Abstract
We report a method for high-throughput, cost-efficient empirical discovery of optimal proteotypic peptides and fragment ions for targeted proteomics applications using in vitro-synthesized proteins. We demonstrate the approach using human transcription factors – which are typically difficult, low-abundance – targets with an overall success rate of 98%. We show further that targeted proteomic assays developed using our approach facilitate robust in vivo quantification of human transcription factors.
Targeted proteomics is a powerful approach that enables quantitative analysis of tryptic peptides from complex biological samples with high sensitivity and specificity1,2. However, a major bottleneck limiting wider application of targeted proteomics has been the identification of optimal proteotypic peptides that are readily detectable by the mass spectrometer, as well as the characteristic fragmentation patterns of these peptides.
Because of differences in physiochemical properties, different peptides from the same protein can produce drastically different signal intensities when measured with a mass spectrometer1. Peptides are referred to as ‘proteotypic’ if they (i) are unique to a given protein, (ii) have good response characteristics in the mass spectrometer, and (iii) have a fragmentation pattern with salient features to accurately detect and quantify. Traditional strategies for identifying proteotypic peptides and their fragmentation patterns have relied on the combination of experimental data with bioinformatic analyses. A common approach has been to use peptides catalogued in the course of ‘shotgun’ proteomic experiments conducted by data-dependent acquisition3,4. This approach assumes that the peptides most frequently identified in shotgun experiments will produce the best response in a targeted proteomics setting. This assumption also underlies the application of machine learning methods, which aim to predict proteotypic peptides (but not their fragmentation spectra) de novo5,6. Complicating these efforts, a large subset of the human proteome is absent from fragmentation spectra databases, and this deficit is particularly acute for low abundance proteins such as transcription factors and kinases. To generate such peptide fragmentation data, large-scale efforts aim to synthesize predicted proteotypic peptides and empirically determine their fragmentation patterns7. However, which, if any, of these approaches is best suited for sensitive targeted proteomic analyses is unknown.
Here we report an empirically-driven approach for generating both optimal proteotypic peptides and their fragmentation patterns in a scalable, economical, and generalizable fashion. Rather than relying on sparsely populated spectral databases3,4, prediction algorithms5,6, costly peptide synthesis7 or the costly purchase of full-length proteins8, we leveraged the rich collection of tagged cDNA clones that are currently available for most human and model organism proteins9,10 to generate in vitro-synthesized full-length protein samples, followed by tryptic digestion and mass spectrometry analysis using selected reaction monitoring (SRM). Because all monitored tryptic peptides for each protein originate from the same full-length protein molecules, we are able to compare the relative intensities of different peptides to identify those that provide the most sensitive proxy for the target protein. In addition to the relative peptide response, we are able to identify in parallel the fragmentation patterns of these peptides in a triple-quadruple mass spectrometer using SRM (Fig. 1).
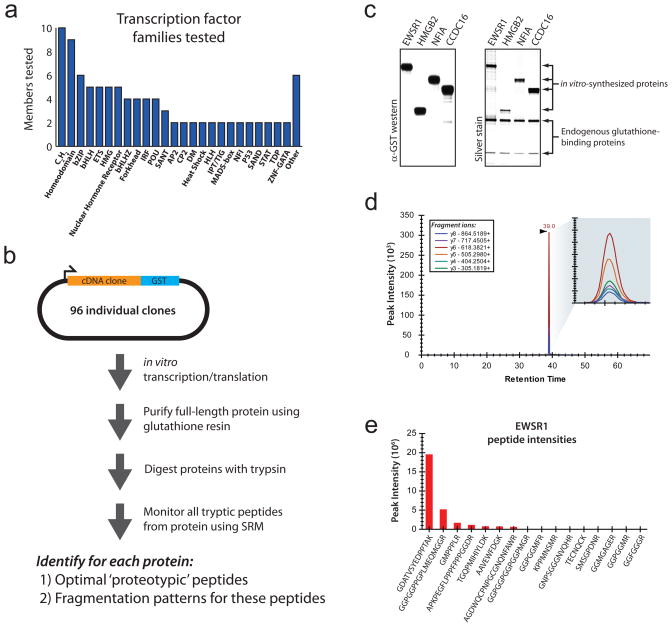
Figure 1. Development of targeted proteomics assays using enriched in vitro synthesized full length proteins.
(a) Proteins for which targeted assays were built. (b) Schematic of the synthesis, enrichment, digestion and analysis of proteins to identify proteotypic peptides and their fragmentation patterns. (c) Protein samples were highly-enriched and full-length, as detected by silver-staining and immunodetection with an anti-schistosomal GST antibody. (d) SRM chromatographic traces from the NFIA peptide EDFVLTVTGK were readily detected over background. (e) Proteotypic peptides for EWSR1 were identified by comparing the signal intensity of all of the tryptic peptides monitored.
To demonstrate our approach, we studied transcription factors, a diverse class of low-abundance proteins with a paucity of spectral data in public databases (Supplementary Fig. 1). We selected 96 human transcription factor proteins spanning all major structural families11 (Fig. 1a). For each of these proteins, we obtained full-length cDNA clones contained within an in vitro transcription/translation compatible vector with an in-frame c-terminal Schistosoma japonicum glutathione S-transferase (GST) tag12 (Supplementary Data 1). We then optimized in vitro protein production and purification in a 96-well plate format. We tested different protein production conditions, capture conditions, wash conditions and digestion conditions to develop a protocol that gave maximal protein yield at the highest possible purity (Methods). To verify that enriched full-length proteins were produced, we performed silver-staining and western blotting analyses for 46 of the 96 proteins (Fig. 1c and Supplementary Fig. 2). For nearly all of the tested proteins, the target protein and the two endogenous glutathione-binding proteins GSTM3 and EEF1G were the top three most intense bands on silver staining, indicating that SRM signal contamination should be minimal. In total, 96% (44/46) of the tested clones produced highly enriched proteins with the correct molecular weight. The remaining two samples produced multiple species of different molecular weights, likely originating from alternative methionine start codons.
For each protein, we selected peptides and fragment ions to measure using the software package Skyline13,, an open source application for building SRM methods and analyzing the resulting mass spectrometry data. We focused our analysis on predicted fully tryptic peptides with lengths between 7 and 23 amino acids. For each doubly charged monoisotopic precursor, we monitored singly charged monoisotopic y3 to yn-1 product ions using a TSQ-Vantage triple-quadrupole mass spectrometer. These measurements were imported into Skyline to identify the relative peptide responses and their fragmentation patterns (Fig. 1d,e). An annotated Skyline file containing the measured peptides and fragment ions for all 96 proteins can be found at http://proteome.gs.washington.edu/supplementary_data/IVT_SRM/.
To quantify the amount of each protein synthesized, heavy forms of the schistosomal GST peptides LLLEYLEEK and IEAIPQIDK were spiked into each in vitro synthesis reaction. The light-to-heavy ratio of these two peptides was measured and this ratio was calibrated to generate an absolute quantification curve containing the same amount of the heavy peptides but different known quantities of the light peptide (Supplementary Fig. 3 and Supplementary Note). Using this approach we determined that all of the 96 tested proteins produced at least 0.5 nM of product (Fig. 2a).
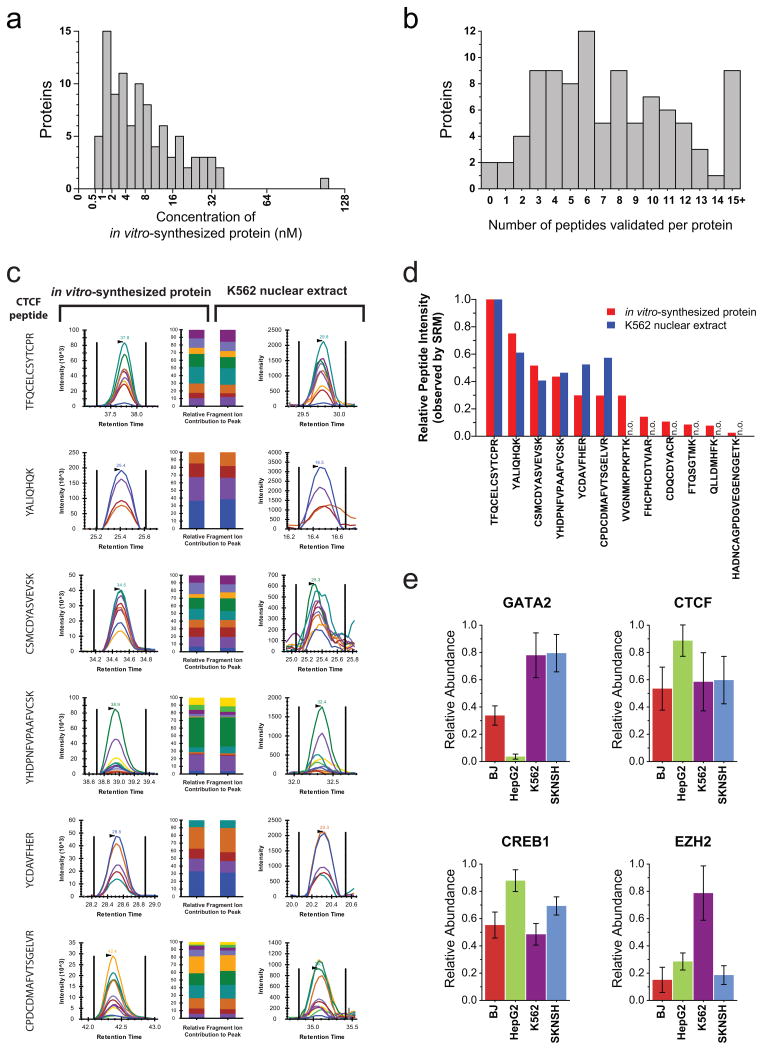
Figure 2. Targeted assays can be efficiently developed using in vitro synthesized proteins and applied to measure proteins in vivo.
(a)The absolute quantity of each in vitro-synthesized protein sample, as measured using a tryptic peptide contained within the c-terminal schistosomal GST tag. (b) The number of peptides per protein empirically assessed with salient features to accurately detect and quantify the target proteins (peptides with a quality score of either 1 or 2). (c) Proteotypic peptides identified using in vitro-synthesized CTCF were monitored in K562 nuclear extracts. The relative contribution of each fragment ion to each peptide peak is displayed as different colors. (d)For each proteotypic peptide from CTCF, the relative signal intensity observed using in vitro synthesized protein is displayed alongside the relative signal intensity observed using K562 nuclear extract Peptides not observed (n.o.) in K562 nuclear extracts are indicated. (e) The measured relative abundance of four transcription factors between the fibroblast (BJ), hepatic carcinoma (HepG2), erythroleukemia (K562) and neuroblastoma (SKNSH) human cell lines. Data points are mean ± s. d. (n = 6).
Chromatographic data from each peptide was manually analyzed to determine the quality of the peptide signal. Each peptide was given a quality score between 1 and 4, with 1 being the highest quality (Methods). Only peptides with a quality score of either 1 or 2 were considered for further analysis. On average we were able to identify eight peptides per protein with a quality score of 1 or 2. Additionally, all but two of the proteins assayed had at least one peptide with a quality score of 1 or 2 (Fig. 2b and Supplementary Data 1). Of note, although sufficient quantities of both CEBPG and HMGA1 protein were produced using our in vitro approach (Supplementary Fig. 2) and the proteins were sufficiently digested as indicated by the mass spectrometry responses of the GST peptides, none of the monitored tryptic peptides from these two proteins gave a good response in the mass spectrometer. This suggests that a small minority of transcription factor proteins may not be amenable to proteomic analysis using trypsin-based digestion.
To determine the quality of our fragmentation patterns, we compared our observed peptide fragmentation patterns with those contained in the National Institute of Standards and Technology (NIST) spectral database. Of the 760 peptides in our dataset with a quality score of either 1 or 2, only 18% (136) were represented in the NIST database (Methods). Of these, all had high spectral similarity scores, with 93% having dot-products greater than 0.85 (Supplementary Fig. 4). This finding mutually corroborates both our data and the NIST database and further highlights the scarcity of proteotypic peptides within large spectral databases.
We next determined the utility of predictor algorithms and shotgun analyses to identify optimal proteotypic peptides. A comparison of our empirical ranking of proteotypic peptides with peptide rank predictions from the ESPP redictor algorithm6 revealed spearman correlations ranging from −0.45 to 0.85 with an average correlation of 0.47 (Supplementary Data 2 and Supplementary Fig. 5). Similarly, roughly half of the optimal proteotypic peptides from our experiments were undetected by shotgun analyses of the identical samples (Supplementary Fig. 6). While these approaches are better than selecting proteotypic peptides at random, our results suggest that current predictor algorithms and spectral counting approaches provide imperfect ranking and identification of optimal proteotypic peptides – potentially limiting the utility of large-scale peptide synthesis efforts that rely on such approaches as a first round filter7.
Finally, we sought to confirm the utility of proteotypic peptides identified using our approach for in vivo analyses, and how the in vitro-derived intensity rankings compared with those from complex biological samples. To test this, we first monitored all 12 of the quality score 1 and 2 peptides from the genomic master regulatory transcription factor CTCF in trypsin-digested nuclear lysate from erythrolukemia cells (K562). Using the fragmentation patterns identified in vitro, we identified corresponding chromatographic peaks for six of these CTCF peptides in K562 nuclear extract (Fig. 2c). The relative intensity of these peptides in vitro and in vivo closely matched, confirming the relevance of the rank order of peptides identified empirically using in vitro-synthesized protein (Fig. 2d). Next, we selected top-ranking peptides from four transcription factors and used these to generate nuclear abundance measurements of these factors across four distinct cell types (Fig. 2e). The relative abundance measurements are consistent with previous reports on the tissue distribution of these transcription factors using RNA abundance14,15.
In summary, we demonstrate and validate a rapid and cost-efficient method for empirical identification of optimal proteotypic peptides and their fragmentation patterns using in vitro-synthesized proteins. Our method can be readily applied to generate assays to identify and quantify structurally diverse low-abundance proteins, such as human transcription factors, in unfractionated cellular extracts.
Supplementary Material
Supplementary Figure 1. Poor coverage of target transcription factor proteins in NIST database
Supplementary Figure 2. In vitro-synthesized proteins are enriched full-length proteins
Supplementary Figure 3. Calibration curve for the schistosomal GST peptide IEAIPQIDK
Supplementary Figure 4. Histogram of dot-products for quality score 1 and 2 peptides
Supplementary Figure 5. Spearman correlation of our empirical peptide ranking with ESPP redactor rankings
Supplementary Figure 6. Peptide MS/MS spectrum counts are a poor predictor of targeted peptide signal intensity using selected reaction monitoring-mass spectrometry
Supplementary Figure 7. Reagent cost for generating in vitro-synthesized proteins from plasmids
Supplementary Data 1. Rank order of proteotypic peptides and fragment ions for targeted proteins
Supplementary Data 2. Comparison of our empirical rankings with peptide rank predictions from the ESPP redictor algorithm
Supplementary Note. Skyline tutorial on protein absolute quantification using a heavy isotope internal standard and an external calibration curve
Acknowledgments
We thank Priska von Haller (University of Washington), Michael Bereman (University of Washington), Eric Hommema (Thermo Scientific) and John Rogers (Thermo Scientific) for their discussions and technical assistance. This work was supported in part by the University of Washington’s Proteomics Resource (UWPR95794), the Thermo Scientific Pierce Human In Vitro Translation Research Grant, and NIH grants P41RR011823 (M.J.M.) and U54HG004592 (J.A.S.).
Footnotes
AUTHORS CONTRIBUTIONS
A.B.S., J.A.S. and M.J.M. conceived and designed the experiments and wrote the paper. A.B.S. and K.L. performed the wet laboratory experiments. A.B.S. and B.M. analyzed the data. All authors read and approved the final manuscript.
COMPETING FINANCIAL INTERESTS
The authors acknowledge financial support from Thermo Fisher Scientific.
References
- 1.Lange V, Picotti P, Domon B, Aebersold R. Mol Syst Biol. 2008;4:222. doi: 10.1038/msb.2008.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carr SA, Anderson L. Clin Chem. 2008;54:1749–1752. doi: 10.1373/clinchem.2008.114686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Picotti P, et al. Nat Methods. 2008;5:913–914. doi: 10.1038/nmeth1108-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Prakash A, et al. J Proteome Res. 2009;8:2733–2739. doi: 10.1021/pr801028b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallick P, et al. Nat Biotechnol. 2006;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 6.Fusaro VA, Mani DR, Mesirov JP, Carr SA. Nat Biotechnol. 2009;27:190–198. doi: 10.1038/nbt.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picotti P, et al. Nat Methods. 2010;7:43–46. doi: 10.1038/nmeth.1408. [DOI] [PubMed] [Google Scholar]
- 8.Keshishian H, Addona T, Burgess M, Kuhn E, Carr SA. Mol Cell Proteomics. 2007;6:2212–2229. doi: 10.1074/mcp.M700354-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goshima N, et al. Nat Methods. 2008;5:1011–1017. doi: 10.1038/nmeth.1273. [DOI] [PubMed] [Google Scholar]
- 10.Ramachandran N, et al. Nat Methods. 2008;5:535–538. doi: 10.1038/nmeth.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaquerizas JM, Kummerfeld SK, Teichmann SA, Luscombe NM. Nat Rev Genet. 2009;10:252–263. doi: 10.1038/nrg2538. [DOI] [PubMed] [Google Scholar]
- 12.Rolfs A, et al. Proc Natl Acad Sci USA. 2008;105:4364–4369. doi: 10.1073/pnas.0712049105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacLean B, et al. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee ME, Temizer DH, Clifford JA, Quertermous T. J Biol Chem. 1991;266:16188–16192. [PubMed] [Google Scholar]
- 15.Klenova EM, et al. Mol Cell Biol. 1993;13:7612–7624. doi: 10.1128/mcb.13.12.7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dorschner MO, et al. Nat Methods. 2004;1:219–225. doi: 10.1038/nmeth721. [DOI] [PubMed] [Google Scholar]
- 17.Maclean B, et al. Anal Chem. 2010;82:10116–10124. doi: 10.1021/ac102179j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eng JK, McCormack AL, Yates JR. J Am Soc Mass Spectr. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 19.Kall L, Canterbury JD, Weston J, Noble WS, MacCoss MJ. Nat Methods. 2007;4:923–925. doi: 10.1038/nmeth1113. [DOI] [PubMed] [Google Scholar]
- 20.Stein SE, Rudnick PA, editors. Human Peptide Mass Spectral Reference DataH. sapiens, ion trap. National Institute of Standards and Technology; Gaithersburg, MD: Jan 14, 2010. NIST Peptide Tandem Mass Spectral Libraries; p. 20899. Downloaded from http://peptide.nist.gov on September 12, 2011. [Google Scholar]
- 21.Frewen BE, Merrihew GE, Wu CC, Noble WS, MacCoss MJ. Anal Chem. 2006;78:5678–5684. doi: 10.1021/ac060279n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Poor coverage of target transcription factor proteins in NIST database
Supplementary Figure 2. In vitro-synthesized proteins are enriched full-length proteins
Supplementary Figure 3. Calibration curve for the schistosomal GST peptide IEAIPQIDK
Supplementary Figure 4. Histogram of dot-products for quality score 1 and 2 peptides
Supplementary Figure 5. Spearman correlation of our empirical peptide ranking with ESPP redactor rankings
Supplementary Figure 6. Peptide MS/MS spectrum counts are a poor predictor of targeted peptide signal intensity using selected reaction monitoring-mass spectrometry
Supplementary Figure 7. Reagent cost for generating in vitro-synthesized proteins from plasmids
Supplementary Data 1. Rank order of proteotypic peptides and fragment ions for targeted proteins
Supplementary Data 2. Comparison of our empirical rankings with peptide rank predictions from the ESPP redictor algorithm
Supplementary Note. Skyline tutorial on protein absolute quantification using a heavy isotope internal standard and an external calibration curve