Abstract
Members of the ERF transcription-factor family participate in a number of biological processes, viz., responses to hormones, adaptation to biotic and abiotic stress, metabolism regulation, beneficial symbiotic interactions, cell differentiation and developmental processes. So far, no tissue-expression profile of any cucumber ERF protein has been reported in detail. Recent completion of the cucumber full-genome sequence has come to facilitate, not only genome-wide analysis of ERF family members in cucumbers themselves, but also a comparative analysis with those in Arabidopsis and rice. In this study, 103 hypothetical ERF family genes in the cucumber genome were identified, phylogenetic analysis indicating their classification into 10 groups, designated I to X. Motif analysis further indicated that most of the conserved motifs outside the AP2/ERF domain, are selectively distributed among the specific clades in the phylogenetic tree. From chromosomal localization and genome distribution analysis, it appears that tandem-duplication may have contributed to CsERF gene expansion. Intron/exon structure analysis indicated that a few CsERFs still conserved the former intron-position patterns existent in the common ancestor of monocots and eudicots. Expression analysis revealed the widespread distribution of the cucumber ERF gene family within plant tissues, thereby implying the probability of their performing various roles therein. Furthermore, members of some groups presented mutually similar expression patterns that might be related to their phylogenetic groups.
Keywords: Cucumis sativus L., ERF, phylogenetic analysis, transcription factor, genome sequence
Introduction
The AP2/ERF superfamily, one of the largest groups of transcription factors in plants, is characterized by the presence of the AP2/ERF-type DNA-binding domain consisting of from 60 to 70 highly conserved amino acids (Wessler, 2005). Based on sequence similarities and the number of AP2/ERF domains, this superfamily can be classified into three families, viz., AP2, ERF and RAV (Sakuma et al., 2002; Nakano et al., 2006). AP2 family proteins contain two repeated AP2/ERF domains, the ERF, a single AP2/ERF domain, and the RAV one AP2/ERF domain, as well as a B3 domain conserved in other plant-specific transcription factors. The ERF family is usually classified into two major subfamilies, CBF/DREB, and ERF, the latter based on the amino acid sequence of the DNA-binding domain. Both are divisible into I to X groups (Nakano et al., 2006).
ERF family proteins are involved in a series of biological events, such as hormonal signal transduction mediated by ethylene, cytokinin and brassinosteroid (Hu et al., 2004; Rashotte et al., 2006), response to biotic and abiotic stress (Stockinger et al., 1997; Liu et al., 1998), metabolism regulation (van der Fits and Memelink, 2000; Aharoni et al., 2004; Zhang et al., 2005), beneficial symbiotic interaction (Vernie et al., 2008), and cell differentiation (Iwase et al, 2011), as well as developmental processes, such as leaf epidermal cell density (Moose and Sisco, 1996), flower development (Elliott et al., 1996), and embryo development (Boutilier et al., 2002) in various plant species. To date, some ERF family proteins have been identified in various plant species, viz., Arabidopsis (Arabidopsis thaliana) (Sakuma et al., 2002), soybeans (Li et al., 2005), rice (Cao et al., 2006; Sharoni et al., 2011), cotton (Jin and Liu, 2008), Populus trichocarpa (Zhuang et al., 2008), tomato (Sharma et al., 2010) and Vitis vinifera (Licausi et al., 2010). The sequenced Arabidopsis genome contains 147 postulated genes encoding AP2/ERF-type proteins, 122 of which belonging to the ERF family(Nakano et al., 2006). In Arabidopsis, expression of both the DREB1A gene and its two homologs in group III is induced by low-temperature stress, but not by drought or high-salt stress, whereas, expression of both the DREB2A gene and its single homolog in another group, group IV, is induced by dehydration, but not by low-temperature stress (Liu et al., 1998; Gilmour et al., 2000), which suggests the functions of members within the same group in the ERF family are likely related to each other, similar to reported MADS-box and bHLH families (Parenicova et al., 2003; Toledo-Ortiz et al., 2003). Thus, the assessment of structural relationships between all the ERF family proteins in plants, as part of each transcription factor function analysis, would provide a guide for predicting the functions of these genes.
Cucumber (Cucumis sativus L.), belonging to the Cucurbitaceae family, is an economically and nutritionally important vegetable crop cultivated world-wide. Huang et al. (2009) proposed the existence of 110 AP2/ERF family genes in the cucumber genome. However, they did not present any specific information regarding individual genes, and no member of the cucumber ERF family has been characterized so far. Furthermore, the expression patterns of this family, as well as details on phylogenetic relationships with ERF members of other plants, remain poorly understood. Thus, the genome-wide identification, and phylogenetic and expression analysis of the family in cucumbers, as well as the comparative analyses with Arabidopsis and rice ERF members, all undertaken here, could be extremely useful in studies on the biological functions of each gene in the cucumber ERF family.
Material and Methods
Database search for cucumber ERF genes
The AP2/ERF domain of a cucumber ethylene response factor sequence (GenBank number AY792593) was used as a query sequence for TBLASTN (Altschul et al., 1997) searches of AP2/ERF superfamily genes encoded in the cucumber genome. The cucumber genome sequence from Cucumber Genome Initiative (CuGI), obtained and released by The Institute of Vegetables and Flowers, of the Chinese Academy of Agricultural Sciences (IVF-CAAS) was used. Default parameters with the TBLASTN program were wordsize 2 and extension 11. Redundant sequences with the same scaffold or chromosome location were removed from the data set. In addition, we have also obtained the same sequences from the CuGI database using Hidden Markov Model (HMM) analysis with the Pfam number PF00847 cotaining typical AP2/ERF domain.
To further confirm hypothetical AP2/ERF superfamily genes, the cDNA sequences, first translated into amino-acid sequences, were then searched for the AP2/ERF domain using the Simple Modular Architecture Research Tool (SMART)(Letunic et al., 2004).
Multiple sequence alignment, tree building and conserved motif prediction
Multiple sequence alignment, using Clustal X (Larkin et al., 2007) with default parameters, was with predicted cucumber CsERF protein sequences, with sequential manual adjustment. Similar amino acids were highlighted using the GeneDoc tool (Nicholas et al., 1997). Multalin software (Corpet, 1988) was also used as a secondary method for aligning sequences and rechecking results. To compare the evolutionary relationships of cucumber, Arabidopsis, and rice ERF family members, multiple sequence alignment was applied, by way of Clustal X, on already obtained CsERF protein sequences, and 122 Arabidopsis AtERF and 139 rice OsERF members predicted by Nakano et al. (2006), also with posterior manual adjustment of alignments.
A phylogenetic tree was constructed with aligned CsERF protein sequences using MEGA4 (Tamura et al., 2007), and the Neighbor Joining (NJ) method, with Poission correction, pairwise deletion and bootstrap (1,000 replicates; random seeds), as parameters. Simultaneously, the Maximum Parsimony (MP) method of PHYLIP 3.69 software (Felsenstein., 1989) was employed to create a second phylogenetic tree with a bootstrap of 1,000 replicates, to so validate the results from the NJ method. A combined CsERF, AtERF and OsERF phylogenetic tree was then constructed, also with MEGA4, the NJ method and a bootstrap of 1,000 replicates. The subsequent tree file was visualized by the TreeView1.6.6 tool (Page, 1996).
The MEME tool (Bailey et al., 2003) was used in the search for conserved motifs shared by CsERF members, to so identify similar sequences.
Intron/exon structure, genome distribution, and segmental duplication
For intron/exon structure analysis, the DNA and cDNA sequences corresponding to each predicted gene from BLASTN research and CuGI database annotation, were unloaded, and their intron distribution patterns and splicing phases analyzed, using the GSDS web-based bioinformatics tool.
In order to obtain information on CsERF gene location, a map with the distribution of CsERF family members throughout the cucumber genome, was drawn with the MapInspect tool. The 100 kb DNA segments flanking each CsERF gene were analyzed to detect large segment-duplicated events. Regions on the different linkage groups containing six or more homologous pairs, each with less than 25 nonhomologous intervening genes, were defined as duplicated segments. A gene-pair, separated by less than five intervening genes and sharing ≥ 40% sequence similarity at the amino acid level, was considered as tandem-duplicated. BioEdit5.0.6 software (Hall., 1999) was used for analyzing homologs for similarity on the NJ phylogenetic tree of these CsERF genes.
Expression analysis of Cucumber ERF genes
The Expressed Sequence Tag (EST) was used to detect CsERF gene expression patterns. EST data were obtained from 353,941 previously reported high quality EST sequences (Guo et al., 2010), as well as the ~8,210 cucumber EST sequences available in GenBank. An EST was considered as corresponding to its gene on sharing ≥ 95% sequence similarity, E values ≤ 10−10, and the length of matching sequences ≥ 100 bp. Semi-quantitative RT-PCR was also used to detect the expression patterns of two CsERF genes from each group. PCR primers were designed to avoid the conserved region. Information on primer sequences appears in detail in Table S4. Seeds of the ‘Chinese long’ 9930 inbred line, commonly used in modern cucumber breeding (Huang et al., 2009), were germinated and grown in trays containing a soil mixture (peat: sand: pumice, 1:1:1, v/v/v). Plants were adequately watered and grown at day/night temperatures of 24/18 °C with a 16 h photoperiod. Total RNA of root, stem, leaf, and flower of cucumber at the stage of the 20 main-stem nodes was isolated using the TRIzol Reagent (Invitrogen, USA). RT-PCR was carried out according to manufacturer’s recommendations (Tiangen Biotech Co. Ltd, Beijing China). The cucumber actin DNA fragment (161 bp) was employed as inner standard for each gene.
Results and Discussion
Identification of 103 CsERF genes
In order to identify the CsERF genes in cucumber genomes, the AP2/ERF domain of a cucumber ethylene response factor sequence (GenBank number AY792593) was used as BLAST query sequence. 131 genes were identified as possibly encoding proteins containing the AP2/ERF domain (Table 1). The same 131 sequences were also obtained from the Cucumber Genome Initiative (CuGI) database, using HMM analysis with PF00847containing a typical AP2/ERF domain. The individual genes are listed in Table S1. Among these, the 18 predicted to encode proteins containing two AP2/ERF domains, and the 4 to encode one AP2/ERF domain together with one B3 domain, were thus assigned to the AP2 and RAV families, respectively. The remaining 109 genes were all predicted to encode proteins containing a single AP/ERF domain. Among these, 103 were assigned to the ERF family. Of the remaining 6, two, Csa002695 and Csa012456, although also containing a single AP/ERF domain, were distinct from the ERF type and more closely related to the AP2. Hence, they were assigned to the AP2 family. As homology appeared to be quite low in comparison with the others, the remaining 4, viz., Csa020380, Csa005269, Csa013415 and Csa012810, were designated as soloists (Figure S1). Based on the amino acid similarity of AP2/ERF domains, the 103 ERF family members were further classified into two subfamilies, 42 genes encoding CBF/DREB-like proteins were assigned to the CBF/DREB subfamily, and the other 61, encoding ERF-like proteins, to the ERF subfamily. As number designation of the ERF family genes was based on the order of multiple sequence alignments, for study purposes, each was provisionally distinguished by a generic name, viz., CsERF001--CsERF103 (Table S1).
Table 1.
Summary of the structure and size of each group of the AP2/ERF superfamily in cucumber, compared with those in Arabidopsis and rice, as classified by Nakano et al. (2006). Totals for each family are in bold-type.
| Family | Subfamily | Group | Cucumber | Arabidopsis | Rice |
|---|---|---|---|---|---|
| AP2 | 20 | 18 | 29 | ||
| ERF | 103 | 122 | 139 | ||
| DREB | 42 | 57 | 56 | ||
| I | 5 | 10 | 9 | ||
| II | 10 | 16 | 15 | ||
| III | 20 | 22 | 26 | ||
| IV | 7 | 9 | 6 | ||
| ERF | 61 | 65 | 76 | ||
| V | 15 | 12 | 11 | ||
| VI | 8 | 8 | 6 | ||
| VII | 3 | 5 | 15 | ||
| VIII | 11 | 15 | 13 | ||
| IX | 16 | 18 | 18 | ||
| X | 8 | 7 | 13 | ||
| a single group | 7 | ||||
| RAV | 4 | 6 | 5 | ||
| Soloist | 4 | 1 | 1 | ||
| Total | 131 | 147 | 174 |
SMART analysis indicated that the AP2/ERF domain of each of the 103 CsERF genes was typical, thereby certifying to their reliability.
Multiple sequence alignments and tree building
To examine sequence features of 103 CsERF proteins, we performed a multiple sequence alignment using amino acid sequences of the AP2/ERF domain (Figure S2). The alignment indicated that the residues Gly-4, Arg-6, Arg-8, Trp-38, Gly-40, and Ala-48 were completely conserved (Figure S2). Furthermore, more than 95% of CsERF-family members contain Gly-12, Glu-17, Ile-18, Arg-36, Leu-39, Ala-49, Ala-51, and Asp-53 residues (Figure S2).
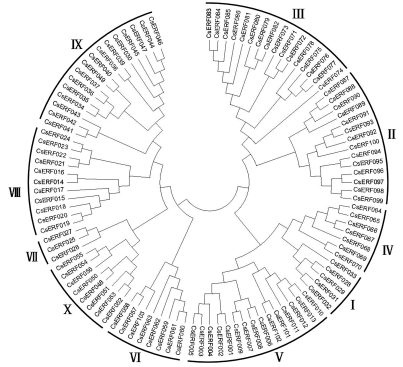
To determine the evolutionary relationship among CsERF proteins, an unrooted NJ phylogenetic tree was constructed, with bootstrap analysis (1000 replicates) based on the multiple sequence alignments of the 103 CsERF proteins (Figure 1). The analysis result showed that the 103 CsERF members were divided into 10 groups, designated I to X, in accordance with the Arabidopsis ERF gene family classification (Nakano et al., 2006). Detailed information appears in Figure 1 and Table S1. The bootstrapping values for the nodes in this phylogenetic tree were not high in every case, similar to the results of the phylogenetic analysis done on Arabidopsis ERF proteins (Nakano et al., 2006). This is most likely due to the AP2/ERF domain being relatively short, and members within the subfamily highly conserved, with relatively few informative-character positions.
Figure 1.
Phylogenetic analysis of 103 cucumber ERF proteins. An unrooted neighbor-joining phylogenetic tree was constructed using MEGA4 for the multiple sequence alignment of 103 cucumber ERF protein sequences. 10 groups are marked I to X, as described by Nakano et al., (2006).
NJ-tree reliability was certified by generating another phylogenetic tree by MP analysis (Figure S3), whereupon it was found that nearly all the CsERF members were placed within the same groups.
Conserved motifs outside the AP2/ERF domain
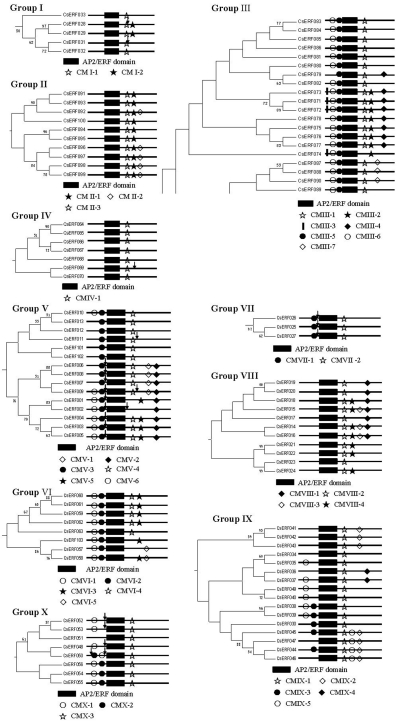
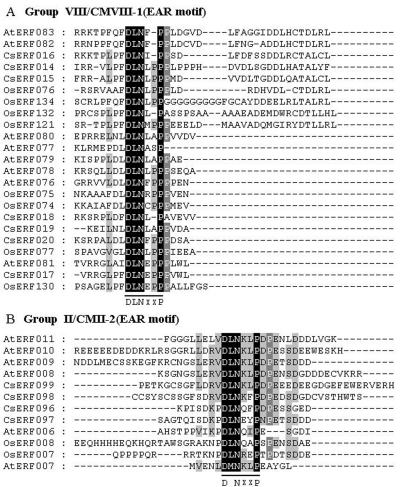
Regions outside the DNA-binding domain in transcription factors generally contain either functionally important domains, or motifs associated with transcription-regulation and nuclear localization (Liu et al., 1999). Proteins within a group that share these domains or motifs in a phylogenetic tree are likely to share similar functions. It has been reported that an ERF-associated amphiphilic repression (EAR) motif (DLNxxP) is a repression domain in the C-terminal regions of the repressor-type ERF proteins playing key roles in several biological functions by negatively regulating genes involved in developmental, hormonal and stress signaling pathways (Fujimoto et al., 2000; Ohta et al., 2001). Motif analysis in this study revealed that the EAR motif was only found among proteins within groups II and VIII, as CMII-2 and CMVIII-1 motifs, respectively (Figure 2, Figure 3, Table S2), thus leading us to suspect their involvement in negative-regulation functions. Previous research showed that the Cys repeat sequence-CX2CX4CX2~4C, possibly a zinc-finger motif, plays a part either in DNA binding, or in protein-protein interactions (Nakano et al., 2006). such a consensus sequence was only found within the CMX-2 motif in the N-terminal region of group X proteins. Liu et al. (1999) believed that regions of acidic amino acid-rich, Gln-rich, Pro-rich, and/or Ser/Thr-rich amino acid sequences, can usually be designated as transcriptional-activation domains (Liu et al., 1999). The conserved motifs identified in this study have similar features, such as Gln-rich in group III as the CMIII-7 motif, Pro-rich in group III as the CMIII-2 motif, and/or Ser/Thr-rich in group VIII as the CMVIII-4 motif (Figure 2; Table S2).
Figure 2.
Phylogenetic relationships among CsERF proteins from the 10 groups, I to X, in the cucumber ERF family. Only > 50% bootstrap values are shown in this phylogenetic tree. The positions of introns are marked with arrowheads. Each black box represents an AP2/ERF domain. Conserved motifs are marked with different signs, as indicated below the tree.
Figure 3.
The EAR motif-like sequences conserved in group VIII and II ERF proteins. (A) Sequence alignment of C-terminal regions in group VIII proteins. (B) Sequence alignment of C-terminal regions in group II proteins. Conserved motifs are underlined. Black and gray shading indicate identical and conserved amino acid residues present in > 50% of the aligned sequences, respectively. Consensus amino acid residues are given below the alignment. The “x” in the sequence indicates no conservation at this position.
In addition, we also found that most of the motifs were selectively distributed among the specific clades in the phylogenetic tree, thereby demonstrating structural similarities among members within the same group (Figure 2, Table S2).
Structure and evolution of CsERF genes
Apparently, most Arabidopsis ERF genes do not possess introns (Sakuma et al., 2002). A similar situation also appeared in this study, among the 103 CsERF genes, 83 (81%) having no intron, the remaining 20, unevenly distributed in groups I, IV, V, VII and X, having only one or two introns (Figure 2). As shown in Figure 2, 17 of the 20 possess only a single intron, and the other three genes CsERF002, CsERF009 and CsERF050 possess two. The presence and position of the introns was highly conserved in each group, thus further validating the reliability of the cucumber ERF-family gene classification in this study.
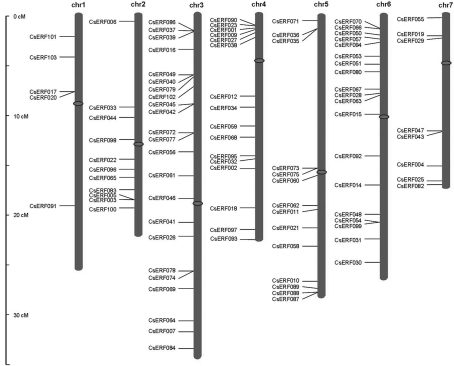
The genomic distribution of the 103 CsERF genes was analyzed, in order to acquire an insight into their evolution. With the exception of the seven genes CsERF008, CsERF013, CsERF024, CsERF052, CsERF076, CsERF081 and CsERF085 lying within unassembled scaffold000393, scaffold000131, scaffold000677, scaffold000111, scaffold000379, scaffold000576 and scaffold000118, respectively, the remainder were found to be unevenly distributed among seven chromosomes (Figure 4, Table S1). As indicated in Figure 4, some, clustered in a large group, within the same small chromosomal region, as, for example, group III members CsERF087, CsERF088 and CsERF089, located in a region close to a telomere on chromosome 5, whereas other members of this group were distributed among different chromosomes, i.e., CsERF083 in chromosome 2 and CsERF090 in chromosome 4. A similar situation also occurred among OsERF and AtERF members (Nakano et al., 2006), thereby indicating that ERF genes are distributed widely within the genome of the common ancestor of monocots and eudicots.
Figure 4.
Chromosomal localization of 103 cucumber ERF genes. The scale is in megabases (Mb). The ovals in the middle of the seven chromosomes show the centromeric positions according to the sequencing results of the cucumber genome (Huang et al., 2009). Seven genes CsERF008, CsERF013, CsERF024, CsERF052, CsERF076, CsERF081, and CsERF085 on scaffold000393, scaffold000131, scaffold000677, scaffold000111, scaffold000379, scaffold000576, and scaffold000118, respectively, could not be anchored onto a specific chromosome.
Although previous research has shown that whole-genome duplication, as a recent event, is not the case with cucumbers, several tandem duplications have in fact actually occurred (Huang et al., 2009), with considerable impact on the increase in the number of family genes in the genome. As the analysis of 100 kb DNA segments flanking each CsERF gene indicated that none could have derived from segment duplication, it is most likely that tandem duplication played a crucial role in gene multiplication. According to previously reported results with Arabidopsis, members of groups III and IX play crucial roles in biotic and/or abiotic stress response. Our analyses indicated both to be the two largest groups, gene multiplication in the two possibly having arisen from the higher frequency of tandem duplication, as a means of adapting to various environment changes.
In silico identification of ERF genes in Arabidopsis and rice, and a comparative analysis of cucumbers
As a previous report indicated there to be 122 and 139 ERF family members distributed within Arabidopsis and rice genomes, respectively (Nakano et al., 2006), Arabidopsis and rice genomes were re-screened for ERF sequences, with the subsequent discovery of a further four AtERF and six OsERF genes, designated as AtERF123~AtERF126 and OsERF140~OsERF145 (Table S3), respectively, based on the names of the previous 122 AtERFs and 139 OsERFs. This discrepancy is probably owing to fresh information on Arabidopsis and rice genome sequences.
To define the evolutionary relationship of cucumber ERF family proteins with those of Arabidopsis and rice, an unrooted neighbor-joining (NJ) phylogenetic tree was generated, based on bootstrap analysis (1000 replicates) of multiple sequence alignment of their respective ERF members. In addition to the ten groups I to X in Arabidopisis and rice, described by Nakano et al. (2006), another group was found, containing four new AtERF members clustering with three new OsERF members, viz., OsERF140, OsERF142 and OsERF144 (Figure S4). A further three new OsERF members, viz., OsERF141, OsERF143 and OsERF145, were placed in groups I, II and VIII, respectively.
Phylogenetic analysis revealed that most CsERF members were closer to eudicot AtERF members than monocot OsERFs in this classification. For example, based on bootstrap values, three group III CsERF members (CsERF071~CsERF073) clustered with six AtERF (AtERF028~AtERF033), whereas another nine OsERFs (OsERF024~OsERF031, OsERF133) were branched into a single clade (Figure S4). On closely examining group IV, it was found that only one member, CsERF070, was clustered with OsERF117 and AtERF052. As previous studies have shown that AtERF052 mainly mediates the effects of exogenous trehalose on Arabidopsis growth and starch breakdown, and vegetative development by sugar, besides repressing endosperm induced seed germination, CsERF070 may also participates in these plant-development processes, the detailed function needs further researched and confirmation. At the same time, CsERF078 was close to AtERF024 in group III and CsERF017 to AtERF081 in group VIII, although the functions of these genes have not, as yet, been rigorously demonstrated.
Usually, the intron/exon position pattern provides clues on evolutionary relationships. Research by Nakano et al. (2006) indicated that the position of the intron was conserved in Arabidopsis ERF groups V, VII, X, with Xb-L containing only one intron. As with Arabidopsis and rice ERF, in the present study, it was revealed that both the presence and position of CsERF introns in groups IV, V, VII and X were highly conserved, with only one or two exceptions (Figure 2). Thus, besides further validating the classification of CsERF family genes, this was an indication of the conservation of intron position patterns that existed in the common ancestor of monocots and eudicots. On the other hand, and in the same group, introns were observed in only one species, but not in others. For example, two CsERF members in group I, namely CsERF028 and CsERF031, possessed one intron at the N-terminal region, although AtERF and OsERF members in the same group possessed none. A similar situation also occured in group II, two OsERF members, OsERF015 and OsERF016, having one intron, and CsERF and AtERF members none, thus possibly indicating intron insertion after the divergence of monocots and eudicots.
As described above, proteins within a group that share conserved domains or motifs outside the DNA-binding domain in transcription factors in a phylogenetic tree, are likely to share similar functions. Cheong et al. (2003) believed that the CMVII-4 motif, as a putative MAP kinase phosphorylation site in OsEREBP1, and the phosphorylation of OsEREBP1, resulted in the enhancement of its binding to the GCC box and GCC box-mediated transcriptional activation. Conserved motif analysis showed that the very CMVII-4 motif (CMVII-3 in this study) has been observed in CsERF025 and CsERF026 in group VII, OsERF070~OsERF072 in rice, and AtERF074 and AtERF075 in Arabidopsis. Whether CsERF025 and CsERF026 share similar functions in transcriptional activation regulation, needs to be confirmed.
Expression analysis of 103 cucumber ERF genes
As gene expression patterns are often correlated with their functions, the ESTs, created by partially sequencing randomly isolated gene transcripts, have proved to be invaluable in discovery through expression pattern analysis.
By using data originating from both 353,941 previously reported high quality ESTs (Guo et al. 2010) and ~8,210 cucumber ESTs available in GenBank, 41 CsERF genes were discovered in at least one tissue among the four investigated, i.e., root, shoot, leaf and flower, the remaining 62 having presented no expression signal. Whereas among the 41 expressed genes, 35, with a high 85% ratio, were found in flowers, two, CsERF067 and CsERF072, of the remaining six were expressed in roots, one, CsERF067, in shoots, and, three, CsERF026, CsERF036, and CsERF045, in leaves. The fact that most sequence tag expression corresponded to CsERF genes in flowers and little in the other tissues, might be due to the 353,941 EST sequences (98%) having all originated from one flower tissue (Guo et al. 2010).
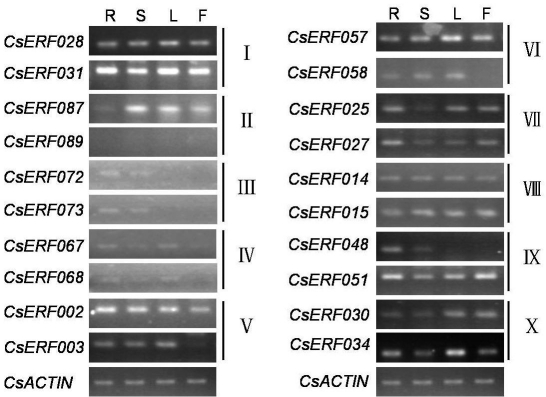
For further study of CsERF gene expression patterns, two members of each group were selected for RT-PCR analysis with RNA from roots, stems, leaves and flowers. In general, patterns were conserved within subfamilies, although expression levels of specific members could change in different organs. Similar expression patterns were observed among members belonging to 6 groups of CsERF genes (I, III, IV, VII, VIII, X). Among these, the members of 4 groups (I, VII, VIII, X) were expressed wherever investigated, thereby implying that these genes could play regulatory roles in various cucumber tissues. As regards the other two groups, CsERF067 and CsERF068 from group IV presented transcript signals in three plant tissues, whereas in group III, CsERF072 and CsERF073 transcript signals were detected only in the root and stem, a possible indication of their taking part in specific biological processes in cucumber vegetative development. Given that similar expression patterns were observed in two members in each group, it is speculated that this similarity might also extend to other group-members, pending corroboration by further experiments.
On the other hand, expression patterns in members of the other 4 groups (II, V, VI and IX) were varied. As indicated in Figure 5, in CsERF02 and CsERF03 in group V, the high transcript signals detected in three of the four vegetative tissues were different from those expressed in flowers. A similar situation was also found in two members, CsERF057 and CsERF058, in group VI. More obvious variation in gene expression among group members could be observed in the remaining groups, II and IX. Group II member CsERF087 presented transcripts in the stem, leaf and flower, whereas for the other member, CsERF089, no detectable signal was observed anywhere. In group IX, CsERF051 was highly expressed in all the tissues, whereas the other member, CsERF048, was expressed only in the root and stem. The different expression patterns among these 4 groups could imply the existence of a probable intragroup functional divergence.
Figure 5.
Expression analysis of 20 cucumber ERF genes in different tissues using RT-PCR. RT-PCR was with primers specific for the 20 CsERF genes. PCR products were run on 1.5% agarose gels. CsACTIN primers giving a 161 bp product were used as inner standard for each gene. Sample identities are as follows: root (R), stem (S), leaf (L) and flower (F).
In summary, after extensive analysis,103 CsERF genes were compared with 126 Arabidopsis and 145 rice ERF genes. The 103 CsERF genes were divided into 10 groups, I to X, thus in general accordance with previous studies (Nakano et al., 2006). This classification was based on the presence and position of the introns and the conserved amino acid sequence motifs outside the AP2/ERF domain. Chromosomal localization and genome distribution revealed that tandem duplication may have contributed to CsERF-gene expansion. Expression data revealed the widespread distribution of this gene family within cucumber plant tissues. Furthermore, in most of the groups, two different members presented similar expression patterns, whereby a possible basis for functional analysis to discover the role of CsERF genes in cucumber development.
Supplementary Material
Acknowledgments
The authors wish to thank Zhonghua Zhang for help with statistical analysis. This work was supported by the National Natural Science Foundation of China (31060262), the Natural Science Foundation of Jiangxi, China (2009GQN0034) and the Foundation of Jiangxi Agricultural University, Jiangxi, China (2009).
Footnotes
Associate Editor: Adriana Silva Hemerly
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. Plant Cell. 2004;16:2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aukerman MJ, Sakai H. Regulation of flowering time and floral organ identity by a MicroRNA and its APE-TALA2-like target genes. Plant Cell. 2003;15:2730–2741. doi: 10.1105/tpc.016238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey PC, Martin C, Toledo-Ortiz G, Quail PH, Huq E, Heim MA, Jakoby M, Werber M, Weisshaar B. Update on the basic helix-loop-helix transcription factor gene family in Arabidopsis thaliana. Plant Cell. 2003;15:2497–2502. doi: 10.1105/tpc.151140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutilier K, Offringa R, Sharma VK, Kieft H, Ouellet T, Zhang L, Hattori J, Liu CM, van Lammeren AA, Miki BL, et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broun P, Poindexter P, Osborne E, Jiang CZ, Riechmann JL. WIN1, a transcriptional activator of epidermal wax accumulation in Arabidopsis. Proc Natl Acad Sci USA. 2004;101:4706–4711. doi: 10.1073/pnas.0305574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RL, Kazan K, McGrath KC, Maclean DJ, Manners JM. A role for the GCC-box in jasmonate-mediated activation of the PDF1.2 gene of Arabidopsis. Plant Physiol. 2003;132:1020–1032. doi: 10.1104/pp.102.017814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Song F, Goodman RM, Zheng Z. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. J Plant Physiol. 2006;163:1167–1178. doi: 10.1016/j.jplph.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Che P, Lall S, Nettleton D, Howell SH. Gene expression programs during shoot, root, and callus development in Arabidopsis tissue culture. Plant Physiol. 2006;141:620–637. doi: 10.1104/pp.106.081240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong YH, Moon BC, Kim JK, Kim CY, Kim MC, Kim IH, Park CY, Kim JC, Park BO, Koo SC, et al. BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 2003;132:1961–1972. doi: 10.1104/pp.103.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong CJ, Liu JY. The Arabidopsis EAR-motif-containing protein RAP2.1 functions as an active transcriptional repressor to keep stress responses under tight control. BMC Plant Biol. 2010;10:e47. doi: 10.1186/1471-2229-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. AINTEGU-MENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP: Phylogeny Inference Package. Cladistics. 1989;5:164–166. [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Zheng Y, Joung JG, Liu S, Zhang Z, Crasta OR, Sobral BW, Xu Y, Huang S, Fei Z. Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genomics. 2010;11:e384. doi: 10.1186/1471-2164-11-384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall TA. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999;41:95–98. [Google Scholar]
- Hinz M, Wilson IW, Yang J, Buerstenbinder K, Llewellyn D, Dennis ES, Sauter M, Dolferus R. Arabidopsis RAP2.2: An ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 2010;153:757–772. doi: 10.1104/pp.110.155077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu YX, Wang YX, Liu XF, Li JY. Arabidopsis RAV1 is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Res. 2004;14:8–15. doi: 10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, et al. The genome of the cucumber, Cucumis sativus L. Nat Genet. 2009;41:1275–1281. doi: 10.1038/ng.475. [DOI] [PubMed] [Google Scholar]
- Ikeda Y, Banno H, Niu QW, Howell SH, Chua NH. The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 2006;47:1443–1456. doi: 10.1093/pcp/pcl023. [DOI] [PubMed] [Google Scholar]
- Iwase A, Mitsuda N, Koyama T, Hiratsu K, Kojima M, Arai T, Inoue Y, Seki M, Sakakibara H, Sugimoto K, et al. The AP2/ERF transcription factor WIND1 controls cell dedifferentiation in Arabidopsis. Curr Biol. 2011;21:508–514. doi: 10.1016/j.cub.2011.02.020. [DOI] [PubMed] [Google Scholar]
- Jin LG, Liu JY. Molecular cloning, expression profile and promoter analysis of a novel ethylene responsive transcription factor gene GhERF4 from cotton (Gossypium hirstum) Plant Physiol Biochem. 2008;46:46–53. doi: 10.1016/j.plaphy.2007.10.004. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X v. 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Letunic I, Copley RR, Schmidt S, Ciccarelli FD, Doerks T, Schultz J, Ponting CP, Bork P. SMART 4.0: Towards genomic data integration. Nucleic Acids Res. 2004;32:D142–D144. doi: 10.1093/nar/gkh088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XP, Tian AG, Luo GZ, Gong ZZ, Zhang JS, Chen SY. Soybean DRE-binding transcription factors that are responsive to abiotic stresses. Theor Appl Genet. 2005;110:1355–1362. doi: 10.1007/s00122-004-1867-6. [DOI] [PubMed] [Google Scholar]
- Licausi F, Giorgi FM, Zenoni S, Osti F, Pezzotti M, Perata P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics. 2010;11:e719. doi: 10.1186/1471-2164-11-719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim CJ, Hwang JE, Chen H, Hong JK, Yang KA, Choi MS, Lee KO, Chung WS, Lee SY, Lim CO. Over-expression of the Arabidopsis DRE/CRT-binding transcription factor DREB2C enhances thermotolerance. Biochem Biophys Res Commun. 2007;362:431–436. doi: 10.1016/j.bbrc.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Lin RC, Park HJ, Wang HY. Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol Plant. 2008;1:42–57. doi: 10.1093/mp/ssm004. [DOI] [PubMed] [Google Scholar]
- Liu L, White MJ, MacRae TH. Transcription factors and their genes in higher plants functional domains, evolution and regulation. Eur J Biochem. 1999;262:247–257. doi: 10.1046/j.1432-1327.1999.00349.x. [DOI] [PubMed] [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moose SP, Sisco PH. Glossy15, an APETALA2-like gene from maize that regulates leaf epidermal cell identity. Genes Dev. 1996;10:3018–3027. doi: 10.1101/gad.10.23.3018. [DOI] [PubMed] [Google Scholar]
- Nag A, Yang Y, Jack T. DORNROSCHEN-LIKE, an AP2 gene, is necessary for stamen emergence in Arabidopsis. Plant Mol Biol. 2007;65:219–232. doi: 10.1007/s11103-007-9210-7. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima K, Shinwari ZK, Sakuma Y, Seki M, Miura S, Shinozaki K, Yamaguchi-Shinozaki K. Organization and expression of two Arabidopsis DREB2 genes encoding DRE-binding proteins involved in dehydration- and high-salinity-responsive gene expression. Plant Mol Biol. 2000;42:657–665. doi: 10.1023/a:1006321900483. [DOI] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Jr, Deerfield DW., II GeneDoc: Analysis and visualization of genetic variation. Embnew News. 1997;4:14. [Google Scholar]
- Ogawa T, Pan L, Kawai-Yamada M, Yu LH, Yamamura S, Koyama T, Kitajima S, Ohme-Takagi M, Sato F, Uchimiya H. Functional analysis of Arabidopsis ethylene-responsive element binding protein conferring resistance to Bax and abiotic stress-induced plant cell death. Plant Physiol. 2005;138:1436–1445. doi: 10.1104/pp.105.063586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell. 2001;13:1959–1968. doi: 10.1105/TPC.010127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RD. TreeView: An application to display phylogenetic trees on personal computers. Comput Appl Biosci. 1996;12:357–358. doi: 10.1093/bioinformatics/12.4.357. [DOI] [PubMed] [Google Scholar]
- Parenicova L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell. 2003;15:1538–1551. doi: 10.1105/tpc.011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pre M, Atallah M, Champion A, De Vos M, Pieterse CM, Memelink J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008;147:1347–1357. doi: 10.1104/pp.108.117523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, Fujita M, Umezawa T, Sawano Y, Miyazono K, et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell. 2008;20:1693–1707. doi: 10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Mason MG, Hutchison CE, Ferreira FJ, Schaller GE, Kieber JJ. A subset of Arabidopsis AP2 transcription factors mediates cytokinin responses in concert with a two-component pathway. Proc Natl Acad Sci USA. 2006;103:11081–11085. doi: 10.1073/pnas.0602038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, Reuber L, Jiang C, Keddie J, Adam L, Pineda O, Ratcliffe OJ, Samaha RR, et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Schmid M, Uhlenhaut NH, Godard F, Demar M, Bressan R, Weigel D, Lohmann JU. Dissection of floral induction pathways using global expression analysis. Development. 2003;130:6001–6012. doi: 10.1242/dev.00842. [DOI] [PubMed] [Google Scholar]
- Shaikhali J, Heiber I, Seidel T, Stroher E, Hiltscher H, Birkmann S, Dietz KJ, Baier M. The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 2008;8:e48. doi: 10.1186/1471-2229-8-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma MK, Kumar R, Solanke AU, Sharma R, Tyagi AK, Sharma AK. Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Mol Genet Genomics. 2010;284:455–475. doi: 10.1007/s00438-010-0580-1. [DOI] [PubMed] [Google Scholar]
- Sharoni AM, Nuruzzaman M, Satoh K, Shimizu T, Kondoh H, Sasaya T, Choi IR, Omura T, Kikuchi S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011;52:344–360. doi: 10.1093/pcp/pcq196. [DOI] [PubMed] [Google Scholar]
- Solano R, Stepanova A, Chao Q, Ecker JR. Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 1998;12:3703–3714. doi: 10.1101/gad.12.23.3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger EJ, Gilmour SJ, Thomashow MF. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc Natl Acad Sci USA. 1997;94:1035–1040. doi: 10.1073/pnas.94.3.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software v. 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15:1749–1770. doi: 10.1105/tpc.013839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- Vernie T, Moreau S, de Billy F, Plet J, Combier JP, Rogers C, Oldroyd G, Frugier F, Niebel A, Gamas P. EFD Is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell. 2008;20:2696–2713. doi: 10.1105/tpc.108.059857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch R, Maass D, Voegel T, Dellapenna D, Beyer P. Transcription factor RAP2.2 and its interacting partner SINAT2: Stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 2007;145:1073–1085. doi: 10.1104/pp.107.104828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR. Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci. 2005;10:54–56. doi: 10.1016/j.tplants.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Xu H, Wang X, Chen J. Overexpression of the Rap2.4f transcriptional factor in Arabidopsis promotes leaf senescence. Sci China Life Sci. 2010;53:1221–1226. doi: 10.1007/s11427-010-4068-3. [DOI] [PubMed] [Google Scholar]
- Zhang JY, Broeckling CD, Blancaflor EB, Sledge MK, Sumner LW, Wang ZY. Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa) Plant J. 2005;42:689–707. doi: 10.1111/j.1365-313X.2005.02405.x. [DOI] [PubMed] [Google Scholar]
- Zhuang J, Cai B, Peng RH, Zhu B, Jin XF, Xue Y, Gao F, Fu XY, Tian YS, Zhao W, et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem Biophys Res Commun. 2008;371:468–474. doi: 10.1016/j.bbrc.2008.04.087. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, Zhang H. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene. 2010;457:1–12. doi: 10.1016/j.gene.2010.02.011. [DOI] [PubMed] [Google Scholar]
Internet Resources
- Cucumber Genome Initiative (CuGI) http://cucumber.genomics.org.cn(October 15, 2010)
- Clustal X. http://www.clustal.org(October 20, 2010)
- GeneDoc tool. http://www.nrbsc.org/gfx/genedoc/(October 20, 2010).
- Multalin software. http://multalin.toulouse.inra.fr/multalin/multalin.html(October 20, 2010)
- MEGA4. http://www.megasoftware.net/index.html(October 20, 2010)
- PHYLIP 3.69. http://evolution.genetics.washington.edu/phylip.html(October 20, 2010)
- TreeView1.6.6. http://taxonomy.zoology.gla.ac.uk/rod/treeview.html(October 20, 2010).
- MEME. http://meme.sdsc.edu/meme4_4_0/cgi-bin/meme.cgi(October 20, 2010).
- GSDS. http://gsds.cbi.pku.edu.cn/(October 20, 2010).
- MapInspect. http://www.plantbreeding.wur.nl/UK/software_mapinspect.html(October 20, 2010).
- BioEdit5.0.6. http://www.mbio.ncsu.edu/BioEdit/bioedit.html(October 20, 2010).
- TIGR Arabidopsis annotation deduced protein database. http://www.tigr.org/tdb/e2k1/ath1/(October 15, 2010).
- Rice genome, release version 5 of TIGR pseudomolecules. http://rice.plantbiology.msu.edu/(October 15, 2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.