Abstract
A detailed study of putative recombination events and their evolution frequency in the whole genome of the currently known members of the family Tombusviridae, comprising 79 accessions retrieved from the international databases, was carried out by using the RECCO and RDP version 3.31β algorithms. The first program allowed the detection of potential recombination sites in seven out of eight virus genera (Aureusvirus, Avenavirus, Carmovirus, Dianthovirus, Necrovirus, Panicovirus, and Tombusvirus), the second program provided the same results except for genus Dianthovirus. On the other hand, both methods failed to detect recombination breakpoints in the genome of members of genus Machlomovirus. Furthermore, based on Fisher’s Exact Test of Neutrality, positive selection exerted on protein-coding genes was detected in 17 accession pairs involving 15 different lineages. Except genera Machlomovirus, and Panicovirus along with unclassified Tombusviridae, all the other taxonomical genera and the unassigned Tombusviridae encompassed representatives under positive selection. The evolutionary history of all members of the Tombusviridae family showed that they segregated into eight distinct groups corresponding to the eight genera which constitute this family. The inferred phylogeny reshuffled the classification currently adopted by the International Committee on Taxonomy of Viruses. A reclassification was proposed.
Keywords: bioinformatics, phylogeny, recombination, positive selection, Tombusviridae, sequence, taxonomy
Introduction
RNA recombination is one of the major factors responsible for the generation of new RNA viruses and retroviruses. The biological mechanisms of recombination differ across organisms, but in broad terms recombination results in the creation of mosaic sequences where the evolutionary history at each site may be different. Recombination, defined as the exchange of genetic information between two nucleotide sequences, is an important process that influences biological evolution at many different levels. Recombination explains a considerable amount of genetic diversity in natural populations and, in general, genes located in regions of the genome with low levels of recombination have low levels of polymorphism (Posada and Crandall, 2001). Recombination reshuffles existing variation and even creates new variants. It has been shown that RNA recombination enables the exchange of genetic material, not only between the same or similar viruses but also between distinctly different viruses (Worobey and Holmes, 1999). Sometimes, it also permits crossovers between viral and host RNA (Greene and Allison, 1994; Aaziz and Tepfer, 1999; Baroth et al., 2000; Nagai et al., 2003). Taking into account the structure of viral genomic molecules and the location of crossover sites, three basic types of RNA recombination were distinguished: homologous, aberrant homologous and non-homologous (Lai, 1992; Alejska et al., 2001). The former two occur between two identical or similar RNAs (or between molecules displaying local homology), while the latter involves two different molecules. Most of the collected data suggest that RNA recombinants are formed according to a copy choice model (Alejska et al., 2001). A viral replication complex starts nascent RNA strand synthesis on one template, called RNA donor, and then switches to another template, called RNA acceptor. Accordingly, two main factors are thought to affect RNA recombination: the structure of recombining molecules and the ability of the viral replicase to switch templates. Through generations, viral populations evolve under various selective forces at different regions and sites that display different functional constraints. A stringent and robust criterion for detecting adaptive evolution in a protein-coding gene is an accelerated nonsynonymous (dN, amino acid replacing) rate relative to the synonymous (dS, silent) rate of substitutions, with the rate ratio ω = dN/dS > 1. As silent mutations do not change the amino acid whereas replacement mutations do, the difference in their fixation rates provides a measure of selective pressure on the protein.
Amongst positive-strand plant RNA viruses, the family Tombusviridae encompasses several viruses with an important economical impact. According to the 8th ICTV (International Committee on Taxonomy of Viruses) report (Fauquet et al., 2005), the family Tombusviridae includes the following genera: Tombusvirus, Carmovirus, Necrovirus, Dianthovirus, Machlomovirus, Avenavirus, Aureusvirus and Panicovirus. According to the Baltimore classification, the viruses in this family are classified as Type IV viruses, and are part of the luteovirus supergroup (Habili and Symons, 1989). The RNA is contained in an icosahedral (T = 3) capsid, composed of 180 units of a single coat protein 27–42 kDa in size; the virion measures 28–35 nm in diameter, and is not enveloped. All Tombusviridae have a positive- sense, single-stranded linear genome, with the exception of dianthoviruses, whose genome is bipartite. The genome is approximately 4–5.4 kb in length, depending on the genus. The 3′ terminus is not polyadenylated. The 5′ terminus is capped only in Carnation mottle carmovirus, Red clover necrotic mosaic dianthovirus and Maize chlorotic mottle machlomovirus. The genome encodes 4–6 ORFs. The polymerase ORF encodes an amber stop codon that is the site of a readthrough event within ORF 1 (except in dianthoviruses, where readthrough occurs via a frameshift), producing two products necessary for replication. There is no helicase encoded by the virus. The replication process of members of family Tombusviridae comprises the following steps: (i) the virus penetrates into the host cell, (ii) the viral genomic RNA is uncoated and released into the cytoplasm, (iii) the viral RNA is translated to produce the two proteins necessary for RNA synthesis (replication and transcription), (iv) a negative-sense complementary ssRNA is synthesized using the genome RNA as a template, (v) a new genomic RNA is synthesized using the negative-sense RNA as a template, (vi) the RNA-dependant RNA polymerase (RdRp) recognizes internal subgenomic promoters on the negative-sense RNA, to transcribe the 3′ co-terminal subgenomic RNAs that will generate the capsid and movement protein, (vii) new virus particles are formed (White and Nagy, 2004).
The main objective of this work was to determine and characterize virus evolution mechanisms of the Tombusviridae based on the occurrence of putative recombination events and positive selection in their full-length genome. This was achieved by the analysis of 79 accessions obtained from GenBank. As a result, we propose a reclassification according to their predicted evolutionary history.
Material and Methods
The sequences of the entire genome of 79 accessions cataloged in GenBank were used in this study (Table 1).
Table 1.
Current taxonomic status of the members of the family Tombusviridae included in the study and their accession numbers.
| Genus (in bold), unclassified and unassigned viruses | Virus/Isolate | GenBank accession number |
|---|---|---|
| Aureusvirus | Cucumber leaf spot virus (CLSV) | NC_007816 |
| Cucumber leaf spot virus/Canada (CLSV/Canada) | EU127904 | |
| Pothos latent virus/Pigeonpea (PoLV/Pigeonpea) | NC_000939 | |
| Johnsongrass chlorotic stripe mosaic virus/Iran (JCSMV/Iran) | NC_005287 | |
| Maize white line mosaic virus/USA (MaWLMV/USA) | NC_009533 | |
| Avenavirus | Oat chlorotic stunt virus (OCSV) | NC_003633 |
| Carmovirus | Cardamine chlorotic fleck virus (CCFV) | NC_001600 |
| Carnation mottle virus/China (CarMoV/China) | NC_001265 | |
| Carnation mottle virus (CarMoV) | X02986 | |
| Carnation mottle virus/Indian (CarMoV/Indian) | AJ811998 | |
| Cowpea mottle virus (CPMoV) | NC_003535 | |
| Hibiscus chlorotic ringspot virus (HCRSV) | NC_003608 | |
| Hibiscus chlorotic ringspot virus.Tw (HCRSV.Tw) | DQ392986 | |
| Japanese iris necrotic ring virus (JINRV) | NC_002187 | |
| Melon necrotic spot virus/Yamaguchi (MeNSV/Yamaguchi) | AB250687 | |
| Melon necrotic spot virus/Nagasaki (MeNSV/Nagasaki) | AB250686 | |
| Melon necrotic spot virus/Kochi (MeNSV/Kochi) | AB250685 | |
| Melon necrotic spot virus/Chiba (MeNSV/Chiba) | AB250684 | |
| Melon necrotic spot virus/Tottori (MeNSV/Tottori) | AB232925 | |
| Melon necrotic spot virus/Kochi2 (MeNSV/Kochi2) | AB232926 | |
| Melon necrotic spot virus (MeNSV) | NC_001504 | |
| Melon necrotic spot virus/MNSV-ISR (MeNSV/MNSV-ISR) | DQ922807 | |
| Melon necrotic spot virus/MNSV-Al (MeNSV/MNSV-Al) | DQ339157 | |
| Melon necrotic spot virus/MNSV264 (MeNSV/MNSV264) | AY330700 | |
| Melon necrotic spot virus/nK (MeNSV/nK) | AB044292 | |
| Melon necrotic spot virus/NH (MeNSV/NH) | AB044291 | |
| Melon necrotic spot virus/Malfa5 (MeNSV/Malfa5) | AY122286 | |
| Pea stem necrosis virus/Japan (PSNV/Japan) | NC_004995 | |
| Pelargonium flower break virus/MZ10 (PFBV/MZ10) | NC_005286 | |
| Pelargonium flower break virus/SP18 (PFBV/SP18) | DQ256073 | |
| Saguaro cactus virus (SCV) | NC_001780 | |
| Turnip crinkle virus (TCV) | NC_003821 | |
| Turnip crinkle virus/UK (TCV/UK) | AY312063 | |
| Unclassified | Angelonia flower break virus/Florida (AFBV/Florida) | NC_007733 |
| Carmoviruses | Soybean yellow mottle mosaic virus/MS1-USA (SYMoMV/MS1-USA) | FJ707484 |
| Soybean yellow mottle mosaic virus/South Korea (SYMoMV/s.Korea) | NC_011643 | |
| Dianthovirus | Carnation ringspot virus RNA 1 (CarRSV-RNA 1) | NC_003530 |
| Carnation ringspot virus RNA 2 (CarRSV-RNA 2) | NC_003531 | |
| Red clover necrotic mosaic virus RNA 1 (RCNMV-RNA 1) | NC_003756 | |
| Red clover necrotic mosaic virus RNA 2 (RCNMV-RNA 2) | NC_003775 | |
| Red clover necrotic mosaic virus RNA 1/Can (RCNMV-RNA 1/Can) | AB034916 | |
| Red clover necrotic mosaic virus RNA 2/Can (RCNMV-RNA 2/Can) | AB034917 | |
| Sweet clover necrotic mosaic virus RNA 1/59 (SCNMV-RNA 1/59) | NC_003806 | |
| Sweet clover necrotic mosaic virus RNA 2/59 (SCNMV-RNA 2/59) | NC_003807 | |
| Sweet clover necrotic mosaic virus RNA 2/38 (SCNMV-RNA 2/38) | S46027 | |
| Unclassified dianthovirus | Rice virus X RNA 1 (RVX-RNA 1) | AB033715 |
| Machlomovirus | Maize chlorotic mottle virus (MCMoV) | NC_003627 |
| Maize chlorotic mottle virus/Nebraska (MCMoV/Nebraska) | EU358605 | |
| Necrovirus | Beet black scorch virus (BBSV) | NC_004452 |
| Beet black scorch virus/Val25-Iran (BBSV/Val25-Iran) | EU545828 | |
| Beet black scorch virus/CO-USA (BBSV/CO-USA) | EF153268 | |
| Beet black scorch virus/Xinjiang (BBSV/Xinjiang) | AY626780 | |
| Leek white stripe virus (LWSV) | NC_001822 | |
| Olive latent virus 1/Citrus (OLV-1/Citrus) | NC_001721 | |
| Olive latent virus 1/GM6-Portugal (OLV-1/GM6-Portugal) | DQ083996 | |
| Tobacco necrosis virus A/FM1B (TNV-A/FM1B) | NC_001777 | |
| Tobacco necrosis virus A/C (TNV-A/C) | AY546104 | |
| Tobacco necrosis virus D/Hungarian (TNV-D/Hungarian) | NC_003487 | |
| Tobacco necrosis virus D/Rhotamsted (TNV-D/Rhotamsted) | D00942 | |
| Unclassified necrovirus | Olive mild mosaic virus/GP-POrtugal (OMMV/GP-Portugal) | NC_006939 |
| Panicovirus | Panicum mosaic virus (PMV) | NC_002598 |
| Tombusvirus | Artichoke mottled crinkle virus/Bari (AMoCV/Bari) | NC_001339 |
| Carnation Italian ringspot virus (CarIRSV) | NC_003500 | |
| Cucumber bulgarian latent virus (CBLV) | NC_004725 | |
| Cucumber necrosis virus (CNV) | NC_001469 | |
| Cymbidium ringspot virus (CymRSV) | NC_003532 | |
| Grapevine algerian latent virus/nipplefruit (GALV/nipplefruit) | NC_011535 | |
| Pear latent virus (PeLV) | NC_004723 | |
| Tomato bushy stunt virus/Statice (TBSV/Statice) | AJ249740 | |
| Tomato bushy stunt virus/Nipplefruit (TBSV/Nipplefruit) | AY579432 | |
| Tomato bushy stunt virus/Pepper (TBSV/Pepper) | U80935 | |
| Tomato bushy stunt virus/Cherry (TBSV/Cherry) | M21958 | |
| Unclassified Tombusviruses | Lisianthus necrosis virus/L (LNV/L) | NC_007983 |
| Lisianthus necrosis virus/Zantedeschia (LNV/Zantedeschia) | AM711119 | |
| Pelargonium necrotic spot virus (PNSV) | NC_005285 | |
| Unassigned Tombusviridae | Maize necrotic streak virus (MaNSV) | NC_007729 |
| Pelargonium line pattern virus/PV-0193 (PLPV/PV-0193) | NC_007017 | |
| Unclassified Tombusviridae | Nootka lupine vein clearing virus/Alaska (NLVCV/Alaska) | NC_009017 |
| Pelargonium chlorotic ring pattern virus/GR 57 (PCRPV/GR 57) | NC_005985 | |
The nucleotide sequences were aligned using programs CLUSTALW 2.0.9 and CLUSTALX 2.0.9 (Larkin et al., 2007) with default configuration. Their phylogenetic relationships were determined with the Maximum-likelihood (ML) algorithm incorporated in the MEGA version 5 program (Tamura et al., 2011) under assumption of the substitution models proposed by Jukes and Cantor (1969) (JC), Hasegawa et al. (1985) (HKY85), and Tamura and Nei (1993) (TN93). Bootstrap analyses with 500 replicates were performed to assess the robustness of the branches.
Using the MEGA4.1β program (Kumar et al., 2008), positive selection was inferred by the counting method described by Nei and Gojobori (1986) and, later on, by Suzuki and Gojobori (1999). According to this method, the phylogenetic tree of sequences analyzed was used. For the parsimony method, the total numbers of synonymous (cS) and nonsynonymous (cN) substitutions as well as the average numbers of synonymous (sS) and nonsynonymous (sN) sites per codon over the phylogenetic tree for each codon site were computed according to the maximum parsimony principle (Fitch, 1971; Hartigan, 1973). The null hypothesis of selective neutrality (rS = rN or ω = 1) was tested for each site by computing the probability (p) of obtaining the observed or more biased values for cS and cN, which were assumed to follow a binomial distribution with the probabilities of occurrence of synonymous and nonsynonymous substitutions given by sS/(sS + sN) and sN/(sS + sN), respectively. Positive selection is inferred when p < 0.05 and cN/sN > cS/sS (Suzuki, 2006).
Potential recombination events between diverged nucleotide sequences were explored using two programs: RDP v3.31β (Martin et al., 2005b) and RECCO (Maydt and Lengauer, 2006). RDP incorporates several published recombination detection methods into a single suite of tools: RDP (Martin and Rybicki, 2000), GENECONV (Padidam et al., 1999), BOOTSCAN (Martin et al., 2005a), MAXCHI (Smith, 1992), CHIMAERA (Posada and Crandall, 2001), SISCAN (Gibbs et al., 2000), and 3SEQ (Boni et al., 2007). In all cases, default parameters were used. Only events predicted by more than half of the methods are considered as significant. The algorithm developed and described by Maydt and Lengauer (2006) as being a fast, simple and sensitive method for detecting recombination in a set of sequences and locating putative recombination breakpoints is based on cost minimization. This method has only two tunable parameters, recombination and mutation cost. In practice the only parameter considered is α, representing the cost of mutation relative to recombination. When α changes from 0 to 1, the cost of mutation weighted by α increases, and the cost for recombination weighted by 1 - α decreases. In other words, parameter α controls the ambiguity between mutation and recombination.
Results
Recombination events during Tombusviridae evolution
Examination of the RECCO program output regarding the occurrence of recombination events in the complete genome of the Tombusviridae family, revealed that three out of five aureusviruses were putative recombinants (PoLV.Pigeonpea, JCSMV.Iran, MaWLMV.USA). In contrast, CLSV (unknown isolate) and CLSV.Canada did not show any recombinant signal (Table 2). Within the genus Aureusvirus, the most frequently recombining virus was PoLV.Pigeonpea (33 putative recombination sites), whereas only 28 possible recombination signals were detected in the genome of viruses JCSMV.Iran and MaWLMV.USA. Similarly, the only representative of the genus Avenavirus (OCSV) was a potential recombinant with 175 putative sites. The RDP package confirmed these results for both genera. Among the carmoviruses, 14 out of 30 members were possible recombinants. According to RECCO, the most frequently recombining virus was JINRSV with 134 putative events, while MeNSV.Nagasaki and MeNSV.NK had the lowest number of recombination sites (two putative sites). The RDP v3.31β algorithm confirmed the occurrence of possible recombination events only for accessions MeNSV.NK and MeNSV.NH. Recombination investigations of genus Dianthovirus based on RECCO analysis showed that only 80% of them were possible recombinants (CarRSV-RNA 1 and 2, RCNMV-RNA 1 and 2, RCNMV.Can. RNA 1 and 2, SCNMV.59.RNA 1, and RVX.RNA 1). in contrast, SCNMV.59.RNA 2 and SCNMV.38.RNA did not show any putative recombination signals. However, the RDP package did not predict recombination in the dianthoviruses (Table 3). Although the most frequently recombining necrovirus was RVX (166 putative sites), RCNMV.RNA 2 had only two putative sites. Based on RECCO analysis, 50% of the necroviruses (BBSV.Val25.Iran, LWSV, TNV-A.FMB, TNV-A.C, TNV-D.Hungarian, and OMMV-GP.Portugal,) were possible recombinants. Conversely, BBSV, BBSV.CO.USA, BBSV.Xinjiang, OLV-1.Citrus, OLV-1.GM6.Portugal, and TNV-D.Rhotamsted were not possible recombinants. These results were congruent with those obtained with the RDP package. While the most frequently recombining virus was LWSV (39 sites), TNV.A.C recombined into two sites. Regarding the sole representative of genus Panicovirus (PMV), the results obtained by the two methods (RECCO and RDP v3.31β) were incongruent. Indeed, with RECCO, 108 possible sites were detected, whereas no recombination signals were found with the RDP package. A similar situation was observed with regard to the newly proposed carmoviruses (NLVCV.Alaska, PCRPV.GR 57, PLPV.PV-0193). According to RECCO analysis, although NLVCV.Alaska was the most frequently recombining virus (65 sites), PLPV.PV-0193 recombined only into 38 sites (Table 4). Regarding the members of genus Tombusvirus, there was an agreement between the two methods indicating that 80% of the analyzed accessions were putative recombinants. While CBLV had the highest number of putative recombination signals (67 sites), TBSV.Cherry had only two recombination sites. Furthermore, it is noteworthy that the two representatives of genus Machlomovirus (MCMoV, and MCMoV.Nebraska) were not recombinants as assessed by the two methods of analysis used in this study. Seeking for the recombination frequency in the genome of the Tombusviridae, two-thirds of the aureusviruses (JCSMV.Iran, and MaWLMV.USA) showed that in most cases, their breakpoint length was a single residue. In contrast, the breakpoint length of most putative recombination sites of PoLV.Pigeonpea was between three and 37 nucleotides (Table 2). Also, the breakpoint length of the major recombination sites of the single representative of genus Avenavirus (OCSV) consisted of a single residue. In about 50% of the members of the genus Carmovirus, the length of their most detected recombination sites was a single residue. As opposed to that, the breakpoint interval of the remaining members exceeded three residues reaching a size as long as 82 residues (MeNSV.NH). In 62% of the investigated dianthoviruses, the breakpoint length exceeded three nucleotides reaching 100 residues (CarRSV.RNA 1) (Table 3). In the necroviruses, the breakpoint interval distribution was similar i.e., 50% of the breakpoints consisted of a single residue, while the remaining breakpoints were between three and 77 nucleotides. For the sole member of the genus Panicovirus (PMV), most of the recombination sites had a breakpoint length of a single residue (45) (Table 3). As for the tombusviruses, 75% showed a breakpoint length exceeding three residues up to 161 nucleotides (AMoCV.Bari) (Table 4).
Table 2.
Determination of inferred putative recombination events and their frequency along the sequences of the entire genome of aureusviruses, one avenavirus and carmoviruses. Algorithm RDP v3.31β showed that only events supported by more than half of the different methods are reported. Nucleotide numbering corresponds to the aligned sequences. Abbreviations: NRS: -number of recombination sites, GIRE: -genomic interval of recombination events (the span of sequences in the viral genome where recombination events were predicted).
| Recombination determined by RECCO | Recombination determined by RDP v3.31β | |||||||
|---|---|---|---|---|---|---|---|---|
| Virus.isolate | NRS | GIRE (nt) | Length of breakpoint | Genomic position of the longest breakpoint (size in nucleotide) | Putative parental (Major × Minor) | |||
| 1 residue | 2 residues | 3 residues | > 3 residues | |||||
| PoLV.Pigeonpea | 33 | 607–5320 | 10 | 8 | 3 | 12 | 2148–2184 (37) | OCSV × MeNSV/MNSV-264 CarRSV RNA 1 × MeNSV-Tottori CarRSV RNA 1 × MeNSV-Kochi 2 |
| JCSMV.Iran | 28 | 2457–5288 | 11 | 6 | 3 | 8 | 3790–3802 (13) | CarRSV RNA 1 × MeNSV-Tottori CarRSV RNA 1 × MeNSV-Kochi 2 |
| MaWLMV.USA | 28 | 3212–4043 | 18 | 3 | 1 | 6 | 3836–3853 (18) | OCSV × MeNSV/MNSV-264 CarRSV RNA 1 × MeNSV-Tottori |
| OCSV | 175 | 766–5052 | 65 | 33 | 23 | 54 | 1295–1306 (12) 5036–5047 (12) |
TCV.UK × MaWLMV.USA |
| CCFV | 20 | 894–4681 | 7 | 2 | 3 | 8 | 1896–1908 (13) | / |
| CarMoV | 5 | 1349–2124 | 0 | 0 | 1 | 4 | 1986–2045 (60) | / |
| CPMoV | 14 | 1316–3143 | 4 | 6 | 2 | 2 | 2850–2854 (5) | / |
| JINRSV | 134 | 689–5098 | 58 | 30 | 20 | 26 | 1736–1760 (25) | / |
| MeNSV.Nagasaki | 2 | 623–4892 | 0 | 0 | 0 | 2 | 4847–4892 (46) | / |
| MeNSV.Kochi | 4 | 1843–4694 | 1 | 0 | 0 | 3 | 3866–3907 (42) | / |
| MeNSV.MNSV-Al | 3 | 619–4908 | 0 | 0 | 0 | 3 | 4864–4908 (45) | / |
| MeNSV.MNSV-264 | 5 | 4940–5131 | 3 | 0 | 2 | 0 | 4952–4954 (3) 5129–5131 (3) |
/ |
| MeNSV.NK | 2 | 615–4908 | 0 | 1 | 0 | 1 | 615–874 (60) | SYMoMV/MS1-USA × TBSV.Statice |
| MeNSV.NH | 5 | 615–4983 | 1 | 1 | 0 | 3 | 2070–2151 (82) | YMoMV/MS1-USA × TBSV.Statice |
| MeNSV.Malfa5 | 13 | 677–4831 | 0 | 0 | 2 | 11 | 1736–1796 (61) | / |
| PSNV.Japan | 80 | 645–5174 | 34 | 15 | 9 | 22 | 4225–4266 (42) | / |
| SCV | 61 | 665–4506 | 28 | 6 | 13 | 14 | 1887–1898 (12) 2761–2772 (12) |
/ |
| AFBV.Florida | 87 | 665–5046 | 34 | 15 | 15 | 23 | 2151–2166 (16) | / |
Table 3.
Determination of inferred putative recombination events and their frequency along the sequences of necroviruses, one panicovirus and dianthoviruses. Algorithm RDP v3.31β showed that only events supported by more than half of the different methods are reported. Nucleotide numbering corresponds to the aligned sequences. Abbreviations: NRS: -number of recombination sites, GIRE: -genomic interval of recombination events (the span of sequences in the viral genome where recombination events were predicted).
| Recombination determined by RECCO | Recombination determined by RDP v3.31β | |||||||
|---|---|---|---|---|---|---|---|---|
| Virus.isolate | NRS | GIRE (nt) | Length of breakpoint | Genomic position of the longest breakpoint (size in nucleotide) | Putative parental (Major × Minor) | |||
| 1 residue | 2 residues | 3 residues | > 3 residues | |||||
| CarRSV-RNA 1 | 19 | 709–4561 | 7 | 3 | 2 | 7 | 2581–2680 (100) | / |
| CarRSV-RNA 2 | 3 | 1826–2811 | 0 | 0 | 0 | 3 | 1826–1835 (10) 2802–2811 (10) |
/ |
| RCNMV-RNA 1 | 8 | 1297–4754 | 0 | 1 | 2 | 5 | 1479–1533 (55) | / |
| RCNMV-RNA 2 | 2 | 2696–2812 | 1 | 0 | 0 | 1 | 2803–2812 (10) | / |
| RCNMV-RNA1.Can | 7 | 720–4218 | 2 | 0 | 0 | 5 | 1556–1644 (89) | / |
| RCNMV-RNA 2.Can | 10 | 1730–2827 | 3 | 1 | 2 | 4 | 1730–1765 (36) | / |
| SCNMV-RNA 1.59 | 12 | 858–4443 | 2 | 2 | 1 | 7 | 3126–3193 (68) | / |
| RVX-RNA 1 | 166 | 439–5107 | 65 | 39 | 19 | 43 | 840–855 (16) | / |
| BBSV-Val25.Iran | 4 | 800–4881 | 2 | 0 | 0 | 2 | 1435–1468 (34) | MeNSV.NH × LNV.Zantedeschia MeNSV.Malfa5 × MaNSV SCV × CNV MeNSV.Yamaguchi × CNV |
| LWSV | 39 | 1027–4853 | 17 | 9 | 5 | 8 | 4846–4853 (8) | MeNSV.NH × LNV.Zantedeschia SCV × CNV |
| TNV-A.FM1B | 3 | 2419–2789 | 3 | 0 | 2 | 1 | 2419–2496 (77) | OLV1.GM6-Portugal × BBSV.Xinjiang |
| TNV-A.C | 2 | 2275–2619 | 0 | 0 | 0 | 2 | 2275–2619 (10) | OLV1.GM6-Portugal × BBSV.Xinjiang LWSV × TNV.D.Rhotamsted |
| TNV-D.Hungarian | 4 | 2323–4287 | 2 | 0 | 0 | 2 | 3959–3964 (6) | MeNSV.NH × LNV.Zantedeschia MeNSV.Malfa5 × MaNSV BBSV × OMMV.GP.Portugal SCV × CNV |
| OMMV-GP.Portugal | 20 | 935–4857 | 6 | 3 | 4 | 7 | 2116–2145 (30) | TNV.A.C × TNV.D.Hungarian TNV.A.C × TNV.D.Rhotamsted TNV.D.Rhotamsted × TNV.A.FM1B OLV1.GM6-Portugal × TNV.D.Hungarian |
| PMV | 108 | 166–4751 | 45 | 28 | 12 | 23 | 1491–1505 (15) | / |
Table 4.
Determination of inferred putative recombination events and their frequency along the sequences of the tombusviruses and tentative new members of genus Carmovirus. Algorithm RDP v3.31β showed that only events supported by more than half of the different methods are reported. Nucleotide numbering corresponds to the aligned sequences. Abbreviations: NRS: -number of recombination sites, GIRE: -genomic interval of recombination events (the span of sequences in the viral genome where recombination events were predicted).
| Recombination determined by RECCO | Recombination determined by RDP v3.31β | |||||||
|---|---|---|---|---|---|---|---|---|
| Virus.isolate | NRS | GIRE (nt) | Length of breakpoint | Genomic position of the longest breakpoint (size in nucleotide) | Putative parental (Major × Minor) | |||
| 1 residue | 2 residues | 3 residues | > 3 residues | |||||
| AMoCV.Bari | 27 | 1074–4774 | 2 | 4 | 3 | 18 | 4614–4774 (161) | CymRSV × CBLV GALV.nipplefruit × MeNSV.Yamaguchi |
| CarIRSV | 28 | 1998–5345 | 9 | 5 | 8 | 6 | 4775–4842 (68) | TBSV.pepper × PNSV TBSV.Statice × MeNSV.Tottori AMoCV.Bari × PNSV LNV.Zantedeschia × MaNSV SCV × CBLV PNSV × TBSV.Pepper CBLV × MeNSV.MNSV-ISR LNV.Zantedeschia × TBSV.Statice |
| CBLV | 67 | 343–5390 | 16 | 17 | 10 | 24 | 1804–1836 (33) | TBSV.Pepper × JCSMV.Iran CarRSV.RNA 1 × MeNSV.Tottori GALV.nipplefruit × MaWLMV.USA PeLV × MaWLMV.USA LNV.Zantedeschia × MaNSV CymRSV × MaNSV |
| CNV | 34 | 417–5280 | 8 | 2 | 6 | 18 | 2681–2714 (34) | CymRSV × MeNSV TBSV.Pepper × PNSV CymRSV × SCV LNV.L × TBSV.Pepper CymRSV × CBLV TBSV.Statice × LNV.L |
| CymRSV | 44 | 675–5066 | 9 | 9 | 4 | 22 | 1719–1767 (49) | PNSV × TBSV.Pepper CarIRSV × CLSV.Israel TBSV.Statice × TCV.UK CNV × CLSV.Canada CNV × TCV.UK MeNSV.MNSV.ISR × PeLV |
| GALV.nipplefruit | 45 | 369–5299 | 7 | 4 | 11 | 23 | 4481–4532 (52) | TBSV.Statice × CNV TBSV.nipplefruit × MaNSV TBSV.nipplefruit × TBSV.Pepper TBSV.Statice × PeLV CymRSV × CBLV CBLV × PNSV CymRSV × PeLV AMoCV.Bari × TBSN.Pepper TBSV.nipplefruit × PeLV PeLV × TBSV.Pepper TBSV.Pepper × CymRSV |
| TBSV.statice | 11 | 379–2743 | 1 | 0 | 1 | 9 | 1902–1931 (30) | GALV.nipplefruit × LNV.Zantedeschia CymRSV × CBLV CymRSV × PeLV TBSV.nipplefruit × TBSV.Pepper |
| TBSV.nipplefruit | 7 | 618–1931 | 0 | 0 | 1 | 6 | 1550–1651 (102) | GALV.nipplefruit × LNV.Zantedeschia AMoCV.Bari × GALV.nipplefruit |
| TBSV.pepper | 7 | 2167–4890 | 0 | 0 | 1 | 5 | 4852–4890 (39) | CymRSV × CBLV PeLV × AMoCV.Bari GALV.nipplefruit × LNV.Zantedeschia |
| TBSV.cherry | 2 | 5324–5345 | 0 | 0 | 2 | 0 | 5324–5326 (3) 5343–5345 (3) |
CymRSV × CBLV CarIRSV × AMoCV.Bari CymRSV × PeLV GALV.nipplefruit × LNV.Zantedeschia |
| MaNSV | 54 | 547–5334 | 29 | 4 | 7 | 14 | 1932–1955 (24) | GALV.nipplefruit × MaWLMV.USA PeLV × MaWLMV.USA |
| PNSV | 24 | 2025–5032 | 7 | 2 | 3 | 12 | 5009–5032 (23) | TBSV.Statice × MeNSV.Tottori CNV × CLSV.Canada AMoCV.Bari × TBSV.Cherry TBSV.nipplefruit × PeLV LNV.Zantedeschia × MaNSV TBSV.Statice × CNV CBLV × MeNSV.MNSV.ISR LNV.Zantedeschia × TBSV.Statice |
| NLVCV.alaska | 65 | 771–5301 | 28 | 13 | 7 | 17 | 771–782 (12) | / |
| PCRPV.GR 57 | 45 | 1109–5104 | 11 | 17 | 4 | 13 | 2307–2316 (20) | / |
| PLPV.PV-0193 | 38 | 991–4908 | 20 | 7 | 2 | 9 | 2813–2821 (9) | / |
Nucleotide sequence analysis
Maximum composite likelihood estimate of the nucleotide substitution pattern were made using the MEGA4.1β program. The results for Tombusviridae showed that the rates of different transitional substitutions varied from 3.18 to 14.61, and those of transversional substitutions varied from 6.6 to 8.57. The nucleotide frequencies were: 0.269 (A), 0.258 (T/U), 0.207 (C), and 0.266 (G). The transition/transversion rate ratios were k1 = 1.705 (purines) and k2 = 0.482 (pyrimidines). The overall transition/transversion bias was R = 0.547, where R = [AGk1 + TCk2]/[(A+G)(T+C)]. There were a total of 1218 positions in the final dataset. In all these analyses, the codon positions included were first + second + third + noncoding. All positions containing gaps and missing data were excluded from the dataset (complete deletion option).
The MEGA4.1β program also incorporates the Tajima’s Neutrality Test. The purpose of this test is to indentify sequences which do not fit the neutral theory model at equilibrium between mutation and genetic drift. Tajima’s test compares a standardized measure of the total number of segregating sites (the polymorphic DNA sites) in the sampled DNA and the average number of mutations between pairs in the sample. Tajima’s D was determined (D = 5.280926).
Positive selection
The high genetic stability of viruses can be attributed to negative or purifying selection to maintain the functional integrity of the viral genome. The degree of negative selection in genes, or the degree of functional constraint for the maintenance of the encoded protein sequence, can be estimated, as mentioned above, by the ratio between the nucleotide diversities in nonsynonymous and synonymous positions (dN/dS). For most coding genes the dN/dS ratio is < 1 which is consistent with negative selection against protein change. In contrast, a dN/dS ratio > 1 may be an indication that adaptive or positive selection is driving gene divergence. In this study, pairwise comparisons of all screened accessions showed that, none of the members of the genera Machlomovirus and Panicovirus, and unclassified Tombusviridae was under positive selection. On the contrary, the genera Aureusvirus (JCMSV.Iran), Avenavirus (OCSV), Carmovirus (CarMoV.China, CarMoV.Indian), Dianthovirus (CarRSV-RNA 2, RCNMV-RNA 2, SCNMV-RNA 2.59, SCNMV-RNA 2.38), Necrovirus (BBSV, BBSV.Val25.Iran), Tombuvirus (GALV.nipplefruit, PeLV, TBSV.Statice, PNSV) along with the unassigned Tombusviridae (PLPV.PV.0193) were under positive selection (Table 5). It is worth pointing out that, in the viruses with a segmented genome, positive selection was detected only in RNA 2, suggesting that probably reassortment events occurred. All these results were obtained by testing neutrality in sequence pairs with Fisher’s Exact Test. The probability of rejecting the null hypothesis of strict-neutrality (dN = dS) in favor of positive selection for each sequence pair was determined. Values of p less than 0.05 were considered significant at the 5% level. The variance of the difference (dN - dS) was computed using the bootstrap method (500 replicates). All analyses were made using the Nei-Gojobori method incorporated in the MEGA program. All positions containing gaps and missing data were excluded from the dataset (complete deletion option). The final dataset comprised a total of 234 positions.
Table 5.
Accession pairs in family Tombusviridae under positive selection with probability determined at the 5% level, based on Fisher’s Exact Test of Neutrality and calculated dN - dS.
| Accession pairs | Virus.isolate pairs | p value at 5% level | dN - dS |
|---|---|---|---|
| NC_007017/NC_003775 | PLPV/PV-0193/RCNMV-RNA 2 | 0.028 | 0.923 |
| AB034917/NC_003633 | RCNMV-RNA 2.Can/OCSV | 0.016 | 2.325 |
| NC_003531/NC_003633 | CarRSV-RNA 2/OCSV | 0.039 | 1.820 |
| S46027/NC_003633 | SCNMV-RNA 2.38/OCSV | 0.041 | 1.847 |
| NC_003775/NC_003633 | PLPV.PV-0193/OCSV | 0.048 | 1.975 |
| NC_003807/NC_003633 | SCNMV-RNA 2.59/OCSV | 0.033 | 1.999 |
| AB034917/NC_001265 | RCNMV-RNA 2.Can/CarMoV.China | 0.034 | 1.620 |
| AB034917/AJ249740 | RCNMV-RNA 2.Can/TBSV.Statice | 0.029 | 1.921 |
| NC_003531/NC_005287 | CarRSV-RNA 2/JCSMV.Iran | 0.037 | 1.790 |
| S46027/EU545828 | SCNMV-RNA 2.38/BBSV.Val25-Iran | 0.045 | 1.682 |
| AB034917/AJ811998 | RCNMV-RNA 2/CarMoV.Indian | 0.031 | 1.679 |
| AB034917/NC_011535 | RCNMV-RNA 2/GALV.nipplefruit | 0.041 | 1.933 |
| NC_003775/NC_011535 | PLPV/PV-0193/GALV.nipplefruit | 0.033 | 1.921 |
| NC_003807/NC_005285 | SCNMV-RNA 2.59/PNSV | 0.049 | 1.773 |
| S46027/NC_004723 | SCNMV-RNA 2.38/PeLV | 0.047 | 1.735 |
| S46027/NC_004452 | SCNMV-RNA 2.38/BBSV | 0.043 | 1.749 |
| NC_003807/NC_004452 | SCNMV-RNA 2.59/BBSV | 0.050 | 1.654 |
Phylogenetic relationships
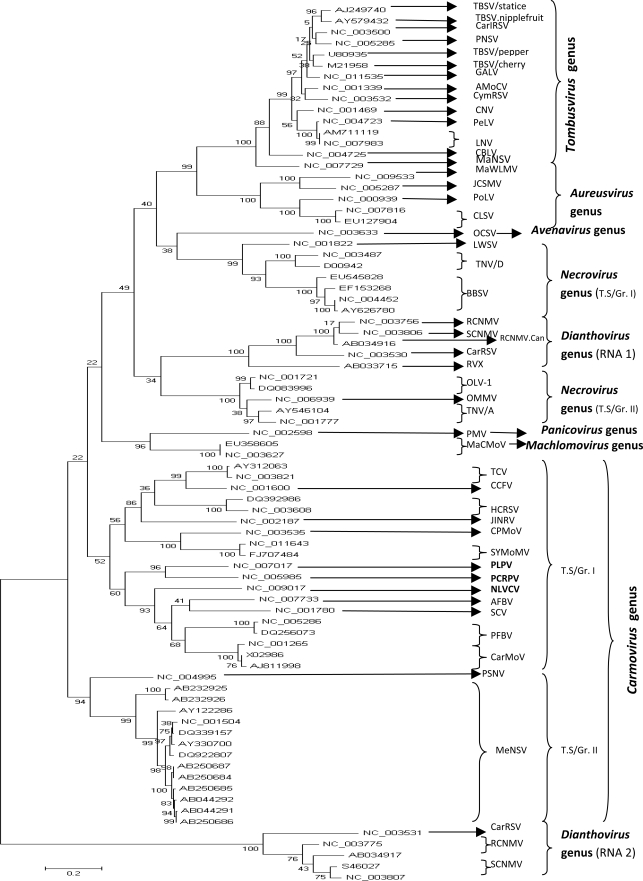
The phylogenetic relationships among members of the family Tombusviridae, based on the sequences of their complete genome, were inferred using a Maximum Likelihood algorithm under the assumption of three models of substitution (JC, HKY85, TN93). The topologies of the constructed trees were identical. The inferred phylogeny showed that each taxonomical genus in the family Tombusviridae constituted a homogenous group clearly distinct from the others. However, the results obtained in this study evidenced a few differences in terms of virus species composition within each taxonomical genus compared to the current classification adopted by the ICTV. In fact, three viruses considered by the ICTV as unassigned (PLPV.PV-0193) and unclassified Tombusviridae (NLVCV.Alaska, PCRPV.GR 57) showed a close phylogenetic relationship to known members of the genus Carmovirus. Moreover, the viruses belonging to this genus were divided into two distinct subgroups. The first subgroup comprised viruses: TCV, CCFV, JINRV, HCRSV, PLPV, PCRPV, NLVCV, SCV, AFBV, PFBV, CPMoV, SYMoMV and CarMoV, and the second subgroup encompassed viruses: MeNSV, and PSNV. Furthermore, it was proposed that genus Necrovirus should be constituted by two distinct subgroups named tentative Subgroup I (BBSV, TNV.D, LWSV) and tentative subgroup II (OMMV, TNV.A, OLV-1) (Figure 1). It should be noted that here OMMV is an integral part of subgroup I rather than an unclassified Necrovirus. In contrast, genus Aureusvirus encompassed members that evolved in a homogenous manner: CLSV, PoLV, MaWLMV, and JCSMV. Similarly, the following members of genus Tombusvirus also formed a coherent ensemble: MaNSV, CBLV, LNV.L, LNV.Zantedeschia, PeLV, CNV, CymRSV, AMoCV, TBSV.Statice, TBSV.Nipplefruit, TBSV.Pepper, TBSV.Cherry, GALV, PNSV, and CarIRSV. Their evolutionary history reshuffled the existing classification adopted by the ICTV since 2009. In fact, according to this classification, MaNSV was considered as an unassigned Tombusviridae, whereas LNV and PNSV were included in the unclassified Tombusvirus group. Concerning genus Dianthovirus which clearly was not monophyletic, the clustering pattern showed two distinct clades representing their RNAs 1 and 2, as illustrated in Figure 1. Originally, RVX was considered as an unclassified virus within genus Dianthovirus.
Figure 1.
Dendrogram depicting phylogenetic relationships among the studied members of the family Tombusviridae, based on their complete genome sequences. Eight clusters representing the eight genera were clearly delineated. While members of genera Carmovirus, Necrovirus, and Dianthovirus fell into two tentative distinct subgroups (T.S/Gr I, T.SGr. II), those of genera Aureusvirus and Tombusvirus constituted a homogenous ensemble. The tree was produced using the ML algorithm option of MEGA5.03. The numbers above the branches indicate the bootstrap confidence value. The scale bar shows the number of substitutions per nucleotide.
Discussion
This study evidenced the prediction of putative recombination events in the genome of several members of the family Tombusviridae and demonstrated that tombusviruses and carmoviruses are highly recombinant compared to viruses of the other genera. For this purpose, two methods were chosen (RECCO and RDP v.3.31β), based on the fact that they are appropriate for the mosaic structure of viruses as reported in previous works (Boulila, 2009; 2010). In this study, using the RECCO algorithm, it was demonstrated that the viruses belonging to the following genera contained putative recombination signals in their genome: Aureusvirus, Avenavirus, Carmovirus, Dianthovirus, Necrovirus, Panicovirus, and Tombusvirus. These results were in good agreement with those obtained by the RDP package except for members of genus Diantovirus. By both methods, the two representatives of genus Machlomovirus (MCMoV, MCMoV.Nebraska) were found to be non-recombinant. As revealed by RECCO, the most frequently recombining viruses were: OCSV, RVX.RNA 1, JINRSV, and PMV with 175, 166, 134, and 108 putative recombination sites, respectively. All of these recombination signals were constituted by a single residue. MeNSV.Nagasaki, MeNSV.NK, RCNMV.RNA 2, TNV-A.C, and TBSV.Cherry (2 sites), MeNSV.MNSV-Al, CarRSV-RNA 2, and TNV-A.FM1B (3 sites), MeNSV.Kochi, BBSV-Val25.Iran, and TNV-D.Hungarian (4 sites), CarMoV, MeNSV.MNSV-264, and MeNSV.NH (5 sites), RCNMV.RNA 1.Can, TBSV.nipplefruit, and TBSV.pepper (7 sites), and RCNMV.RNA 1 (8 sites) showed the lowest frequency of recombination breakpoints. In contrast, most of these breakpoints had an interval exceeding three nucleotides. Furthermore, this study showed that recombination may occur between viruses belonging to different genera. For example: Oat chlorotic stunt avenavirus (OCSV) and Melon necrotic spot carmovirus (MeNSV) may give rise to Pothos latent aureusvirus (PoLV). Similarly, OCSV itself may result from a recombination between Turnip crinkle carmovirus (TCV) and Maize white line mosaic aureusvirus (MaWLMV) (Table 2). Seemingly, these viruses could contain part of their sequences particularly in the coat protein-encoding gene of each other. Such an event was largely studied for Cucumber necrosis tombusvirus (CNV) and Melon necrotic spot carmovirus (MeNSV) (Riviere and Rochon, 1990).
On the other hand, investigations of selective pressure acting on protein expression of virus genes led to the identification of positive selection in 17 accession pairs involving 15 different lineages. It is worth mentioning that numerous viruses: JCSMV.Iran, OCSV, CarRSV-RNA 2, RCNMV-RNA 2.Can, BBSV.Val25.Iran, GALV.nipplefruit, TBSV.Statice, PNSV, and PLPV.PV.0193 evolved under both mechanisms: recombination and positive selection between which synergism might be occurring. Such a synergism between recombination and natural selection may have played a major role in Darwinian molecular evolution.
The evolutionary history of the Tombusviridae has shown that the 79 accessions split into eight clearly separated clusters representing the eight genera of the Tombusviridae family. From the present phylogenetic study, at least two taxonomic implications can be drawn: (i) three viruses (NLVCV.Alaska, PCRPV.GR 57, PLPV.PV-0193) currently considered by the ICTV as: one unassigned Tombusviridae (PLPV.PV-0193), and two unclassified Tombusviridae (NLVCV.Alaska and PCRPV.GR 57). All of them should be included in genus Carmovirus; (ii) In addition to the viruses belonging to genus Carmovirus which have formed two separated subgroups, the members of genera Necrovirus, and Dianthovirus evolved separately and divided into two distinct subgroups as shown in Figure 1. In contrast, members of genera Aureusvirus, and Tombusvirus formed separately a single ensemble. The evolutionary relationships among viruses are a reliable approach for classification. As stated by Stuart et al. (2004) (who reported similar results regarding the genetic divergence of components of genus Necrovirus), the comparison of complete genomes is a more balanced approach that should provide a more precise scheme of relatedness. On the other hand, it should be pointed out that, in genus Dianthovirus, the genetic divergence between RNAs 1 and 2 is correlated to the final products synthesized and their use by the virus to survive. For example: RNA silencing is a small RNA-guided sequence- specific gene activation mechanism in eukaryotes that is involved in different biological phenomena (e.g. development, heterochromatin formation and defense against molecular parasites such as viruses). Many viruses express suppressors to counteract RNA-silencing-mediated antiviral defenses. These RNA silencing suppressors have been identified in the following genera: Aureusvirus, Carmovirus, Tombusvirus, and Dianthovirus (Voinnet e al., 1999; Qu et al., 2003; Mérai et al., 2005; Takeda et al., 2005). Dianthovirus uses a unique strategy to suppress RNA silencing. The dianthoviral suppressor consists of multiple components including P27, P88 (encoded by two ORFs in RNA 1) and viral RNA (Takeda et al., 2005). Moreover, sequence variability of the coat protein-coding gene (RNA 1) may be linked to the interaction between this structural protein and the host and vector which themselves show a major diversity among diathoviruses. In contrast, the ORF in RNA 2 encodes the movement protein. All these factors can influence the divergence between the two RNAs.
Finally, to the author’s best knowledge, this is the largest study in the literature so far on recombination potentially occurring in the entire genome of all currently known members of the family Tombusviridae as well as positive selection operating on protein expression and their phylogenetic reconstruction. In addition, a reclassification based on their predicted evolutionary history, is proposed.
Footnotes
Associate Editor: Carlos F.M. Menck
References
- Aaziz R, Tepfer M. Recombination in RNA viruses and in virus-resistant transgenic plants. J Gen Virol. 1999;80:1339–1346. doi: 10.1099/0022-1317-80-6-1339. [DOI] [PubMed] [Google Scholar]
- Alejska M, Kurzyniska-Kokorniak A, Broda M, Kierzek R, Figlerowicz M. How RNA viruses exchange their genetic material. Acta Bioch Polon. 2001;48:391–407. [PubMed] [Google Scholar]
- Baroth M, Orlich M, Thiel HJ, Becher P. Insertion of cellular NEDD8 coding sequences in a pestivirus. Virology. 2000;278:456–466. doi: 10.1006/viro.2000.0644. [DOI] [PubMed] [Google Scholar]
- Boni MF, Posada D, Feldman MW. An exact non-parametric method for inferring mosaic structure in sequence triplets. Genetics. 2007;176:1035–1047. doi: 10.1534/genetics.106.068874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulila M. Recombination structure and genetic relatedness among members of the family Bromoviridae based on their RNAs 1 and 2 sequence analyses. Virus Genes. 2009;38:435–444. doi: 10.1007/s11262-009-0340-7. [DOI] [PubMed] [Google Scholar]
- Boulila M. Putative recombination events and evolutionary history of five economically important viruses of fruit trees based on the coat protein-encoding gene sequence analysis. Biochem Genet. 2010;48:357–375. doi: 10.1007/s10528-009-9317-4. [DOI] [PubMed] [Google Scholar]
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press; London: 2005. Virus taxonomy: Classification and nomenclature of viruses; p. 1259. [Google Scholar]
- Fitch WN. Toward defining the course of evolution minimum change for a specific tree topology. Syst Zool. 1971;20:406–416. [Google Scholar]
- Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-scanning: A Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000;16:573–582. doi: 10.1093/bioinformatics/16.7.573. [DOI] [PubMed] [Google Scholar]
- Greene AE, Allison RF. Recombination between viral RNA and transgenic plants transcripts. Science. 1994;263:1423–1425. doi: 10.1126/science.8128222. [DOI] [PubMed] [Google Scholar]
- Habili N, Symons RH. Evolutionary relationship between luteoviruses and other RNA plant viruses based on sequence motifs in their putative RNA polymerase and nucleic acid helicases. Nucleic Acids Res. 1989;17:9543–9555. doi: 10.1093/nar/17.23.9543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartigan JA. Minimum mutation fits to a given tree. Biometrics. 1973;29:53–63. [Google Scholar]
- Hasegawa M, Kishino H, Yano T. Dating of human-ape splitting by a molecular clock of mitochondrial DNA. J Mol Evol. 1985;22:160–174. doi: 10.1007/BF02101694. [DOI] [PubMed] [Google Scholar]
- Jukes T, Cantor C. Evolution of protein molecules. In: Munro HN, editor. Mammalian Protein Metabolism. Academic Press; New York: 1969. pp. 21–132. [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: A biologist-centric software for evolutionary analysis of DNA and protein sequences. Brief Bioinfor. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai MMC. RNA recombination in animal and plant viruses. Microbiol Rev. 1992;56:61–79. doi: 10.1128/mr.56.1.61-79.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshileds G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X v.2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Martin D, Rybicki E. RDP: Detection of recombination amongst aligned sequences. Bioinformatics. 2000;16:562–563. doi: 10.1093/bioinformatics/16.6.562. [DOI] [PubMed] [Google Scholar]
- Martin DP, Posada D, Crandall KA, Williamson C. A modified bootscan algorithm for automated identification of recombination sequences and recombination breakpoints. AIDS Res H Retrovir. 2005a;21:98–102. doi: 10.1089/aid.2005.21.98. [DOI] [PubMed] [Google Scholar]
- Martin DP, Williamson C, Posada D. RDP2: Recombination detection and analysis from sequence alignments. Bioinformatics. 2005b;21:260–262. doi: 10.1093/bioinformatics/bth490. [DOI] [PubMed] [Google Scholar]
- Maydt J, Lengauer T. Recco: Recombination analysis using cost optimization. Bioinformatics. 2006;22:1064–1071. doi: 10.1093/bioinformatics/btl057. [DOI] [PubMed] [Google Scholar]
- Mérai Z, Kerényi Z, Molnar A, Barta E, Valoczi A, Bisztray G, Havelda Z, Burgyan J, Silhavy D. Aureuvirus P14 is an efficient RNA silencing suppressor that binds double-stranded RNAs without size specificity. J Virol. 2005;79:7217–7226. doi: 10.1128/JVI.79.11.7217-7226.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai M, Sakoda Y, Mori M, Hayashi M, Kida H, Akashi H. Insertion of a cellular sequence and RNA recombination in the structural protein coding region of cytopathogenic bovine viral diarrhea virus. J Gen Virol. 2003;84Pt2:447–452. doi: 10.1099/vir.0.18773-0. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–225. doi: 10.1006/viro.1999.0056. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall K. Evaluation of methods for detecting recombination from DNA sequences. Computer simulation. Proc Natl Acad Sci USA. 2001;98:13757–13762. doi: 10.1073/pnas.241370698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu F, Ren T, Morris TJ. The coat protein of Turnip crinkle virus suppresses post transcriptional gene silencing at an early initiation step. J Virol. 2003;77:511–522. doi: 10.1128/JVI.77.1.511-522.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riviere CJ, Rochon DM. Nucleotide sequence and genomic organization of melon necrotic spot virus. J Gen Virol. 1990;71:1887–1896. doi: 10.1099/0022-1317-71-9-1887. [DOI] [PubMed] [Google Scholar]
- Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–129. doi: 10.1007/BF00182389. [DOI] [PubMed] [Google Scholar]
- Stuart G, Moffet K, Bozarth RF. A whole genome perspective on the phylogeny of the plant virus family Tombusviridae. Arch Virol. 2004;149:1595–1610. doi: 10.1007/s00705-004-0298-7. [DOI] [PubMed] [Google Scholar]
- Suzuki Y. Statistical properties of the methods for detecting positively selected amino acid sites. Gene. 2006;365:125–129. doi: 10.1016/j.gene.2005.09.002. [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Gojobori T. A method for detecting positive selection at single amino acid sites. Mol Biol Evol. 1999;16:1315–1328. doi: 10.1093/oxfordjournals.molbev.a026042. [DOI] [PubMed] [Google Scholar]
- Tajima F. Statistical-method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585–595. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Tsukuda M, Mizumoto H, Okamoto K, Kaido M, Mise K, Okuno T. A plant RNA virus suppresses RNA silencing through viral RNA replication. EMBO J. 2005;24:3147–3157. doi: 10.1038/sj.emboj.7600776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Nei M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol. 1993;10:512–526. doi: 10.1093/oxfordjournals.molbev.a040023. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular Biology and Evolution. 2011 doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet O, Pinto YM, Baulcombe DC. Suppression of gene silencing: A general strategy used by diverse DNA and RNA viruses of plants. Proc Natl Acad Sci USA. 1999;96:14147–14152. doi: 10.1073/pnas.96.24.14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White KA, Nagy PD. Advances in the molecular biology of tombusviruses: Gene expression, genome replication and recombination. Prog Nucleic Acid Res Mol Biol. 2004;78:187–226. doi: 10.1016/S0079-6603(04)78005-8. [DOI] [PubMed] [Google Scholar]
- Worobey M, Holmes EC. Evolutionary aspects of recombination in RNA viruses. J Gen Virol. 1999;80:2535–2543. doi: 10.1099/0022-1317-80-10-2535. [DOI] [PubMed] [Google Scholar]