Abstract
Unlike the dominant role of one class II invariant chain peptide (CLIP) in blocking MHC class II, comparative lipidomics analysis shows that human cluster of differentiation (CD) proteins CD1a, CD1b, CD1c, and CD1d bind lipids corresponding to hundreds of diverse accurate mass retention time values. Although most ions were observed in association with several CD1 proteins, ligands binding selectively to one CD1 isoform allowed the study of how differing antigen-binding grooves influence lipid capture. Although the CD1b groove is distinguished by its unusually large volume (2,200 Å3) and the T′ tunnel, the average mass of compounds eluted from CD1b was similar to that of lipids from CD1 proteins with smaller grooves. Elution of small ligands from the large CD1b groove might be explained if two small lipids bind simultaneously in the groove. Crystal structures indicate that all CD1 proteins can capture one antigen with its hydrophilic head group exposed for T-cell recognition, but CD1b structures show scaffold lipids seated below the antigen. We found that ligands selectively associated with CD1b lacked the hydrophilic head group that is generally needed for antigen recognition but interferes with scaffold function. Furthermore, we identified the scaffolds as deoxyceramides and diacylglycerols and directly demonstrate a function in augmenting presentation of a small glycolipid antigen to T cells. Thus, unlike MHC class II, CD1 proteins capture highly diverse ligands in the secretory pathway. CD1b has a mechanism for presenting either two small or one large lipid, allowing presentation of antigens with an unusually broad range of chain lengths.
Keywords: antigen presentation, Mycobacterium tuberculosis, cluster of differentiation 1
Most mammals have preserved large cluster of differentiation 1 (CD1) gene families, comprised of orthologs of human CD1a, CD1b, CD1c, and CD1d antigen-presenting molecules. Each of these CD1 protein types, known as isoforms, differs in sequence and 3D structure, so that their heavy chains form antigen-binding grooves of differing size and shape. CD1 proteins initially fold in the endoplasmic reticulum (ER), exit through the secretory pathway, and traffic to endosomes via direct or surface mechanisms that require the binding of cytoplasmic sequences to adaptor proteins (1). These subcellular trafficking pathways expose nascent CD1 proteins to diverse classes of endogenous cellular lipids in ER, Golgi, and endosomal compartments. However, whether CD1 proteins, especially the less-studied group 1 CD1 proteins (CD1a, CD1b, and CD1c), initially capture one or many types of groove-blocking lipids is unknown. CD1 proteins might capture a dominant lipid class that is functionally equivalent with the class II invariant chain peptide (CLIP) (2), or instead broadly survey various endogenous lipids.
Cellular pH strongly influences the capture and release of lipids. In particular, CD1b and CD1d are relatively resistant to exogenous antigen loading at neutral pH conditions normally found in the secretory pathway (3–6). They more rapidly exchange antigens at pH 4–6, which is characteristic of the late endosomal or lysosomal environment (7, 8). The differing pH requirements for antigen loading, combined with enrichment of endogenous lipids in the secretory pathway and exogenous lipids endosomes, are coalescing into a two-step model of lipid antigen presentation. First, newly translated CD1 proteins, aided by the microsomal triglyceride transport protein (MTP) (9), capture endogenous self-lipids during or after folding at neutral pH in the ER or secretory pathway. Second, upon reaching the endosomes by recycling or direct routes, the acidic pH facilitates the unfolding or untethering of the parallel α-helices that normally restrict ligand access to the groove (3–6). Increased groove access promotes both unloading of the initially bound endogenous lipids and subsequent capture of the exogenous lipids delivered by infection, lipid transfer proteins, or receptor-mediated uptake (8, 10).
Whereas later antigen exchange events within endosomes are extensively characterized, the early events in which newly folded CD1 proteins load self-lipids are less well understood. The folding of nascent CD1 heavy chains is thought to involve cotranslational binding of self lipids in the ER (11). This expectation is supported by experiments that detect self-diacylglycerols, monoacylglycerols, and sphingolipids eluted from CD1 proteins that have ER retention signals or lack endosomal recycling motifs (2, 12, 13). Some of these endogenous lipids activate T cells, so are true autoantigens (14). Others are more likely nonantigenic place-holding lipids that provide a hydrophobic substrate for the initial folding reactions and serve a groove-blocking function until antigenic exogenous lipids are encountered in acidic endosomes. Therefore, whether the human CD1 family uses one or many types of place-holding lipids, or whether individual CD1 proteins bind the same or different lipids, is largely unknown. CD1a (1,280 Å3), CD1b (2,200 Å3), CD1c (1,780 Å3), and CD1d (1,650 Å3) grooves differ in volume and the number of pockets, known as A′, F′, C′, and the T′ tunnel, located at the bottom of the groove. All grooves have an F′ and an A′ pocket, but only CD1b forms the T′ tunnel and C′ pocket (15–18). These structural studies predict that each CD1 protein might capture somewhat distinct classes of lipids, with the strongest prediction being that CD1b captures lipids with long alkyl chains and higher mass.
After deleting transmembrane and cytoplasmic tail sequences needed for endosomal trafficking, we captured cellular CD1a, CD1b, CD1c, and CD1d complexes and eluted ligands for entry into a newly validated comparative lipidomics platform. As contrasted with conventional MS, this platform rapidly generates lipidomes, derived from triplicate datasets generated back-to-back under rigorously comparable conditions. Validated protocols measure equivalent input lipids, lack of detector saturation, broad detection of many lipid classes, and computer-based analysis with data filters, so that comparative analyses of thousands of molecular features in one lipidome are possible (19). We found that the human CD1 system surveys hundreds of distinct endogenous lipids that differ in polarity, in contrast to the CLIP mechanism whereby one ligand dominates in the initial loading of MHC II. Contrary to the simple predictions arising from the large volume of CD1b groove, the spectrum of cellular lipids binding selectively to CD1b is dominated by ligands with relatively low mass and no hydrophilic head groups. These distinct chemical features suggest that such small, headless lipids might not be antigens that contact the T-cell receptor (TCR), but instead are the long-sought scaffold lipids binding to CD1b in conjunction with antigens. We identified the candidate scaffold lipids as deoxyceramides and diacylglycerols, and demonstrate their cofactor function in antigen presentation to T cells, defining a natural cellular mechanism for presenting lipids with extraordinary lipid length discrepancy.
Results
Linked and Zippered CD1 Proteins Capture Similar Lipids.
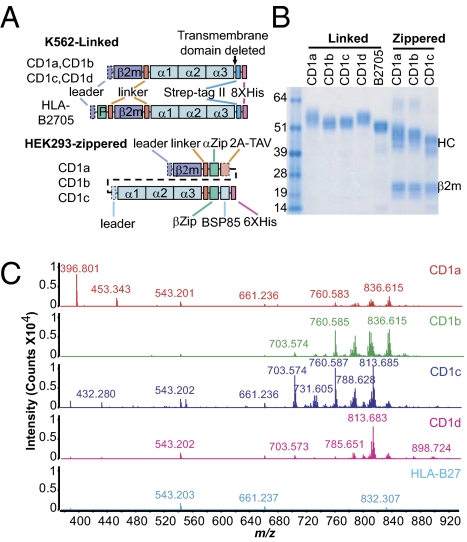
Elution of peptides from MHC proteins was key to understanding the differing antigen motifs for MHC I and II proteins and the discovery of groove-blocking peptides (20–22). Similarly, broad comparisons of lipids eluted from group 1 (CD1a, CD1b, CD1c) or group 2 (CD1d) CD1 proteins might lead to insights into isoform-specific CD1 functions. We expressed β-2 microglobulin (β2m)-linked CD1 molecules in human hematopoietic cell K562 and zippered CD1 proteins in the human HEK293 cells. Deletion of the transmembrane and cytoplasmic sequences limits or abolishes endosomal recycling, so that lipids are loaded predominantly in early compartments (Fig. 1A and Fig. S1A) (3, 6). Transmembrane sequence deletion allows capture of soluble molecules for lipid elution without the need for detergents, which can exchange for endogenous lipids and hamper MS detection. These two sets of proteins were purified through polyhistidine, anti-zipper, and strep II tags (Fig. 1 A and B). These various expression tags allowed differing approaches to CD1–lipid complex purification, which was later proven to be an important design consideration. Linked CD1 complexes purified with polyhistidine/nickel methods or zippered complexes treated with base generated high-quality eluents with good lipid ligand yield and few contaminants observable by MS. In contrast, harsher methods that require acidic buffers resulted in premature unloading or chemical alteration of lipids during complex purification (Fig. 1C). Generation of two sets of proteins with differing construct designs and expressed in distinct cell types also offered the opportunity to measure effects of cell type and possible artifacts generated by protein engineering. We used human MHC I (HLA-B2705) and MHC I of rhesus macaque (MamuA01) lacking a hydrophobic groove as negative control proteins because their overall structure is similar to CD1, but they lack hydrophobic grooves. Proteins were recovered with milligram yields, high purity, and expected apparent masses of linked monomers or zippered dimers (Fig. 1B). The recovered proteins were recognized by isoform-specific anti-CD1 antibodies, confirming a proper folding (Fig. S1B). These CD1 proteins provided eluents that yielded strong and diverse mass spectral signals in HPLC-MS, allowing identification of many ions not seen with MHC I, extracted buffers, or solvent blanks, especially at the late retention time period (Fig. 1C and Fig. S1C). These two sets of human CD1 proteins provide an unambiguous and consistent demonstration of CD1-associated ions for comparative determination of isoform-specific ligands through lipidomic analyses.
Fig. 1.
Two sets of human CD1 proteins bind cellular lipids. (A) Covalently linked molecules were constructed by linking β2m to the heavy chains of CD1 or HLA-B2705 molecules through a flexible linker. The HLA-B2705 sequence contains a groove-binding peptide (P) with linker. Zippered CD1 molecules consist of zipper-encoding regions (αZip and βZip) at the C termini of heavy chains and human β2m sequences, which are linked by cleavable 2A peptide from Thosea asigna virus (2A-TAV). Regions cleaved upon maturation (dashed line) and biotinylation site (BSP85) are also indicated. (B) One microgram of purified human CD1 proteins estimated with spectrophotometry was separated by SDS/PAGE under denatured and reduced conditions and visualized with Coomassie blue. HC, heavy chain. (C) Lipid ligands from purified single-chain CD1 proteins were eluted and profiled with HPLC-MS in comparison with the HLA-B2705 control. Coeluting ions (18–42 min) in solvents and buffer were subtracted from the CD1 spectra.
Comparative Lipidomics of CD1 Ligands.
Comparative lipidomics is a computer-based analysis that provides broad comparisons of datasets generated before and after altering one biological variable, such as the CD1 isoform from which lipids are eluted. Unlike conventional MS analysis (Fig. 1C and Fig. S1C), comparative lipidomics converts each ion seen in two or more replicates into one feature, defined as a 3D data point of linked intensity, retention time (RT), and m/z values. After aligning features with equivalent accurate mass-RT across samples, fold change and statistical significance are calculated for each pairwise comparison for identification of all features that selectively eluted from one but not other human CD1 isoforms (Fig. 2B). This method provides a broad overview of CD1 isoform-specific lipid capture (Fig. 2 A and B). Like all high-throughput methods, broad comparisons contain individual errors, so key features emerging from the global analysis are further tested through focused reanalysis of one variable (Fig. 2C), raw (Fig. 2D) or collisional mass spectra (Figs. 3 and 4 and Figs. S2–S5). Unlike conventional MS analysis, the comparative and quantitative nature of this analysis imposes specialized design and performance criteria for HPLC-MS. We recently validated the normalization of input lipids, reproducibility of intensity measurements, avoidance of detector saturation through detection of a wide dynamic range, automated methods for quantitation of thousands of chromatograms, software protocols for aligning large datasets, and data filters for censoring features with high variance (19).
Fig. 2.
Human CD1b captures small and nonpolar lipids. (A) CD1-associated and isoform-specific features are plotted with embedded mass values, with n indicating the number of validated features associated with each isoform. (B) CD1b-associated features are shown in a volcano plot in comparison with the ions with highest intensity from isoforms CD1a, CD1c, or CD1d. (C) All CD1b-associated (Left) and CD1b-specific (Right) features eluted from linked and zippered CD1b are plotted. The numbers of features are indicated and two early eluting ions (m/z 584.523 and 612.555) from CD1b are annotated with arrows (B and C). (D) The early eluting, CD1b-specific ions emerging from lipidomic comparisons are validated with raw mass spectra extracted at 3.1 min. The depicted isoform-specific lipids are shown by subtracting the ions in solvent, buffer, MHC class I controls, or any other CD1 proteins. CD1b-associated ions (green) are shown with and without subtraction.
Fig. 3.
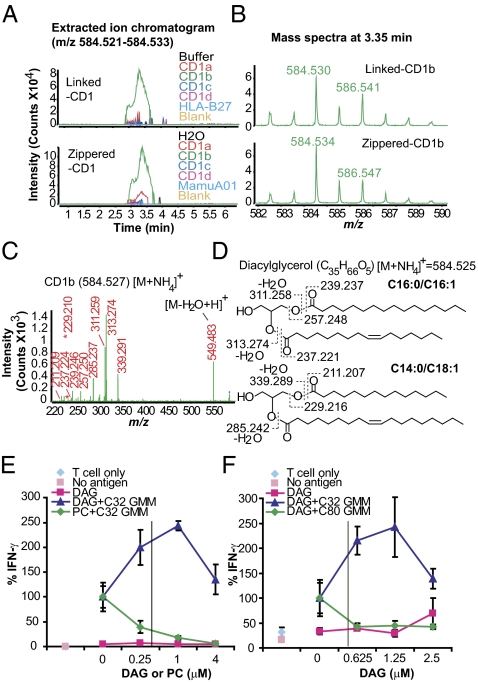
Identification of a deoxydihydroceramide as a CD1b ligand. Chromatograms (A) and mass spectra (B) highlight the target ion (m/z 524.540) corresponding to a candidate spacer detected in association with CD1b. (C) Collision-induced dissociation (CID) of precursor ion 524.540 displays a collisional pattern well-matched to that of a standard lipid 1-deoxydihydroceramide (d18:0/16:0). (D) Calculated m/z values of the protonated collision fragments from the standard lipid.
Fig. 4.
Identification of diacylglycerol series as CD1b ligands and its regulatory function in T-cell activation. Chromatograms (A) and mass spectra (B) of ions from zippered and linked CD1 proteins highlight m/z 584.527 as an ion corresponding to a second candidate spacer lipid. CID (C) of precursor ion 584.527 is interpreted as diacylglycerol isomers with calculated m/z values of protonated collision fragments (D). A detailed comparison with the standard is shown in Fig. S3. LDN5 T cells were activated with plate-bound CD1b protein after acid stripping of endogenous ligands. Pilot experiments determined the concentration of C32 GMM (0.5 μm) and C80 GMM (0.5 μm) that give nearly half-maximal activation. DAG (18:1/18:1 or 16:0/18:1) or PC was then added simultaneously with GMM before measuring T-cell activation (E and F). The concentration of GMM at 0.5 μm was annotated with vertical lines in comparison with the concentration of DAG or PC. The percent of IFN-γ production mediated by GMM upon the presence of DAG or PC was plotted in comparison with that in absence of DAG or PC. The vertical line represents equal mass of added antigen and spacing cofactor. The percent of background IFN-γ production with DAG or PC was obtained by comparing to that with C32 GMM. This result is representative of three or more experiments.
To assess reproducibility of intensity measurements, input lipids are normalized to CD1 concentration and monitored by total ion current with continuously infused calibrants. To optimize the reproducibility of chromatography, we used a universal normal-phase chromatographic system that generates one lipidome within 50 min, allowing consecutive generation of lipidomes in triplicate from many conditions in <1 d (19). Data filters remove the background features present in calibrants, solvent blanks, extracted buffers, and the features in MHC I eluents. We determined the CD1-associated features as those with intensity >2,000 counts, 10-fold above the background, fivefold higher than the MHC class I control. We obtained hundreds of features for each human CD1 isoform (Fig. 2A, Left). The large absolute number of ligands (Fig. 2A) and diverse RT (Fig. 2C) reflect the somewhat nonspecific nature of hydrophobic interactions, as contrasted with the more specific hydrogen bonding mechanisms used by MHC–peptide complexes. Therefore, the global survey of lipids eluted from human CD1 proteins suggests that unlike MHC II–CLIP complexes, all four CD1 isoforms naturally capture diverse endogenous lipids. Because these ions are not seen in MHC I homologs, these ligands are likely associated with hydrophobic CD1 grooves, rather than the exposed surface of β2m heavy-chain complexes. The smaller number of isoform-specific features among total features (Fig. 2A, Left and Center) illustrates that many features were seen in association with two or more CD1 isoforms. This finding is in agreement with others that suggest that the sulfatide glycolipid can be presented by several CD1 isoforms (23). Also, the global similarities in the profiles of lipids from two protein designs and two cell types (Figs. 1C, 2D, and 4A and Fig. S1C), supported the reproducibility of CD1 lipidomic analyses and indicated that tags, linkers, and cell types did not fundamentally alter the number and types of ligands bound, with some exceptions (Fig. 2D).
CD1b Captures Low-Mass Lipids.
A second stage of data analyses highlight ions selectively appearing in eluents of individual CD1 isoforms. Features were considered specific to one CD1 isoform when they showed more than fivefold-higher intensity values, compared with the highest-intensity value detected with any other isoform (Fig. 2 A, Center, and B). Although many features are detected with more than one or all CD1 proteins (23), many others associate only with one human CD1 isoform. For example, comparison of all features associated with CD1b with all mass-aligned features associated with any other CD1 protein yielded 35 CD1b-specific features (Fig. 2B). We focused on CD1b, because clear, but unexpected patterns were observed. CD1a has the smallest groove volume (1,280 Å3) among human CD1 proteins and generates the smallest average mass, supporting a correlation of groove volume with ligand size (Fig. 2A). In contrast, the CD1b groove, which can bind exogenous C70–C86 mycolyl or sulfoglycolipids in endosomes (24–26), greatly exceeds the volume of other CD1 isoforms, which mainly present antigens with C36–C46 in overall lipid length. However, average m/z calculated from all endogenously acquired CD1b ligands was similar to that of other isoforms (Fig. 2A, Left).
We reasoned that highly available cellular lipids might promiscuously bind to multiple CD1 isoforms and therefore mask any isoform-specific function of CD1b to capture larger lipids. After censoring features associated with two or more CD1 proteins (Fig. 2A, Center), reanalysis showed that the average m/z value of CD1-specific features was similar among isoforms and not higher for CD1b. In electrospray ionization with positive mode detection, lipids typically ionize as monomers, [M + H]+, but certain zwitterionic lipids form dimeric complexes of [2M + H]+. In the latter case, the detected m/z overestimates the mass of monomeric lipids. For example, sphingomyelin can be detected as two ion chromatograms with the same retention time, but appearing in two mass windows corresponding to [2M + H]+ (m/z 1,626.362) and [M + H]+ (m/z 813.685; Fig. S1D). To avoid any systematic bias of average mass of CD1 ligands, a third analysis censors features in the mass range in which this artifact can be detected (m/z 1,200–1,900). This analysis again returned a similar average mass for features associated with each CD1 isoform (Fig. 2A, Right). It was surprising that the CD1 isoform with a groove capacity that is at least 400 Å3 larger than other CD1 grooves did not release larger ligands, when measured as monomers. This conclusion was reinforced by an apparent mismatch of the calculated groove volume with the average mass of eluted ligands (Fig. 2A). Molecular models indicate that the C′ pocket and combined A′T′F channel of CD1b would be fully occupied by a C74 lipid with a mass in the range of 1,037, plus the mass of any head group extending out of the portal (16, 25). Nearly all CD1b-specific features showed lower mass values, suggesting that CD1b uniquely captures lipids with alkyl chains smaller than the available capacity of the groove (Fig. 2A).
Elution of small lipids from a large groove could be explained if CD1b simultaneously captures two endogenous lipids that act together to fill the internal volume. This possibility was first suggested by crystallography (16) and later by MS (27) of intact CD1b–phospholipid complexes. For example, the CD1b–phosphatidylinositol (PI) crystal first revealed that the inositol head group protrudes upward for TCR contact, but two aliphatic hydrocarbon units of PI bind together with small lipids, presumably detergents used for refolding, seated deeply within the F′ and T′ pockets. The unknown deeply bound lipids might be considered scaffold lipids because they bind beneath the antigen, occupying space within the groove so that PI is positioned upward and ectopically toward the F′ portal and the plane of TCR contact. The scaffold hypothesis could solve the observed length mismatch problem because the combined length of the aliphatic alkyl chains of one scaffold and one antigen could approximate C74, fully occupying all four pockets. The endogenous antigen and scaffold might be subsequently exchanged for one large exogenous antigen such as one ∼C80 mycolyl glycolipid, imported into endosomes (1, 28). This proposed scaffold function is distinct from the place-holding function of CLIP, which fully occupies the groove and is exchanged one to one for endosomal peptides. This model predicts three general features that guided lipidomic analysis: spacers should show CD1b specificity, m/z values <1,000, and unusual hydrophobicity due to the lack of a hydrophilic head group.
CD1b Captures Nonpolar Lipids with Early Retention Time.
Because RT correlates directly with polarity, display of RT values embedded in all CD1b-associated features showed that CD1b binds molecules that span the full range of low- (3–14 min) and high-polarity lipids (14–42 min), which are enriched for neutral lipids and phosphoglycolipids, respectively. However, among CD1b-specific features, most were early eluting with m/z <1,000, suggesting that CD1b is structurally specialized to capture certain small and unusually hydrophobic lipids. This automated lipidomic analysis was supported by manual validation of the mass spectra for these early eluting lipids (3.1 min), which confirmed the many CD1b-specific ions lacking from CD1a, CD1c, or CD1d (Fig. 2D). These mass spectra further provided a detailed display of isotope windows and mass intervals corresponding with lipid length and unsaturation (Fig. 2D). For both zippered and linked CD1b proteins, we detected ions at m/z 584.527, 612.557, and 640.587 corresponding to an alkane series of molecules differing by C2H4. Other ions corresponded to the expected unsaturated forms and isotopes. For zippered CD1b proteins from HEK293 cells, we detected this series, as well as a second set of weaker ions near m/z 524.541. Because these low-mass, highly hydrophobic, CD1b-specific lipids fulfilled all criteria for spacers, we determined their structures by collisional MS.
Deoxyceramides Eluted from CD1b.
Extracted ion chromatograms at m/z 524.544 confirmed that this ion was associated with CD1b, but was absent or present in trace levels with other CD1 or MHC proteins (Fig. 3 A and B). Potential matches included an ammoniated fatty ester or a pronated deoxysphingolipid, [C34H70NO2]+, with m/z 524.540. We first tested oleyl palmitate because such fatty acyl esters were suggested to be a candidate CD1b ligand based on a prior report (27). However, the collisional spectrum of the wax ester standard did not match the CD1b ligand with regard to the [M − H2O + H]+ ion (m/z 506.525) and other features (Fig. 3C). Surprisingly, the CD1b ligands did match all ions in the collision spectrum of the recently described 1-deoxy form of dihydroceramide (29), including those interpreted as the dehydration product, pronated fatty amide chain (m/z 256.264), and sphinganine (m/z 268.300; Fig. 3D). Independently, the CD1b-associated ion of m/z 522.528 was also determined as a deoxyceramide whose fatty acid contained one additional methylene group and one saturation (Fig. S2).
Diacylglycerols Eluted from CD1b.
The ion at m/z 584.530, which was detected with zippered and linked forms of CD1b (Fig. 2 B and C), matched the expected mass of an ammonium adduct of diacylglycerol (DAG), C35H72O5N, within 4 ppm. Collisional MS (Fig. 4 C and D) demonstrated fragments corresponding to the loss of C16, C16:1, or both fatty acyl units, and the isobars of C14, C18:1 of DAG. Nearly coeluting ions with m/z 612.557 and 640.587 were additional members of alkane series, whereas m/z 586.538 and 638.574 were saturated or unsaturated analogs as demonstrated by collisional MS in comparison with DAG standards (Figs. S4 and S5). Thus, we identified nine molecular species of diacylglycerols and two deoxyceramides in association with CD1b, which account for many but not all of the CD1b-specific lipids (Fig. 2).
Spacers Influence Antigen Recognition.
The identified DAG and deoxyceramides are low-mass, hydrophobic, CD1b-specific ligands that meet the criteria for scaffold lipids. Candidate scaffold lipids observed in the CD1b-PI structure were expected to be two C16 detergents plus the size of the head group (16). The mass of unknown scaffold lipids in the CD1b-PC structure were reported to have m/z values varying from 587.4 to 643.5 (27). The two types of natural cellular ligands reported here range in mass from 522 to 640 amu and length from C34 to C38. Therefore, the overall size of the natural cellular ligands reported here matches well to the size of densities seen in CD1b–lipid complexes studied by crystallography (16, 27). However, direct evidence for a functional role of any molecule as a scaffold required testing in lipid-mediated assays of T-cell activation. We focused on diacylglycerols (DAG) because this lipid is common in cells, observed with high intensity, and detected in association with both CD1b protein preparations. To bypass confounding effects of DAG on antigen-presenting cells and focus on the interactions with the glycolipid antigen, glucose monomycolate (GMM), we used cell-free assays. These T cells respond to GMM with an average alkyl chain length of C80 (C80 GMM) or C32 (C32 GMM) based on the ability of GMM to mediate a physical interaction of CD1b with the clonotypic TCR (28, 30). Therefore, we measured IFN-γ release by the CD1b-restricted, GMM-reactive T-cell line LDN5 as a measure of antigen presentation. C80 GMM matches or slightly exceeds the CD1b groove volume, so should be nonpermissive for binding with a scaffold lipid. In contrast, C32 GMM is predicted to occupy less than half of the groove volume, and so might be unaffected by or even stabilized by pretreatment with a scaffold lipid.
The recombinant biotinylated CD1b from which DAG was originally eluted was bound to streptavidin plates and washed with acid to strip endogenous ligands (25). Next we titrated in C32 or C80 GMM ligands to yield nearly half-maximal levels of stimulation, allowing measurement of both positive and negative effects of concomitantly added lipid cofactors, measured as percent change from the baseline condition of no cofactor. Controls showed that the T-cell response was not affected by DAG, when antigen was absent (Fig. 4 E and F). Importantly, coloading with DAG improved the T-cell recognition C32 GMM and suppressed the recognition of C80 GMM, fulfilling both aspects of predictions related to the effect of scaffold lipids on short- and long-chain antigens (Fig. 4 E and F). When DAG was added in large molar excess to either antigen, T-cell response was suppressed, consistent with the conclusion that this nonantigen lipid binds within the groove but does not activate T cells. As an additional specificity control for the scaffold function, we determined whether coaddition of C32 GMM with phosphatidylcholine (PC) could augment recognition. PC is a known groove-binding CD1b ligand with a chain length similar to DAG, but is predicted not to function as a scaffold, because its large zwitterionic head group likely prevents interaction with hydrophobic residues deeply seated in the groove (27). PC failed to augment C32 GMM recognition, but instead inhibited responses, indicating that it likely interacted with the groove in competing with the coloaded C32 GMM. These results are most consistent with CD1b capture of heterologous C32 GMM–DAG complexes, demonstrating a cofactor role of DAG in antigen recognition.
Discussion
Both CD1 and MHC II have a two-step antigen capture mechanism whereby endogenous lipids are initially bound in the ER or a secretory compartment until exchanged for exogenous antigens imported into endosomes. CLIP peptides broadly occupy the grooves of most MHC II proteins, owing to positioning of the CLIP sequence in proximity to the folding event by the physical association of invariant chain to MHC II (22). In vitro, various lipids stabilize and promote CD1 folding (16, 24, 31), implying that cellular lipid loading of CD1 might be a cotranslational event that could be supported by several lipid types. In contrast to MHC II–CLIP, there is no known cellular mechanism to position one kind of lipid onto nascent CD1 heavy chains during folding. The one well-established early acting cofactor for loading, MTP, shows promiscuity for lipids and affects all human CD1 isoforms (9, 32). Actual measurements of mouse CD1d complexes formed in cells first suggested that cellular ligands were dominated by one class of phosphatidylinositol-containing molecule (2), and CD1b was reported to bind PC in a selective way (27). However, subsequent study of mouse and human CD1d detected several types of phospho- and sphingolipid ligands (12–14), suggesting greater diversity of lipid ligands for CD1d. Using cellular CD1 proteins lacking endosomal targeting motifs and captured under two different conditions that avoid the acid-mediated unloading effects, our comparative lipidomics approach provides evidence for substantial diversity of CD1 ligands for each of the four human CD1 proteins, detecting hundreds of molecular features whose retention times vary greatly. These results support a model, contrasting starkly with MHC II–CLIP, in which early loading events capture broadly diverse lipids to which CD1 proteins are exposed in cells.
By aligning features based on mass across the various CD1 proteins, many ligands were promiscuously detected in response to two or more CD1 isoforms, in agreement with a study showing that sulfatide can be presented by multiple CD1 isoforms (23). However, isoform-specific features occurred as well. Here we focus on CD1b-specific features to provide biochemical and functional evidence for a CD1b-specific model of antigen capture that allows presentation of a particularly diverse size range of lipid antigens. The structural evidence for concomitant binding of antigen and scaffold lipids comes from crystallography (16, 27), and these studies identify the molecular structures of candidate scaffolds as deoxyceramides and diacylglycerols and provide functional evidence for a cofactor role in T-cell recognition. The pattern of T-cell activation observed is highly specific, because it is seen only with a headless cofactor and only when the two lipid ligands' total length matches but does not exceed the groove volume (16, 27). Although we cannot directly measure the amount of GMM and CD1b in the groove, optimized T-cell activation is observed when adding similar absolute molar concentration of antigen and scaffold. Antigens bind in the groove, and the ability of excess DAG to suppress recognition suggests that it also binds to the groove. These data, size considerations, and prior crystallography data all point strongly to the conclusion that one antigen and one scaffold bind simultaneously.
Among cellular molecules, lipids are hydrophobic. Among lipids, the scaffolds are at the extreme of hydrophobicity. MS analysis directly demonstrated that these scaffolds lack the peptide, phosphate, or carbohydrate moieties that normally interact with the TCR. In fact, it is notable that the deoxyceramide moieties are a recently discovered sphingolipid variant, whose deoxy state renders it even less polar than more common sphingosines (29). Thus, CD1b captures molecules that are among most highly hydrophobic lipids present in cells. The unusually hydrophobic structure likely allows these lipids to settle deeply in the hydrophobic groove in ways that are not possible if a charged or bulky head group is present.
A model whereby two or three short-chain lipids are initially loaded and subsequently exchanged for one long (∼C80) foreign lipid in endosomes has been previously proposed (33), and the current data provide specific insights into how this could occur. We speculate that spacer lipids are bound to bridge the discrepancy in the length between most naturally occurring endogenous lipids and long-chain foreign antigens, likely mycolyl lipids. Most endogenous sphingolipids or phospholipids contain two alkyl tails, whose combined lengths of C32–C46 match well to the groove volumes of CD1a, CD1c, or CD1d, but are smaller than the CD1b groove. The CD1b groove is larger than needed for most naturally produced endogenous lipids. Triacylgycerols (C51–C66) and dolichols (C90–C95) might more closely match the CD1b groove volume. However, the former still has a length mismatch and lacks one long lipid for the A′T′F′ superchannel, and the latter is large, branched, and semirigid, owing to polyunsaturation. Therefore, no single endogenous lipid class provides an obvious hand-in-glove fit to CD1b (33). This tentative conclusion supported by our data showing that the average mass of nearly all CD1b-associated features is smaller than that predicted by groove volume, so that groove volume mismatch with endogenous lipids is likely a general phenomenon. These considerations suggest two functions for DAG and deoxyceramide lipids in cellular antigen processing: (i) they might have a scaffold function in pushing short-chain self-lipids upward toward the TCR interface, so that they can be recognized; and (ii) they might be like a hemi-CLIP, holding space in the CD1b groove until large ∼C80 mycolyl or sulfolipids are first encountered in endosomes.
Materials and Methods
Expression and Purification of CD1 Proteins.
Linked CD1 proteins were constructed by covalently linking β2m to α1–3 domains of the heavy chain together with Strep-tag II and polyhistidine tags at C termini. Zippered CD1 proteins were generated by stabilizing β2m and the heavy chain with C-terminal zipper sequences. The linked and zippered CD1 proteins together with MHC class I controls HLA-B2705 and MamuA01 were expressed in K562 (34) and HEK293 (4, 5), respectively. The secreted proteins were purified as described (24). Strategies for CD1 expression and purification are detailed in SI Materials and Methods.
HPLC-MS and Lipidomics Analyses of Lipids.
The lipid ligands from CD1 proteins were extracted (25) and tested with an Agilent Technologies 6520 Accurate-Mass Q-TOF coupled with an Agilent 1200 series HPLC system controlled by MassHunter software as described (19). MS data were initially visualized and manually compared using MassHunter software and then converted into mzData format. XCMS software (version 1.24; http://metlin.scripps.edu/xcms/index.php) carried out filtering, peak picking, ion intensity quantification, and ion alignment across samples. CD1-associated features were assigned when ion counts were >2,000, 10-fold higher than buffer control and fivefold higher than MHC class I control. CD1 isoform-specific features met the additional criteria of fivefold-higher intensity than any aligned features from all other CD1 proteins and a corrected P < 0.05, as calculated with Student's paired t test and Benjamini and Hochberg multiple testing correction. Detailed criteria for data acquisition and analyses are described in SI Materials and Methods.
T-Cell Assays.
Influence of antigen loading by diacylglycerol was tested using the T-cell line restricted by C32 and C80 glucose monomycolates as described (23). The lipid compounds used and detailed procedures are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Rick Willis for technical advice and Dr. Ted Hansen for providing protein expression vector and the HLA-B2705 gene template. This work was supported by The Burroughs Wellcome Fund Program in Translational Science; National Institutes of Health Grants AR R01 048632 and AI R01 049313; and National Institutes of Health Tetramer Facility Grant N01 AI25456.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112969108/-/DCSupplemental.
References
- 1.Moody DB, Porcelli SA. Intracellular pathways of CD1 antigen presentation. Nat Rev Immunol. 2003;3(1):11–22. doi: 10.1038/nri979. [DOI] [PubMed] [Google Scholar]
- 2.Joyce S, et al. Natural ligand of mouse CD1d1: Cellular glycosylphosphatidylinositol. Science. 1998;279:1541–1544. doi: 10.1126/science.279.5356.1541. [DOI] [PubMed] [Google Scholar]
- 3.Jackman RM, et al. The tyrosine-containing cytoplasmic tail of CD1b is essential for its efficient presentation of bacterial lipid antigens. Immunity. 1998;8:341–351. doi: 10.1016/s1074-7613(00)80539-7. [DOI] [PubMed] [Google Scholar]
- 4.Jayawardena-Wolf J, Benlagha K, Chiu YH, Mehr R, Bendelac A. CD1d endosomal trafficking is independently regulated by an intrinsic CD1d-encoded tyrosine motif and by the invariant chain. Immunity. 2001;15:897–908. doi: 10.1016/s1074-7613(01)00240-0. [DOI] [PubMed] [Google Scholar]
- 5.Kang SJ, Cresswell P. Regulation of intracellular trafficking of human CD1d by association with MHC class II molecules. EMBO J. 2002;21:1650–1660. doi: 10.1093/emboj/21.7.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sugita M, et al. Failure of trafficking and antigen presentation by CD1 in AP-3-deficient cells. Immunity. 2002;16:697–706. doi: 10.1016/s1074-7613(02)00311-4. [DOI] [PubMed] [Google Scholar]
- 7.Relloso M, et al. pH-dependent interdomain tethers of CD1b regulate its antigen capture. Immunity. 2008;28:774–786. doi: 10.1016/j.immuni.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Im JS, et al. Kinetics and cellular site of glycolipid loading control the outcome of natural killer T cell activation. Immunity. 2009;30:888–898. doi: 10.1016/j.immuni.2009.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brozovic S, et al. CD1d function is regulated by microsomal triglyceride transfer protein. Nat Med. 2004;10:535–539. doi: 10.1038/nm1043. [DOI] [PubMed] [Google Scholar]
- 10.Prigozy TI, et al. The mannose receptor delivers lipoglycan antigens to endosomes for presentation to T cells by CD1b molecules. Immunity. 1997;6:187–197. doi: 10.1016/s1074-7613(00)80425-2. [DOI] [PubMed] [Google Scholar]
- 11.Sugita M, Porcelli SA, Brenner MB. Assembly and retention of CD1b heavy chains in the endoplasmic reticulum. J Immunol. 1997;159:2358–2365. [PubMed] [Google Scholar]
- 12.Cox D, et al. Determination of cellular lipids bound to human CD1d molecules. PLoS ONE. 2009;4:e5325. doi: 10.1371/journal.pone.0005325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yuan W, Kang SJ, Evans JE, Cresswell P. Natural lipid ligands associated with human CD1d targeted to different subcellular compartments. J Immunol. 2009;182:4784–4791. doi: 10.4049/jimmunol.0803981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox LM, et al. Recognition of lyso-phospholipids by human natural killer T lymphocytes. PLoS Biol. 2009;7:e1000228. doi: 10.1371/journal.pbio.1000228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zajonc DM, Elsliger MA, Teyton L, Wilson IA. Crystal structure of CD1a in complex with a sulfatide self antigen at a resolution of 2.15 A. Nat Immunol. 2003;4:808–815. doi: 10.1038/ni948. [DOI] [PubMed] [Google Scholar]
- 16.Gadola SD, et al. Structure of human CD1b with bound ligands at 2.3 A, a maze for alkyl chains. Nat Immunol. 2002;3:721–726. doi: 10.1038/ni821. [DOI] [PubMed] [Google Scholar]
- 17.Zeng Z, et al. Crystal structure of mouse CD1: An MHC-like fold with a large hydrophobic binding groove. Science. 1997;277:339–345. doi: 10.1126/science.277.5324.339. [DOI] [PubMed] [Google Scholar]
- 18.Scharf L, et al. The 2.5 Å structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation. Immunity. 2010;33:853–862. doi: 10.1016/j.immuni.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Layre E, et al. A comparative lipidomics platform for chemotaxonomic analysis of Mycobacterium tuberculosis. Chemistry & Biology. 2011 doi: 10.1016/j.chembiol.2011.10.013. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rötzschke O, et al. Isolation and analysis of naturally processed viral peptides as recognized by cytotoxic T cells. Nature. 1990;348:252–254. doi: 10.1038/348252a0. [DOI] [PubMed] [Google Scholar]
- 21.Rudensky AYu, Preston-Hurlburt P, Hong SC, Barlow A, Janeway CA., Jr Sequence analysis of peptides bound to MHC class II molecules. Nature. 1991;353:622–627. [PubMed] [Google Scholar]
- 22.Cresswell P. Invariant chain structure and MHC class II function. Cell. 1996;84:505–507. doi: 10.1016/s0092-8674(00)81025-9. [DOI] [PubMed] [Google Scholar]
- 23.Shamshiev A, et al. Presentation of the same glycolipid by different CD1 molecules. J Exp Med. 2002;195:1013–1021. doi: 10.1084/jem.20011963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batuwangala T, et al. The crystal structure of human CD1b with a bound bacterial glycolipid. J Immunol. 2003;170:2382–2388. doi: 10.4049/jimmunol.172.4.2382. [DOI] [PubMed] [Google Scholar]
- 25.Cheng TY, et al. Role of lipid trimming and CD1 groove size in cellular antigen presentation. EMBO J. 2006;25:2989–2999. doi: 10.1038/sj.emboj.7601185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gilleron M, et al. Diacylated sulfoglycolipids are novel mycobacterial antigens stimulating CD1-restricted T cells during infection with Mycobacterium tuberculosis. J Exp Med. 2004;199:649–659. doi: 10.1084/jem.20031097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Alles LF, et al. Endogenous phosphatidylcholine and a long spacer ligand stabilize the lipid-binding groove of CD1b. EMBO J. 2006;25:3684–3692. doi: 10.1038/sj.emboj.7601244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moody DB, et al. Lipid length controls antigen entry into endosomal and nonendosomal pathways for CD1b presentation. Nat Immunol. 2002;3:435–442. doi: 10.1038/ni780. [DOI] [PubMed] [Google Scholar]
- 29.Zitomer NC, et al. Ceramide synthase inhibition by fumonisin B1 causes accumulation of 1-deoxysphinganine: A novel category of bioactive 1-deoxysphingoid bases and 1-deoxydihydroceramides biosynthesized by mammalian cell lines and animals. J Biol Chem. 2009;284:4786–4795. doi: 10.1074/jbc.M808798200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kasmar AG, et al. CD1b tetramers bind αβ. T cell receptors to identify a mycobacterial glycolipid-reactive T cell repertoire in humans. J Exp Med. 2011;208:1741–1747. doi: 10.1084/jem.20110665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Karadimitris A, et al. Human CD1d-glycolipid tetramers generated by in vitro oxidative refolding chromatography. Proc Natl Acad Sci USA. 2001;98:3294–3298. doi: 10.1073/pnas.051604498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaser A, et al. Microsomal triglyceride transfer protein regulates endogenous and exogenous antigen presentation by group 1 CD1 molecules. Eur J Immunol. 2008;38:2351–2359. doi: 10.1002/eji.200738102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moody DB, Zajonc DM, Wilson IA. Anatomy of CD1-lipid antigen complexes. Nat Rev Immunol. 2005;5:387–399. doi: 10.1038/nri1605. [DOI] [PubMed] [Google Scholar]
- 34.de Jong A, et al. CD1a-autoreactive T cells are a normal component of the human αβ T cell repertoire. Nat Immunol. 2010;11:1102–1109. doi: 10.1038/ni.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.