Summary
Protein arginylation and arginine methylation are two posttranslational modifications of emerging importance that involve Arg residues and their modifications. To test a hypothesis that posttranslationally added arginines can be methylated, we used high precision mass spectrometry and metabolic labeling to find whether posttranslationally added arginines can serve as methyation sites. We identified a number of proteins in vivo, on which posttranslationally added Arg have undergone mono- and dimethylation. This double modification predominantly affects the chromatin-containing nuclear fraction and likely plays an important regulatory role in chromatin-associated proteins. Moreover, inhibition of arginylation and Arg methylation results in a significant reduction of the nucleus size in cultured cells, suggesting changes in chromatin compaction and nuclear architecture. Our findings suggest a functional link between protein regulation by arginylation and methylation that affects nuclear structure in vivo.
Introduction
Posttranslational modifications regulate the majority of protein functions and physiological pathways and add an extra level of complexity to the structural and functional diversity of the proteome. Many modifications that have been discovered decades ago are now coming into the spotlight, as new studies show evidence of their major physiological role. One such modification is arginine methylation, recently demonstrated to regulate a variety of processes, including protein-protein interactions, RNA binding, transcription, signal transduction, and DNA repair (see (Bedford and Richard, 2005) for review). Another modification, whose role has emerged in recent studies, is arginylation – posttranslational addition of Arg onto proteins that has been shown to affect many proteins in vivo (Wong et al., 2007) and modulate critical physiological processes such as cell motility and cardiovascular development (Karakozova et al., 2006; Kwon et al., 2002).
While no link between the two modifications has been previously suggested, a natural question arises of whether posttranslationally added Arg can serve as a site for the action of protein Arg-methyltransferases. Here we addressed this question by examining posttranslational arginylation and methylation in subcellular fractions and found that posttranslationally arginylated proteins can be methylated on added arginines and that this double modification occurs with higher frequency in the nuclear compared to the cytosolic proteins, due likely to the higher nuclear activity of arginylation and methylation enzymes. The identity of these proteins and the location of arginylation and arginylation/methylation sites suggest that these modifications in the nucleus are involved in the global regulation of chromatin structure, alternative splicing, and gene expression. Further, our studies in cultured cells showed that inhibition of arginylation and/or Arg-methylation results in a significant reduction of the nuclear size, suggesting that these two modifications are essential for maintaining normal chromatin compaction and nuclear architecture. Our results are the first demonstration of posttranslational modifications doubled up on the same sites to regulate an important subset of proteins in vivo.
Results
Posttranslationally added arginines can be methylated in vivo
To address the possible relationship between posttranslational arginylation and Arg-methylation, we examined the samples isolated from subcellular fractions of mouse tissues and cultured cells by high precision mass spectrometry to find out whether any of the posttranslationally added Arg are found in the methylated state (i.e., contain a mass addition of Arg (+156.1011) with an extra mass of either one (+14.0157) or two (+28.0314) methyl groups).
Analysis of the cytosolic fractions derived from cultured embryonic fibroblasts and mouse heart tissue revealed two peptides in different samples, in which an internal site within the molecule contained dimethylated Arg (mass addition +184.1325). One peptide belongs to alpha cardiac actin (identified in a 2D gel spot corresponding to alpha actin from the mouse heart Fig. 1) and one is common between heat shock proteins 90 and 84b (identified by six different scans in a protein sample isolated by immunoaffinity chromatography with N-terminal Arg-specific antibody (Wong et al., 2007) from mouse heart tissue) (Fig.1, S1 and Supplemental Table S1).
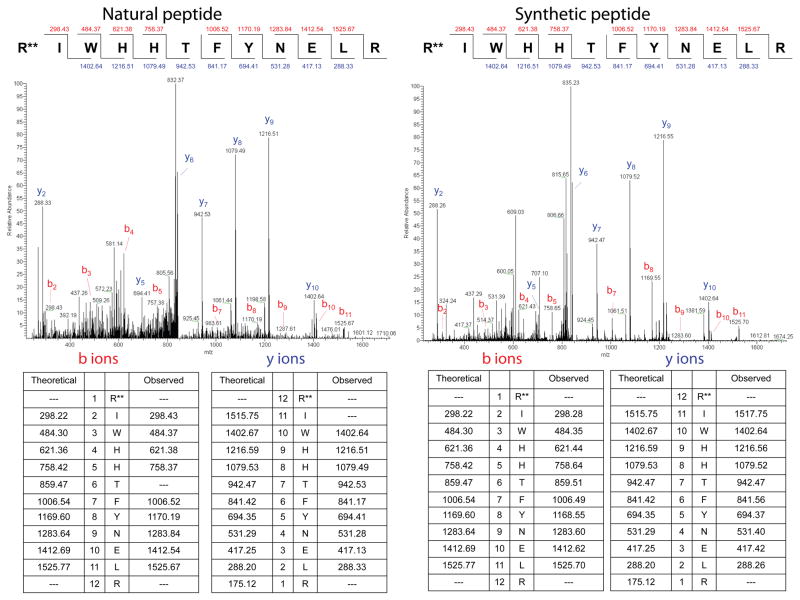
Figure 1. MS/MS spectra of the experimentally identified dimethyl-arginylated actin peptide (NP-033736.1) (left, natural peptide) and synthetic standard peptide containing N-terminal dimethylated Arg (right, dimethyl groups in both are denoted with double stars).
Peptide sequences and b and y ion masses are shown on the top right for each spectrum and in the tables underneath. See Supplemental Table S1 for the search parameters. See also Figure S1.
To confirm that the identified modification indeed represents Arg methylation and does not arise from an ambiguity that mimics a mass addition of +184.1325, we synthesized both peptides with a dimethyl-Arg residue in the N-terminal position, and compared the tandem mass spectra of these peptides with the tandem mass spectra of the arginylated/methylatied peptides identified in the real sample. We found that the spectra between the standard peptides and the in vivo samples matched, both in the precursor masses and in the fragmentation patterns (see Fig.1 and S1 for mass spectra and Supplemental Table S1 for the precursor and ion fragment masses), confirming that the peptides identified by our analysis were truly arginylated/methylated.
Posttranslational arginylation and methylation regulates nuclear and chromatin-bound proteins
Since Arg-methylation is known to affect a large number of chromatin-associated proteins, it appears likely that if posttranslational arginylation and methylation indeed have a functional relationship, arginylation should also prominently affect chromatin-associated proteins, and such proteins could then be methylated on the added Arg to a higher extent than proteins in the cytosol. To test this hypothesis, we performed metabolic labeling assays to compare the overall patterns of posttranslational Arg and methyl incorporation in different subcellular fractions (Fig. 2A), by incubating cycloheximide and chloramphenicol-treated cells (to inhibit cotranslational incorporation of amino acids) with radioactively labeled 3H-Arg or 3H-Met (the source of methyl groups). As a negative control in these experiments we used Arginyl-tRNA-protein transferase 1 knockout (Ate1 KO) fibroblasts, in which arginylation does not occur and protein methylation happens without the contribution of posttranslationally added Arg.
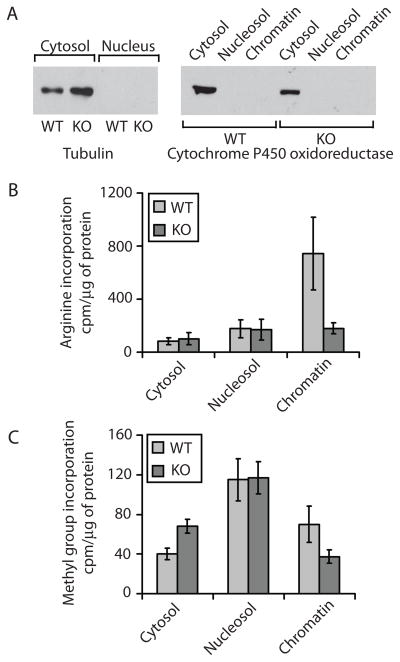
Figure 2. Arg and methyl group incorporation into different subcellular fractions.
A. Western blot of the cytosolic and nuclear fractions with the cytoplasmic marker tubulin (left) and the ER marker cytochrome P450 oxidoreductase indicate that purified nuclear fractions are devoid of cytosolic and ER contaminants. All the fractions for Western blotting were normalized by total protein amount. B. In vivo incorporation of 3H-Arg into cycloheximide and chloramphenicol -treated wild type (WT) and Ate1 knockout (KO) cells presented as c.p.m. per μg of protein. Error bars +/− SEM, n = 4. Chromatin fraction p value is 0.0874. C. In vivo incorporation of 3H-Met (the source of the methyl groups) into cycloheximide/chloramphenicol-treated WT and Ate1 KO cells presented as c.p.m. per μg of protein. Error bars +/− SEM, n = 3. Cytosol and chromatin fraction p values are 0.016 and 0.1, respectively. See also Figures S2 and S3.
During both treatments, residual incorporation of radioactively labeled amino acids occurred due to incompletely inhibited protein synthesis, producing a background level of radioactive label incorporation (as seen from the Ate1 KObars in Fig. 2B). Surprisingly, Arg incorporation into the cytosolic and nucleosolic protein fractions in wild type (WT) cells did not exceed this background and did not significantly differ from that in the Ate1 knockout, suggesting that the overall stable level of posttranslational arginylation under these conditions is below the detection level for this assay. However, the amount of Arg incorporated into the chromatin-associated fraction in WT cells significantly exceeded that in the arginylation knockout (the rightmost set of bars in Fig. 2B), indicating that posttranslational arginylation level is very significant in the chromatin fraction. Control experiments using cells without cycloheximide treatment (that incorporate Arg both during translation and posttranslationally) showed that this incorporation is also comparatively higher in the nucleosolic and chromatin fractions of WT cells, compared to the cytosolic protein fraction and to the Ate1 KO cells (Fig. S3A).
Comparison of methylation levels in different subcellular fractions from wild type and arginylation knockout cells revealed the same trend (Fig. 2C). While the nucleosol fraction from WT and Ate1 KO cells had similar amount of methyl groups incorporation after 4 hr of metabolic labeling, the chromatin fraction in Ate1 KO cells had significantly less methylation than wild type. At the same time methylation levels in the cytosol in Ate1 KO cells were higher than in WT cells, suggesting that absence of arginylation may lead to specific changes in protein Arg-methylation in the cells. To test this and to evaluate the total methyl-Arg content in the protein-incorporated and free amino acid state, we performed amino acid analysis to detect methylated Arg derivatives in hydrolyzed total protein samples and in protein-free low molecular weight pools obtained from total cell lysates containing both the nuclear and the cytosolic fraction. We found that total methylation, as well as the amount of methylated Arg both in the total protein and in the free amino acid pool was higher in Ate1 KO than in wild type (Fig. S2), suggesting that in these cells the overall Arg-methylation was increased via arginylation-independent mechanisms. Given these results, the increased arginylation and methylation in the chromatin fraction of wild type appears even more striking and strongly suggests that arginylation-dependent methylation contributes significantly to the overall methyl incorporation specifically in the chromatin. Control experiments suggested that this methyl-arginylation was likely not mediated by arginyltransferase itself, since purified ATE1 mixed in vitro with methylated Arg, tRNA, and Arg-tRNA-synthethase was unable to transfer methylated Arg onto the test substrate. Control experiments also suggested that inhibition of Arg-methylation using the commercially available protein arginine methyltransferase inhibitor AMI-1, resulted in a reduction of methyl incorporation into the chromatin proteins in WT cells but not in the Ate1 KO cells, suggesting that only the Arg-methyltransferases specifically affected by this inhibitor are likely responsible for arginylation-dependent Arg-methylation, while the overall increase of Arg methylation in Ate1 KO is likely mediated by other type of Arg methyltransferases. In agreement with this, AMI 1 treatment had negligible and somewhat opposite effect on the cytosolic and nucleosolic Arg methylation in WT and Ate1 KO cells (Fig. S3B). Overall, these data suggest that posttranslational arginylation/methylation requires both the activity of arginyltransferase and the activity of specific type(s) of Arg-methyltransferase(s).
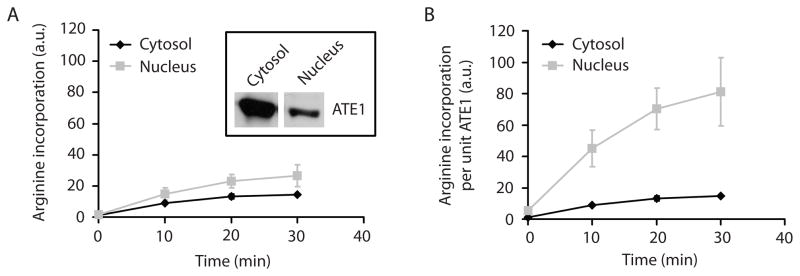
To test whether the higher incorporation of Arg into the nuclear proteins is related to the higher levels and/or activity of nuclear ATE1, we first detected the levels of ATE1 in the cytosol and the nucleus by immunoblotting and found that while ATE1 is present in both subcellular fractions, its level in the nucleus is significantly lower than that in the cytosol (inset in Fig. 3A). We then used the cytosolic and the nuclear fractions in an in vitro arginylation assay and found that in the nuclear fraction, arginylation activity was higher than in the cytosol (Fig. 3A). When normalized to the comparative ATE1 levels in the two fractions, this activity was several times higher in the nucleus compared to the cytosol (Fig. 3B). Control experiments showed that RNase A treatment abolished this Arg incorporation activity both in the nucleus and in the cytosol, confirming that the activity was RNA dependent (Fig. S4). Therefore, increased arginylation of nuclear proteins is associated with a significantly higher arginyltransferase activity in the nucleus compared to the cytosol.
Figure 3. ATE1 is less abundant but more active in the nucleus.
A. Inset: Western blot showing ATE1 levels in the cytosol and nucleus in the samples loaded at the equal protein level. Chart: 3H-Arg incorporation by endogenous ATE1 into cellular proteins in cytosolic and nuclear extracts at different time points. Error bars +/− SEM, n = 2. B. The same curves as in A normalized to the ATE1 levels detected by the Western blots shown in A, inset. ATE1 activity per molecule is significantly higher in the nucleus than in the cytosol. See also Figure S4.
Identification of arginylated and arginylated/methylated proteins in the nucleus
To identify proteins that are arginylated and arginylated/methylated in the nuclear subfractions, we analyzed total proteins found in the fractionated nuclear samples from mouse embryonic fibroblasts by mass spectrometry. This analysis revealed a large number of proteins that were arginylated in the nuclear subfractions. A significant subset of these proteins was found to contain posttranslationally added methyl-Arg, suggesting that these proteins have undergone methylation upon arginylation (see Tables 1 and 2 for the lists of proteins, and Datasets S1 and S2 for the mass spectra of arginylated/methylated and arginylated peptides, respectively). Some of these proteins contained dimethylated Arg, similarly to those previously identified in the cytosol. Others contained monomethylated Arg, suggesting that methylation of posttranslationally added Arg occurs in steps and is likely mediated by the same enzymes as regular Arg-methylation. Remarkably, these proteins were identified without any special enrichment in the total nuclear pool, suggesting that the extent of arginylation and arginylation/methylation on these proteins is relatively high.
Table 1. Posttranslationally arginylated/methylated proteins.
(see Supplemental Table 2 for the search parameters and peptide sequences, and Fig. 1, Fig. S1, and Dataset S1 for the corresponding mass spectra)
| Accession No. | Name | Modified Residue | Modification | Cellular Fraction | Function |
|---|---|---|---|---|---|
| AAA37866 | HSP90-α/HSP84b | S587 | DMA | Cytosol | Heat shock |
| NP_033736.1 | Actin, alpha skeletal muscle | I87 | DMA | Cytosol | Actin cytoskeleton |
| NP_058557.2 | PDZ and LIM domain protein 1 | I30 | MMA | Nucleosol | Actin cytoskeleton |
| NP_062752.2 | Zinc finger and BTB domain containing 20 isoform L | I69 | MMA | Nucleosol | Transcriptional repressor |
| NP_001104761.1 | Cytoplasmic activation/proliferation-associated protein 1 isoform c | L170 | DMA | Nucleosol | Cell proliferation, translation regulation, RNA binding |
| NP_598432 | Splicing factor U2AF 65 kDa subunit | G248 | MMA | Nucleosol | Pre-mRNA splicing, 3′-end mRNA processing |
| NP_035561.1 | 116 kDa U5 sn RNP component isoform a | E689 | DMA | Nucleosol | Pre-mRNA splicing |
| NP_598841.1 | Filamin B | S1911 | MMA | Chromatin | Actin cytoskeleton |
| CAM16973.1 | Spectrin beta 2 | E741 | MMA | Chromatin | Actin cytoskeleton |
| NP_835509.2 | Histone H2B F | K47 | MMA | Chromatin | Chromatin structure and organization |
| NP_033736.1 | Actin, alpha skeletal muscle | N299 | MMA | Chromatin | Actin cytoskeleton |
| NP_032017.2 | Fibrillarin | Q160 | MMA | Chromatin | snoRNP particle component, pre-rRNA processing, pre-rRNA methylation, ribosome assembly |
| NP_035897.2 | Zinc finger RNA binding protein | A444 | DMA | Chromatin | RNA transport and localization |
Table 2. Proteins arginylated in the nucleus.
(see Supplemental Table 2 for the search parameters and peptide sequences and Dataset S2 for the corresponding mass spectra)
| Accession No. | Name | Modified Residue | Cellular Fraction | Function |
|---|---|---|---|---|
| NP_034928.1 | Macrophage migration inhibitory factor | A35 | Nucleosol | Pro-inflammatory cytokine |
| CAM24508.1 | Far upstream element binding protein 3 | I111 | Nucleosol | ssDNA and RNA binding, transcriptional and posttranscriptional regulation |
| P11499.2 | HSP 90β | L439 | Nucleosol | Heat shock |
| NP_034368.2 | Bromodomain- containing protein 2 isoform a | L10 | Nucleosol | Transcriptional regulation, chromatin remodeling, signal transduction, nuclear kinase |
| EDL34349.1 | Similar to glyceraldehyde-3- phoSphate dehydrogenase | V121 | Nucleosol/Chromatin | Glycolysis, transcription activation, initiation of apoptosis, and ER to golgi vesicle transport. |
| NP_035931.1 | craniofacial development protein 1 | K104 | Nucleosol | Transcriptional developmental signaling |
| CAI24328.1 | RAB34 | D75 | Nucleosol | Lysosomal positioning, protein transport and secretion |
| NP_034063.1 | Collagen alpha-1(VI) chain precursor | E576 | Nucleosol | Extracellular matrix |
| NP_084206.1 | Hypothetical protein LOC77574 | G633 | Nucleosol | Unknown |
| NP_663600.2 | Eukaryotic translation initiation factor 4B | G138 | Nucleosol | Translation |
| BAC36440.1 | Histone H2B homolog | A59 | Chromatin | Chromatin structure and organization |
| NP_783597.2 | Histone H2B type 2-B | Q48; A59 | Chromatin | Chromatin structure and organization |
| XP_981474.1 | Histone H4 | T136; V116 | Chromatin | Chromatin structure and organization |
| AAA37763.1 | Histone H2A.1 | S41 | Chromatin | Chromatin structure and organization |
| EDK99240.1 | Ribophorin I, isoform CRA_a | V468 | Chromatin | N-glycosylation, ER chaperone |
| NP_031419.1 | Actin, beta | D51 | Chromatin | Actin cytoskeleton, transcriptional regulation |
| XP_001476183.1 | Similar to ribosomal protein L23a | L140 | Chromatin | Ribosomal organization |
| NP_035831.2 | Vimentin | V224 | Chromatin | Intermediate filament, cytoskeleton |
| NP_080447.1 | Nuclear valosin- containing protein-like | T628 | Chromatin | Nuclear ATPase |
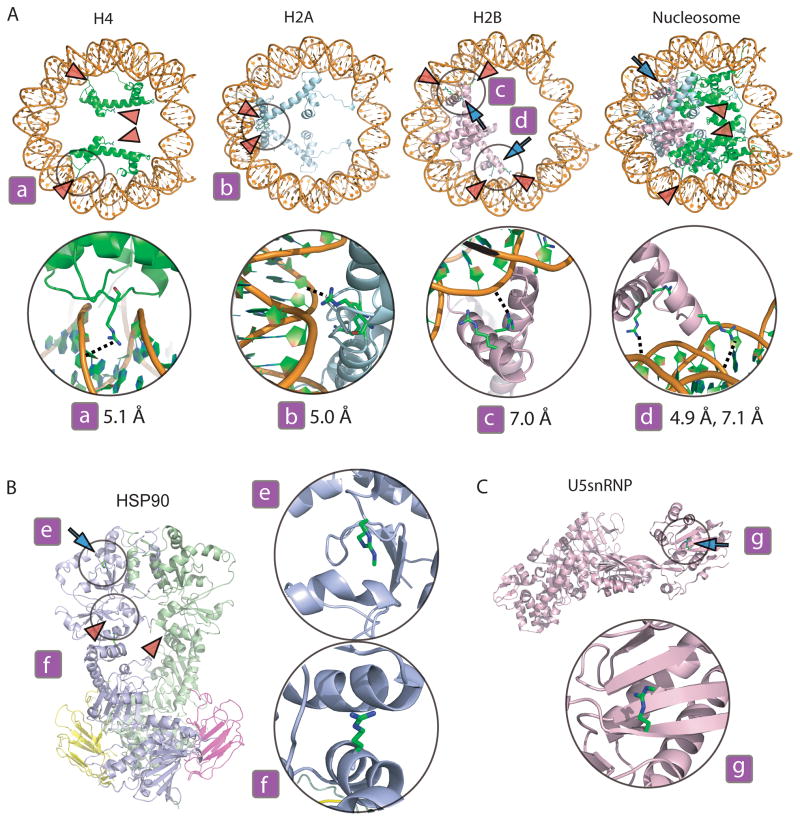
Overall, the arginylated and arginylated/methylated proteins identified by this analysis, fell into distinct functional groups. A prominent subset of these proteins included histones, which were arginylated and in some cases further methylated on the sites involved in DNA binding, ion interaction, and association with other histones in the nucleosome complex (Fig. 4A). On these proteins, arginylated sites were identified in multiple scans in different analyzed subsets, suggesting that the overall abundance of the arginylated form of these proteins was significant. Another subset of arginylated proteins have roles in gene expression and RNA splicing, including RNA-binding proteins and related factors. While, similar to the previous studies (Wong et al., 2007), no arginylation consensus was found at or around these sites, a Lys residue was prevalent in the (−1) position to the arginylated site. This finding suggests that in the nucleus, unlike in the cytosol, recognition of proteins by the arginylation machinery may be mediated by Lys-containing primary sequence motifs. Structural analysis using the 3D structures available in PDB database for these proteins or their close homologues revealed that in all cases arginylation sites were located on the protein surface (Fig. 4, Fig. S5) and in many cases fell onto functionally significant regulatory regions of these proteins. Structural modeling of the arginylated histone proteins and the nucleosome indicated that some of the added Arg and methyl-Arg have a distance of 5–7 Å from the nearest phosphate in the DNA backbone which, given the opposite charges in these groups, can potentially facilitate the interaction of the histones, with DNA (Fig 4A). These results confirm that arginylation and arginylation/methylation occurs to a significant extent in the nucleus compared to the cytosol, and that these modifications in the nucleus likely play an important regulatory role.
Figure 4. Structures of some of the arginylated and arginylated/methylated proteins identified in this study.
The position of the postranslationally added Arg or methyl-Arg residues in the represented structures are modeled by incorporating the Arg or methyl-Arg in the primary structure of the protein. Postranslationally added arginines are marked with green and the side chain amino groups are in blue. The methyl groups in arginine are marked by green spots attached to the blue amino group. Arginylation sites are shown with red arrowheads and arginylation/methylation sites – with blue arrows. A. Nucleosome (structures shown are selected from the complex between nucleosome core particle (H3, H4, H2a, H2b) and 146 Bp Long DNA fragment, PDB identifier: 1AOI). The position of individual histones (marked on top) and the entire nucleosome assembly (marked ‘Nucleosome in the top rightmost panel) are shown in the topmost panels. The modified sites are located on the DNA binding and histone interaction surfaces likely to affect nucleosome assembly. The distances between the nitrogen atoms of added Arg or methyl-Arg and the nearest oxygen atoms of phosphate backbone of the DNA molecule are represented in the inset images a, b, c, and d and marked by black dotted lines. B. Hsp90 dimer (structure shown is of S. cerevisiae Hsp90, PDB identifier: 2CG9). Modified sites are located on the protein surface and near subunit interaction sites. Inset images e and f show a detailed image of the modified regions. C. U5snRNP specific protein (structure shown is of a homologus S. cerevisiae protein in complex with rRNA obtained by selecting the relevant structures from the S. cerevisiae ribosomal 80s-Eef2-sordarin complex, PDB identifier: 1S1H). Inset image g shows a detailed image of the modified regions. See also Supplemental Figures S4 for additional structures. See also Figure S5, Supplemental Table 2 and Supplemental Datasets S1 and S2.
Arginylation/methylation regulates nuclear size in cultured cells
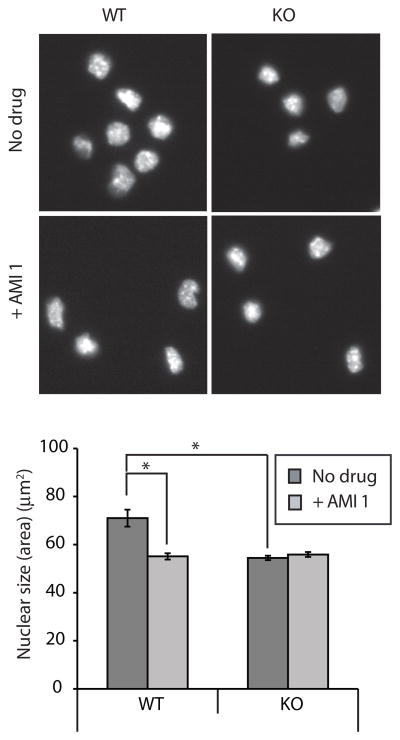
To assess the biological role of arginylation/methylation of chromatin proteins, we analyzed the identity of these proteins (Tables 1 and 2) and noticed that a large fraction of these proteins had prominent roles in DNA compaction and architecture. To test the hypothesis that arginylation/methylation regulates nuclear architecture, we compared the nucleus size in cultured wild type and Ate1 KO mouse embryonic fibroblasts and found that Ate1 KO nuclei were ~20% smaller compared to WT nuclei (Fig 5A, B). Strikingly, treatment of WT and Ate1 KO cells with AMI1 reduced the nuclear size of the WT MEFs to the same extent (by approximately 20%), without affecting the nuclear size in Ate1 KO cells (Fig 5A, B). Thus, inhibition of both arginylation and Arg-methylation lead to exactly the same effect on the nuclear size, clearly indicating that arginylation combined with methylation regulates nuclear size in vivo. As nuclear size can change due to change in chromatin compaction or other structural change in nucleus, exact mechanism of arginylation/methylation mediated regulation of nuclear size remains to be investigated.
Fig. 5. Arginylation methylation regulates nuclear size.
A. DAPI stained untreated or AMI 1 treated WT and Ate1 KO MEF nucleus. Bar 25 μm. B. Quantification of nuclear size observed in the experiment presented in panel A from following number of nucleus analyzed. WT, n= 41; Ate1 KO, n=96; WT+ AMI 1, n= 84; Ate1 KO+ AMI 1, n=150. Error bar +/− SEM. t-test p value <0.0001 for WT vs Ate1 KO and WT+ AMI 1.
Discussion
This work is the first demonstration that two posttranslational modifications can affect the same sites in a protein, and the first comprehensive analysis of arginylation in the nucleus that reveals the identity of a significant number of previously unknown protein arginylation targets and outlines a likely function for protein arginylation in chromatin structure and gene expression. Our result that ATE1 is less abundant and more active in the nucleus suggest that this enzyme undergoes an additional level of regulation that could increase or silence its activity dependent on its intracellular localization. Such regulation could be achieved by modulating the interaction of ATE1 with different binding partners in the cytoplasm and the nucleus that may form either activatory or inhibitory complexes with this enzyme and/or regulate its activity by changing its conformation or posttranslationally modified state. Different chemical environments in these two intracellular compartments may also play a role, by creating favorable or unfavorable ionic conditions for arginylation.
It has been previously shown that some ATE1 isoforms showed preferential nuclear localization under certain conditions, suggesting that this localization depends on the cell’s physiological state (Kwon et al., 1999; Rai and Kashina, 2005; Wang et al., 2011). It is possible that higher arginylation activity in nucleus could be necessary because of the existence of higher number and/or amount of natural substrates of ATE1 in nucleus. It was previously found that partially purified ATE1 preparation can arginylate nuclear proteins in vitro (Kaji, 1976). Our data demonstrates for the first time that ATE1 preferentially arginylates nuclear proteins in vivo and that the activity of this enzyme can be differentially regulated in different intracellular compartments.
Many of the proteins identified in the cytosolic and chromatin fractions have a demonstrated role in the corresponding compartments, such as HSP90 in the cytosol and DNA- and RNA-binding proteins involved in chromatin structure, gene expression, and RNA processing in vivo. For such proteins, regulation by arginylation with or without subsequent methylation can conceivably facilitate their structural interactions and affect their in vivo functions. For nucleic acid-binding proteins, the modifications likely affect their ability to interact with the nucleic acids (which should be facilitated by the addition of the positively charged Arg), or other proteins, which could be repelled or attracted by the chemical groups on the matching surfaces of the interaction partners. A number of proteins found in the nucleosolic and chromatin fractions, however, have been previously characterized as cytoplasmic proteins. For some of those, including actin, GAPDH, filamin, and spectrin, recent data demonstrate their involvement in gene expression and transcriptional regulation (Grummt, 2006; Loy et al., 2003; Sawa et al., 1997; Tang et al., 2003). It is likely that arginylation and arginylation/methylation of these proteins in the nucleus can facilitate this specific function. For others, such as collagen, no such function has been previously demonstrated. It is possible that their interaction with the chromatin is a non-specific effect of the purification, resulting from the presence of the surface Arg residues that may induce their interaction with DNA after the cell and nuclear lysis. It is also possible, however, that their presence in the nuclear fractions found in this study reflects their previously unknown function in the chromatin and/or transcriptional control.
Structural and sequence analysis shows that arginylated sites for most of the identified proteins are located in the middle of their polypeptide chains, in conserved domains predicted to affect key molecular interactions (Fig. 4, S5). It has been previously found that for many arginylated proteins sites for the posttranslational addition of Arg are located in the middle of the polypeptides, far away from the N-terminus, suggesting a novel regulatory mechanism that couples arginylation with partial regulatory proteolysis without disrupting the protein’s quarternary structure (Wong et al., 2007). In agreement with this, all the available structural data presented in our current study (Fig. 4, Fig. S4) show that arginylated sites are exposed within the structurally important conserved domains on the protein surface, suggesting that arginylation on these sites affects fully synthesized and folded proteins and plays an important regulatory role. The measured distances of the added Arg or methyl-Arg residues in histones to the phosphate groups in the DNA backbone are particularly encouraging, as these distances with the added Arg are short enough to significantly affect the interaction of histones with DNA, and thus can affect DNA packaging and/or transcriptional regulation. Conceivably, addition of methyl groups onto Arg in these positions should stabilize the arginylation sites and ensure their longer-term action in the nucleosome. Further studies of the individual arginylation sites identified in this study will reveal the functional significance of their regulation by arginylation and advance the knowledge about this poorly understood posttranslational modification to the next level of understanding.
Our finding that posttranslationally added Arg can be methylated constitutes a proof of principle and the first demonstration that posttranslational modifications can double on the same sites within the proteins. It has been previously shown that a negative posttranslational regulation of methylation by phosphorylation can occur by modifying the neighboring residues to change the charge and conformation of the methylation sites (Bedford and Richard, 2005). We show for the first time a positive regulation by a posttranslational modification that can facilitate methylation by creating an acceptor site for methyl groups. While many of the functions of such double modifications remain to be determined, our study shows a clear correlation between arginylation/methylation and the nuclear size. As a smaller nucleus implies a higher compaction, which can affect nuclear functions significantly, it is reasonable to suggest that arginylation/methylation will have a significant effect on nuclear function. It also appears likely that arginylation with subsequent methylation of the added Arg residues can serve as a second-order regulatory mechanism that could affect those major metabolic processes in which arginylation and methylation have been implicated. Methylation may also act to protect posttranslationally added Arg from chemical modifications in vivo that could destroy or inactivate them (for example, by methylglyoxal – a cytotoxic agent linked to disease), in line with a recently proposed model that methylation acts as a ‘guardian’ of Arg (Fackelmayer, 2005). Finally, methylation may be necessary to protect proteins against degradation by the N-end rule pathway, shown to affect some proteins with Arg in the N-terminal position (Varshavsky, 1997). It appears likely that the posttranslationally added Arg residues within a protein that further serve as targets of methylation should be especially important and/or long-lived to allow their further modification by Arg methyltransferases, an additional level of regulation or protection of these proteins and their important functional sites in vivo.
Significance
The current study demonstrates that posttranslationally added arginines on proteins can be further modified by arginine methylation and that this double posttranslational modification can regulate DNA arrangement in the chromatin and nuclear architecture. This is the first evidence that posttranslationally modified sites of proteins can undergo subsequent posttranslational modifications, and the first functional link between protein arginylation and Arg-methylation, which has been long known to regulate chromatin and nuclear proteins in vivo. It appears likely that such double modifications can not only increase the complexity of the signaling through Arg and the methylation pathways, but also exert other regulatory functions through a limited number of protein sites, and even provide a unique type of protein code. Our study demonstrated that the interaction of the two important protein modifications, arginylation and methylation, affects the nuclear size, suggesting its possible role in nuclear architecture and function.
Experimental Procedures
Nuclear fractionation and nuclear protein isolation for mass spectrometry
Cells were lysed in the lysis buffer (0.3 M sucrose, 4 mM Mg acetate, 12.5 mM KCl, 50 mM Tris pH: 8.0, 0.5 mM arginine, 1 mM DTT, 0.2 mM PMSF, protease inhibitor cocktail) and nuclear and cytosolic fractions were separated by centrifugation and further purification of nuclei by centrifugation through sucrose cushion. Pure nuclei were lysed in 400 mM NaCl-containing lysis buffer, and the soluble part was collected as nucleosol. The insoluble chromatin part was further extracted with 2 M NaCl containing buffer, and finally by DNase I treatment (see Supplement for the detailed procedure). Proteins from each fraction were precipitated by 20% TCA and the pellets were analyzed by mass spectrometry.
Mass spectrometry and database searches
Extracts from subcellular fractions of adult mouse heart (cytoplasmic fractions) and immortalized mouse embryonic fibroblasts (cytoplasmic and nuclear fractions) were used throughout this study. For the initial identification of arginylation/methylation, samples obtained during the previous study on the global analysis of protein arginylation (Wong et al., 2007) were subjected to a new search to detect the N-terminal mass addition of Arg plus one or two methyl groups as described below and in (Xu et al., 2009). In addition, one protein spot excised from 2D gel of mouse cardiac actin (Rai et al., 2008) was analyzed. For the analysis of nuclear proteins, samples were obtained as described above and the TCA-precipitated protein pellets were dissolved in invitrosolTM LC/MS protein solubilizer following the manufacturer’s protocol (Invitrogen), reduced and alkylated with 10 mM Tris(2-carboxyethyl)phosphine hydrochloride (TCEP, Roche Applied Science, Palo Alto, CA) and 55 mM iodoacetamide (IAM, Sigma) in 100 mM ammonium bicarbonate. Digestion was performed in the presence of 50 mM ammonium bicarbonate and 5 mM calcium chloride (Sigma) using sequencing grade soluble trypsin (Promega, Madison, WI). The resulting peptides were extracted by 5% formic acid, re-dissolved into buffer A (5% acetonitrile with 0.1% formic acid), and were pressure-loaded onto a reversed-phase (RP) or strong cation exchange (SCX)/reversed-phase MudPIT column and eluted with a linear gradient of acetonitrile or pulses of ammonium acetate salt (5–100%). Synthetic peptides were dissolved in water and supplemented with 1% formic acid to the final concentration of 100 fmol μl−1 and analyzed under the same LC-MS/MS conditions as those used for the digested protein samples. Data-dependent tandem mass spectrometry (MS/MS) analysis was performed in a LTQ-Orbitrap mass spectrometer (Thermo Electron, San Jose, CA). Full MS spectra were acquired by the precursor ion scan using the Orbitrap analyzer with resolution set at 60,000, followed by nine MS/MS events in the linear ion trap (LTQ), sequentially generated on the first, second and third most intense ions selected from the full MS spectrum. MS scan functions and HPLC solvent gradients were controlled by the Xcalibur data system (Thermo Electron). Tandem mass spectra were searched against EBI-IPI and NCBI mouse protein databases concatenated with reversed sequences to estimate false positive rate, using the ProLuCID (Xu, 2006) protein database search algorithm with peptide mass tolerance of +/− 3 amu, fragment ion mass tolerance of 0.4 amu and a static modification of 57.0215 on Cys due to carboxyamidomethylation. In order to identify N-terminal arginylated peptides, differential N-terminal modification searches for a mass addition of Arg (+156.1011) with an extra mass of either one (+14.0157) or two (+28.0314) methyl groups were performed with no enzymatic cleavage conditions imposed on the database search. ProLuCID search results were then filtered with DTASelect 2.0 (Tabb et al., 2002) using XCorr and DeltaCN at a false positive rate of 0.1% (--fp 0.001) estimated by number of reverse hits. Half or fully tryptic (-y 1) peptides only with N-terminal arginylation and Arg-methylation (-m 0) and a DeltaMass <= 5ppm (-DM 5) were accepted, the minimum number of peptides to identify a protein was set to 1(-p 1). All spectra were verified by automated filtering and manual filtering against the mass ambiguities shown in Supplemental Tables S3 and S4. The final proteins with arginylation modifications that passed these filtering criteria and manual validation are listed in Tables 1 and 2 and the corresponding mass spectra are shown in Fig. 1, S1, and Datasets 1 and 2.
In vivo methylation assay
1.2 million of immortalized mouse embryonic fibroblasts were seeded in 100 mm tissue culture dishes and grown for 15 hr. The plates were washed once with 5 ml of Met- and Cys-free DMEM without serum and incubated with 5 ml of the same media for 1 hr 30 min to induce amino acid starvation. After the initial starvation period, cycloheximide (100 μg/ml) and chloroamphenicol (40 μg/ml) (as well as arginine methyltransferase inhibitor AMI-1 (100 μM) in case of AMI-1 treatment) were added to the medium and the incubation continued for an additional 1 hr, followed by addition of 30 μCi of L-[methyl-3H]-Methionine (GE Amersham) for a 4 hr incorporation. At the end of the incorporation period, cells were washed in PBS and harvested by scraping and centrifugation. Cell pellets were lysed by pipetting in buffer A (50 mM Tris pH: 8, 0.3 M Sucrose, 4 mM Mg-acetate, 12.5 mM KCl, 10 mM β-ME, 0.5% NP 40, 1 mM DTT, 0.2 mM PMSF, protease inhibitor cocktail). Nucleus and cytosol were separated using the protocol adapted from (Kaji, 1976), by spinning at 4°C for 5 min at 2,000 × g. The supernatant (cytosol) was collected and the nuclear pellet was resuspended in buffer B (50 mM Tris pH: 7.5, 25 mM KCl, 5 mM MgCl2 0.5% NP 40, 1 mM DTT, 0.2 mM PMSF, protease inhibitor cocktail) and layered onto an equal volume of 0.88 M sucrose in buffer B, followed by centrifugation at 4°C for 5 min at 2,000 × g. The pellet containing pure nuclei was collected and lysed by pipetting in hypertonic buffer C (50 mM Tris pH:8.0, 25 mM KCl, 400 mM NaCl, 1 mM DTT, 0.2 mM PMSF, protease inhibitor cocktail), followed by centrifugation at 4°C for 10 min at 16,000 × g. The supernatant was collected as nucleosol and the pellet -- as chromatin. Protein concentrations in the cytosol and nucleosol, were estimated by the Bradford assay. Protein concentration in the chromatin fractions was estimated by running known amounts of nucleosolic proteins and 2X Laemmli – buffer-solubilized chromatin pellets from similar preparations in SDS PAGE, followed by densitometric analysis of the whole lane of Coomassie-stained gel. Equal amounts of protein from cytosol and nucleosol were precipitated by TCA, and incorporation into the pelleted protein was monitored by liquid scintillation. The chromatin pellet was washed three times with buffer B and incorporation was monitored by liquid scintillation.
In vivo arginylation assay
This assay was performed similar to the in vivo methylation assay, except for the following changes. Instead of Met- and Cys-free DMEM, custom-made Arg-free DMEM (Hyclone) was used. 60 μCi of L-[2,3,4-3H] Arginine (Perkin Elmer) was added for 4 hrs to each dish containing 5 ml of Arg-free DMEM without serum. For in vivo arginylation in the absence of translation inhibitors, the assay was performed in the same way except cycloheximide/chloramphenicol treatment step was omitted.
Estimation of intracellular methyl-Arg concentrations in the protein and free metabolite pools
Protein fractions prepared as described in the Supplement were precipitated by 10% TCA and the supernatant was analyzed by HPLC without acid hydrolysis. The pure methyl arginines (MMA, SDMA, and ADMA) were used as the standards in HPLC analysis (see Supplement for standard concentrations). HPLC, amino acid hydrolysis, and Arg determination was performed by the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University. See Supplement for the detailed procedure.
In vitro arginylation assay to estimate endogenous ATE1 activity in cellular fractions
This assay was performed using purified charged Arg-tRNA and protein fractions as the source of Arg-transferase as described in (Wang et al., 2011) with some modifications. Cytosol and nuclei were purified in the same way as described in the nucleus fractionation section except the pure nuclei were lysed by sonication followed by clarification by centrifugation at 16,000 × g and used for the assay. After the reaction, protein was precipitated with 10% cold TCA and counted in a liquid scintillation counter (see Supplement for the detailed procedure).
Estimation of ATE1 in level in cytosol and nucleus fractions
Cells were fractionated in the same way as described in the section ‘in vivo methylation assay’. Proteins in the cytosol and nucleosol fractions were estimated by Bradford assay. Equal amount of proteins were analyzed by Western blot using an ATE1 rat monoclonal antibody (Wang et al., 2011) to estimate the difference in the level of ATE1 in these two fractions.
Estimation of the nuclear size
WT or Ate1 KO MEF cells, untreated or treated with 100 μM AMI 1 for 24 hr, were harvested by trypsinization. Cells were washed once with PBS and fixed for 20min with 4% paraformaldehyde in PBS at room temperature. Post fixation, cells were washed thrice with PBS and stained with 1μg/ml DAPI for 10 min and spotted on poly lysine coated coverslips and mounted with aqua-polymount (Polysciences Inc). 5 randomly chosen fields from each preparation were captured at 40X and analyzed using Metamorph software. The pixel area covered by each nucleus was scored as nuclear size and converted to μm2 using the Metamorph internal calibration tool.
Arginylated and methyl-arginylated protein structure models and the measurement of distances between arginyl and methylarginyl residues and the neighboring phosphate atoms of DNA in the histone complexes were performed using the program Coot (Emsley and Cowtan, 2004). Illustrations of structures were prepared with the program PyMOL (Schrödinger).
Supplementary Material
Highlights.
Methylation of posttranslationally added arginine (arginylation/methylation)
Arginylation/methylation of nuclear proteins
Arginylation/methylation regulate nuclear size
Acknowledgments
We thank members of the Kashina lab for obtaining samples for the mass spectrometry analysis, J.M. Crawford and the W.M. Keck Foundation Biotechnology Resource Laboratory at Yale University for amino acid analysis, and Dr. Y.I. Wolf for helpful discussions and analysis of the arginylation consensus site using on-line Weblogo tool (http://weblogo.berkeley.edu/). This work was supported by NIH grant 1R01HL084419 to A.K., and by NIH grants P41 RR11823-09 and 5R01 MH067880 to J.R.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bedford MT, Richard S. Arginine Methylation: An Emerging Regulator of Protein Function. Molecular Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Fackelmayer FO. Protein arginine methyltransferases: guardians of the Arg? Trends Biochem Sci. 2005;30:666–671. doi: 10.1016/j.tibs.2005.10.002. [DOI] [PubMed] [Google Scholar]
- Grummt I. Actin and myosin as transcription factors. Curr Opin Genet Dev. 2006;16:191–196. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- Kaji H. Amino-terminal arginylation of chromosomal proteins by arginyl-tRNA. Biochemistry. 1976;15:5121–5125. doi: 10.1021/bi00668a027. [DOI] [PubMed] [Google Scholar]
- Karakozova M, Kozak M, Wong CC, Bailey AO, Yates JR, 3rd, Mogilner A, Zebroski H, Kashina A. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science. 2006;313:192–196. doi: 10.1126/science.1129344. [DOI] [PubMed] [Google Scholar]
- Kwon YT, Kashina AS, Davydov IV, Hu RG, An JY, Seo JW, Du F, Varshavsky A. An essential role of N-terminal arginylation in cardiovascular development. Science. 2002;297:96–99. doi: 10.1126/science.1069531. [DOI] [PubMed] [Google Scholar]
- Kwon YT, Kashina AS, Varshavsky A. Alternative splicing results in differential expression, activity, and localization of the two forms of arginyl-tRNA-protein transferase, a component of the N-end rule pathway. Mol Cell Biol. 1999;19:182–193. doi: 10.1128/mcb.19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loy CJ, Sim KS, Yong EL. Filamin-A fragment localizes to the nucleus to regulate androgen receptor and coactivator functions. Proc Natl Acad Sci U S A. 2003;100:4562–4567. doi: 10.1073/pnas.0736237100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Kashina A. Identification of mammalian arginyltransferases that modify a specific subset of protein substrates. Proc Natl Acad Sci U S A. 2005;102:10123–10128. doi: 10.1073/pnas.0504500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai R, Wong CC, Xu T, Leu NA, Dong DW, Guo C, McLaughlin KJ, Yates JR, 3rd, Kashina A. Arginyltransferase regulates alpha cardiac actin function, myofibril formation and contractility during heart development. Development. 2008;135:3881–3889. doi: 10.1242/dev.022723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa A, Khan AA, Hester LD, Snyder SH. Glyceraldehyde-3-phosphate dehydrogenase: nuclear translocation participates in neuronal and nonneuronal cell death. Proc Natl Acad Sci U S A. 1997;94:11669–11674. doi: 10.1073/pnas.94.21.11669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., 3rd DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y, Katuri V, Dillner A, Mishra B, Deng CX, Mishra L. Disruption of transforming growth factor-beta signaling in ELF beta-spectrin-deficient mice. Science. 2003;299:574–577. doi: 10.1126/science.1075994. [DOI] [PubMed] [Google Scholar]
- Varshavsky A. The N-end rule pathway of protein degradation. Genes Cells. 1997;2:13–28. doi: 10.1046/j.1365-2443.1997.1020301.x. [DOI] [PubMed] [Google Scholar]
- Wang J, Han X, Saha S, Xu T, Rai R, Zhang F, Wolf YI, Wolfson A, Yates JR, III, Kashina A. Arginyltransferase is an ATP-independent Self-Regulating Enzyme that Forms Distinct Functional Complexes in vivo. Chemistry and Biology. 2011;18:121–130. doi: 10.1016/j.chembiol.2010.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong CCL, Xu T, Rai R, Bailey AO, Yates JR, Wolf YI, Zebroski H, Kashina A. Global Analysis of Posttranslational Protein Arginylation. PLoS Biology. 2007;5:e258. doi: 10.1371/journal.pbio.0050258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Venable JD, Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR., III ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Molecular & Cellular Proteomics. 2006;5:S174. [Google Scholar]
- Xu T, Wong CC, Kashina A, Yates JR., 3rd Identification of N-terminally arginylated proteins and peptides by mass spectrometry. Nat Protoc. 2009;4:325–332. doi: 10.1038/nprot.2008.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.