Abstract
Smad proteins are cytoplasmic signaling effectors of transforming growth factor-β (TGF-β) family cytokines and regulate gene transcription in the nucleus. Receptor-activated Smads (R-Smads) become phosphorylated by the TGF-β type I receptor. Rapid and precise transport of R-Smads to the nucleus is of crucial importance for signal transduction. By focusing on the R-Smad Smad3 we demonstrate that 1) only activated Smad3 efficiently enters the nucleus of permeabilized cells in an energy- and cytosol-dependent manner. 2) Smad3, via its N-terminal domain, interacts specifically with importin-β1 and only after activation by receptor. In contrast, the unique insert of exon3 in the N-terminal domain of Smad2 prevents its association with importin-β1. 3) Nuclear import of Smad3 in vivo requires the action of the Ran GTPase, which mediates release of Smad3 from the complex with importin-β1. 4) Importin-β1, Ran, and p10/NTF2 are sufficient to mediate import of activated Smad3. The data describe a pathway whereby Smad3 phosphorylation by the TGF-β receptor leads to enhanced interaction with importin-β1 and Ran-dependent import and release into the nucleus. The import mechanism of Smad3 shows distinct features from that of the related Smad2 and the structural basis for this difference maps to the divergent sequences of their N-terminal domains.
INTRODUCTION
Transforming growth factor-βs (TGF-βs) are multifunctional peptide growth factors that regulate cell proliferation, differentiation, and death, and are involved in normal development and several disease conditions (Roberts and Sporn, 1990; Massaguéet al., 2000). TGF-β signals via plasma membrane receptor serine/threonine kinases that activate, by phosphorylation, the receptor-activated Smad (R-Smad) proteins at their C-terminal conserved SSXS motifs (Massagué and Chen, 2000; ten Dijke et al., 2000). The activated R-Smads rapidly translocate to the nucleus and simultaneously carry along the common mediator (Co)-Smad. The nuclear complex of Smads associates directly with specific DNA sequences and with a large number of transcription factors, leading to target gene regulation (Attisano and Wrana, 2000; Massagué and Wotton, 2000; ten Dijke et al., 2000). Although the nuclear import of R-Smads is of crucial importance to this signaling pathway, its nature and details have just started to surface.
The pathway of regulated nuclear import of proteins is well-explored (Mattaj and Englmeier, 1998; Görlich and Kutay, 1999). Accordingly, protein cargoes associate with importin carriers, which traverse the nuclear pores via nucleoporin-mediated associations (Rout et al., 2000). In the nucleoplasm, the protein cargo is released by the action of the small GTPase Ran that interacts with the importin carrier and disrupts the cargo–importin complex. Nuclear import is energetically driven by a precise gradient of Ran-GTP, which increases from the cytoplasmic toward the nuclear side of the pore (Görlich and Kutay, 1999; Yoneda et al., 1999). For many proteins, regulated nuclear import depends on a nuclear localization signal (NLS) rich in basic amino acids, which interacts specifically with importin-α, an adaptor molecule that mediates interaction with importin-β, the primary carrier that associates with various nucleoporins and Ran (Mattaj and Englmeier, 1998; Görlich and Kutay, 1999). In other cases, the cargo protein interacts directly with importin-β or other members of this family of carriers via a positively charged NLS-like motif or via uncharacterized protein domains of the cargo (Görlich and Kutay, 1999; Yoneda et al., 1999). Finally, certain proteins do not require importins for their nuclear import but seem to associate directly with nucleoporins (Görlich and Kutay, 1999).
Smad proteins consist of conserved N-terminal, Mad-homology (MH)1, and C-terminal, MH2 domains that associate with each other within the same Smad molecule conferring an autoinhibitory property to the protein (Heldin et al., 1997; Massaguéet al., 2000). Phosphorylation of the carboxyl-terminal serines of the R-Smad SSXS motif by the type I receptor serine/threonine kinase leads to conformational changes of the MH1 and MH2 domains and functional activation (Heldin et al., 1997). One primary function of the MH1 domain of most Smads is to bind DNA sequences via its β-hairpin loop (Shi et al., 1998; Massagué and Wotton, 2000). A conserved lysine-rich motif resides in the N-terminal half of the MH1 domain of most Smads (Shi et al., 1998; Xiao et al., 2000a). This motif resembles the classic simian virus 40 large T antigen NLS and it was recently shown to function as an NLS in Smad3 (Xiao et al., 2000a). In contrast, another recent report described the Smad2 import mechanism as NLS-independent (Xu et al., 2000). These reports provide contradictory evidence for Smad nuclear import and leave the precise import mechanism still open for investigation. Of great impact is the question whether the Ran GTPase regulates Smad import. To address these fundamental issues we focused on Smad3, a central effector of the TGF-β signaling pathway. We describe a mechanism of regulated Smad3 nuclear import via the nonclassical importin-β/Ran pathway. We also provide molecular evidence for the difference of import mechanisms between Smad2 and Smad3.
MATERIALS AND METHODS
Reagent and Antibodies
TGF-β1 was from Dr. N. Ferrara (Genentech, San Francisco, CA). Bone morphogenetic protein 7 (BMP-7) was from Dr. K. Sampath (Creative Biomolecules, Hopkinton, MA). Purified baculoviral Smad3 and TGF-β type I receptor-phosphorylated Smad3 proteins were from Drs. F.M. Hoffman and A. Comer (Johnson et al., 1999). Restriction enzymes and DNA-modifying enzymes were from New England Biolabs (Beverly, MA) or MBI Fermentas Inc. (St. Leou-Rot, Germany). The glutathione S-transferase (GST) purification kit was from Amersham Pharmacia Biotech (Uppsala, Sweden). Isopropyl-β-d-thiogalactopyranoside and digitonin were from Calbiochem (San Diego, CA). All reagents for cell culture (DMEM, fetal bovine serum [FBS], trypsin-EDTA, and phosphate-buffered saline) were from Life Technologies (Gaithersburg, MD). Monoclonal anti-Flag (M2, F-3165, and M5, F-4042) antibodies and 4′,6-diamidino-2-phenylindole were from Sigma (St. Louis, MO). Monoclonal anti-myc antibody was produced by the 9E10 hybridoma cell clone. Anti-histidine antibody (Penta-His) was from Qiagen (Chatsworth, CA). Anti-phospho-serine rabbit polyclonal antibody (Poly-Z-PS1) was from Zymed Laboratories (South San Francisco, CA). Anti-Smad2/3 antibody (clone H-2) was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-phospho-Smad2/3 rabbit polyclonal antisera were as described (Piek et al., 1999). Anti-mouse horseradish peroxidase-, anti-mouse Cy3-, and anti-mouse tetramethylrhodamine isothiocyanate-conjugated secondary antibodies from Amersham Pharmacia Biotech, Molecular Probes (Eugene, OR), and DAKO (Carpinteria, CA), respectively.
Plasmid Constructions
The expression vector pcDNA3 encoding the nontagged Smad2, nontagged Smad3, 6myc-tagged human Smad3, the deletion mutants of Smad3 MH1 plus linker (1–229), Smad3 linker plus MH2 (135–424), Smad3 MH1 (1–134), Smad3 linker (135–229), Smad3 MH2 (230–424) were generously provided by Dr S. Itoh of our institute. The expression vector pcDNA3-HA-CA-ALK5 was described earlier (Nakao et al., 1997). The three MH1 domain Smad3-Smad2 chimeras in pcDNA3.1 (Smad2ΔTID, Smad2ΔGAG, and Smad2 ΔGAG ΔTID) were from Dr. J.-M. Gauthier exactly as described (Dennler et al., 1999).
To construct pGEX4T-1-Flag-Smad3 plasmid, the BamHI-XhoI fragment encoding Flag-tagged human Smad3 was obtained from pcDNA3-FlagSmad3 (Morén et al., 2000) and subcloned into the BamHI-XhoI sites of pGEX-4T-1 (Amersham Pharmacia Biotech). To generate pGEX-FlagSmad3D, the BglII-NotI fragment (amino acids 342–425) of pMX-IRES-GFP-FlagSmad3 (Liu et al., 1997) was subcloned into the BglII-NotI site of pGEX-FlagSmad3. To generate pGEX-GFP-Smad3 and its deletion mutants, the EcoRI-NotI fragments of pcDNA3-6myc-Smad3 and its mutants were subcloned into the EcoRI-NotI site of pGEX-6P-2-hGFP, which carries the S65A/Y145F humanized green fluorescent protein (GFP) gene at the multicloning site (provided by Dr. S. Kuroda, Institute of Scientific and Industrial Research, Osaka, Japan).
Bacterial Expression and Purification of Proteins
The GST fusion proteins were expressed in Escherichia coli BL21(DE3) or BL21(DE3) carrying the pT-Trx plasmid (Yasukawa et al., 1995). To induce proteins, bacteria were grown at 37°C to a density of 0.7 (OD550), and then cultured in LB medium with 0.1 mM isopropyl-β-d-thiogalactopyranoside at 20°C overnight. Cells were harvested and disrupted by sonication in a lysis buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, and 1% Nonidet P-40). After centrifugation, the supernatant was incubated with glutathione-Sepharose (Amersham Pharmacia Biotech) at 4°C. GST-fusion proteins bound on beads were washed extensively with washing buffer (20 mM Tris-HCl pH 7.4 and 150 mM NaCl) and used for pull-down experiments. For in vitro import assays and microinjection, where noted, the GST moiety was cleaved by thrombin or precision protease according to the manufacturer's (Amersham Pharmacia Biotech) protocols. The soluble protein passed through a PD10 column (Amersham Pharmacia Biotech) equilibrated with transport buffer (20 mM HEPES, pH 7.3, 110 mM potassium acetate, 2 mM magnesium acetate, 5 mM sodium acetate, and 0.5 mM EGTA) containing 2 mM DTT and protease inhibitor mixture, followed by concentration by Centricon 30 or Centricon 50 (Amicon, Beverly, MA). All primary protein species were detectable without significant degradation products as determined by SDS-PAGE (not shown).
Expression and purification of mouse importin β (PTAC 97) was performed as described (Kose et al., 1997), as were the purification of GST-mouse importin α (GST-PTAC 58) and GST-importin β (GST-PTAC 97) (Imamoto et al., 1995). Recombinant human p10/NTF2 protein was expressed and purified as described (Tachibana et al., 1996). Recombinant wild-type Ran and RanQ69L were expressed, purified, and charged with GDP or GTP, respectively, as described previously (Hieda et al., 1999). The GST-NLS-GFP fusion protein was expressed and purified to homogeneity using glutathione-Sepharose as described previously (Nagoshi et al., 1999). Aliquots of each recombinant protein were frozen in liquid nitrogen and stored at −80°C.
Cell Culture, Transient Transfections, and Viral Infections
Mink lung cells CCL64, the human colorectal cancer cell line SW480.7 cells, and human embryonic kidney 293T cells were cultured in DMEM supplemented with 10% FBS, l-glutamine, and penicillin/streptomycin at 37°C, in a 5% CO2 atmosphere. HeLa cells were cultured in the same medium but supplemented with 5% FBS. Transient transfections of 293T cells were performed as described (Morén et al., 2000; Pardali et al., 2000). Transient adenoviral infections of SW480.7 cells or CCL64 cells were performed as described (Piek et al., 1999; Pardali et al., 2000). In brief, cells seeded at a density of 1 × 106 in 100-mm dishes were infected the next day with the appropriate adenovirus and cultured for 48 h. The following day cells were washed and fed fresh DMEM containing 10% FBS, and then treated with or without 10 ng/ml TGF-β1 for 1 h, and total cell lysates were prepared.
In Vitro Import Assays in Digitonin-permeabilized Cells
Transport assays were performed essentially as described (Adam et al., 1990; Imamoto et al., 1995). Briefly, 1 × 105 HeLa cells/ml were plated on an eight-well multitest slide (ICN, Costa Mesa, CA) 24–48 h before use. Cells were rinsed twice with transport buffer and permeabilized for 5 min in ice-cold transport buffer containing 2 mM DTT, protease inhibitor mixture, and 40 μg/ml digitonin. After washing twice the slides were immersed in transport buffer containing 2 mM DTT and protease inhibitor mixture at room temperature for 6 min. Excess buffer was removed and cells were incubated with 10 μl/well of reaction mixture for 12 min at room temperature (25–26°C). All reactions contained an ATP regeneration system (1 mM ATP, 5 mM creatine phosphate, and 20 U/ml creatine phosphokinase; Sigma), 2% bovine serum albumin, 2 mM DTT, and protease inhibitor mixture in transport buffer. For in vitro import assays in the absence of ATP, 1.8 U/ml hexokinase (Toyobo, Osaka, Japan) and 5 mM glucose were added to the reaction mixture in the absence of the ATP regeneration system. For wheat germ agglutinin (WGA) treatment, permeabilized cells were incubated with 0.5 mg/ml WGA (E.Y. Laboratories, San Mateo, CA) in transport buffer containing 2 mM DTT and protease inhibitors for 5 min on ice before the import reaction. For import assays in the presence of cytosol, Ehrlich ascites tumor cell total cytosol was prepared as described (Imamoto et al., 1995). For import reconstitution assays, 0.4 μM importin-β1, 1 μM p10, and 3 μM Ran-GDP or 3 μM RanQ69L-GTP were used with 0.5 μM cargo proteins. After incubation, the cells were rinsed with transport buffer and fixed with 3.7% formaldehyde in transport buffer for 15 min at room temperature. After rinsing with transport buffer, GFP-fusion proteins were detected by autofluorescence microscopy. To examine the import of FlagSmad3D, fixed cells were permeabilized with 0.5% Triton X-100 in transport buffer for 5 min at room temperature, blocked with 3% skim milk for 30 min, and then subjected to indirect immunofluorescence using a monoclonal anti-FLAG M2 antibody as the first antibody and Cy3-labeled goat anti-mouse IgG as secondary antibody. All photomicrographs were obtained in a Zeiss Axioplan microscope equipped with a Hammamatsu digital camera.
In Vivo Import Assays in Microinjected Cells
For microinjection of recombinant Flag-tagged Smad proteins, 1 × 105 SW480.7 or HeLa cells were plated on coverslips in 35-mm tissue culture dishes. The next day, the medium was changed to normal medium containing 50 mM HEPES (pH 7.3), and cultured for 3 h. Flag-Smad3 (3 mg/ml), Flag-Smad3D (1 mg/ml), GFPSmad3 (1 mg/ml), or GFPSmad3MH1 (1 mg/ml) protein was injected through a glass capillary into the cytoplasm by using an Eppendorf (model 5246) automatic microinjector, and the cells were cultured for 1 h in normal medium. In the case of Flag-Smad3, cells were cultured with or without 10 ng/ml TGF-β1 for 1 h. GFP-tagged Smad proteins were microscopically detected after cell fixation with 3.7% formaldehyde in phosphate-buffered saline. Flag-tagged Smad proteins were detected by indirect immunofluorescence as described above. To observe the effect of dominant negative Ran on the nuclear import of Smad3, cells were treated as described above and the control protein GSTNLSGFP with or without RanQ69L-GTP was injected into the cytoplasm, and the cells were analyzed as described above.
GST Protein Interaction Assays
Interaction assays of GST-importin β1 with nontagged, 6myc-tagged, and chimeric Smad proteins expressed in 293T cells were performed as described (Pardali et al., 2000). For interaction assays of GST-importins with adenovirally expressed Flag-Smad3 proteins, SW480.7 or CCL64 cells were infected as described above and GST pull-down assays of cell extracts were performed as described (Sekimoto et al., 1997).
Dissociation assays of GST-importin-β1 with Flag-Smad3D or maltose-binding protein (MBP)-importin-β-binding domain (IBB) were performed by incubating 3 or 15 μg of the latter two purified proteins, respectively, with 25 μg of GST-importin-β1 at 4°C for 2 h followed by glutathione-Sepharose bead incubation for 1 h and extensive washing with transport buffer. Then, 10 μM of purified Ran-GDP or RanQ69L-GTP were added to the bound complexes followed by incubation at 4°C for 1 h, centrifugation, and SDS-PAGE analysis of the obtained pellet and supernatant. Proteins were detected by Coomassie Brilliant Blue staining and quantified densitometrically using a luminescent image analyzer LAS-1000plus and the integrated advanced image data analyzer (Fuji Photo Film, Stockholm, Sweden).
Interaction assays of baculoviral 6-his-Smad3 or receptor-phosphorylated 6-his-Smad3 (6-his-Smad3-P) with GST-importin-β1 were performed by mixing 2.5 μg of pure Smad proteins with COS-7 total cell lysate prepared as previously described (Sekimoto et al., 1997), and diluted in transport buffer to simulate nuclear transport conditions. The protein–cell extract mixture was precleared with Sepharose beads at 4°C for 30 min, and then 1 nmol of GST or GST-importin-β1 was added and incubation continued at 4°C for 2 h, followed by exhaustive washing with transport buffer and SDS-PAGE analysis. Smad proteins were detected by immunoblotting with penta-histidine antibody and the phosphorylation status of the 6-his-Smad3-P preparation was verified by anti-phosphoserine antibody blotting.
RESULTS
Establishment of In Vitro and In Vivo Nuclear Import Assays Point to an Active Transport Mechanism for Smad3
An in vitro Smad3 import assay was developed using the established digitonin-permeabilized HeLa cell-free transport system (Figure 1) (Adam et al., 1990). In this system and in many other TGF-β–responsive or not responsive cell lines, permeabilization led to quantitative loss of all cellular Smad3 (our unpublished results). As a source of exogenous Smad3 protein, N terminally Flag-tagged Smad3 was isolated from E. coli as a GST-fusion that was subsequently cleaved with thrombin and purified (Figure 1A, lane 1). To obtain activated Smad3 in the in vitro import assay (where the signaling pathway is disrupted by digitonin treatment), we used a triple point mutant of Smad3, Smad3D, in which the three carboxyl terminal serine residues of the SSXS motif are mutated to aspartate residues. The substitution of these serine residues to negatively charged residues has been shown to mimic phosphorylation by receptor and results in constitutive activation of the protein (Liu et al., 1997). The Smad3D protein was also purified from E. coli (Figure 1A, lane 2).
Figure 1.
In vitro import assay for Smad3. (A) Profile of bacterially purified Smad3 (lane 1) and Smad3D proteins (lane 2) after SDS-PAGE and Coomassie Brilliant Blue staining. (B) Microinjection of bacterial Smad3 together with bacterial GSTNLSGFP protein in the cytoplasm of human colon carcinoma SW480.7 cells. The cells were previously transiently infected with adenoviruses that encode for LacZ (control) or caALK-5. The caALK-5–infected cells were also treated with TGF-β1 (+TGF-β) immediately after microinjection and for 1 h. (C) In vitro import of Smad3, Smad3D, and GSTNLSGFP in the absence (−) and presence (+) of cytosol in digitonin-permeabilized HeLa cells. In this assay, each protein was added at 0.5 μM. Smad proteins were detected by anti-Flag/Cy3 immunofluorescence and GSTNLSGFP by green autofluorescence. Nuclear morphology was assessed by phase contrast microscopy.
The functional integrity of the recombinant Smad3 proteins was tested by microinjection into the cytoplasm of live human colon carcinoma SW480.7 cells together with a control GSTNLSGFP protein that contains the potent simian virus 40 virus large T antigen NLS fused between the GST and GFP moieties (Figure 1B). To obtain ligand-stimulated conditions the cells were transiently infected with an adenovirus expressing the constitutively active (ca) type I receptor of TGF-β (activin receptor-like kinase [ALK]-5) and treated with extracellular TGF-β1 to achieve maximal level of Smad activation and nuclear translocation. The corresponding nonstimulated condition was obtained by transiently infecting cells with an adenovirus expressing the LacZ gene and treatment with vehicle. The results demonstrate that the recombinant Smad3 behaved physiologically, because it exhibited diffuse cytoplasmic distribution in the absence of stimulation and translocated quantitatively to the nucleus after stimulation with TGF-β1 (Figure 1B). Smad3D localized in the nucleus in the absence or presence of stimulation (our unpublished results). As expected, the coinjected control GSTNLSGFP protein constitutively localized to the nuclei of the microinjected cells and its distribution was not affected by activation of TGF-β receptors.
In vitro import assays in the absence of added cytosol followed by anti-Flag immunofluorescence revealed that Smad3 accumulated around the nuclear envelope (Figure 1C). When cytosol was added, the concentrated perinuclear staining was replaced by a weak and diffuse nuclear staining of Smad3, suggesting inefficient nuclear import (Figure 1C). Thus, Smad3 that has not been activated by TGF-β receptors cannot accumulate efficiently in the nucleus as expected. When Smad3D was tested, it exhibited ring-like perinuclear staining like Smad3 in the absence of cytosol (Figure 1C). In contrast to wild-type Smad3, Smad3D efficiently entered the nucleus when cytosol was added (Figure 1C). The behavior of Smad3D in this assay was identical to that of the positive control, the GSTNLSGFP (Figure 1C). We conclude that Smad3 activated by the TGF-β signal is transported to the nucleus by an active mechanism that requires cytosolic factors, as is the case for most other NLS-containing proteins.
Smad3 Interacts with Importin-β1 in a TGF-β Pathway-specific Manner
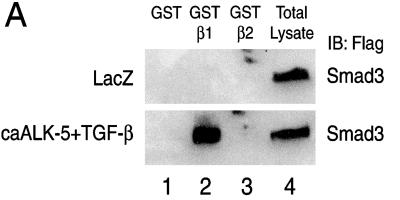
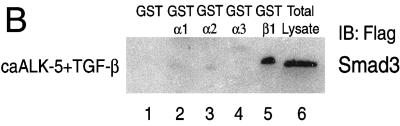
The cytosol-dependent import of Smad3D suggests that carriers of the importin-β family may be involved in activated Smad3 transport. To test this hypothesis, we performed protein–protein interaction assays (Figure 2A). We prepared total cell extracts of SW480.7 or mink lung CCL64 cells transiently infected with an adenovirus that expresses wild-type Smad3, after stimulation or not with TGF-β1 and ca-ALK-5. These extracts were incubated with GST-fusions of importin-β1 and -β2, bound proteins were isolated and analyzed by SDS-PAGE, and Smad3 was detected by anti-Flag immunoblotting (Figure 2A). Importin-β1 demonstrated positive and specific interaction with Smad3, and only after stimulation with TGF-β1 and/or ca-ALK-5. Less efficient association was obtained when cells were either treated with TGF-β1 alone or infected with ca-ALK-5 alone (our unpublished results). The control GST protein did not interact. We also tested whether adaptors of the importin-α family can interact with Smad3. For this, we repeated the GST-interaction assays using GST-importin-α1, -α2, and -α3 (Figure 2B). None of these proteins showed any specific interaction with Smad3.
Figure 2.
Smad3 interacts with importin-β1 in an activation/phosphorylation-dependent manner. (A-C) SW480.7 and CCL64 cells were used giving essentially identical results. Representative immunoblots are shown in this figure. (A) In vitro interaction assay between GST (lane 1), GST-importin-β1 (GST-β1, lane 2), GST-importin-β2 (GST-β2, lane 3) and Smad3. Detergent extracts from transiently coinfected cells with adenoviruses encoding LacZ and Smad3 or caALK-5 and Smad3 and treated (+TGF-β) or not (LacZ) with TGF-β1 were isolated. Extracts were incubated with the three GST protein affinity columns and the bound Smad3 was analyzed by immunoblotting (IB) with anti-Flag antibody. In lane 4 an aliquot of the total cell lysate was analyzed by immunoblotting directly. (B) In vitro interaction assay between GST (lane 1), GST-importin-α1 (GST-α1, lane 2), GST-importin-α2 (GST-α2, lane 3), GST-importin-α3 (GST-α3, lane 4) and GST-importin-β1 (GST-β1, lane 5) and Smad3. Cell extracts were prepared and analyzed as in A. The total lysate control is in lane 6. (C) Specificity of the Smad3–importin-β1 interaction. In vitro interaction assay between GST-importin-β1 (GST-β1, all lanes) and Smad3. Cell extracts treated as indicated on top of the panel were prepared and analyzed as in A by using anti-Flag (first panel) or anti-phospho-Smad (second panel) immunoblotting. Analysis of total lysate aliquots (Total) with the same two antibodies is shown in the third and fourth panels, respectively. Smad7 was also detected by anti-Flag immunoblotting (fifth panel). Asterisks on the right of the second and fourth panels indicate nonspecific bands resulting from the rabbit anti-phospho-Smad serum. (D) C-Terminal phosphorylation-dependent interaction between Smad3 and importin-β1. In vitro interaction assay between GST (lanes 1 and 2), GST-importin-β1 (GST-β1, lanes 3 and 4) and histidine-tagged baculoviral Smad3 (S3, lanes 1 and 3), or C terminally phosphorylated Smad3 (S3P, lanes 2 and 4). Bound Smad3 proteins were analyzed by immunoblotting by using anti-histidine (His) antibody. Aliquots of the baculoviral proteins were separately analyzed by sequential anti-histidine (lanes 5 and 6) and anti-phospho-serine (P-Ser, lanes 7 and 8) immunoblotting. (A–D) Position of the relevant protein bands is indicated on the sides of the panels.
To further analyze the specificity of the Smad3-importin-β1 interaction, we used similar cell extracts that contained in addition to Smad3 and ca-ALK-5, the inhibitory Smad7 that blocks R-Smad phosphorylation and activation by the type I receptor (Figure 2C) (ten Dijke et al., 2000). Whereas activated and phosphorylated Smad3 avidly bound to importin-β1, Smad7 efficiently blocked the Smad3–importin-β1 interaction, confirming that this protein complex forms only when the signaling pathway is activated. Probing for phosphorylated Smad3 in these experiments, we demonstrated that Smad7 inhibited Smad3 phosphorylation by receptor, thus resulting in weak Smad3 association with importin-β1 (Figure 2C). As a negative control, we tested the Smad3–importin-β1 association under conditions where the cells were stimulated with another TGF-β superfamily member, BMP-7, and the constitutively active BMP type IB receptor (ca-ALK-6). This treatment, which results in Smad1, Smad5, and Smad8 activation (our unpublished results), but not Smad3 activation, failed to induce Smad3 phosphorylation or Smad3–importin-β1 association, as expected (Figure 2C).
Finally, to confirm that the strict ligand dependency of the Smad3–importin-β1 association depends on the C-terminal SSXS phosphorylation status, we performed interaction assays with 1) histidine (his)-tagged Smad3 (S3) purified from baculovirus-infected insect cells, or with 2) C terminally phosphorylated Smad3 (S3P) that was produced in insect cells coinfected with Smad3 and ca-ALK-5 receptor (Figure 2D) (Johnson et al., 1999). Whereas nonphosphorylated Smad3 interacted weakly with importin-β1, phospho-Smad3 reproducibly exhibited significantly stronger interaction. Control GST could not support any specific association with the Smad proteins used, and control immunoblot analysis with anti-histidine and anti-phospho-serine antibodies verified the levels and fidelity of phosphorylation of the baculoviral Smad3 preparations (Figure 2D). We therefore conclude that Smad3, after phosphorylation by the TGF-β type I receptor, can specifically interact with importin-β1 but not with importin-α adaptors. The same conclusion has been recently reported by another group (Xiao et al., 2000b).
Dominant Negative Mutant RanQ69L-GTP Quantitatively Inhibits the TGF-β and Receptor-activated Import of Smad3 into the Nucleus
If importin-β1 indeed mediates nuclear transport of activated Smad3, then the Ran GTPase could possibly regulate this process. To test this hypothesis we performed in vivo experiments in SW480.7 cells whose cytoplasm was microinjected with the internal control GSTNLSGFP protein together with bacterially purified dominant negative RanQ69L-GTP (which binds GTP strongly but cannot hydrolyze it) (Görlich and Kutay, 1999). To monitor Smad3, the same cells were infected with the Smad3 adenovirus before microinjection and were treated or not with TGF-β1 for 1 h before immunostaining (Figure 3A). Control uninjected cells demonstrated the correct ligand-dependent translocation of Smad3 to the nucleus (top panels) confirming that the conditions of adenoviral infection preserve the physiological behavior of the signaling pathway as previously described (Piek et al., 1999). When GSTNLSGFP plus buffer alone were microinjected into the infected cells, Smad3 exhibited the same normal localization as in the uninjected controls, demonstrating that the microinjection process also preserves the physiology of the signaling pathway (middle panels). Under these conditions, GSTNLSGFP showed constitutive nuclear accumulation and treatment with TGF-β1 did not alter the nuclear transport of GSTNLSGFP, as expected. Finally, microinjection of RanQ69L-GTP into the cytoplasm of the infected cells resulted in the expected inhibition of the control GSTNLSGFP in both untreated cells and in cells treated with TGF-β1 (bottom panels). RanQ69L-GTP potently inhibited the nuclear translocation of Smad3 in the TGF-β1–stimulated cells and led to partial nuclear accumulation of Smad3 in nonstimulated cells. These experiments demonstrate that the Ran GTPase regulates the active process of Smad3 import after stimulation with ligand. They also raise the possibility that Smad3 is localized to the cytoplasm of unstimulated cells due to an active export mechanism (see DISCUSSION).
Figure 3.
Ran GTPase mediates the nuclear import of Smad3. (A) Dominant negative RanQ69L-GTP inhibits the ligand-dependent nuclear import of Smad3. SW480.7 cells transiently infected with an adenovirus expressing Smad3 were left intact (−), microinjected with GSTNLSGFP plus buffer (Buffer), or with GSTNLSGFP plus RanQ69L-GTP (RanQ69L-GTP). Immediately after microinjection cells were treated with vehicle (−TGF-β) or treated with TGF-β1 (+TGF-β) for 1 h. The proteins were detected by GFP autofluorescence (NLS), anti-Flag/tetramethylrhodamine isothiocyanate immunofluorescence (Smad3), and nuclei were scored by 4′,6-diamidino-2-phenylindole fluorescence. Each three-panel row of photographs represents the same microscopic field. (B) Ran-GTP dissociates Smad3 from the complex with importin-β1. In vitro dissociation assay of purified Smad3D bound to GST–importin-β1 (GST-β1) affinity columns (lanes 1–4). A similar assay was performed between purified MBP-IBB fusion protein and GST–importin-β1 (GST-β1) affinity columns (lanes 5–8). Purified proteins were bound to the affinity columns and eluted with buffer containing Ran-GDP (lanes 1, 2, 5, and 6) or Ran-GTP (lanes 3, 4, 7, and 8). The proteins remaining bound (lanes 1, 3, 5, and 7) to the affinity columns and those eluted (lanes 2, 4, 6, and 8) off the columns were analyzed by SDS-PAGE and Coomassie Brilliant Blue staining. The position of the relevant protein bands is indicated on the sides of the panel. Densitometric quantification of the eluted Smad3D and MBP-IBB protein bands of the gel is presented as a bar graph. The eluted protein band intensity is graphed as percentage of the total band intensity (bound plus eluted) in each of the four binding reactions. Numbers corresponding to the eluted sample lanes of the gel described above are indicated below the bar graph.
To biochemically link the action of the dominant negative Ran with the observed Smad3–importin-β1 complexes, we attempted to dissociate Smad3 from the in vitro complex with importin-β1 (Figure 3B). Incubation of the Smad3D–importin-β1 complex with Ran-GDP primarily resulted in retention of the complex on the glutathione-Sepharose column via the immobilized GST-importin-β1. Under these conditions only 2.5% of Smad3D dissociated from the column. However, incubation of the immobilized importin-β1–Smad3D complex with recombinant RanQ69L-GTP resulted in measurable elution (21.5%) of Smad3D (Figure 3B). As a positive control, a similar experiment was performed with a complex of importin-β1 and the domain of importin-α that specifically binds to importin-β1 (IBB) (Görlich and Kutay, 1999). These two proteins also formed a strong complex that resulted in only 1.7% elution after incubation with Ran-GDP but was partially dissociated (21%) by RanQ69L-GTP. The dissociation profiles of IBB and Smad3D are similar, suggesting that binding of Ran-GTP to importin-β1 induces dissociation of the importin-β1–Smad3D complex as it does for the IBB fragment. Thus, upon stimulation with TGF-β, Smad3 is imported into the nucleus by importin-β1, after which Ran mediates Smad3 release from the importin complex.
MH1 Domain of Smad3 but not Smad2 Interacts Directly with Importin-β1
We then dissected the domain of Smad3 that is required for interaction with importin-β1 and nuclear import (Figure 4A). Using a panel of Smad3 deletion mutants transiently overexpressed in 293T cells, which were stimulated with TGF-β1, we performed in vitro interaction assays by passing extracts from the transfected cells over the GST-importin-β1 column or a GST column as negative control. The bound Smad3 proteins were detected via their N-terminal 6-myc tag by immunoblotting analysis (Figure 4B). Full-length Smad3, the MH1-linker, and the MH1 domains all specifically bound to the importin-β1 affinity column. In contrast, the linker-MH2, the linker, and the MH2 domains did not associate with importin-β1. Interestingly deletion of the MH2 domain resulted in relative enhancement of Smad3 interaction with importin-β1 compared with the full-length protein. Further deletion of the linker domain, leaving only the isolated MH1 domain, enhanced the interaction even more (Figure 4B). This is not due to differential protein expression of the various Smad3 domains (Figure 4D). As a negative control, the interaction of the Smad3 mutants with GST was tested and no specific associations were detected (Figure 4C). We conclude that the MH1 domain of Smad3 is the primary determinant for the specific association with importin-β1 and that this interaction requires conformational exposure of the MH1 domain that is induced by receptor phosphorylation, negatively charged amino acid substitution in the SSXS motif (Smad3D), or C-terminal truncation.
Figure 4.
Importin-β1 interacts specifically with the Smad3 MH1 domain but not with Smad2. (A) A diagram of Smad3 with its three domains (MH1, linker [L] and MH2) is shown. N and C denote the amino and carboxyl termini. The conserved lysine-rich motif in MH1 is indicated as a striped box. (B) 293T cells were transiently transfected with empty vector (Mock) or the indicated 6-myc-tagged Smad3 (S3) domain mutants together with ca-ALK-5 to stimulate the pathway. Cell extracts were incubated with GST-importin-β1 (GST-β1) and the bound Smad3 proteins were analyzed by immunoblotting (IB) with anti-myc antibody. (C) Aliquots of the same extracts were incubated with GST alone as negative control and analyzed as in B. (D) Aliquots of the same extracts (Total) were analyzed by direct immunoblotting with anti-myc antibody to monitor the expression levels of the different Smad3 mutants in the transfected cell extracts. (E) Diagrams of wild-type Smad2 and the three Smad2-Smad3 chimeras are shown with their three domains (MH1, L, and MH2). N and C denote the amino and carboxyl termini. The conserved lysine-rich motif in MH1 is indicated as a striped box. The two unique inserts of Smad2 are shown as highlighted boxes labeled GAG and TID. Smad2 sequences are shown as black and Smad3 sequences as white boxes. The positions in Smad3 MH1 sequences that mark the absence of the Smad2 unique inserts are also depicted with straight vertical lines and are interconnected with the two inserts between constructs. (F) 293T cells were transiently transfected with the indicated nontagged wild-type Smad2 (lanes 1 and 5) and three Smad2-Smad3 chimeras (lanes 2–4 and 6–8) and then left unstimulated (−, lanes 1–4) or were stimulated (+, lanes 5–8) with TGF-β1. Cell extracts were incubated with GST-importin-β1 (pull-down) and the bound Smad2 proteins were analyzed by IB with the anti-Smad2/3 antibody. Aliquots of the same extracts (total) were analyzed by direct immunoblotting with the anti-Smad2/3 antibody to monitor the expression levels of the different Smad2 variants in the transfected cell extracts. The closely migrating positions of the four Smad2 variants are shown by bracket on the right side of the panels.
In contrast, a recent report demonstrated the role of the MH2 domain on the cytosolic factor-independent import mechanism of Smad2 (Xu et al., 2000). On the other hand, another group emphasized the importance of an NLS-like sequence in the MH1 domain of Smad3 for nuclear import and interaction with importin-β1, in agreement with the above-described data (Xiao et al., 2000a,b). This NLS-like motif is identical in both Smad2 and Smad3. To resolve the question of possible differences between the two related R-Smads of the TGF-β/activin pathways, we focused on the strength of interaction between Smads and importin-β1 as a measure of specificity of nuclear import mechanism. Smad2 and Smad3 are highly related in primary amino acid sequence (Heldin et al., 1997). However, the MH1 domain of Smad2 contains two inserts that are specifically absent from Smad3 and have been termed GAG and TID based on the central amino acid triplet of each insert (Dennler et al., 1999). These two inserts localize on either side of the lysine-rich NLS-like motif of Smad2 (Shi et al., 1998). Insert TID has been shown to render Smad2 unable to recognize the Smad binding element on DNA, whereas an alternatively spliced form of Smad2 that specifically lacks the insert TID binds to DNA as efficiently as Smad3 (Dennler et al., 1999; Yagi et al., 1999). We therefore focused on the importance of these inserts by analyzing a series of chimeras between the MH1 domains of Smad2 and Smad3 (Figure 4E). Using the same 293T overexpression protocol followed by GST-importin-β1 pull-down assay described in Figure 4A, we demonstrated that Smad2 cannot associate with importin-β1 in the absence or presence of stimulation by TGF-β1 (Figure 4F). The chimera that lacks both MH1 inserts (Smad2-ΔGAG-ΔTID) exhibited weak constitutive association with importin-β1 that was strongly enhanced by treatment with TGF-β1, thus resembling partially Smad3 (Figure 4F). The critical sequence affecting the association with importin-β1 maps to the insert TID because removal of this insert only makes Smad2 capable of interaction in a ligand-dependent manner, like Smad3, whereas removal of the insert GAG had no effect compared with wild-type Smad2 (Figure 4F). It must be noted that the chimeric proteins contain a small number of amino acid substitutions in the nondeleted portions of the MH1 domain, most of which are conservative and thus are not expected to contribute significantly to the observed associations with importin-β1. Using the same assay we observed that addition of an N-terminal epitope tag on Smad2 also resulted in efficient association of Smad2 with importin-β1 (our unpublished results). We conclude that Smad2 and Smad3 indeed differ in their capacity to associate with importin-β1 and this specificity is defined by the unique Smad2 insert TID encoded by exon 3. Thus, the mechanisms of nuclear import of these two related Smads rely on different components that must be dictated by the differential folded structures of their activated MH1 and MH2 domains. The fully conserved NLS-like motif may be important but is not the primary determinant of specificity.
MH1 Domain Mimics Signal-dependent Wild-Type Smad3 Import, and the MH2 Domain Exhibits Only Weak Constitutive Import Activity
To examine the role of the various Smad3 domains in nuclear import we performed in vitro import assays in permeabilized HeLa cells by using a panel of Smad3 deletion mutants whose N termini were now fused to GFP to facilitate detection (Figure 5). The deletion mutants were compared with Smad3D, which was also fused N terminally to GFP. GSTNLSGFP served as the positive control. As anticipated, GFP-Smad3D exhibited cytosol-dependent nuclear import, confirming the data of Figure 1C. GFP-Smad3MH1 showed a perinuclear ring in the absence of cytosol and strong nuclear accumulation in the presence of cytosol. The cytosol-dependent import of Smad3D and Smad3 MH1 in vitro was blocked 1) when ATP was depleted from the cytosol by the addition of hexokinase and glucose; 2) when the permeabilized cells were pretreated with the lectin WGA, which binds to the cytoplasmic sugars of the nuclear pore complex; or 3) when the system was incubated on ice (Figure 5). All such blocking treatments resulted in the perinuclear, ring-like accumulation of the exogenous Smads. GFP-Smad3Linker (L) performed rather inefficiently in in vitro import assays, exhibiting both weak constitutive and cytosol-enhanced import activity (Figure 5). GFP-Smad3MH2 showed weak but distinct nuclear localization in the absence of cytosol (our unpublished results). The efficiency of constitutive and cytosol-independent nuclear import of the Smad3 MH2 domain was under all conditions tested much lower than the efficiency of cytosol-dependent import of the full length or MH1 domain of Smad3. A second chimera encoding GSTGFP-Smad3MH2, which is substantially larger than the GFP-Smad3MH2 fusion protein, exhibited almost no nuclear import under all conditions tested (Figure 5). The cytosol-dependent and energy-dependent import of the GSTNLSGFP control also confirms the quality and fidelity of the digitonin-permeabilized HeLa cell system. We conclude that the MH1 domain specifies the active nuclear import of Smad3. However, in certain fusion constructs, the nuclear import activity of the Smad3 MH2 domain resembles the constitutive import function of the Smad2 MH2 domain described recently (Xu et al., 2000).
Figure 5.
In vitro nuclear import assay of various Smad3 domains. In vitro import of N-terminal GFP fusions of Smad3D (GFPS3D, A–E), Smad3 (S3) domain mutants MH1 (GFPS3MH1, F–J), linker (GFPS3L, panels K–O), N-terminal GSTGFP fusion of Smad3 MH2 (GSTGFPS3MH2, P–T), and control GSTNLSGFP (U–Y) proteins in digitonin-permeabilized HeLa cells. Each protein cargo was added at 0.5 μM. The in vitro import conditions are indicated to the left of the panels and included: the absence (−) and presence (+) of cytosol, ATP depletion by hexokinase plus glucose treatment (−ATP), wheat germ agglutinin blocking of the nuclear pores (+WGA), and incubation on ice in the presence of cytosol. Protein detection was obtained by green autofluorescence. Nuclear staining was assessed with phase contrast microscopy.
In Vitro Reconstitution of Smad3 Import by Importin-β1, p10/NTF2, and Ran
To demonstrate that the nuclear transport components importin-β1 and Ran are sufficient for Smad3 nuclear transport, we designed reconstitution experiments of in vitro import by using only purified proteins as reaction components. As demonstrated in Figure 6, importin-β1 alone was capable of only limited nuclear import of Smad3D. Ran-GDP in the presence of its nuclear carrier p10/NTF2 showed no nuclear translocation. Finally, combination of importin-β1 and Ran-GDP plus p10/NTF2 gave strong nuclear accumulation, to the same level as that seen when cytosol was added. Importantly, replacement of Ran-GDP with RanQ69L-GTP led to perinuclear accumulation and complete blockade of Smad3D import. The same results were obtained when the GFP-MH1 protein was used as cargo (Figure 6). The results demonstrate that importin-β1 and Ran, in association with p10/NTF2, are all the components required to mediate the active nuclear import of phosphorylated Smad3, or an isolated MH1 domain.
Figure 6.
In vitro reconstitution of Smad3 nuclear import. In vitro import of Smad3D, N-terminal GFP fusion of Smad3 MH1 domain (GFPS3MH1) and control GSTNLSGFP proteins in the presence or absence of cytosol in digitonin-permeabilized HeLa cells. For Smad3D and GFPS3MH1, additional import reactions are shown with purified protein components (no cytosol): importin-β1 alone (+imp β1), Ran-GDP plus p10, importin-β1 plus Ran-GDP plus p10, or importin-β1 plus RanQ69L-GTP plus p10. Smad3D was detected via anti-Flag/Cy3 immunofluorescence and GFPS3MH1 and GSTNLSGFP via their green autofluorescence.
DISCUSSION
Active Nuclear Import of the Receptor-activated Smad3
The in vitro import assay developed for Smad3 in digitonin-permeabilized HeLa cells enabled us to demonstrate that receptor-activated Smad3, represented by the point mutant Smad3D, is transported into the nucleus in an active manner that requires soluble cytosolic factors and metabolic energy (Figures 1 and 5). By contrast, nonactivated Smad3 protein is inefficiently retained in the nuclei of permeabilized cells, suggesting a slow rate of constitutive import in the presence of cytosolic factors.
The fundamental difference between nonactivated and activated Smad3 is consistent with the previously proposed model of Smad autoinhibition, whereby the intramolecular interaction of the MH1 and MH2 domains results in cytoplasmic retention and inactivity (Hata et al., 1997). Smad cytoplasmic retention could possibly involve the specific membrane anchor of Smads, SARA, the receptor-associated scaffold protein ARIP, and/or microtubules (Tsukazaki et al., 1998; Dong et al., 2000; Shoji et al., 2000). We propose that in addition to release from autoinhibition, C-terminal serine phosphorylation by the type I receptor kinase must result in specific conformational changes that modify the structure of the MH1 domain (Shi et al., 1997, 1998; Qin et al., 1999), resulting in exposure of the importin-β1–associating interface. These predictions also deserve further biochemical and biophysical experimentation.
Smad3 MH1 Domain and Importin-β1 Define the Specificity of Smad3 Import
The active import of receptor-activated Smad3 could be explained by a classic transport pathway involving the adaptor importin-α as a cargo-interacting component and importin-β as the carrier mediating shuttling through the nuclear pore (Görlich and Kutay, 1999). However, the screen we performed for various importin members that could specifically interact with Smad3 scored positively only for importin-β1 (Figure 2). Thus, Smad3 is capable of direct interaction with the major carrier component and does not require the adaptor importin-α. In this sense Smad3 joins a growing list of proteins whose regulated nuclear import is mediated solely by importin-β. Such proteins include cyclin B1, the retroviral Tat, Rev, and Rex proteins, sterol-regulated element binding protein-2, insulin-like growth factor binding protein-3 and -5, and more (Moore et al., 1999; Nagoshi et al., 1999; Palmeri and Malim, 1999; Takizawa et al., 1999; Truant and Cullen, 1999; Schedlich et al., 2000). Shuttling through the nuclear pore occurs by a series of association-dissociation events of the importin–cargo complex to nucleoporins (Görlich and Kutay, 1999; Allen et al., 2000; Rout et al., 2000). Our preliminary evidence suggests that the Smad3–importin-β1 complex can associate with nucleoporins containing phenylalanine-glycine repeats (Kurisaki and Moustakas, unpublished observations).
The specificity assays of Figure 2C further support the evidence that the importin-β1–Smad3 association, and by extension, Smad3 import, depend on an active TGF-β signaling pathway that cannot be phenocopied by overexpression of a related but distinct signaling pathway such as the BMP pathway. In addition, Smad7 was shown to efficiently block the TGF-β–induced Smad3 phosphorylation and association with importin-β1, presumably due to Smad7's ability to block the kinase activity of the type I receptor (Hayashi et al., 1997; Nakao et al., 1997).
The Smad3 structural determinant(s) required for the interaction with importin-β1 mapped to the N-terminal conserved MH1 domain (Figure 4). Furthermore, the isolated MH1 domain exhibited cytosol- and energy-dependent import that was similar to full-length Smad3, but independent from activation by ligand (Figure 5). This is in contrast to the other two domains, the linker and MH2, of Smad3. We postulate that the MH1 domain must include specific structural determinants that mediate the importin-β1 interaction and efficient Smad3 import. The short motif, K40KLKK44, which is part of a relatively unexposed-to-solvent α-helix, as proposed by the Smad3 MH1-DNA complex structure, is conserved in all the R-Smads, the CoSmads Smad4 and 4β, and the inhibitory Smad7 (Shi et al., 1998; Masuyama et al., 1999). Consistent with our findings, Xiao et al. (2000a,b) recently reported that this specific Smad3 sequence indeed functions as a true NLS and mediates interaction with importin-β1. However, our analysis of Smad2–importin-β1 interaction (Figure 4B) demonstrated that additional structural determinants in the MH1 domain regulate the specificity of this association. Thus, it is possible that a higher order structure is the specific structural determinant of the Smad3–importin-β1 interface and the defined NLS sequence may be important but not the only critical determinant.
The Smad3 MH2 domain in certain fusion constructs was found to spontaneously and partially accumulate in the nuclei of permeabilized cells in the absence of cytosol (our unpublished results), whereas in other constructs not (Figure 5). It is possible that the MH2 domain may exhibit weak but significant affinity for nucleoporins and thus enters the nucleus constitutively. Constitutive but strong nuclear accumulation of the isolated MH2 domain of Smad2 was recently reported and this finding led to a model whereby Smad2 nuclear import is receptor phosphorylation-independent, cytosol-independent but energy-dependent and mediated by the MH2 domain (Xu et al., 2000). Although our in vitro import data for the Smad3 MH2 domain partially agree with the Smad2 model, both in vitro and in vivo import experiments of full-length Smad3 point to the distinct nuclear import mechanisms of these two related Smads. This suggested to us that the mechanism of Smad2 import might differ from that of Smad3, despite the conservation of the putative MH1 NLS motif. In agreement with this hypothesis we demonstrated that Smad2 could not interact with importin-β1 due to a unique sequence insert termed TID in its MH1 domain (Figure 4B). Interestingly, the same sequence determinant also results in Smad2's inefficient binding to the Smad binding element (Dennler et al., 1999; Yagi et al., 1999). Thus, two important functions, nuclear import and DNA binding are coregulated by the same structural motifs in the MH1 domain of Smad2. These results allow us to postulate that in Smad2, importin-mediated nuclear import is deficient and thus the intrinsic import activity of the MH2 domain prevails as recently described (Xu et al., 2000). In addition, our findings predict that the alternatively spliced form of Smad2 that lacks exon 3 could associate with importin-β1 and thus be imported by a different mechanism than Smad2.
Ran-dependence of Smad3 Import
Both our in vitro and in vivo import approaches establish a requirement for the small GTPase Ran in the regulation of Smad3 nuclear import (Figures 3 and 6). Ran-GTP is important at the nuclear face of the pore for release of the cargo protein to the nucleoplasm (Görlich and Kutay, 1999). This is achieved by the specific interaction of Ran-GTP with importin-β. In agreement with this mechanism, we demonstrate that the mutant RanQ69L-GTP, which is constitutively locked in the GTP form and thus mimics endogenous Ran-GTP, can cause dissociation of Smad3D from the importin-β1 complex under in vitro conditions (Figure 3B). Thus, when RanQ69L-GTP is injected into the cytoplasm, it inhibits binding of importin-β1 to activated Smad3 and so inhibits Smad3 nuclear import (Figure 3A). Consistent with this notion, in the in vitro reconstitution system where only purified components are added, RanQ69L-GTP efficiently blocked the import of Smad3D (Figure 6). This result proves that the mechanism of regulation of Smad3 import by Ran operates at the level of Smad3–importin-β1 complex dissociation. The efficient inhibition of TGF-β–stimulated Smad3 nuclear import by RanQ69L-GTP in the microinjection experiments supports the notion that the dominant import pathway for Smad3 is Ran-dependent. Therefore, a cytosolic factor-independent nuclear import pathway mediated by the MH2 domain may be a minor route for Smad3 translocation to the nucleus.
An unexpected result was that RanQ69L-GTP microinjection in live cells consistently resulted in partial nuclear accumulation of Smad3 in the nonligand-stimulated state (Figure 3A). This can be interpreted by RanQ69L-GTP squelching the available cytosolic importin-β1, thus enhancing the weak constitutive import activity of Smad3 driven by its MH2 domain. Alternatively, because the Ran GTPase is involved in active nuclear export of macromolecules (Mattaj and Englmeier, 1998; Yoneda et al., 1999), we propose that nuclear accumulation of Smad3 under these conditions can also be explained by a change in the Ran-GTP equilibrium across the nuclear pore, which leads to nuclear accumulation of the exported cargo, as previously demonstrated (Nachury and Weis, 1999).
Generalization of the Import Mechanism for Smad Proteins
This report describes a nuclear transport pathway for a member of the Smad family of tumor suppressors. The import process is active and regulated by Ran and thus guarantees rapid, efficient, and regulated nuclear import. In this report we have focused on a single Smad, Smad3, to develop useful import assays for this family of proteins. However, this report and those recently describing the Smad3 NLS and the Smad2 import mechanism (Xiao et al., 2000a,b; Xu et al., 2000) pose the question of how general is the import mechanism of Smad proteins. In contrast to the previous reports we provide evidence that the two related Smads of the TGF-β/activin pathways are imported via distinct mechanisms due to the differential ability of their MH1 domains to interact with importin-β1, which possibly leads to a differential utilization of the constitutive nuclear import function of their MH2 domains. Thus, we propose that the nuclear import mechanisms of all other family members deserves further experimentation and cannot be safely extrapolated by the findings on a single Smad protein. It is also of interest to examine whether other Smad members, including Smad2, are imported to the nucleus in response to ligand and receptor activation by a Ran-dependent mechanism. Finally, Smad2, Smad3, and Smad4 have been proposed to translocate as a single oligomeric complex (Heldin et al., 1997). The import mechanism for such a complex must be characterized to define whether it behaves like any of its components or independently. Thus, the paradigm of Smad3 nuclear import established here offers an important reference point for future investigations of the mechanisms of Smad protein nucleo-cytoplasmic shuttling.
ACKNOWLEDGMENTS
We are grateful to N. Ferrara and K. Sampath for providing TGF-β1 and BMP-7, respectively, and to G. Blobel, J.-M. Gauthier, D. Görlich, S. Itoh, S. Kuroda, X. Liu, H. F. Lodish, P. ten Dijke, and N. Yaseen for various bacterial and mammalian expression plasmids. We thank K. Miyazono for the adenoviral stocks, M. F. Hoffman, and A. Comer for the baculoviral 6-his-Smad3 proteins, and D.O. Morgan for advice and a thorough review of this manuscript. This research was partly supported by grants from the Human Frontier Science Program (to A.M. and Y.Y.). A.K. is recipient of a postdoctoral fellowship from the Swedish Foundation for International Cooperation in Research and High Education.
REFERENCES
- Adam SA, Marr RS, Gerace L. Nuclear protein import in permeabilized mammalian cells requires soluble cytoplasmic factors. J Cell Biol. 1990;111:807–816. doi: 10.1083/jcb.111.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113:1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- Attisano L, Wrana JL. Smads as transcriptional co-modulators. Curr Opin Cell Biol. 2000;12:235–243. doi: 10.1016/s0955-0674(99)00081-2. [DOI] [PubMed] [Google Scholar]
- Dennler S, Huet S, Gauthier J-M. A short amino-acid sequence in MH1 domain is responsible for functional differences between Smad2 and Smad3. Oncogene. 1999;18:1643–1648. doi: 10.1038/sj.onc.1202729. [DOI] [PubMed] [Google Scholar]
- Dong C, Li Z, Alvarez R, Jr, Feng XH, Goldschmidt-Clermont PJ. Microtubule binding to Smads may regulate TGF β activity. Mol Cell. 2000;5:27–34. doi: 10.1016/s1097-2765(00)80400-1. [DOI] [PubMed] [Google Scholar]
- Görlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Hata A, Lo RS, Wotton D, Lagna G, Massagué J. Mutations increasing autoinhibition inactivate tumor suppressors Smad2 and Smad4. Nature. 1997;388:82–87. doi: 10.1038/40424. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, Richardson MA, Topper JN, Gimbrone MA, Jr, Wrana JL, Falb D. The MAD-related protein Smad7 associates with the TGFβ receptor and functions as an antagonist of TGFβ signaling. Cell. 1997;89:1165–1173. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Heldin C-H, Miyazono K, ten Dijke P. TGf-β“ § 12 signaling from cell membrane to nucleus through SMAD proteins. Nature. 1997;390:465–471. doi: 10.1038/37284. [DOI] [PubMed] [Google Scholar]
- Hieda M, Tachibana T, Yokoya F, Kose S, Imamoto N, Yoneda Y. A monoclonal antibody to the COOH-terminal acidic portion of Ran inhibits both the recycling of Ran and nuclear protein import in living cells. J Cell Biol. 1999;144:645–655. doi: 10.1083/jcb.144.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamoto N, Shimamoto T, Takao T, Tachibana T, Kose S, Matsubae M, Sekimoto T, Shimonishi Y, Yoneda Y. In vivo evidence for involvement of a 58 kDa component of nuclear pore-targeting complex in nuclear protein import. EMBO J. 1995;14:3617–3626. doi: 10.1002/j.1460-2075.1995.tb00031.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K, Kirkpatrick H, Comer A, Hoffmann FM, Laughon A. Interaction of Smad complexes with tripartite DNA-binding sites. J Biol Chem. 1999;274:20709–20716. doi: 10.1074/jbc.274.29.20709. [DOI] [PubMed] [Google Scholar]
- Kose S, Imamoto N, Tachibana T, Shimamoto T, Yoneda Y. Ran-unassisted nuclear migration of a 97-kD component of nuclear pore-targeting complex. J Cell Biol. 1997;139:841–849. doi: 10.1083/jcb.139.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Sun Y, Constantinescu SN, Karam E, Weinberg RA, Lodish HF. Transforming growth factor β-induced phosphorylation of Smad3 is required for growth inhibition and transcriptional induction in epithelial cells. Proc Natl Acad Sci USA. 1997;94:10669–10674. doi: 10.1073/pnas.94.20.10669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J, Blain SW, Lo RS. TGFβ signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Massagué J, Chen YG. Controlling TGF-β signaling. Genes Dev. 2000;14:627–644. [PubMed] [Google Scholar]
- Massagué J, Wotton D. Transcriptional control by the TGF-β/Smad signaling system. EMBO J. 2000;19:1745–1754. doi: 10.1093/emboj/19.8.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuyama N, Hanafusa H, Kusakabe M, Shibuya H, Nishida E. Identification of two Smad4 proteins in Xenopus. Their common and distinct properties. J Biol Chem. 1999;274:12163–12170. doi: 10.1074/jbc.274.17.12163. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- Moore JD, Yang J, Truant R, Kornbluth S. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J Cell Biol. 1999;144:213–224. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morén A, Itoh S, Moustakas A, ten Dijke P, Heldin C-H. Functional consequences of tumorigenic missense mutations in the amino-terminal domain of Smad4. Oncogene. 2000;19:4396–4404. doi: 10.1038/sj.onc.1203798. [DOI] [PubMed] [Google Scholar]
- Nagoshi E, Imamoto N, Sato R, Yoneda Y. Nuclear import of sterol regulatory element-binding protein-2, a basic helix-loop-helix-leucine zipper (bHLH-Zip)-containing transcription factor, occurs through the direct interaction of importin beta with HLH-Zip. Mol Biol Cell. 1999;10:2221–2233. doi: 10.1091/mbc.10.7.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Morén A, Nakayama T, Christian JL, Heuchel R, Itoh S, Kawabata M, Heldin NE, Heldin C-H, ten Dijke P. Identification of Smad7, a TGFβ-inducible antagonist of TGFf-β“ § 12 signaling. Nature. 1997;389:631–635. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Nachury MV, Weis K. The direction of transport through the nuclear pore can be inverted. Proc Natl Acad Sci USA. 1999;96:9622–9627. doi: 10.1073/pnas.96.17.9622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmeri D, Malim MH. Importin β can mediate the nuclear import of an arginine-rich nuclear localization signal in the absence of importin α. Mol Cell Biol. 1999;19:1218–1225. doi: 10.1128/mcb.19.2.1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardali K, Kurisaki A, Morén A, ten Dijke P, Kardassis D, Moustakas A. Role of Smad proteins and transcription factor Sp1 in p21WAF-1/Cip-1 regulation by transforming growth factor-β. J Biol Chem. 2000;275:29244–29256. doi: 10.1074/jbc.M909467199. [DOI] [PubMed] [Google Scholar]
- Piek E, Moustakas A, Kurisaki A, Heldin C-H, ten Dijke P. TGF-β type I receptor/ALK-5 and Smad proteins mediate epithelial to mesenchymal transdifferentiation in NMuMG breast epithelial cells. J Cell Sci. 1999;122:4557–4568. doi: 10.1242/jcs.112.24.4557. [DOI] [PubMed] [Google Scholar]
- Qin B, Lam SS, Lin K. Crystal structure of a transcriptionally active Smad4 fragment. Structure Fold Des. 1999;7:1493–1503. doi: 10.1016/s0969-2126(00)88340-9. [DOI] [PubMed] [Google Scholar]
- Roberts AB, Sporn MB. The transforming growth factors βs. In: Sporn MB, Roberts AB, editors. Peptide Growth Factors and Their Receptors. Heidelberg, Germany: Springer Verlag; 1990. pp. 419–472. [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schedlich LJ, Le Page SL, Firth SM, Briggs LJ, Jans DA, Baxter RC. Nuclear import of insulin-like growth factor-binding protein-3 and -5 is mediated by the importin β subunit. J Biol Chem. 2000;275:23462–23470. doi: 10.1074/jbc.M002208200. [DOI] [PubMed] [Google Scholar]
- Sekimoto T, Imamoto N, Nakajima K, Hirano T, Yoneda Y. Extracellular signal-dependent nuclear import of Stat1 is mediated by nuclear pore-targeting complex formation with NPI-1, but not Rch1. EMBO J. 1997;16:7067–7077. doi: 10.1093/emboj/16.23.7067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y, Hata A, Lo RS, Massagué J, Pavletich NP. A structural basis for mutational inactivation of the tumor suppressor Smad4. Nature. 1997;388:87–93. doi: 10.1038/40431. [DOI] [PubMed] [Google Scholar]
- Shi Y, Wang YF, Jayaraman L, Yang H, Massagué J, Pavletich NP. Crystal structure of a Smad MH1 domain bound to DNA: insights on DNA binding in TGF-β signaling. Cell. 1998;94:585–594. doi: 10.1016/s0092-8674(00)81600-1. [DOI] [PubMed] [Google Scholar]
- Shoji H, Tsuchida K, Kishi H, Yamakawa N, Matsuzaki T, Liu Z, Nakamura T, Sugino H. Identification and characterization of a PDZ protein that interacts with activin type II receptors. J Biol Chem. 2000;275:5485–5492. doi: 10.1074/jbc.275.8.5485. [DOI] [PubMed] [Google Scholar]
- Tachibana T, Hieda M, Sekimoto T, Yoneda Y. Exogenously injected nuclear import factor p10/NTF2 inhibits signal-mediated nuclear import and export of proteins in living cells. FEBS Lett. 1996;397:177–182. doi: 10.1016/s0014-5793(96)01180-5. [DOI] [PubMed] [Google Scholar]
- Takizawa CG, Weis K, Morgan DO. Ran-independent nuclear import of cyclin B1-Cdc2 by importin β. Proc Natl Acad Sci USA. 1999;96:7938–7943. doi: 10.1073/pnas.96.14.7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ten Dijke P, Miyazono K, Heldin C-H. Signaling inputs converge on nuclear effectors in TGFβ signaling. Trends Biochem Sci. 2000;25:64–70. doi: 10.1016/s0968-0004(99)01519-4. [DOI] [PubMed] [Google Scholar]
- Truant R, Cullen BR. The arginine-rich domains present in human immunodeficiency virus type 1 Tat and Rev function as direct importin β-dependent nuclear localization signals. Mol Cell Biol. 1999;19:1210–1217. doi: 10.1128/mcb.19.2.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukazaki T, Chiang TA, Davison AF, Attisano L, Wrana JL. SARA, a Fyre domain protein that recruits Smad2 to the TGF-β receptor. Cell. 1998;95:779–791. doi: 10.1016/s0092-8674(00)81701-8. [DOI] [PubMed] [Google Scholar]
- Xiao Z, Liu X, Henis YI, Lodish HF. A distinct nuclear localization signal in the N terminus of Smad 3 determines its ligand-induced nuclear translocation. Proc Natl Acad Sci USA. 2000a;97:7853–7858. doi: 10.1073/pnas.97.14.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Liu X, Lodish HF. Importin β mediates nuclear translocation of Smad 3. J Biol Chem. 2000b;275:23425–23428. doi: 10.1074/jbc.C000345200. [DOI] [PubMed] [Google Scholar]
- Xu L, Chen YG, Massagué J. The nuclear import function of Smad2 is masked by SARA and unmasked by TGFβ-dependent phosphorylation. Nat Cell Biol. 2000;2:559–562. doi: 10.1038/35019649. [DOI] [PubMed] [Google Scholar]
- Yagi K, Goto D, Hamamoto T, Takenoshita S, Kato M, Miyazono K. Alternatively spliced variant of Smad2 lacking exon 3. Comparison with wild-type Smad2 and Smad3. J Biol Chem. 1999;274:703–709. doi: 10.1074/jbc.274.2.703. [DOI] [PubMed] [Google Scholar]
- Yasukawa T, Kanei-Ishii C, Maekawa T, Fujimoto J, Yamamoto T, Ishii S. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. J Biol Chem. 1995;270:25328–25331. doi: 10.1074/jbc.270.43.25328. [DOI] [PubMed] [Google Scholar]
- Yoneda Y, Hieda M, Nagoshi E, Miyamoto Y. Nucleocytoplasmic protein transport and recycling of Ran. Cell Struct Funct. 1999;24:425–433. doi: 10.1247/csf.24.425. [DOI] [PubMed] [Google Scholar]