Abstract
Despite the importance of migratory birds in the ecology and evolution of avian influenza virus (AIV), there is a lack of information on the patterns of AIV spread at the intra-continental scale. We applied a variety of statistical phylogeographic techniques to a plethora of viral genome sequence data to determine the strength, pattern, and determinants of gene flow in AIV sampled from wild birds in North America. These analyses revealed a clear isolation-by-distance of AIV among sampling localities. In addition, we show that phylogeographic models incorporating information on the avian flyway of sampling proved a better fit to the observed sequence data than those specifying homogeneous or random rates of gene flow among localities. In sum, these data strongly suggest that the intra-continental spread of AIV by migratory birds is subject to major ecological barriers, including spatial distance and avian flyway.
Keywords: avian influenza, phylogeography, evolution, gene flow, ecological barriers, flyways, spatial distance
INTRODUCTION
Avian influenza virus (AIV) has been isolated from over a hundred species of wild bird, with Anseriformes (e.g. ducks, geese, and swans) and Charadriiformes (e.g. gulls, terns, and waders) constituting its major natural reservoir (Webster et al. 1992; Munster et al. 2007). AIV prevalence in wild birds is generally high, but varies between seasons, geographic regions and bird species (0.25 – 8%) (Olsen et al. 2006), and can exceed 30% in juveniles in some pre-migration staging areas (Hinshaw et al. 1985). AIV in wild birds is also usually characterized by a low pathogenicity. However, the occasional inter-species transmission of AIV may lead to the emergence of highly pathogenic forms of the virus, reflected in high mortality rates in poultry (Swayne & Suarez 2000), sometimes in wild birds (Chen et al. 2005), and in humans (Uyeki 2009).
A diverse array of antigenic and genetic variants of AIV are maintained in wild birds, and through a combination of point mutation and reassortment generate novel genotypes, some of which occasionally adapt to new species (Parrish et al. 2008). Because of the potential health burden to humans and avian species that might result from AIV, as well as the necessity to better understand its ecology and evolution, it is critical to determine the factors that shape the spread of AIV in wild birds. On a global scale AIV is separated into two major lineages, representing those viruses that circulate in the Western and Eastern hemispheres, respectively (Fig. 1A). Although such a phylogenetic pattern is indicative of the long-term ecological separation of the bird species in these hemispheres, there is some evidence of inter-hemispheric AIV gene flow (Krauss et al. 2004; Koehler et al. 2008). Alaska and Nunavut (Canada) are likely localities for such inter-hemispheric transmission as many birds from the Western and Eastern hemispheric flyways congregate there for nesting and breeding. However, despite the potential importance of bird migration for the inter-hemisphere spread of AIV (Si et al. 2009), how viral genetic diversity is shaped by avian flyway at the intra-continental scale is less clear (Causey & Edwards 2008).
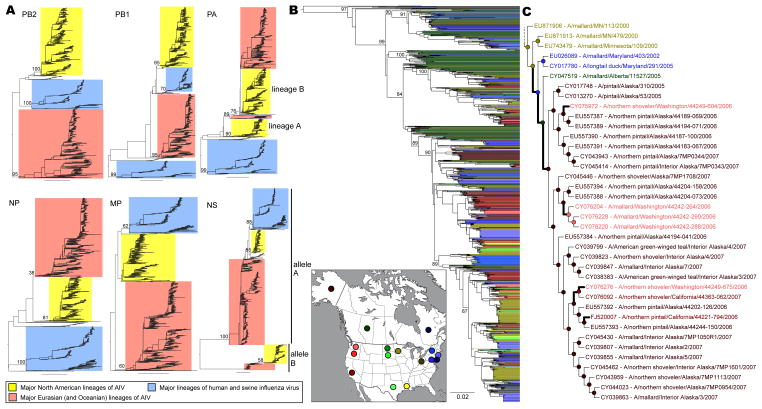
Figure 1.
Phylogenetic trees of the internal genome segments of influenza A virus. (A) ‘Panoramic’ phylogenies of the PB2, PB1, PA, NP, MP, and NS segments. Bootstrap support values are shown adjacent to selected nodes, and the scale bar represents 0.1 substitutions/site. Major North American lineages of AIV are highlighted in yellow. (B) Maximum likelihood phylogeny of the major North American wild bird AIV (NA-WB-AIV) lineage inferred from the PB2 gene. Terminal branches are extended and colored according to the place of sampling as shown in the inset map (14 US states and two Canadian provinces; others shown in grey). The scale bar is 0.02 substitutions/site. (C) A sub-lineage of NA-WB-AIV in the PB2 phylogeny, shown as a cladogram. Geographical states of the ancestral nodes (circles) were estimated, using parsimony, from taxon localities, and their colors are the same as those in panel A. Changes in geographical state that occurred during evolutionary history are indicated by bold branches.
Many Anseriformes and Charadriiformes species undergo regular long-distance migration (del Hoyo et al. 1996). Hence, it is commonly thought that influenza virus and other avian pathogens, especially those that cause mild disease, are dispersed over long distances during the journeys made by avian migrants (Causey & Edwards 2008). There is a large body of literature investigating the ecology and evolution of AIV in different wild bird species, some of which suggests a role for migratory birds in dispersing AIV (Romvary et al. 1980; Deibel et al. 1985; Kawaoka et al. 1988; Krauss et al. 2004; Munster et al. 2007; Wallace et al. 2007). However, this widely assumed premise has not been formally tested. In addition, most studies undertaken to date have focused solely on the spread of highly pathogenic H5N1 AIV in Eurasia (Wallace et al. 2007; Si et al. 2009). As such, the ecological factors shaping AIV transmission in North American wild birds remain poorly understood.
Herein we determined the extent and structure of AIV gene flow among wild birds resident in North America by analyzing an extensive data set of AIV genome sequences (~1,000) with a suite of statistical phylogeographic methods. In particular, we assessed the influence of spatial distance and avian flyway on the frequency and pattern of AIV gene flow.
MATERIAL AND METHODS
Sample collection
100 AIV from the USGS National Wildlife Health Center’s collection were selected with the aim of providing a broad survey of the viruses present on the US west coast (Pacific flyway). The viruses originated from samples collected between 2005–2008 from 15 species of migratory birds in 16 counties in California, Oregon, and Washington as part of coordinated surveillance for avian influenza using live bird, hunter-killed, and mortality monitoring techniques (http://www.pacificflyway.gov/Documents/AIS_plan.pdf). Cloacal and/or oropharyngeal swab samples were collected, and inoculated in embryonated specific pathogen-free chicken eggs. Viruses were recovered and viral RNA were extracted from the allantoic fluid following incubation as previously described (Ip et al. 2008).
Viral genome sequencing
All samples were first sequenced using the traditional PCR/Sanger high-throughput sequencing pipeline at the JCVI as described previously (Dugan et al. 2008). Sequences were then processed and assembled. Difficulties associated with the high level of genetic variability in the sample set, likely a function of frequent mixed serotype infections (Dugan et al. 2008), led to the processing of all samples using the next generation sequencing pipeline at JCVI that includes the 454/Roche GS-FLX and the Illumina Genome Analyzer II (Djikeng et al. 2008; Zhou et al. 2009). A final mapping of all next generation sequences to the updated reference sequences was performed using CLC Bio software. More details of the sequencing procedure are available in Appendix S1 in Supporting Information. All sequences generated here have been assigned GenBank accession numbers (Table S1).
GenBank sequence collection and alignment
The 100 AIV sequences generated here were combined with nucleotide sequences (excluding partial sequences <70% of full-length) of all influenza A viruses available on GenBank (Bao et al. 2008). An alignment of each segment was constructed using the default settings in MUSCLE v3.5 (Edgar 2004). Due to a large imbalance in the number of sequences, human viruses were randomly sampled to a total of 100–300 per subtype. Total data set sizes were as follows: PB2, N = 5389 sequences (alignment length of 2277 nucleotides); PB1, n=5744 (2271 nt); PA, n=5712 (2148 nt); HA(H3), n=1270 (1698 nt); HA(H4), n=334 (1692 nt); NP, n=6028 (1494 nt); NA(N6), n=278 (1410 nt); NA(N8), n=426 (1410nt); MP, n=8336 (982 nt); and NS, n=7626 (838 nt). The final number of sequences from the major North American lineage(s) of AIV in wild birds (NA-WB-AIV; defined in Results) are provided in Table S2.
Phylogenetic analysis
A complete ‘panoramic’ maximum likelihood phylogeny for each of the eight genome segments of AIV was inferred using a rapid tree searching algorithm implemented in RAxML v7.04 program (Stamatakis 2006), and employing the GTRGAMMA substitution model which was most often the best-fit as determined by MODELTEST (Posada & Crandall 1998). The highest likelihood phylogenies were collated from 200 independent tree searches (Fig. 1A). A total of 100 pseudo-replicates were generated for bootstrap analysis using the neighbor-joining clustering method and employing the maximum likelihood distance criterion.
Sequences falling into the NA-WB-AIV lineage were extracted for further analyses (Tables S1 and S2). A small number (<5%) of non-wild bird sequences (mainly poultry and swine) that fell within the NA-WB-AIV lineage were excluded. For the HA and NA gene segments, only the H3, H4, N6, and N8 gene data sets were studied because of insufficient sequences and/or geographical diversity in the other subtypes. To obtain more accurate maximum likelihood phylogenies for the NA-WB-AIV sequences we used the PhyML v3.14 program (Guindon et al. 2009), employing an heuristic search and the GTR+I+Γ4 model of nucleotide substitution. These trees were highly congruent to those produced by RAxML above. Finally, 1,000 bootstrap maximum likelihood trees were estimated using PhyML. More details of the phylogenetic methods used are provided in Appendix S1.
Geographical and avian species information
Each AIV sequence was assigned a discrete geographical state according to its province/state of isolation (Table S2). Although the earliest AIV was sampled in 1968, the majority (>75%) were isolated during the period 1998–2008. Therefore, sequences prior to 1998, as well as those from poorly sampled or unknown provinces/states (n ≤ 5) were either removed from analysis or labeled as an ‘other’ state. Each US state and Canadian province was categorized into a specific North American flyway; the Atlantic flyway (AF), Mississippi flyway (MF), Central flyway (CF), or Pacific flyway (PF), according to the United States Fish and Wildlife Service and Flyway Councils (Lincoln 1979). The spatial distance between each pair of states/provinces was calculated as the great circle distance between the average latitude and longitude of each state/province.
Six major species groups of waterfowl and shorebirds were present in our AIV data set: mallard (Anas platyrhynchos), northern pintail (Anas acuta), northern shoveler (Anas clypeata), blue-winged teal (Anas discors), green-winged teal (Anas carolinensis), and ruddy turnstone (Arenaria interpres). Viral sequences were labeled according to their host species for the analysis of the species contribution to AIV gene flow. Species for which we had only a small number of sequences, including Aythya americana, Aythya collaris, Aythya valisineria, Anas rubripes, Clangula hyemalis, were excluded.
Testing panmixia in AIV
Slatkin-Maddison tests (Slatkin & Maddison 1989; Maddison & Slatkin 1990) were used to assess the extent of panmixia in the AIV sequence data. Ancestral geographical states were inferred from the maximum likelihood phylogenies and geographically coded taxa using a parsimony procedure (Fitch 1971). The number of changes (S) in ancestral and taxon geographical states was then counted. The mean value of S was obtained by analyzing 200 randomly selected maximum likelihood bootstrap trees. To simulate the expected number of state changes (Sp) assuming panmixis, the geographical states of the taxa were shuffled randomly and the ancestral geographical states inferred as described above. A total of 100,000 randomizations (500 per tree) were performed for the 200 bootstrap phylogenies. Sp was then counted and compared to S. Panmixis was rejected if S was significantly less than Sp (at the 5% significance level).
Estimating levels of gene flow between North American localities
We used three summary statistics to determine the extent of NA-WB-AIV gene flow: (i) The fixation index (FST), which examines the degree of genetic differentiation between the viral populations of two localities (Nei 1982; Lynch & Crease 1990). We employed the FST generalized by Hudson et al. (1992) and the GTR+I+Γ4 substitution model to estimate pairwise genetic distances between sequences. (ii) A modified version of Slatkin-Maddison’s phylogeny-trait association test, in which the minimum number of geographical state changes in the phylogeny (s) provides an estimate of the extent of gene flow among populations (Slatkin & Maddison 1989). To correct for differences in sampling frequency among localities we scaled s by the maximum possible frequency of gene flow among each pair of localities that could occur under panmixia. This scaled index was denoted as σ; (iii) The rate of state transition (q) among localities estimated using a maximum likelihood method (Pagel 1994), and which assumes that the geographical state of each sequence is a trait that has co-evolved with the virus. More details of these indices are given in Appendix S1.
The influence of flyway on AIV gene flow
To assess the extent to which North American avian flyways influence the level of AIV gene flow we compared, within a likelihood framework, the fit to the data of four phylogeographic models: (i) a homogenous rate model (HRM), which specifies the same rate of transition among localities; (ii) a 4-flyway rate model (4-FRM) in which each flyway is characterized by its own specific transition rate, and where transition rates are the same from localities within a flyway to those in another flyway; (iii) a 3-flyway rate model (3-FRM), the same as 4-FRM but where the CF and MF are merged since they overlap in central North America; and (iv) a randomly-categorized rate model (RRM), in which localities were randomly categorized into any of the flyways, regardless of their real sampling locations. The rate parameters (q) for the extent of gene flow between any two localities were then categorized as those for their flyway of assignment (e.g. qCF↔AF, qAF↔PF, qPF↔MF) (Table S2). Hence, there are 10 reversible transition rates in the 4-flyway model (and six in the 3-flyway model). Transition rates were estimated using Pagel’s maximum likelihood method (Pagel 1994) as implemented in the APE library (Paradis et al. 2004) running R (version 2.11.1). Various starting values (20, 10, 5, 1, 0.1 and 0.01) of transition rate were employed, and estimates with the highest likelihoods were retained. Likelihood ratio and randomization tests were used to assess the goodness-of-fit of the FRM to the genetic and geographic data. The likelihood ratio test assesses whether the 4-FRM and 3-FRM fitted the data significantly better than the HRM, while in the randomization test the distribution of likelihoods was computed by fitting 500 RRMs to the geographically coded phylogeny which are then compared to the 4-FRM and 3-FRM.
Inter-hemisphere transmission of AIV
Instances of Eurasian sequences falling within the North American major lineage and North American sequences falling within the Eurasian lineages in the panoramic phylogeny (Fig. 1A) were considered as evidence for the inter-hemisphere transmission of AIV. The number of such occurrences was then counted. The potential origins (place and flyway) of each introduction was inferred from the geographic locations of their closest neighboring sequences (Table S4).
The species contribution to patterns of AIV gene flow
To determine the role played by different bird species in AIV gene flow we co-estimated the species and geographical state of each node in each phylogeny using parsimony. Specifically, an avian species was considered responsible for a particular gene flow event between two localities if it was inferred to be the ancestral species at a node connecting the branches in question.
RESULTS
Major lineages of AIV circulating in North American wild birds
Our panoramic phylogenetic analyses of all influenza A virus sequences identified a major lineage of North American wild bird viruses (the ‘NA-WB-AIV lineage’) that was apparent as a well-supported monophyletic group (most bootstrap support values > 70%) (Fig. 1A, yellow boxes). Most AIV gene segments depicted a single such NA-WB-AIV lineage, although two lineages were identified in the PA (‘PA-A’ and ‘PA-B’) and NS (‘NS-A’ and ‘NS-B’) gene segments. The NA-WB-AIV lineage was comprised of approximately 1,000 AIV sequences that together constituted >90% of avian influenza strains isolated in the North American wild bird population during the period 1968–2009 (>75% of which were Anseriformes species). Since a majority (>75%) of samples were collected between 1998 and 2008, the genome sequences of these NA-WB-AIV, including 100 full genomes newly sequenced as part of this study (Table S1), were analyzed further. Each of these sequences was coded with a discrete geographic state according to its sampling location (US state or Canadian province; Fig. 1B, Table S2).
Spatial structure of NA-WB-AIV
Slatkin-Maddison tests (Slatkin & Maddison 1989) were performed to determine whether there was significant geographical structure within the NA-WB-AIV lineage. In all gene data sets (n=12), the observed frequency (S) of AIV gene flow did not overlap with the expected frequency (Sp) simulated under the assumption of panmixia (Fig. S1), thereby significantly (p < 0.00001) rejecting random mixing and suggesting that there are barriers to viral gene flow in North America.
Gene flow of NA-WB-AIV between localities
The extent of gene flow between localities inferred by plotting FST against spatial distance revealed significant and positive correlations in a majority of gene segments (8 out of 12; Fig. 2A and S2; slopes = 0.107 ×10−4– 0.942 ×10−4; R = 0.243 – 0.561; p < 0.05 in Mantel tests). For example, PA (lineage B), NS (allele A), NP, and HA (H4) had Pearson’s correlation coefficients (R) of 0.381, 0.323, 0.561, and 0.410 in the linear regression analyses, respectively. Since higher FST values indicate greater genetic differentiation between populations, these positive correlations reveal that gene flow decreases with increasing spatial distance, such that there is effectively isolation-by-distance.
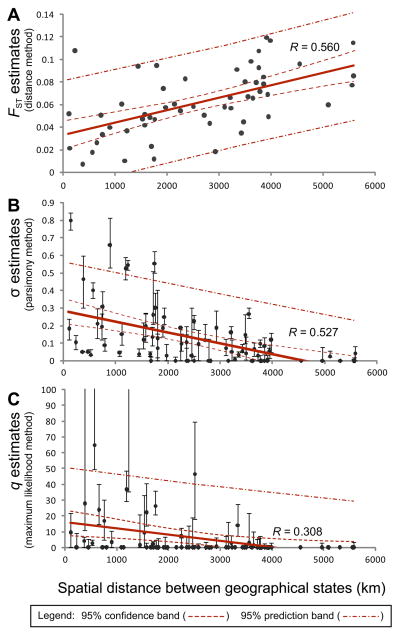
Figure 2.
The level of AIV gene flow between two geographical states plotted against their spatial distance. Estimates of viral gene flow (n=91) were inferred using three approaches: (A) FST (a distance method), (B) modified Slatkin-Maddison’s s (σ; a parsimony method), and (C) state transition rate (q; a maximum likelihood method). For illustration, we show the results from the NP gene data set only. The thick solid straight lines are linear regressions of gene flow rates and spatial distances (in units of km). 95% confidence and prediction ranges of regression are shown as dashed lines. Pearson’s coefficients (R) of the regressions are also shown. Error bars represent 95% confidence intervals of the estimates from 200 bootstrap phylogenies.
Because FST is solely based on genetic distances it is in sensitive to varying rates of evolutionary change (Neigel 2002). In this context it is notable that in our NP, MP and NS-B data sets the AIV sequences sampled from New York state formed clusters separated from other viruses by long branches (Fig. S2). As these New York viruses cluster with those from a poultry outbreak, such a phylogenetic pattern is suggestive of rapid viral evolution following a period of circulation in poultry. Including these outlying samples in our analyses (grey circles in Fig. S2) resulted in substantially weaker correlations in the linear regression (e.g. R decreased from 0.469 to 0.222 (NS-B), from 0.561 to 0.001 (NP), and from 0.292 to 0.074 (MP)). As a consequence, phylogeny-based estimates of the extent of AIV gene flow (σ and q) are likely to be more robust in this case.
We similarly found consistent evidence of strong spatial structure using the parsimony-based σ statistic. Higher values of σ signify more abundant gene flow, so that their negative correlations with spatial distance are again indicative of isolation-by-distance (Fig. 2B, 2C and S3; slope for σ = −1.066×10−4 – −0.454×10−4; R for σ = 0.295–0.527; p < 0.05 in Mantel tests). Although broadly equivalent results were observed using the maximum likelihood q statistics (Fig. 2B), there was a larger estimation uncertainty, suggesting that the phylogeographic model may be over-parameterized (data not shown). It is also noteworthy that there was greater variation in σ and q for pairs of localities separated by short spatial distances than those separated by long distances (Fig. S3). This suggests that factors other than spatial distance affect the rate of AIV gene flow among localities in close proximity.
Avian flyways as determinants for NA-WB-AIV gene flow
Each of the 14 US states and two Canadian provinces was categorized into one of four North American avian flyways (Fig. 3 and Table S2). Likelihood ratio tests revealed that in all data sets both the 3-FRM and 4-FRM, which take into account the flyway of sampling, significantly (p < 0.05) outperformed the HRM where no flyway information is incorporated (Table 1). Likewise, randomization tests revealed that the 4-FRM and 3-FRM were significantly (p < 0.05) better than the RRM in 9 and 8 of the 12 NA-WB-AIV gene data sets, respectively (Fig. S4 and Table 1). Hence, incorporating information on avian flyway greatly improves phylogeographic descriptions of the pattern of AIV gene flow.
Figure 3.
Flyway-specific rates of AIV gene flow. (A) Maximum likelihood estimates (q) of the average level of AIV internal gene flow (excluding the NS gene which has large confidence intervals) within and between the four flyways; Pacific flyway (PF; red), Central flyway (CF; green), Mississippi flyway (MF; yellow) and Atlantic flyway (AF; blue). The 4×4 rate matrices were projected onto the map. The color of the bar and the geographical region where the bar is located denote the flyway where gene flow is measured. For example, the red bar in the yellow terrestrial region represents the extent of gene flow between the PF and MF, while the red bar in the red terrestrial region represents the gene flow within PF. The rate of AIV gene flow within PF is shown next to the bar. Average 95% confidence intervals for the rate estimates are indicated by error bars. (B) The rate matrices (3×3) in a 3-flyway model in which CF and MF are merged.
Table 1.
Likelihood ratio test (LRT) and randomization test (RT) results for the flyway contribution to patterns of AIV gene flow. FRM denotes the flyway-specific rate models and RRM denotes the randomly-categorized rate model.
| Gene data set | 4-Flyway Model (4-FRM) | 3-Flyway Model (3-FRM) | Homogeneous Rate Model (HRM) | 4-FRM vs. RRM | 3-FRM vs. RRM | HRM vs. 4-FRM (3-FRM) | 4-FRM vs. 3- FRM | |||
|---|---|---|---|---|---|---|---|---|---|---|
| -lnL* | Para# | -lnL | Para# | -lnL | Para#‡ | RT p-value | RT p-value | LRT p-value | LRT p-value | |
| PB2 (n=1,316) | −1628.10 | 11 | −1630.51 | 7 | −1692.44 | 2 | 0.034¶ | <0.002¶ | <0.00001 (<0.00001) | 0.30676 |
| PB1 (n=1,346) | −1613.76 | 11 | −1621.97 | 7 | −1702.13 | 2 | <0.002¶ | <0.002¶ | <0.00001 (<0.00001) | 0.00250§ |
| PA-B (n=862) | −916.58 | 11 | −924.84 | 7 | −970.08 | 2 | 0.018¶ | <0.002¶ | <0.00001 (<0.00001) | 0.00239§ |
| PA-A (n=466) | −622.80 | 11 | −624.84 | 7 | −660.83 | 2 | 0.016¶ | <0.002¶ | <0.00001 (<0.00001) | 0.39414 |
| NP (n=1,002) | −1186.56 | 11 | −1203.45 | 7 | −1278.01 | 2 | 0.028¶ | 0.006¶ | <0.00001 (<0.00001) | <0.00001§ |
| MP (n=1,281) | −1642.58 | 11 | −1650.53 | 7 | −1715.02 | 2 | 0.024¶ | <0.002¶ | <0.00001 (<0.00001) | 0.00315§ |
| NS-A (n=735)† | −794.84 | 10 | −797.60 | 7 | −851.10 | 2 | 0.020¶ | 0.024¶ | <0.00001 (<0.00001) | 0.137 |
| NS-B (n=386) | −470.87 | 11 | −474.96 | 7 | −491.52 | 2 | 0.260 | 0.160 | <0.00001 (<0.00001) | 0.08540 |
| H3 (n=294) | −279.23 | 11 | −288.72 | 7 | −297.49 | 2 | 0.002¶ | 0.044¶ | <0.00001 (0.00358) | 0.00079§ |
| H4 (n=286) | −326.78 | 11 | −336.10 | 7 | −354.45 | 2 | 0.122 | 0.204 | <0.00001 (<0.00001) | 0.00092§ |
| N6 (n=239)† | −276.43 | 10 | −281.27 | 6 | −290.07 | 2 | 0.042¶ | 0.056 | 0.00063 (0.00148) | 0.04618§ |
| N8 (n=270)† | −241.47 | 10 | −246.83 | 6 | −256.86 | 2 | 0.092 | 0.082 | 0.00015 (0.00049) | 0.02990§ |
Log likelihood.
Because theN6 and N8 data sets had only one geographical state in the Atlantic flyway (AF), and NS-A had only one geographical state in the Central flyway (CF), it was impossible to estimate the level of gene flow in these cases. As a consequence they have one less parameter than the other data sets (N6 and N8 only).
Both the FRM and HRM contain the same extra rate parameter accommodating transitions involving the ‘other’ geographical state (e.g. taxa collected before 1998, having an unknown geographical state, or state frequency ≤ 5).
Data sets in which the 4-FRM is significantly better than the 3-FRM.
Data sets in which the 4-FRM (or 3-FRM) is significantly better than the RR
To measure the level of viral gene flow within and between the four flyways we conducted further analyses using both the maximum likelihood (q) and parsimony (σ) methods. These yielded largely congruent results: levels of gene flow within a flyway were usually higher than those between flyways, and the average level of viral gene flow was lower between the more distantly separated flyways (Fig. 3 and Fig. S5). For example, the average level of gene flow decreased progressively from that in the AF, to that between the AF and CF, more so between the AF and MF, and reaching its lowest level between the most distant AF and PF (Fig. 3, Fig. S5 and Table S3). Such a pattern is again indicative of isolation-by-distance, and consistent with the fact that neighboring flyways experience more frequent crossing of avian migration routes than those that are more distant.
Increased AIV gene flow between the Central and Mississippi flyways
The pattern of a higher level of gene flow within than among flyways was most apparent for the Pacific and Atlantic flyways, and less so for the Central and Mississippi flyways. As an example of the latter, gene flow between CF and MF (qCF↔MF = 15.7; average σCF↔MF = 0.324) was more frequent than within CF (qCF↔CF = 3.99; average σCF↔CF = 0.077), and almost as frequent as within MF (qMF↔MF = 23.0; average σMF↔MF = 0.359). Similarly, viruses from Texas, South Dakota, and North Dakota (CF) had more gene flow with Minnesota and Louisiana (MF) than with other localities in their own flyway (Fig. 4A). However, despite the apparent mixing of AIV between CF and MF, the 4-flyway model was a better fit to more gene data sets than the 3-flyway model that merges CF and MF (8 out of 12 comparisons).
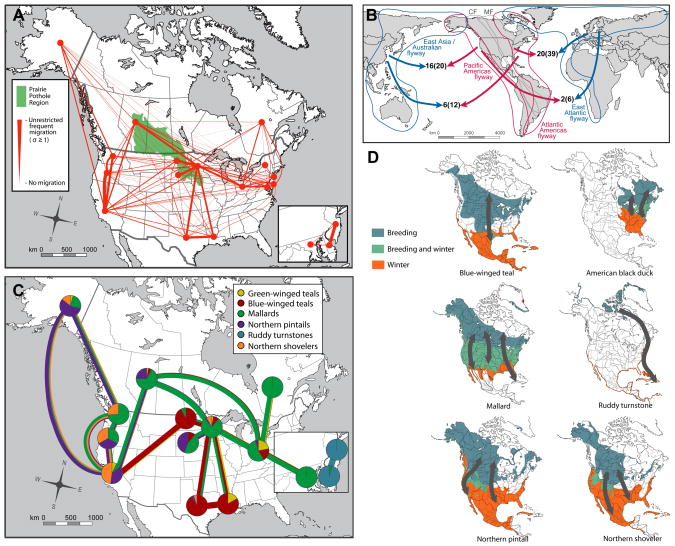
Figure 4.
Overall patterns of gene flow in AIV and the contribution of individual avian species. (A) Parsimony estimates (σ) of the average level of AIV gene flow in internal genes (n=120) between 14 US states and two Canada provinces (red circles). The thickness of the red lines represents the magnitude of σ estimates relative to panmixis (shown in the legend box). Gene flow between Maryland, New Jersey, and Delaware are shown in a zoom-in inset for clarity. (B) The putative number of inter-hemispheric AIV gene flow events during 1998–2008 (and 1927–2008, shown in parentheses) inferred from the panoramic phylogenies of the eight gene segments. (C) Species prevalence in the AIV data set (indicated by the pie-charts) and their contribution (indicated by the colored thick lines, in which the thickness denotes percentage) to AIV gene flow between selected localities where the extent of gene flow is above the 80th percentile of that seen in all localities. New York State was not displayed due to the small sample sizes of the duck species found in this region. (D) Distribution of bird species in North America during breeding and wintering periods. The approximate migration pathways observed in these bird species are shown by grey arrows. The illustrations were adapted from Birds of North America Online (http://bna.birds.cornell.edu).
Inter-hemisphere transmission of AIV
Although the inter-hemisphere transmission of AIV is generally thought to be a rare event (Krauss et al. 2004; Koehler et al. 2008; Pearce et al. 2010), we observed a number of occurrences in our study. Most notably, US states falling in the Pacific flyway experienced more gene flow with localities in the East Asia/Australasian flyway than with localities in the East Atlantic flyway (16 and two transferred lineages, respectively; Fig. 4B and Table S4). This is compatible with the fact that the PF is more geographically distant to the East Atlantic flyway, and that birds in the PF share a stopover region in Alaska with those of the East Asia/Australasian flyway. A similar observation was seen in the Atlantic flyway, which exhibited more gene flow with the adjacent East Atlantic than with the distant East Asia/Australasian flyway (20 and 6 lineages, respectively).
The contribution of different avian species to AIV gene flow
Six avian species groups – mallards, northern pintails, northern shovelers, blue-winged teals, green-winged teals, and ruddy turnstones – were commonly observed in our AIV data. These species have specific ranges of breeding, migration and wintering (Fig. 4D), and which is also apparent in our sample of AIV sequences (Fig. 4C). For example, shovelers were more commonly found in the western part of North America, as were they in our sequence data (orange in the pie-charts in Fig. 4C). The thickness of the colored lines connecting localities in Fig. 4C indicates the relative contribution of different avian species groups (represented by different colors) to the extent of AIV gene flow. This analysis revealed that mallards, blue-winged teals and pintails were the major contributors to AIV gene flow among the selected localities. Mallards were particularly important contributors to long-range viral gene flow from Canada (through Alberta) to different parts of the US (e.g. to California, Minnesota and Ohio). In contrast, teals play a more important role in the central part of North America, including the grasslands of the Prairie Pothole Region (see below), and down to Gulf Coast areas in Texas and Louisiana. Lastly, while both pintails and shovelers move through the western part of the continent, with pintails playing a greater role in long distance gene flow between Alaska and California, short distance gene flow within the PF was mainly due to shovelers, mallards and teals.
DISCUSSION
Our phylogeographic analysis reveals that avian influenza virus exhibits a strongly spatially structured population in North America, with relatively infrequent gene flow among localities and especially between those that are spatially distant or belong to different flyways. Of particular note was the association between the extent of AIV gene flow and avian flyway. Although avian flyways in North America are primarily administrative constructs used to simplify waterfowl management, they are commonly conceived as areas in which related migration routes occur within a defined geographic region, representing aggregations of avian populations that are spatially and temporally isolated from those elsewhere in the continent. Flyways can therefore be thought of as broader highways within which the avian migration routes are tributary, largely based on the migration patterns observed in banding studies (Lincoln 1979; Davidson et al. 1995; Olsen et al. 2006). Our observation that flyways have a marked influence on the extent of AIV gene flow provides additional support for their importance in avian ecology. In this context biotelemetry studies, which provide a detailed picture of the migration trajectories of wild birds using GPS satellite transmitters (Miller et al. 2010; Prosser et al. 2011), constitute a useful complement to phylogeography, providing a more precise picture of the association between the pattern of AIV gene flow and the flight routes used by different bird species.
As might be expected, the influence of flyway is less apparent for geographic regions in which the flyways overlap. Indeed, our broad-scale information on the place of sampling entailed that we categorized each US state and Canadian province into a single flyway, even though some localities are overlaid by multiple flyways. Most notably, we observed an increased level of gene flow in Minnesota, within which AIV gene flow occurs frequently with North and South Dakota, Texas, Louisiana, Alberta and Ohio (above the 85th percentile of all gene flow events determined here). Much of this gene flow occurs within the Prairie Pothole Region that straddles the U.S.-Canadian border (Fig. 4A). The Prairie Pothole Region covers a large portion of land encompassed by the Central and Mississippi flyways, and contains an abundance of shallow wetlands that are of critical importance to migratory birds as stopover points to replenish nutrients and energy, and as breeding habitats for species such as pintails, mallards, gadwall, teals, and shovelers. These wetlands allow migratory aquatic birds from the Central and Mississippi flyways to congregate, and the transmission of influenza virus can occur through either direct contact or virally contaminated water (Stallknecht et al. 1990; Roche et al. 2009). In addition, these mid-continent flyways have more similarity in bird populations and habitats than the coastal flyways, which are isolated by geophysical barriers such as the Rocky and Appalachian Mountains (Lincoln 1979).
We also observed that species of wild bird play varying roles in the geographic dispersal of AIV. For example, pintails are associated with the long distance movement of viruses along the west coast of North America, and which is consistent with their migratory routes as tracked by satellite telemetry (Miller et al. 2010). These differing species contributions to AIV gene flow are to some extent consistent with the migration patterns of bird species deduced from banding studies (USGS 2011). However, our estimates may also be biased by differences in species prevalence; the more AIV sequences of a specific bird species included in the analysis, the more likely this species will contribute to AIV gene flow by chance alone. Despite this, we observed that pintails made a major contribution to AIV gene flow between Alaska and Washington state even though no pintail sequences were present in our Washington state data set. Indeed, pintails are highly cosmopolitan birds, traveling long-distances, and a portion of their population breed in Siberia and winter in North America (and there are records of pintails banded in Japan or Hawaii being shot in locations in North America). In contrast, mallards and shovelers tend to have more restricted in long distance movements, both within North America and between continents.
Although we have necessarily focused on low pathogenic avian influenza virus in wild birds, there is contact between these avian species and domestic poultry populations. In particular, wild birds may serve as a conduit for AIV to enter poultry. This has been particularly well demonstrated in the case of highly pathogenic H5N1 influenza virus (Swayne & Suarez 2000; Si et al. 2009; Ward et al. 2009). The pattern of AIV spread in wild birds may therefore influence the risk of outbreaks in poultry farms, and which is demonstrated to some extent by biotelemetry studies of wild bird migration across the Qinghai-Tibetan plateau (Prosser et al. 2011). It is possible that wild birds also influence the pattern of AIV spread in poultry in North America, although this will require analysis of a larger data set of domestic bird viruses.
We suggest that the strongly spatially dependent pattern of AIV gene flow observed here may serve as a useful estimator of the risk of disease spread when a novel AIV lineage enters the wild bird populations of North America. In particular, we would expect any newly invading virus to be similarly structured by flyway and geographical distance, and therefore follow a route of dispersal that is in some aspects predictable. This may also apply to the emergence of highly pathogenic AIV, although other factors may come into place in this circumstance. For instance, spatial distance may constitute a stronger barrier if highly pathogenic AIV reduces migratory capability (Feare & Yasue 2006; Gauthier-Clerc et al. 2007), and high density poultry populations may represent more efficient vectors for the transmission of highly virulent viruses (Lebarbenchon et al. 2010).
Compared to the wealth of knowledge on the ecology of many bird species in North America, the available AIV genetic data is limited, and subject to over-sampling from some localities and species. Indeed, some regions of North America, such as northern Canada and the central US, have not been extensively sampled for AIV, reducing the resolution of the inferred gene flow network. Such limitations result in a ‘closed system’ in which all phylogeographic results should be interpreted conservatively. To achieve an integrated statistical framework that fully collates geographical, temporal and host species information will require a major increase in the amount of genome sequence data from a diverse array of bird species, and a more even sampling in time and geographic space. Development of statistical models that simultaneously account for factors such as missing data and observation error may also be beneficial.
Although the analytical indices used here are not absolute measures of rates of gene flow per unit time, they do provide a powerful relative index of the average level of viral traffic among different geographical regions during a specific time period. However, it is important to note that rates of AIV gene flow between localities may vary over time as a result of changing migration patterns and/or viral prevalence in different avian species and locations. Such temporal patterns could not be resolved here due to the insufficient AIV samples for each month in our study period. This issue clearly merits further investigation, as information of this kind could help determine how the spread of avian diseases is influenced by the changing anthropogenic impact on bird habitats and behaviors.
It is generally thought, although rarely tested, that AIV evolution is shaped by a combination of ecological factors including the migratory pathways of birds, their susceptibility to influenza, the distance between habitats, and the availability of habitats shared by different bird species. Although our phylogeographic study only considered avian flyway and spatial distance as possible ecological constraints to AIV, we believe that the statistical techniques utilized here could be used to study the influence of other ecological factors when appropriate data becomes available. Indeed, there is accumulating information on the migration routes of specific bird species, and which will enable analysis of how species-specific migration behavior shapes the dispersal of AIV. Perhaps of more importance, the future surveillance of AIV by more frequent (e.g. monthly), consistent (e.g. samples collected regularly from various species, locations and times), and geographically extensive (e.g. Canada and central US) sampling will increase the power of phylogeography, ultimately providing a higher resolution picture of the spread of AIV.
Supplementary Material
Appendix S1. More details of the genome sequencing procedure and the indices used to measure gene flow.
Figure S1. Distribution of the observed and expected numbers of changes between geographical states in the U.S. and Canada.
Figure S2. FST versus spatial distance.
Figure S3. Modified Slatkin-Maddison’s s (σ) plotted against the spatial distance between two localities.
Figure S4. Frequency distribution of log-likelihoods for fitting the randomly categorized rate model (RRM) to the data.
Figure S5. Parsimony estimates (σ) of the flyway-specific rates of AIV gene flow.
Table S1. Sequences used in this study.
Table S2. Summary of NA-WB-AIV nucleotide sequences used in this study.
Table S3. Levels of gene flow within and between the four American flyways.
Table S4. Inter-hemisphere AIV gene flow.
Acknowledgments
This research was funded in part by grant R01 GM080533 from the National Institutes of Health, by the US Department of the Interior and U.S. Fish and Wildlife Service’s Avian Health Program, and by the U.S. Geological Survey, Biological Resources Discipline. We thank the US Fish and Wildlife Service, as well as State and Tribal agencies. It was also in part funded with federal funds from the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract numbers HHSN272200900007C. The samples from the Pacific Flyway were collected under the ‘Surveillance for Early detection of Highly Pathogenic Avian Influenza H5N1 in Wild Migratory Birds: a Strategy for the Pacific Flyway’ Pacific Flyway Council, 2006. We thank the field personnel who sampled birds throughout the flyway, and gratefully acknowledge the contributions of many biologists who have sequenced the influenza genomes published in GenBank. We also thank members of the Diagnostic Virology Laboratory at the National Wildlife Health Center, especially Katy Griffin, Jessica Montez, Samantha Scott, and Kim Kooiman. Additional bioinformatic and computational resources were provided by the Computer Centre at The University of Hong Kong (HKU) and TWGrid in Taiwan. We also thank W. K. Kwan and Frankie Cheung (HKU) for technical support. Any use of trade, product, or firm names is for descriptive purposes only and does not imply endorsement by the US government.
Footnotes
Statement of authorship: All authors agree to the submission of this manuscript. TT-YL, HSI and ECH conceived the study. HSI, RJD, JBB, JH, BDB and DRY contributed the virus samples, and advised on aspects of avian and virus ecology. EG, DEW, RAH, TBS and DJS generated the genome sequence data. TT-YL performed the data analysis. TT-YL, HSI, EG, RJD, JBB and ECH wrote the paper with assistance from the other authors.
References
- Bao Y, Bolotov P, Dernovoy D, Kiryutin B, Zaslavsky L, Tatusova T, Ostell J, Lipman D. The influenza virus resource at the National Center for Biotechnology Information. J Virol. 2008;82:596–601. doi: 10.1128/JVI.02005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causey D, Edwards SV. Ecology of avian influenza virus in birds. J Infect Dis. 2008;197:S29–33. doi: 10.1086/524991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Smith GJ, Zhang SY, Qin K, Wang J, Li KS, Webster RG, Peiris JS, Guan Y. Avian flu: H5N1 virus outbreak in migratory waterfowl. Nature. 2005;436:191–2. doi: 10.1038/nature03974. [DOI] [PubMed] [Google Scholar]
- Davidson NC, Rothwell PI, Pienkowski MW. Towards a flyway conservation strategy for waders. Wader Study Group Bull. 1995:70–81. [Google Scholar]
- Deibel R, Emord DE, Dukelow W, Hinshaw VS, Wood JM. Influenza viruses and paramyxoviruses in ducks in the Atlantic flyway, 1977–1983, including an H5N2 isolate related to the virulent chicken virus. Avian Dis. 1985;29:970–85. [PubMed] [Google Scholar]
- del Hoyo J, Elliot A, Sargatal J. Handbook of the Birds of the World. Lynx Edicions; Barcelona: 1996. [Google Scholar]
- Djikeng A, Halpin R, Kuzmickas R, Depasse J, Feldblyum J, Sengamalay N, Afonso C, Zhang X, Anderson NG, Ghedin E, Spiro DJ. Viral genome sequencing by random priming methods. BMC Genomics. 2008;9:5. doi: 10.1186/1471-2164-9-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugan VG, Chen R, Spiro DJ, Sengamalay N, Zaborsky J, Ghedin E, Nolting J, Swayne DE, Runstadler JA, Happ GM, Senne DA, Wang R, Slemons RD, Holmes EC, Taubenberger JK. The evolutionary genetics and emergence of avian influenza viruses in wild birds. PLoS Pathog. 2008;4:e1000076. doi: 10.1371/journal.ppat.1000076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nuc Acids Res. 2004;32:1792–7. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feare CJ, Yasue M. Asymptomatic infection with highly pathogenic avian influenza H5N1 in wild birds: how sound is the evidence? Virol J. 2006;3:96. doi: 10.1186/1743-422X-3-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch WM. Toward defining the course of evolution: minimum change for a specified tree topology. Syst Zool. 1971;20:406–16. [Google Scholar]
- Gauthier-Clerc M, Lebarbenchon C, Thomas F. Recent expansion of highly pathogenic avian in uenza H5N1: a critical review. Ibis. 2007:202–214. [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol. 2009;537:113–37. doi: 10.1007/978-1-59745-251-9_6. [DOI] [PubMed] [Google Scholar]
- Hinshaw VS, Wood JM, Webster RG, Deibel R, Turner B. Circulation of influenza viruses and paramyxoviruses in waterfowl originating from two different areas of North America. Bull World Health Org. 1985;63:711–9. [PMC free article] [PubMed] [Google Scholar]
- Ip HS, Flint PL, Franson JC, Dusek RJ, Derksen DV, Gill RE, Jr, Ely CR, Pearce JM, Lanctot RB, Matsuoka SM, Irons DB, Fischer JB, Oates RM, Petersen MR, Fondell TF, Rocque DA, Pedersen JC, Rothe TC. Prevalence of Influenza A viruses in wild migratory birds in Alaska: patterns of variation in detection at a crossroads of intercontinental flyways. Virol J. 2008;5:71. doi: 10.1186/1743-422X-5-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaoka Y, Chambers TM, Sladen WL, Webster RG. Is the gene pool of influenza viruses in shorebirds and gulls different from that in wild ducks? Virology. 1988;163:247–50. doi: 10.1016/0042-6822(88)90260-7. [DOI] [PubMed] [Google Scholar]
- Koehler AV, Pearce JM, Flint PL, Franson JC, Ip HS. Genetic evidence of intercontinental movement of avian influenza in a migratory bird: the northern pintail (Anas acuta) Mol Ecol. 2008;17:4754–62. doi: 10.1111/j.1365-294X.2008.03953.x. [DOI] [PubMed] [Google Scholar]
- Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, Hinshaw VS, Webster RG. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis. 2004;4:177–89. doi: 10.1089/vbz.2004.4.177. [DOI] [PubMed] [Google Scholar]
- Lebarbenchon C, Feare CJ, Renaud F, Thomas F, Gauthier-Clerc M. Persistence of highly pathogenic avian influenza viruses in natural ecosystems. Emer Infect Dis. 2010;16:1057–62. doi: 10.3201/eid1607.090389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln FC. Circular. Vol. 16. U.S. Fish and Wildlife Service; Washington, D.C: 1979. Migration of birds. [Google Scholar]
- Lynch M, Crease TJ. The analysis of population survey data on DNA sequence variation. Mol Biol Evol. 1990;7:377–94. doi: 10.1093/oxfordjournals.molbev.a040607. [DOI] [PubMed] [Google Scholar]
- Maddison WP, Slatkin M. Null models for the number of evolutionary steps in a character on a phylogenetic tree. Evolution. 1990;45:1184–97. doi: 10.1111/j.1558-5646.1991.tb04385.x. [DOI] [PubMed] [Google Scholar]
- Miller MR, Takekawa JY, Battaglia DS, Golightly RT, Perry WM. Spring migration and summer destinations of northern pintails from the coast of southern California. The Southwestern Naturalist. 2010;55:501–509. [Google Scholar]
- Munster VJ, Baas C, Lexmond P, Waldenstrom J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WE, Schutten M, Olsen B, Osterhaus AD, Fouchier RA. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog. 2007;3:e61. doi: 10.1371/journal.ppat.0030061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Evolution of human races at the gene level. Prog Clin Biol Res. 1982;103:167–81. [PubMed] [Google Scholar]
- Neigel JE. Is FST obsolete? Conservation Genet. 2002:167–173. [Google Scholar]
- Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–8. doi: 10.1126/science.1122438. [DOI] [PubMed] [Google Scholar]
- Pagel M. Detecting correlated evolution on phylogenies: a general method for the comparative analysis of discrete characters. Proc R Soc Lond B. 1994;255:37–45. [Google Scholar]
- Paradis E, Claude J, Strimmer K. APE: Analyses of Phylogenetics and Evolution in R language. Bioinformatics. 2004;20:289–90. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- Parrish CR, Holmes EC, Morens DM, Park EC, Burke DS, Calisher CH, Laughlin CA, Saif LJ, Daszak P. Cross-species virus transmission and the emergence of new epidemic diseases. Microbiol Mol Biol Rev. 2008;72:457–70. doi: 10.1128/MMBR.00004-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce JM, Ramey AM, Ip HS, Gill RE., Jr Limited evidence of trans-hemispheric movement of avian influenza viruses among contemporary North American shorebird isolates. Virus Res. 2010;148:44–50. doi: 10.1016/j.virusres.2009.12.002. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–8. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Prosser DJ, Cui P, Takekawa JY, Tang M, Hou Y, Collins BM, Yan B, Hill NJ, Li T, Li Y, Lei F, Guo S, Xing Z, He Y, Zhou Y, Douglas DC, Perry WM, Newman SH. Wild bird migration across the Qinghai-Tibetan plateau: a transmission route for highly pathogenic H5N1. PLoS One. 2011;6:e17622. doi: 10.1371/journal.pone.0017622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche B, Lebarbenchon C, Gauthier-Clerc M, Chang CM, Thomas F, Renaud F, van der Werf S, Guegan JF. Water-borne transmission drives avian influenza dynamics in wild birds: the case of the 2005–2006 epidemics in the Camargue area. Infect Genet Evol. 2009;9:800–5. doi: 10.1016/j.meegid.2009.04.009. [DOI] [PubMed] [Google Scholar]
- Romvary J, Meszaros J, Barb K, Matskasi I. The role of wild birds in the spread of influenza viruses. Acta Microbiol Acad Sci Hung. 1980;27:269–77. [PubMed] [Google Scholar]
- Si Y, Skidmore AK, Wang T, de Boer WF, Debba P, Toxopeus AG, Li L, Prins HH. Spatio-temporal dynamics of global H5N1 outbreaks match bird migration patterns. Geospat Health. 2009;4:65–78. doi: 10.4081/gh.2009.211. [DOI] [PubMed] [Google Scholar]
- Slatkin M, Maddison WP. A cladistic measure of gene flow inferred from the phylogenies of alleles. Genetics. 1989;123:603–13. doi: 10.1093/genetics/123.3.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallknecht DE, Shane SM, Kearney MT, Zwank PJ. Persistence of avian influenza viruses in water. Avian Dis. 1990;34:406–11. [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–90. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Swayne DE, Suarez DL. Highly pathogenic avian influenza. Rev Sci Tech. 2000;19:463–82. doi: 10.20506/rst.19.2.1230. [DOI] [PubMed] [Google Scholar]
- USGS. The North American Bird Banding Program. 2011 URL http://www.pwrc.usgs.gov/bbl/
- Uyeki TM. Human infection with highly pathogenic avian influenza A (H5N1) virus: review of clinical issues. Clin Infect Dis. 2009;49:279–90. doi: 10.1086/600035. [DOI] [PubMed] [Google Scholar]
- Wallace RG, Hodac H, Lathrop RH, Fitch WM. A statistical phylogeography of influenza A H5N1. Proc Natl Acad Sci U S A. 2007;104:4473–8. doi: 10.1073/pnas.0700435104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward MP, Maftei DN, Apostu CL, Suru AR. Association between outbreaks of highly pathogenic avian influenza subtype H5N1 and migratory waterfowl (family Anatidae) populations. Zoonoses and Public Health. 2009;56:1–9. doi: 10.1111/j.1863-2378.2008.01150.x. [DOI] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152–79. doi: 10.1128/mr.56.1.152-179.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B, Donnelly ME, Scholes DT, St George K, Hatta M, Kawaoka Y, Wentworth DE. Single-reaction genomic amplification accelerates sequencing and vaccine production for classical and Swine origin human influenza a viruses. J Virol. 2009;83:10309–13. doi: 10.1128/JVI.01109-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. More details of the genome sequencing procedure and the indices used to measure gene flow.
Figure S1. Distribution of the observed and expected numbers of changes between geographical states in the U.S. and Canada.
Figure S2. FST versus spatial distance.
Figure S3. Modified Slatkin-Maddison’s s (σ) plotted against the spatial distance between two localities.
Figure S4. Frequency distribution of log-likelihoods for fitting the randomly categorized rate model (RRM) to the data.
Figure S5. Parsimony estimates (σ) of the flyway-specific rates of AIV gene flow.
Table S1. Sequences used in this study.
Table S2. Summary of NA-WB-AIV nucleotide sequences used in this study.
Table S3. Levels of gene flow within and between the four American flyways.
Table S4. Inter-hemisphere AIV gene flow.