | 0.27 | 0.21 | 0.28 | 0.06 | C | |
| CAC2497 | Hypothetical protein | 0.17 | 0.28 | 0.19 | 0.22 | 0.21 | 0.05 | – | |
| CAC2517 | nrpE | Extracellular neutral metalloprotease. NPRE | 0.06 | 0.06 | 0.06 | 0.04 | 0.05 | 0.01 | E |
| CAC2606 | Hypothetical protein | 0.18 | 0.19 | 0.37 | 0.36 | 0.28 | 0.10 | G | |
| CAC2607 | Gluconate 5-dehydrogenase | 0.20 | 0.19 | 0.52 | 0.28 | 0.30 | 0.15 | I,Q,R | |
| CAC2650 | pyrD | Dihydroorotate dehydrogenase | 0.15 | 0.14 | 0.53 | 0.38 | 0.30 | 0.19 | F |
| CAC2651 | pyrZ | Dihydroorotate dehydrogenase electron transfer subunit | 0.15 | 0.13 | 0.56 | 0.32 | 0.29 | 0.20 | H,C |
| CAC2652 | pyrF | Orotidine 5′-phosphate decarboxylase | 0.16 | 0.12 | 0.63 | 0.30 | 0.30 | 0.23 | F |
| CAC2653 | pyrI | Aspartate carbamoyltransferase regulatory subunit | 0.21 | 0.21 | 0.44 | 0.33 | 0.30 | 0.11 | F |
| CAC2654 | pyrB | Aspartate carbamoyltransferase catalytic subunit | 0.15 | 0.12 | 0.34 | 0.34 | 0.24 | 0.12 | F |
| CAC2920 | tenI | Thiamine monophosphate synthase | 0.18 | 0.16 | 0.52 | 0.36 | 0.31 | 0.17 | H |
| CAC2921 | thiH | Thiamine biosynthesis protein ThiH | 0.15 | 0.13 | 0.56 | 0.33 | 0.29 | 0.20 | H,R |
| CAC2922 | thiG | Thiazole synthase | 0.15 | 0.14 | 0.39 | 0.24 | 0.23 | 0.11 | H |
| CAC2923 | Thiamine biosynthesis protein ThiF | 0.15 | 0.14 | 0.37 | 0.24 | 0.23 | 0.11 | H | |
| CAC2924 | thiS | Hypothetical protein | 0.15 | 0.15 | 0.35 | 0.21 | 0.22 | 0.10 | H |
| CAC2928 | Predicted membrane protein | 0.22 | 0.15 | 0.10 | 0.06 | 0.13 | 0.07 | S | |
| CAC3166 | Predicted DNA-binding protein | 0.32 | 0.27 | 0.37 | – | 0.32 | 0.05 | R | |
| CAC3167 | Hypothetical protein | 0.26 | 0.20 | 0.44 | 0.24 | 0.29 | 0.11 | S | |
| CAC3276 | nrdF | Ribonucleotide-diphosphate reductase beta-subunit | 0.38 | 0.49 | 0.17 | 0.14 | 0.30 | 0.17 | F |
| CAC3279 | ChW repeat-containing protein | 0.26 | 0.28 | 0.33 | 0.20 | 0.27 | 0.05 | N | |
| CAC3280 | ChW repeat-containing protein | 0.33 | 0.30 | 0.34 | 0.21 | 0.29 | 0.06 | N | |
| CAC3387 | Pectate lyase | 0.16 | 0.17 | 0.06 | 0.07 | 0.11 | 0.06 | – | |
| CAC3469 | Endoglucanase family protein | 0.05 | 0.04 | 0.10 | 0.06 | 0.06 | 0.03 | G | |
| CAC3526 | FMN-binding protein | 0.30 | 0.22 | 0.40 | 0.37 | 0.32 | 0.08 | – | |
| CAC3612 | Hypothetical protein | 0.27 | 0.23 | 0.22 | 0.26 | 0.25 | 0.03 | – | |
| CAC3684 | Polygalacturonase | 0.21 | 0.20 | 0.42 | 0.27 | 0.27 | 0.10 | M | |
| CAC3693 | Hypothetical protein | 0.30 | 0.41 | 0.24 | 0.31 | 0.31 | 0.07 | – | |
| CAC3694 | TPR-repeat-containing protein | 0.32 | 0.37 | 0.30 | 0.31 | 0.32 | 0.03 | R | |
| CAC3695 | Transcriptional regulator, containing DNA-binding domain of xre family | 0.27 | 0.30 | 0.31 | 0.28 | 0.29 | 0.02 | – | |
| CAC3696 | Hypothetical protein | 0.43 | 0.31 | 0.30 | 0.29 | 0.33 | 0.07 | S | |
| CA_P0004 | Cysteine protease | 0.02 | 0.02 | 0.04 | 0.04 | 0.03 | 0.01 | O | |
| CA_P0009 | Response regulator | 0.25 | 0.28 | 0.51 | 0.24 | 0.32 | 0.13 | T | |
| CA_P0010 | bgla | Beta-glucosidase | 0.30 | 0.32 | 0.35 | 0.33 | 0.32 | 0.02 | G |
| CA_P0040 | Xre family DNA-binding domain and TPR repeats containing protein | 0.21 | 0.19 | – | 0.26 | 0.22 | 0.03 | – | |
| CA_P0044 | Hypothetical protein | 0.18 | 0.17 | 0.29 | 0.19 | 0.21 | 0.05 | – | |
| CA_P0045 | Glycosyl transferase | 0.19 | 0.18 | 0.23 | 0.14 | 0.18 | 0.04 | G,C | |
| CA_P0053 | xynb | Xylanase | 0.06 | 0.07 | 0.05 | 0.05 | 0.06 | 0.01 | G |
| CA_P0054 | Xylanase/chitin deacetylase family protein | 0.06 | 0.06 | 0.06 | 0.04 | 0.05 | 0.01 | G | |
| CA_P0056 | pell | Pectate lyase | 0.10 | 0.09 | 0.16 | 0.11 | 0.12 | 0.03 | – |
| CA_P0065 | Secreted metalloprotease | 0.07 | 0.07 | 0.07 | 0.06 | 0.06 | 0.004 | – | |
| CA_P0102 | Membrane protein | 0.11 | 0.10 | 0.12 | 0.08 | 0.10 | 0.02 | R | |
| CA_P0112 | Hypothetical protein | 0.27 | 0.25 | 0.18 | 0.17 | 0.22 | 0.05 | – | |
| CA_P0116 | Xylanase | 0.17 | 0.17 | 0.28 | 0.28 | 0.22 | 0.06 | G | |
| CA_P0117 | Beta-xylosidase | 0.12 | 0.12 | 0.22 | 0.19 | 0.17 | 0.05 | – | |
| CA_P0118 | Xylan degradation enzyme | 0.14 | 0.16 | 0.18 | 0.16 | 0.16 | 0.02 | M | |
| CA_P0119 | Xylan degradation enzyme | 0.09 | 0.09 | 0.15 | 0.12 | 0.11 | 0.03 | M | |
| CA_P0120 | Xylan degradation enzyme | 0.09 | 0.10 | 0.16 | 0.13 | 0.12 | 0.03 | G | |
| CA_P0128 | Permease, MDR related | 0.30 | 0.25 | 0.05 | 0.04 | 0.16 | 0.13 | G | |
| CA_P0129 | Glycogen-binding regulatory subunit of S/T protein phosphatase I | 0.19 | 0.21 | 0.22 | 0.25 | 0.22 | 0.02 | – | |
| CA_P0162 | adhe1 | Aldehyde dehydrogenase | 0.012 | 0.009 | 0.008 | 0.004 | 0.008 | 0.003 | C |
| CA_P0163 | ctfa | Butyrate-acetoacetate CoA-transferase subunit A | 0.009 | 0.010 | 0.009 | 0.004 | 0.008 | 0.003 | I |
| CA_P0164 | ctfb | Butyrate-acetoacetate CoA-transferase subunit B | 0.014 | 0.010 | 0.007 | 0.004 | 0.008 | 0.004 | I |
| CA_P0168 | amyA | Alpha-amylase | 0.11 | 0.09 | 0.16 | 0.09 | 0.11 | 0.03 | G |
Another protein involved in solvent production, NADH-dependent butanol dehydrogenase B (CAC3298, BdhB), was also significantly increased with an infinite ratio as its spot was not detected at pH 5.7 (Table 2). In contrast to the proteins mentioned above, its transcript level was only slightly (~1.7-fold) elevated (SD1, Table S2).
The role of the other proteins found in significantly higher amounts (Table 2) for cells growing at pH 4.5 remains unknown. Interesting examples are carbon monoxide dehydrogenase (CAC0116) which increased up to ~2.1-fold and flagellin FlaC (or hook-associated flagellin protein Hag; CAC2203) which appeared in multiple spots. One of them, spot number 445, was only observed at pH 4.5, indicating a smaller apparent molecular weight and higher pI than expected (SD1, Fig. S2; SD3). In contrast, the two main spots of FlaC revealed considerable higher molecular masses of ~42 kDa with respect to the calculated mass of 29.5 kDa. This phenomenon might be explained by multiple glycosylation (Lyristis et al. 2000).
Other proteins listed in Table 2 represent typical extracellular proteins like an extracellular neutral metalloprotease (CAC2517) or a processive endoglucanase (CAC0911). Increased production at pH 4.5 was in agreement with upregulated transcript levels of ~20-fold and ~111-fold, respectively (Table 4).
Transcriptome analysis
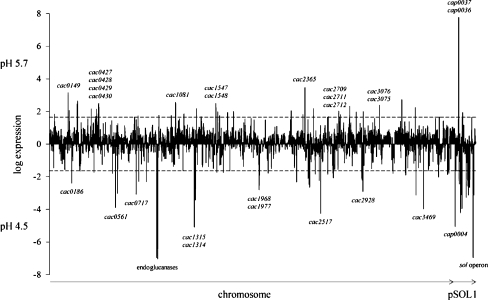
Acidogenic cells and solventogenic cells of a master fermentation were comprehensively compared by DNA microarray analyses. The time points of sampling were identical to those of the proteome analyses (Fig. 1). Complete results are listed in the supplementary data files SD5 and SD6 whereas in Fig. 4 striking differences are highlighted. Accordingly, transcription of 53 genes (1.4%) in acidogenic cells and 95 genes (2.5%) in solventogenic cells was found to be highly upregulated, each in comparison to the status at the other pH value. The majority (66.3%) of the genes did not show significant alterations and further 29.8% showed no transcripts matching the filter criteria (Hillmann et al. 2009).
Fig. 4.
Overview of the transcript levels during acidogenesis at pH 5.7 and solventogenesis at pH 4.5. Log expression ratios of acidogenesis to solventogenesis are shown. All genes with log values (as logarithms to the basis of 2) higher than 1.6 are significantly induced at pH 5.7, and genes with a negative log of less than −1.6 were significantly induced at pH 4.5. According to this definition, all genes between the dashed lines were expected to be not significantly influenced
Genes with increased transcript amounts in acidogenic cells
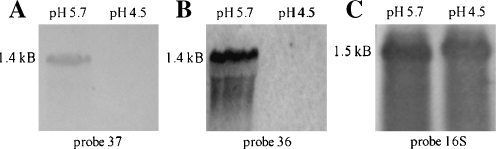
The detailed results of the 53 open reading frames (ORFs) that revealed at least ≥3.0-fold increase in their transcript levels at pH 5.7 are given in Table 3. Seven of them are located on the megaplasmid pSOL1. Two of them showed particularly high scores of more than 140-fold increase each, cap0036 and cap0037. This finding was in good accordance with the proteome data and prompted us to verify their RNA levels by Northern blot analysis. The cap0036- and cap0037-specific mRNA transcripts were exclusively detected in acidogenic chemostat cells of C. acetobutylicum (Fig. 5). Individual probes against both genes (Fig. 5a, b) revealed signals of the same size of about 1.4 kb, indicating a common transcription unit of a bicistronic operon. More intense hybridization signals using the cap0036 probe may suggest higher mRNA quantities, but a higher hybridization efficiency due to specific base composition and individual nucleotide sequence might also explain the differences.
Fig. 5.
Northern blot analyses of cap0037 and cap0036. Total RNA samples (15 μg per lane in a and b; 1 μg in c) of C. acetobutylicum cells grown at pH 5.7 and pH 4.5 (as indicated) were hybridized with DIG-labeled probes against cap0037 (a), cap0036 (b), or 16S rRNA transcript (c) as control of RNA integrity
The other five genes of pSOL1 with increased transcript levels (approximately threefold to fourfold) comprised the putative operon cap0073 and cap0074 and the putative orphan genes cap0072, cap0038, and cap0149 (Karp et al. 2005). The functions of their gene products are “hypothetical or uncharacterized” although some similarities to a protein of an ABC transporter (CAP0073) or to a DNA-binding domain (CAP0149) have been postulated (Nölling et al. 2001).
Among the chromosomal open reading frames, cac2365 showed the highest transcript increase of ~11-fold at pH 5.7. This gene putatively encodes an SspA-like protein, annotated as a small acid-soluble DNA-binding spore protein which might protect the spore genome. SspA is known to be upregulated in batch cultures of C. acetobutylicum and Clostridium beijerinckii in cells entering stationary growth phase (Jones et al. 2008; Shi and Blaschek 2008). However, C. acetobutylicum did not form spores in exponentially growing cells of continuous chemostat cultures, and its function under these conditions is not clear. Transcriptional induction of SspA might be explained as a consequence of the simultaneous increase (~2.0-fold; SD5) of sigG (CAC1696) because σG regulated the synthesis of SspA in Bacillus subtilis (Jones et al. 2008).
Not surprisingly, genes directly responsible for butyrate formation like ptb (CAC3076) and buk (CAC3075) (Hartmanis 1987; Walter et al. 1993; Wiesenborn et al. 1989a) were also strongly induced (approximately fivefold) in acidogenic cells (Table 3, SD1, Table S2). Similarly increased induction was found for genes of enzymes involved in the conversion of acetyl-CoA to butyryl-CoA, e.g., thiolase A (thlA, CAC2873), crotonase (crt, CAC2712), butyryl-CoA dehydrogenase (bcd, CAC2711), and the α subunit of the electron transfer flavoprotein (etfA, CAC2709). Otherwise, 3-hydroxybutyryl-CoA dehydrogenase (hbd, CAC2708) and the second subunit β of the electron transfer flavoprotein (etfB, CAC2710) did not show significantly elevated transcript levels (SD1, Table S2).
Furthermore, seven genes of four different ABC transporter systems were induced at pH 5.7, e.g., a putative ATP binding protein involved in sugar transport (msmX, CAC3237) and the genes cac0427, cac0428, and cac0429 of a glycerol-3-phosphate ABC transporter system which are organized in a common operon together with glycerophosphoryl diester phosphodiesterase (cac0430; Karp et al. 2005). Additionally, ten chromosomal open reading frames (Table 3) encoding unknown “hypothetical proteins” revealed significant higher transcript levels of approximately threefold to ninefold.
Genes with increased transcript amounts in solventogenic cells
The transcript pattern confirmed the proteome studies that in solventogenic cells more genes were found to be upregulated than in acidogenic cells. The mRNA transcripts of 95 genes matched the significance criterion of ≥3.0-fold induction (pH 4.5 versus pH 5.7; Table 4). Particularly, high values (~125-fold) were documented for the genes of the sol operon (cap0162–cap0164) due to the requirement of AdhE1, and the subunits of CoA-transferase are necessary for the production of solvents after the metabolic shift (Fischer et al. 1993; Nair et al. 1994; Wiesenborn et al. 1989b). In addition, similar high levels of the mRNA transcripts of the chromosomal genes cac0910–cac0915 and cac0918–cac0919 were detected. Although glucose represented the single carbohydrate source in the medium, their deduced gene products are likely involved in cellulose degradation (Nölling et al. 2001). Gene products with related functions in xylan degradation (cap0053–cap0054, cap0118–cap0120), a pectate lyase (cap0056), a cellulose CelE-like protein (cac0561), and other endoglucanases (cac0826, cac3469) also showed clearly increased gene expression (~5–20-fold).
Furthermore, transcription of several putative proteolytic enzymes was found to be upregulated between ~5- and 30-fold, e.g., a cysteine protease (cap0004), a secreted metalloprotease (cap0065), a secreted metal-dependent protease (cac0746), and an extracellular neutral metalloprotease NPRE (cac2517). Also, several ORFs with significantly increased transcription at pH 4.5 encode proteins predicted as hypothetical (at least 12) or membrane proteins (at least five) which are encoded by the plasmid pSOL1 (e.g., CAP0044, CAP0102, CAP0112) or the chromosome (e.g., CAC0717, CAC1324, CAC2053, CAC2497), respectively.
Discussion
Several investigations have been conducted to provide useful information on the transcriptome of C. acetobutylicum ATCC824, and limited proteome data are also available (Alsaker and Papoutsakis 2005; Jones et al. 2008; Sullivan and Bennett 2006; Tomas et al. 2003). However, all studies published so far were based on batch cultures with the focus on regulatory aspects of solvent formation and sporulation linked to the transition phase of cell growth. In contrast, exponentially growing cells of a chemostat culture have not been analyzed yet on the molecular level, which constituted the motivation of this study. To accomplish analyses of steady-state cells of C. acetobutylicum reproducibly adapted to the two distinct physiological phases, advantage was taken of continuous cultivation states of C. acetobutylicum for the comparison of acidogenic and solventogenic metabolism as a function of the external pH.
Protein patterns were analyzed by 2-D gel electrophoresis, covering more than 21% of the cytosolic proteins with a pI range of 4–7. This was in good accordance with other proteome reference maps, e.g., 4.6% protein coverage of Corynebacterium glutamicum (Li et al. 2007), 9.8% of Deinococcus geothermalis (Liedert et al. 2009), 16.9% of Corynebacterium jeikeium K411 (Hansmeier et al. 2006), and 21.4% of Bifidobacterium longum (Yuan et al. 2006). However, with respect to a total number of 3,847 annotated protein coding genes of C. acetobutylicum (Nölling et al. 2001), a relatively small fraction of 251 different proteins (6.5%) was identified since only the intracellular fraction of soluble proteins was analyzed and extracellular and insoluble membrane polypeptides were not considered. Furthermore, proteins of a general low abundance might not be detectable due to the limited sensitivity of colloidal Coomassie-stained 2-D gels.
The most interesting proteins identified here were the gene products of cap0036 and cap0037, which appeared as multiple spots on 2-D gels of acidogenic cells. In agreement with the transcriptome data, their high abundance could simply be explained by considerably increased amounts of mRNA at pH 5.7. It is noteworthy that no other transcript in the DNA microarrays showed a similar high induction of more than 200-fold. Northern blot analyses revealed that cap0036 and cap0037 constitute a bicistronic transcription unit with a length of ~1.4 kb, corresponding to the size of both genes plus an 18-b intergenic region and a short 5′ untranslated region (630 b [cap0037] + 729 b [cap0036] + 18 b + UTR = 1,377 b + UTR).
In previous batch cultures, mRNA-transcripts of cap0037/cap0036 were identified in negligible amounts which may be associated with late stationary growth (Alsaker and Papoutsakis 2005; Jones et al. 2008). In 2-D gels presented by Schaffer et al. (2002), the spots of CAP037 and CAP0036 were visible in acidogenic cells as well as in a cell sample referred to as “onset of solventogenesis” which was taken 6 h after the drop of the pH in the chemostat culture of C. acetobutylicum DSM792. Obviously, the time period after the drop of the pH was too short to achieve steady-state solventogenic growth, and CAP0037/CAP0036 still appeared in the protein patterns, preventing a clearly distinguished assignment to acidogenesis.
Unfortunately, the physiological function of CAP0037/CAP0036 could not be deduced. Database searches proposed a transmembrane domain (TMHMM2 program; Krogh et al. 2001) at the C terminus of CAP0037, and alignments with CAP0036 predicted a conserved domain of unknown function belonging to the DUF583 superfamily (Nölling et al. 2001). Orthologous genes seemed to be restricted to Gram-positive bacteria (Karp et al. 2005), and only one clostridial strain, Clostridium difficile, a nonsolvent producer, revealed a homologous gene locus. Orthologs might exist in B. subtilis (ygaS, formerly yhbD), Bacillus thuringiensis, Bacillus cereus, and Bacillus licheniformis. Usually, genes orthologous to cap0037 are accompanied by genes resembling cap0036 (in B. subtilis ygaT, formerly yhbE) found in the same gene order at adjacent positions (exception C. difficile: interchanged gene positions). In contrast to C. acetobutylicum, other bacteria showed one (C. difficile, B. cereus) or up to six genes present in the respective operon (Bacillus anthracis, B. subtilis, Geobacillus thermodenitrificans, B. thuringiensis, Bacillus weihenstephanensis). For example, in B. subtilis, orthologs are located in a three-gene operon (yhbD–yhbE–yhbF; Flórez et al. 2009). The genes yhbD, yhbE, and yhbF were only found among the bacilli and clostridia, which indicates a role in sporulation (Lai et al. 2003). The limited distribution of CAP0037 and CAP0036 suggests a distinct function in C. acetobutylicum. Direct involvement in endospore formation seems questionable because the cultivation conditions applied here explicitly uncoupled solventogenesis and sporulation. This might also be the reason why these proteins have not been noticed in earlier studies employing transient batch cultivation conditions. However, the role of CAP0037/CAP0036 during acidogenic growth and their clear repression during solventogenesis remains to be elucidated.
Regarding significantly induced proteins of steady-state solventogenic cells, the gene products of the sol operon, aldehyde alcohol dehydrogenase (AdhE1) and the subunits of the butyrate-acetoacetate CoA-transferase (CtfA, CtfB) were identified. This correlated well with previous results because the transcription of the sol operon was known to be highly upregulated during batch cultivation at the onset of solvent formation in the stationary growth phase (Alsaker and Papoutsakis 2005; Harris et al. 2002; Jones et al. 2008), and similar expression profiles were also shown in chemostat experiments (Fischer et al. 1993; Fontaine et al. 2002; Sauer and Dürre 1995). On the other hand, further genes coding for enzymes directly involved in solvent production like the NADH-dependent butanol dehydrogenase A (bdhA) and the acetoacetate decarboxylase (adc) did not reveal elevated transcript levels in this study. These genes might show a temporarily limited transcriptional increase coupled to the metabolic switch, corresponding to phase III of the master fermentation (Fig. 1). Accordingly, their expression patterns in steady-state cells before and after the metabolic shift were almost in the same range. Another explanation would be a higher mRNA turnover pushing their amounts below the significance criteria. Several other genes of different functions reported earlier, e.g., genes involved in stress response, branched chain amino acid synthesis, cell motility, and lipid metabolism revealed only marginal differences between acidogenic and solventogenic growth in continuous cultures. It seems likely that their expression maxima during the cell cycle of C. acetobutylicum can be found within a specific time window of the transition phase, i.e., at the onset of solventogenesis (Alsaker and Papoutsakis 2005; Jones et al. 2008; Sauer and Dürre 1995).
The proteins Adc, Bcd, GroEL, GroES, and Hsp18 were found to be highly expressed during the onset of solventogenesis in previous proteome analyses (Schaffer et al. 2002; Sullivan and Bennett 2006), but we did not detect significant differences between the two phases of the chemostat cells growing at pH 5.7 and pH 4.5, respectively, for similar reasons as mentioned above.
Another group of genes induced during the onset of solventogenesis (Alsaker and Papoutsakis 2005; Jones et al. 2008) maintained high expression levels during solventogenic growth of chemostat cultures. Examples are enzymes of xylan and starch metabolism as well as some glycosyltransferases. In theory, glycosyltransferases can be necessary for regulatory purposes, but the increased expression levels of enzymes for xylan and starch metabolism cannot be explained at this point because the synthetic medium used in the chemostat cultures contained a surplus of glucose as single carbon and energy source. Furthermore, referring to the increased expression of putative gene products involved in cellulose degradation (cac0910–cac0915 and cac0918–cac0919), it must be mentioned that C. acetobutylicum cannot utilize cellulose (Lee et al. 1985a) although the genome comprises several genes encoding parts of the cellulosome and its assembly (López-Contreras et al. 2003, 2004; Sabathé et al. 2002). On the other hand, xylanolytic activity of C. acetobutylicum is well known (Lee et al. 1985b), and xylan and xylooligosaccharide hydrolyzing activities were demonstrated for the xylanases CAP0053 (formerly Xyn10A) and CAP0116 (formerly Xyn10B) (Mursheda et al. 2004, 2005).
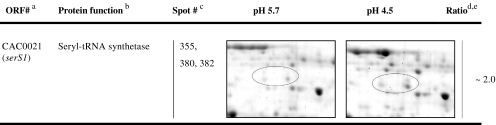
In general, elaborate proteome analyses require much more time and effort than transcriptome analyses; thus, more DNA microarray data are available for different perturbations affecting the life cycle and solvent production of C. acetobutylicum. Since proteins are more closely linked to the metabolism in the broadest sense than the respective genes, proteomics can be regarded as a more meaningful approach. Nevertheless, both proteome and transcriptome analyses of steady-state cells representing acidogenic and solventogenic growth were used in this study. The general understanding that increased mRNA amounts led to increased protein abundances was confirmed for the majority of the proteins, with CAP0036/CAP0037, the alcohol aldehyde dehydrogenase AdhE1, the CoA-transferase CtfA/B, a neutral metalloprotease (CAC2517), and a processive endoglucanase (CAC0911) as the most significant candidates. However, the global approach including a large number of proteins also revealed several exceptions, e.g., the flagellin FlaC, the seryl-tRNA synthetase SerS1, a Zn-dependent peptidase (CAC3006), and a response regulator (CAC3220) exhibited higher protein abundances at pH 4.5, but the respective transcript levels did not show substantial alteration. Possibly due to increased translation efficiency and/or higher protein stability, similar observations have been reported earlier, e.g., for the C. acetobutylicum glyceraldehyde 3-phosphate dehydrogenase Gap (Schaffer et al. 2002). Contrariwise, some genes revealed significant higher mRNA levels but no or only marginal increases in protein abundance (~1.1-fold to ~1.3-fold). Examples of acidogenic proteins were the acetyl-CoA acetyltransferase (thlA), the enoyl-CoA hydratase (crt), the butyryl-CoA dehydrogenase (bcd), the electron transfer flavoprotein alpha-subunit (etfA), the enoyl-CoA hydratase (buk), and the phosphate butyryltransferase (ptb).
Finally, the protein profiles of the central glycolytic enzymes glucose-6-phosphate isomerase (Pgi), triosephosphate isomerase (Tpi), fructose-bisphosphate aldolase (Fba), glyceraldehyde 3-phosphate dehydrogenase (GapC), phosphoglycerate kinase (Pgk), phosphoglyceromutase (Pgm), and pyruvate kinase (PykA) showed no significant differences in acidogenesis and solventogenesis and correlated well with their mRNA levels.
In conclusion, novel reference maps of C. acetobutylicum acidogenic and solventogenic proteins with a maximum degree of reproducibility were generated from steady-state cells of chemostat cultures. The proteome maps and accompanying transcriptome analyses constitute a reliable basis for various future investigations to specifically alter endogenous (e.g., certain mutant strains) or exogenous (e.g., defined nutrients) parameters. Another clear advantage of the approach presented here was the strict uncoupling of solventogenesis and sporulation which allows the identification of candidate proteins distinctly related to the metabolic shift. Whether the striking acidogenic proteins CAP0037 and CAP0036 play a major role in the switch between acidogenic and solventogenic metabolism remains to be investigated. Moreover, the generated data sets provide a useful fundamental for systems biology projects such as the European COSMIC project (C. acetobutylicum systems microbiology, http://www.sysmo.net) to assess the biotechnological impact of C. acetobutylicum as a superior solvent production strain.
Electronic supplementary material
Below is the link to the electronic supplementary material.
pH 5.7 acidogenesis (XLS 124 kb)
pH 4.5 solventogenesis (XLS 132 kb)
Table of influenced spots (XLS 48 kb)
Transcriptome data (XLS 552 kb)
Transcript amounts (XLS 984 kb)
(PDF 343 kb)
(PDF 283 kb)
Acknowledgement
This work was supported by the German Federal Ministry of Education and Research (BMBF) through the projects COSMIC-SysMO (0313981D) and BaCell-SysMO (0313978A) (http://www.sysmo.net). We are grateful to C. Voigt for experimental support. Furthermore, we thank T. Lütke-Eversloh for critical reviewing of the manuscript.
References
- Alsaker KV, Papoutsakis ET. Transcriptional program of early sporulation and stationary-phase events in Clostridium acetobutylicum. J Bacteriol. 2005;187:7103–7118. doi: 10.1128/JB.187.20.7103-7118.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahl H, Andersch W, Braun K, Gottschalk G. Effect of pH and butyrate concentration on the production of acetone and butanol by Clostridium acetobutylicum grown in continuous culture. Eur J Appl Microbiol Biotechnol. 1982;14:17–20. doi: 10.1007/BF00507998. [DOI] [Google Scholar]
- Bahl H, Andersch W, Gottschalk G. Continuous production of acetone and butanol by Clostridium acetobutylicum in a two-stage phosphate limited chemostat. Eur J Appl Microbiol Biotechnol. 1982;15:201–205. doi: 10.1007/BF00499955. [DOI] [Google Scholar]
- Balodimos IA, Rapaport E, Kashket ER. Protein phosphorylation in response to stress in Clostridium acetobutylicum. Appl Environ Microbiol. 1990;56:2170–2173. doi: 10.1128/aem.56.7.2170-2173.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornillot E, Nair RV, Papoutsakis ET, Soucaille P. The genes for butanol and acetone formation in Clostridium acetobutylicum ATCC 824 reside on a large plasmid whose loss leads to degeneration of the strain. J Bacteriol. 1997;179:5442–5447. doi: 10.1128/jb.179.17.5442-5447.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürre P. Biobutanol: an attractive biofuel. Biotechnol J. 2007;2:1525–1534. doi: 10.1002/biot.200700168. [DOI] [PubMed] [Google Scholar]
- Dürre P. Fermentative butanol production: bulk chemical and biofuel. Ann N Y Acad Sci. 2008;1125:353–362. doi: 10.1196/annals.1419.009. [DOI] [PubMed] [Google Scholar]
- Fiedler T, Mix M, Meyer U, Mikkat S, Glocker MO, Bahl H, Fischer RJ. The two-component system PhoPR of Clostridium acetobutylicum is involved in phosphate-dependent gene regulation. J Bacteriol. 2008;190:6559–6567. doi: 10.1128/JB.00574-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RJ, Helms J, Dürre P. Cloning, sequencing, and molecular analysis of the sol operon of Clostridium acetobutylicum, a chromosomal locus involved in solventogenesis. J Bacteriol. 1993;175:6959–6969. doi: 10.1128/jb.175.21.6959-6969.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer RJ, Oehmcke S, Meyer U, Mix M, Schwarz K, Fiedler T, Bahl H. Transcription of the pst operon of Clostridium acetobutylicum is dependent on phosphate concentration and pH. J Bacteriol. 2006;188:5469–5478. doi: 10.1128/JB.00491-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flórez LA, Roppel SF, Schmeisky AG, Lammers CR, Stülke J. A community-curated consensual annotation that is continuously updated: the Bacillus subtilis centred wiki SubtiWiki. Database (Oxford) 2009;2009:bap12. doi: 10.1093/database/bap012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine L, Meynial-Salles I, Girbal L, Yang X, Croux C, Soucaille P. Molecular characterization and transcriptional analysis of adhE2, the gene encoding the NADH-dependent aldehyde/alcohol dehydrogenase responsible for butanol production in alcohologenic cultures of Clostridium acetobutylicum ATCC 824. J Bacteriol. 2002;184:821–830. doi: 10.1128/JB.184.3.821-830.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansmeier N, Chao TC, Daschkey S, Müsken M, Kalinowski J, Pühler A, Tauch A. A comprehensive proteome map of the lipid-requiring nosocomial pathogen Corynebacterium jeikeium K411. Proteomics. 2006;7:1076–1096. doi: 10.1002/pmic.200600833. [DOI] [PubMed] [Google Scholar]
- Harris LM, Welker NE, Papoutsakis ET. Northern, morphological, and fermentation analysis of spo0A inactivation and overexpression in Clostridium acetobutylicum ATCC 824. J Bacteriol. 2002;184:3586–3597. doi: 10.1128/JB.184.13.3586-3597.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmanis MGN. Butyrate kinase from Clostridium acetobutylicum. J Biol Chem. 1987;262:617–621. [PubMed] [Google Scholar]
- Hiller K, Grote A, Maneck M, Münch R, Jahn D. JVirGel 2.0: computational prediction of proteomes separated via two-dimensional gel electrophoresis under consideration of membrane and secreted proteins. Bioinformatics. 2006;22:2441–2443. doi: 10.1093/bioinformatics/btl409. [DOI] [PubMed] [Google Scholar]
- Hillmann F, Döring C, Riebe O, Ehrenreich A, Fischer RJ, Bahl H. The role of PerR in O2-affected gene expression of Clostridium acetobutylicum. J Bacteriol. 2009;191:6082–6093. doi: 10.1128/JB.00351-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT. Applied acetone–butanol fermentation. In: Bahl H, Dürre P, editors. Clostridia. Biotechnological and medical applications. Weinheim: Wiley-VCH; 2001. pp. 125–168. [Google Scholar]
- Jones DT, Woods DR. Acetone–butanol fermentation revisited. Microbiol Rev. 1986;50:484–524. doi: 10.1128/mr.50.4.484-524.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DT, Westhuizen A, Long S, Allcock ER, Reid SJ, Woods DR. Solvent production and morphological changes in Clostridium acetobutylicum. Appl Environ Microbiol. 1982;43:1434–1439. doi: 10.1128/aem.43.6.1434-1439.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SW, Paredes CJ, Tracy B, Cheng N, Sillers R, Senger RS, Papoutsakis ET. The transcriptional program underlying the physiology of clostridial sporulation. Genome Biol. 2008;7:R114. doi: 10.1186/gb-2008-9-7-r114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahrén D, Tsoka S, Darzentas N, Kunin V, López-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Research. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, Heijne G, Sonnhammer E. Predicting transmembrane protein topology with a hidden Markov model. Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Lai EM, Phadke ND, Kachman MT, Giorno R, Vazquez S, Vazquez JA, Maddock JR, Driks A. Proteomic analysis of the spore coats of Bacillus subtilis and Bacillus anthracis. J Bacteriol. 2003;185:1443–1454. doi: 10.1128/JB.185.4.1443-1454.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF, Forsberg CW, Gibbins LN. Cellulolytic activity of Clostridium acetobutylicum. Appl Environ Microbiol. 1985;50:220–228. doi: 10.1128/aem.50.2.220-228.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SF, Forsberg CW, Gibbins LN. Xylanolytic activity of Clostridium acetobutylicum. Appl Environ Microbiol. 1985;50:1068–1076. doi: 10.1128/aem.50.4.1068-1076.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Park JH, Jang SH, Nielsen LK, Kim J, Jung KS. Fermentative butanol production by clostridia. Biotechnol Bioeng. 2008;101:209–228. doi: 10.1002/bit.22003. [DOI] [PubMed] [Google Scholar]
- Li L, Wada M, Yokota A. Cytoplasmic proteome reference map for a glutamic acid-producing Corynebacterium glutamicum ATCC 14067. Proteomics. 2007;7:4317–4322. doi: 10.1002/pmic.200700269. [DOI] [PubMed] [Google Scholar]
- Liedert C, Bernhardt J, Albrecht D, Voigt B, Hecker M, Salkinoja-Salonen M, Neubauer P. Two-dimensional proteome reference map for the radiation-resistant bacterium Deinococcus geothermalis. Proteomics. 2009;10:555–563. doi: 10.1002/pmic.200800657. [DOI] [PubMed] [Google Scholar]
- López-Contreras AM, Martens AA, Szijarto N, Mooibroek H, Claassen PAM, Oost J, Vos WM. Production by Clostridium acetobutylicum ATCC 824 of CelG, a cellulosomal glycoside hydrolase from family 9. Appl Environ Microbiol. 2003;69:869–877. doi: 10.1128/AEM.69.2.869-877.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Contreras AM, Gabor K, Martens AA, Renckens BAM, Claassen PAM, Oost J, Vos WM. Substrate-induced production and secretion of cellulases by Clostridium acetobutylicum. Appl Environ Microbiol. 2004;70:5238–5243. doi: 10.1128/AEM.70.9.5238-5243.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyristis M, Boynton ZL, Petersen D, Kann Z, Bennett GN, Rudolph FB. Cloning, sequencing, and characterization of the gene encoding flagellin, flaC, and the posttranslational modification of flagellin, FlaC, from Clostridium acetobutylicum ATCC824. Anaerobe. 2000;6:69–79. doi: 10.1006/anae.1999.0311. [DOI] [Google Scholar]
- Mursheda KA, Rudolph FB, Bennett GN. Thermostable xylanase10B from Clostridium acetobutylicum ATCC824. J Ind Microbiol Biotechnol. 2004;31:229–234. doi: 10.1007/s10295-004-0143-8. [DOI] [PubMed] [Google Scholar]
- Mursheda KA, Rudolph FB, Bennett GN. Characterization of thermostable Xyn10A enzyme from mesophilic Clostridium acetobutylicum ATCC 824. J Ind Microbiol Biotechnol. 2005;32:12–18. doi: 10.1007/s10295-004-0192-z. [DOI] [PubMed] [Google Scholar]
- Nair RV, Bennett GN, Papoutsakis ET. Molecular characterization of an aldehyde/alcohol dehydrogenase gene from Clostridium acetobutylicum ATCC 824. J Bacteriol. 1994;176:871–885. doi: 10.1128/jb.176.3.871-885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Y, Sun Z. Recent progress on industrial fermentative production of acetone-butanol-ethanol by Clostridium acetobutylicum in China. Appl Microbiol Biotechnol. 2009;83:415–423. doi: 10.1007/s00253-009-2003-y. [DOI] [PubMed] [Google Scholar]
- Nölling J, Breton G, Omelchenko MV, Makarova KS, Zeng Q, Gibson R, Lee HM, Dubois JA, Qiu D, Hitti J, GTC Sequencing Center Production, Finishing, and Bioinformatics Teams. Wolf YI, Tatusov RL, Sabathe F, Doucette-Stamm L, Soucaille P, Daly MJ, Bennett GN, Koonin EV, Smith DR. Genome sequence and comparative analysis of the solvent-producing bacterium Clostridium acetobutylicum. J Bacteriol. 2001;183:4823–4838. doi: 10.1128/JB.183.16.4823-4838.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen R, Ron EZ. Proteome analysis in the study of the bacterial heat-shock response. Mass Spec Rev. 2002;21:244–265. doi: 10.1002/mas.10031. [DOI] [PubMed] [Google Scholar]
- Rosen R, Becher D, Büttner K, Biran D, Hecker M, Ron EZ. Highly phosphorylated bacterial proteins. Proteomics. 2004;4:3068–3077. doi: 10.1002/pmic.200400890. [DOI] [PubMed] [Google Scholar]
- Sabathé F, Bélaïch A, Soucaille P. Characterization of the cellulolytic complex (cellulosome) of Clostridium acetobutylicum. FEMS Microbiol Lett. 2002;217:15–22. doi: 10.1111/j.1574-6968.2002.tb11450.x. [DOI] [PubMed] [Google Scholar]
- Sauer U, Dürre P. Differential induction of genes related to solvent formation during the shift from acidogenesis to solventogenesis in continuous culture of Clostridium acetobutylicum. FEMS Microbiol Lett. 1995;125:115–120. doi: 10.1111/j.1574-6968.1995.tb07344.x. [DOI] [Google Scholar]
- Schaffer S, Isci N, Zickner B, Dürre P. Changes in protein synthesis and identification of proteins specifically induced during solventogenesis in Clostridium acetobutylicum. Electrophoresis. 2002;23:110–121. doi: 10.1002/1522-2683(200201)23:1<110::AID-ELPS110>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Schwarz K, Fiedler T, Fischer RJ, Bahl H. A standard operating procedure (SOP) for preparation of intra- and extracellular proteins of Clostridium acetobutylicum for proteome analysis. J Microbiol Methods. 2007;68:396–402. doi: 10.1016/j.mimet.2006.09.018. [DOI] [PubMed] [Google Scholar]
- Shi Z, Blaschek HP. Transcriptional analysis of Clostridium beijerinckii NCIMB 8052 and the hyper-butanol-producing mutant BA101 during the shift from acidogenesis to solventogenesis. Appl Environ Microbiol. 2008;74:7709–7714. doi: 10.1128/AEM.01948-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan L, Bennett GN. Proteome analysis and comparison of Clostridium acetobutylicum ATCC 824 and Spo0A strain variants. J Ind Microbiol Biotechnol. 2006;33:298–308. doi: 10.1007/s10295-005-0050-7. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Galperin MY, Natale DA, Koonin EV. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomas CA, Alsaker KV, Bonarius HPJ, Hendriksen WT, Yang H, Beamish JA, Parades CJ, Papoutsakis ET. DNA-array based transcriptional analysis of asporogenous, non-solventogenic Clostridium acetobutylicum strains SKO1 and M5. J Bacteriol. 2003;185:4539–4547. doi: 10.1128/JB.185.15.4539-4547.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt B, Schweder T, Sibbald MJ, Albrecht D, Ehrenreich A, Bernhardt J, Feesche J, Maurer KH, Gottschalk G, Dijl JM, Hecker M. The extracellular proteome of Bacillus licheniformis grown in different media and under different nutrient starvation conditions. Proteomics. 2006;6:268–281. doi: 10.1002/pmic.200500091. [DOI] [PubMed] [Google Scholar]
- Walter KA, Nair RV, Cary JW, Bennett GN, Papoutsakis ET. Sequence and arrangement of two genes of the butyrate-synthesis pathway of Clostridium acetobutylicum ATCC 824. Gene. 1993;134:107–111. doi: 10.1016/0378-1119(93)90182-3. [DOI] [PubMed] [Google Scholar]
- Wiesenborn DP, Rudolph FB, Papoutsakis ET. Phosphotransbutyrylase from Clostridium acetobutylicum ATCC 824 and its role in acidogenesis. Appl Environ Microbiol. 1989;55:317–322. doi: 10.1128/aem.55.2.317-322.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesenborn DP, Rudolph FB, Papoutsakis ET. Coenzyme A transferase from Clostridium acetobutylicum ATCC 824 and its role in the uptake of acids. Appl Environ Microbiol. 1989;55:323–329. doi: 10.1128/aem.55.2.323-329.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Zhu L, Liu X, Li T, Zhang Y, Ying T, Wang B, Wang J, Dong H, Feng E, Li Q, Wang J, Wang H, Wei K, Zhang X, Huang C, Huang P, Huang L, Zeng M, Wang H. A proteome reference map and proteomic analysis of Bifidobacterium longum NCC2705. Mol Cell Proteomics. 2006;5:1105–1118. doi: 10.1074/mcp.M500410-MCP200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Below is the link to the electronic supplementary material.
pH 5.7 acidogenesis (XLS 124 kb)
pH 4.5 solventogenesis (XLS 132 kb)
Table of influenced spots (XLS 48 kb)
Transcriptome data (XLS 552 kb)
Transcript amounts (XLS 984 kb)
(PDF 343 kb)
(PDF 283 kb)