Abstract
Brefeldin A-inhibited guanine nucleotide-exchange protein (BIG) 1 activates class I ADP ribosylation factors (ARFs) by accelerating the replacement of bound GDP with GTP to initiate recruitment of coat proteins for membrane vesicle formation. Among proteins that interact with BIG1, kinesin family member 21A (KIF21A), a plus-end-directed motor protein, moves cargo away from the microtubule-organizing center (MTOC) on microtubules. Because KANK1, a protein containing N-terminal KN, C-terminal ankyrin-repeat, and intervening coiled-coil domains, has multiple actions in cells and also interacts with KIF21A, we explored a possible interaction between it and BIG1. We obtained evidence for a functional and physical association between these proteins, and found that the effects of BIG1 and KANK1 depletion on cell migration in wound-healing assays were remarkably similar. Treatment of cells with BIG1- or KANK1-specific siRNA interfered significantly with directed cell migration and initial orientation of Golgi/MTOC toward the leading edge, which was not mimicked by KIF21A depletion. Although colocalization of overexpressed KANK1 and endogenous BIG1 in HeLa cells was not clear microscopically, their reciprocal immunoprecipitation (IP) is compatible with the presence of small percentages of each protein in the same complexes. Depletion or overexpression of BIG1 protein appeared not to affect KANK1 distribution. Our data identify actions of both BIG1 and KANK1 in regulating cell polarity during directed migration; these actions are consistent with the presence of both BIG1 and KANK1 in dynamic multimolecular complexes that maintain Golgi/MTOC orientation, differ from those that might contain all three proteins (BIG1, KIF21A, and KANK1), and function in directed transport along microtubules.
Cell motility is crucial in diverse biological events, including embryonic development, immune surveillance, and wound healing (1, 2). Directed migration of cells growing on dishes, usually in response to external chemical or mechanical cues (3), requires complex and precisely coordinated actions, from initial polarization and extension of protrusions in the direction of movement to formation of adhesions at the leading edge, translocation of the cell body, and detachment with retraction from an extracellular substratum at the trailing edge (1, 4). Generation and maintenance of cell polarity, which are essential for directional migration, result from asymmetric membrane traffic achieved by cytoskeletal reorganization to direct secretory transport and delivery of additional membrane to the leading edge (5, 6). Positioning of Golgi and microtubule-organizing center (MTOC) structures anterior to the nucleus, facing the direction of forward movement, and supplementation of content at the cell front are important for coordination of intracellular traffic (7, 8). As elucidation of mechanisms of polarization and migration continues, more molecular participants are identified (3, 9, 10).
Brefeldin A (BFA)-inhibited guanine nucleotide-exchange protein (BIG) 1 (∼200 kDa), originally purified with BIG2 (∼190 kDa) in ∼670-kDa multiprotein complexes from bovine brain cytosol (11), activates class I ARFs (human ARF1 and 3) by catalyzing the replacement of ARF-bound GDP with GTP to enable critical vesicular transport (12–15). BIG1 is often found at trans-Golgi membranes (16, 17); after BIG1 depletion, membranes of the HepG2 cell Golgi trans face appeared less smooth by electron microscopy with more vesicle-like structures (18). Boal and Stephens also reported that Golgi structure was altered after BIG1 depletion (19). In addition to its Golgi association, BIG1 was accumulated in nuclei of serum-deprived HepG2 cells (20) or after their incubation with the immunosuppressive FK506 (21) or 8-Br-cAMP (22). In other conditions, BIG2 was associated with the trans-Golgi network and recycling endosomes (17, 23).
In addition to ARF activation by specific sequence in the central Sec7 domain (24), other parts of BIG1 molecules participate in multimolecular complexes with nucleolin, U3 small nucleolar RNA, and U3-binding protein fibrillarin or the RNA-binding protein La in different nuclear complexes (25). An A kinase-anchoring protein (AKAP) sequence in BIG1 is identical to one of three such sequences first identified in BIG2, and antibodies against RI or RII, regulatory subunits of cAMP-dependent protein kinase, coprecipitated endogenous BIG1 (26). ARF activation by BIG1 was decreased after its phosphorylation by PKA in vitro (27). Reported interactions of the C-terminal region of BIG1 with myosin IXb (28) and with kinesin KIF21A (29) suggest that BIG1 also plays a role in cargo movement along actin fibers or microtubules.
Kakinuma and Kiyama reported an interaction of KIF21A and KANK1 that influenced membrane localization of KANK1 (30). The human KANK1 gene was first described as a growth suppressor in renal carcinoma cells (31). Two KANK1 proteins, KANK1-L (∼175 kDa) and KANK1-S (∼160 kDa), produced by alternative splicing of exon 1 are present in many human tissues, and levels of KANK1-L in kidney tumors were lower than those in normal tissue (32). Predominantly cytoplasmic KANK1 (31, 33) interfered with actin polymerization by blocking interaction of insulin receptor substrate p53 (IRSp53) and Rac1 (34). Inhibition of RhoA activity via PI3K-Akt signaling regulated by KANK1 binding to 14-3-3 decreased actin stress fiber formation and cell migration (35). Participation of KANK1, with its nuclear export (NES) and nuclear localization (NLS) signals, in nucleocytoplasmic shuttling relevant to the activation of β-catenin-dependent transcription was also reported (36). Here, we describe the coimmunoprecipitation of endogenous BIG1 and KANK1 from HeLa cells and subsequent exploration of actions of the two proteins revealing that depletion of either one interfered seriously with regulation of HeLa cell polarity during directed cell migration in wound-healing assays. BIG1 and KANK1 apparently use, at least in part, the same pathway to produce those effects, although no evidence of their direct physical interaction in cells was obtained.
Results
Interaction of BIG1 with KANK1.
To begin to assess KANK1 interaction with BIG1, we demonstrated its presence among proteins immunoprecipitated from HepG2 cells with antibodies against BIG1. The finding was confirmed in HeLa cells, which we then used instead of HepG2 cells because of their higher transfection efficiency and faster proliferation. BIG1 or BIG2 was partially coprecipitated from cell lysates by antibodies against the other protein. KIF21A was also among proteins precipitated with BIG1 antibodies (29, 37). Endogenous KANK1 specifically coprecipitated with BIG1 antibodies, but not BIG2 or control IgG (Fig. 1A). Immunoprecipitation (IP) of KANK1 yielded also BIG1 and KIF21A (Fig. 1B), and again, no BIG2 was detected; this result is consistent with the presence of BIG1 and KANK1 together in some multimolecular complexes that differ from those containing both BIG1 and BIG2.
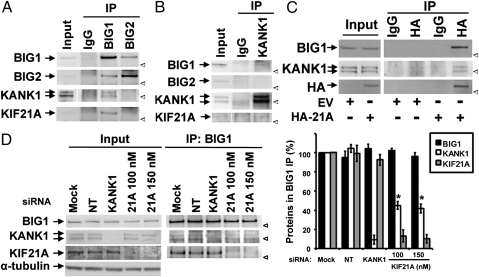
Fig. 1.
Immunoprecipitation of KANK1 with BIG1. (A and B) Samples of Input (2.5% of total) and 25% of proteins from IP with antibodies against BIG1, BIG2, or KANK1, or control IgG from extracts of HeLa cells (1 mg) were separated by SDS/PAGE before reaction of Western blots with indicated antibodies. (A and B are enlarged in Fig. S1.) (C) Samples (50%) of proteins precipitated with anti-HA antibodies or control IgG from 200 μg of extracts prepared 24 h after transfection of cells with HA-KIF21A (HA-21A) or empty vector (EV) were used for Western blotting with indicated antibodies. Input (20 μg) was 10% of amount used for IP. (D) Cells transfected with 100 nM nontargeted (NT) or KANK1-specific siRNA or 100 or 150 nM KIF21A-specific siRNA or with vehicle alone (Mock) were lysed 48 h later. Proteins precipitated with antibodies against BIG1 were analyzed by Western blotting and densitometric quantification. Amounts of proteins from BIG1 IP in three experiments expressed relative to that of the same protein in Mock cells (=100%) are means ± SEM *P < 0.005 vs. Mock. Arrow: protein band. Arrowhead: position of 160-kDa marker.
Because we did not have KIF21A antibodies specific enough for IP of endogenous protein, we transiently overexpressed HA-KIF21A, which enabled co-IP of BIG1 and KANK1 along with HA-tagged KIF21A from those cells but not from cells expressing empty vector (Fig. 1C). These results were in agreement with earlier reports of BIG1-KIF21A (29) and KANK1-KIF21A associations (30), and also with the possibility of KANK1 as a component of some BIG1 complexes. In further experiments, we used siRNA that decreased endogenous KANK1 to 4 ± 3% and KIF21A to 9 ± 5% of control cells (Fig. S1C). Co-IP of endogenous KIF21A with BIG1 was not altered after KANK1 depletion, but BIG1 IP after KIF21A depletion yielded significantly less KANK1 than it did from control cells (Fig. 1D), consistent with the notion that co-IP of KANK1 with BIG1 was due, in some part, to its interaction with KIF21A (30). The small amounts of KANK1 in BIG1 IP from cells after ∼90% depletion of KIF21A were still ∼40% of that from control cells (Fig. 1D); however, this result is compatible with the existence of BIG1 and KANK1 in complexes different from those that include KIF21A.
Because the C-terminal amino acids 886–1849 of BIG1 had interacted with KIF21A (29), we used anti-tag antibodies to investigate co-IP of KANK1-myc and HA-BIG1 or its fragments coexpressed in HeLa cells (Fig. S2B). Despite differences in levels of expression of the BIG1 constructs, reciprocal IP of both N- and C-terminal fragments (but not Sec7) with KANK1-myc was found. We concluded that any direct interaction of BIG1 and KANK1 molecules could involve only very small fractions of both proteins and cannot involve those complexes of BIG1 that include BIG2.
Intracellular Localizations of BIG1 and KANK1.
KANK1 is widely viewed as a cytosolic protein, whereas BIG1 is most often associated with Golgi structures (29, 37) identified by specific Golgi or TGN markers (Fig. S3). However, the presence of small amounts of each protein at other intracellular loci has been clearly shown. Our questions about direct interaction of BIG1 and KANK1 molecules prompted a search for clues to previously unrecognized loci of one or the other. We assessed endogenous protein distributions by iodixanol density gradient fractionation, confirming very little overlap between largely cytosolic KANK1 and the essentially single peak of BIG1 in fractions 7–10 (Fig. S4). Amounts of endogenous BIG1 or KANK1 were markedly reduced in cells treated, respectively, with BIG1- or KANK1-specific siRNAs, although distributions of the proteins and endogenous βCOP, BiP, and α-tubulin were similar to those in control cells transfected with nonspecific siRNA.
Microscopically, endogenous BIG1 was prominently clustered in a perinuclear region and scattered through the cytoplasm (29, 37). Insufficient specificity and sensitivity of our antibodies precluded identification of endogenous KANK1. Overexpressed untagged KANK1, in about sevenfold the endogenous amount, was seen at membrane ruffles and throughout the cytoplasm (Fig. 2). To further explore possible effects of BIG1 levels on distribution of KANK1, we overexpressed HA-BIG1 plus untagged KANK1 in HeLa cells (Fig. S5). Like endogenous BIG1, HA-BIG1 was seen in a punctate distribution with perinuclear concentration (Fig. S5B), and its amount did not apparently affect KANK1 localization (Fig. S5C and Fig. 2). Overall, none of our experiments provided unequivocal evidence of direct interactions or reciprocal effects of total cell content of BIG1 or KANK1.
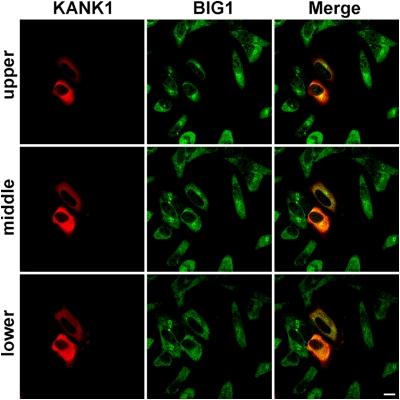
Fig. 2.
Endogenous BIG1 and overexpressed KANK1 in HeLa cells. After transfection (24 h) with cDNA encoding untagged KANK1, cells were fixed, reacted with rabbit anti-BIG1 and mouse anti-KANK1 antibodies, and prepared for confocal immunofluorescence microscopy. In three Z-stack images of 1-μm planes (upper, middle, and lower), overexpressed KANK1 (red) did not apparently coincide with or alter the distribution of endogenous BIG1 (green), which is scattered in punctate collections through cytoplasm of all cells, with greatest perinuclear concentration in the upper plane. (Scale bar, 10 μm.)
Effects of BIG1 and KANK1 on Wound Healing.
Depletion of endogenous BIG1 interfered with HepG2 cell attachment to collagen as well as COS7 cell transwell migration (18). Kiyama and coworkers showed that KANK1 influenced actin cytoskeleton reorganization and lamellipodium formation via RhoA and Rac activation (34, 35). To assess potential roles for BIG1–KANK1 interaction in cell motility, we evaluated migration in wound-healing assays after treatment of cells with vehicle alone (Mock) or control nontargeted or specific siRNAs (Fig. 3A). Endogenous BIG1, BIG2, KANK1, and KIF21A levels were only 14 ± 6%, 9 ± 3%, 14 ± 5%, and 11 ± 2%, respectively, of control (Mock) cells (mean ± SEM, n = 3) 48 h after the addition of specific cognate siRNAs. Wound area covered by migrating monolayer cells was quantified 6 h after wounding. BIG1, BIG2, or KANK1 siRNA treatments each delayed wound closure (Fig. 3 B and C), but KIF21A depletion did not significantly alter any of the migration characteristics quantified in wound-healing experiments.
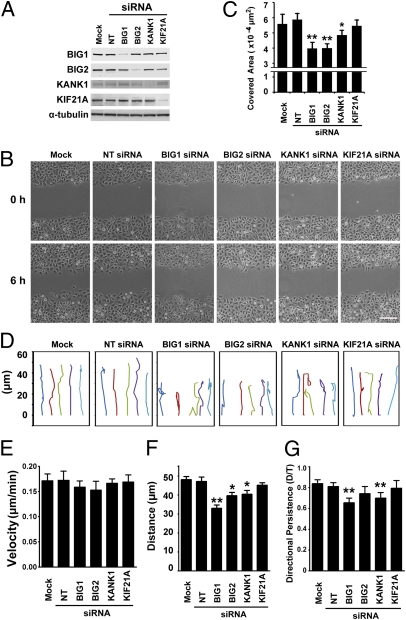
Fig. 3.
Effects of BIG1, BIG2, KANK1, or KIF21A depletion on wound healing. (A) Cells were lysed 48 h after transfection with nontargeted (NT), BIG1, BIG2, KANK1, or KIF21A siRNA or vehicle alone (Mock) and amounts of proteins quantified by densitometry of Western blots. (B) Images of cells at 0 and 6 h after wounding as in A. (Scale bar, 70 μm.) (C) Covered area is the difference between wound areas 0 and 6 h after wounding. Data are means ± SEM of values from five experiments. **P < 0.005 *; P < 0.05, (two-tailed t test) for difference from cells transfected with nontargeted siRNA. (D) Migration paths of five representative cells at wound edges during 6-h assays (B). Initiation of cell migration = 0. (E–G) Motility characteristics in B and C were calculated for individual cells as described in Materials and Methods. Data are means ± SEM of values from three experiments for at least 120–150 cells in each group. **P < 0.005; *P < 0.05, (ANOVA test) for difference from cells transfected with vehicle alone (Mock).
Individual cell paths (Fig. 3D) recorded by tracing the centers of cell nuclei from time-lapse images and analyzing trajectories of individual cells at the wound edge over a 6-h period are especially useful for seeing flaws in directed migration. We found that velocity (total length of single cell path in 6 h) was not significantly altered by BIG1, BIG2, KANK1, or KIF21A depletion (Fig. 3E), suggesting an absence of effects on cell motility.
Net translocation toward wound closure of cells transfected with BIG1-, BIG2-, or KANK1-specific siRNA was significantly less than that of control cells (Fig. 3F), but only cells depleted of BIG1 or KANK1 made directional changes significantly more often than did control cells. Depletion of BIG1 or KANK1 appeared to delay wound healing by interfering with directional persistence, defined as the distance (D) between cell start and end points divided by total migration path length (T) during 6-h assays (Fig. 3G) of cell migration. Although some data in Fig. 3 seem to show effects of KIF21A depletion, none of these reached statistical significance; taken together, the data indicate that delayed wound healing by cells depleted of BIG1 or KANK1 resulted from defects in persistence of polarization, rather than velocity of cell movement.
BIG1 and KANK1 Contribute to Cell Polarization During Directed Migration.
Polarized morphology of wound-edge cells is achieved by translocation of MTOC and Golgi to precede the nucleus in cells oriented to the direction of migration (7, 8). This enhances microtubule growth to the lamella, enabling vesicle transport to extend protrusions at the leading edge (2, 5) and establish polarity. To assess these actions, the positions of Golgi (Fig. 4 A and C) and MTOC (Fig. 4 B and C) in wound-edge cells were recorded 6 h after wounding. Only in cells depleted of BIG1 or KANK1 were positions of Golgi marker GM130 and MTOC (immunoreactive γ-tubulin in the perinuclear region) significantly different from those in controls (Mock or nontargeted siRNA). Aberrant Golgi positions appeared correlated with extent of BIG1 or KANK1 depletion (Fig. S6E), and effects of maximal depletion of either one were not greater with additional depletion of the other (Fig. S6 F and G).
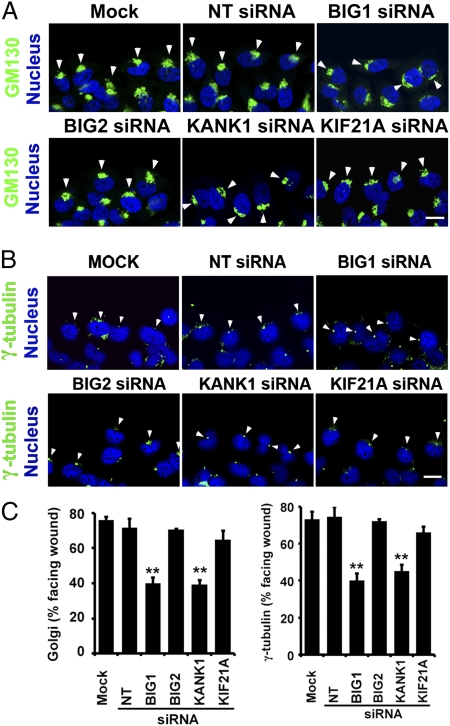
Fig. 4.
Depletion of BIG1 or KANK1 interfered with cell polarization during wound healing. Confluent monolayers of HeLa cells transfected 48 h before with indicated siRNA or vehicle alone (Mock), as in Fig.3, were wounded and fixed 6 h later for staining with DAPI and anti-GM130 (A) or anti-γ-tubulin antibodies (B) and confocal immunofluorescence microscopy. Arrowheads indicate GM130 (A) or γ-tubulin (B). (Scale bar, 10 μm.) (C) Percentage of wound-edge cells with Golgi or γ-tubulin structures in forward-facing 120° sector between nucleus and wound was recorded for at least 100 cells of each population for Golgi localization and 30 cells for γ-tubulin in each experiment. Data are means ± SEM of values from six experiments. **P < 0.02.
Polarization of Golgi and MTOC persisted in cells depleted of BIG2 or KIF21A, but was grossly disturbed by BIG1 or KANK1 siRNA treatment. BIG1 and KANK1 were each important for establishing cell polarity and preserving correctly directed cell movement during wound healing. The lack of significant effects of KIF21A knockdown on those characteristics means there was no evidence that KIF21A was critical for effects of BIG1 or KANK1 on orientation of Golgi/MTOC and cell polarity (Fig. 4C).
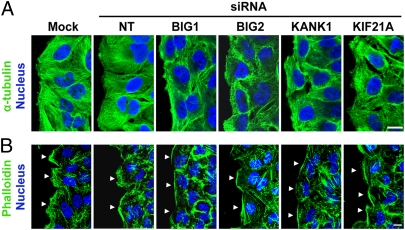
Effects of BIG1 or KANK1 depletion on microtubule (α-tubulin) and actin (phalloidin) cytoskeletons were also evaluated (Fig. 5 A and B). Microtubules in control cells were aligned perpendicular to the leading edge and extended well behind it, but were short and failed to form organized networks in cells depleted of BIG1 or KANK1. Effects of BIG2 or KIF21A siRNAs, although similar to one another, were clearly different from the phenotype of cells depleted of BIG1 or KANK1 (Fig. 5A). Stress fibers are less prominent in HeLa cells than in fibroblasts, but differences in F-actin patterns at leading edges of control cells (densely populated with actin-rich membrane ruffles) and BIG1- or KANK1-depleted cells, without evidence of F-actin-based ruffles and with nonuniform, irregular phalloidin staining, were obvious (Fig. 5B). Microscopically, both actin and microtubule skeletons differed after BIG2 or KIF21A depletion from those of control cells (Mock or nontargeted siRNA). However, they were also quite distinct from patterns in BIG1- and KANK1-depleted cells. The BIG2- and KIF21A-depleted cells still exhibited protrusions toward the wound edge and F-actin-based ruffles within the leading edge of migrating cells.
Fig. 5.
Effects of BIG1, BIG2, KANK1, or KIF21A depletion on intracellular microtubule and actin morphology 6 h after wounding. HeLa cells treated as in Figs. 3 and 4 were fixed 6 h after wounding and reacted with anti-α-tubulin antibodies to mark microtubules (A) or Alexa Fluor 488-conjugated phalloidin for F-actin (B). Patterns of microtubules and F-actin were altered in cells depleted of any of the four proteins, but effects on F-actin were most obvious and prominent in cells treated with BIG1 or KANK1 siRNA. (Scale bar, 10 μm.) Arrowheads indicate wound-edge membrane.
Although most of the events quantified in wound-healing assays were not significantly altered, at least some effects of BIG2 or KIF21A depletion may be recognized as functionally relevant when we better understand the mechanisms through which these and numerous additional molecules act to achieve directed cell migration via integrated operations of diverse multimolecular machines throughout the cell.
Discussion
Participation in macromolecular complexes is required for many BIG1 and KANK1 functions. Copurification of BIG1 and BIG2 in ∼670-kDa complexes (37) and formation of homo- or heterodimers in multimolecular complexes (38) was suggested by structures of specific domains of the two molecules. However, redundancy of their actions at TGN (39) seems rather less likely in light of the apparently long evolutionary histories and evident functional differentiation of the two BIG molecules, along with those of their specific substrates, such as human ARF1 and ARF3, which differ by only 7 of 181 amino acids. Ishizaki et al. (39) found that depletion of BIG1 alone had no obvious effects on the location of Golgi or endosomal proteins that were examined, whereas depletion of BIG2 alone induced extension from recycling endosomes of tubular structures that reacted with antibodies against TfnR, AP-1, and Rab11. However, depletion of both BIG1 and BIG2 led to redistribution of proteins that were otherwise seen in TGN (e.g., TGN46, AP-1) or recycling endosomes and blocked retrograde trafficking of furin from late endosomes to the TGN, consistent with, as Ishizaki et al. wrote, “redundant roles of BIG1 and BIG2 in membrane traffic between the trans-Golgi network and endosomes” (39). Several other functions of BIG1 and BIG2 in cell organization and membrane trafficking seem clearly not redundant; for example, BIG1 was required to maintain usual morphology of the TGN (18), and BIG2 was important for endosome integrity (17, 39). Perhaps observations like those of Ishizaki et al. (39) are more likely due to the extent of “cross-talk” among pathways involving these molecules as seen after siRNA-knockdown of pairs of different ARF molecules rather than the effects of single siRNAs (40).
Here, we failed to demonstrate direct interaction of BIG1 with KANK1, and concluded that BIG1 and KANK1 might exist together in multimolecular complexes different from those containing both BIG1 and BIG2. Similarly, the lack of effect of BIG2 depletion on Golgi/MTOC orientation in wound healing likely reflects differing functional interactions of BIG1 and BIG2. Kakinuma and Kiyama reported that KIF21A interacted directly with KANK1 (30), and the level of KIF21A protein significantly influenced the extent of KANK1 co-IP with BIG1 antibodies (Fig. 1D), compatible with the existence of complexes containing BIG1, KIF21A, and KANK1. Depletion of KIF21A altered BIG1 and KANK1 distributions without changing the microscopic appearance of intrinsic Golgi proteins (29, 30), suggesting that BIG1 might act as a scaffold as well as a partner in KIF21A-powered intracellular transport (29). We found no change in KANK1 distribution after BIG1 depletion, although our methodology (Fig. S4) may have been inadequate for detection of differences in associated molecules or location of any small fraction of the total KANK1. The effect of KIF21A depletion on migration in wound healing did differ, at least quantitatively, from that of BIG1 or KANK1 depletion. Molecular assemblies containing BIG1, KIF21A, and KANK1 could exist during transport, without being responsible for effects of BIG1 or KANK1 depletion on Golgi/MTOC orientation. We showed that the percentage of cells with distorted Golgi orientation during wound healing migration was no greater after depletion of both BIG1 and KANK1 than after either one alone, suggesting that BIG1 and KANK1 affect Golgi polarity via the same pathways. However, further studies are required to exclude the possibility that either one might use an additional mechanism to influence cell polarity.
We described here a previously unrecognized role for BIG1 in the regulation of Golgi/MTOC orientation. Further studies are needed to learn whether or how signaling upstream of BIG1 or KANK1 controls cell polarity and to understand mechanisms used by both proteins in maintenance or remodeling of Golgi structure. Actin network action in Golgi structure and function has been demonstrated (41). Cdc42 and ARF1 effects on short actin filament dynamics in a Golgi complex pool could contribute to the polarization of intracellular trafficking (42, 43). BIG1 interaction with myosin IXb (28) would be consistent with participation of the motor molecule in maintenance of Golgi morphology via function of the actin cytoskeleton. The report of myosin IXb control of murine macrophage shape and motility via its RhoGAP activity (44) appears directly relevant to the demonstrated effects of BIG1, and perhaps KANK1, depletion on directional persistence of HeLa cell movement.
KANK1 is known to influence the actin cytoskeleton, but the extent to which it contributes to cytoskeleton function in maintaining Golgi structure and protein translocation remains to be established. Recently, Arai et al. (45) described an intrinsic phosphoinositide signaling system in Dictyostelium that is responsible and required for random cell migration. Stochastic variations in activities of phosphoinositide-3-kinase (PI3K) and PTEN (phosphatase and tensin homolog) produced temporally and spatially confined waves of individual phosphoinositide concentrations. Mechanisms that coordinate these phenomena with actin-based cell movement remain to be elucidated, as do the similarities between these systems in HeLa and Dictyostelium cells.
Regulation of actin polymerization is among KANK1 functions seemingly involved in cell migration (33). Potential routes of KANK1 influence on actin remodeling include PI3K/Akt signaling via 14-3-3 in RhoA regulation (35) and effects of lower Rac1 activity after interaction with IRSp53 (34). Depletion of KANK1 increased RhoA activity and enhanced insulin-stimulated single cell movement in transwell filters coated with entactin-collagen IV-laminin (35), whereas overexpression of GFP-KANK1 inhibited random migration of NIH 3T3 cells (33). It will be important to learn why our data regarding effects of KANK1 depletion on cell migration appear inconsistent with such reports (33–35). In our wound-healing experiments, KANK1 depletion clearly interfered with persistence of directional migration, which requires mechanisms more complicated than those of random or single cell migration (46, 47) for which self-organized, intrinsic phosphoinositide signaling may suffice (45). Continuity of directional translocation and lamellipodium structure may involve numerous regulatory influences; for example, the nature of extracellular matrix, stability of cell polarity, and multiple, diverse intra- and intercell signals (3, 46, 47). Whether parallel differences in cell mobility and KANK1 content reflect differences in cell type, study conditions, or the cues guiding migration remain to be determined. Our present fragmentary understanding of specific molecular forms of KANK1 (and other proteins) with their posttranslation modifications is probably equally important.
Inhibition of cell migration by depletion of BIG2 could be related to effects of BIG2 depletion on transferrin receptor recycling (17, 23) or its interaction with Exo70, a component of the exocyst complex required for exocytosis (48). Exo70 interacted with the Arp2/3 complex, a key regulator of actin polymerization, and its inhibition blocked formation of actin-based membrane protrusions plus other aspects of cell locomotion (49). Information on exocyst participation in returning internalized cycling proteins to the cell surface is also needed. Similarly, understanding mechanisms through which cAMP/PKA signals control or coordinate BIG1, BIG2, and/or KANK1 function in actin-based cell migration will surely be important. One or more AKAP sequences in both BIG1 and BIG2 (26) enable them to compartmentalize PKA with other molecular complexes relevant to cAMP metabolism and action, thereby contributing to the critical spatial and temporal specificity of cAMP signaling and/or action (27, 50, 51). Reversible PKA-catalyzed phosphorylation of BIG1 and BIG2 inhibits their ARF GEF activity and might be relevant to a KANK1-14-3-3 interaction (27, 35).
Roles for both BIG1 and KANK1, which appear to act in part via the same pathway(s) to regulate HeLa cell polarity during oriented motion, seem clear. Direct interaction of the two molecules was not proven, although co-IP of in vitro synthesized KANK1-myc and HA-BIG1 was consistent with both BIG1 N and C termini binding to KANK1. Unequivocal characterization in cells of such interactions, involving only small fractions of total endogenous protein, will likely require the remarkable spatial and temporal resolution of microscopic techniques like those now used by Waterman and Lippincott-Schwartz (52–54). Identification of additional components of BIG1/KANK1 functional complexes and elucidation of detailed molecular mechanisms that accomplish directional migration, including potential BIG1/KANK1 interactions, require further studies.
Materials and Methods
Sources of antibodies and other specific reagents not otherwise identified are reported in SI Materials and Methods, along with details of immunoprecipitation and procedures for prior, reversible cross-linking of protein complexes. HeLa cells (from American Type Culture Collection) were grown, and wound-healing assays for analyses of directed cell migration as well as confocal immunofluorescence microscopy experiments were performed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We are grateful to members of the Cell Biology and Physiology Center, National Heart, Lung and Blood Institute (NHLBI), National Institutes of Health (NIH) for invaluable help in time-lapse imaging and to Drs. Daniela Malide and Christian Combs (Light Microscopy Core Facility, NHLBI) for their much appreciated assistance in confocal microscopy. This research was supported by the Intramural Research Program of the NIH, NHLBI.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1117011108/-/DCSupplemental.
References
- 1.Ridley AJ, et al. Cell migration: Integrating signals from front to back. Science. 2003;302:1704–1709. doi: 10.1126/science.1092053. [DOI] [PubMed] [Google Scholar]
- 2.Raftopoulou M, Hall A. Cell migration: Rho GTPases lead the way. Dev Biol. 2004;265:23–32. doi: 10.1016/j.ydbio.2003.06.003. [DOI] [PubMed] [Google Scholar]
- 3.Petrie RJ, Doyle AD, Yamada KM. Random versus directionally persistent cell migration. Nat Rev Mol Cell Biol. 2009;10:538–549. doi: 10.1038/nrm2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weijer CJ. Collective cell migration in development. J Cell Sci. 2009;122:3215–3223. doi: 10.1242/jcs.036517. [DOI] [PubMed] [Google Scholar]
- 5.Etienne-Manneville S. Cdc42—the centre of polarity. J Cell Sci. 2004;117:1291–1300. doi: 10.1242/jcs.01115. [DOI] [PubMed] [Google Scholar]
- 6.Etienne-Manneville S, Hall A. Cell polarity: Par6, aPKC and cytoskeletal crosstalk. Curr Opin Cell Biol. 2003;15:67–72. doi: 10.1016/s0955-0674(02)00005-4. [DOI] [PubMed] [Google Scholar]
- 7.Kupfer A, Louvard D, Singer SJ. Polarization of the Golgi apparatus and the microtubule-organizing center in cultured fibroblasts at the edge of an experimental wound. Proc Natl Acad Sci USA. 1982;79:2603–2607. doi: 10.1073/pnas.79.8.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes ER, Jani S, Gundersen GG. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 9.Sütterlin C, Colanzi A. The Golgi and the centrosome: Building a functional partnership. J Cell Biol. 2010;188:621–628. doi: 10.1083/jcb.200910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iden S, Collard JG. Crosstalk between small GTPases and polarity proteins in cell polarization. Nat Rev Mol Cell Biol. 2008;9:846–859. doi: 10.1038/nrm2521. [DOI] [PubMed] [Google Scholar]
- 11.Morinaga N, Tsai SC, Moss J, Vaughan M. Isolation of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP ribosylation factor (ARF) 1 and ARF3 that contains a Sec7-like domain. Proc Natl Acad Sci USA. 1996;93:12856–12860. doi: 10.1073/pnas.93.23.12856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jackson CL, Casanova JE. Turning on ARF: The Sec7 family of guanine-nucleotide-exchange factors. Trends Cell Biol. 2000;10:60–67. doi: 10.1016/s0962-8924(99)01699-2. [DOI] [PubMed] [Google Scholar]
- 13.Morinaga N, Adamik R, Moss J, Vaughan M. Brefeldin A inhibited activity of the sec7 domain of p200, a mammalian guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem. 1999;274:17417–17423. doi: 10.1074/jbc.274.25.17417. [DOI] [PubMed] [Google Scholar]
- 14.Mansour SJ, et al. p200 ARF-GEP1: A Golgi-localized guanine nucleotide exchange protein whose Sec7 domain is targeted by the drug brefeldin A. Proc Natl Acad Sci USA. 1999;96:7968–7973. doi: 10.1073/pnas.96.14.7968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moss J, Vaughan M. Molecules in the ARF orbit. J Biol Chem. 1998;273:21431–21434. doi: 10.1074/jbc.273.34.21431. [DOI] [PubMed] [Google Scholar]
- 16.Zhao X, Lasell TK, Melançon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: Evidence for distinct functions in protein traffic. Mol Biol Cell. 2002;13:119–133. doi: 10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shen X, et al. Association of brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) with recycling endosomes during transferrin uptake. Proc Natl Acad Sci USA. 2006;103:2635–2640. doi: 10.1073/pnas.0510599103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shen X, Hong MS, Moss J, Vaughan M. BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein, is required for correct glycosylation and function of integrin beta1. Proc Natl Acad Sci USA. 2007;104:1230–1235. doi: 10.1073/pnas.0610535104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Boal F, Stephens DJ. Specific functions of BIG1 and BIG2 in endomembrane organization. PLoS ONE. 2010;5:e9898. doi: 10.1371/journal.pone.0009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Padilla PI, Pacheco-Rodriguez G, Moss J, Vaughan M. Nuclear localization and molecular partners of BIG1, a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. Proc Natl Acad Sci USA. 2004;101:2752–2757. doi: 10.1073/pnas.0307345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Padilla PI, et al. Interaction of FK506-binding protein 13 with brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1): Effects of FK506. Proc Natl Acad Sci USA. 2003;100:2322–2327. doi: 10.1073/pnas.2628047100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Citterio C, et al. Effect of protein kinase A on accumulation of brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) in HepG2 cell nuclei. Proc Natl Acad Sci USA. 2006;103:2683–2688. doi: 10.1073/pnas.0510571103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin HW, Morinaga N, Noda M, Nakayama K. BIG2, a guanine nucleotide exchange factor for ADP-ribosylation factors: Its localization to recycling endosomes and implication in the endosome integrity. Mol Biol Cell. 2004;15:5283–5294. doi: 10.1091/mbc.E04-05-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Togawa A, Morinaga N, Ogasawara M, Moss J, Vaughan M. Purification and cloning of a brefeldin A-inhibited guanine nucleotide-exchange protein for ADP-ribosylation factors. J Biol Chem. 1999;274:12308–12315. doi: 10.1074/jbc.274.18.12308. [DOI] [PubMed] [Google Scholar]
- 25.Padilla PI, et al. Association of guanine nucleotide-exchange protein BIG1 in HepG2 cell nuclei with nucleolin, U3 snoRNA, and fibrillarin. Proc Natl Acad Sci USA. 2008;105:3357–3361. doi: 10.1073/pnas.0712387105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Adamik R, Pacheco-Rodriguez G, Moss J, Vaughan M. Protein kinase A-anchoring (AKAP) domains in brefeldin A-inhibited guanine nucleotide-exchange protein 2 (BIG2) Proc Natl Acad Sci USA. 2003;100:1627–1632. doi: 10.1073/pnas.0337678100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuroda F, Moss J, Vaughan M. Regulation of brefeldin A-inhibited guanine nucleotide-exchange protein 1 (BIG1) and BIG2 activity via PKA and protein phosphatase 1gamma. Proc Natl Acad Sci USA. 2007;104:3201–3206. doi: 10.1073/pnas.0611696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saeki N, Tokuo H, Ikebe M. BIG1 is a binding partner of myosin IXb and regulates its Rho-GTPase activating protein activity. J Biol Chem. 2005;280:10128–10134. doi: 10.1074/jbc.M413415200. [DOI] [PubMed] [Google Scholar]
- 29.Shen X, et al. Interaction of brefeldin A-inhibited guanine nucleotide-exchange protein (BIG) 1 and kinesin motor protein KIF21A. Proc Natl Acad Sci USA. 2008;105:18788–18793. doi: 10.1073/pnas.0810104105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kakinuma N, Kiyama R. A major mutation of KIF21A associated with congenital fibrosis of the extraocular muscles type 1 (CFEOM1) enhances translocation of Kank1 to the membrane. Biochem Biophys Res Commun. 2009;386:639–644. doi: 10.1016/j.bbrc.2009.06.109. [DOI] [PubMed] [Google Scholar]
- 31.Sarkar S, et al. A novel ankyrin repeat-containing gene (Kank) located at 9p24 is a growth suppressor of renal cell carcinoma. J Biol Chem. 2002;277:36585–36591. doi: 10.1074/jbc.M204244200. [DOI] [PubMed] [Google Scholar]
- 32.Wang Y, et al. Alternative splicing of the human Kank gene produces two types of Kank protein. Biochem Biophys Res Commun. 2005;330:1247–1253. doi: 10.1016/j.bbrc.2005.03.106. [DOI] [PubMed] [Google Scholar]
- 33.Kakinuma N, Zhu Y, Wang Y, Roy BC, Kiyama R. Kank proteins: Structure, functions and diseases. Cell Mol Life Sci. 2009;66:2651–2659. doi: 10.1007/s00018-009-0038-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roy BC, Kakinuma N, Kiyama R. Kank attenuates actin remodeling by preventing interaction between IRSp53 and Rac1. J Cell Biol. 2009;184:253–267. doi: 10.1083/jcb.200805147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kakinuma N, Roy BC, Zhu Y, Wang Y, Kiyama R. Kank regulates RhoA-dependent formation of actin stress fibers and cell migration via 14-3-3 in PI3K-Akt signaling. J Cell Biol. 2008;181:537–549. doi: 10.1083/jcb.200707022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y, Kakinuma N, Zhu Y, Kiyama R. Nucleo-cytoplasmic shuttling of human Kank protein accompanies intracellular translocation of beta-catenin. J Cell Sci. 2006;119:4002–4010. doi: 10.1242/jcs.03169. [DOI] [PubMed] [Google Scholar]
- 37.Yamaji R, et al. Identification and localization of two brefeldin A-inhibited guanine nucleotide-exchange proteins for ADP-ribosylation factors in a macromolecular complex. Proc Natl Acad Sci USA. 2000;97:2567–2572. doi: 10.1073/pnas.97.6.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramaen O, et al. Interactions between conserved domains within homodimers in the BIG1, BIG2, and GBF1 Arf guanine nucleotide exchange factors. J Biol Chem. 2007;282:28834–28842. doi: 10.1074/jbc.M705525200. [DOI] [PubMed] [Google Scholar]
- 39.Ishizaki R, Shin HW, Mitsuhashi H, Nakayama K. Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol Biol Cell. 2008;19:2650–2660. doi: 10.1091/mbc.E07-10-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Volpicelli-Daley LA, Li Y, Zhang CJ, Kahn RA. Isoform-selective effects of the depletion of ADP-ribosylation factors 1-5 on membrane traffic. Mol Biol Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.De Matteis MA, Luini A. Exiting the Golgi complex. Nat Rev Mol Cell Biol. 2008;9:273–284. doi: 10.1038/nrm2378. [DOI] [PubMed] [Google Scholar]
- 42.Cao H, et al. Actin and Arf1-dependent recruitment of a cortactin-dynamin complex to the Golgi regulates post-Golgi transport. Nat Cell Biol. 2005;7:483–492. doi: 10.1038/ncb1246. [DOI] [PubMed] [Google Scholar]
- 43.Dubois T, et al. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat Cell Biol. 2005;7:353–364. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
- 44.Hanley PJ, et al. Motorized RhoGAP myosin IXb (Myo9b) controls cell shape and motility. Proc Natl Acad Sci USA. 2010;107:12145–12150. doi: 10.1073/pnas.0911986107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Arai Y, et al. Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proc Natl Acad Sci USA. 2010;107:12399–12404. doi: 10.1073/pnas.0908278107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Friedl P, Gilmour D. Collective cell migration in morphogenesis, regeneration and cancer. Nat Rev Mol Cell Biol. 2009;10:445–457. doi: 10.1038/nrm2720. [DOI] [PubMed] [Google Scholar]
- 47.Mayor R, Carmona-Fontaine C. Keeping in touch with contact inhibition of locomotion. Trends Cell Biol. 2010;20:319–328. doi: 10.1016/j.tcb.2010.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu KF, et al. Interaction of BIG2, a brefeldin A-inhibited guanine nucleotide-exchange protein, with exocyst protein Exo70. Proc Natl Acad Sci USA. 2005;102:2784–2789. doi: 10.1073/pnas.0409871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuo X, et al. Exo70 interacts with the Arp2/3 complex and regulates cell migration. Nat Cell Biol. 2006;8:1383–1388. doi: 10.1038/ncb1505. [DOI] [PubMed] [Google Scholar]
- 50.Wong W, Scott JD. AKAP signalling complexes: Focal points in space and time. Nat Rev Mol Cell Biol. 2004;5:959–970. doi: 10.1038/nrm1527. [DOI] [PubMed] [Google Scholar]
- 51.Carnegie GK, Means CK, Scott JD. A-kinase anchoring proteins: From protein complexes to physiology and disease. IUBMB Life. 2009;61:394–406. doi: 10.1002/iub.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanchanawong P, et al. Nanoscale architecture of integrin-based cell adhesions. Nature. 2010;468:580–584. doi: 10.1038/nature09621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Manley S, et al. High-density mapping of single-molecule trajectories with photoactivated localization microscopy. Nat Methods. 2008;5:155–157. doi: 10.1038/nmeth.1176. [DOI] [PubMed] [Google Scholar]
- 54.Betzig E, et al. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.