Abstract
Structured RNAs encode native conformations that are more stable than the vast ensembles of alternative conformations, but how this specificity is evolved is incompletely understood. Here we show that a variant of the Tetrahymena group I intron ribozyme that was generated previously by in vitro selection for enhanced thermostability also displays modestly enhanced specificity against a stable misfolded structure that is globally similar to the native state, despite the absence of selective pressure to increase the energy gap between these structures. The enhanced specificity for native folding arises from mutations in two nucleotides that are close together in space in the native structure, and additional experiments show that these two mutations do not affect the stability of the misfolded conformation relative to the largely unstructured transition state ensemble for interconversion between the native and misfolded conformers. Thus, they selectively stabilize the native state, presumably by strengthening a local tertiary contact network that cannot form in the misfolded conformation. The stabilization is larger in the presence of the peripheral element P5abc, suggesting that cooperative tertiary structure formation plays a key role in the enhanced stability. The increased specificity in the absence of explicit selection suggests that the large energy gap in the wild-type RNA may have arisen analogously, a consequence of selective pressure for stability of the functional structure. More generally, the structural rigidity and intricate networks of contacts in structured RNAs may allow them to evolve substantial structural specificity without explicit negative selection, even against closely-related alternative structures.
To be functional, structured RNAs are faced with the challenge that they must selectively populate conformations representing a very small portion of the energy landscape. An important component of this challenge is to generate active conformations that are more stable than the most stable alternative ones, such that these functional structures remain populated at equilibrium (1-4).
How this specificity is achieved during evolution is not clear. In general, two scenarios can be envisioned. In one, the RNA is under selective pressure to stabilize the native structure relative to partially folded or unfolded forms, and a sufficiently large fraction of mutations stabilize contacts that cannot form in any given alternative structure. Thus, selection for native stability alone produces enough specificity against all viable alternative structures. Alternatively, it is possible that negative selection is necessary. In this process, some mutations would become fixed in the RNA population not because they stabilize the native state relative to unfolded structures, but instead because they destabilize a particular family of misfolded structures that would otherwise be sufficiently populated as to interfere with the functions of the RNA (5). There is increasing evidence that some proteins have also evolved in this latter scenario, with negative selection to reduce the ability to form extended beta sheet structures or other misfolded structures (6, 7), and this principle has been incorporated into protein design efforts (8-10).
Although it is difficult to disentangle the complex and varied selective pressures experienced by RNA molecules in natural environments, artificial selection can be a powerful tool because it can define explicitly the selective pressures. An in vitro selection scheme was used previously to generate a mutant of the Tetrahymena group I intron ribozyme with substantially enhanced thermostability, as demonstrated by its ability to migrate rapidly at higher temperatures in temperature-gradient gel electrophoresis (TGGE)1 (11). This mutant has nine single-nucleotide substitutions that are localized predominantly away from helical elements, and further work showed that the effects of individual mutations are strikingly non-additive, suggesting that the enhanced stability arises largely from cooperative effects of the mutations (12). Analogously, an RNase P RNA of thermophilic origin was also shown to be more stable than its mesophilic counterpart and to display increased cooperativity in tertiary structure formation (4, 13).
In addition to forming a native structure, the Tetrahymena ribozyme folds in vitro to a long-lived misfolded intermediate under standard conditions (14-20). This misfolded conformation shares extensive similarity with the native structure, including a network of long-range peripheral contacts (19). Although it is defective for the standard ribozyme catalytic reaction, cleavage of an oligonucleotide that mimics the 5′-splice site, the misfolded ribozyme catalyzes efficient cleavage at the 3′-splice site, underscoring the similarity between the two structures (19). Despite this similarity, the native state is much more stable, with a robust energy gap of 6 kcal/mol (21).
The previous in vitro selection scheme included a step in each cycle that required the RNA to remain active, ensuring that the native state remained the most stable form. However, there was no selective pressure to increase or even to maintain the substantial energy gap between the native and misfolded structures. Here, we measure the relative stability of these two conformations for the evolved ribozyme mutant and find that the energy gap is increased by 1.2 kcal/mol (~10-fold) to >7 kcal/mol. Analysis of subsets of the mutations shows that the enhanced specificity against misfolding arises exclusively from two of the nine point mutations, which are close in space and cooperatively stabilize the native state (12). The environment of these nucleotides differs in the misfolded conformation (19), suggesting that the enhanced specificity against misfolding arose from fortuitous strengthening of a contact that cannot form in the misfolded conformation. Similar processes in nature may allow structured RNAs to achieve large energy gaps between their native and misfolded structures, even closely related ones, without explicit negative selection.
MATERIALS AND METHODS
Preparation of RNA
The R14CΔP5abc ribozyme was constructed by Quickchange PCR (Stratagene) from the gene encoding the full-length R14C using oligonucleotides encoding the 76-nucleotide P5abc deletion (21). Individual substitutions were also made using Quickchange, and the complete sequences of all mutant ribozymes were verified by DNA sequencing. Catalytic activity of all mutants was measured and found to be within 3-fold of the wild-type ribozyme (data not shown). Full-length ribozymes, P5abc-deleted versions, and P5abc RNA were prepared by in vitro transcription using T7 RNA polymerase and purified by Qiagen RNeasy columns (22). Oligonucleotides (Dharmacon, Lafayette, C O) were 5 ′-end-labeled with [γ-32P]ATP using T4 polynucleotide kinase and purified by non-denaturing polyacrylamide gel electrophoresis (PAGE) (23). R14C ribozyme was 3′-end-labeled with [α-32P]ATP using the Klenow Fragment of DNA Polymerase I and a complementary DNA template (24).
Catalytic activity assay to monitor native ribozyme formation
The refolding of misfolded ribozyme was followed by catalytic activity. P5abc-deleted ribozymes (200 nM) were folded to a mixture of the native and misfolded conformations by adding 10 mM Mg2+ at 25°C (50 mM Na-MOPS, pH 7.0) (see Figure 2A). At various times thereafter, aliquots were mixed with a ‘folding quench’ solution containing guanosine and P5abc (600 nM final concentration). This solution was incubated for 5 min, sufficient time for P5abc to bind and activate the P5abc-deleted ribozymes for substrate cleavage but not enough time to allow interconversion of the native and misfolded conformers (17, 19). Refolding of full-length ribozymes was measured in the same way except that the folding quench did not include P5abc. Radiolabeled substrate CCCUCUA5 (S*) was added and aliquots were quenched after 1-2 min by adding 2 volumes of 90% formamide, 20 mM EDTA (1 min for the wild-type ribozyme and 1.75 min for mutants with modestly reduced cleavage rates). Labeled substrate and product were separated by 20% denaturing PAGE and quantitated by a phosphorimager. The fraction of native ribozyme was determined by the fraction of S* that was cleaved to the shorter product CCCUCU (P*) (17, 25). To correct for a small fraction of damaged and inactive ribozyme (<20%), data are shown after normalization against an equivalent reaction in which the ribozyme was incubated at 50 °C in the presence of P5abc for 30 min to give essentially 100% native ribozyme (17). For P5abc-deleted ribozymes, observed rate constants for the approach to equilibrium (kobs) were converted to rate constants for refolding from M to N (kM-->N) by using the formula kM-->N = kobs (KM<-->N/KM<-->N + 1), where KM<-->N is the equilibrium between the native and misfolded forms.
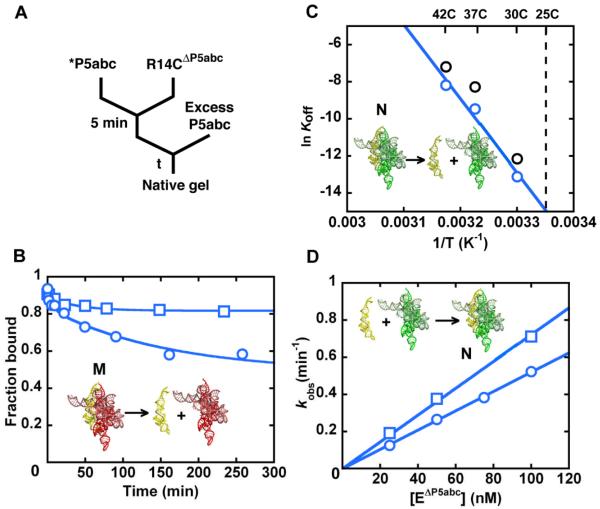
Figure 2.
Increased specificity for native folding in the R14CΔP5abc ribozyme. (A) RNA was folded in Mg2+ for time t1, and then P5abc was added, followed by S* and guanosine (G). Reactions were incubated long enough to give complete S* cleavage by the native ribozyme (time t2, 1 min for the wild-type ribozyme and 1.75 min for mutants with modestly reduced cleavage rates; see Methods). (B) Gel showing approach to equilibrium of native and misfolded conformations for the wild-type EΔP5abc (top) and R14CΔP5abc (bottom) ribozymes. Time t1 was varied from 1 min to >250 min. (C) Folding progress curves for R14CΔP5abc (blue) and EΔP5abc (black) in side-by-side reactions. Rate constants for the approach to equilibrium were 0.25 ± 0.01 min−1 and 0.018 ± 0.001 min−1 for R14CΔP5abc and EΔP5abc, respectively (average ± standard error). The latter result is the same within error as reported previously (21) Fractions of native ribozyme in this and other figure s are calculated by dividing the fraction. of S* cleaved during t2 by the corresponding fraction for fully-native ribozyme formed by extended incubation with P5abc (>0.8, see Methods). (D) Approach to equilibrium of R14CΔP5abc upon folding at 50 mM Mg2+ and subsequent dilution to standard conditions (10 mM Mg2+).
An analogous activity assay was used to measure the Mg2+ dependence of native ribozyme formation as described previously (26). Briefly, the ribozyme or its variants were incubated with varying Mg2+ concentrations (0-1 mM for full-length ribozymes and 0-20 mM for P5abc-deleted ribozymes) at 25°C (50 mM Na-MOPS, pH 7.0) for 2 hr to allow equilibration of folded and unfolded conformers (see Figure 6A). For EΔP5abc, P5abc (500 nM) and additional Mg2+ (50 mM) and guanosine (500 μM) were then added to restore catalytic activity of ribozyme that was in the native state in the initial incubation, whereas the majority of the ribozyme that was non-native was subsequently trapped in the misfolded conformation. After 10 min to allow full binding of P5abc, labeled substrate was added to initiate reactions. Substrate cleavage reactions with full-length ribozyme were performed similarly, except that Mg2+ and S was added, followed by guanosine to initiate the cleavage reaction. Reactions were quenched after 1 min for wild-type ribozyme or after 1.75 min for R14C ribozyme variants, sufficient time for complete substrate cleavage by the native ribozyme. To compare visually the Mg2+ dependences of different ribozyme variants, the results were normalized by subtracting the small fraction that avoided misfolding after addition of 50 mM Mg2+ and dividing by the maximal fraction of native ribozyme. For example, the R14CΔP5abc ribozyme began at a higher value and progressed to a higher endpoint value, the former reflecting a more favorable kinetic partitioning during the folding process and the latter reflecting the increased specificity for folding to the native state (0.91 and 0.56 for R14CΔP5abc and EΔP5abc, respectively).
Figure 6.
Mg2+ dependence of native ribozyme formation. (A) Reaction scheme for measuring the Mg2+ dependence for the EΔP5abc ribozymes (25, 26). (B) Mg2+ dependences for the EΔP5abc ribozymes. The A269G/A304GΔP5abc ribozyme (magenta) gives a reduced K1/2 value relative to the wild-type EΔP5abc ribozyme (black, 0.86 mM vs. 2.3 mM Mg2+). R14CΔP5abc (blue) displays the same Mg2+ requirement within error as the wild-type EΔP5abc (K1/2 = 2.2 mM Mg2+). To facilitate comparison, each data set is normalized to the maximum and minimum values (see Methods). (C) Mg2+ dependence of native state formation for the full-length R14C (blue) and wild-type (black) ribozymes. The R14C ribozyme gives a modest but readily detectable decrease in K1/2 value for native state formation (0.2 mM vs. 0.3 mM Mg2+).
P5abc association and dissociation
To measure dissociation of P5abc, wild-type or mutant EΔP5abc (100 nM) was incubated for 15 s or 1 h (50 mM Na-MOPS, 10 mM Mg2+, 25 °C) to generate predominantly misfolded or native ribozyme, respectively. A trace amount of radiolabeled P5abc was added for 8 min to allow full binding, and then excess unlabeled P5abc (1 μM) was added. The fraction of labeled P5abc remaining bound over time was determined by loading aliquots on a continuously-running 12% native PAGE at 4°C and analyzed by using a phosphorimager.
P5abc association kinetics were measured analogously. EΔP5abc ribozyme was incubated as above to generate populations of predominantly native ribozyme or as much misfolded ribozyme as possible, and then labeled P5abc was added. At various times thereafter, excess unlabeled P5abc (2 μM) was added to block further binding of labeled P5abc, and aliquots were processed as above. Rate constants for P5abc association with the native and misfolded conformations were calculated from the observed rate constants with these two mixtures as described previously (21).
Calculation of native specificity in the full-length ribozyme
The equilibrium values relating the stabilities of the native and misfolded conformations for the R14C ribozyme and other variants were calculated as described previously (21). Briefly, the corresponding equilibrium value for each P5abc-deleted ribozyme was multiplied by the affinity difference of P5abc for the native vs the misfolded core. This analysis makes the assumption that the behavior of the trans complex with P5abc is the same as that of the corresponding full-length ribozyme, as suggested by equivalent catalytic activity and structure probing results (21, 27, 28). Because the equilibrium values for the full-length ribozymes are calculated from experiments in which incubations were chosen to give extensive population of the native or misfolded conformation as desired, the values are not subject to errors that could arise from identifying exceedingly minor populations (i.e. establishing that 1 in 105 molecules are misfolded rather than being non-functional as a result of damage).
Thermal denaturation
RNA denaturation was followed on a DU7400 Beckman-Coulter UV spectrophotometer (260 nM). Ribozyme was incubated in 50 mM Na-MOPS, pH 7.0 (25 °C), 5 mM MgCl2 (80 °C, 3 min), slowly cooled to room temperature and incubated for 5 min. Absorbance data were collected as the ribozyme was heated from 25 °C to 90 °C (0.5 °C per min).
RESULTS
To measure the relative stability of the native and misfolded conformations of the thermostable ribozyme mutant, termed R14C (11), we used a thermodynamic cycle developed previously with the wild-type ribozyme (21) (Figure 1A). The energetic preference for native state formation, relative to the misfolded conformation, has been defined as the structural specificity of folding (21), and this terminology or the abbreviated term ‘specificity’ is used herein to refer specifically to the equilibrium between the native and misfolded conformations. Our earlier work showed that the equilibrium between the native and misfolded conformations for a version of the ribozyme that lacks the peripheral element P5abc (EΔP5abc) is near unity (Keq(M<->N) = 1.4, as determined directly by catalytic activity (21)). P5abc was shown to bind to the native core nearly 105-fold tighter than to the misfolded core. Completion of the thermodynamic cycle led to a calculated value of approximately 105 for the equilibrium between the native and misfolded conformations of the complex. It is not possible to measure the equilibrium directly for the full-length ribozyme because the native state is so strongly favored, but we infer that the equilibrium for the full-length ribozyme is similar or equivalent to that of the complex because P5abc assembles with the EΔP5abc ribozyme to form a complex that is fully active and recapitulates the global footprinting pattern of the full-length ribozyme (21, 28-30), indicating comparable structural and energetic properties.
Figure 1.
Experimental system for measuring the relative stability of the native (N) and misfolded (M) conformations of the R14C ribozyme. (A) Thermodynamic cycle used to calculate the equilibrium between the native and misfolded forms. Here and in other figures, the native and misfolded conformations are depicted schematically, in green and red respectively, by showing the ribozyme structural model (43). Equilibrium constants obtained herein for the R14C ribozyme are blue and corresponding values for the wild-type ribozyme are black (21). (B) The two-piece ribozyme construct. The mutations in the R14C ribozyme are shown in large letters. The two substitutions shown herein to increase native specificity are red, and the other seven are blue. (C) Ribozyme stereoview showing the nine substitutions of the R14C ribozyme. Colors are the same as in panel B.
To perform analogous measurements of the R14C ribozyme, we constructed a variant that includes all nine mutations and lacks P5abc (R14CΔP5abc; Figure 1B and C). Below, we describe experiments using R14CΔP5abc and P5abc added separately to measure the values of this thermodynamic cycle, and then we dissect contributions from individual and pairwise mutations.
Increased native specificity in the thermostable R14C mutant
We first measured the equilibrium of the R14CΔP5abc ribozyme between the native and misfolded conformations using catalytic activity (17, 25). To determine the fraction of native R14CΔP5abc ribozyme during Mg2+-induced folding, we added radiolabeled substrate (CCCUCUA5, S*) to aliquots of a folding reaction and measured the fraction of S* that was cleaved rapidly (Figure 2A and B). As expected, much of the R14CΔP5abc ribozyme did not reach the native state on the time scale of seconds, although the fraction that avoided misfolding was larger than that for the wild-type EΔP5abc ribozyme (Figure 2C).2 Hydroxyl radical footprinting of the misfolded R14C ribozyme revealed structural signatures of the misfolded wild-type ribozyme (Figure S1), suggesting a common origin of misfolding. Unlike the wild-type EΔP5abc ribozyme, however, R14CΔP5abc then refolded to give predominantly native ribozyme (Keq(M<->N)= 8.4, Figure 2C). We confirmed that this endpoint reflected an equilibrium by folding at higher Mg2+ concentration, which gave a further shift toward the native state as observed previously for the EΔP5abc ribozyme (21), and then diluting back to 10 mM Mg2+ (Figure 2D). Upon dilution, the native fraction of R14CΔP5abc ribozyme decreased to the same endpoint as upon initial folding at 10 mM Mg2+. Thus, compared to the wild-type EΔP5abc ribozyme, R14CΔP5abc displays increased specificity for native state formation relative to the misfolded conformation, shifting the equilibrium between these two conformations by a factor of six (Figure 2C and Table 1).
Table 1.
Kinetic and thermodynamic constants of P5abc-deleted ribozymes1
| Number and description of ribozyme2 |
kobs (min−1)3 |
kM->N (min−1)4 |
Equilibrium w/o P5abc KM <-->N |
kon, (M) P5abc (M−1 min−1) |
koff, (M) P5abc (min−1) |
Kd, (M) P5abc (nM) |
kon, (N) P5abc (M−1 min−1) |
koff, (N) P5abc (min−1) |
Kd, (N) P5abc (fM) |
Equilibrium w/ P5abc KM <-->N |
|
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | WT5 | 0.018 ± 0.001 | (1.1 ± 0.1) × 10−2 | 1.4 ± 0.1 | (8.0 ± 2.0) × 106 | 0.026 ± 0.006 | 3.1 ± 1.0 | (1.9 ± 0.5) × 107 | (7.2 ± 2.4) × 10−7 | 38 ± 16 | (1.1 ± 0.6) × 105 |
| 2 | R14C | 0.25 ± 0.01 | 0.22 ± 0.03 | 8.4 ± 1.2 | (2.8 ± 0.6) × 106 | (7.2 ± 0.3) × 10−3 | 2.6 ± 0.8 | (8.6 ± 1.3) × 106 | (2.4 ± 0.8) × 10−7 | 28 ± 10 | (8 ± 4) × 105 |
| 3 | WT A269G/A304G |
(9.2 ± 0.6) × 10−3 | (8.0 ± 1.0) × 10−3 | 6.8 ± 0.7 | (1.1 ± 0.3) × 107 | 0.021 ± 0.002 | 1.9 ± 0.5 | (4.4 ± 0.2) × 107 | (2.7 ± 0.9) × 10−7 | 6.1 ± 2.0 | (2.1 ± 0.9) × 106 |
| 4 | R14C G269A/G304A |
0.14 ± 0.02 | 0.08 ± 0.02 | 1.3 ± 0.3 | (9.9 ± 1.6) × 106 | 0.026 ± 0.006 | 2.6 ± 0.7 | (2.8 ± 0.6) × 107 | (1.4 ± 0.5) × 10−6 | 50 ± 20 | (7 ± 4) × 104 |
| 5 | WT U277C |
0.20 ± 0.01 | 0.11 ± 0.01 | 1.3 ± 0.1 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 6 | R14C C277U |
(6.0 ± 0.3) × 10−3 | (5.2 ± 2.0) × 10−3 | 6.5 ± 2.5 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 7 | WT U277C/ A269G/304G |
0.70 ± 0.20 | 0.60 ± 0.20 | 6.2 ± 0.7 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 8 | R14C C277U/ G269A/G304A |
0.018 ± 0.006 | 0.011 ± 0.003 | 1.7 ± 0.1 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 9 | WT A269G |
0.016 ± 0.001 | 0.013 ± 0.001 | 3.6 ± 0.1 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 10 | WT A304G |
(6.9 ± 0.6) × 10−3 | (4.9 ± 0.9) × 10−3 | 2.4 ± 0.4 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 11 | WT U259A |
0.018 ± 0.001 | (1.1 ± 0.1) × 10−2 | 1.4 ± 0.2 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
| 12 | WT U340A6 |
0.016 | 1.1 × 10−2 | 2.1 | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. | N.D. |
All values are shown as the average of multiple determinations. For experiments that were performed twice, the uncertainty is expressed as the range of the two measurements. For those that were performed more than twice, the uncertainty reflects the standard error of the set of measurements.
Mutants are named by first indicating either the WT EΔP5abc or R14CΔP5abc background and then indicating the nucleotide substitutions.
Values are the observed rate constant for equilibration of the native and misfolded species (25 °C, 50 mM Na-MOPS, pH 7.0, 10 mM Mg2+).
The value of kM->N is calculated from the rate and equilibrium values for exchange of the native and misfolded conformations (kobs and KM<--->N).
Values for the wild-type ribozyme are from side-by-side measurements except for P5abc binding rate constants, which are reproduced from previous work (21). All values from the side-by-side measurements were in good agreement with the earlier work (21).
Values for the U340A ribozyme reflect a single determination.
N.D., not determined.
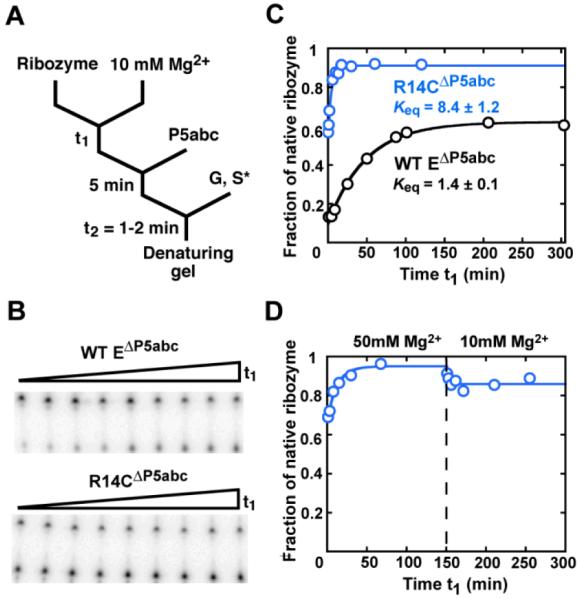
With knowledge of the rate and equilibrium constants for interconversion of the native and misfolded conformations of R14CΔP5abc, we used a pulse-chase native gel shift approach to determine the affinity of P5abc for each conformation by measuring the kinetics of binding and dissociation (21). Dissociation of P5abc from a mixture of native and misfolded R14CΔP5abc was biphasic, with a major phase of 0.0072 min−1 (Figure 3A and 3B). This phase was much less prominent in the reaction with predominantly native R14CΔP5abc ribozyme, indicating that it reflects dissociation from misfolded R14CΔP5abc ribozyme. In contrast, dissociation of P5abc from the native R14CΔP5abc ribozyme was extremely slow, as observed previously for the wild-type EΔP5abc ribozyme (21). Extrapolation from higher temperature (Figure 3C) gave a rate constant of 2.4 × 10−7 min−1. An analogous pulse-chase approach was used to measure the binding rate of P5abc. From two sets of binding reactions, one with largely native R14CΔP5abc ribozyme and one with a mixture of native and misfolded R14CΔP5abc ribozyme, the association rate constants were determined to be 8.6 × 106 M−1 min−1 and 2.8 × 106 M−1 min−1 for the native and misfolded ribozyme respectively (Figure 3D).
Figure 3.
Preferential binding of P5abc to the native R14CΔP5abc ribozyme. (A) To measure dissociation, radiolabeled P5abc (*P5abc) was allowed to bind to the ribozyme and then chased with unlabeled P5abc. (B) Dissociation kinetics of P5abc from a population of predominantly native R14CΔP5abc ribozyme (blue squares) or a mixture of native and misfolded R14CΔP5abc (blue circles). The mixture gave a major phase of dissociation with a rate constant of 0.0072 ± 0.0003 min−1, whereas most of the P5abc remained bound to the native ribozyme on the observable time scale (up to 5 days, additional data points not shown). (C) Determination of the rate constant for P5abc dissociation from the native R14CΔP5abc ribozyme by extrapolation from measurements at higher temperatures (blue). Data for the EΔP5abc ribozyme obtained side-by-side are shown in black and were consistent with previous results (21). (D) Association kinetics of P5abc to predominantly native R14CΔP5abc ribozyme (blue squares) or a mixture of native and misfolded R14CΔP5abc ribozyme (blue circles).
The much slower dissociation, coupled with the modestly faster binding, indicates that P5abc binds to the native R14CΔP5abc ribozyme 90,000-fold tighter than to the misfolded conformation (Figure 1A), the same difference within error as for the wild-type EΔP5abc ribozyme (Figure 1A and ref. 21). Completing the thermodynamic cycle gives a Keq(M<->N) value for the complex, and presumably for the full-length R14C ribozyme, of 8 × 105, about 7-8-fold larger than for the wild-type ribozyme. Strikingly, the in vitro selection produced a ribozyme that is better able to discriminate between the native and misfolded conformations despite the absence of selective pressure to increase or even to maintain this energy gap.
Two mutations are responsible for the increased specificity
We next probed individual mutations to identify changes that gave rise to increased specificity for native folding. We identified A269G and A304G as strong candidates because these mutations were shown previously to stabilize the native state when made in tandem (12) and because structure probing indicated that the limited differences between the native and misfolded conformations in both wild-type and R14C mutant ribozymes are concentrated in this region (19).
Therefore, we utilized the thermodynamic cycle again to determine the effects on specificity of these mutations (Figure 4A). In the absence of added P5abc, the P5abc-deleted, A269G/A304G ribozyme displayed enhanced specificity against the misfolded form (Figure 4B), with an equilibrium value indistinguishable from that of R14CΔP5abc (Keq(M<->N) = 6.8, Figure 4A). This enhancement required both mutations, with the individual substitutions giving smaller effects (Table 1, ribozymes 9 and 10). The preferential binding of P5abc to the native form was maintained and even modestly enhanced in the A269G/A304G EΔP5abc mutant (Figure 4C and 4D), with an affinity difference of 3 × 105-fold. Thus, completion of the thermodynamic cycle indicates that the A269G and A304G mutations impart at least as much specificity to the full-length ribozyme as the entire set of R14C mutations (Keq(M<->N) = 2.1 × 106, Figure 4A). (Individual mutations in other selected nucleotides did not give effects; see Table 1, ribozymes 11-12 and Table 2, ribozymes 21-22). This conclusion was confirmed using a variant in which 269 and 304 were reverted to their wild-type identities in the background of R14CΔP5abc. This ribozyme displayed a complete loss of the enhanced specificity against the misfolded state with bound P5abc, giving a value indistinguishable from the wild-type ribozyme (Figure S2; Table 1, ribozyme 4).
Figure 4.
Enhanced specificity for native folding from A269G and A304G substitutions. (A) Thermodynamic cycle for the A269G/A304G ribozyme variant. Both in the absence and presence of P5abc (top and bottom, respectively), the equilibrium is shifted toward the native state by at least as large an amount as for the R14C variants (Figure 1A, see text). (B) Equilibrium folding of the A269G/A304GΔP5abc ribozyme. The observed rate constant is 0.008 ± 0.001 min−1 and the equilibrium value is 6.8 ± 0.7. (C) Dissociation of P5abc from the A269G/A304GΔP5abc ribozyme. As in Figure 3B, dissociation was followed from a solution of predominantly native ribozyme (magenta squares) or a mixture of native and misfolded ribozyme (magenta circles). Data from wild-type EΔP5abc are shown in black for comparison. (D) P5abc association kinetics. P5abc association was measured for a population of largely native A269G/A304GΔP5abc ribozyme (magenta squares) or a population of predominantly misfolded A269G/A304GΔP5abc ribozyme (magenta circles)
Table 2.
Kinetics and thermodynamic constants of full-length mutant ribozymes
| Number and description of ribozyme |
kobs (min−1)1 |
kobs (relative)2 |
ΔΔGM (kcal/mol)3 |
KM <-->N4 | ΔΔGN (kcal/mol)5 |
|||
|---|---|---|---|---|---|---|---|---|
| 50 °C | 37 °C | 25 °C | ||||||
| 13 | WT | 0.075 ± 0.017 | 1.4 × 10−3 | 2.1 × 10−4 | (1) | (0) | (1.1 ± 0.6) × 105 | (0) |
| 14 | R14C | 0.017 ± 0.004 | 4.0 × 10−3 | 5.1 × 10−5 | 0.22 ± 0.07 | −0.95 ± 0.25 | (8 ± 4) × 105 | −2.1 ± 0.7 |
| 15 | WT A269G/A304G | 0.043 | 8.1 × 10−3 | 1.3 × 10−4 | 0.57 | −0.36 | (2.1 ± 0.9) × 106 | −2.1 ± 0.7 |
| 16 | R14C G269A/G304A | 0.012 | N.D. | N.D. | 0.16 | −1.2 | (7 ± 4) × 104 | −0.9 ± 0.9 |
| 17 | WT U277C | 0.041 | 1.5 × 10−3 | 2.5 × 10−4 | 0.53 | −0.39 | N.D. | N.D. |
| 18 | R14C C277U | 8.2 × 10−3 | 1.2 × 10−4 | N.D. | 0.11 | −1.4 | N.D. | N.D. |
| 19 | WT U277C/A269G/304G | 0.027 | 1.6 × 10−3 | N.D. | 0.40 | −0.59 | N.D. | N.D. |
| 20 | R14C C277U/G269A/G304A | 6.0 × 10−3 | 1.3 × 10−4 | N.D. | 0.070 | −1.7 | N.D. | N.D. |
| 21 | WT U259A | 0.12 | N.D | N.D | 1.6 | 0.30 | N.D | N.D |
| 22 | WTU340A | 0.046 | 1.3 × 10−3 | N.D | 0.61 | −0.32 | N.D | N.D |
Values are rate constants for refolding of the misfolded ribozyme to the native state. For the wild-type and R14C ribozymes, values are the average and range from two independent determinations. Results from single determinations are reported for the other mutant ribozymes.
Relative values were obtained from measurements at 50 °C.
ΔΔGM values reflect the change in stability of the misfolded conformation for the indicated mutant, relative to the transition state for refolding. These values were calculated from the relative refolding rates as ΔΔGM = −RT ln (1/kobs (relative)). Values are based on measurements at 50 °C. Similar effects of mutations were observed at 25 °C for all of the mutants for which measaurements were made at 25 °C.
KM <-->N values are reproduced from Table 1. These values were determined using the thermodynamic cycle of P5abc binding to the native and misfolded conformers of the corresponding P5abc-deleted ribozymes.
ΔΔGN values reflect the change in stability of the native conformation for the indicated mutant, relative to the transition state for refolding, and were calculated from ΔΔGM and the change in KM <-->N as ΔΔGN = ΔΔGM + (−RT ln(KM<-->N)Mutant/(KM<-->N)WT) (see Figure 3).
N.D., not determined.
Mutation of A269 and A304 do not affect stability of the misfolded conformation
To probe further the mechanism of these mutations in conferring specificity for native folding, we were interested in determining whether the mutations affect the stability of the misfolded conformation relative to unstructured conformations. Although this question cannot be addressed directly because we do not have a means of populating only the misfolded and unfolded conformations to determine their relative stabilities, we used the kinetics of refolding. This measurement is useful because the refolding transition proceeds through a transition state ensemble that is much less structured than the misfolded conformation, as earlier work showed strong increases in the refolding rate from urea and tertiary contact disruptions (19, 32), and the refolding rate provides a relative measure of the free energy difference between the misfolded conformation and this less structured transition state ensemble. The full-length A269G/A304G mutant gave a refolding rate constant within 2-fold of that of the wild-type ribozyme (Figure 5A, Table 2, ribozyme 15), indicating that these substitutions have minimal effect on the stability of the misfolded conformation relative to the transition state ensemble. Therefore, the enhanced specificity for native state folding indicates that these mutations stabilize the native state relative to the less structured transition state, consistent with previous TGGE gel data indicating that this double-mutant increased the melting temperature of the wild-type ribozyme (12).
Figure 5.

The A269G and A304G mutations specifically stabilize the native state without stabilizing the misfolded conformer. (A) Refolding of full-length ribozyme variants. Refolding progress curves at 50 °C and 10 mM Mg2+ are shown for the A269G/A304G variant (magenta, kobs = 0.043 min−1) with the wild-type ribozyme as a reference (black, kobs = 0.075 ± 0.017 min−1), and for the G269A/G304A reversion mutant (violet, kobs = 0.012 min−1) with the R14C mutant as a reference (blue, kobs = 0.017 ± 0.004 min−1). (B) Free energy profile of the A269G/A304G mutant in the wild-type background. The reference states are the relatively unstructured transition states for exchange between the native (N) and misfolded (M) conformations. The change in relative free energy of M (vertical position) is determined from the refolding rate constant of the A269G/A304G variant (magenta) relative to the wild-type ribozyme (black). The relative free energy of N is then calculated from the equilibrium value between the native and misfolded conformers, determined using the thermodynamic cycle of P5abc binding to the native and misfolded species of the corresponding P5abc-deleted ribozymes. Whereas N is substantially stabilized by the A269G/A304G mutations (2.1 kcal/mol), M is at most marginally stabilized (0.36 kcal/mol). (C) Stabilization from the A269G/A304G mutations in the R14C background. The mutant R14C G269A/G304A (violet) contains the other seven mutations. From this background, the forward mutations of 269 and 304 from A to G specifically stabilize N by 1.2 kcal/mol (R14C mutant, blue). Comparing the R14C G269A/G304A (violet) with the wild-type ribozyme (black) shows that the other seven mutations stabilize the native and misfolded conformers equally.
To probe possible context-dependent effects, we also tested a full-length mutant that contains all of the R14C mutations except nucleotides 269 and 304, which are reverted to the wild-type sequence. This mutant refolded with the same rate constant within error as the full-length R14C ribozyme (Figure 5A and Table 2, ribozyme 16). Thus, in either background the substitutions at nucleotides 269 and 304 have little or no effect on the stability of the misfolded conformation, instead specifically stabilizing the native state. These results and the conclusion are depicted in the free energy profiles in Figure 5B and 5C, in which the reference state is defined as being the transition state ensemble for refolding. Although there may be limits to the assumption that the R14C ribozyme variant refold with the same transition state ensemble, the R14C ribozyme adopts a highly-related misfolded conformation (Figure S1) and is accelerated for refolding with the same urea dependence as the corresponding wild-type ribozymes, indicating at least substantial similarity of the refolding processes and likely of the transition states (Figure S3A).
With the free energy profiles, it is apparent that the 269/304 mutations stabilize the native state but have relatively little effect on the misfolded conformation (Figure 5B). In contrast, when the mutant with reversion only of 269 and 304 is compared to the wild-type ribozyme, it is apparent that the other seven mutations together stabilize both conformations by about 1 kcal/mol (Figure 5C and Table 2, ribozyme 16). The simplest explanation for these results is that 269G and 304G stabilize the native state by participating in contacts that are unable to form in the misfolded conformation (see Discussion).
Peripheral element P5abc enhances effects of stabilizing mutations
The use of the thermodynamic cycle required that we measure folding of the P5abc-deleted versions of the ribozyme. As described below, in the course of these analyses we found that the stabilizing effects of the mutations are enhanced or even dependent on the presence of P5abc. P5abc forms extensive tertiary connections with the core and other peripheral elements, completing a ring of tertiary connections that encircles the core, and these contacts are known or proposed to form cooperatively (21, 33; T. Johnson and R.R., unpublished results). Thus, we suggest that tertiary structure cooperativity plays a key role in the stabilizing effects of the mutations, consistent with the interpretation of earlier work (12).
For the P5abc-deleted ribozymes, catalytic activity assays showed that the mutations in nucleotides 269 and 304 increase specificity for native folding, as indicated by the shift in endpoint (Figure 4B), with at most a small effect on the stability of the misfolded conformation, as indicated by the same refolding rate as the wild-type EΔP5abc ribozyme. (Analogous to the full-length ribozymes, similar urea dependences for refolding kinetics of the R14C and wild-type EΔP5abc ribozymes suggest similar extents of unfolding; Figure S3B). Thus, this analysis indicates that these two mutations stabilize the native conformation of EΔP5abc relative to unstructured conformations by ~1 kcal/mol (5-fold, compare ribozymes 1 and 3 in Table 1). To confirm this conclusion, we wanted to measure this stabilization directly. Although the most direct method of measuring stability might be imagined to be thermal unfolding as monitored by UV absorbance or TGGE, these methods appeared to be largely insensitive to tertiary structure loss in the P5abc-deleted ribozymes, instead reporting on loss of secondary structure (see Supporting Information). We therefore turned to the Mg2+ dependence, with catalytic activity providing a readout for native state formation (25, 26). It was shown previously that the Mg2+ dependence for native ribozyme formation of EΔP5abc mirrors that for formation of tertiary structure monitored by hydroxyl radical footprinting (26). Indeed, using this approach we found that the A269G/A304GΔP5abc mutant gave a reduced Mg2+ requirement for native state formation (Figure 6A and B), directly demonstrating that the mutations stabilize the native conformation even in the absence of P5abc. However, P5abc binds several-fold more tightly to the native A269G/A304GΔP5abc mutant than to the wild-type EΔP5abc, without a corresponding increase in binding affinity for the misfolded conformer (see Figure 4 and Table 1, ribozymes 1 and 3), leading to a calculated equilibrium value between the native and misfolded states of the full-length A269G/A304G mutant that is 20-fold larger than that of the wild-type ribozyme. Thus, P5abc contributes to the preferential stabilization of the native state by the mutations, presumably by tightening contacts that are specific to the mutated nucleotides.
A more complicated picture emerged when considering the full set of R14C mutations. First, the Mg2+ dependence of native state formation for R14CΔP5abc showed no stabilization relative to the wild-type EΔP5abc ribozyme (Figure 6B). (The control experiment shown in Figure 6C confirmed that the full-length ribozyme is stabilized by the R14C mutations, consistent with the earlier results (12).) This result could appear paradoxical given that the R14C mutations enhance the specificity for native state of EΔP5abc, but further analysis of the refolding curves revealed that the R14C mutations increase the rate of refolding from the misfolded conformation for EΔP5abc (see Figure 2C and Table 1, ribozyme 2), indicating a decrease of 1.7 kcal/mol in the stability of the misfolded EΔP5abc relative to the transition state (Figure 7). This destabilization roughly compensates for the enhanced specificity, leaving no net stabilization of the native state of the R14CΔP5abc ribozyme.
Figure 7.
Only three substitutions affect stability and specificity in the absence of P5abc. A cycle of mutations is depicted starting from EΔP5abc (top left). Free energy profiles analogous to those in Figure 5 are shown, along with the refolding data used to calculate the profiles (insets, k represents kM→N), for the A269G/A304GΔP5abc variant (bottom left) and the U277CΔP5abc variant (top right). At the bottom right, the free energy profile and data are shown for R14CΔP5abc (blue). This ribozyme gives the same results within a factor of three from values calculated as the energetic sum of the A269G/A304G and U277C mutations. A variant of EΔP5abc with the three mutations, A269G/A304G/U277CΔP5abc gives essentially the same behavior (bottom right, orange curve) but with modestly accelerated kinetics.3 Each ribozyme is labeled with its corresponding number from Table 1.
The behavior of the R14CΔP5abc mutant could be explained by a mutation that destabilized the native and misfolded states equivalently and acted additively with the stabilization by the A269G and A304G mutations. A candidate was the U277C mutation, which disrupts a base pair in the P3 helix, because previous work suggested that P3 is formed in both the native and misfolded conformations (19) but not in the unfolded ensemble (31, 34). Supporting this hypothesis, we found that a U277C variant of EΔP5abc destabilized the native and misfolded states equally, giving a refolding rate equal to that of the R14CΔP5abc ribozyme without shifting the equilibrium toward the native state (Figure 7, top right). Therefore, U277C destabilizes the native and misfolded EΔP5abcconformations equally (Figure 7, top right). In contrast, in the full-length ribozyme, the U277C mutation slowed refolding by a factor of 2 (Table 2, ribozyme 17), and previous work showed that this substitution stabilizes the native state modestly (12).
In the P5abc-deleted background, the additive effects of 277 and 269/304 would be predicted to mirror the overall behavior of the R14CΔP5abc mutant (Figure 7). To test this hypothesis, we constructed triple mutants (A269G/A304G/U277C) in the wild-type EΔP5abc and R14CΔP5abc backgrounds. The triple mutant in the wild-type EΔP5abc background displayed the native specificity associated with the A269G and A304G mutations, the faster refolding rate associated with U277C, and gave results similar to the R14CΔP5abc ribozyme (Figure 7, bottom right). Analogously, the triple reversion mutant from the R14CΔP5abc background gave additive effects and overall behavior that was indistinguishable from that of the wild type EΔP5abc ribozyme (Figure S4).
Two important conclusions arise from these results. First, the stabilization by the R14C mutations is enhanced by the presence of P5abc, and most of the mutations do not give detectable effects on stability in its absence.3 These nucleotides may not form base-specific tertiary contacts in the absence of P5abc and its tertiary connections (33). Second, the mutations that do give effects do so in an energetically independent manner to a first approximation, most likely reflecting a loss of global cooperativity in the absence of P5abc.
DISCUSSION
In the in vitro selection experiment that led to the generation of the R14C Tetrahymena ribozyme, the dominant selective pressure was for enhanced thermostability, specifically the ability to migrate rapidly through a gel at increased temperature (11). Here we showed that, in adition to being more therostable, the R14C variant is better able to discriminate between the native and misfolded conformations than the wild-type ribozyme. As described below, this striking result indicates that structured RNAs are capable of evolving very large energy gaps between their native states and alternative structures, even closely-related structures, without negative selection against the alternative structures.
An important question is whether the original in vitro selection included ‘hidden’ selective pressure to destabilize the misfolded conformation relative to the native state (11). Within each cycle of the selection, there were two parts, and in principle, selection against misfolding could have been introduced in either part. First, after a heating/cooling cycle, Mg2+ was added to 1 mM and the RNA was loaded on a temperature gradient gel with 0.2 – 0.5 mM Mg2+. Although much of the ribozyme was presumably misfolded when it was loaded into the gel well, at these low Mg2+ concentrations and the elevated temperature of the relevant portions of the gel (i.e. near the ribozyme denaturation temperature), interconversion of the native and misfolded conformations is likely on the time scale of seconds or faster (19), presumably allowing equilibration in the well prior to entering the gel. Thus, it is unlikely that the misfolded conformation remained populated during the gel separation, and therefore unlikely that there was selective pressure against its stability. Second, RNA extracted from the gel was checked for catalytic activity, and only those molecules that retained catalytic activity were taken on to the next round. In this step, there is clearly selective pressure against stable misfolding; that is, the misfolded form could not have become more stable than the native state. However, there was no selective pressure to increase or even maintain the large energy gap between the native and misfolded conformations, provided that the native state remained the dominant form. It is also important to point out that the catalytic activity test was performed after only a brief incubation in 5 mM Mg2+ (≤30 min at room temperature), conditions that would be expected to trap both the wild-type and selected ribozymes largely in the misfolded conformation (16, 35; data herein). Thus, there may have been selective pressure for the ability to avoid the misfolded conformer during folding, and indeed the R14C ribozyme partitions more favorably than the wild-type ribozyme toward the native state (Figure 5A). However, this pressure is exerted on upstream intermediates, not on the misfolded structure, and indeed there appears to be no correlation between mutations that change the likelihood of misfolding and those that modulate the stability of the misfolded conformation (17, 31). Thus, we conclude that the increased energy gap between the native and misfolded conformations arose by in vitro selection without selective pressure against stability of the misfolded conformation.
Although the magnitude of the increase in energy gap was modest, ~10-fold, it suggests that the very large gap in the wild-type ribozyme may also have arisen in the absence of explicit negative selection against stability of the misfolded conformation. To the extent that the specific stabilization of the native structure arises from mutations that strengthen tertiary contacts in the native state but not in the misfolded state (see below), the simplest expectation is that there would be more opportunities for such improvement early in the evolutionary process because it would be possible to take advantage of all regions that differ in structure between the two conformations. As the energy gap increases, it would most likely be increasingly difficult to generate further specificity, as the regions of structural difference would already be optimized for differential stability, leaving dwindling opportunities for further improvement.
Physical origin of enhanced structural specificity
A further dissection of the effects of individual and pair-wise mutations has given physical insight into how the native state was stabilized relative to the misfolded conformation. The selective stabilization arose from just two of the nine mutations, A269G and A304G. These two nucleotides are close in space in the native structure of the wild-type ribozyme (36), and are brought even closer, to within 5 Å, in the native crystal structure of the R14C ribozyme (37; see Figure 1C). Thus, the mutations to G may stabilize the native state by stabilizing a network of contacts involving both nucleotides (12). It is also possible that in the native structure in solution, these nucleotides contact each other directly. An analogous contact is observed by equivalent guanosine nucleotides in the Azoarcus ribozyme (38, 39). Although the misfolded and native conformations share a high degree of global similarity, structure-probing results indicate that local differences include and surround these nucleotides (Figure S1; refs. 19, 32). Thus, the simplest model is that the connection that is formed or strengthened by the mutations is unable to form in the misfolded structure (Figure 8), most likely because local differences between the native and misfolded conformations result in re-positioning of one or both structural elements such that they cannot interact with each other. These results therefore provide a specific structural constraint that should be useful for future modeling of the misfolded conformation (19, 32).
Figure 8.
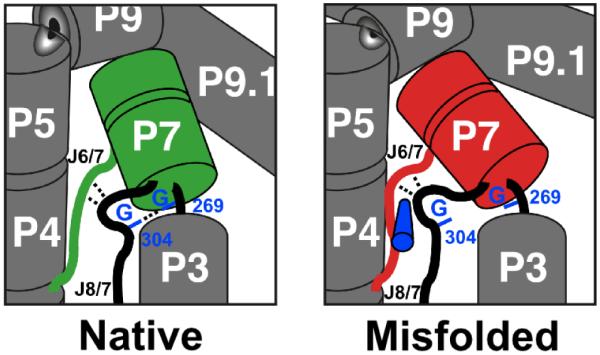
Physical model for the enhanced specificity from the A269G and A304G mutations. In the native state, guanosines at these positions can form base-specific contacts (dashed line between labeled nucleotides), analogous to contacts formed in the Azoarcus ribozyme between the equivalent bases (37, 39). In the misfolded conformation, local rearrangements or re-orientations of P7 and J6/7 (red), as indicated by changes in footprinting patterns of these elements (19, 32), change the relative positions of nucleotides 269 and 304 such that they do not contact each other. The structural differences within the core have been suggested to include a topological change (19), which may block contacts in the misfolded ribozyme by creating steric barriers to local rearrangements, as indicated schematically by the blue cylinder adjacent to J6/7.
The requirement for P5abc for enhanced stability from these mutations suggests that their contacts form cooperatively with global tertiary structure. For A269 and A304, all of the data are consistent with a simple picture in which the mutations to G stabilize the native state without affecting the misfolded state, and therefore the enhanced stability calculated from the thermodynamic cycle is equivalent to the enhanced binding of P5abc to the native core. Interestingly, this relationship does not hold when considering the other seven mutations, which stabilize the native state only in the presence of P5abc but do not lead to tighter binding of P5abc to the native core (see Table 1, mutant 4, and Table 2, mutant 14). This paradoxical result may reflect changes in the unfolded ground state or the transition state for refolding induced by these mutations, or may reflect uncertainties associated with the P5abc binding measurements or limitations in the assumption that the full-length ribozymes mirror the trans complexes with P5abc. Further physical studies on the folding transitions of these ribozymes will be necessary to resolve this question. Nevertheless, the results overall suggest that the effects of mutations are larger with the network of tertiary interactions that require P5abc (33), and these additional contacts would then be expected to contribute to the overall level of cooperativity, which is much larger in the presence of P5abc (12, 30; data herein).
Highlighting the dependence on global tertiary structure of stability contributed from the R14C mutations, we found that the U277C mutation is destabilizing for both the native and misfolded ribozyme in the P5abc-deleted ribozymes, presumably because it disrupts a base pair in P3. On the other hand, this substitution is not destabilizing in the full-length context, and the crystal structure of the R14C ribozyme suggested that mutation of U277 may allow a new tertiary contact to form between A97 and U300, compensating for the loss of the base pair (12). Thus, this mutation would be expected to enhance the cooperativity of folding in the R14C ribozyme by more closely linking secondary and tertiary structure and making folding less hierarchical.
Implications for evolution of structured RNAs
Previous results have suggested that structured RNAs can achieve enhanced stability through evolution by stabilizing networks of tertiary connections that form cooperatively (4, 12, 13), and our results suggest that these processes can generate substantial specificity against alternative structures, without explicit negative selection, because the alternative structures do not have all of the structural elements positioned to form the same networks of contacts. A key factor may be the relative rigidity of the helical elements that come together to form structured RNAs, which may prevent small-scale rearrangements and thereby enforce local differences between conformations (22). Rigidity is also likely to contribute by reducing the entropic penalties for fixation of helical elements, allowing even a single hydrogen bond to provide measureable stabilization relative to conformations in which the partners presumably form hydrogen bonds with water (40, 41). The presence of a large and robust energy gap between the native structures and inactive conformations has been suggested to be critical for ensuring that native RNA structures remain populated in the complex cellular environment (4, 42), and the results here demonstrate that this specificity can be evolved and maintained under selective pressure for thermostability of the native structure.
Supplementary Material
ACKNOWLEDGMENTS
We thank Anne Gooding and Tom Cech for the plasmid encoding the R14C ribozyme and Anne Gooding for TGGE experiments, Paul Paukstelis and Alan Lambowitz for assistance with thermal denaturation experiments, and Dan Herschlag for helpful comments on the manuscript. This work was supported by a grant to R.R. from the Welch Foundation (F-1563).
Footnotes
Abbreviations: EDTA, ethylene-dinitrilo-tetraacetic acid; EΔP5abc, Tetrahymena ribozyme variant lacking the P5abc peripheral element; R14C, the full-length ribozyme variant with enhanced thermostability; R14CΔP5abc, thermostable variant ribozyme with deletion of P5abc; PAGE, polyacrylamide gel electrophoresis; MOPS, 3-(n-morpholino) propanesulfonic acid; P, the oligonucleotide product CCCUCU; P*, (5′-32P)-labeled P; S, the oligonucleotide substrate CCCUCUA5; S*, (5′-32P)-labeled S; TGGE, temperature-gradient gel electrophoresis.
The greater accumulation of native R14CΔP5abc ribozyme early in folding reflects a shift in the kinetic partitioning between different folding pathways and does not correlate directly with the relative stabilities of the native and misfolded conformers (17, 31).
Refolding is modestly faster (3-fold) for the A269G/A304G/U277C mutant than for the R14C mutant, implying that the other six mutations together give a small stabilization of both the native and misfolded structures in the background of A269G/A304G/U277C. In the wild-type background, the six mutations give no detectable stabilization of the native or misfolded structures relative to the transition state that separates them (Figure S4).
SUPPORTING INFORMATION AVAILABLE: Hydroxyl radical footprinting data of native and misfolded R14C ribozyme (Figure S1), measurement of native specificity of the G269A/G304A mutant (Figure S2), urea-dependent refolding of full-length and P5abc-deleted R14C ribozymes (Figure S3), and effects of mutations of nucleotides 269/304 and 277 in the R14CΔP5abc background (Figure S4). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Sigler PB. An analysis of the structure of tRNA. Annu Rev Biophys Bioeng. 1975;4:477–527. doi: 10.1146/annurev.bb.04.060175.002401. [DOI] [PubMed] [Google Scholar]
- 2.Herschlag D. RNA chaperones and the RNA folding problem. J Biol Chem. 1995;270:20871–20874. doi: 10.1074/jbc.270.36.20871. [DOI] [PubMed] [Google Scholar]
- 3.Tinoco I, Jr., Bustamante C. How RNA folds. J Mol Biol. 1999;293:271–281. doi: 10.1006/jmbi.1999.3001. [DOI] [PubMed] [Google Scholar]
- 4.Fang XW, Golden BL, Littrell K, Shelton V, Thiyagarajan P, Pan T, Sosnick TR. The thermodynamic origin of the stability of a thermophilic ribozyme. Proc Natl Acad Sci U S A. 2001;98:4355–4360. doi: 10.1073/pnas.071050698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brown TS, Chadalavada DM, Bevilacqua PC. Design of a highly reactive HDV ribozyme sequence uncovers facilitation of RNA folding by alternative pairings and physiological ionic strength. J Mol Biol. 2004;341:695–712. doi: 10.1016/j.jmb.2004.05.071. [DOI] [PubMed] [Google Scholar]
- 6.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci U S A. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berezovsky IN, Zeldovich KB, Shakhnovich EI. Positive and negative design in stability and thermal adaptation of natural proteins. PLoS Comput Biol. 2007;3:e52. doi: 10.1371/journal.pcbi.0030052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Havranek JJ, Harbury PB. Automated design of specificity in molecular recognition. Nat Struct Biol. 2003;10:45–52. doi: 10.1038/nsb877. [DOI] [PubMed] [Google Scholar]
- 9.Kortemme T, Joachimiak LA, Bullock AN, Schuler AD, Stoddard BL, Baker D. Computational redesign of protein-protein interaction specificity. Nat Struct Mol Biol. 2004;11:371–379. doi: 10.1038/nsmb749. [DOI] [PubMed] [Google Scholar]
- 10.Bolon DN, Grant RA, Baker TA, Sauer RT. Specificity versus stability in computational protein design. Proc Natl Acad Sci U S A. 2005;102:12724–12729. doi: 10.1073/pnas.0506124102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo F, Cech TR. Evolution of Tetrahymena ribozyme mutants with increased structural stability. Nat Struct Biol. 2002;9:855–861. doi: 10.1038/nsb850. [DOI] [PubMed] [Google Scholar]
- 12.Guo F, Gooding AR, Cech TR. Comparison of crystal structure interactions and thermodynamics for stabilizing mutations in the Tetrahymena ribozyme. RNA. 2006;12:387–395. doi: 10.1261/rna.2198206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang XW, Srividya N, Golden BL, Sosnick TR, Pan T. Stepwise conversion of a mesophilic to a thermophilic ribozyme. J Mol Biol. 2003;330:177–183. doi: 10.1016/s0022-2836(03)00582-5. [DOI] [PubMed] [Google Scholar]
- 14.Walstrum SA, Uhlenbeck OC. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990;29:10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- 15.Pan J, Woodson SA. Folding intermediates of a self-splicing RNA: mispairing of the catalytic core. J Mol Biol. 1998;280:597–609. doi: 10.1006/jmbi.1998.1901. [DOI] [PubMed] [Google Scholar]
- 16.Russell R, Herschlag D. New pathways in folding of the Tetrahymena group I RNA enzyme. J Mol Biol. 1999;291:1155–1167. doi: 10.1006/jmbi.1999.3026. [DOI] [PubMed] [Google Scholar]
- 17.Russell R, Herschlag D. Probing the folding landscape of the Tetrahymena ribozyme: commitment to form the native conformation is late in the folding pathway. J Mol Biol. 2001;308:839–851. doi: 10.1006/jmbi.2001.4751. [DOI] [PubMed] [Google Scholar]
- 18.Treiber DK, Williamson JR. Concerted kinetic folding of a multidomain ribozyme with a disrupted loop-receptor interaction. J Mol Biol. 2001;305:11–21. doi: 10.1006/jmbi.2000.4253. [DOI] [PubMed] [Google Scholar]
- 19.Russell R, Das R, Suh H, Travers KJ, Laederach A, Engelhardt MA, Herschlag D. The paradoxical behavior of a highly structured misfolded intermediate in RNA folding. J Mol Biol. 2006;363:531–544. doi: 10.1016/j.jmb.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 20.Russell R. RNA misfolding and the action of chaperones. Front Biosci. 2008;13:1–20. doi: 10.2741/2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson TH, Tijerina P, Chadee AB, Herschlag D, Russell R. Structural specificity conferred by a group I RNA peripheral element. Proc Natl Acad Sci U S A. 2005;102:10176–10181. doi: 10.1073/pnas.0501498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Russell R, Herschlag D. Specificity from steric restrictions in the guanosine binding pocket of a group I ribozyme. RNA. 1999;5:158–166. doi: 10.1017/s1355838299981839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaug AJ, Grosshans CA, Cech TR. Sequence-specific endoribonuclease activity of the Tetrahymena ribozyme: enhanced cleavage of certain oligonucleotide substrates that form mismatched ribozyme-substrate complexes. Biochemistry. 1988;27:8924–8931. doi: 10.1021/bi00425a008. [DOI] [PubMed] [Google Scholar]
- 24.Huang Z, Szostak JW. A simple method for 3′-labeling of RNA. Nucleic Acids Res. 1996;24:4360–4361. doi: 10.1093/nar/24.21.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wan Y, Mitchell D, 3rd, Russell R. Catalytic activity as a probe of native RNA folding. Methods Enzymol. 2009;468:195–218. doi: 10.1016/S0076-6879(09)68010-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Russell R, Tijerina P, Chadee AB, Bhaskaran H. Deletion of the P5abc peripheral element accelerates early and late folding steps of the Tetrahymena group I ribozyme. Biochemistry. 2007;46:4951–4961. doi: 10.1021/bi0620149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Doherty EA, Doudna JA. The P4-P6 domain directs higher order folding of the Tetrahymena ribozyme core. Biochemistry. 1997;36:3159–3169. doi: 10.1021/bi962428+. [DOI] [PubMed] [Google Scholar]
- 28.Engelhardt MA, Doherty EA, Knitt DS, Doudna JA, Herschlag D. The P5abc peripheral element facilitates preorganization of the tetrahymena group I ribozyme for catalysis. Biochemistry. 2000;39:2639–2651. doi: 10.1021/bi992313g. [DOI] [PubMed] [Google Scholar]
- 29.van der Horst G, Christian A, Inoue T. Reconstitution of a group I intron self-splicing reaction with an activator RNA. Proc Natl Acad Sci U S A. 1991;88:184–188. doi: 10.1073/pnas.88.1.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doherty EA, Herschlag D, Doudna JA. Assembly of an exceptionally stable RNA tertiary interface in a group I ribozyme. Biochemistry. 1999;38:2982–2990. doi: 10.1021/bi982113p. [DOI] [PubMed] [Google Scholar]
- 31.Russell R, Zhuang X, Babcock HP, Millett IS, Doniach S, Chu S, Herschlag D. Exploring the folding landscape of a structured RNA. Proc Natl Acad Sci U S A. 2002;99:155–160. doi: 10.1073/pnas.221593598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wan Y, Suh H, Russell R, Herschlag D. Multiple unfolding events during native folding of the Tetrahymena group I ribozyme. J Mol Biol. 2010;400:1067–1077. doi: 10.1016/j.jmb.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sattin BD, Zhao W, Travers K, Chu S, Herschlag D. Direct measurement of tertiary contact cooperativity in RNA folding. J Am Chem Soc. 2008;130:6085–6087. doi: 10.1021/ja800919q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zarrinkar PP, Williamson JR. Kinetic intermediates in RNA folding. Science. 1994;265:918–924. doi: 10.1126/science.8052848. [DOI] [PubMed] [Google Scholar]
- 35.Herschlag D, Cech TR. Catalysis of RNA cleavage by the Tetrahymena thermophila ribozyme. 1. Kinetic description of the reaction of an RNA substrate complementary to the active site. Biochemistry. 1990;29:10159–10171. doi: 10.1021/bi00496a003. [DOI] [PubMed] [Google Scholar]
- 36.Golden BL, Gooding AR, Podell ER, Cech TR. A preorganized active site in the crystal structure of the Tetrahymena ribozyme. Science. 1998;282:259–264. doi: 10.1126/science.282.5387.259. [DOI] [PubMed] [Google Scholar]
- 37.Guo F, Gooding AR, Cech TR. Structure of the Tetrahymena ribozyme: base triple sandwich and metal ion at the active site. Mol Cell. 2004;16:351–362. doi: 10.1016/j.molcel.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 38.Adams PL, Stahley MR, Gill ML, Kosek AB, Wang J, Strobel SA. Crystal structure of a group I intron splicing intermediate. RNA. 2004;10:1867–1887. doi: 10.1261/rna.7140504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adams PL, Stahley MR, Kosek AB, Wang J, Strobel SA. Crystal structure of a self-splicing group I intron with both exons. Nature. 2004;430:45–50. doi: 10.1038/nature02642. [DOI] [PubMed] [Google Scholar]
- 40.Silverman SK, Cech TR. Energetics and cooperativity of tertiary hydrogen bonds in RNA structure. Biochemistry. 1999;38:8691–8702. doi: 10.1021/bi9906118. [DOI] [PubMed] [Google Scholar]
- 41.Blose JM, Silverman SK, Bevilacqua PC. A simple molecular model for thermophilic adaptation of functional nucleic acids. Biochemistry. 2007;46:4232–4240. doi: 10.1021/bi0620003. [DOI] [PubMed] [Google Scholar]
- 42.Bhaskaran H, Russell R. Kinetic redistribution of native and misfolded RNAs by a DEAD-box chaperone. Nature. 2007;449:1014–1018. doi: 10.1038/nature06235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lehnert V, Jaeger L, Michel F, Westhof E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.