Abstract
It is challenging to effectively deprotect hydroxyl groups of acid-or-base sensitive bio-macromolecules without causing even minor defects and compromising high quality of final products. We report here a mild detritylation strategy in mildly acidic buffers to remove the DMTr protection from the 5’-hydroxyl groups of synthetic nucleic acids. The DMTr-groups can be easily and effectively removed at pH 4.5 or 5.0 with slight warming-up (40 °C), offering virtually quantitative deprotection. This warming-up strategy is particularly useful for deprotection of the modified nucleic acids that are sensitive to the conventional acid deprotection. As a first step towards our long-term goal of synthesizing defect-free nucleic acids, our novel and simple strategy further increases the quality of synthetic nucleic acids.
Keywords: DNA, RNA, detritylation, mild deprotection condition, modified nucleic acid
INTRODUCTION
Protection of hydroxyl groups is constantly required in the syntheses of bio-macromolecules, such as nucleic acids and saccharides. Due to the presence of multiple functionalities in a bio-macromolecule, selective deprotection of the hydroxyl groups without causing even minor molecular decompositions (defects) is challenging. The deprotection is a critical step of the synthesis, such as chemical synthesis of modified and acid-labile nucleic acids. Because of the easy access and convenient synthesis of nucleic acids on solid phase, synthetic nucleic acids and their analogs have found a myriad of applications in genetic engineering, therapeutics, structural biology, nucleic acid-protein interactions, forensics, bioinformatics, DNA nanotechnology, and history and anthropology.[1–7] Clearly, nucleic acids synthesized by the well-established approach have dramatically advanced nucleic acid research field. However, the chemically synthesized nucleic acids suffer from minor defects (less than 1%), such as depurination, deamination and abasic-site formation. These defects in synthesized longer nucleic acids are difficult to detect. Fortunately, these minor defects do not cause obvious disruptions in most of the current applications. Since DNA and RNA molecules are fully protected during the solid-phase synthesis, minor defects mostly likely occur during the deprotection steps, where relatively strong bases and acids are used.
Mild deprotection is critical for preparing defect-free nucleic acids, especially those used in applications sensitive to the molecular defects, such as DNA and RNA crystallization and structure determination, which require defect-free molecules for perfect molecular packing. So far, we have developed a mild detritylation strategy as a first step towards our long-term goal of synthesizing defect-free nucleic acids. In the conventional 5’-hydroxyl deprotection (the detritylation step), the hydrophobic 5’-DMTr protecting group is removed predominantly by a relatively strong acid treatment. In the synthesis, the nucleic acids (or oligonucleotides) of interest are often prepared on a DNA/RNA synthesizer in the DMTr-on mode, which allows convenient separation of the full-length oligonucleotide from failure sequences by HPLC.[8]Isolation and purification are usually accomplished by reversed-phase HPLC (RP-HPLC) or by purification cartridges (Glen-Pak, Fluoro-pak or Clarity). In order to purify oligonucleotides, the DMTr protection on 5’-HO groups is normally kept to facilitate reversed-phase HPLC purification. Finally, this 5’-hydroxyl protection is removed by a relatively strong acid treatment. Treating nucleic acids with a strong acid can cause minor depurination.[9–13]
RESULTS AND DISCUSSION
In order to avoid a strong acid treatment, herein we report a mild strategy (at pH 4.5 or 5.0 and 40 °C) to remove the DMTr protection from the 5’-hydroxyl groups of nucleic acids without the depurination and backbone cleavage. Our warming-up strategy avoids the strong acid treatment and base neutralization, minimizes consumption of extra reagents, and significantly simplifies the deprotection procedure. This warming-up approach is useful to deprotect the hydroxyl groups from nucleic acids, especially modified nucleic acids sensitive to the conventional acid treatment, and to simplify preparation of siRNAs, microRNAs and antisense DNAs with potential therapeutic applications. This novel and simple strategy is an important step toward high-quality and defect-free nucleic acids, which are extremely useful for microchip fabrication, disease detection, nucleic acid derivatization, and X-ray crystal structure study of nucleic acids and protein-nucleic acid complexes.[14–16] Furthermore, this mild warming-up approach will be useful in synthesis of many other bio-molecules that are sensitive to a strong acid treatment.
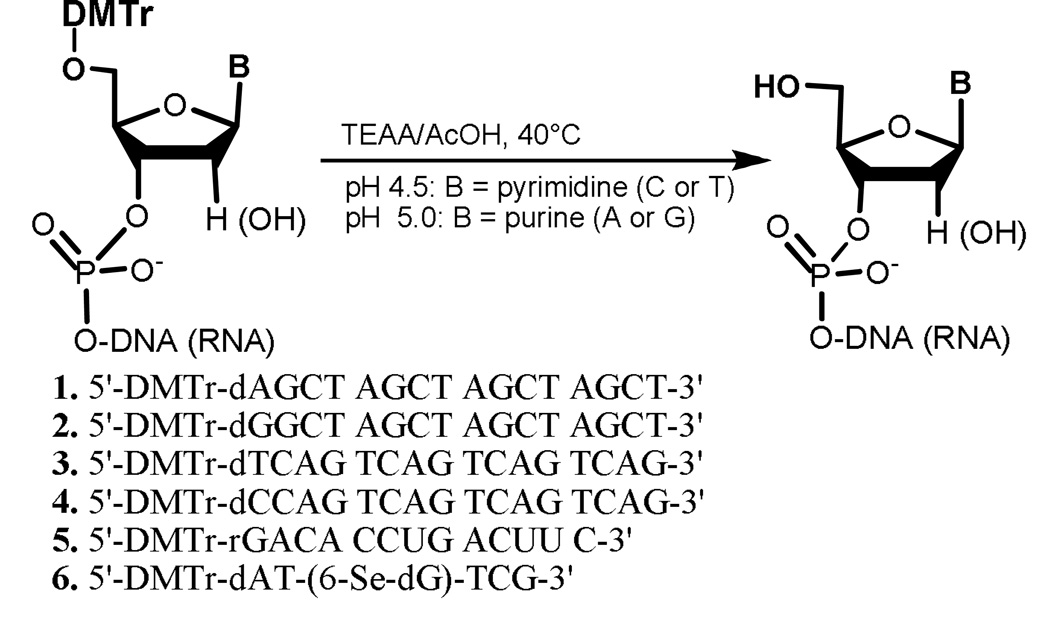
The standard DMTr deprotection from the 5’-hydroxyl groups of synthetic nucleic acids[17–18] requires the use of 80% acetic acid (pKa = 4.8),[8] dichloroacetic acid (DCA, pKa = 1.5), trichloroacetic acid (TCA, pKa = 0.7),[19] or trifluoroacetic (TFA, pKa = 0.5), resulting in a strongly acidic solution. The acidic solution may cause minor depurination and backbone cleavage.[11] To avoid these problems, we decided to investigate the possibility of removing DMTr from the 5’-hydroxyl groups of oligonucleotides (Scheme 1) under a mildly acidic condition, such as using 50 mM triethylammonium acetate (TEAA) buffer at a pH ranging from 4.5 to 6. Oligonucleotides 1–6 were chosen as model DNA or RNA sequences with different 5’-terminal nucleotides. Use of the mildly acidic TEAA solution with low ionic strength (50 mM) allowed us to easily adjust the pH for the detritylation reaction, monitored by RP-HPLC (Figure 1).
SCHEME 1.
Mild removal of DMTr protection from the 5’-hydroxyl groups of nucleic acids.
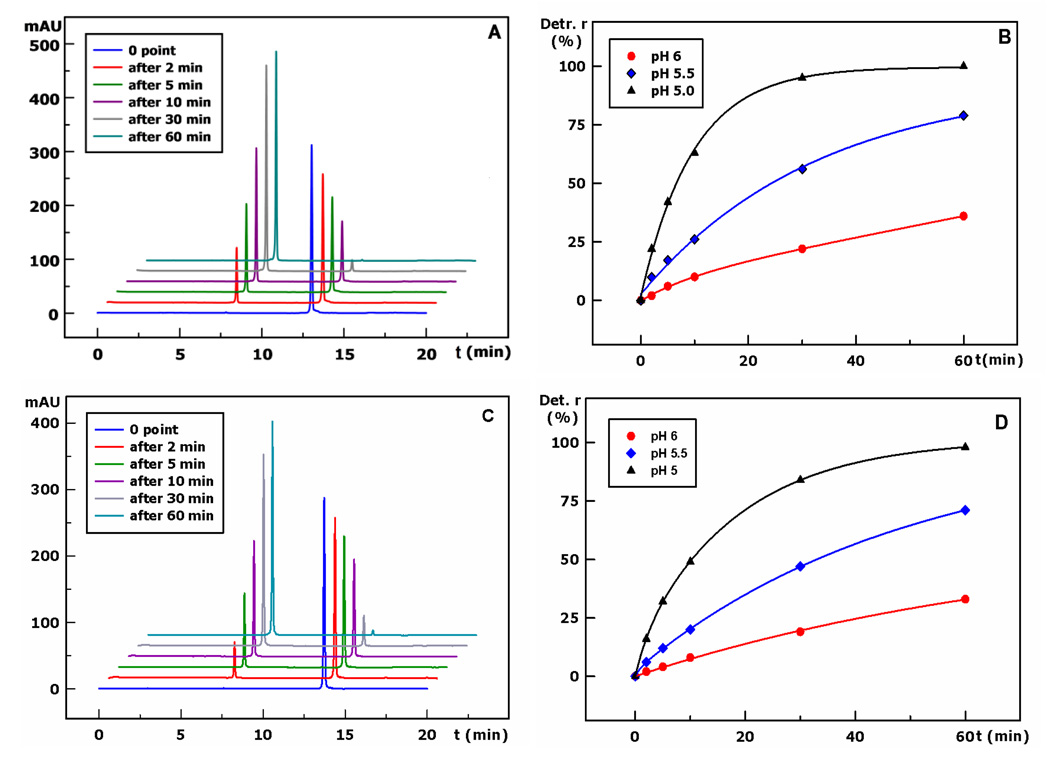
FIGURE 1.
DMTr deprotection study (at 40 °C) at different time and pH. A) The detritylation progress of the 16-mer DNA (5’-DMTr-dAGCTAGCTAGCTAGCT) over a period of 0, 2, 5, 10, 30, and 60 min (pH 5.0); HPLC gradient: from 5 to 40% of buffer B in 15 min; B) the detritylation as a function of time at pH 5.0, 5.5, and 6.0; C) the detritylation progress of the 13-mer RNA (5’-DMTr-rGACACCUGACUUC) over a period of 0, 2, 5, 10, 30, and 60 min (pH 5.0); HPLC gradient: from 5 to 40% of buffer B in 15 min; D) the detritylation as a function of time at pH 5.0, 5.5, and 6.0.
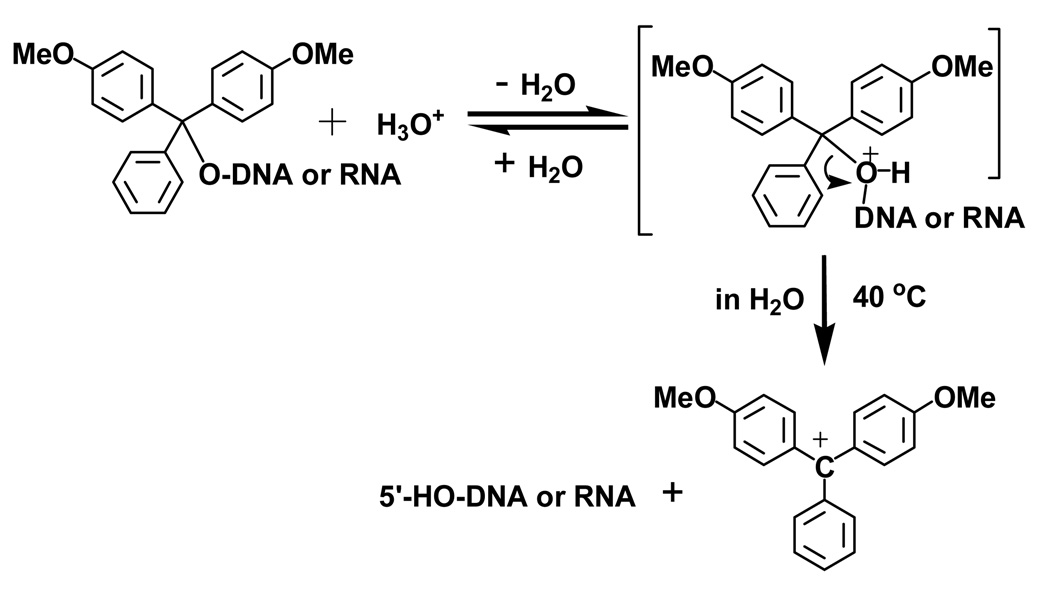
Since the DMTr cation is stable (Scheme 2), we reasoned that in aqueous solution, the rate of the O-C bond [5’-O-C(Ar)3] cleavage is increased at an elevated temperature, resulting in the stable DMTr carbocation and the desired 5’-HO-oligonucleotide (R-OH).[19–21]Our experiments indicated that the acid-dependent detritylation reaction is also sensitive to higher temperature, especially in an aqueous medium. The rate of detritylation reaction increases when the pH decreases. Moreover, an increase in temperature facilitates the detritylation reaction rate. As we expected, the detritylation reaction is slow at the room temperature, when solution of a DMTr-on nucleic acid is just slightly acidic. However, a temperature increase (such as up to 40 °C) can largely facilitate this deprotection reaction (Figure 1). Oligonucleotides (1–6) were examined under various pH values and temperatures in order to study their detritylation. The results are summarized in Table 1. Consistent with our hypothesis, our experimental results indicate that the pH (such as 5.0) and temperature (such as 40 °C) play critical roles in this deprotection reaction (Figure 1). Our experimental data indicated that the nature of the 5’-terminal nucleotides can also have minor effects on the detritylation reaction; the deprotection rate: dA>dG>dC>T (Table 1 and Figure 1). Furthermore, under the same conditions, the detritylation of DNA is slightly faster than that of RNA (Table 1 and Figure 1). Since the detritylation reactions were performed under conditions of low salt (less than 2 mM) and elevated temperature (40 °C), where the duplexes were not formed, the sequence self-complementarity did not play a role in the detritylation. This general acid-catalyzed reaction shows a large increase of detritylated products when the pH decreases (Figure 1). Moreover, the depurination and backbone cleavage were not detected under the mild conditions for removing the DMTr groups.
SCHEME 2.
The warming-up mechanism of the DMTr removal from the 5’-hydroxyl groups of nucleic acids. The DMTr cation is rapidly quenched in aqueous solution and the red color of the cation shown here as the end product of the reaction is not observed.
TABLE 1.
Mild removal of the 5’-O-DMTr group from the native and modified oligonucleotides (1–6) in Scheme 1a
| Oligo -entry |
pH b |
t1/2c (min) |
Td (min) |
Yielde (%) |
|---|---|---|---|---|
| 1 | 5.0 | 6.5 | 45 | 99 |
| 2 | 5.0 | 10 | 60 | 99 |
| 3 | 4.5 | 9.4 | 60 | 98 |
| 4 | 4.5 | 8.6 | 60 | 98 |
| 5 | 5.0 | 11 | 60 | 98 |
| 6 | 4.5 | 7.8 | 60 | 97 |
All experiments were carried out at 40 °C (± 1 °C);
pH measurement error (σ = ± 0.04);
the half-life time for the DMTr removal: it was calculated from the following equation t1/2 = ln (0.5) / ln (%DMTr-on/100) × t;
the reaction time of detritylation reaction;
the yields of the generated DMTr-off oligonucleotides (confirmed by MS analysis), which were measured via the peak area.
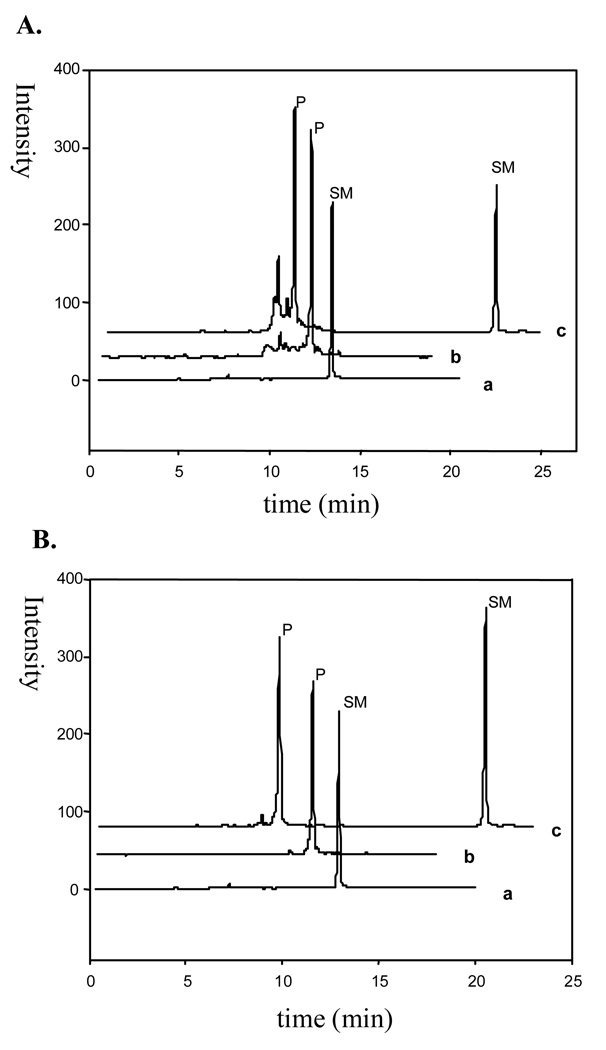
We have also directly compared the conventional approach with our novel strategy in the 5’-OH deprotection reaction. Thought no significant difference was observed in the detritylation of native oligonucleotides using either the conventional approach or our warming-up strategy, we observed the significant difference in the detritylation of the modified DNA using these two strategies (Figure 2). The 6-Se-dG-modified DNA (6, 5'-DMTr-AT-6-SeG-TCG-3')[22] developed previously was examined using these two detritylation strategies. The difference is obvious: the warming-up strategy led to quantitative formation of the desired product without significant decomposition, while the conventional strategy (the acid treatment) caused significant by-product formation. Our results indicate the convenience and advantage of this novel warming-up strategy in the detritylation and modified nucleic acid synthesis for nucleic acid drug discovery.[23–25]
FIGURE 2.
Comparison of these two DMTr-deprotection strategies on 5'-DMTr-AT-6-SeG-TCG-3' (SM). Profile a: SM; Profile b: product (P) after the deprotection; Profile c: co-injection. HPLC gradient for SM: runs from 5 to 60% buffer B in 8 min and then from 60 to 100% buffer B in 2 min. Gradient for P: from 5 to 40% buffer B in 20 min and then from 40 to 100% buffer B in 3 min. Gradient for co-injection: from 5 to 40% buffer B in 17 min and then from 40 to 100% buffer B in 3 min. A) using the conventional strategy (treatment with 3% CCl3COOH for 3 min before neutralization); B) using the mild detritylation strategy (heating at 40 °C and pH 4.5 for 1 hr).
CONCLUSION
In summary, we have developed a mild method for removal of the DMTr protection from the 5’-hydroxyl groups of native and modified DNA and RNA. This is a first step towards the synthesis of defect-free oligonucleotides and nucleic acids. This strategy is useful to ensure high purity of synthetic nucleic acids for investigations in function, structure, diagnosis, and therapeutic discovery. Furthermore, this simple and effective approach is critical for synthesizing the modified nucleic acids and other bio-molecules that are sensitive to the conventional acid treatment. Moreover, this useful strategy is essential to prepare high-purity DNAs and RNAs for defect-free derivatization, crystallization, and X-ray crystal structure studies of nucleic acids and their protein complexes. Our novel strategy offers a great opportunity to significantly increase the high quality of synthetic nucleic acids and oligonucleotides, such as siRNAs, microRNAs and antisense DNAs with potential therapeutic applications.
EXPERIMENTAL
Oligonucleotide Synthesis
Six oligonucleotides (1–6 in Scheme S1) were prepared using the phosphoramidites (dA, Ac-dC, dG, dT, or 2’-O-TOM ribo-series ones), and the standard dT and dG-lcaa-CPG supports (500 Å) or 2’-OMe-Ac-C-RNA CPG (500 Å) were purchased from Glen Research. The oligonucleotides were synthesized on an ABI 3400 DNA synthesizer in the DMTr-on mode at a 1 µmol scale. (I) Coupling: phosphoramidites in dry acetonitrile (0.1 M) were activated by 0.3 M benzylthiotetrazole in dry acetonitrile before coupling to the solid support with optimized coupling times (20 s for DNA and 180 s for RNA); (II) Capping: (A) Ac2O/2,6-Lutidine/THF, and (B) 16% 1-Methylimidazole in THF; (III) Oxidation: 0.02 M I2 in THF/Py/H2O; (III) detritylation: 3% CCl3COOH in CH2Cl2. After synthesis, DMTr-on DNAs were removed from the solid support and deprotected by conc. ammonia at 55 °C, while the DMTr-on RNA was handled according to the procedure for the synthesis, deprotection, and isolation of RNA using TOM-protected monomers. Crude DMTr-on DNAs & RNA were purified by reverse phase HPLC on an XB-C18 column (Welch Materials, 21.2 × 250 mm); flow rate: 6 mL/min; buffer A: 30 mM TEAA (pH 7.6); buffer B: 60% CH3CN in 30 mM TEAA (pH 7.6); gradient: 5–100% B in 20 min; single injection (1 mL). Fractions containing DMTr-on DNA or RNA were subsequently adjusted to pH 8.0 using Et3N. All pH measurements were performed using a UB-10 pH/mV meter (Denver Instrument) equipped with a micro-pH combination electrode (Orion 98-03, ± 0.04).
Deprotection of 5’-DMTr-containing Oligonucleotides
Prior to the removal of DMTr from the 5’-termini of oligonucleotides, DNA & RNA fractions were evaporated on the vacufuge concentrator to remove all the organic solvent (CH3CN), and each oligonucleotide was analyzed by RP-HPLC to confirm the purity. An aliquot (200 µL) of each pure oligonucleotide was taken out, its pH was adjusted to the desired value (pH = 4.5, 5.0, 5.5 or 6.0) with a HOAc aqueous solution (5.0 M), and the deprotection was subsequently investigated at various temperatures (40, 50, and 60 °C). Right after this pH adjustment, the aliquot was distributed into five 200 µL tubes (30 µl each). These five tubes were then heated (at such as 40 °C) for 2, 5, 10, 30, and 60 min. At each time point, the corresponding sample was removed from the heating system and added with neat Et3N (0.1 µl) to quench the detritylation. The detritylated samples were analyzed on HPLC by an XB-C18 column (Welchrom, 4.6 × 250 mm) at 25 °C; flow rate: 1 mL/min; buffer A: 20 mM triethylammonium acetate (TEAA, pH 7.6); buffer B: 50 % CH3CN in 20 mM TEAA (pH 7.6); gradient: 5–60% B in first 10 min and then 60–100% B in 5 min (unless otherwise indicated); pressure: 125–135 Bar. After the deprotection, the deprotected DMTr-OH was removed from the oligonucleotides by either ethanol precipitation or extraction with ethyl acetate.
In a typical detritylation reaction, a HPLC-purified DMTr-on DNA or RNA (0.2 µmole) dissolved in water (200 µL) is adjusted to pH 5.0 with acetic acid (just a few microliters of 10% HOAc; pH is monitored by a micro-pH meter.), and it is then heated at 40 °C for 1 hr and neutralized with neat Et3N (a few microliters) to pH 7.6, before removing DMTr-OH by either ethanol precipitation or extraction with ethyl acetate. It is depended on its use whether the detritylated DNA or RNA sample needs to be further purified by HPLC.
Acknowledgment
This work was financially supported by NIH (GM095086) and the Georgia Cancer Coalition (GCC) Distinguished Cancer Clinicians and Scientists.
REFERENCES
- 1.Baisnee PF, Baldi P, Brunak S, Pedersen AG. Flexibility of the genetic code with respect to DNA structure. Bioinformatics. 2001;17:237–248. doi: 10.1093/bioinformatics/17.3.237. [DOI] [PubMed] [Google Scholar]
- 2.Carrasco N, Ginsburg D, Du Q, Huang Z. Synthesis of Selenium-Derivatized Nucleosides and Oligonucleotides for X-ray Crystallography. Nucl. Nucl. Nucl. Acids. 2001;20:1723–1734. doi: 10.1081/NCN-100105907. [DOI] [PubMed] [Google Scholar]
- 3.Collins A, Morton NE. Likelihood ratios for DNA identification. Proc Natl Acad Sci U S A. 1994;91:6007–6011. doi: 10.1073/pnas.91.13.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Du Q, Carrasco N, Teplova M, Wilds CJ, Egli M, Huang Z. Internal derivatization of oligonucleotides with selenium for X-ray crystallography using MAD. J Am Chem Soc. 2002;124:24–25. doi: 10.1021/ja0171097. [DOI] [PubMed] [Google Scholar]
- 5.Goff SP, Berg P. Construction of hybrid viruses containing SV40 and lambda phage DNA segments and their propagation in cultured monkey cells. Cell. 1976;9:695–705. doi: 10.1016/0092-8674(76)90133-1. [DOI] [PubMed] [Google Scholar]
- 6.Rothemund PW. Folding DNA to create nanoscale shapes and patterns. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 7.Sheng J, Huang Z. Selenium Derivatization of Nucleic Acids for X-ray Crystal Structure and Function Studies. Chem. & Biodiver. 2010;7:753–785. doi: 10.1002/cbdv.200900200. [DOI] [PubMed] [Google Scholar]
- 8.Beaucage SL, Iyer RP. Advances in the Synthesis of Oligonucleotides by the Phosphoramidite Approach. Tetrahedron. 1992;48:2223–2311. [Google Scholar]
- 9.Krotz AH, Cole DL, Ravikumar VT. Synthesis of Antisense Oligonucleotides with Minimum Depurination. Nucl. Nucl. Nucl. Acids. 2003;22:129–134. doi: 10.1081/NCN-120019499. [DOI] [PubMed] [Google Scholar]
- 10.Septak M. Kinetic studies on depurination and detritylation of CPG-bound intermediates during oligonucleotide synthesis. Nucleic Acids Res. 1996;24:3053–3058. doi: 10.1093/nar/24.15.3053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Suzuki T, Ohsumi S, Makino K. Mechanistic studies on depurination and apurinic site chain breakage in oligodeoxyribonucleotides. Nucleic Acids Res. 1994;22:4997–5003. doi: 10.1093/nar/22.23.4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka T, Letsinger RL. Syringe method for stepwise chemical synthesis of oligonucleotides. Nucleic Acids Res. 1982;10:3249–3260. doi: 10.1093/nar/10.10.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zon GS, Stec WJ. Oligonucleotides and Analogues. In: Eckstein F, editor. A Practical Approach. 1991. pp. 87–108. [Google Scholar]
- 14.Caton-Williams J, Huang Z. Synthesis and DNA Polymerase Incorporation of Colored 4-Selenothymidine Triphosphate with a Single Atom Substitution. Angew. Chem. Int. Ed. 2008;47:1723–1725. doi: 10.1002/anie.200705213. [DOI] [PubMed] [Google Scholar]
- 15.Hassan AE, Sheng J, Zhang W, Huang Z. High fidelity of base pairing by 2-selenothymidine in DNA. J Am Chem Soc. 2010;132:2120–2121. doi: 10.1021/ja909330m. [DOI] [PubMed] [Google Scholar]
- 16.Salon J, Sheng J, Jiang J, Chen G, Caton-Williams J, Huang Z. Oxygen replacement with selenium at the thymidine 4-position for the Se base pairing and crystal structure studies. J Am Chem Soc. 2007;129:4862–4863. doi: 10.1021/ja0680919. [DOI] [PubMed] [Google Scholar]
- 17.Carrasco N, Buzin Y, Tyson E, Halpert E, Huang Z. Selenium derivatization and crystallization of DNA and RNA oligonucleotides for X-ray crystallography using multiple anomalous dispersion. Nucleic Acids Res. 2004;32:1638–1646. doi: 10.1093/nar/gkh325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reddy MP, Farooqui F, Hanna NB. Methylamine deprotection provides increased yield of oligoribonucleotides. Tetrahedron Lett. 1995;36:8929–8932. [Google Scholar]
- 19.Adams SP, Kavka KS, Wykes EJ, Holder SB, Galluppi GR. Hindered dialkylamino nucleoside phosphite reagents in the synthesis of two DNA 51-mers. J. Am. Chem. Soc. 1983;105:661–663. [Google Scholar]
- 20.Crugeiras HM. Can. J. Chem. 1999;77:530–536. [Google Scholar]
- 21.Crugeiras HM. J. Chem. Soc. Perkin Trans. 2. 2000;2:441–445. [Google Scholar]
- 22.Salon J, Jiang J, Sheng J, Gerlits OO, Huang Z. Derivatization of DNAs with selenium at 6-position of guanine for function and crystal structure studies. Nucleic Acids Res. 2008;36:7009–7018. doi: 10.1093/nar/gkn843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salon J, Sheng J, Gan J, Huang Z. Synthesis and crystal structure of 2'-Se-modified guanosine containing DNA. J Org Chem. 2010;75:637–641. doi: 10.1021/jo902190c. [DOI] [PubMed] [Google Scholar]
- 24.Sheng JS, Gan JH, Huang Z. Synthesis and Crystal Structure Study of 2’-Se-Adenosine-Derivatized DNA. Sciense China: Chemistry. 2010;53:78–85. [Google Scholar]
- 25.Gan JH, Sheng J, Huang Z. Chemical and Structural Biology of Nucleic Acids and Protein-nuclei Acid Complexes for Novel Drug Discovery. Sciense China: Chemistry. 2011;54:3–23. [Google Scholar]