Abstract
μ-Conotoxin KIIIA (μ-KIIIA) blocks mammalian voltage-gated sodium channels (VGSCs) and is a potent analgesic following systemic administration in mice. Previous structure-activity studies of μ-KIIIA identified a helical pharmacophore for VGSC blockade. This suggested a route for designing truncated analogues of μ-KIIIA by incorporating the key residues into an α-helical scaffold. As (i, i+4) lactam bridges constitute a proven approach for stabilizing α-helices, we designed and synthesized six truncated analogues of μ-KIIIA containing single lactam bridges at various locations. The helicity of these lactam analogues was analysed by NMR spectroscopy, and their activities were tested against mammalian VGSC subtypes NaV1.1 through 1.7. Two of the analogues, Ac-cyclo9/13[Asp9,Lys13]KIIIA7–14 and Ac-cyclo9/13[Lys9,Asp13]KIIIA7–14, displayed µM activity against VGSC subtypes NaV1.2 and NaV1.6; importantly, the subtype selectivity profile for these peptides matched that of μ-KIIIA. Our study highlights structure-activity relationships within these helical mimetics and provides a basis for the design of additional truncated peptides as potential analgesics.
Introduction
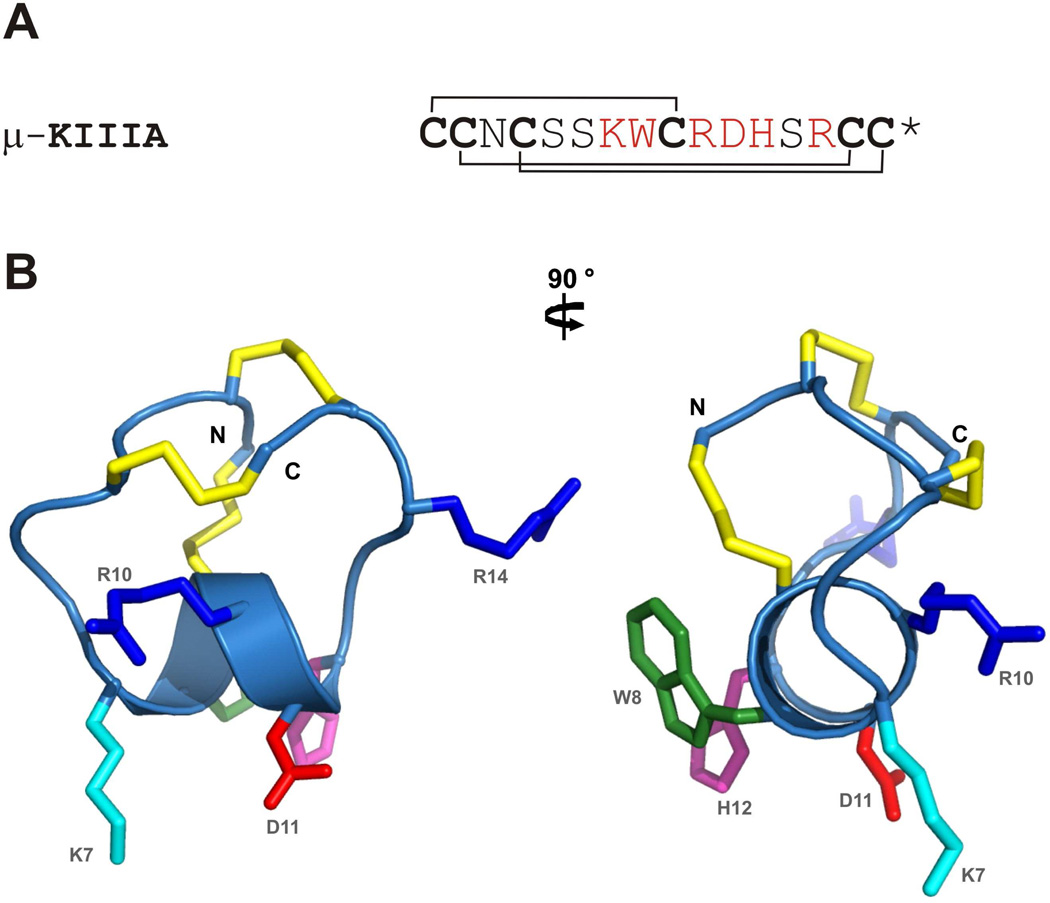
μ-Conotoxin KIIIA (μ-KIIIA), a neuroactive peptide from the marine cone snail Conus kinoshitai, contains 16 amino acid residues and is stabilized by three disulfide bonds.1 In common with previously characterized μ-conotoxins, it blocks voltage-gated sodium channels (VGSCs) by occluding the pore of the channel.2–4 Nine different subtypes of the α-subunit of VGSCs, NaV1.1 through 1.9, have been identified in mammals. NaV1.4 and 1.5 are the skeletal and cardiac muscle-specific subtypes, respectively, while the remaining NaV1 channels are neuronal subtypes. Several of the neuronal subtypes have been implicated in pain perception and identified as potential targets for the treatment of pain because of their involvement in the generation and propagation of action potentials in peripheral pain pathways.5,6 μ-KIIIA has been shown to possess potent analgesic activity following its systemic administration in mice.7 Previous structure-activity studies examining the block of NaV1.2 and 1.4 by μ-KIIIA7 indicated that five of the six residues important for functional activity (K7, W8, R10, D11, H12) occurred in an α-helical region of the peptide and the sixth (R14) was located immediately C-terminal to the helix (Fig.1); moreover, the first disulfide bond could be removed without significant loss of activity.8,9 These findings suggested that the VGSC-blocking activity of μ-KIIIA could be captured in truncated peptides corresponding to the helical region of μ-KIIIA, provided their conformations were stabilized to maintain the helical structure.6
Fig.1.
(A) Amino acid sequence of μ-KIIIA with the disulfide connectivities indicated. Asterisk (*) denotes C-terminal amidation. Key residues important for NaV1.2 and 1.4 blockade are highlighted in red. (B) Closest-to-average solution structure of μ-KIIIA with side chain heavy atoms of the key residues displayed and labelled. Disulfide bonds are shown in gold. Side chains are colored as follows; Arg in blue, Lys in cyan, Asp in red, Trp in green and His in magenta. The two views are related by a 90 ° counter-clockwise rotation about the vertical axis.
Several peptides (ANP, NPY) have been shortened successfully while retaining high potency,10–12 and there are many advantages to such an approach. Smaller peptides are not only less expensive to synthesize but also more amenable to modification, which can be useful for improving specific characteristics to make them more viable as therapeutic leads. In the process of truncating peptides, it is also desirable to remove disulfide bonds as they are susceptible to degradation in some extracellular environments.13,14 In addition, from a synthetic perspective, oxidative folding of multiple-disulfide-bonded peptides can be a time-consuming and inefficient process. Once disulfide bridges are removed, however, it is often necessary to introduce other means of structural stabilization to maintain the bioactive conformation.
In stabilizing α-helices, the use of (i, i+4) lactam bridges has proven to be a successful approach.11, 15–18 Side-chain carboxyl groups of Asp or Glu are linked via an amide bond with side-chain ammonium groups of Lys or Orn (Ornithine) four residues apart, thereby stabilizing the typical 3.6-residue turn of an α-helix. In a study undertaken on helix-stabilizing effects of (i, i+4) side-chain lactam bridges, it was found that a Lys(i) – Asp(i+4) linkage resulted in maximum helicity.19 For a mimetic of μ-KIIIA, the result that Cys9 can be replaced with no significant loss in activity8,9 identifies a position in the helix that can be substituted to form a helix-stabilizing (i, i+4) lactam bridge to either residue 5 or 13, both of which are also non-essential residues.7,8 Alternatively, a bridge can be created between existing Lys7 and Asp11 residues, although the results from a previous alanine walk imply that modifying residues in positions 7 and 11 would reduce activity against the skeletal muscle subtype NaV1.4 more than the neuronal subtype NaV1.2.7
In this study, we have designed and synthesized six truncated analogues of μ-KIIIA (Peptides 1 – 6 in Table 1), each lacking residues at both the N- and C-termini of the native peptide and having the remaining sequence stabilized by a lactam bridge. Both Lys-Asp and Asp-Lys lactam bridges were used to link residues 5–9, 9–13 and 7–11 (using the residue numbering of native μ-KIIIA). The structures of the six lactam analogues were analysed by NMR spectroscopy and their activities were tested against a range of VGSC subtypes. In addition, two analogues lacking a lactam bridge (Peptides 7 and 8 in Table 1) were synthesized to explore the effects of lactam bridge stabilization on the activity against VGSCs.
Table 1.
Amino acid sequences of lactam-bridged and linear analogues of μ-KIIIA. Residues involved in lactam bridge formation are highlighted in bold. Residue numbering follows native μ-KIIIA
| Peptide no. |
Sequence | Lactam position | Peptide name |
|---|---|---|---|
| 1 | Ac-KSKWDRDHSR-NH2 | Lys5-Asp9 | K5D9 |
| 2 | Ac-DSKWKRDHSR-NH2 | Asp5-Lys9 | D5K9 |
| 3 | Ac-KWARDHSR-NH2 | Lys7-Asp11 | K7D11 |
| 4 | Ac-DWARKHSR-NH2 | Asp7-Lys11 | D7K11 |
| 5 | Ac-KWKRDHDR-NH2 | Lys9-Asp13 | K9D13 |
| 6 | Ac-KWDRDHKR-NH2 | Asp9-Lys13 | D9K13 |
| 7 | Ac-KWKRDHDR-NH2 | - | K9D13 linear |
| 8 | Ac-KWDRDHKR-NH2 | - | D9K13 linear |
Results
Synthesis
Peptides 1 – 6 were synthesized by the Boc strategy and cleaved by HF. Purification was accomplished by preparative RP-HPLC. Peptides 1 – 6 were characterized by mass spectrometry, analytical RP-HPLC, and capillary electrophoresis (see Supporting Information).
NMR evidence for α-helicity
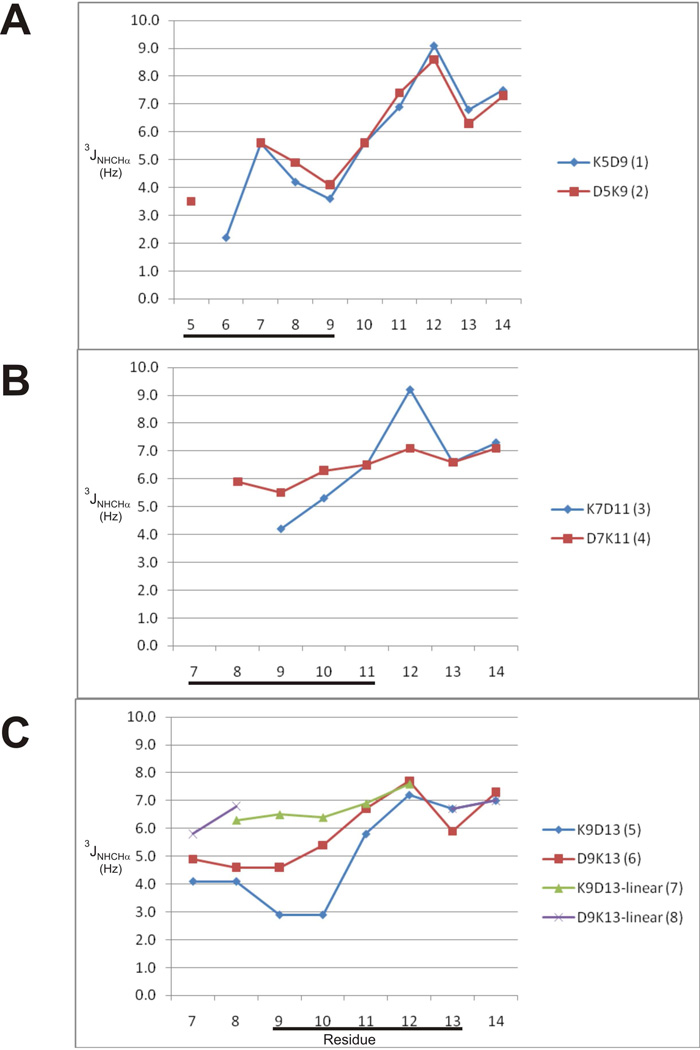
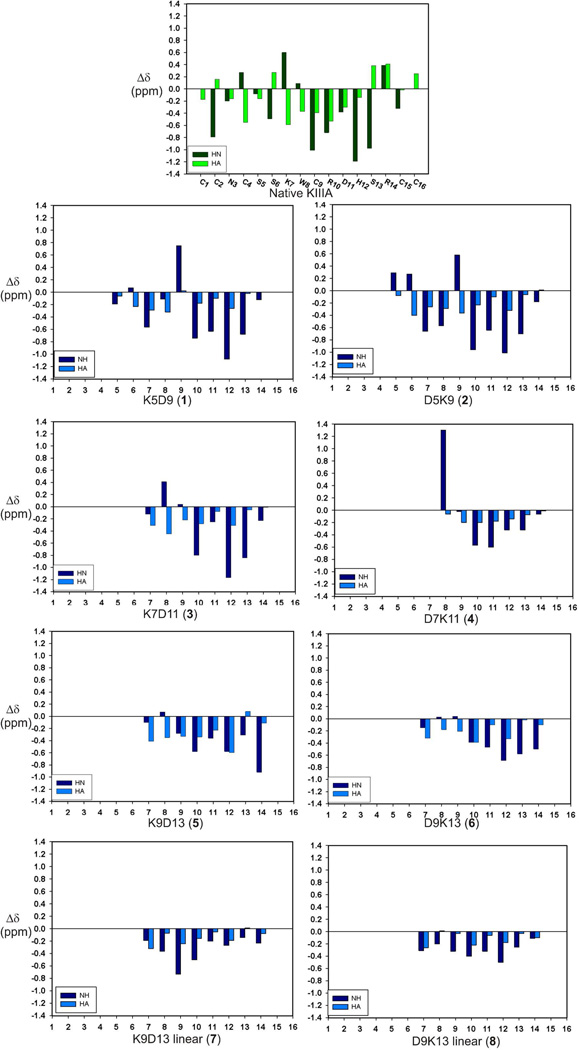
Good quality 1H NMR spectra were obtained for all six lactam analogues. No evidence of multiple conformations was observed, implying that the side-chain lactam bridges were conformationally constrained in all peptides. Inter-chain NOEs observed between the Lys NζH and Asp CβH resonances of the lactam-bridged residues confirmed the presence of the lactam bridge. Sequence-specific resonance assignments for each residue were made using 2D TOCSY and NOESY spectra at 5 °C for lactam analogues 1 – 6. Analysis of measurable temperature coefficients for backbone amide resonances indicated that Arg10 and Asp11 had low values (<4.5 ppb/K) in all lactam analogues except D7K11 (peptide 4). Ser13 in peptides 1 and 3, Asp13 in peptide 5 and Arg14 in peptides 5 and 6 also had small temperature coefficients (<4.0 ppb/K). The low temperature dependence for the amide NH chemical shifts of these residues suggests their possible involvement in hydrogen bonds characterizing an α-helix. Small temperature coefficients (<4.0 ppb/K) were observed for the amide NH chemical shifts of Arg10, Asp11, Ser13 in the native toxin. Several other spectral features pointed to an α-helical structure in these peptides. 3JNHCHα coupling constants < 6 Hz are indicative of α-helical backbone dihedral angles.20 With the exception of the D7K11 lactam analogue, 3JNHCHα coupling constants obtained from 1D and DQF-COSY spectra were < 6 Hz for amide resonances from Lys7 – Arg10 (Fig. 2), indicative of α-helix in this region. Indeed, small coupling constants in this region were comparable to those observed in the corresponding region of the native toxin.8 Coupling constants were also < 6 Hz for residues 5 and 6 in both 5–9 lactam analogues, although in this case the corresponding residues in native μ-KIIIA had large coupling constants (> 7 Hz). In addition, the backbone NH and Hα chemical shifts of the lactam analogues indicate a general upfield shift relative to random coil values, which is characteristic of a helical conformation (Fig. 3).21,22 However, the Hα chemical shifts of the lactam analogues did not deviate as much from random coil values as the Hα chemical shifts in the corresponding helical region (residues 7–12) of native μ-KIIIA (Fig. 3).
Fig.2.
Comparison of 3JNHCαH coupling constants of (A) 5–9 lactam-bridged peptides, (B) 7–11 lactam-bridged peptides and (C) 9–13 lactam-bridged and linear peptides. Coupling constants were measured from 1D and DQF-COSY spectra. Location of lactam bridge is indicated by the black bar.
Fig.3.
Deviation of backbone amide and CαH chemical shifts from random coil values at 5 °C41 for peptides 1 – 8 (blue) compared with that of native μ-KIIIA (green). Respective peptide names are given at the bottom of each graph.
The Hα shifts of the K9D13 lactam analogue displayed the most significant deviations from random coil values, with resonances from residues 7–12 all showing upfield shifts of > 0.2 ppm. With the exception of the terminal residues 7 and 14, the chemical shift deviation pattern of the K9D13 analogue was similar to the corresponding region of native μ-KIIIA. A similar pattern was also observed for the D9K13 analogue with the exception of the NH chemical shift of residue 9 and the Hα chemical shift of residue 13, both of which are involved in lactam bridge formation. In the D7K11 analogue, the Hα chemical shifts of residues 8–14 all showed upfield deviations < 0.2 ppm and the NH chemical shift of Trp8 had a relatively large downfield deviation of 1.3 ppm. In the 5–9 lactam analogues, upfield Hα chemical shift deviations of > 0.1 ppm were observed for residues 6–12, with the exception of residue 9 of K5D9. Ser6, encompassed by a lactam bridge in both K5D9 and D5K9, experienced upfield Hα chemical shift deviations, in contrast to Ser6 in the native μ-KIIIA, which experienced a downfield shift. The NH chemical shifts of Trp8 in both K5D9 and D5K9 also showed upfield chemical shift deviations, differing from the downfield chemical shift deviations observed for Trp8 in native μ-KIIIA as well as the other lactam analogues.
NOE patterns were analysed for the D9K13 lactam analogue. The presence of dαN(i,i+3) and dαβ(i,i+3) NOEs was consistent with helical structure (Fig. SI-1). Possible dαβ(i,i+3) NOEs between residues 9 and 12 and 10 and 13, respectively, could not be assigned unambiguously owing to overlap. Based on the observed NOE pattern in D9K13, it seems that the helix is located towards the N-terminus, not necessarily coincident with the position of the lactam bridge. This correlates well with the 3JNHCHα coupling constants observed for this analogue, in which Lys7 and Trp8, which precede the lactam bridge, both had coupling constants < 6 Hz, whereas Asp11 and His12 within the lactam constraint had coupling constants of 6.7 and 7.7 Hz, respectively.
Coupling constants and chemical shift deviations from random coil values were also determined for the linear analogues of D9K13 and K9D13 (Peptides 7 and 8). Measurable coupling constants of the K9D13 linear analogue from Trp8 to His12 were all > 6 Hz. The coupling constant for Trp8 of the D9K13 linear analogue was also > 6 Hz, but those for Asp9 to His12 could not be measured owing to overlap. Comparing Hα chemical shift deviations from random coil values, both linear analogues had smaller deviations than their lactam-stabilized analogues, with several residues within the 7–11 region showing deviations of < 0.1 ppm. In addition, the amide resonances of the D9K13 linear analogue were not particularly well dispersed, with overlap occurring at 8.28 ppm for residues 9–12, consistent with a lack of helical secondary structure.
VGSC-blocking activity
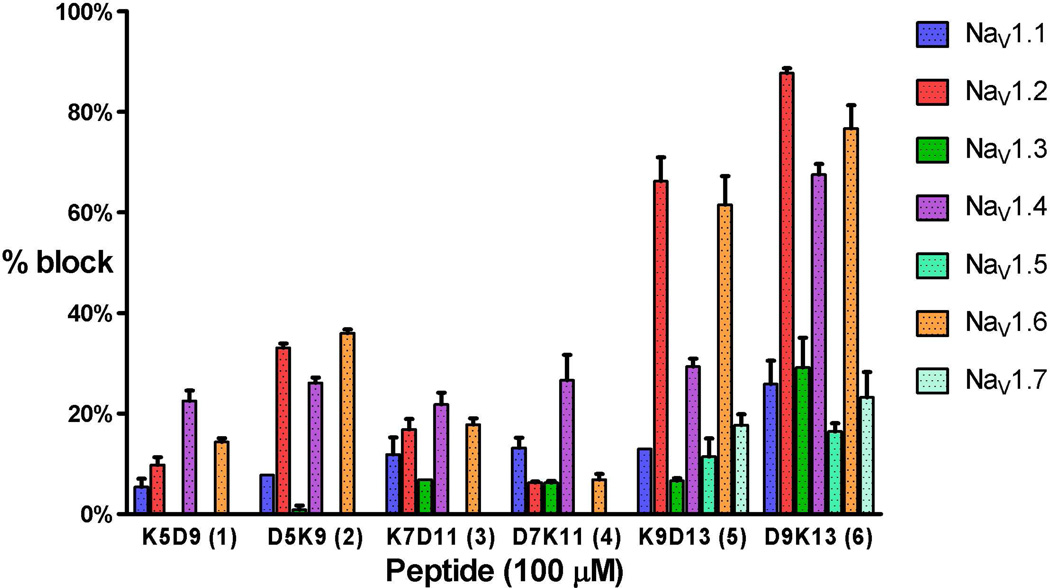
The ability of the truncated μ-KIIIA lactam analogues to block cloned α-subunits of rodent sodium channels expressed in oocytes was assessed by voltage-clamp protocols on seven VGSC subtypes, NaV1.1 through 1.7, as described in Experimental procedures. Based on percentage block at a fixed peptide concentration, the 9–13 lactam analogues were the most active (Fig. 4). At 100 µM, K9D13 displayed strong activity against neuronal subtypes NaV1.2 (66.2 ± 4.7% block) and NaV1.6 (61.5 ± 5.7% block) and modest activity against the skeletal muscle subtype NaV1.4 (29.4 ± 1.6% block). The D9K13 peptide had strong activity against NaV1.2 (87.7 ± 1.0% block), NaV1.4 (67.5 ± 2.1% block) and NaV1.6 (76.7 ± 4.6% block), and relatively strong activity against NaV1.3 (29.1 ± 5.9% block) compared to the other peptides. Only the 9–13 lactam peptides showed measurable activity against NaV1.5 and NaV1.7, albeit < 25%. All lactam-stabilized peptides displayed at least 20% block of NaV1.4. The D5K9 lactam peptide also had moderate activity against NaV1.2 (33.1 ± 0.9% block) and NaV1.6 (36.0 ± 0.7% block). In all cases blockade of VGSC subtypes was reversible (koff values are given in Fig.5).
Fig.4.
Block of NaV1.1 through 1.7 by the lactam-stabilized analogues at a concentration of 100 µM. Functional activities were tested on voltage-clamped Xenopus oocytes expressing the indicated NaV1 subtypes as described in Experimental procedures. NaV1.1, 1.2, 1.3, 1.6 and 1.7 are neuronal subtypes, while NaV1.4 and NaV1.5 are the major isoforms found in skeletal and cardiac muscles, respectively. The most active peptides (K9D13, D9K13, and D5K9) favored the block of NaV1.2, 1.4, and 1.6, while NaV1.4 was preferentially blocked by the remaining peptides. Values represent mean ± S.E.M. (N ≥ 3 oocytes).
Fig.5.
Dose-response curves for K9D13 (left) and D9K13 (right) tested against NaV1.1 through 1.7 expressed in Xenopus oocytes. Curves represent fits of data to the Hill equation, % Block = 100 × 1/(1 + (IC50/[peptide])nH), where % Block was measured by voltage clamp protocols described in Experimental procedures. Data could not be fit for K9D13 with NaV1.1, 1.3 and 1.7; instead their IC50 values (">300 µM") were estimated by visual inspection. koff was determined from single-exponential fits of the time course of recovery from block following toxin washout. Values represent mean ± S.E.M (N ≥ 3 oocytes). In those instances where the kinetics of peptide washout were too fast to be accurately measured, an estimated lower limit for koff (>3.0 min−1) is indicated.
Of the 9–13 lactam analogues, D9K13 was the more potent, with an IC50 of 13.3 ± 0.2 µM against NaV1.2 (Fig. 5). The K9D13 lactam analogue had an IC50 of 52.3 ± 2.8 µM against this subtype. Comparing IC50 values across the VGSC subtypes, the K9D13 analogue seems to be selective for the neuronal subtypes NaV1.2 (IC50 = 52.3 ± 2.8 µM) and NaV1.6 (IC50 = 54.3 ± 9.0 µM).
Linear peptides of the 9–13 lactam analogues were also assayed. At a concentration of 300 µM, the D9K13 linear analogue had little, if any, activity against NaV1.1 through 1.7. In contrast, the K9D13 linear peptide, when tested at 100 and 300 µM, blocked NaV1.2 by 35.9 ± 4.1% and 73.1 ± 3.3%, respectively, with an IC50 of 144 ± 1.1 µM.
Molecular dynamics
Molecular dynamics simulations on each of the truncated analogues of μ-KIIIA, with and without lactam bridges, were conducted to assess the utility of this approach as a probe of conformation and flexibility. Snapshot trajectories of the truncated analogues at 10 ns intervals reflect the flexibility of each peptide during the MD simulation (Fig.SI-2). The helical content of each peptide was analysed throughout the MD simulation, and the percentage of time during which the residue was helical was plotted as a function of residue number (Fig.SI-3). These analyses suggest that the 5–9 lactam analogues were the most helical throughout the simulation, adopting a helical conformation over residues 7–11 for > 50 % of the time of the simulation. The helix across these residues was similarly stable in the 5–9 analogues without the lactam bridge. Of the 9–13 analogues, the K9D13 analogue without the lactam was highly flexible, with < 10 % helicity throughout the simulation. Addition of the lactam bridge improved helix stability slightly. On the other hand, the results of MD simulations on the D9K13 analogue suggested that the analogue without the lactam bridge had higher helix stability compared to that with the lactam.
In cases where a helix was evident in the MD simulations, that helix generally formed between residues 7–11 regardless of the position of the lactam bridge, although the 5–9 lactam analogues were observed to have a slightly more N-terminally extended helix compared to the other analogues. These observations are reflected in the NMR data, where small coupling constants (< 6 Hz) were measured from residues Lys7 to Arg10 in most of the lactam analogues regardless of where the lactam bridge was positioned. Small coupling constants were also observed in residues 5 and 6 of the 5–9 lactam analogues, indicative of an extended helix as predicted by MD simulations.
Overall, however, the results of MD simulations for these truncated peptides did not correlate well with the helical content observed by NMR. The D9K13 lactam analogue was predicted by MD simulations not to have a stable helical structure whereas NMR data (coupling constants, NOE patterns) suggested otherwise. NMR data also suggest a lack of helical structure in both of the 9–13 linear analogues, even though a helix was predicted by MD simulations in the K9D13 linear analogue.
Discussion
In this study, we have described the design, synthesis, structural analysis and VGSC-blocking activity of lactam-stabilized truncated analogues of μ-KIIIA. Our initial design was based on previous structure-activity relationship studies,7 which showed that five of the six residues important for functional activity (K7, W8, R10, D11, H12) occurred in an α-helical region of the peptide and that the first disulfide bond could be removed without significant loss of activity and structure.8 In shortening μ-KIIIA, we retained key residues on a truncated scaffold and incorporated helix-stabilizing (i, i+4) lactam bridges across the α-helical motif of μ-KIIIA.
Activity of 9–13 lactam analogues
In previous studies we investigated the consequences of removing each of the three native disulfide bonds of μ-KIIIA and incorporating a backbone spacer to replace the two non-essential serine residues at positions 5 and 6 in a truncated analogue of μ-KIIIA.8,9 Disulfide-deficient scaffolds of μ-KIIIA have also been used as a more efficient strategy in synthesizing derivatives of μ-KIIIA to study their efficacy and potency in blocking the VGSCs.23 In these studies at least two of the three native disulfide bonds were present, whereas the lactam analogues in the present study represent the first truncated peptides in which all of the native disulfide bonds of μ-KIIIA have been eliminated.
Based on our electrophysiology experiments, the 9–13 lactam analogues displayed significantly higher potency than the other tested lactam analogues, with the most potent analogue, D9K13, achieving an IC50 of 13.3 ± 0.2 µM on the NaV1.2 subtype. This analogue was significantly less potent than wild-type μ-KIIIA, for which an IC50 value of 0.061 µM against NaV1.2 has been reported recently.24 It is clear that removing the disulfide bonds and truncating the peptide compromised bioactivity, which was not unexpected since previous analogues lacking just the Cys4–Cys16 disulfide bond reduced the ability to block VGSCs dramatically.9 Nonetheless, it is encouraging that micromolar activity was achievable in the most active lactam analogue (D9K13). More importantly, the subtype selectivity profile of the 9–13 lactam-bridged peptides mimicked native μ-KIIIA well, and we believe that these smaller peptides, which do not have any requirement for oxidative refolding, represent a valuable platform for defining the structure-activity relationships associated with NaV1 subtype blockade. For the lactam-bridged peptides to be useful as therapeutic leads, further modifications will be required to improve their potency and selectivity.
Effect of lactam position
Our initial designs explored lactam bridges across three positions in truncated analogues of μ-KIIIA: 5–9, 7–11 and 9–13. The results indicate that, regardless of the position of the lactam bridge, residues in the region Lys7 – Arg10 in all six lactam-stabilized analogues displayed helical propensity, similar to that of the parent peptide. Although our NMR studies suggest that these lactam analogues adopted a helical conformation, electrophysiology results indicated that only those analogues with a lactam bridge linking residues 9 and 13 retained significant activity against the VGSCs. It seems that a local constraint towards the C-terminal end of the peptide is more effective than stabilizing either the middle or the N-terminal end, although the NMR data do not suggest any clear evidence of a shift in the location of the helix.
The lactam bridge linking positions 7 and 11 essentially replaces two of the key residues (Lys7 and Asp11) identified as being more important for targeting NaV1.4 than NaV1.2.7 Our electrophysiology results indicated that the ability of the 7–11 lactam analogues to block VGSCs was reduced significantly across all VGSC subtypes, including NaV1.2. It is not known how Lys7 and Asp11 contribute to VGSC blockade; they may be involved in electrostatic interactions with the channel or steric blockade of the channel, or they may form a (i,i+4) salt bridge involved in stabilizing the helix bearing the other key residues.
Effect of lactam orientation
Several studies have investigated the effects of lactam ring size, orientation and location on stabilizing an α-helical conformation in short peptides, although the results of optimal bridge size and orientation have been inconclusive.18–19, 25 Our initial designs investigated the effects of lactam bridge orientation by evaluating both Lys-Asp and Asp-Lys orientations. Analogues with Asp-Lys lactam bridges in the 9–13 and 5–9 lactam analogues were more potent than their oppositely-oriented counterparts. The fact that potency is affected simply by reversing the orientation of the lactam bridge implies that the binding site in the VGSC pore is quite sensitive to changes in the binding peptide. It is possible that the carbonyl group in the lactam bridge interferes with interactions between the key residues and the channel in one orientation more than the other owing to the different lengths of Asp and Lys of the lactam bridge. This could reflect a direct steric interference or an indirect interference that prevents the proper orientation of key side chains interacting with the receptor.
Linear analogues
Linear homologues of the active 9–13 lactam analogues were synthesized to establish whether the lactam bridge indeed stabilizes the helix and is essential for retaining biological activity. While the D9K13 linear peptide had little activity, the K9D13 linear peptide, somewhat surprisingly, displayed significant activity against NaV1.2. This could be explained by the possible formation of a salt bridge between K9 and D13 to stabilize a helical formation. Presumably the reverse orientation may have resulted in a destabilizing interaction between the charged side chains and the helix dipole.26 Measurable 3JNHCHα coupling constants from Trp8 to Asp11 of the K9D13 linear analogue, however, were slightly larger than 6 Hz. It remains to be investigated if eliminating the possibility of salt bridge formation would give a different result. This could be carried out by acetylating the amino group of Lys9 and replacing Asp13 with an Asn residue to neutralize the charge.
Helical conformation and binding
The design of the lactam analogues described here was based on the helical pharmacophore predicted by previous structure-activity relationship studies of μ-KIIIA.7,8 However, it must be borne in mind that the helical region of the toxin may experience some distortion in the bound state.27 For the active 9–13 lactam analogues, measured koff values were much higher than for native μ-KIIIA. The shorter peptides presumably make fewer key binding interactions with the channel, and their greater flexibility may also contribute to the weaker binding and increased dissociation rate. For these truncated peptides, a helix spanning residues 7–11 and a constraint stabilizing the C-terminal end seemed to be the most effective in retaining activity against the VGSCs. Such a constraint clearly stabilizes a helical conformation, as evident from comparing NMR results of the 9–13 lactam analogues and their linear counterparts. However, it is possible that a canonical α-helix is not required for activity.
Conclusions and further work
Both the NaV1.3 and NaV1.7 subtypes have been implicated in transmitting pain signals to the spinal cord. NaV1.3, normally expressed in embryonic neurons, is up-regulated following nerve injury and inflammation, as well as in painful disorders such as trigeminal neuralgia.28,29 The role of NaV1.7 in pain perception has been well studied in families with mutations in this subtype, who are unable to perceive pain.30 Analogues that target these subtypes selectively could be ideal therapeutics to treat pain, although loss of the sense of smell could be a significant side effect from targeting NaV1.7.31
The lactam-stabilized peptide D9K13 exhibited the greatest activity against NaV1.2, 1.4 and 1.6, as well as NaV1.3 and 1.7. The peptides containing 5–9 and 7–11 lactam bridges showed no activity against NaV1.7 and those containing 5–9 bridges also showed none against NaV1.3. Further work is now needed to modify the sequence of D9K13 in order to reduce activity against NaV1.2, 1.4 and 1.6 and enhance activity against NaV1.3 and 1.7. Alternatively, peptides with little activity against NaV1.2, 1.4 and 1.6 could be modified to target the NaV1.3 and NaV1.7 subtypes. As the original Ala scan analogues of μ-KIIIA were tested against only NaV1.2 and 1.4,7 we do not have an ideal basis for creating analogues selective for NaV1.3 and/or 1.7, so this will have to be pursued by creating a limited library of analogues of D9K13 and perhaps K9D13. Recently, an R14A substitution was demonstrated to improve the subtype selectivity for NaV1.7 over NaV1.2 and NaV1.4.32 Other studies have also shown that single residue mutations of positions 7 or 8 can lead to more selective blockers for the neuronal sodium channels.23,24 Our finding that activity was retained in the 9–13 lactam-stabilized analogues establishes this as a possible truncated scaffold to explore subtype selectivity, as compared to the disulfide-containing full-length peptide, which needs to be refolded. In addition to their potential as therapeutics, subtype-selective lactam analogues should be valuable experimental tools in probing the role of VGSCs in pain and other physiological conditions.
Experimental procedures
Synthesis
Peptides 1 – 6 were synthesized manually by Boc solid phase utilizing MBHA resin (0.5 g). Amino acid derivatives, Boc-Ala-OH, Boc-Asp(OFm)-OH, Boc-Arg(Tos)-OH, Boc-His(DNP)-OH, Boc-Lys(Fmoc)-OH, Boc-Ser(Bzl)-OH, and Boc-Trp-OH, were obtained from Bachem Inc. (Torrance, CA), Chem-Impex International (Wood Dale, IL), Novabiochem (San Diego, CA), Reanal (Budapest, Hungary), and AApptec (Louisville, KY). All solvents were reagent grade or better.
Four equivalents of Boc-amino acid (0.8 mmol) based on the original substitution of the MBHA resin (0.4 mmol/ g) were used for each coupling. Peptide couplings were mediated by N,N'-diisopropylcarbodiimide/1-hydroxy-benzotriazole (DIC/HOBt) (0.8 mmol /1.6 mmol) in NMP (4 mL) for 1 h. Boc removal was achieved with neat trifluoroacetic acid (TFA containing 0.5% m-cresol, 15 mL) for 20 min. A methanol wash followed the TFA treatment and then successive washes (23 mL) with triethylamine (TEA) solution (10% in DCM), methanol, and DCM completed the neutralization sequence. After completion of the synthesis, the Boc group of the terminal amino acid was removed with neat TFA for 20 min and then N-terminal acetylation was accomplished by addition of neat acetic anhydride (10 mL) and pyridine (5 drops) for 10 min. Removal of the DNP group of the side chain of His was achieved using thiophenol (10% in NMP, 15 mL) for 2–3 h. Removal of the Fmoc and OFm side chains of Lys and Asp was accomplished by piperidine (20% in NMP) for 20 min. The lactam bridge was formed by treatment with PyBOP/HOBt/DIEA (0.8 mmol/1.6 mmol/2.4 mmol) in NMP for 1.5 h. Peptide-resin (0.75 g) was deprotected and cleaved from the resin by anhydrous HF (10 mL) containing the scavengers anisole (10% v/v) and methyl sulfide (5% v/v) for 60 min at 0°C. The cold diethyl ether-precipitated crude peptide was dissolved in TEAP buffer at pH 2.25 and purified using a preparative Shimadzu RP-HPLC system (two Shimadzu LC 8A pumps, an SCL-10A controller, and an SDP 10A UV detector), a Waters PrepPak cartridge (4.7 × 30 cm) packed with Grace Vydac C4 (15–20 µm particle size), and the TEAP solvent system (Eluent A = TEAP at pH 2.25, eluent B = 60% CH3CN, 40% A). A linear gradient of 0.5% B/min increased from 0% B with a flow rate of 100 mL/min; detection was at 220 nm. Analytical HPLC screening was performed on a Grace Vydac C18 column (0.46 × 25 cm, 5 µm particle size, 300 Å pore size) using a Shimadzu analytical HPLC system (two Shimadzu LC 10AT pumps, an SCL-10A controller, and an SDP 10A UV detector). A linear gradient of 1% B/min increased from 0%B to 50%B (Eluent A = 0.05% TFA/H2O, Eluent B = 0.05% TFA/CH3CN) at a flow rate of 1.2 mL/min, with detection at 210 nm. Fractions containing the desired product were pooled and desalted in a second purification step on the same column using a linear gradient of 0.5% B/min which increased from 0% B (Eluent A = 0.1% TFA, eluent B = 60% CH3CN, 40% A). The desired product was lyophilized. See Supporting Information (Table SI-2) for yields.
Peptides 7 and 8 were purchased from GLBiochem (Shanghai, China) and were synthesized with purities of 98.6 % and 98.9 %, respectively. Identities were confirmed by mass spectrometry. All peptides were determined to be ≥95 % pure by analytical RP-HPLC.
NMR spectroscopy
NMR samples were prepared by dissolving 5–10 mg of peptide in 550 µL of 94% H2O / 6% 2H2O. For each peptide, a series of one-dimensional spectra at 5 °C intervals was collected over the temperature range 5–25 °C. Two-dimensional homonuclear total correlation (TOCSY) spectra with a spin-lock time of 70 ms, nuclear Overhauser enhancement (NOESY) spectra with mixing times of 50 and 250 ms, and double quantum filtered correlation (DQF-COSY) spectra were acquired at 600 MHz on a Bruker DRX-600 spectrometer. The water resonance was suppressed using the WATERGATE pulse sequence.33 All spectra were acquired at 5 °C and pH 3.0 unless otherwise stated and were referenced via the water resonance. Spectra were processed using TOPSPIN (Version 1.3, Bruker Biospin) and analysed using XEASY (Version 1.3.13).34
Molecular dynamics
Models of the lactam analogues were generated from the solution structure of μ-KIIIA (BMRB entry 20048) by homology modelling methods using Modeller (8v2) software.35 The 20 experimental NMR structures of μ-KIIIA were each used to generate 25 initial models. From each of the 25 initial models the model with the lowest Modeller Objective Function was selected as the representative structure for each of the 20 experimental templates. The representative structure that was most similar to the other 19 models, selected based on a Principal Component Analysis of the root-mean-square deviation of the superimposition of backbone atoms, was used in further modelling. Lactam bridges were built into the models using InsightII (Accelrys, 2005). The representative model was subjected to molecular dynamics simulations using the GROMACS version 3.3.1 package of programs36 and the OPLS-aa force field.37 The peptides were solvated in a cubic box of water with dimensions of 40 Å. The ionisable residues Arg, His and Lys each carried a +1 formal charge, while Asp was modelled with a −1 charge; the total charge of the system was made neutral by replacing water molecules with chloride ions. The LINCS algorithm was used to constrain bond lengths.38 Peptide, water, and ions were coupled separately to a thermal bath at 300 K using velocity rescaling39 applied with a coupling time of 0.1 ps, while the pressure was maintained at a reference pressure of 1 bar through coupling to an isotropic barostat using a time constant of 0.5 ps and compressibility of 4.5 × 10−5 bar−1. All simulations were performed with a single non-bonded cutoff of 10 Å, applying a neighbor-list update frequency of 10 steps (20 fs). The Particle-Mesh Ewald method was applied to deal with long-range electrostatics with a grid width of 1.2 Å and fourth-order spline interpolation. All simulations consisted of an initial minimization to prevent close contacts, followed by 10 ps of positional restrained MD to equilibrate the water molecules with the polypeptide fixed. The time step used in all the simulations was 2 fs and each MD simulation was run for a total period of 50 ns. The helical contents of the conformations sampled throughout the simulation were analysed using the g_helix function in GROMACS.
Electrophysiology of mammalian NaV1 clones expressed in Xenopus laevis oocytes
Oocytes expressing one of the α-subunits NaV1.1 through 1.7 (all from rat except NaV1.6, which was from mouse) were prepared and two-electrode voltage-clamped essentially as described previously.7, 40 Briefly, oocytes were placed in a 30 µl chamber containing ND96 and two-electrode voltage-clamped at a holding potential of −100 mV. To activate VGSCs, the membrane potential was stepped to a value between −20 and 0 mV (depending on NaV1 subtype) for a 50 ms period every 20 sec. To apply peptide, the perfusion was halted, 3 µL of peptide solution (at ten times the final concentration) was added to the 30 µL bath, and the bath was manually stirred for about 5 s by gently aspirating and expelling a few µL of the bath fluid several times with a pipettor. Peptide exposures were in static baths to conserve material. All recordings were made at room temperature (~21 °C). Steady-state IC50 values were obtained by fitting block percentage versus [peptide] data to the Hill equation, Y = 100 × 1/(1 + (IC50/[peptide])nH), where Y is the % block of the peak of the voltage-activated sodium current. Values of koff were determined from single-exponential fits of the time course of recovery from block following toxin washout. Curve fittings were done with KaleidaGraph (Synergy Software), Prism (GraphPad Software), or in-house software written in LabVIEW (National Instruments).
Supplementary Material
Acknowledgements
We thank Prof. Alan A. Goldin for the NaV1.1 through 1.6 clones, Prof. Gail Mandel for the NaV1.7 clone, and Dr. Layla Azam for the cRNA from these clones. This work was supported in part by an Australian Research Council grant DP1094212 (to R.S.N), NIH grant GM 48677 (to J.G., J.E.R., G.B. and D.Y.) as well as NHMRC IRIISS Grant 361646 and a Victorian State Government OIS grant (B.J.S.). R.S.N. acknowledges fellowship support from the Australian National Health and Medical Research Council.
Abbreviations
- Boc
t-Butoxycarbonyl
- Bzl
benzyl
- DCM
dichloromethane
- DIC
N,N'-diisopropylcarbodiimide
- DIEA
N,N-diisopropylethylamine
- Fmoc
Fluorenylmethyloxycarbonyl chloride
- HOBt
1-hydroxy-benzotriazole
- μ-KIIIA
μ-conotoxin KIIIA from Conus kinoshitai
- MBHA
4-methylbenzhydrylamine
- MD
molecular dynamics
- NaV1.1, NaV1.2 etc.
α-subunit of the voltage-gated sodium channel subtype 1.1, 1.2, etc.
- NMP
N-methylpyrrolidinone
- OcHex
cyclohexyl ester
- OFm
9-fluorenylmethyl ester
- PyBOP
Benzotriazol-1-yloxy)tripyrrolidinophosphonium hexafluorophosphate
- RMS
root mean square
- TEA
triethylamine
- TEAP
triethylammonium phosphate
- TFA
trifluoroacetic acid
- VGSC
voltage-gated sodium channel
The abbreviations for the common amino acids are in accordance with the recommendations of the IUPAC-IUB Joint Commission on Biochemical Nomenclature (Eur. J. Biochem. 1984, 138:9–37); the symbols represent the L-isomers.
Footnotes
Supporting Information Available: one table containing assigned NH and Hα chemical shifts and 3JNHCHα for Peptides 1 – 8, one table showing characterization of Peptides 1 – 8 by HPLC, capillary electrophoresis and mass spectroscopy, one figure showing NOE connectivity for D9K13 and two figures showing MD results. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bulaj G, West PJ, Garrett JE, Watkins M, Zhang MM, Norton RS, Smith BJ, Yoshikami D, Olivera BM. Novel conotoxins from Conus striatus and Conus kinoshitai selectively block TTX-resistant sodium channels. Biochemistry. 2005;44:7259–7265. doi: 10.1021/bi0473408. [DOI] [PubMed] [Google Scholar]
- 2.Norton RS, Olivera BM. Conotoxins down under. Toxicon. 2006;48:780–798. doi: 10.1016/j.toxicon.2006.07.022. [DOI] [PubMed] [Google Scholar]
- 3.Terlau H, Olivera BM. Conus venoms: a rich source of novel ion channel-targeted peptides. Physiol. Rev. 2004;84:41–68. doi: 10.1152/physrev.00020.2003. [DOI] [PubMed] [Google Scholar]
- 4.Zhang MM, McArthur JR, Azam L, Bulaj G, Olivera BM, French RJ, Yoshikami D. Synergistic and antagonistic interactions between tetrodotoxin and μ-conotoxin in blocking voltage-gated sodium channels. Channels (Austin) 2009;3:32–38. doi: 10.4161/chan.3.1.7500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Momin A, Wood JN. Sensory neuron voltage-gated sodium channels as analgesic drug targets. Curr. Opin. Neurobiol. 2008;18:383–388. doi: 10.1016/j.conb.2008.08.017. [DOI] [PubMed] [Google Scholar]
- 6.Norton RS. μ-conotoxins as leads in the development of new analgesics. Molecules. 2010;15:2825–2844. doi: 10.3390/molecules15042825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang MM, Green BR, Catlin P, Fiedler B, Azam L, Chadwick A, Terlau H, McArthur JR, French RJ, Gulyas J, Rivier JE, Smith BJ, Norton RS, Olivera BM, Yoshikami D, Bulaj G. Structure/function characterization of μ-conotoxin KIIIA, an analgesic, nearly irreversible blocker of mammalian neuronal sodium channels. J. Biol. Chem. 2007;282:30699–30706. doi: 10.1074/jbc.M704616200. [DOI] [PubMed] [Google Scholar]
- 8.Khoo KK, Feng ZP, Smith BJ, Zhang MM, Yoshikami D, Olivera BM, Bulaj G, Norton RS. Structure of the analgesic μ-conotoxin KIIIA and effects on the structure and function of disulfide deletion. Biochemistry. 2009;48:1210–1219. doi: 10.1021/bi801998a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han TS, Zhang MM, Walewska A, Gruszczynski P, Robertson CR, Cheatham TE, 3rd, Yoshikami D, Olivera BM, Bulaj G. Structurally minimized μ-conotoxin analogues as sodium channel blockers: implications for designing conopeptide-based therapeutics. Chemmedchem. 2009;4:406–414. doi: 10.1002/cmdc.200800292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li B, Tom JY, Oare D, Yen R, Fairbrother WJ, Wells JA, Cunningham BC. Minimization of a polypeptide hormone. Science. 1995;270:1657–1660. doi: 10.1126/science.270.5242.1657. [DOI] [PubMed] [Google Scholar]
- 11.Yao S, Smith-White MA, Potter EK, Norton RS. Stabilization of the helical structure of Y2-selective analogues of neuropeptide Y by lactam bridges. J. Med. Chem. 2002;45:2310–2318. doi: 10.1021/jm010543z. [DOI] [PubMed] [Google Scholar]
- 12.Moellering RE, Cornejo M, Davis TN, Del Bianco C, Aster JC, Blacklow SC, Kung AL, Gilliland DG, Verdine GL, Bradner JE. Direct inhibition of the NOTCH transcription factor complex. Nature. 2009;462:182–188. doi: 10.1038/nature08543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khoo KK, Norton RS. Role of disulfide bonds in peptide and protein conformation. In: Hughes AB, editor. Amino Acids, Peptides and Proteins in Organic Chemistry. Vol. 5. Weinheim, Germany: Wiley-VCH; 2010. p in press. [Google Scholar]
- 14.Trivedi MV, Laurence JS, Siahaan TJ. The role of thiols and disulfides on protein stability. Curr. Protein Pept. Sci. 2009;10:614–625. doi: 10.2174/138920309789630534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirby DA, Britton KT, Aubert ML, Rivier JE. Identification of high-potency neuropeptide Y analogues through systematic lactamization. J. Med. Chem. 1997;40:210–215. doi: 10.1021/jm960593h. [DOI] [PubMed] [Google Scholar]
- 16.Andrews MJI, Tabor AB. Forming Stable Helical Peptides Using Natural and Artificial Amino Acids. Tetrahedron. 1999;55:11711–11743. [Google Scholar]
- 17.Lanigan MD, Pennington MW, Lefievre Y, Rauer H, Norton RS. Designed peptide analogues of the potassium channel blocker ShK toxin. Biochemistry. 2001;40:15528–15537. doi: 10.1021/bi011300b. [DOI] [PubMed] [Google Scholar]
- 18.Taylor JW. The synthesis and study of side-chain lactam-bridged peptides. Biopolymers. 2002;66:49–75. doi: 10.1002/bip.10203. [DOI] [PubMed] [Google Scholar]
- 19.Shepherd NE, Hoang HN, Abbenante G, Fairlie DP. Single turn peptide alpha helices with exceptional stability in water. J. Am. Chem. Soc. 2005;127:2974–2983. doi: 10.1021/ja0456003. [DOI] [PubMed] [Google Scholar]
- 20.Pardi A, Billeter M, Wuthrich K. Calibration of the angular dependence of the amide proton-C alpha proton coupling constants, 3JHNHα, in a globular protein. Use of 3JHNHα for identification of helical secondary structure. J. Mol. Biol. 1984;180:741–751. doi: 10.1016/0022-2836(84)90035-4. [DOI] [PubMed] [Google Scholar]
- 21.Wishart DS, Sykes BD, Richards FM. Relationship between nuclear magnetic resonance chemical shift and protein secondary structure. J. Mol. Biol. 1991;222:311–333. doi: 10.1016/0022-2836(91)90214-q. [DOI] [PubMed] [Google Scholar]
- 22.Wishart DS, Sykes BD, Richards FM. The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy. Biochemistry. 1992;31:1647–1651. doi: 10.1021/bi00121a010. [DOI] [PubMed] [Google Scholar]
- 23.Zhang MM, Han TS, Olivera BM, Bulaj G, Yoshikami D. μ-conotoxin KIIIA derivatives with divergent affinities versus efficacies in blocking voltage-gated sodium channels. Biochemistry. 2010;49:4804–4812. doi: 10.1021/bi100207k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Van Der Haegen A, Peigneur S, Tytgat J. Importance of position 8 in μ-conotoxin KIIIA for voltage-gated sodium channel selectivity. FEBS J. 2011;278:3408–3418. doi: 10.1111/j.1742-4658.2011.08264.x. [DOI] [PubMed] [Google Scholar]
- 25.Houston ME, Jr, Gannon CL, Kay CM, Hodges RS. Lactam bridge stabilization of alpha-helical peptides: ring size, orientation and positional effects. J. Pept. Sci. 1995;1:274–282. doi: 10.1002/psc.310010408. [DOI] [PubMed] [Google Scholar]
- 26.Marqusee S, Baldwin RL. Helix stabilization by Glu-…Lys+ salt bridges in short peptides of de novo design. Proc. Natl. Acad. Sci. U S A. 1987;84:8898–8902. doi: 10.1073/pnas.84.24.8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yao S, Zhang MM, Yoshikami D, Azam L, Olivera BM, Bulaj G, Norton RS. Structure, dynamics, and selectivity of the sodium channel blocker μ-conotoxin SIIIA. Biochemistry. 2008;47:10940–10949. doi: 10.1021/bi801010u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cummins TR, Sheets PL, Waxman SG. The roles of sodium channels in nociception: Implications for mechanisms of pain. Pain. 2007;131:243–257. doi: 10.1016/j.pain.2007.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siqueira SR, Alves B, Malpartida HM, Teixeira MJ, Siqueira JT. Abnormal expression of voltage-gated sodium channels NaV1.7, NaV1.3 and NaV1.8 in trigeminal neuralgia. Neuroscience. 2009;164:573–577. doi: 10.1016/j.neuroscience.2009.08.037. [DOI] [PubMed] [Google Scholar]
- 30.Cox JJ, Reimann F, Nicholas AK, Thornton G, Roberts E, Springell K, Karbani G, Jafri H, Mannan J, Raashid Y, Al-Gazali L, Hamamy H, Valente EM, Gorman S, Williams R, McHale DP, Wood JN, Gribble FM, Woods CG. An SCN9A channelopathy causes congenital inability to experience pain. Nature. 2006;444:894–898. doi: 10.1038/nature05413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weiss J, Pyrski M, Jacobi E, Bufe B, Willnecker V, Schick B, Zizzari P, Gossage SJ, Greer CA, Leinders-Zufall T, Woods CG, Wood JN, Zufall F. Loss-of-function mutations in sodium channel NaV1.7 cause anosmia. Nature. 2011;472:186–190. doi: 10.1038/nature09975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McArthur JR, Singh G, McMaster D, Winkfein R, Tieleman DP, French RJ. Interactions of key charged residues contributing to selective block of neuronal sodium channels by μ-conotoxin KIIIA. Mol. Pharmacol. 2011;80:573–584. doi: 10.1124/mol.111.073460. [DOI] [PubMed] [Google Scholar]
- 33.Piotto M, Saudek V, Sklenar V. Gradient-tailored excitation for single-quantum NMR spectroscopy of aqueous solutions. J. Biomol. NMR. 1992;2:661–665. doi: 10.1007/BF02192855. [DOI] [PubMed] [Google Scholar]
- 34.Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral-analysis of biological macromolecules. J. Biomol. NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 35.Fiser A, Sali A. Modeller: generation and refinement of homology-based protein structure models. Method. Enzymol. 2003;374:461–491. doi: 10.1016/S0076-6879(03)74020-8. [DOI] [PubMed] [Google Scholar]
- 36.Lindahl E, Hess B, van der Spoel D. Gromacs 3.0: A package for molecular simulation and trajectory analysis. J. Mol. Model. 2001;7:306–317. [Google Scholar]
- 37.Jorgensen WL, Tirado-Rives J. The OPLS potential functions for proteins. Energy minimizations for crystals of cyclic peptides and crambin. J. Am. Chem. Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 38.Hess B, Bekker H, Berendsen HJC. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 1977;18:1463–1472. [Google Scholar]
- 39.Bussi G, Donadio D, Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 40.Wilson MJ, Yoshikami D, Azam L, Gajewiak J, Olivera BM, Bulaj G, Zhang MM. μ-Conotoxins that differentially block sodium channels NaV1.1 through 1.8 identify those responsible for action potentials in sciatic nerve. Proc. Natl. Acad. Sci. U S A. 2011;108:10302–10307. doi: 10.1073/pnas.1107027108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Merutka G, Dyson HJ, Wright PE. 'Random coil' 1H chemical shifts obtained as a function of temperature and trifluoroethanol concentration for the peptide series GGXGG. J. Biomol. NMR. 1995;5:14–24. doi: 10.1007/BF00227466. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.